Abstract
Pichia pastoris PEX17 was cloned by complementation of a peroxisome-deficient strain obtained from a novel screen for mutants disrupted in the localization of a peroxisomal membrane protein (PMP) reporter. PEX17 encodes a 267-amino-acid protein with low identity (18%) to the previously characterized Saccharomyces cerevisiae Pex17p. Like ScPex17p, PpPex17p contains a putative transmembrane domain near the amino terminus and two carboxyl-terminal coiled-coil regions. PpPex17p behaves as an integral PMP with a cytosolic carboxyl-terminal domain. pex17Δ mutants accumulate peroxisomal matrix proteins and certain integral PMPs in the cytosol, suggesting a critical role for Pex17p in their localization. Peroxisome remnants were observed in the pex17Δ mutant by morphological and biochemical means, suggesting that Pex17p is not absolutely required for remnant formation. Yeast two-hybrid analysis demonstrated that the carboxyl terminus of Pex19p was required for interaction with Pex17p lacking the carboxyl-terminal coiled-coil domains. Biochemical evidence confirmed the interaction between Pex19p and Pex17p. Additionally, Pex17p cross-linked to components of the peroxisome targeting signal–receptor docking complex, which unexpectedly contained Pex3p. Our evidence suggests the existence of distinct subcomplexes that contain separable pools of Pex3p, Pex19p, Pex17p, Pex14p, and the peroxisome targeting signal receptors. These distinct pools may serve different purposes for the import of matrix proteins or PMPs.
INTRODUCTION
The prevailing model for peroxisome biogenesis posits that peroxisomes arise by growth and division of preexisting peroxisomes (Lazarow and Fujiki, 1985). The coordinate regulation of these processes is essential for the proper functioning of the organelle and consequently the organism. Peroxisomes are absolutely required in multicellular organisms, as evidenced by the numerous human genetic diseases caused by peroxisomal abnormalities (Subramani, 1997). In lower eukaryotes, such as the yeast Pichia pastoris, peroxisomes are required specifically for the use of methanol and oleate as carbon sources. Because many fundamental peroxisomal functions are conserved at the molecular level between yeast and humans, yeast has been heavily exploited as a model system to gain insights into peroxisome biogenesis.
The bulk of our understanding of peroxisome biogenesis concerns the identification of the targeting signals that direct matrix proteins from the cytosol to the peroxisome and the proteins essential for this process. The peroxisome targeting signals (PTSs) for import of peroxisomal matrix proteins are PTS1 and PTS2 (Gould et al., 1987, 1989; Osumi et al., 1991; Swinkels et al., 1991; Elgersma et al., 1996b). By isolating and characterizing mutants defective for the localization of the peroxisome matrix proteins (pex mutants), 22 peroxins have been identified from various species (Distel et al., 1996; Götte et al., 1998; Purdue et al., 1998; Subramani, 1998; Titorenko et al., 1998; Koller et al., 1999). Most of these peroxins are membrane proteins, but a few of them have a predominantly cytosolic localization. For example, Pex5p and Pex7p are predominantly cytosolic proteins that interact specifically with PTS1 and PTS2, respectively (Marzioch et al., 1994; Dodt and Gould, 1996; Rehling et al., 1996; Elgersma et al., 1998). These PTS receptors function to bring newly synthesized cargo from the cytosol to docking sites at the peroxisome membrane. The docking sites have been defined by protein–protein interactions between the receptors and docking proteins at the peroxisome membrane. It has been demonstrated that Pex5p and Pex7p bind to Pex14p (Albertini et al., 1997; Brocard et al., 1997; Fransen et al., 1998; Schliebs et al., 1999; Shimizu et al., 1999; Will et al., 1999) and Pex13p (Elgersma et al., 1996a; Erdmann and Blobel, 1996; Gould et al., 1996; Girzalsky et al., 1999; Shimozawa et al., 1999) to allow docking at the peroxisome membrane. Pex17p (Huhse et al., 1998), identified previously only in Saccharomyces cerevisiae, binds Pex14p and is therefore part of the receptor docking complex.
Despite our understanding of the early stages of matrix protein import, very little is known about the growth of the peroxisome membrane or the targeting and insertion of peroxisomal membrane proteins (PMPs). Several PMPs are synthesized on free polysomes (Fujiki et al., 1984; Suzuki et al., 1987; Bodnar and Rachubinski, 1991) and targeted directly from the cytosol to the peroxisome (Lazarow and Fujiki, 1985). The targeting signal for integral PMPs (mPTS) has been identified for Pex3p from several species (Höhfeld et al., 1992; Baerends et al., 1996; Wiemer et al., 1996; Kammerer et al., 1998). In addition, the mPTSs for S. cerevisiae Pex15p (Elgersma et al., 1997), Candida boidinii PMP47 (Dyer et al., 1996), and P. pastoris Pex22p (Koller et al., 1999) have been characterized. The factors that bind these mPTS sequences to mediate targeting are unknown.
The phenotypes of several pex mutant strains suggest their involvement in the biogenesis of the peroxisome membrane or in membrane protein import. In all pex mutants examined, with the exception of human pex16 mutants (South and Gould, 1999), pex3 mutants (Höhfeld et al., 1991; Baerends et al., 1996; Wiemer et al., 1996), and, in some organisms, pex19 mutants (Götte et al., 1998; Matsuzono et al., 1999), PMPs accumulate in membranous remnants, whereas matrix proteins are mislocalized in the cytosol (Crookes and Olsen, 1999). Therefore, in most pex mutants, with the exceptions noted above, the machinery for the propagation of the peroxisome membrane and the machinery that targets PMPs to those membranes remain intact. In all pex3 mutants and in the human pex16-deficient cell lines, no peroxisome remnants have been detected. Human and S. cerevisiae pex19 mutant strains were reported to lack remnant structures. However, in P. pastoris positive evidence has been presented for membranous remnants that contain Pex3p (Snyder et al., 1999), and additional evidence is accumulating that other PMPs are also in remnants of Pppex19Δ mutants (our unpublished results). The morphology of the remnants in Pppex19Δ mutants suggests that PpPex19p is involved in the maturation of a tubulovesicular, early preperoxisome compartment to the late preperoxisome structures, corresponding to late remnant structures observed in other pex mutant strains (Snyder et al., 1999). It is noteworthy that in P. pastoris and S. cerevisiae, PEX16 has not been identified. The predominant players for peroxisome membrane biogenesis and PMP localization in S. cerevisiae and P. pastoris would therefore be Pex3p and Pex19p.
Recent evidence that could explain the source and mechanism of deposition of membrane lipids to growing peroxisomes is provided by studies that suggest that a vesicular trafficking pathway exists between the endoplasmic reticulum and peroxisomes (for review, see Kunau and Erdmann, 1998; Titorenko and Rachubinski, 1998).
We decided to take a new approach to the understanding of PMP localization in P. pastoris by designing a novel genetic screen for mutants disrupted in the targeting of an mPTS-green fluorescent protein (GFP) reporter protein. This reporter efficiently localizes to peroxisomes in wild-type cells (Wiemer et al., 1996), and to punctate remnant structures in most pex mutants. However, in pex3Δ and pex19Δ mutants the mPTS-GFP appeared diffuse. Differences between these localization patterns correlated with a useable fluorescence-activated cell sorter (FACS) phenotype. Our FACS-based enrichment procedure identified one new complementation group of P. pastoris pex mutant, namely PEX17. Pex17p has only been previously identified in S. cerevisiae as a component of the PTS–receptor docking complex (see above). We provide evidence that PpPex17p is part of the receptor docking complex required for the localization of matrix proteins but is also required for efficient PMP localization. This requirement for PpPex17p in PMP localization is related to functional interactions with the two main players in PMP biogenesis, Pex3p and Pex19p.
MATERIALS AND METHODS
Strains and Growth Conditions
Media and growth conditions used are described elsewhere (Snyder et al., 1999).
Molecular Biological Techniques
P. pastoris strains are listed in Table 1. All plasmids used in this study are listed in Table 2. All DNA oligonucleotide primers used are listed in Table 3.
Table 1.
P. pastoris strain list
| Strain | Relevant genotype | Source |
|---|---|---|
| PPY12 | his4 arg4 | Gould et al., 1992 |
| PPY4 | his4 | Gould et al., 1992 |
| SKF1 | arg4 his4∷pKNSD77 (PAOXPEX31–40-GFP) | Wiemer et al., 1996 |
| MUT9 | SKF1 pex17-1 | This study |
| SWS19DM | PPY12 pex19Δhis4∷pKNSD77 (PAOXPEX31–40-GFP) | This study |
| SWS1DM | PPY12 pex1Δhis4∷pKNSD77 (PAOXPEX31–40-GFP) | This study |
| SWS3DM | PPY12 pex3Δhis4∷pKNSD77 (PAOXPEX31–40-GFP) | This study |
| SWS8DM | PPY12 pex8Δhis4∷pKNSD77 (PAOXPEX31–40-GFP) | This study |
| SMD1163 | his4 pep4 prb | Invitrogen |
| SWS17D | SMD1163 pex17∷KanMX | This study |
| SWS17HA | S17D his4∷p17HA | This study |
Table 2.
Plasmids used in this study
| Name | Relevant elements | Reference |
|---|---|---|
| pKNSD77 | PAOXPEX31–40-GFP | Wiemer et al., 1996 |
| pMut9 | pYM8 2.4 kB genomic PEX17 | This study |
| pBL17 | 900 bp genomic PEX17′ | This study |
| p17HA | pIB1 PEX17-HA | This study |
| Two-hybrid plasmids | ||
| pKNSD123 | pKNSD52 PEX19 | Snyder et al., 1999 |
| pKNSD124 | pKNSD55 PEX19 | Snyder et al., 1999 |
| pKNSD127 | pKNSD52 PEX19[1–232] | Snyder et al., 1999 |
| pKNSD125 | pKNSD52 PEX19[1–75] | Snyder et al., 1999 |
| pKNSD135 | pKNSD52 PEX19[1–42] | Snyder et al., 1999 |
| p2H17 | pKNSD55 PEX17[1–267] | This study |
| p2H17NB | pKNSD55 PEX17[1–124] | This study |
| p2H17lum | pKNSD55 PEX17[1–59] | This study |
| p2H17cyt | pKNSD55 PEX17[52–267] | This study |
Table 3.
