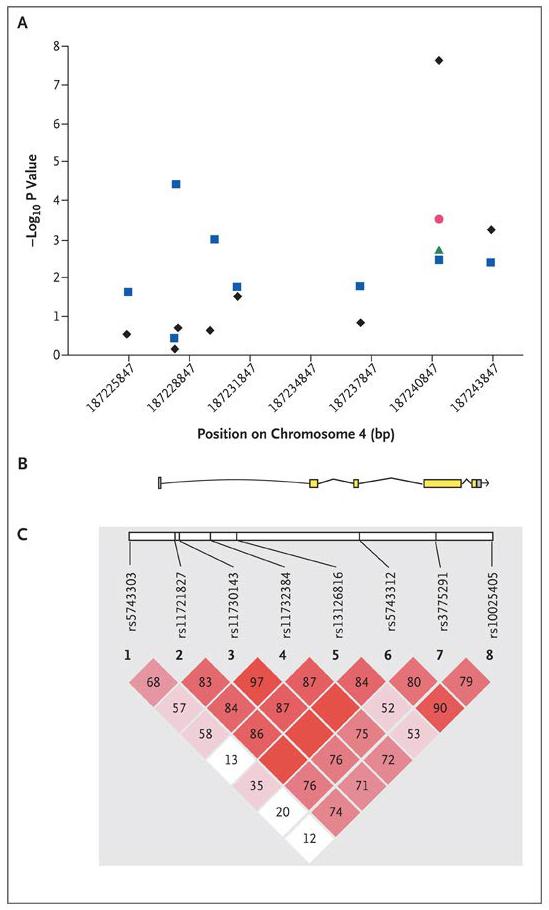
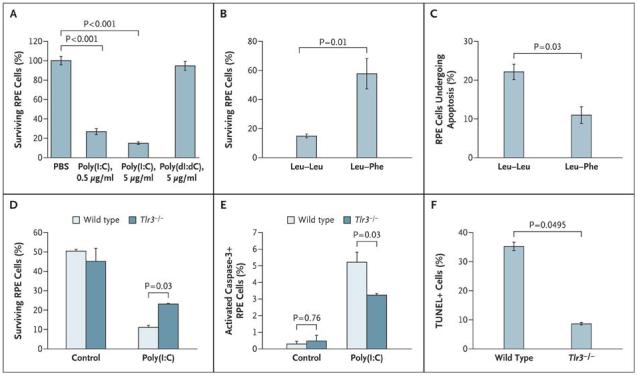
Zhenglin Yang
Zhenglin Yang, M.D.
1Center for Human Molecular Biology and Genetics, Sichuan Academy of Medical Sciences and Sichuan Provincial People's Hospital, Chengdu, China (Z.Y.); Department of Ophthalmology & Visual Sciences, Moran Eye Center, University of Utah School of Medicine, Salt Lake City, UT (Z.Y., C.S., D.G., Z.T., H.C., R.C., X.Y., Y. C., J.Z., X.M., V.S.H., J.H., J.B., E.P., S.P., Y.K., L.L., N.A.Z., P.S.B., A.S., B.Y., K.Z.); Program in Human Molecular Biology & Genetics, Eccles Institute of Human Genetics, University of Utah School of Medicine, Salt Lake City, UT (C.S., D.G., Z.T., H.C., R.C., X.Y., H.C., J.Z., X.M., J.H., J.B., E.P., S.P., Y.K., L.L., B.Y., K.Z.); Department of Ophthalmology, Shiley Eye Center, University of California San Diego, San Diego, CA (Z.Y., Y.C., J.Z., K.Z.); Macular Degeneration Center, Casey Eye Institute, Oregon Health & Science University, Portland, OR (P.J.F., M.L.K., E.S.); Department of Ophthalmology & Visual Sciences, University of Kentucky, Lexington, KY (M.E.K., W.C., J.A.); Department of Human Genetics, University of Utah School of Medicine, Salt Lake City, UT (S.W., L.J.); Institute of Genetic Medicine (P.L.T., L.D., N.K.), Department of Molecular Biology and Genetics (N.K.), Department of Oncology (G.P.), Department of Biostatistics (C.W., G.P.), and Wilmer Eye Institute (J.S.S., P.C., B.C., D.J.Z., N.K.), Johns Hopkins University, Baltimore, MD; Richard E. Hoover Rehabilitation Services for Low Vision and Blindness, Greater Baltimore Medical Center, Baltimore, MD (J.S.S.); Departments of Pathology and Ophthalmology, Keck School of Medicine of the University of Southern California, Doheny Eye Institute, Los Angeles, CA (D.R.H.); Laboratory of Statistical Genetics, Rockefeller University, 1230 York Avenue, New York, NY (S.C.H.).
1,*,
Charity Stratton
Charity Stratton, B.S.
1Center for Human Molecular Biology and Genetics, Sichuan Academy of Medical Sciences and Sichuan Provincial People's Hospital, Chengdu, China (Z.Y.); Department of Ophthalmology & Visual Sciences, Moran Eye Center, University of Utah School of Medicine, Salt Lake City, UT (Z.Y., C.S., D.G., Z.T., H.C., R.C., X.Y., Y. C., J.Z., X.M., V.S.H., J.H., J.B., E.P., S.P., Y.K., L.L., N.A.Z., P.S.B., A.S., B.Y., K.Z.); Program in Human Molecular Biology & Genetics, Eccles Institute of Human Genetics, University of Utah School of Medicine, Salt Lake City, UT (C.S., D.G., Z.T., H.C., R.C., X.Y., H.C., J.Z., X.M., J.H., J.B., E.P., S.P., Y.K., L.L., B.Y., K.Z.); Department of Ophthalmology, Shiley Eye Center, University of California San Diego, San Diego, CA (Z.Y., Y.C., J.Z., K.Z.); Macular Degeneration Center, Casey Eye Institute, Oregon Health & Science University, Portland, OR (P.J.F., M.L.K., E.S.); Department of Ophthalmology & Visual Sciences, University of Kentucky, Lexington, KY (M.E.K., W.C., J.A.); Department of Human Genetics, University of Utah School of Medicine, Salt Lake City, UT (S.W., L.J.); Institute of Genetic Medicine (P.L.T., L.D., N.K.), Department of Molecular Biology and Genetics (N.K.), Department of Oncology (G.P.), Department of Biostatistics (C.W., G.P.), and Wilmer Eye Institute (J.S.S., P.C., B.C., D.J.Z., N.K.), Johns Hopkins University, Baltimore, MD; Richard E. Hoover Rehabilitation Services for Low Vision and Blindness, Greater Baltimore Medical Center, Baltimore, MD (J.S.S.); Departments of Pathology and Ophthalmology, Keck School of Medicine of the University of Southern California, Doheny Eye Institute, Los Angeles, CA (D.R.H.); Laboratory of Statistical Genetics, Rockefeller University, 1230 York Avenue, New York, NY (S.C.H.).
1,*,
Peter J Francis
Peter J Francis, M.D., Ph.D.
1Center for Human Molecular Biology and Genetics, Sichuan Academy of Medical Sciences and Sichuan Provincial People's Hospital, Chengdu, China (Z.Y.); Department of Ophthalmology & Visual Sciences, Moran Eye Center, University of Utah School of Medicine, Salt Lake City, UT (Z.Y., C.S., D.G., Z.T., H.C., R.C., X.Y., Y. C., J.Z., X.M., V.S.H., J.H., J.B., E.P., S.P., Y.K., L.L., N.A.Z., P.S.B., A.S., B.Y., K.Z.); Program in Human Molecular Biology & Genetics, Eccles Institute of Human Genetics, University of Utah School of Medicine, Salt Lake City, UT (C.S., D.G., Z.T., H.C., R.C., X.Y., H.C., J.Z., X.M., J.H., J.B., E.P., S.P., Y.K., L.L., B.Y., K.Z.); Department of Ophthalmology, Shiley Eye Center, University of California San Diego, San Diego, CA (Z.Y., Y.C., J.Z., K.Z.); Macular Degeneration Center, Casey Eye Institute, Oregon Health & Science University, Portland, OR (P.J.F., M.L.K., E.S.); Department of Ophthalmology & Visual Sciences, University of Kentucky, Lexington, KY (M.E.K., W.C., J.A.); Department of Human Genetics, University of Utah School of Medicine, Salt Lake City, UT (S.W., L.J.); Institute of Genetic Medicine (P.L.T., L.D., N.K.), Department of Molecular Biology and Genetics (N.K.), Department of Oncology (G.P.), Department of Biostatistics (C.W., G.P.), and Wilmer Eye Institute (J.S.S., P.C., B.C., D.J.Z., N.K.), Johns Hopkins University, Baltimore, MD; Richard E. Hoover Rehabilitation Services for Low Vision and Blindness, Greater Baltimore Medical Center, Baltimore, MD (J.S.S.); Departments of Pathology and Ophthalmology, Keck School of Medicine of the University of Southern California, Doheny Eye Institute, Los Angeles, CA (D.R.H.); Laboratory of Statistical Genetics, Rockefeller University, 1230 York Avenue, New York, NY (S.C.H.).
1,*,
Mark E Kleinman
Mark E Kleinman, M.D.