Primers
| Name | DNA sequence (5′→3′) |
|---|---|
| TAG17u | GCGAATTCGATCTAGCTGCTCTGGAG |
| HApstD | GCGCCTGCAAGGTCGACTTTTAGAGGATCCC |
| TAG17uL | GAAAAGAAATAGGTCTAGTACCCCCGGGCGCATCTTTTAC |
| TAG17dL | GTAAAAGATGCGCCCGGGGGTACTAGACCTATTTCTTTTC |
| 2h17u | GTCCAGATCTATGTCGTCAAGGCGCAACG |
| 2h17d | GGAATTCGGTACTAGACCTATTTCTTTTC |
| 2h17NB | GAATTCTTAAAACTTGATCGTCTGTCTTCC |
| 2h17lumD | GCGAATTCGCTTCACATAGGTCGAATCAG |
| 2h17cytU | GCGCAGATCTCGACCTATGTTGAAGCTTC |
| P17up | GATCTAGCTGCTCTGGAGAAC |
| M9SEQ8 | GTTACCAGCCTTGAAGGTGG |
| P17P5L | GGGGATCCGTCGACCTGCAGCGTACCATGGTTTGGGAGTTACCAGCC |
| P17P3L | AACGAGCTCGAATTCATCGATGATATGATAATGACTTTCATTTTGATGGC |
Restriction enzyme digestion, cloning, plasmid isolation, and PCRs were performed by standard methods (Sambrook et al., 1989). DNA sequencing was performed by the method of Sanger et al. (1977).
P. pastoris transformations, mating, sporulation, and random spore analysis were performed as described (Gould et al., 1992).
FACS Isolation of Peroxisome Assembly (pex) Mutants
N-Methyl-N-nitro-N-nitrosoguanidine (NTG) mutagenesis of a wild-type strain expressing the mPTS(Pex3p)-GFP, strain SKF1, was performed as described (Elgersma et al., 1998). Cells treated with 150 μg/ml NTG had 41% killing and were grown overnight in YPD to an A600 of 1. These cells were then induced in methanol-containing media for 6 h and subjected to a sterile sort on a FACStar fluorescence-activated sorter (Becton Dickinson, Mountain View, CA) to collect bright cells. This collection pool was plated on YPD. Eighty-six colonies appeared after plating and were individually streaked onto methanol, oleate, and glycerol media. Nine of these colonies were unable to grow on methanol or oleate but were able to grow on glycerol and glucose medium. These strains were back-crossed to wild type (PPY4) and used for complementation analysis.
Cloning and Sequencing of PEX17
A genomic library constructed in plasmid pYM8 (Liu et al., 1995) was used to transform the novel mutant strain (MUT9). A 2.4-kb clone (pMut9) was identified, which complemented the mutant strain for growth on methanol. Approximately 1.5 kb of the genomic insert were sequenced, revealing a 801-bp open reading frame (ORF). A 900-bp subclone was generated by cutting the original pMut9 plasmid with BamHI and religating; there was a BamHI site flanking the genomic insert on one side and another BamHI inside the genomic insert, 600 bp from the other one. This removed 600 bp of the genomic insert and left 900 bp. This plasmid, pBL17, could only express a carboxyl-terminal truncated form of Pex17p but nonetheless complemented the pex17 mutant strains for growth on methanol and oleate media, confirming that this region comprised the essential portion of the PEX17 ORF and the required regulatory elements.
Two-Hybrid Analysis
Cloning vectors, tester strains, and screening by two-hybrid analysis have been described (Faber et al., 1998). Two-hybrid clones containing PEX19 and subdomains were described previously (Snyder et al., 1999). A full-length clone of PEX17 was amplified by PCR (primers 2h17u and 2h17d) and inserted as an EcoRI–BglII fragment into pKNSD55 cut with EcoRI and BamHI, creating p2H17. Fragments of PEX17, amplified by PCR, were introduced in pKNSD55 as follows: PEX17[1–124] (primers 2h17u and 2h17NB) was cut with EcoRI and BglII and inserted into pKNSD55 cut with EcoRI and BamHI, creating p2H17NB; PEX17[1–59] (primers 2h17u and 2h17lumD) was cut with EcoRI and BglII and cloned into pKNSD55 cut with EcoRI and BamHI, creating p2H17lum; PEX17[52–267] (primers 2h17cytU and 2h17d) was cut with EcoRI and BglII and cloned into pKNSD55 cut with EcoRI and BamHI, creating p2H17cyt.
Construction of the pex17Δ Strain
The 5′ and 3′ flanking regions of the PEX17 ORF were amplified by overlap extension PCR (primers P17up, M9SEQ8, P17P5L, and P17P3L), creating a Geneticin resistance cassette between the flanking regions as described (Wach et al., 1994). This PCR product was used to transform strain SMD1163. Transformants were selected on YPD plates containing 200 μg/ml Geneticin, and the expected genomic alteration in the pex17 deletion strain (SWS17D), which was unable to grow on methanol or oleate medium, was confirmed by PCR.
Biochemical Techniques
Crude cell-free extracts were made as described previously (Babst et al., 1997). SDS-PAGE (Laemmli, 1970) and Western blot analyses (Towbin et al., 1979) were performed as described. Goat-anti-rabbit conjugated HRP, goat-anti-rabbit conjugated alkaline phosphatase (Bio-Rad, Hercules, CA), and goat-anti-rat conjugated HRP (Jackson ImmunoResearch, West Grove, PA) were used as secondary antibodies which were detected by ECL (Amersham, Arlington Heights, IL) or 5-bromo-4-chloro-3-indolyl phosphate/nitro blue tetrazolium (Kirkland & Perry, Gaithersburg, MD) according to the manufacturer’s protocols. Primary antibodies and the dilutions used were as follows: α-Pex19p (1:4000), α-Pex3p (1:10,000), α-Pex22p (1:2000), α-ScTHIO (1:10,000), α-CAT (1:10,000), α-Pex4p (1:1000), α-Sc-glucose-6-phospate dehydrogenase (1:2000), and rat-α-hemagglutinin (HA; 1:2000).
Immunoprecipitation and cross-linking with dithiobis(succinimidyl propionate) (Pierce, Rockford, IL) was performed from 5 A600 units of oleate-grown cells as described previously (Rieder and Emr, 1997). One microliter of antisera was used per each immunoprecipitation.
Protease protection and organelle membrane extraction assays are described elsewhere (Koller et al., 1999).
Subcellular Fractionation Experiments
Differential centrifugation of oleate-grown cells was performed as described (Faber et al., 1998). For floatation, all sucrose stocks contained the lysis buffer lacking sorbitol, and the gradients were prepared as follows: 0.375 ml of postnuclear supernatant (PNS) from the methanol-grown cells was mixed with 1.625 ml of 80% (wt/vol) sucrose in a 5-ml ultracentrifuge tube; this was layered with 1.5 ml of 50% (wt/vol) sucrose and 1.5 ml of 35% (wt/vol) sucrose. The gradients were centrifuged in a Beckman Instruments (Palo Alto, CA) SW50.1 rotor for 20 h at 40,000 rpm. Five hundred-microliter fractions were collected from the top and adjusted to 5% trichloroacetic acid (TCA). The material pelleted on the bottom of the tube was resuspended in 1 ml of 5% TCA and transferred to an Eppendorf tube. The TCA precipitates were incubated on ice for >20 min and washed once with 5% TCA and three times with cold acetone. The acetone was evaporated, and the pellets were resuspended in 100 μl of SDS-PAGE sample buffer.
Construction of a Strain Expressing Pex17HAp
The PEX17-HA construct was generated by overlap extension PCR. PEX17 was amplified by PCR from pMut9 with primers TAG17u and TAG17dL; HA was amplified by PCR with primers TAG17uL and HApstD from a triple-HA construct in pBlusescript (a gift from Markus Babst, University of California, San Diego, CA). These products were gel purified and mixed as template for PCR with primers TAG17u and HApstD to generate the PEX17-HA. This fragment was cut with EcoRI and PstI and cloned into pIB1 (Sears et al., 1998) cut with the same enzymes, creating p17HA. This plasmid was linearized with SalI and integrated at the his4 locus of strain SWS17D creating SWS17HA.
Fluorescence and Electron Microscopy
Samples for immunofluorescence were prepared from methanol- or oleate-induced cells spheroplasted as described for biochemical fractionation and then fixed and prepared as described previously (Babst et al., 1998). Pex3p, thiolase and catalase antibodies were used at dilutions of 1:10,000. Microscopy for immunofluorescence was as described (Odorizzi et al., 1998). Preparation and analysis of cells expressing GFP constructs were as described (Monosov et al., 1996). Cells for electron microscopy were prepared as described previously (Sakai et al., 1998).
RESULTS
A FACS-based Screen Yields New pex Mutants
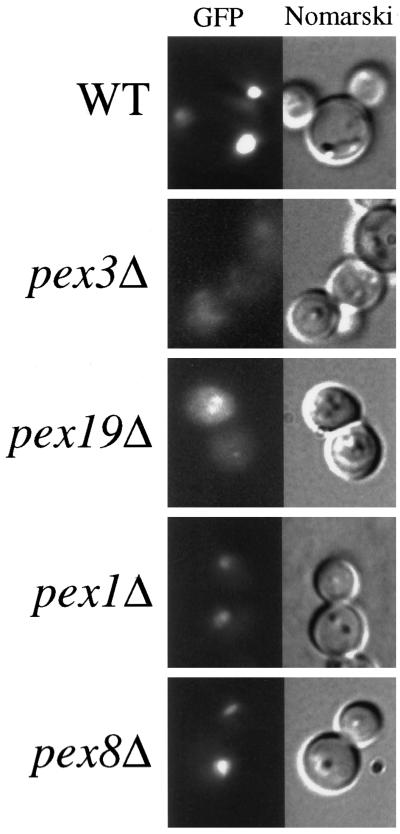
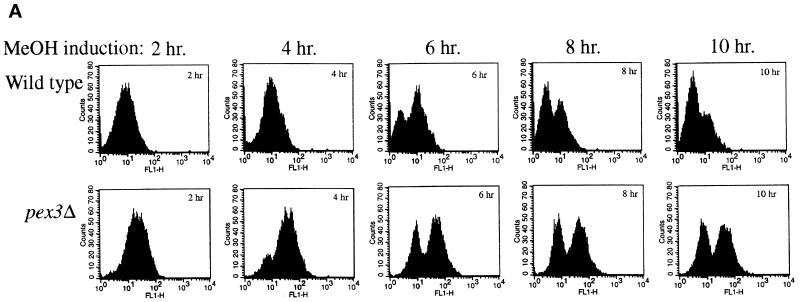
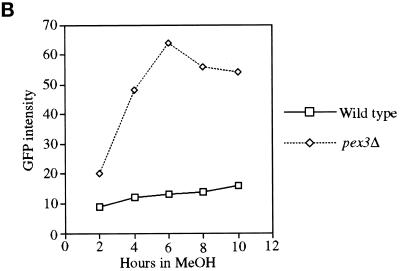
To obtain genes encoding components required for the localization of PMPs, we developed a FACS-based enrichment procedure for pex mutants. Using the 40-amino-acid mPTS of Pex3p fused to GFP [mPTS(Pex3p)-GFP] to follow membrane protein targeting, we observed normal, mature peroxisomes in wild-type cells (Figure 1; Wiemer et al., 1996). In addition, similar peroxisome remnants, or ghosts, were observed in 11 typical pexΔ mutants including pex1, 2, 4, 5, 6, 7, 8, 10, 12, 13, and 22 (Figure 1; our unpublished results). In contrast, the pex3Δ and pex19Δ mutants showed diffuse staining with the mPTS(Pex3p)-GFP that could represent true cytosolic localization and/or small, vesicular structures (Figure 1). To quantitate differences between cells containing mPTS(Pex3p)-GFP in peroxisomes and those containing the reporter in the diffuse, cytosolic pattern, we analyzed wild-type and pex3Δ cells by FACS. A modest increase was noted in fluorescence intensity of the population of wild-type cells after induction of the mPTS(Pex3p)-GFP reporter in methanol-containing media (Figure 2). As the population grew, a small, low-intensity peak was seen after 6 h (Figure 2A), which is likely to correspond to newly formed daughter cells in the population that are smaller than the original mother cells inoculated into the culture. The number of cells in this low-intensity peak increased with time as the larger mother cells from the original inoculum were diluted out in the culture. By analysis of the forward light scattering of the cells, which is proportional to the cell size, we indeed observed a population of low-GFP intensity, small cells that accumulated in the culture (our unpublished results). In contrast, pex3Δ cells showed a dramatic increase in GFP intensity after growth on methanol (Figure 2). The population of daughter cells did not outgrow the original mothers in the pex3Δ culture, because after one doubling these cells stop growing in methanol medium. The difference in fluorescence intensity of the mPTS(Pex3p)-GFP is easily quantitated by plotting the mean intensity of the bright peaks from wild-type and pex3Δ cells versus time (Figure 2B). After a 6-h induction in methanol, the fluorescence intensity of the mPTS(Pex3p)-GFP detected by FACS in pex3Δ cells was sixfold higher than in wild-type cells. Although we cannot definitively explain this difference in intensity, it correlates with the localization of the mPTS(Pex3p)-GFP in punctate structures versus the diffuse pattern in the two cell populations. Furthermore, typical pex mutants, those containing punctate remnants, showed a fluorescence intensity similar to that of wild-type cells (our unpublished results).
Figure 1.
Fluorescence microscopy of mPTS(Pex3p)-GFP in wild-type and pex mutant cells. Methanol-grown wild-type (PPY12), pex3Δ (SWS3DM), pex19Δ (SKF13), pex1Δ (SWS1DM), and pex8Δ (SWS8DM) strains expressing the mPTS(Pex3p)-GFP were visualized by fluorescence microscopy and Nomarski optics.
Figure 2.
FACS analysis of fluorescence intensity of mPTS(Pex3p)-GFP in wild-type and pex3Δ cells. (A) Time course analysis of mPTS(Pex3p)-GFP fluorescence intensity (FL1-H) versus cell number (Counts) from a population of wild-type (SKF1) and pex3Δ (SWS3DM) cells at the indicated times after induction in methanol medium. (B) Mean fluorescence intensity of mPTS(Pex3p)-GFP in the high fluorescence intensity peak of the population plotted versus time.
We exploited the difference in mPTS(Pex3p)-GFP intensity between wild-type and pex3Δ cells for the identification of pex mutant cells that accumulate the mPTS(Pex3p)-GFP in a diffuse pattern. Wild-type cells containing mPTS(Pex3p)-GFP were mutagenized with NTG, and after 6 h of growth in methanol medium, mutants were enriched by collecting a pool of bright cells (intensity >120) by FACS. From 2.5 million cells analyzed, only 86 cells were collected into our sort pool. Nine of these 86 cells were unable to grow with methanol or oleate as the sole carbon source but grew normally on glucose- or glycerol-containing media, suggesting that the enrichment was efficient for obtaining pex mutants. All nine of these mutants contained diffuse, cytosolic mPTS(Pex3p)-GFP. In contrast, brute-force screening of 300 colonies from the mutagenized pool without FACS enrichment did not reveal any strains containing diffuse, cytosolic mPTS(Pex3p)-GFP. The mutant strains were then crossed with all known pex mutants. Three of the mutants belonged to the pex3 complementation group, and five more mapped to known complementation groups. However, null mutants from these previously identified complementation groups contain Pex3p in typical, punctate peroxisome remnants and not in the cytosol. One of the mutants did not correspond to any previously known pex complementation group and, accordingly, was chosen for further analysis.
Cloning of P. pastoris PEX17
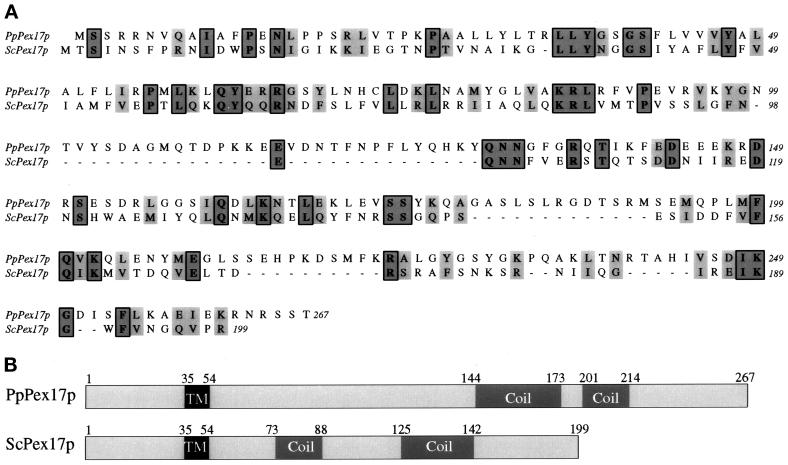
Complementation of the methanol growth defect of the novel mutant by a genomic DNA library identified a 2.4-kb clone that complemented the oleate growth defect as well as restoring the localization of mPTS(Pex3p)-GFP to punctate structures (our unpublished results). Subcloning and sequence analysis revealed an ORF of 801 nucleotides encoding a 267-amino-acid protein with a predicted molecular mass of 30.5 kDa (Figure 3A). This protein is predicted to contain a transmembrane domain near the amino terminus (aa 35–54) and contains two regions predicted to form coiled-coil domains (Lupas, 1996; Figure 3B). Comparison of this ORF with the databases identified one protein with significant homology, the S. cerevisiae Pex17p. Although the sequence identity between ScPex17p and our ORF is only 18%, there are conserved regions throughout the alignment (Figure 3A). In addition, the conservation of the putative transmembrane domain near the amino terminus of both proteins and the carboxyl-terminal coiled-coil domains further suggested that the ORF represents the P. pastoris Pex17p (Figure 3B). However, the ScPEX17 was unable to complement our mutant strain for growth on methanol media (our unpublished results). Additional protein–protein interaction data (see below) further suggested that the ORF is indeed the PpPex17p.
Figure 3.
Sequence alignment and features of P. pastoris and S. cerevisiae Pex17p orthologues. The amino acid sequences were aligned using the ClustalW program (A). Identical residues (boxed) and similar residues (gray) are shaded. Similarity rules: G = A = S, A = V, V = I = L = M, I = L = M = F = Y = W, K = R = H, D = E = Q = N, and S = T = Q = N. Dashes represent gaps. (B) Sequence features and relative positions. TM, putative transmembrane domain; Coil, putative coiled-coil domain.
Peroxisome Membrane and Matrix Protein Localization Defects in pex17Δ Mutants
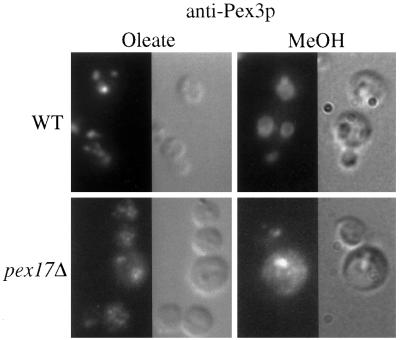
Strains of P. pastoris deleted for PEX17 were constructed as described in MATERIALS AND METHODS. The pex17Δ mutants were unable to grow on methanol or oleate as the sole carbon source. Because the original mutant accumulated the mPTS(Pex3p)-GFP in the cytosol or on small vesicular structures, we wished to visualize where Pex3p accumulates in the pex17Δ mutant. In oleate-grown, wild-type cells, the typical peroxisome staining pattern was observed for Pex3p (Figure 4). The pex17Δ cells, in contrast, exhibited diffuse staining for Pex3p, as well as some brighter structures. Examination of methanol-grown, wild-type cells by anti-Pex3p immunofluorescence revealed the large peroxisome clusters that are typical for methanol-grown P. pastoris. By contrast, the pex17Δ cells contained a few bright, punctate structures, but the Pex3p also appeared to be diffuse. This localization pattern for Pex3p in oleate- and methanol-grown pex17Δ cells is consistent with the conclusion that Pex3p accumulated in large peroxisome remnant structures, small vesicular structures, and, perhaps also, the cytosol.