1Center for Human Molecular Biology and Genetics, Sichuan Academy of Medical Sciences and Sichuan Provincial People's Hospital, Chengdu, China (Z.Y.); Department of Ophthalmology & Visual Sciences, Moran Eye Center, University of Utah School of Medicine, Salt Lake City, UT (Z.Y., C.S., D.G., Z.T., H.C., R.C., X.Y., Y. C., J.Z., X.M., V.S.H., J.H., J.B., E.P., S.P., Y.K., L.L., N.A.Z., P.S.B., A.S., B.Y., K.Z.); Program in Human Molecular Biology & Genetics, Eccles Institute of Human Genetics, University of Utah School of Medicine, Salt Lake City, UT (C.S., D.G., Z.T., H.C., R.C., X.Y., H.C., J.Z., X.M., J.H., J.B., E.P., S.P., Y.K., L.L., B.Y., K.Z.); Department of Ophthalmology, Shiley Eye Center, University of California San Diego, San Diego, CA (Z.Y., Y.C., J.Z., K.Z.); Macular Degeneration Center, Casey Eye Institute, Oregon Health & Science University, Portland, OR (P.J.F., M.L.K., E.S.); Department of Ophthalmology & Visual Sciences, University of Kentucky, Lexington, KY (M.E.K., W.C., J.A.); Department of Human Genetics, University of Utah School of Medicine, Salt Lake City, UT (S.W., L.J.); Institute of Genetic Medicine (P.L.T., L.D., N.K.), Department of Molecular Biology and Genetics (N.K.), Department of Oncology (G.P.), Department of Biostatistics (C.W., G.P.), and Wilmer Eye Institute (J.S.S., P.C., B.C., D.J.Z., N.K.), Johns Hopkins University, Baltimore, MD; Richard E. Hoover Rehabilitation Services for Low Vision and Blindness, Greater Baltimore Medical Center, Baltimore, MD (J.S.S.); Departments of Pathology and Ophthalmology, Keck School of Medicine of the University of Southern California, Doheny Eye Institute, Los Angeles, CA (D.R.H.); Laboratory of Statistical Genetics, Rockefeller University, 1230 York Avenue, New York, NY (S.C.H.).
1,*,
Perciliz L Tan
Perciliz L Tan, B.S.
1Center for Human Molecular Biology and Genetics, Sichuan Academy of Medical Sciences and Sichuan Provincial People's Hospital, Chengdu, China (Z.Y.); Department of Ophthalmology & Visual Sciences, Moran Eye Center, University of Utah School of Medicine, Salt Lake City, UT (Z.Y., C.S., D.G., Z.T., H.C., R.C., X.Y., Y. C., J.Z., X.M., V.S.H., J.H., J.B., E.P., S.P., Y.K., L.L., N.A.Z., P.S.B., A.S., B.Y., K.Z.); Program in Human Molecular Biology & Genetics, Eccles Institute of Human Genetics, University of Utah School of Medicine, Salt Lake City, UT (C.S., D.G., Z.T., H.C., R.C., X.Y., H.C., J.Z., X.M., J.H., J.B., E.P., S.P., Y.K., L.L., B.Y., K.Z.); Department of Ophthalmology, Shiley Eye Center, University of California San Diego, San Diego, CA (Z.Y., Y.C., J.Z., K.Z.); Macular Degeneration Center, Casey Eye Institute, Oregon Health & Science University, Portland, OR (P.J.F., M.L.K., E.S.); Department of Ophthalmology & Visual Sciences, University of Kentucky, Lexington, KY (M.E.K., W.C., J.A.); Department of Human Genetics, University of Utah School of Medicine, Salt Lake City, UT (S.W., L.J.); Institute of Genetic Medicine (P.L.T., L.D., N.K.), Department of Molecular Biology and Genetics (N.K.), Department of Oncology (G.P.), Department of Biostatistics (C.W., G.P.), and Wilmer Eye Institute (J.S.S., P.C., B.C., D.J.Z., N.K.), Johns Hopkins University, Baltimore, MD; Richard E. Hoover Rehabilitation Services for Low Vision and Blindness, Greater Baltimore Medical Center, Baltimore, MD (J.S.S.); Departments of Pathology and Ophthalmology, Keck School of Medicine of the University of Southern California, Doheny Eye Institute, Los Angeles, CA (D.R.H.); Laboratory of Statistical Genetics, Rockefeller University, 1230 York Avenue, New York, NY (S.C.H.).
1,
Daniel Gibbs
Daniel Gibbs, BA
1Center for Human Molecular Biology and Genetics, Sichuan Academy of Medical Sciences and Sichuan Provincial People's Hospital, Chengdu, China (Z.Y.); Department of Ophthalmology & Visual Sciences, Moran Eye Center, University of Utah School of Medicine, Salt Lake City, UT (Z.Y., C.S., D.G., Z.T., H.C., R.C., X.Y., Y. C., J.Z., X.M., V.S.H., J.H., J.B., E.P., S.P., Y.K., L.L., N.A.Z., P.S.B., A.S., B.Y., K.Z.); Program in Human Molecular Biology & Genetics, Eccles Institute of Human Genetics, University of Utah School of Medicine, Salt Lake City, UT (C.S., D.G., Z.T., H.C., R.C., X.Y., H.C., J.Z., X.M., J.H., J.B., E.P., S.P., Y.K., L.L., B.Y., K.Z.); Department of Ophthalmology, Shiley Eye Center, University of California San Diego, San Diego, CA (Z.Y., Y.C., J.Z., K.Z.); Macular Degeneration Center, Casey Eye Institute, Oregon Health & Science University, Portland, OR (P.J.F., M.L.K., E.S.); Department of Ophthalmology & Visual Sciences, University of Kentucky, Lexington, KY (M.E.K., W.C., J.A.); Department of Human Genetics, University of Utah School of Medicine, Salt Lake City, UT (S.W., L.J.); Institute of Genetic Medicine (P.L.T., L.D., N.K.), Department of Molecular Biology and Genetics (N.K.), Department of Oncology (G.P.), Department of Biostatistics (C.W., G.P.), and Wilmer Eye Institute (J.S.S., P.C., B.C., D.J.Z., N.K.), Johns Hopkins University, Baltimore, MD; Richard E. Hoover Rehabilitation Services for Low Vision and Blindness, Greater Baltimore Medical Center, Baltimore, MD (J.S.S.); Departments of Pathology and Ophthalmology, Keck School of Medicine of the University of Southern California, Doheny Eye Institute, Los Angeles, CA (D.R.H.); Laboratory of Statistical Genetics, Rockefeller University, 1230 York Avenue, New York, NY (S.C.H.).
1,
Zongzhong Tong
Zongzhong Tong, Ph.D.
1Center for Human Molecular Biology and Genetics, Sichuan Academy of Medical Sciences and Sichuan Provincial People's Hospital, Chengdu, China (Z.Y.); Department of Ophthalmology & Visual Sciences, Moran Eye Center, University of Utah School of Medicine, Salt Lake City, UT (Z.Y., C.S., D.G., Z.T., H.C., R.C., X.Y., Y. C., J.Z., X.M., V.S.H., J.H., J.B., E.P., S.P., Y.K., L.L., N.A.Z., P.S.B., A.S., B.Y., K.Z.); Program in Human Molecular Biology & Genetics, Eccles Institute of Human Genetics, University of Utah School of Medicine, Salt Lake City, UT (C.S., D.G., Z.T., H.C., R.C., X.Y., H.C., J.Z., X.M., J.H., J.B., E.P., S.P., Y.K., L.L., B.Y., K.Z.); Department of Ophthalmology, Shiley Eye Center, University of California San Diego, San Diego, CA (Z.Y., Y.C., J.Z., K.Z.); Macular Degeneration Center, Casey Eye Institute, Oregon Health & Science University, Portland, OR (P.J.F., M.L.K., E.S.); Department of Ophthalmology & Visual Sciences, University of Kentucky, Lexington, KY (M.E.K., W.C., J.A.); Department of Human Genetics, University of Utah School of Medicine, Salt Lake City, UT (S.W., L.J.); Institute of Genetic Medicine (P.L.T., L.D., N.K.), Department of Molecular Biology and Genetics (N.K.), Department of Oncology (G.P.), Department of Biostatistics (C.W., G.P.), and Wilmer Eye Institute (J.S.S., P.C., B.C., D.J.Z., N.K.), Johns Hopkins University, Baltimore, MD; Richard E. Hoover Rehabilitation Services for Low Vision and Blindness, Greater Baltimore Medical Center, Baltimore, MD (J.S.S.); Departments of Pathology and Ophthalmology, Keck School of Medicine of the University of Southern California, Doheny Eye Institute, Los Angeles, CA (D.R.H.); Laboratory of Statistical Genetics, Rockefeller University, 1230 York Avenue, New York, NY (S.C.H.).
1,
Haoyu Chen
Haoyu Chen, M.D.
1Center for Human Molecular Biology and Genetics, Sichuan Academy of Medical Sciences and Sichuan Provincial People's Hospital, Chengdu, China (Z.Y.); Department of Ophthalmology & Visual Sciences, Moran Eye Center, University of Utah School of Medicine, Salt Lake City, UT (Z.Y., C.S., D.G., Z.T., H.C., R.C., X.Y., Y. C., J.Z., X.M., V.S.H., J.H., J.B., E.P., S.P., Y.K., L.L., N.A.Z., P.S.B., A.S., B.Y., K.Z.); Program in Human Molecular Biology & Genetics, Eccles Institute of Human Genetics, University of Utah School of Medicine, Salt Lake City, UT (C.S., D.G., Z.T., H.C., R.C., X.Y., H.C., J.Z., X.M., J.H., J.B., E.P., S.P., Y.K., L.L., B.Y., K.Z.); Department of Ophthalmology, Shiley Eye Center, University of California San Diego, San Diego, CA (Z.Y., Y.C., J.Z., K.Z.); Macular Degeneration Center, Casey Eye Institute, Oregon Health & Science University, Portland, OR (P.J.F., M.L.K., E.S.); Department of Ophthalmology & Visual Sciences, University of Kentucky, Lexington, KY (M.E.K., W.C., J.A.); Department of Human Genetics, University of Utah School of Medicine, Salt Lake City, UT (S.W., L.J.); Institute of Genetic Medicine (P.L.T., L.D., N.K.), Department of Molecular Biology and Genetics (N.K.), Department of Oncology (G.P.), Department of Biostatistics (C.W., G.P.), and Wilmer Eye Institute (J.S.S., P.C., B.C., D.J.Z., N.K.), Johns Hopkins University, Baltimore, MD; Richard E. Hoover Rehabilitation Services for Low Vision and Blindness, Greater Baltimore Medical Center, Baltimore, MD (J.S.S.); Departments of Pathology and Ophthalmology, Keck School of Medicine of the University of Southern California, Doheny Eye Institute, Los Angeles, CA (D.R.H.); Laboratory of Statistical Genetics, Rockefeller University, 1230 York Avenue, New York, NY (S.C.H.).