Figure 4.
Immunofluorescence microscopy of Pex3p in wild-type and pex17Δ cells. Oleate- and methanol-grown wild-type (SMD1163) and pex17Δ (SWS17D) spheroplasts were indirectly labeled with the anti-Pex3p antibody and visualized by fluorescence microscopy and Nomarski optics.
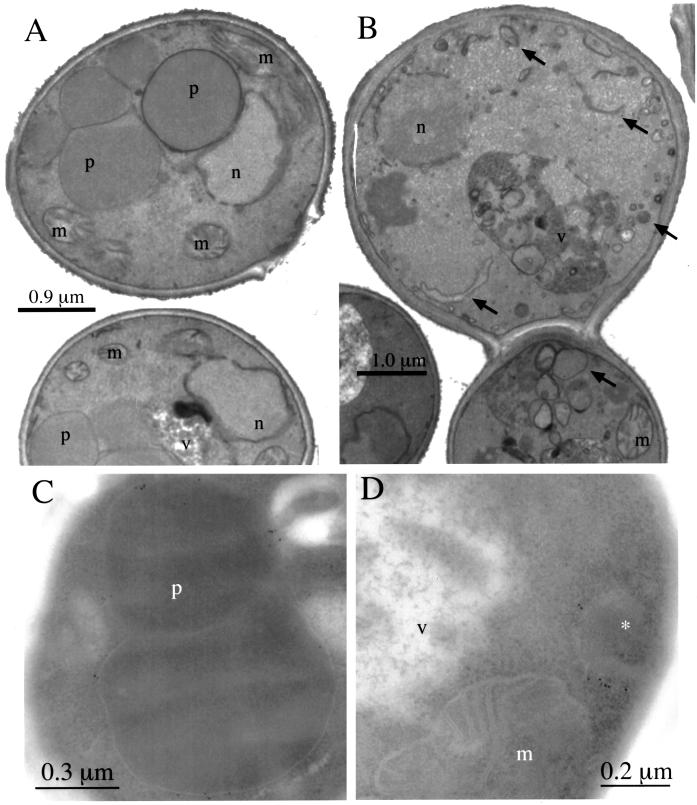
The morphology of peroxisome remnant structures in the pex17Δ mutants was observed by electron microscopy. Methanol-grown wild-type cells contained the usual, large clusters of peroxisomes (Figure 5A). In contrast, methanol-grown pex17Δ mutants contained no normal peroxisomes. Instead the pex17Δ mutants contained vesicular and tubular structures of varying size (Figure 5B) that are not normally seen in methanol-grown P. pastoris. To identify conclusively the peroxisome remnant structures in the pex17Δ mutant, immunoelectron microscopy was performed using the anti-Pex3p antibody. In methanol-grown wild-type cells Pex3p was detected on the normal peroxisome clusters (Figure 5C). In the pex17Δ cells, the Pex3p was detected on the membrane of smaller, single-membrane–bound compartments (Figure 5D). These structures are likely to represent the peroxisome remnants in the pex17Δ mutant. No Pex3p was detected in the vacuole or mitochondria of the pex17Δ cells.
Figure 5.
Electron microscopy of wild-type and pex17Δ cells. Methanol-grown wild-type (SMD1163; A and C) and pex17Δ (SWS17D; B and D) cells were prepared as described in MATERIALS AND METHODS for membrane staining (A and B) and immunoelectron microscopy with anti-Pex3p antibodies (C and D). Arrows point to the novel structures in pex17Δ cells (A and B). n, nucleus; m, mitochondria; v, vacuole; p, peroxisome; *, peroxisme remnant.
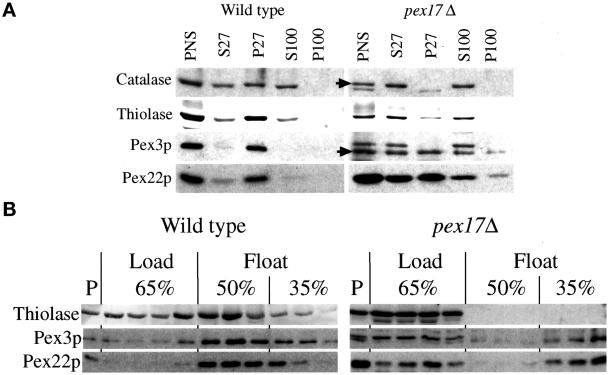
To further characterize the protein localization defects of pex17Δ cells, a subcellular fractionation was performed (Figure 6A). A whole-cell lysate after a low-speed spin to remove unlysed cells and nuclei is called a PNS and is the starting material for the fractionation experiment. Catalase and thiolase are commonly used as markers for lumenal protein import via the PTS1 and PTS2 pathway, respectively. The PNS contained catalase, thiolase, Pex3p, and another integral PMP, Pex22p (Figure 6A). Centrifugation of the PNS at 27,000 × g created a pellet fraction (P27), which contains organelles, including peroxisomes. In wild-type cells, the majority of catalase, thiolase, Pex3p, and Pex22p was found in the P27 fraction. Consistent with previous reports (Kalish et al., 1996; Waterham et al., 1996; Elgersma et al., 1998; Faber et al., 1998; Snyder et al., 1999), some of catalase and thiolase leaked from the organelle during the procedure and was found in the 27,000 × g supernatants (S27). Further centrifugation of the S27 at 100,000 × g left this catalase and thiolase in the supernatant fractions (S100), consistent with a cytosolic localization. The cytosolic marker, glucose-6-phosphate dehydrogenase, was found only in the supernatant fractions from both strains (our unpublished results). In contrast to wild type, the pex17Δ cells contained catalase exclusively in the supernatant fractions, indicative of a cytosolic localization. Some thiolase was found in the P27 of pex17Δ cells but likely represents large aggregates (see below); the majority was found in the supernatant fractions. In pex17Δ cells, the Pex3p and Pex22p were found equally in the P27 and S27 fractions, and only a small fraction of the Pex3p and Pex22p in the S27 fraction could be further pelleted at 100,000 × g (P100). The majority of the Pex3p and Pex22p from the S27 fraction remained in the supernatant (S100), consistent with a cytosolic localization, but some was also found in the P100 fraction.
Figure 6.
Mislocalization of peroxisomal proteins by pex17Δ mutants. (A) Oleate-grown spheroplasts of wild-type and pex17Δ cells (SMD1163 and SWS17D) were lysed and subjected to sequential differential centrifugation. Equivalent amounts of the PNS, 27,000 × g supernatant (S27), 27,000 × g pellet (P27), 100,000 × g supernatant (S100), and 100,000 × g pellet (P100) were resolved by SDS-PAGE, transferred to nitrocellulose, and probed with the indicated antibodies. (B) The PNS from oleate-grown cells was adjusted to 65% sucrose, layered with 50 and 35% sucrose, and centrifuged to allow floatation of membranes into the lighter fractions as described in MATERIALS AND METHODS. P, pelleted material at the bottom of the tube. Arrows point to the normal, full-length catalase and Pex3p.
To provide additional evidence that pex17Δ mutants accumulate membrane-bound and non–membrane-bound pools of integral PMPs, sucrose floatation gradients were used with the PNS fractions (Figure 6B). Proteins able to migrate from the bottom of the sucrose gradient, containing high concentrations of sucrose, to lower-density fractions are membrane associated, whereas those remaining in the high-density sucrose at the bottom of the gradient are non-membrane bound and likely to be cytosolic. In such gradients, the majority of the integral membrane proteins Pex3p and Pex22p from wild-type cells left the load fraction and migrated into the 50 and 35% fractions (Figure 6B). Glucose-6-phosphate dehydrogenase, a cytosolic protein, never leaves the load fraction (our unpublished results). In wild-type cells, a portion of thiolase floated into the fractions containing Pex3p and Pex22p. However, some of the thiolase remained in the load fraction and likely represents the portion that leaks from the peroxisomes during the procedure or non–membrane-bound aggregates that were pelleted in the differential centrifugation experiment. The thiolase from pex17Δ mutants did not float from the load fractions, consistent with the conclusion that it is in the cytosol and not a membrane-bound compartment. Approximately half of the Pex3p and Pex22p from the pex17Δ mutants remained in the load fractions but the remainder floated to the 35% sucrose fractions. These data are consistent with the notion that Pex3p and Pex22p accumulate in the cytosol of pex17Δ mutants but also in membrane-bound compartments of lower density than peroxisomes. Furthermore, the data from immunofluorescence microscopy, differential centrifugation, and sucrose density floatation taken together support the conclusion that integral PMPs accumulate in the cytosol and on membrane-bound remnants in pex17Δ mutants.
Pex17p Is a Peroxisomal Integral Membrane Protein with a C-terminal Cytosolic Domain
A triple-HA epitope tag was fused to the carboxyl terminus of Pex17p, and the tagged protein expressed from the PEX17 promoter was integrated into the chromosome of pex17Δ cells (see MATERIALS AND METHODS). These cells grew similar to wild-type cells on methanol or oleate media, demonstrating that the Pex17HAp was at least partially functional. The Pex17HAp had an apparent molecular mass of 40 kDa, which is slightly higher than the predicted molecular mass of 35 kDa. Pex17HAp was detected in glucose-grown cells, and its expression was not induced upon shift from glucose to oleate or methanol media (our unpublished results).
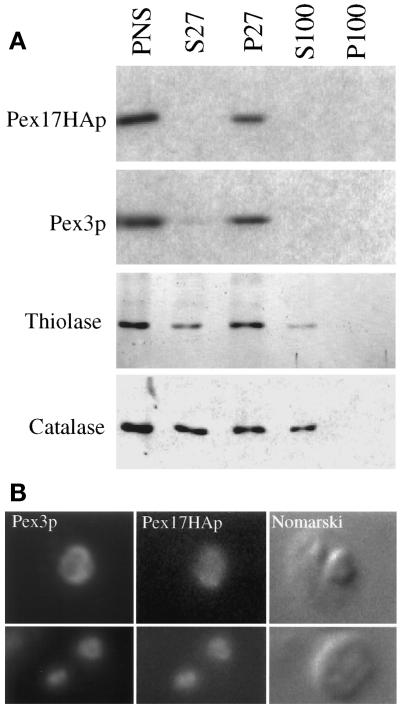
The subcellular localization of Pex17HAp and its ability to fully complement pex17Δ cells was determined by differential centrifugation. In pex17Δ cells expressing Pex17HAp, at least half of the thiolase and catalase was in the P27 fraction (Figure 7A), which was similar to wild type (Figure 6A). Pex3p and Pex17HAp were found almost exclusively in the pellet (Figure 7A). These data show that Pex17HAp complements the peroxisome protein import defects of pex17Δ cells and suggest that Pex17HAp is organelle associated.
Figure 7.
Subcellular localization of Pex17HAp. (A) Oleate-grown spheroplasts of pex17Δ cells expressing Pex17HAp (SWS17HA) were lysed and subjected to sequential differential centrifugation. Equivalent amounts of the PNS, 27,000 × g supernatant (S27), 27,000 × g pellet (P27), 100,000 × g supernatant (S100), and 100,000 × g pellet (P100) were resolved by SDS-PAGE, transferred to nitrocellulose, and probed with the indicated antibodies. (B) Methanol-grown spheroplasts of pex17Δ cells expressing Pex17HAp (SWS17HA) were indirectly labeled using anti-Pex3p or anti-HA antibodies as described in MATERIALS AND METHODS and visualized by fluorescence microscopy (Pex3p and Pex17HAp) and Nomarski optics.
Evidence for peroxisomal localization of Pex17HAp comes from indirect, double-labeling immunofluorescence experiments using anti-HA and anti-Pex3p antibodies. In methanol-grown cells expressing Pex17HAp, Pex3p staining overlapped with the Pex17HAp staining (Figure 7B) in the typical, large peroxisome clusters. These results and the differential centrifugation data show that Pex17HAp localizes to the peroxisome.
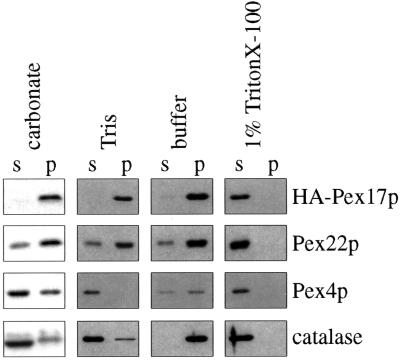
Because Pex17p contains a putative transmembrane domain, we tested its membrane association properties. A 27,000 × g pellet fraction from Pex17HAp-expressing cells was extracted with various buffers to test the strength of interaction with the membrane (see MATERIALS AND METHODS). Extraction with buffer alone left the majority of peroxisomal proteins, Pex22p, Pex4p, catalase, and Pex17HAp, in the pellet fraction (Figure 8). Treatment with low-ionic-strength Tris caused organelle rupture, which released catalase from the lumen and the peripheral membrane protein, Pex4p, from the membrane. However, Pex22p and Pex17HAp were resistant to extraction with Tris (Figure 8). Pex22p was resistant to extraction with sodium carbonate, as observed previously (Koller et al., 1999), as was Pex17HAp, consistent with the conclusion that Pex17HAp is an integral membrane protein. In the presence of TritonX-100, all peroxisomal proteins were released into the supernatant fractions as expected. These data, together with the prediction that Pex17p contains one membrane-spanning domain, indicate strongly that Pex17p is an integral PMP.
Figure 8.
Membrane extraction of Pex17HAp. Organelle membranes from oleate-grown cells expressing Pex17HAp (SWS17HA) were extracted with the indicated buffers and repelleted. The supernatant (s) and pellet (p) fractions were analyzed by immunoblotting with the indicated antibodies. carbonate, 0.1 M sodium carbonate, pH 11; Tris, 10 mM Tris-HCl, pH 8.0; buffer, original lysis buffer only; 1% Triton X-100, original lysis buffer with 1% Triton X-100.
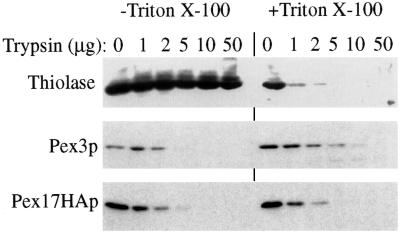
The topology of Pex17HAp at the peroxisome membrane was examined using protease protection experiments. In the absence of detergent, the matrix protein thiolase was completely resistant to exogenously added protease (Figure 9). Pex3p, which contains a large, cytosolic domain, was sensitive to exogenous protease, as observed previously (Koller et al., 1999). Likewise, Pex17HAp was sensitive to protease in the absence of detergent, suggesting that the HA tag was exposed on the cytosolic side of the peroxisome and not in the lumen. In the presence of detergent, all marker proteins, including thiolase, were degraded by exogenous protease, as expected.
Figure 9.
Protease sensitivity of Pex17HAp. Organelle pellet fractions from oleate-grown cells expressing Pex17HAp (SWS17HA) were incubated with the indicated amount of trypsin in the presence (+) or absence (−) of Triton X-100. The samples were analyzed by immunoblotting with the indicated antibodies.
Pex17p Interacts with the PTS–Receptor Docking Complex and Pex19p
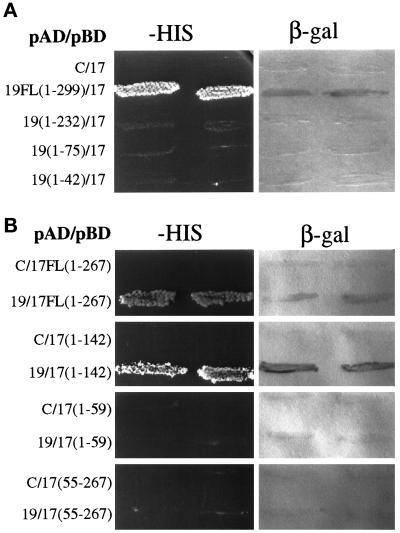
Protein–protein interactions of Pex17p were determined by the yeast two-hybrid system. When expressed as a DNA-binding domain fusion, Pex17p interacted with Pex19p fused to the transcriptional activation domain of LexA, leading to transcriptional activation of the HIS and LacZ/β-galactosidase reporter genes in the yeast tester strain (Figure 10A). The Pex17p DNA-binding domain fusion alone did not activate transcription of the reporter genes (Figure 10A), but Pex19p DNA-binding domain fusions autoactivated the reporter gene (Snyder et al., 1999), so these interactions were only tested with the DNA-binding domain constructs of Pex17p. Subdomains of Pex19p lacking increasing amounts of the Pex19p carboxyl terminus did not interact with Pex17p by the two-hybrid test (Figure 10A), suggesting that the carboxyl terminus of Pex19p (aa 232–299) is required for the interaction with Pex17p. All of these activation domain fusions to Pex19p subdomains were active for interaction with Pex3p (Snyder et al., 1999), suggesting that all of them are expressed and stable.
Figure 10.
Two-hybrid analysis of Pex17p and Pex19p. The indicated Pex17p and Pex19p hybrid protein constructs were tested for trans-activation of the HIS3 gene, resulting in growth on media lacking histidine, or LacZ, resulting in the production of β-galactosidase assayed as described in MATERIALS AND METHODS. Numbers refer to amino acids from Pex19p (A) or Pex17p (B). pAD, transcriptional activation domain fusion constructs; pBD, DNA-binding domain fusion constructs; C, empty DNA-binding domain plasmid (pKNSD55).
To determine the region of Pex17p that interacts with Pex19p, three subdomains of Pex17p were created as DNA-binding domain fusions (Figure 10B). The first subdomain, amino acids 1–142, which stops before the first coiled-coil domain of Pex17p, showed a positive interaction with Pex19p. The smallest, amino-terminal fragment of Pex17p, which includes the amino terminus through the transmembrane domain (aa 1–59), did not interact with Pex19p. The carboxyl-terminal fragment of Pex17p, amino acids 55–267, which is predicted to be the entire cytosolic domain, did not interact with Pex19p. We conclude that the interaction between Pex19p and Pex17p requires the extreme carboxyl terminus of Pex19p but does not require the carboxyl-terminal coiled-coil domain of Pex17p. We did observe a two-hybrid interaction between Pex17p and Pex14p (our unpublished observations), as described previously in S. cerevisiae (Huhse et al., 1998). No other two-hybrid interactions were observed between Pex17p and all other known P. pastoris peroxins (Pex1p, 2, 3, 4, 5, 6, 7, 8, 10, 12, 13, and 22).
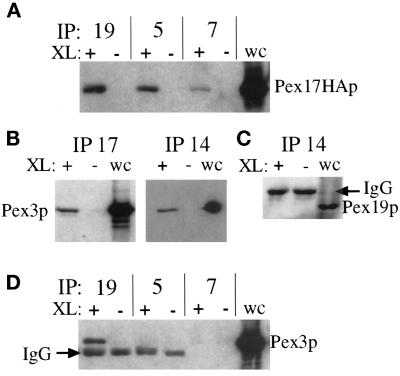
To confirm the interactions we observed in the two-hybrid system and further characterize components of the Pex17p protein complex, we performed coimmunoprecipitation experiments. For these experiments we found it useful to include a cleavable cross-linker, dithiobis(succinimidyl propionate), to covalently link the protein complexes, which were then immunoprecipitated under denaturing conditions. The cross-linked material was dissociated by the addition of reducing agent, which cleaves the cross-linker, before SDS-PAGE and immunoblotting to identify the individual members of the complexes. Immunoprecipitation with Pex19p antisera brought down Pex17HAp in a cross-linker–dependent manner (Figure 11A). The coimmunoprecipitation of Pex17HAp with Pex19p only in the presence of the cross-linker proves the specificity of the coimmunoprecipitation. Likewise, immunoprecipitations with Pex5p and Pex7p antisera also brought down Pex17HAp in a cross-linker–dependent manner. Pex5p and Pex7p have been shown in other species to dock at the peroxisome by binding to Pex14p (Albertini et al., 1997; Brocard et al., 1997; Fransen et al., 1998), and this is thought to mediate the interactions with Pex17p (Huhse et al., 1998). This was true in P. pastoris as well and will be described elsewhere (our unpublished results).