1,
Ryan Constantine
Ryan Constantine, B.A.
1Center for Human Molecular Biology and Genetics, Sichuan Academy of Medical Sciences and Sichuan Provincial People's Hospital, Chengdu, China (Z.Y.); Department of Ophthalmology & Visual Sciences, Moran Eye Center, University of Utah School of Medicine, Salt Lake City, UT (Z.Y., C.S., D.G., Z.T., H.C., R.C., X.Y., Y. C., J.Z., X.M., V.S.H., J.H., J.B., E.P., S.P., Y.K., L.L., N.A.Z., P.S.B., A.S., B.Y., K.Z.); Program in Human Molecular Biology & Genetics, Eccles Institute of Human Genetics, University of Utah School of Medicine, Salt Lake City, UT (C.S., D.G., Z.T., H.C., R.C., X.Y., H.C., J.Z., X.M., J.H., J.B., E.P., S.P., Y.K., L.L., B.Y., K.Z.); Department of Ophthalmology, Shiley Eye Center, University of California San Diego, San Diego, CA (Z.Y., Y.C., J.Z., K.Z.); Macular Degeneration Center, Casey Eye Institute, Oregon Health & Science University, Portland, OR (P.J.F., M.L.K., E.S.); Department of Ophthalmology & Visual Sciences, University of Kentucky, Lexington, KY (M.E.K., W.C., J.A.); Department of Human Genetics, University of Utah School of Medicine, Salt Lake City, UT (S.W., L.J.); Institute of Genetic Medicine (P.L.T., L.D., N.K.), Department of Molecular Biology and Genetics (N.K.), Department of Oncology (G.P.), Department of Biostatistics (C.W., G.P.), and Wilmer Eye Institute (J.S.S., P.C., B.C., D.J.Z., N.K.), Johns Hopkins University, Baltimore, MD; Richard E. Hoover Rehabilitation Services for Low Vision and Blindness, Greater Baltimore Medical Center, Baltimore, MD (J.S.S.); Departments of Pathology and Ophthalmology, Keck School of Medicine of the University of Southern California, Doheny Eye Institute, Los Angeles, CA (D.R.H.); Laboratory of Statistical Genetics, Rockefeller University, 1230 York Avenue, New York, NY (S.C.H.).
1,
Xian Yang
Xian Yang, M.D., Ph.D.
1Center for Human Molecular Biology and Genetics, Sichuan Academy of Medical Sciences and Sichuan Provincial People's Hospital, Chengdu, China (Z.Y.); Department of Ophthalmology & Visual Sciences, Moran Eye Center, University of Utah School of Medicine, Salt Lake City, UT (Z.Y., C.S., D.G., Z.T., H.C., R.C., X.Y., Y. C., J.Z., X.M., V.S.H., J.H., J.B., E.P., S.P., Y.K., L.L., N.A.Z., P.S.B., A.S., B.Y., K.Z.); Program in Human Molecular Biology & Genetics, Eccles Institute of Human Genetics, University of Utah School of Medicine, Salt Lake City, UT (C.S., D.G., Z.T., H.C., R.C., X.Y., H.C., J.Z., X.M., J.H., J.B., E.P., S.P., Y.K., L.L., B.Y., K.Z.); Department of Ophthalmology, Shiley Eye Center, University of California San Diego, San Diego, CA (Z.Y., Y.C., J.Z., K.Z.); Macular Degeneration Center, Casey Eye Institute, Oregon Health & Science University, Portland, OR (P.J.F., M.L.K., E.S.); Department of Ophthalmology & Visual Sciences, University of Kentucky, Lexington, KY (M.E.K., W.C., J.A.); Department of Human Genetics, University of Utah School of Medicine, Salt Lake City, UT (S.W., L.J.); Institute of Genetic Medicine (P.L.T., L.D., N.K.), Department of Molecular Biology and Genetics (N.K.), Department of Oncology (G.P.), Department of Biostatistics (C.W., G.P.), and Wilmer Eye Institute (J.S.S., P.C., B.C., D.J.Z., N.K.), Johns Hopkins University, Baltimore, MD; Richard E. Hoover Rehabilitation Services for Low Vision and Blindness, Greater Baltimore Medical Center, Baltimore, MD (J.S.S.); Departments of Pathology and Ophthalmology, Keck School of Medicine of the University of Southern California, Doheny Eye Institute, Los Angeles, CA (D.R.H.); Laboratory of Statistical Genetics, Rockefeller University, 1230 York Avenue, New York, NY (S.C.H.).
1,
Yuhong Chen
Yuhong Chen, M.D., Ph.D
1Center for Human Molecular Biology and Genetics, Sichuan Academy of Medical Sciences and Sichuan Provincial People's Hospital, Chengdu, China (Z.Y.); Department of Ophthalmology & Visual Sciences, Moran Eye Center, University of Utah School of Medicine, Salt Lake City, UT (Z.Y., C.S., D.G., Z.T., H.C., R.C., X.Y., Y. C., J.Z., X.M., V.S.H., J.H., J.B., E.P., S.P., Y.K., L.L., N.A.Z., P.S.B., A.S., B.Y., K.Z.); Program in Human Molecular Biology & Genetics, Eccles Institute of Human Genetics, University of Utah School of Medicine, Salt Lake City, UT (C.S., D.G., Z.T., H.C., R.C., X.Y., H.C., J.Z., X.M., J.H., J.B., E.P., S.P., Y.K., L.L., B.Y., K.Z.); Department of Ophthalmology, Shiley Eye Center, University of California San Diego, San Diego, CA (Z.Y., Y.C., J.Z., K.Z.); Macular Degeneration Center, Casey Eye Institute, Oregon Health & Science University, Portland, OR (P.J.F., M.L.K., E.S.); Department of Ophthalmology & Visual Sciences, University of Kentucky, Lexington, KY (M.E.K., W.C., J.A.); Department of Human Genetics, University of Utah School of Medicine, Salt Lake City, UT (S.W., L.J.); Institute of Genetic Medicine (P.L.T., L.D., N.K.), Department of Molecular Biology and Genetics (N.K.), Department of Oncology (G.P.), Department of Biostatistics (C.W., G.P.), and Wilmer Eye Institute (J.S.S., P.C., B.C., D.J.Z., N.K.), Johns Hopkins University, Baltimore, MD; Richard E. Hoover Rehabilitation Services for Low Vision and Blindness, Greater Baltimore Medical Center, Baltimore, MD (J.S.S.); Departments of Pathology and Ophthalmology, Keck School of Medicine of the University of Southern California, Doheny Eye Institute, Los Angeles, CA (D.R.H.); Laboratory of Statistical Genetics, Rockefeller University, 1230 York Avenue, New York, NY (S.C.H.).
1,
Jiexi Zeng
Jiexi Zeng, M.D.
1Center for Human Molecular Biology and Genetics, Sichuan Academy of Medical Sciences and Sichuan Provincial People's Hospital, Chengdu, China (Z.Y.); Department of Ophthalmology & Visual Sciences, Moran Eye Center, University of Utah School of Medicine, Salt Lake City, UT (Z.Y., C.S., D.G., Z.T., H.C., R.C., X.Y., Y. C., J.Z., X.M., V.S.H., J.H., J.B., E.P., S.P., Y.K., L.L., N.A.Z., P.S.B., A.S., B.Y., K.Z.); Program in Human Molecular Biology & Genetics, Eccles Institute of Human Genetics, University of Utah School of Medicine, Salt Lake City, UT (C.S., D.G., Z.T., H.C., R.C., X.Y., H.C., J.Z., X.M., J.H., J.B., E.P., S.P., Y.K., L.L., B.Y., K.Z.); Department of Ophthalmology, Shiley Eye Center, University of California San Diego, San Diego, CA (Z.Y., Y.C., J.Z., K.Z.); Macular Degeneration Center, Casey Eye Institute, Oregon Health & Science University, Portland, OR (P.J.F., M.L.K., E.S.); Department of Ophthalmology & Visual Sciences, University of Kentucky, Lexington, KY (M.E.K., W.C., J.A.); Department of Human Genetics, University of Utah School of Medicine, Salt Lake City, UT (S.W., L.J.); Institute of Genetic Medicine (P.L.T., L.D., N.K.), Department of Molecular Biology and Genetics (N.K.), Department of Oncology (G.P.), Department of Biostatistics (C.W., G.P.), and Wilmer Eye Institute (J.S.S., P.C., B.C., D.J.Z., N.K.), Johns Hopkins University, Baltimore, MD; Richard E. Hoover Rehabilitation Services for Low Vision and Blindness, Greater Baltimore Medical Center, Baltimore, MD (J.S.S.); Departments of Pathology and Ophthalmology, Keck School of Medicine of the University of Southern California, Doheny Eye Institute, Los Angeles, CA (D.R.H.); Laboratory of Statistical Genetics, Rockefeller University, 1230 York Avenue, New York, NY (S.C.H.).
1,
Lisa Davey
Lisa Davey, M.S.