Figure 11.
Cross-linking and coimmunoprecipitation of Pex17HAp with the PTS–receptor docking complex and Pex19p. Immunoprecipitations from cross-linked (+) or non–cross-linked (−) extracts of the Pex17HAp-expressing strain (SWS17HA) were analyzed by immunoblotting. (A) Pex19p, Pex5p, and Pex7p were immunoprecipitated and immunoblotted with anti-HA. (B) Pex17HAp and Pex14p were immunoprecipitated and immunoblotted with anti-Pex3p. (C) Pex14p was immunoprecipitated and immunoblotted with anti-Pex19p. (D) Pex19p, Pex5p, and Pex7p were immunoprecipitated and immunoblotted with anti-Pex3p. Whole-cell lysates (wc) were loaded (0.033 A600) as a control for immunoblotting.
Unexpectedly, immunoprecipitations with the HA antibody, to precipitate Pex17HAp, and with the anti-Pex14p antisera brought down Pex3p (Figure 11B). Pex3p has not previously been seen as part of the receptor docking complex composed of Pex13p, Pex14p, and Pex17p, but these studies were done in S. cerevisiae using immunoprecipitations of Pex7p (Albertini et al., 1997; Huhse et al., 1998). We did not see Pex3p in coimmunoprecipitation experiments with Pex5p or Pex7p antisera in P. pastoris as well (Figure 11D). Moreover, we did not see Pex19p in coimmunoprecipitation experiments with Pex14p antisera (Figure 11C), suggesting that the linkage between Pex14p and Pex3p is not mediated by Pex19p, because it was previously shown that Pex19p interacts with Pex3p (see DISCUSSION). These results confirmed the interactions observed by two-hybrid analysis and suggest that Pex3p is part of the Pex14p–Pex17p complex.
DISCUSSION
We have identified the P. pastoris PEX17 by functional complementation of a pex17 mutant strain obtained from a novel, FACS-based screen for mutants impaired in the ability to localize a PMP reporter, mPTS(Pex3p)-GFP. Although, the amino acid identity between the previously characterized S. cerevisiae Pex17p and PpPex17p is extremely low, the conservation of sequence features such as a putative transmembrane domain and coiled-coil regions, as well as the conservation of protein–protein interactions, supports the conclusion that we have identified PpPex17p. The fact that the sequence identity between PpPex17p and ScPex17p is low, and that PpPex17p is significantly larger, suggests that PpPex17p could have different, or additional, functions beyond that of ScPex17p. PpPex17p behaves as an integral PMP (Figures 7 and 8) with its large carboxyl-terminal domain in the cytosol (Figure 9), whereas ScPex17p, despite having a sequence predicted to form a transmembrane domain, behaves as a peripheral membrane protein on the cytosolic side of the peroxisome membrane (Huhse et al., 1998). In both species, however, the majority of the protein would be facing the cytosol, poised to carry out its function(s).
Role of Pex17p in Peroxisome Biogenesis
Previous studies in S. cerevisiae have implicated Pex17p as a component of unknown function in the receptor docking complex comprising Pex5p, Pex7p, Pex13p, Pex14p, and Pex17p at the peroxisomal membrane (Huhse et al., 1998). This complex functions for the import of proteins via the PTS1 and PTS2 pathways, but there were no data to suggest an involvement of this complex in the biogenesis of PMPs. Our studies clearly show that Pppex17Δ strains are deficient not only in the import of PTS1- and PTS2-containing matrix proteins, as described previously for S. cerevisiae, but also for the localization of integral PMPs, such as Pex3p and Pex22p. This suggests that PpPex17p has an additional role in PMP localization that ScPex17p lacks, or the role for ScPex17p in PMP import was missed. In S. cerevisiae, pex17Δ mutants localize two PMPs, Pex11p and Pex3p, to peroxisome remnants (Huhse et al., 1998), but we are not able to test the localization of Pex11p in Pppex17 mutants because it has not been discovered in P. pastoris.
Because pex17Δ mutants can still partially localize Pex3p to peroxisome remnants, it must not be absolutely required for the import of PMPs to membranous remnants. However, the significant amounts of Pex3p and Pex22p in the cytosol of pex17Δ mutants suggest a major role of Pex17p in membrane protein localization. It should be noted that we do not yet know whether this function for Pex17p would start with the formation of a cytosolic subcomplex containing newly synthesized Pex17p and other integral PMPs, which is then recruited to sites of insertion on peroxisomal or preperoxisomal membranes. Alternatively, Pex17p may be a stable component of an mPTS receptor docking site and/or translocation machinery. In the yeast two-hybrid system, no interactions were detected between P. pastoris Pex17p and either Pex3p or Pex22p. However, in immunoprecipitates of Pex17p, Pex3p was indeed present (Figure 11B), but we do not know whether this complex is found at the peroxisome membrane or formed in the cytosol from newly synthesized polypeptides.
Pex17p interacts with another key component, Pex19p, which was proposed to be required for the conversion of early preperoxisomes to late preperoxisomes (Snyder et al., 1999). We found evidence for this interaction using the yeast two-hybrid system (Figure 10) as well as by coimmunoprecipitation (Figure 11). As noted in earlier studies, Pex19p is primarily cytosolic with a small pool on peroxisomes (Götte et al., 1998; Snyder et al., 1999). Pex19p is known to interact with two peroxisomal integral membrane proteins, Pex3p and Pex10p (Götte et al., 1998; Snyder et al., 1999). Interestingly, Pex19p also interacts with several other integral PMPs such as Pex2p, Pex13p, and Pex22p (our unpublished results). This ability of Pex19p to interact with several different integral PMPs (Pex2p, Pex3p, Pex10p, Pex13p, Pex17p, and Pex22p) while remaining predominantly cytosolic and only partially associated with the peroxisome membrane suggests that Pex19p may bind to newly synthesized, integral PMPs in the cytosol and chaperone their insertion into the peroxisomal membranes. In the absence of Pex19p, these integral PMPs would be inserted, but in such a way as to prevent maturation of early preperoxisomes, the predominant phenotype of pex19Δ mutants (Snyder et al., 1999). Alternatively, Pex19p may interact with these integral PMPs transiently at the peroxisomal membrane to facilitate their assembly into multimeric complexes, which could then lead to remnant maturation. In this respect it is significant that Pex19p interacts with the cytosolic domain of Pex3p and not the mPTS (Snyder et al., 1999). We have not yet defined the topological domains of other PMPs that interact with Pex19p, but this clearly needs to be addressed.
In addition to the role of Pex17p in the biogenesis of PMPs, it also appears to be part of the complex involved in matrix protein import, as described previously in S. cerevisiae (Albertini et al., 1997; Huhse et al., 1998; Girzalsky et al., 1999). Although Pex17p did not interact in the yeast two-hybrid system with the PTS receptors Pex5p and Pex7p, it did interact with Pex14p, which does appear to interact directly with the PTS receptors (our unpublished results), and Pex17p and Pex14p were found together in coimmunoprecipitates (our unpublished results). This confirms that in P. pastoris Pex17p is a component of the PTS–receptor docking complex. Interestingly, immunoprecipitation of either Pex17p or Pex14p brought down Pex3p specifically (Figure 11B), making Pex3p a component of this receptor docking complex that had not been recognized earlier.
In previous studies, Pex3p may have been missed as a member of the import–receptor docking complex, because the components were defined using immunoprecipitations of Pex7p (Huhse et al., 1998; Girzalsky et al., 1999). We were also unable to detect Pex3p in similar immunoprecipitations (Figure 11D). The presence of Pex3p and Pex17p in the matrix protein import complex (consisting of Pex13p, Pex14p, Pex3p, and Pex17p) and in the peroxisome biogenesis complex (Pex19p, Pex3p, and Pex17p) could explain the impairment of both membrane and matrix protein import in the pex17Δ strains, as well as in pex3Δ strains. Previously no PMP-containing remnants have been observed in pex3Δ mutants, which led to the conclusion that there was a strong peroxisome biogenesis defect in the absence of Pex3p (Wiemer et al., 1996).
Evidence for Distinct Peroxin Subcomplexes
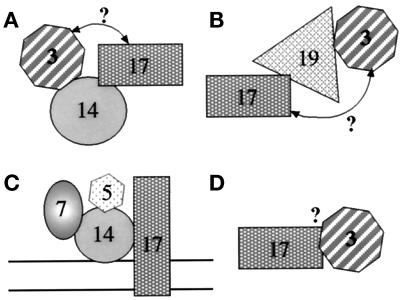
The immunoprecipitation data presented here allow us to start defining subcomplexes that contain separable pools of peroxins (Figure 12). Because Pex14p–Pex17p, Pex17p–Pex19p, and Pex19p–Pex3p interact, it was a formal possibility that the Pex14p–Pex3p complex detected by coimmunoprecipitation (Figure 11B) might have been bridged by Pex19p–Pex17p. This cannot be true, because no Pex19p was found in the immunoprecipitate with Pex14p (Figure 11C) under the same conditions in which Pex19p and Pex3p form a complex (Figure 11D). This argues that separable pools of Pex3p are in a complex with Pex14p (Figure 12A) and Pex19p (Figure 12B). Likewise, the Pex17p that forms a complex with Pex19p must be a separate pool from the Pex17p that forms a complex with Pex14p (Figure 12, A and B). In addition, the pool of Pex17p that forms a complex with Pex5p and Pex7p (Figure 12C) is separate from the pool of Pex17p that complexes with Pex3p and/or Pex19p (Figure 12B), because no Pex3p (Figure 11D) or Pex19p (our unpublished results) has been observed in the Pex5p and Pex7p immunoprecipitations. The mere suggestion of these separable pools hints that complex formation between Pex19p and Pex17p may have a separate role in peroxisome biogenesis from the complex of Pex17p and Pex14p, perhaps the former for membrane protein import and the latter for matrix protein import. We cannot determine from these experiments whether the pool of Pex3p that is in a complex with Pex17p is bridged by Pex19p (Figure 12B) or Pex14p (Figure 12A) or a direct interaction (Figure 12D). Nonetheless, these data point to the diversity of peroxin subcomplexes. We must stress that these separable pools of a peroxin may be physically separated from each other, or they may represent different conformational states of the peroxin, because cross-linking depends on spacing between primary amine residues of the polypeptide chain. Moreover, because the same cross-linker and identical conditions were used in all the immunoprecipitations, the identification or absence of certain protein–protein interactions within a complex depends on the conformational states of the proteins in that complex.
Figure 12.
Schematic representation of various putative Pex protein subcomplexes. The rationale for the definition of these complexes is explained in DISCUSSION. The question mark by the arrows represents a putative bridged interaction, which is not proven or disproven by the data. Because complex formation may occur on the membrane or in the cytosol, no membrane bilayer was included in the diagram for A, B, and D.
Future work will be aimed at determining whether the role of Pex17p in membrane protein import begins in the cytosol (with Pex3p and/or Pex19p) or as a stable, catalytic component of the peroxisome membrane and matrix protein import machinery. In addition, it will be crucial to determine whether certain subcomplexes are formed only during new protein synthesis, suggestive of the dynamics of biogenesis, or whether other complexes are stable components of the peroxisome membrane in the absence of new protein synthesis. Furthermore, the requirement for ATP hydrolysis in the formation and stability of these subcomplexes needs to be addressed, as well as the site of formation of these subcomplexes in either the cytosol or after insertion into the peroxisome membrane. Finally, as additional antibodies and reagents for studying P. pastoris integral PMPs become available, we should be able to address whether Pex17p plays a general role in the insertion of all integral PMPs in the membrane.
ACKNOWLEDGMENTS
We thank Su Hua for technical assistance and Scott Emr for continued access to his microscope. We appreciate the advice of Peter Rehling and Markus Babst. This work was supported by fellowships from the American Cancer Society (to W.B.S.) and the Swiss National Funds (to A.K.). Funding was provided by National Institutes of Health grant DK-41737 (to S.S.) and National Institutes of Health grant DK-43698 (to J.M.C.).
Abbreviations used:
- FACS
fluorescence-activated cell sorter
- GFP
green fluorescent protein
- HA
hemagglutinin
- mPTS
integral peroxisomal membrane protein targeting signal
- NTG
N-methyl-N-nitro-N-nitrosoguanidine
- ORF
open reading frame, P, pellet
- PMP
peroxisomal membrane protein
- PNS
postnuclear supernatant
- PTS
peroxisome targeting signal
- S
supernatant
- TCA
trichloroacetic acid
Footnotes
The nucleotide sequence of PpPEX17 has been submitted to GenBank with accession number AF179352.
REFERENCES
- Albertini M, Rehling P, Erdmann R, Girzalsky W, Kiel JA, Veenhuis M, Kunau WH. Pex14p, a peroxisomal membrane protein binding both receptors of the two PTS-dependent import pathways. Cell. 1997;89:83–92. doi: 10.1016/s0092-8674(00)80185-3. [DOI] [PubMed] [Google Scholar]
- Babst M, Sato TK, Banta LM, Emr SD. Endosomal transport function in yeast requires a novel AAA-type ATPase, Vps4p. EMBO J. 1997;16:1820–1831. doi: 10.1093/emboj/16.8.1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babst M, Wendland B, Estepa EJ, Emr SD. The Vps4p AAA ATPase regulates membrane association of a Vps protein complex required for normal endosome function. EMBO J. 1998;17:2982–2993. doi: 10.1093/emboj/17.11.2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baerends RJS, Rasmussen SW, Hilbrands RE, van der Heide M, Faber KN, Reuvekamp PTW, Kiel JAKW, Cregg JM, van der Klei I, Veenhuis M. The Hansenula polymorpha PER9 gene encodes a peroxisomal membrane protein essential for peroxisome assembly and integrity. J Biol Chem. 1996;271:8887–8894. doi: 10.1074/jbc.271.15.8887. [DOI] [PubMed] [Google Scholar]
- Bodnar AG, Rachubinski RA. Characterization of the integral membrane polypeptides of rat liver peroxisomes isolated from untreated and clofibrate-treated rats. Biochem Cell Biol. 1991;69:499–508. doi: 10.1139/o91-074. [DOI] [PubMed] [Google Scholar]
- Brocard C, Lametschwandtner G, Koudelka R, Hartig A. Pex14p is a member of the protein linkage map of Pex5p. EMBO J. 1997;16:5491–5500. doi: 10.1093/emboj/16.18.5491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crookes WJ, Olsen LJ. Peroxin puzzles and folded freight: peroxisomal protein import in review. Naturwissenschaften. 1999;86:51–61. doi: 10.1007/s001140050572. [DOI] [PubMed] [Google Scholar]
- Distel B, et al. A unified nomenclature for peroxisome biogenesis factors. J Cell Biol. 1996;135:1–3. doi: 10.1083/jcb.135.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodt G, Gould SJ. Multiple PEX genes are required for proper subcellular distribution and stability of Pex5p, the PTS1 receptor: evidence that PTS1 protein import is mediated by a cycling receptor. J Cell Biol. 1996;135:1763–1774. doi: 10.1083/jcb.135.6.1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyer JM, McNew JA, Goodman JM. The sorting sequence of the peroxisomal integral membrane protein PMP47 is contained within a short hydrophilic loop. J Cell Biol. 1996;133:269–280. doi: 10.1083/jcb.133.2.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elgersma Y, Elgersma HM, Wenzel T, McCaffery JM, Farquhar MG, Subramani S. A mobile PTS2 receptor for peroxisomal protein import in Pichia pastoris. J Cell Biol. 1998;140:807–820. doi: 10.1083/jcb.140.4.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elgersma Y, Kwast L, Klein A, Voorn-Brouwer T, van den Berg M, Metzig B, America T, Tabak H, Distel B. The SH3 domain of the peroxisomal membrane protein Pex13p functions as a docking site for Pex5p, a mobile receptor for peroxisomal proteins. J Cell Biol. 1996a;135:97–109. doi: 10.1083/jcb.135.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elgersma Y, Kwast L, van den Berg M, Snyder WB, Distel B, Subramani S, Tabak HF. Overexpression of Pex15p, a phosphorylated peroxisomal integral membrane protein required for peroxisome assembly in S. cerevisiae, causes proliferation of the endoplasmic reticulum membrane. EMBO J. 1997;16:7326–7341. doi: 10.1093/emboj/16.24.7326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elgersma Y, Vos A, van den Berg M, van Roermund CW, van der Sluijs P, Distel B, Tabak HF. Analysis of the carboxyl-terminal peroxisomal targeting signal 1 in a homologous context in Saccharomyces cerevisiae. J Biol Chem. 1996b;271:26375–26382. doi: 10.1074/jbc.271.42.26375. [DOI] [PubMed] [Google Scholar]
- Erdmann R, Blobel G. Identification of Pex13p a peroxisomal membrane receptor for the PTS1 recognition factor. J Cell Biol. 1996;135:111–121. doi: 10.1083/jcb.135.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faber KN, Heyman JA, Subramani S. Two AAA family peroxins, PpPex1p and PpPex6p, interact with each other in an ATP-dependent manner and are associated with different subcellular membranous structures distinct from peroxisomes. Mol Cell Biol. 1998;18:936–943. doi: 10.1128/mcb.18.2.936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fransen M, Terlecky SR, Subramani S. Identification of a human PTS1 receptor docking protein directly required for peroxisomal protein import. Proc Natl Acad Sci USA. 1998;95:8087–8092. doi: 10.1073/pnas.95.14.8087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiki Y, Rachubinski RA, Lazarow PB. Synthesis of a major integral membrane polypeptide of rat liver peroxisomes on free polysomes. Proc Natl Acad Sci USA. 1984;81:7127–7131. doi: 10.1073/pnas.81.22.7127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girzalsky W, Rehling P, Stein K, Kipper J, Blank L, Kunau WH, Erdmann R. Involvement of Pex13p in Pex14p localization and peroxisomal targeting signal 2-dependent protein import into peroxisomes. J Cell Biol. 1999;144:1151–1162. doi: 10.1083/jcb.144.6.1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Götte K, Girzalsky W, Linkert M, Baumgart E, Kammerer S, Kunau WH, Erdmann R. Pex19p, a farnesylated protein essential for peroxisome biogenesis. Mol Cell Biol. 1998;18:616–628. doi: 10.1128/mcb.18.1.616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould SJ, Kalish JE, Morrell JC, Bjorkman J, Urquhart AJ, Crane DI. Pex13p is an SH3 protein of the peroxisome membrane and a docking factor for the predominantly cytoplasmic PTS1 receptor. J Cell Biol. 1996;135:85–95. doi: 10.1083/jcb.135.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould SJ, Keller G-A, Subramani S. Identification of a peroxisomal targeting signal at the carboxy terminus of firefly luciferase. J Cell Biol. 1987;105:2923–2931. doi: 10.1083/jcb.105.6.2923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould SJ, Keller GA, Hosken N, Wilkinson J, Subramani S. A conserved tripeptide sorts proteins to peroxisomes. J Cell Biol. 1989;108:1657–1664. doi: 10.1083/jcb.108.5.1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould SJ, McCollum D, Spong AP, Heyman JA, Subramani S. Development of the yeast Pichia pastoris as a model organism for a genetic and molecular analysis of peroxisome assembly. Yeast. 1992;8:613–628. doi: 10.1002/yea.320080805. [DOI] [PubMed] [Google Scholar]