1Center for Human Molecular Biology and Genetics, Sichuan Academy of Medical Sciences and Sichuan Provincial People's Hospital, Chengdu, China (Z.Y.); Department of Ophthalmology & Visual Sciences, Moran Eye Center, University of Utah School of Medicine, Salt Lake City, UT (Z.Y., C.S., D.G., Z.T., H.C., R.C., X.Y., Y. C., J.Z., X.M., V.S.H., J.H., J.B., E.P., S.P., Y.K., L.L., N.A.Z., P.S.B., A.S., B.Y., K.Z.); Program in Human Molecular Biology & Genetics, Eccles Institute of Human Genetics, University of Utah School of Medicine, Salt Lake City, UT (C.S., D.G., Z.T., H.C., R.C., X.Y., H.C., J.Z., X.M., J.H., J.B., E.P., S.P., Y.K., L.L., B.Y., K.Z.); Department of Ophthalmology, Shiley Eye Center, University of California San Diego, San Diego, CA (Z.Y., Y.C., J.Z., K.Z.); Macular Degeneration Center, Casey Eye Institute, Oregon Health & Science University, Portland, OR (P.J.F., M.L.K., E.S.); Department of Ophthalmology & Visual Sciences, University of Kentucky, Lexington, KY (M.E.K., W.C., J.A.); Department of Human Genetics, University of Utah School of Medicine, Salt Lake City, UT (S.W., L.J.); Institute of Genetic Medicine (P.L.T., L.D., N.K.), Department of Molecular Biology and Genetics (N.K.), Department of Oncology (G.P.), Department of Biostatistics (C.W., G.P.), and Wilmer Eye Institute (J.S.S., P.C., B.C., D.J.Z., N.K.), Johns Hopkins University, Baltimore, MD; Richard E. Hoover Rehabilitation Services for Low Vision and Blindness, Greater Baltimore Medical Center, Baltimore, MD (J.S.S.); Departments of Pathology and Ophthalmology, Keck School of Medicine of the University of Southern California, Doheny Eye Institute, Los Angeles, CA (D.R.H.); Laboratory of Statistical Genetics, Rockefeller University, 1230 York Avenue, New York, NY (S.C.H.).
1,
Xiang Ma
Xiang Ma, B.S.
1Center for Human Molecular Biology and Genetics, Sichuan Academy of Medical Sciences and Sichuan Provincial People's Hospital, Chengdu, China (Z.Y.); Department of Ophthalmology & Visual Sciences, Moran Eye Center, University of Utah School of Medicine, Salt Lake City, UT (Z.Y., C.S., D.G., Z.T., H.C., R.C., X.Y., Y. C., J.Z., X.M., V.S.H., J.H., J.B., E.P., S.P., Y.K., L.L., N.A.Z., P.S.B., A.S., B.Y., K.Z.); Program in Human Molecular Biology & Genetics, Eccles Institute of Human Genetics, University of Utah School of Medicine, Salt Lake City, UT (C.S., D.G., Z.T., H.C., R.C., X.Y., H.C., J.Z., X.M., J.H., J.B., E.P., S.P., Y.K., L.L., B.Y., K.Z.); Department of Ophthalmology, Shiley Eye Center, University of California San Diego, San Diego, CA (Z.Y., Y.C., J.Z., K.Z.); Macular Degeneration Center, Casey Eye Institute, Oregon Health & Science University, Portland, OR (P.J.F., M.L.K., E.S.); Department of Ophthalmology & Visual Sciences, University of Kentucky, Lexington, KY (M.E.K., W.C., J.A.); Department of Human Genetics, University of Utah School of Medicine, Salt Lake City, UT (S.W., L.J.); Institute of Genetic Medicine (P.L.T., L.D., N.K.), Department of Molecular Biology and Genetics (N.K.), Department of Oncology (G.P.), Department of Biostatistics (C.W., G.P.), and Wilmer Eye Institute (J.S.S., P.C., B.C., D.J.Z., N.K.), Johns Hopkins University, Baltimore, MD; Richard E. Hoover Rehabilitation Services for Low Vision and Blindness, Greater Baltimore Medical Center, Baltimore, MD (J.S.S.); Departments of Pathology and Ophthalmology, Keck School of Medicine of the University of Southern California, Doheny Eye Institute, Los Angeles, CA (D.R.H.); Laboratory of Statistical Genetics, Rockefeller University, 1230 York Avenue, New York, NY (S.C.H.).
1,
Vincent S Hau
Vincent S Hau, M.D. Ph.D.
1Center for Human Molecular Biology and Genetics, Sichuan Academy of Medical Sciences and Sichuan Provincial People's Hospital, Chengdu, China (Z.Y.); Department of Ophthalmology & Visual Sciences, Moran Eye Center, University of Utah School of Medicine, Salt Lake City, UT (Z.Y., C.S., D.G., Z.T., H.C., R.C., X.Y., Y. C., J.Z., X.M., V.S.H., J.H., J.B., E.P., S.P., Y.K., L.L., N.A.Z., P.S.B., A.S., B.Y., K.Z.); Program in Human Molecular Biology & Genetics, Eccles Institute of Human Genetics, University of Utah School of Medicine, Salt Lake City, UT (C.S., D.G., Z.T., H.C., R.C., X.Y., H.C., J.Z., X.M., J.H., J.B., E.P., S.P., Y.K., L.L., B.Y., K.Z.); Department of Ophthalmology, Shiley Eye Center, University of California San Diego, San Diego, CA (Z.Y., Y.C., J.Z., K.Z.); Macular Degeneration Center, Casey Eye Institute, Oregon Health & Science University, Portland, OR (P.J.F., M.L.K., E.S.); Department of Ophthalmology & Visual Sciences, University of Kentucky, Lexington, KY (M.E.K., W.C., J.A.); Department of Human Genetics, University of Utah School of Medicine, Salt Lake City, UT (S.W., L.J.); Institute of Genetic Medicine (P.L.T., L.D., N.K.), Department of Molecular Biology and Genetics (N.K.), Department of Oncology (G.P.), Department of Biostatistics (C.W., G.P.), and Wilmer Eye Institute (J.S.S., P.C., B.C., D.J.Z., N.K.), Johns Hopkins University, Baltimore, MD; Richard E. Hoover Rehabilitation Services for Low Vision and Blindness, Greater Baltimore Medical Center, Baltimore, MD (J.S.S.); Departments of Pathology and Ophthalmology, Keck School of Medicine of the University of Southern California, Doheny Eye Institute, Los Angeles, CA (D.R.H.); Laboratory of Statistical Genetics, Rockefeller University, 1230 York Avenue, New York, NY (S.C.H.).
1,
Chi Wang
Chi Wang, B.S.
1Center for Human Molecular Biology and Genetics, Sichuan Academy of Medical Sciences and Sichuan Provincial People's Hospital, Chengdu, China (Z.Y.); Department of Ophthalmology & Visual Sciences, Moran Eye Center, University of Utah School of Medicine, Salt Lake City, UT (Z.Y., C.S., D.G., Z.T., H.C., R.C., X.Y., Y. C., J.Z., X.M., V.S.H., J.H., J.B., E.P., S.P., Y.K., L.L., N.A.Z., P.S.B., A.S., B.Y., K.Z.); Program in Human Molecular Biology & Genetics, Eccles Institute of Human Genetics, University of Utah School of Medicine, Salt Lake City, UT (C.S., D.G., Z.T., H.C., R.C., X.Y., H.C., J.Z., X.M., J.H., J.B., E.P., S.P., Y.K., L.L., B.Y., K.Z.); Department of Ophthalmology, Shiley Eye Center, University of California San Diego, San Diego, CA (Z.Y., Y.C., J.Z., K.Z.); Macular Degeneration Center, Casey Eye Institute, Oregon Health & Science University, Portland, OR (P.J.F., M.L.K., E.S.); Department of Ophthalmology & Visual Sciences, University of Kentucky, Lexington, KY (M.E.K., W.C., J.A.); Department of Human Genetics, University of Utah School of Medicine, Salt Lake City, UT (S.W., L.J.); Institute of Genetic Medicine (P.L.T., L.D., N.K.), Department of Molecular Biology and Genetics (N.K.), Department of Oncology (G.P.), Department of Biostatistics (C.W., G.P.), and Wilmer Eye Institute (J.S.S., P.C., B.C., D.J.Z., N.K.), Johns Hopkins University, Baltimore, MD; Richard E. Hoover Rehabilitation Services for Low Vision and Blindness, Greater Baltimore Medical Center, Baltimore, MD (J.S.S.); Departments of Pathology and Ophthalmology, Keck School of Medicine of the University of Southern California, Doheny Eye Institute, Los Angeles, CA (D.R.H.); Laboratory of Statistical Genetics, Rockefeller University, 1230 York Avenue, New York, NY (S.C.H.).
1,
Jennifer Harmon
Jennifer Harmon
1Center for Human Molecular Biology and Genetics, Sichuan Academy of Medical Sciences and Sichuan Provincial People's Hospital, Chengdu, China (Z.Y.); Department of Ophthalmology & Visual Sciences, Moran Eye Center, University of Utah School of Medicine, Salt Lake City, UT (Z.Y., C.S., D.G., Z.T., H.C., R.C., X.Y., Y. C., J.Z., X.M., V.S.H., J.H., J.B., E.P., S.P., Y.K., L.L., N.A.Z., P.S.B., A.S., B.Y., K.Z.); Program in Human Molecular Biology & Genetics, Eccles Institute of Human Genetics, University of Utah School of Medicine, Salt Lake City, UT (C.S., D.G., Z.T., H.C., R.C., X.Y., H.C., J.Z., X.M., J.H., J.B., E.P., S.P., Y.K., L.L., B.Y., K.Z.); Department of Ophthalmology, Shiley Eye Center, University of California San Diego, San Diego, CA (Z.Y., Y.C., J.Z., K.Z.); Macular Degeneration Center, Casey Eye Institute, Oregon Health & Science University, Portland, OR (P.J.F., M.L.K., E.S.); Department of Ophthalmology & Visual Sciences, University of Kentucky, Lexington, KY (M.E.K., W.C., J.A.); Department of Human Genetics, University of Utah School of Medicine, Salt Lake City, UT (S.W., L.J.); Institute of Genetic Medicine (P.L.T., L.D., N.K.), Department of Molecular Biology and Genetics (N.K.), Department of Oncology (G.P.), Department of Biostatistics (C.W., G.P.), and Wilmer Eye Institute (J.S.S., P.C., B.C., D.J.Z., N.K.), Johns Hopkins University, Baltimore, MD; Richard E. Hoover Rehabilitation Services for Low Vision and Blindness, Greater Baltimore Medical Center, Baltimore, MD (J.S.S.); Departments of Pathology and Ophthalmology, Keck School of Medicine of the University of Southern California, Doheny Eye Institute, Los Angeles, CA (D.R.H.); Laboratory of Statistical Genetics, Rockefeller University, 1230 York Avenue, New York, NY (S.C.H.).