- Höhfeld J, Martens D, Franziska FF, Kunau WH. Defining components required for peroxisome assembly in Saccharomyces cerevisiae. In: Neupert W, Lill R, editors. Membrane Biogenesis and Protein Targeting, New Comprehensive Biochemistry. Vol. 22. New York: Elsevier; 1992. pp. 185–207. [Google Scholar]
- Höhfeld J, Veenhuis M, Kunau WH. PAS3, a Saccharomyces cerevisiae gene encoding a peroxisomal integral membrane protein essential for peroxisome biogenesis. J Cell Biol. 1991;114:1167–1178. doi: 10.1083/jcb.114.6.1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huhse B, Rehling P, Albertini M, Blank L, Meller K, Kunau WH. Pex17p of Saccharomyces cerevisiae is a novel peroxin and component of the peroxisomal protein translocation machinery. J Cell Biol. 1998;140:49–60. doi: 10.1083/jcb.140.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalish JE, Keller GA, Morrell JC, Mihalik SJ, Smith B, Cregg JM, Gould SJ. Characterization of a novel component of the peroxisomal protein import apparatus using fluorescent peroxisomal proteins. EMBO J. 1996;15:3275–3285. [PMC free article] [PubMed] [Google Scholar]
- Kammerer S, Holzinger A, Welsch U, Roscher AA. Cloning and characterization of the gene encoding the human peroxisomal assembly protein Pex3p. FEBS Lett. 1998;429:53–60. doi: 10.1016/s0014-5793(98)00557-2. [DOI] [PubMed] [Google Scholar]
- Koller A, Snyder WB, Faber KN, Wenzel TJ, Rangell L, Keller GA, Subramani S. Pex22p of Pichia pastoris, essential for peroxisome matrix protein import, anchors the ubiquitin-conjugating enzyme, Pex4p, on the peroxisome membrane. J Cell Biol. 1999;146:99–112. doi: 10.1083/jcb.146.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunau WH, Erdmann R. Peroxisome biogenesis: back to the endoplasmic reticulum? Curr Biol. 1998;8:299–302. doi: 10.1016/s0960-9822(98)70191-5. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lazarow PB, Fujiki Y. Biogenesis of peroxisomes. Annu Rev Cell Biol. 1985;1:489–530. doi: 10.1146/annurev.cb.01.110185.002421. [DOI] [PubMed] [Google Scholar]
- Liu H, Tan X, Russell KA, Veenhuis M, Cregg JM. PER3, a gene required for peroxisome biogenesis in Pichia pastoris, encodes a peroxisomal membrane protein involved in protein import. J Biol Chem. 1995;270:10940–10951. doi: 10.1074/jbc.270.18.10940. [DOI] [PubMed] [Google Scholar]
- Lupas A. Prediction and analysis of coiled-coil structures. Methods Enzymol. 1996;266:513–525. doi: 10.1016/s0076-6879(96)66032-7. [DOI] [PubMed] [Google Scholar]
- Marzioch M, Erdmann R, Veenhuis M, Kunau WH. PAS7 encodes a novel yeast member of the WD-40 protein family essential for import of 3-oxoacyl-CoA thiolase, a PTS2-containing protein, into peroxisomes. EMBO J. 1994;13:4908–4918. doi: 10.1002/j.1460-2075.1994.tb06818.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzono Y, Kinoshita N, Tamura S, Shimozawa N, Hamasaki M, Ghaedi K, Wanders RJ, Suzuki Y, Kondo N, Fujiki Y. Human PEX19: cDNA cloning by functional complementation, mutation analysis in a patient with Zellweger syndrome, and potential role in peroxisomal membrane assembly. Proc Natl Acad Sci USA. 1999;96:2116–2121. doi: 10.1073/pnas.96.5.2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monosov EZ, Wenzel TJ, Luers GH, Heyman JA, Subramani S. Labeling of peroxisomes with green fluorescent protein in living P. pastoris cells. J Histochem Cytochem. 1996;44:581–589. doi: 10.1177/44.6.8666743. [DOI] [PubMed] [Google Scholar]
- Odorizzi G, Babst M, Emr SD. Fab1p PtdIns(3)P5-kinase function essential for protein sorting in the multivesicular body. Cell. 1998;95:847–858. doi: 10.1016/s0092-8674(00)81707-9. [DOI] [PubMed] [Google Scholar]
- Osumi T, Tsukamoto T, Hata S, Yokota S, Miura S, Fujiki Y, Hijikata M, Miyazawa S, Hashimoto T. Amino-terminal presequence of the precursor of peroxisomal 3-ketoacyl-CoA thiolase is a cleavable signal peptide for peroxisomal targeting. Biochem Biophys Res Commun. 1991;181:947–954. doi: 10.1016/0006-291x(91)92028-i. [DOI] [PubMed] [Google Scholar]
- Purdue PE, Yang X, Lazarow PB. Pex18p and Pex21p, a novel pair of peroxins essential for peroxisomal targeting by the PTS2 pathway. J Cell Biol. 1998;143:1859–1869. doi: 10.1083/jcb.143.7.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehling P, Marzioch M, Niesen F, Wittke E, Veenhuis M, Kunau WH. The import receptor for the peroxisomal targeting signal 2 (PTS2) in Saccharomyces cerevisiae is encoded by the PAS7 gene. EMBO J. 1996;15:2901–2913. [PMC free article] [PubMed] [Google Scholar]
- Rieder SE, Emr SD. A novel RING finger protein complex essential for a late step in protein transport to the yeast vacuole. Mol Biol Cell. 1997;8:2307–2327. doi: 10.1091/mbc.8.11.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai Y, Koller A, Rangell LK, Keller GA, Subramani S. Peroxisome degradation by microautophagy in Pichia pastoris: identification of specific steps and morphological intermediates. J Cell Biol. 1998;141:625–636. doi: 10.1083/jcb.141.3.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schliebs W, Saidowsky J, Agianian B, Dodt G, Herberg FW, Kunau WH. Recombinant human peroxisomal targeting signal receptor PEX5. Structural basis for interaction of PEX5 with PEX14. J Biol Chem. 1999;274:5666–5673. doi: 10.1074/jbc.274.9.5666. [DOI] [PubMed] [Google Scholar]
- Sears IB, O’Connor J, Rossanese OW, Glick BS. A versatile set of vectors for constitutive and regulated gene expression in Pichia pastoris. Yeast. 1998;14:783–790. doi: 10.1002/(SICI)1097-0061(19980615)14:8<783::AID-YEA272>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Shimizu N, et al. The peroxin Pex14p. cDNA cloning by functional complementation on a Chinese hamster ovary cell mutant, characterization, and functional analysis. J Biol Chem. 1999;274:12593–12604. doi: 10.1074/jbc.274.18.12593. [DOI] [PubMed] [Google Scholar]
- Shimozawa N, et al. Nonsense and temperature-sensitive mutations in PEX13 are the cause of complementation group H of peroxisome biogenesis disorders. Hum Mol Genet. 1999;8:1077–1083. doi: 10.1093/hmg/8.6.1077. [DOI] [PubMed] [Google Scholar]
- Snyder WB, Faber KN, Wenzel TJ, Koller A, Luers GH, Rangell L, Keller GA, Subramani S. Pex19p interacts with Pex3p and Pex10p and is essential for peroxisome biogenesis in Pichia pastoris. Mol Biol Cell. 1999;10:1745–1761. doi: 10.1091/mbc.10.6.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- South ST, Gould SJ. Peroxisome synthesis in the absence of preexisting peroxisomes. J Cell Biol. 1999;144:255–266. doi: 10.1083/jcb.144.2.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramani S. PEX genes on the rise. Nat Genet. 1997;15:331–333. doi: 10.1038/ng0497-331. [DOI] [PubMed] [Google Scholar]
- Subramani S. Components involved in peroxisome import, biogenesis, proliferation, turnover, and movement. Physiol Rev. 1998;78:171–188. doi: 10.1152/physrev.1998.78.1.171. [DOI] [PubMed] [Google Scholar]
- Suzuki Y, Orii T, Takiguchi M, Mori M, Hijikata M, Osumi T. Biosynthesis of membrane polypeptides of rat liver peroxisomes. J Biochem. 1987;101:491–496. doi: 10.1093/oxfordjournals.jbchem.a121935. [DOI] [PubMed] [Google Scholar]
- Swinkels BW, Gould SJ, Bodnar AG, Rachubinski RA, Subramani S. A novel, cleavable peroxisomal targeting signal at the amino-terminus of the rat 3-ketoacyl-CoA thiolase. EMBO J. 1991;10:3255–3262. doi: 10.1002/j.1460-2075.1991.tb04889.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titorenko VI, Rachubinski RA. The endoplasmic reticulum plays an essential role in peroxisome biogenesis. Trends Biochem Sci. 1998;23:231–233. doi: 10.1016/s0968-0004(98)01226-2. [DOI] [PubMed] [Google Scholar]
- Titorenko VI, Smith JJ, Szilard RK, Rachubinski RA. Pex20p of the yeast Yarrowia lipolytica is required for the oligomerization of thiolase in the cytosol and for its targeting to the peroxisome. J Cell Biol. 1998;142:403–420. doi: 10.1083/jcb.142.2.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wach A, Brachat A, Pohlmann R, Philippsen P. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast. 1994;10:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- Waterham HR, de Vries Y, Russel KA, Xie W, Veenhuis M, Cregg JM. The Pichia pastoris PER6 gene product is a peroxisomal integral membrane protein essential for peroxisome biogenesis and has sequence similarity to the Zellweger syndrome protein PAF-1. Mol Cell Biol. 1996;16:2527–2536. doi: 10.1128/mcb.16.5.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiemer EAC, Luers G, Faber KN, Wenzel T, Veenhuis M, Subramani S. Isolation and characterization of Pas2p, a peroxisomal membrane protein essential for peroxisome biogenesis in the methylotrophic yeast Pichia pastoris. J Biol Chem. 1996;271:18973–18980. doi: 10.1074/jbc.271.31.18973. [DOI] [PubMed] [Google Scholar]
- Will GK, Soukupova M, Hong X, Erdmann KS, Kiel JA, Dodt G, Kunau WH, Erdmann R. Identification and characterization of the human orthologue of yeast Pex14p. Mol Cell Biol. 1999;19:2265–2277. doi: 10.1128/mcb.19.3.2265. [DOI] [PMC free article] [PubMed] [Google Scholar]