1,
Jeanette Buehler
Jeanette Buehler, B.S.
1Center for Human Molecular Biology and Genetics, Sichuan Academy of Medical Sciences and Sichuan Provincial People's Hospital, Chengdu, China (Z.Y.); Department of Ophthalmology & Visual Sciences, Moran Eye Center, University of Utah School of Medicine, Salt Lake City, UT (Z.Y., C.S., D.G., Z.T., H.C., R.C., X.Y., Y. C., J.Z., X.M., V.S.H., J.H., J.B., E.P., S.P., Y.K., L.L., N.A.Z., P.S.B., A.S., B.Y., K.Z.); Program in Human Molecular Biology & Genetics, Eccles Institute of Human Genetics, University of Utah School of Medicine, Salt Lake City, UT (C.S., D.G., Z.T., H.C., R.C., X.Y., H.C., J.Z., X.M., J.H., J.B., E.P., S.P., Y.K., L.L., B.Y., K.Z.); Department of Ophthalmology, Shiley Eye Center, University of California San Diego, San Diego, CA (Z.Y., Y.C., J.Z., K.Z.); Macular Degeneration Center, Casey Eye Institute, Oregon Health & Science University, Portland, OR (P.J.F., M.L.K., E.S.); Department of Ophthalmology & Visual Sciences, University of Kentucky, Lexington, KY (M.E.K., W.C., J.A.); Department of Human Genetics, University of Utah School of Medicine, Salt Lake City, UT (S.W., L.J.); Institute of Genetic Medicine (P.L.T., L.D., N.K.), Department of Molecular Biology and Genetics (N.K.), Department of Oncology (G.P.), Department of Biostatistics (C.W., G.P.), and Wilmer Eye Institute (J.S.S., P.C., B.C., D.J.Z., N.K.), Johns Hopkins University, Baltimore, MD; Richard E. Hoover Rehabilitation Services for Low Vision and Blindness, Greater Baltimore Medical Center, Baltimore, MD (J.S.S.); Departments of Pathology and Ophthalmology, Keck School of Medicine of the University of Southern California, Doheny Eye Institute, Los Angeles, CA (D.R.H.); Laboratory of Statistical Genetics, Rockefeller University, 1230 York Avenue, New York, NY (S.C.H.).
1,
Erik Pearson
Erik Pearson, B.S.
1Center for Human Molecular Biology and Genetics, Sichuan Academy of Medical Sciences and Sichuan Provincial People's Hospital, Chengdu, China (Z.Y.); Department of Ophthalmology & Visual Sciences, Moran Eye Center, University of Utah School of Medicine, Salt Lake City, UT (Z.Y., C.S., D.G., Z.T., H.C., R.C., X.Y., Y. C., J.Z., X.M., V.S.H., J.H., J.B., E.P., S.P., Y.K., L.L., N.A.Z., P.S.B., A.S., B.Y., K.Z.); Program in Human Molecular Biology & Genetics, Eccles Institute of Human Genetics, University of Utah School of Medicine, Salt Lake City, UT (C.S., D.G., Z.T., H.C., R.C., X.Y., H.C., J.Z., X.M., J.H., J.B., E.P., S.P., Y.K., L.L., B.Y., K.Z.); Department of Ophthalmology, Shiley Eye Center, University of California San Diego, San Diego, CA (Z.Y., Y.C., J.Z., K.Z.); Macular Degeneration Center, Casey Eye Institute, Oregon Health & Science University, Portland, OR (P.J.F., M.L.K., E.S.); Department of Ophthalmology & Visual Sciences, University of Kentucky, Lexington, KY (M.E.K., W.C., J.A.); Department of Human Genetics, University of Utah School of Medicine, Salt Lake City, UT (S.W., L.J.); Institute of Genetic Medicine (P.L.T., L.D., N.K.), Department of Molecular Biology and Genetics (N.K.), Department of Oncology (G.P.), Department of Biostatistics (C.W., G.P.), and Wilmer Eye Institute (J.S.S., P.C., B.C., D.J.Z., N.K.), Johns Hopkins University, Baltimore, MD; Richard E. Hoover Rehabilitation Services for Low Vision and Blindness, Greater Baltimore Medical Center, Baltimore, MD (J.S.S.); Departments of Pathology and Ophthalmology, Keck School of Medicine of the University of Southern California, Doheny Eye Institute, Los Angeles, CA (D.R.H.); Laboratory of Statistical Genetics, Rockefeller University, 1230 York Avenue, New York, NY (S.C.H.).
1,
Shrena Patel
Shrena Patel, M.D.
1Center for Human Molecular Biology and Genetics, Sichuan Academy of Medical Sciences and Sichuan Provincial People's Hospital, Chengdu, China (Z.Y.); Department of Ophthalmology & Visual Sciences, Moran Eye Center, University of Utah School of Medicine, Salt Lake City, UT (Z.Y., C.S., D.G., Z.T., H.C., R.C., X.Y., Y. C., J.Z., X.M., V.S.H., J.H., J.B., E.P., S.P., Y.K., L.L., N.A.Z., P.S.B., A.S., B.Y., K.Z.); Program in Human Molecular Biology & Genetics, Eccles Institute of Human Genetics, University of Utah School of Medicine, Salt Lake City, UT (C.S., D.G., Z.T., H.C., R.C., X.Y., H.C., J.Z., X.M., J.H., J.B., E.P., S.P., Y.K., L.L., B.Y., K.Z.); Department of Ophthalmology, Shiley Eye Center, University of California San Diego, San Diego, CA (Z.Y., Y.C., J.Z., K.Z.); Macular Degeneration Center, Casey Eye Institute, Oregon Health & Science University, Portland, OR (P.J.F., M.L.K., E.S.); Department of Ophthalmology & Visual Sciences, University of Kentucky, Lexington, KY (M.E.K., W.C., J.A.); Department of Human Genetics, University of Utah School of Medicine, Salt Lake City, UT (S.W., L.J.); Institute of Genetic Medicine (P.L.T., L.D., N.K.), Department of Molecular Biology and Genetics (N.K.), Department of Oncology (G.P.), Department of Biostatistics (C.W., G.P.), and Wilmer Eye Institute (J.S.S., P.C., B.C., D.J.Z., N.K.), Johns Hopkins University, Baltimore, MD; Richard E. Hoover Rehabilitation Services for Low Vision and Blindness, Greater Baltimore Medical Center, Baltimore, MD (J.S.S.); Departments of Pathology and Ophthalmology, Keck School of Medicine of the University of Southern California, Doheny Eye Institute, Los Angeles, CA (D.R.H.); Laboratory of Statistical Genetics, Rockefeller University, 1230 York Avenue, New York, NY (S.C.H.).
1,
Yuuki Kaminoh
Yuuki Kaminoh, B.S.
1Center for Human Molecular Biology and Genetics, Sichuan Academy of Medical Sciences and Sichuan Provincial People's Hospital, Chengdu, China (Z.Y.); Department of Ophthalmology & Visual Sciences, Moran Eye Center, University of Utah School of Medicine, Salt Lake City, UT (Z.Y., C.S., D.G., Z.T., H.C., R.C., X.Y., Y. C., J.Z., X.M., V.S.H., J.H., J.B., E.P., S.P., Y.K., L.L., N.A.Z., P.S.B., A.S., B.Y., K.Z.); Program in Human Molecular Biology & Genetics, Eccles Institute of Human Genetics, University of Utah School of Medicine, Salt Lake City, UT (C.S., D.G., Z.T., H.C., R.C., X.Y., H.C., J.Z., X.M., J.H., J.B., E.P., S.P., Y.K., L.L., B.Y., K.Z.); Department of Ophthalmology, Shiley Eye Center, University of California San Diego, San Diego, CA (Z.Y., Y.C., J.Z., K.Z.); Macular Degeneration Center, Casey Eye Institute, Oregon Health & Science University, Portland, OR (P.J.F., M.L.K., E.S.); Department of Ophthalmology & Visual Sciences, University of Kentucky, Lexington, KY (M.E.K., W.C., J.A.); Department of Human Genetics, University of Utah School of Medicine, Salt Lake City, UT (S.W., L.J.); Institute of Genetic Medicine (P.L.T., L.D., N.K.), Department of Molecular Biology and Genetics (N.K.), Department of Oncology (G.P.), Department of Biostatistics (C.W., G.P.), and Wilmer Eye Institute (J.S.S., P.C., B.C., D.J.Z., N.K.), Johns Hopkins University, Baltimore, MD; Richard E. Hoover Rehabilitation Services for Low Vision and Blindness, Greater Baltimore Medical Center, Baltimore, MD (J.S.S.); Departments of Pathology and Ophthalmology, Keck School of Medicine of the University of Southern California, Doheny Eye Institute, Los Angeles, CA (D.R.H.); Laboratory of Statistical Genetics, Rockefeller University, 1230 York Avenue, New York, NY (S.C.H.).
1,
Scott Watkins
Scott Watkins, M.S
1Center for Human Molecular Biology and Genetics, Sichuan Academy of Medical Sciences and Sichuan Provincial People's Hospital, Chengdu, China (Z.Y.); Department of Ophthalmology & Visual Sciences, Moran Eye Center, University of Utah School of Medicine, Salt Lake City, UT (Z.Y., C.S., D.G., Z.T., H.C., R.C., X.Y., Y. C., J.Z., X.M., V.S.H., J.H., J.B., E.P., S.P., Y.K., L.L., N.A.Z., P.S.B., A.S., B.Y., K.Z.); Program in Human Molecular Biology & Genetics, Eccles Institute of Human Genetics, University of Utah School of Medicine, Salt Lake City, UT (C.S., D.G., Z.T., H.C., R.C., X.Y., H.C., J.Z., X.M., J.H., J.B., E.P., S.P., Y.K., L.L., B.Y., K.Z.); Department of Ophthalmology, Shiley Eye Center, University of California San Diego, San Diego, CA (Z.Y., Y.C., J.Z., K.Z.); Macular Degeneration Center, Casey Eye Institute, Oregon Health & Science University, Portland, OR (P.J.F., M.L.K., E.S.); Department of Ophthalmology & Visual Sciences, University of Kentucky, Lexington, KY (M.E.K., W.C., J.A.); Department of Human Genetics, University of Utah School of Medicine, Salt Lake City, UT (S.W., L.J.); Institute of Genetic Medicine (P.L.T., L.D., N.K.), Department of Molecular Biology and Genetics (N.K.), Department of Oncology (G.P.), Department of Biostatistics (C.W., G.P.), and Wilmer Eye Institute (J.S.S., P.C., B.C., D.J.Z., N.K.), Johns Hopkins University, Baltimore, MD; Richard E. Hoover Rehabilitation Services for Low Vision and Blindness, Greater Baltimore Medical Center, Baltimore, MD (J.S.S.); Departments of Pathology and Ophthalmology, Keck School of Medicine of the University of Southern California, Doheny Eye Institute, Los Angeles, CA (D.R.H.); Laboratory of Statistical Genetics, Rockefeller University, 1230 York Avenue, New York, NY (S.C.H.).
1,
Ling Luo
Ling Luo, M.D.
1Center for Human Molecular Biology and Genetics, Sichuan Academy of Medical Sciences and Sichuan Provincial People's Hospital, Chengdu, China (Z.Y.); Department of Ophthalmology & Visual Sciences, Moran Eye Center, University of Utah School of Medicine, Salt Lake City, UT (Z.Y., C.S., D.G., Z.T., H.C., R.C., X.Y., Y. C., J.Z., X.M., V.S.H., J.H., J.B., E.P., S.P., Y.K., L.L., N.A.Z., P.S.B., A.S., B.Y., K.Z.); Program in Human Molecular Biology & Genetics, Eccles Institute of Human Genetics, University of Utah School of Medicine, Salt Lake City, UT (C.S., D.G., Z.T., H.C., R.C., X.Y., H.C., J.Z., X.M., J.H., J.B., E.P., S.P., Y.K., L.L., B.Y., K.Z.); Department of Ophthalmology, Shiley Eye Center, University of California San Diego, San Diego, CA (Z.Y., Y.C., J.Z., K.Z.); Macular Degeneration Center, Casey Eye Institute, Oregon Health & Science University, Portland, OR (P.J.F., M.L.K., E.S.); Department of Ophthalmology & Visual Sciences, University of Kentucky, Lexington, KY (M.E.K., W.C., J.A.); Department of Human Genetics, University of Utah School of Medicine, Salt Lake City, UT (S.W., L.J.); Institute of Genetic Medicine (P.L.T., L.D., N.K.), Department of Molecular Biology and Genetics (N.K.), Department of Oncology (G.P.), Department of Biostatistics (C.W., G.P.), and Wilmer Eye Institute (J.S.S., P.C., B.C., D.J.Z., N.K.), Johns Hopkins University, Baltimore, MD; Richard E. Hoover Rehabilitation Services for Low Vision and Blindness, Greater Baltimore Medical Center, Baltimore, MD (J.S.S.); Departments of Pathology and Ophthalmology, Keck School of Medicine of the University of Southern California, Doheny Eye Institute, Los Angeles, CA (D.R.H.); Laboratory of Statistical Genetics, Rockefeller University, 1230 York Avenue, New York, NY (S.C.H.).
1,
Norman A Zabriskie
Norman A Zabriskie, M.D.
1Center for Human Molecular Biology and Genetics, Sichuan Academy of Medical Sciences and Sichuan Provincial People's Hospital, Chengdu, China (Z.Y.); Department of Ophthalmology & Visual Sciences, Moran Eye Center, University of Utah School of Medicine, Salt Lake City, UT (Z.Y., C.S., D.G., Z.T., H.C., R.C., X.Y., Y. C., J.Z., X.M., V.S.H., J.H., J.B., E.P., S.P., Y.K., L.L., N.A.Z., P.S.B., A.S., B.Y., K.Z.); Program in Human Molecular Biology & Genetics, Eccles Institute of Human Genetics, University of Utah School of Medicine, Salt Lake City, UT (C.S., D.G., Z.T., H.C., R.C., X.Y., H.C., J.Z., X.M., J.H., J.B., E.P., S.P., Y.K., L.L., B.Y., K.Z.); Department of Ophthalmology, Shiley Eye Center, University of California San Diego, San Diego, CA (Z.Y., Y.C., J.Z., K.Z.); Macular Degeneration Center, Casey Eye Institute, Oregon Health & Science University, Portland, OR (P.J.F., M.L.K., E.S.); Department of Ophthalmology & Visual Sciences, University of Kentucky, Lexington, KY (M.E.K., W.C., J.A.); Department of Human Genetics, University of Utah School of Medicine, Salt Lake City, UT (S.W., L.J.); Institute of Genetic Medicine (P.L.T., L.D., N.K.), Department of Molecular Biology and Genetics (N.K.), Department of Oncology (G.P.), Department of Biostatistics (C.W., G.P.), and Wilmer Eye Institute (J.S.S., P.C., B.C., D.J.Z., N.K.), Johns Hopkins University, Baltimore, MD; Richard E. Hoover Rehabilitation Services for Low Vision and Blindness, Greater Baltimore Medical Center, Baltimore, MD (J.S.S.); Departments of Pathology and Ophthalmology, Keck School of Medicine of the University of Southern California, Doheny Eye Institute, Los Angeles, CA (D.R.H.); Laboratory of Statistical Genetics, Rockefeller University, 1230 York Avenue, New York, NY (S.C.H.).
1,
Paul S Bernstein
Paul S Bernstein, M.D., Ph.D.
1Center for Human Molecular Biology and Genetics, Sichuan Academy of Medical Sciences and Sichuan Provincial People's Hospital, Chengdu, China (Z.Y.); Department of Ophthalmology & Visual Sciences, Moran Eye Center, University of Utah School of Medicine, Salt Lake City, UT (Z.Y., C.S., D.G., Z.T., H.C., R.C., X.Y., Y. C., J.Z., X.M., V.S.H., J.H., J.B., E.P., S.P., Y.K., L.L., N.A.Z., P.S.B., A.S., B.Y., K.Z.); Program in Human Molecular Biology & Genetics, Eccles Institute of Human Genetics, University of Utah School of Medicine, Salt Lake City, UT (C.S., D.G., Z.T., H.C., R.C., X.Y., H.C., J.Z., X.M., J.H., J.B., E.P., S.P., Y.K., L.L., B.Y., K.Z.); Department of Ophthalmology, Shiley Eye Center, University of California San Diego, San Diego, CA (Z.Y., Y.C., J.Z., K.Z.); Macular Degeneration Center, Casey Eye Institute, Oregon Health & Science University, Portland, OR (P.J.F., M.L.K., E.S.); Department of Ophthalmology & Visual Sciences, University of Kentucky, Lexington, KY (M.E.K., W.C., J.A.); Department of Human Genetics, University of Utah School of Medicine, Salt Lake City, UT (S.W., L.J.); Institute of Genetic Medicine (P.L.T., L.D., N.K.), Department of Molecular Biology and Genetics (N.K.), Department of Oncology (G.P.), Department of Biostatistics (C.W., G.P.), and Wilmer Eye Institute (J.S.S., P.C., B.C., D.J.Z., N.K.), Johns Hopkins University, Baltimore, MD; Richard E. Hoover Rehabilitation Services for Low Vision and Blindness, Greater Baltimore Medical Center, Baltimore, MD (J.S.S.); Departments of Pathology and Ophthalmology, Keck School of Medicine of the University of Southern California, Doheny Eye Institute, Los Angeles, CA (D.R.H.); Laboratory of Statistical Genetics, Rockefeller University, 1230 York Avenue, New York, NY (S.C.H.).
1,
Wongil Cho
Wongil Cho, Ph.D.
1Center for Human Molecular Biology and Genetics, Sichuan Academy of Medical Sciences and Sichuan Provincial People's Hospital, Chengdu, China (Z.Y.); Department of Ophthalmology & Visual Sciences, Moran Eye Center, University of Utah School of Medicine, Salt Lake City, UT (Z.Y., C.S., D.G., Z.T., H.C., R.C., X.Y., Y. C., J.Z., X.M., V.S.H., J.H., J.B., E.P., S.P., Y.K., L.L., N.A.Z., P.S.B., A.S., B.Y., K.Z.); Program in Human Molecular Biology & Genetics, Eccles Institute of Human Genetics, University of Utah School of Medicine, Salt Lake City, UT (C.S., D.G., Z.T., H.C., R.C., X.Y., H.C., J.Z., X.M., J.H., J.B., E.P., S.P., Y.K., L.L., B.Y., K.Z.); Department of Ophthalmology, Shiley Eye Center, University of California San Diego, San Diego, CA (Z.Y., Y.C., J.Z., K.Z.); Macular Degeneration Center, Casey Eye Institute, Oregon Health & Science University, Portland, OR (P.J.F., M.L.K., E.S.); Department of Ophthalmology & Visual Sciences, University of Kentucky, Lexington, KY (M.E.K., W.C., J.A.); Department of Human Genetics, University of Utah School of Medicine, Salt Lake City, UT (S.W., L.J.); Institute of Genetic Medicine (P.L.T., L.D., N.K.), Department of Molecular Biology and Genetics (N.K.), Department of Oncology (G.P.), Department of Biostatistics (C.W., G.P.), and Wilmer Eye Institute (J.S.S., P.C., B.C., D.J.Z., N.K.), Johns Hopkins University, Baltimore, MD; Richard E. Hoover Rehabilitation Services for Low Vision and Blindness, Greater Baltimore Medical Center, Baltimore, MD (J.S.S.); Departments of Pathology and Ophthalmology, Keck School of Medicine of the University of Southern California, Doheny Eye Institute, Los Angeles, CA (D.R.H.); Laboratory of Statistical Genetics, Rockefeller University, 1230 York Avenue, New York, NY (S.C.H.).
1,
Andrea Schwager
Andrea Schwager, B.S.
1Center for Human Molecular Biology and Genetics, Sichuan Academy of Medical Sciences and Sichuan Provincial People's Hospital, Chengdu, China (Z.Y.); Department of Ophthalmology & Visual Sciences, Moran Eye Center, University of Utah School of Medicine, Salt Lake City, UT (Z.Y., C.S., D.G., Z.T., H.C., R.C., X.Y., Y. C., J.Z., X.M., V.S.H., J.H., J.B., E.P., S.P., Y.K., L.L., N.A.Z., P.S.B., A.S., B.Y., K.Z.); Program in Human Molecular Biology & Genetics, Eccles Institute of Human Genetics, University of Utah School of Medicine, Salt Lake City, UT (C.S., D.G., Z.T., H.C., R.C., X.Y., H.C., J.Z., X.M., J.H., J.B., E.P., S.P., Y.K., L.L., B.Y., K.Z.); Department of Ophthalmology, Shiley Eye Center, University of California San Diego, San Diego, CA (Z.Y., Y.C., J.Z., K.Z.); Macular Degeneration Center, Casey Eye Institute, Oregon Health & Science University, Portland, OR (P.J.F., M.L.K., E.S.); Department of Ophthalmology & Visual Sciences, University of Kentucky, Lexington, KY (M.E.K., W.C., J.A.); Department of Human Genetics, University of Utah School of Medicine, Salt Lake City, UT (S.W., L.J.); Institute of Genetic Medicine (P.L.T., L.D., N.K.), Department of Molecular Biology and Genetics (N.K.), Department of Oncology (G.P.), Department of Biostatistics (C.W., G.P.), and Wilmer Eye Institute (J.S.S., P.C., B.C., D.J.Z., N.K.), Johns Hopkins University, Baltimore, MD; Richard E. Hoover Rehabilitation Services for Low Vision and Blindness, Greater Baltimore Medical Center, Baltimore, MD (J.S.S.); Departments of Pathology and Ophthalmology, Keck School of Medicine of the University of Southern California, Doheny Eye Institute, Los Angeles, CA (D.R.H.); Laboratory of Statistical Genetics, Rockefeller University, 1230 York Avenue, New York, NY (S.C.H.).
1,
David R Hinton
David R Hinton, M.D.
1Center for Human Molecular Biology and Genetics, Sichuan Academy of Medical Sciences and Sichuan Provincial People's Hospital, Chengdu, China (Z.Y.); Department of Ophthalmology & Visual Sciences, Moran Eye Center, University of Utah School of Medicine, Salt Lake City, UT (Z.Y., C.S., D.G., Z.T., H.C., R.C., X.Y., Y. C., J.Z., X.M., V.S.H., J.H., J.B., E.P., S.P., Y.K., L.L., N.A.Z., P.S.B., A.S., B.Y., K.Z.); Program in Human Molecular Biology & Genetics, Eccles Institute of Human Genetics, University of Utah School of Medicine, Salt Lake City, UT (C.S., D.G., Z.T., H.C., R.C., X.Y., H.C., J.Z., X.M., J.H., J.B., E.P., S.P., Y.K., L.L., B.Y., K.Z.); Department of Ophthalmology, Shiley Eye Center, University of California San Diego, San Diego, CA (Z.Y., Y.C., J.Z., K.Z.); Macular Degeneration Center, Casey Eye Institute, Oregon Health & Science University, Portland, OR (P.J.F., M.L.K., E.S.); Department of Ophthalmology & Visual Sciences, University of Kentucky, Lexington, KY (M.E.K., W.C., J.A.); Department of Human Genetics, University of Utah School of Medicine, Salt Lake City, UT (S.W., L.J.); Institute of Genetic Medicine (P.L.T., L.D., N.K.), Department of Molecular Biology and Genetics (N.K.), Department of Oncology (G.P.), Department of Biostatistics (C.W., G.P.), and Wilmer Eye Institute (J.S.S., P.C., B.C., D.J.Z., N.K.), Johns Hopkins University, Baltimore, MD; Richard E. Hoover Rehabilitation Services for Low Vision and Blindness, Greater Baltimore Medical Center, Baltimore, MD (J.S.S.); Departments of Pathology and Ophthalmology, Keck School of Medicine of the University of Southern California, Doheny Eye Institute, Los Angeles, CA (D.R.H.); Laboratory of Statistical Genetics, Rockefeller University, 1230 York Avenue, New York, NY (S.C.H.).
1,
Michael L Klein
Michael L Klein, M.D.
1Center for Human Molecular Biology and Genetics, Sichuan Academy of Medical Sciences and Sichuan Provincial People's Hospital, Chengdu, China (Z.Y.); Department of Ophthalmology & Visual Sciences, Moran Eye Center, University of Utah School of Medicine, Salt Lake City, UT (Z.Y., C.S., D.G., Z.T., H.C., R.C., X.Y., Y. C., J.Z., X.M., V.S.H., J.H., J.B., E.P., S.P., Y.K., L.L., N.A.Z., P.S.B., A.S., B.Y., K.Z.); Program in Human Molecular Biology & Genetics, Eccles Institute of Human Genetics, University of Utah School of Medicine, Salt Lake City, UT (C.S., D.G., Z.T., H.C., R.C., X.Y., H.C., J.Z., X.M., J.H., J.B., E.P., S.P., Y.K., L.L., B.Y., K.Z.); Department of Ophthalmology, Shiley Eye Center, University of California San Diego, San Diego, CA (Z.Y., Y.C., J.Z., K.Z.); Macular Degeneration Center, Casey Eye Institute, Oregon Health & Science University, Portland, OR (P.J.F., M.L.K., E.S.); Department of Ophthalmology & Visual Sciences, University of Kentucky, Lexington, KY (M.E.K., W.C., J.A.); Department of Human Genetics, University of Utah School of Medicine, Salt Lake City, UT (S.W., L.J.); Institute of Genetic Medicine (P.L.T., L.D., N.K.), Department of Molecular Biology and Genetics (N.K.), Department of Oncology (G.P.), Department of Biostatistics (C.W., G.P.), and Wilmer Eye Institute (J.S.S., P.C., B.C., D.J.Z., N.K.), Johns Hopkins University, Baltimore, MD; Richard E. Hoover Rehabilitation Services for Low Vision and Blindness, Greater Baltimore Medical Center, Baltimore, MD (J.S.S.); Departments of Pathology and Ophthalmology, Keck School of Medicine of the University of Southern California, Doheny Eye Institute, Los Angeles, CA (D.R.H.); Laboratory of Statistical Genetics, Rockefeller University, 1230 York Avenue, New York, NY (S.C.H.).
1,
Sara C Hamon
Sara C Hamon, Ph.D.
1Center for Human Molecular Biology and Genetics, Sichuan Academy of Medical Sciences and Sichuan Provincial People's Hospital, Chengdu, China (Z.Y.); Department of Ophthalmology & Visual Sciences, Moran Eye Center, University of Utah School of Medicine, Salt Lake City, UT (Z.Y., C.S., D.G., Z.T., H.C., R.C., X.Y., Y. C., J.Z., X.M., V.S.H., J.H., J.B., E.P., S.P., Y.K., L.L., N.A.Z., P.S.B., A.S., B.Y., K.Z.); Program in Human Molecular Biology & Genetics, Eccles Institute of Human Genetics, University of Utah School of Medicine, Salt Lake City, UT (C.S., D.G., Z.T., H.C., R.C., X.Y., H.C., J.Z., X.M., J.H., J.B., E.P., S.P., Y.K., L.L., B.Y., K.Z.); Department of Ophthalmology, Shiley Eye Center, University of California San Diego, San Diego, CA (Z.Y., Y.C., J.Z., K.Z.); Macular Degeneration Center, Casey Eye Institute, Oregon Health & Science University, Portland, OR (P.J.F., M.L.K., E.S.); Department of Ophthalmology & Visual Sciences, University of Kentucky, Lexington, KY (M.E.K., W.C., J.A.); Department of Human Genetics, University of Utah School of Medicine, Salt Lake City, UT (S.W., L.J.); Institute of Genetic Medicine (P.L.T., L.D., N.K.), Department of Molecular Biology and Genetics (N.K.), Department of Oncology (G.P.), Department of Biostatistics (C.W., G.P.), and Wilmer Eye Institute (J.S.S., P.C., B.C., D.J.Z., N.K.), Johns Hopkins University, Baltimore, MD; Richard E. Hoover Rehabilitation Services for Low Vision and Blindness, Greater Baltimore Medical Center, Baltimore, MD (J.S.S.); Departments of Pathology and Ophthalmology, Keck School of Medicine of the University of Southern California, Doheny Eye Institute, Los Angeles, CA (D.R.H.); Laboratory of Statistical Genetics, Rockefeller University, 1230 York Avenue, New York, NY (S.C.H.).
1,
Emily Simmons
Emily Simmons, B.S.
1Center for Human Molecular Biology and Genetics, Sichuan Academy of Medical Sciences and Sichuan Provincial People's Hospital, Chengdu, China (Z.Y.); Department of Ophthalmology & Visual Sciences, Moran Eye Center, University of Utah School of Medicine, Salt Lake City, UT (Z.Y., C.S., D.G., Z.T., H.C., R.C., X.Y., Y. C., J.Z., X.M., V.S.H., J.H., J.B., E.P., S.P., Y.K., L.L., N.A.Z., P.S.B., A.S., B.Y., K.Z.); Program in Human Molecular Biology & Genetics, Eccles Institute of Human Genetics, University of Utah School of Medicine, Salt Lake City, UT (C.S., D.G., Z.T., H.C., R.C., X.Y., H.C., J.Z., X.M., J.H., J.B., E.P., S.P., Y.K., L.L., B.Y., K.Z.); Department of Ophthalmology, Shiley Eye Center, University of California San Diego, San Diego, CA (Z.Y., Y.C., J.Z., K.Z.); Macular Degeneration Center, Casey Eye Institute, Oregon Health & Science University, Portland, OR (P.J.F., M.L.K., E.S.); Department of Ophthalmology & Visual Sciences, University of Kentucky, Lexington, KY (M.E.K., W.C., J.A.); Department of Human Genetics, University of Utah School of Medicine, Salt Lake City, UT (S.W., L.J.); Institute of Genetic Medicine (P.L.T., L.D., N.K.), Department of Molecular Biology and Genetics (N.K.), Department of Oncology (G.P.), Department of Biostatistics (C.W., G.P.), and Wilmer Eye Institute (J.S.S., P.C., B.C., D.J.Z., N.K.), Johns Hopkins University, Baltimore, MD; Richard E. Hoover Rehabilitation Services for Low Vision and Blindness, Greater Baltimore Medical Center, Baltimore, MD (J.S.S.); Departments of Pathology and Ophthalmology, Keck School of Medicine of the University of Southern California, Doheny Eye Institute, Los Angeles, CA (D.R.H.); Laboratory of Statistical Genetics, Rockefeller University, 1230 York Avenue, New York, NY (S.C.H.).
1,
Beifeng Yu
Beifeng Yu, M.D.
1Center for Human Molecular Biology and Genetics, Sichuan Academy of Medical Sciences and Sichuan Provincial People's Hospital, Chengdu, China (Z.Y.); Department of Ophthalmology & Visual Sciences, Moran Eye Center, University of Utah School of Medicine, Salt Lake City, UT (Z.Y., C.S., D.G., Z.T., H.C., R.C., X.Y., Y. C., J.Z., X.M., V.S.H., J.H., J.B., E.P., S.P., Y.K., L.L., N.A.Z., P.S.B., A.S., B.Y., K.Z.); Program in Human Molecular Biology & Genetics, Eccles Institute of Human Genetics, University of Utah School of Medicine, Salt Lake City, UT (C.S., D.G., Z.T., H.C., R.C., X.Y., H.C., J.Z., X.M., J.H., J.B., E.P., S.P., Y.K., L.L., B.Y., K.Z.); Department of Ophthalmology, Shiley Eye Center, University of California San Diego, San Diego, CA (Z.Y., Y.C., J.Z., K.Z.); Macular Degeneration Center, Casey Eye Institute, Oregon Health & Science University, Portland, OR (P.J.F., M.L.K., E.S.); Department of Ophthalmology & Visual Sciences, University of Kentucky, Lexington, KY (M.E.K., W.C., J.A.); Department of Human Genetics, University of Utah School of Medicine, Salt Lake City, UT (S.W., L.J.); Institute of Genetic Medicine (P.L.T., L.D., N.K.), Department of Molecular Biology and Genetics (N.K.), Department of Oncology (G.P.), Department of Biostatistics (C.W., G.P.), and Wilmer Eye Institute (J.S.S., P.C., B.C., D.J.Z., N.K.), Johns Hopkins University, Baltimore, MD; Richard E. Hoover Rehabilitation Services for Low Vision and Blindness, Greater Baltimore Medical Center, Baltimore, MD (J.S.S.); Departments of Pathology and Ophthalmology, Keck School of Medicine of the University of Southern California, Doheny Eye Institute, Los Angeles, CA (D.R.H.); Laboratory of Statistical Genetics, Rockefeller University, 1230 York Avenue, New York, NY (S.C.H.).
1,
Betsy Campochiaro
Betsy Campochiaro, B.S., M.S.W
1Center for Human Molecular Biology and Genetics, Sichuan Academy of Medical Sciences and Sichuan Provincial People's Hospital, Chengdu, China (Z.Y.); Department of Ophthalmology & Visual Sciences, Moran Eye Center, University of Utah School of Medicine, Salt Lake City, UT (Z.Y., C.S., D.G., Z.T., H.C., R.C., X.Y., Y. C., J.Z., X.M., V.S.H., J.H., J.B., E.P., S.P., Y.K., L.L., N.A.Z., P.S.B., A.S., B.Y., K.Z.); Program in Human Molecular Biology & Genetics, Eccles Institute of Human Genetics, University of Utah School of Medicine, Salt Lake City, UT (C.S., D.G., Z.T., H.C., R.C., X.Y., H.C., J.Z., X.M., J.H., J.B., E.P., S.P., Y.K., L.L., B.Y., K.Z.); Department of Ophthalmology, Shiley Eye Center, University of California San Diego, San Diego, CA (Z.Y., Y.C., J.Z., K.Z.); Macular Degeneration Center, Casey Eye Institute, Oregon Health & Science University, Portland, OR (P.J.F., M.L.K., E.S.); Department of Ophthalmology & Visual Sciences, University of Kentucky, Lexington, KY (M.E.K., W.C., J.A.); Department of Human Genetics, University of Utah School of Medicine, Salt Lake City, UT (S.W., L.J.); Institute of Genetic Medicine (P.L.T., L.D., N.K.), Department of Molecular Biology and Genetics (N.K.), Department of Oncology (G.P.), Department of Biostatistics (C.W., G.P.), and Wilmer Eye Institute (J.S.S., P.C., B.C., D.J.Z., N.K.), Johns Hopkins University, Baltimore, MD; Richard E. Hoover Rehabilitation Services for Low Vision and Blindness, Greater Baltimore Medical Center, Baltimore, MD (J.S.S.); Departments of Pathology and Ophthalmology, Keck School of Medicine of the University of Southern California, Doheny Eye Institute, Los Angeles, CA (D.R.H.); Laboratory of Statistical Genetics, Rockefeller University, 1230 York Avenue, New York, NY (S.C.H.).
1,
Janet S Sunness
Janet S Sunness, M.D.
1Center for Human Molecular Biology and Genetics, Sichuan Academy of Medical Sciences and Sichuan Provincial People's Hospital, Chengdu, China (Z.Y.); Department of Ophthalmology & Visual Sciences, Moran Eye Center, University of Utah School of Medicine, Salt Lake City, UT (Z.Y., C.S., D.G., Z.T., H.C., R.C., X.Y., Y. C., J.Z., X.M., V.S.H., J.H., J.B., E.P., S.P., Y.K., L.L., N.A.Z., P.S.B., A.S., B.Y., K.Z.); Program in Human Molecular Biology & Genetics, Eccles Institute of Human Genetics, University of Utah School of Medicine, Salt Lake City, UT (C.S., D.G., Z.T., H.C., R.C., X.Y., H.C., J.Z., X.M., J.H., J.B., E.P., S.P., Y.K., L.L., B.Y., K.Z.); Department of Ophthalmology, Shiley Eye Center, University of California San Diego, San Diego, CA (Z.Y., Y.C., J.Z., K.Z.); Macular Degeneration Center, Casey Eye Institute, Oregon Health & Science University, Portland, OR (P.J.F., M.L.K., E.S.); Department of Ophthalmology & Visual Sciences, University of Kentucky, Lexington, KY (M.E.K., W.C., J.A.); Department of Human Genetics, University of Utah School of Medicine, Salt Lake City, UT (S.W., L.J.); Institute of Genetic Medicine (P.L.T., L.D., N.K.), Department of Molecular Biology and Genetics (N.K.), Department of Oncology (G.P.), Department of Biostatistics (C.W., G.P.), and Wilmer Eye Institute (J.S.S., P.C., B.C., D.J.Z., N.K.), Johns Hopkins University, Baltimore, MD; Richard E. Hoover Rehabilitation Services for Low Vision and Blindness, Greater Baltimore Medical Center, Baltimore, MD (J.S.S.); Departments of Pathology and Ophthalmology, Keck School of Medicine of the University of Southern California, Doheny Eye Institute, Los Angeles, CA (D.R.H.); Laboratory of Statistical Genetics, Rockefeller University, 1230 York Avenue, New York, NY (S.C.H.).
1,
Peter Campochiaro
Peter Campochiaro, M.D.
1Center for Human Molecular Biology and Genetics, Sichuan Academy of Medical Sciences and Sichuan Provincial People's Hospital, Chengdu, China (Z.Y.); Department of Ophthalmology & Visual Sciences, Moran Eye Center, University of Utah School of Medicine, Salt Lake City, UT (Z.Y., C.S., D.G., Z.T., H.C., R.C., X.Y., Y. C., J.Z., X.M., V.S.H., J.H., J.B., E.P., S.P., Y.K., L.L., N.A.Z., P.S.B., A.S., B.Y., K.Z.); Program in Human Molecular Biology & Genetics, Eccles Institute of Human Genetics, University of Utah School of Medicine, Salt Lake City, UT (C.S., D.G., Z.T., H.C., R.C., X.Y., H.C., J.Z., X.M., J.H., J.B., E.P., S.P., Y.K., L.L., B.Y., K.Z.); Department of Ophthalmology, Shiley Eye Center, University of California San Diego, San Diego, CA (Z.Y., Y.C., J.Z., K.Z.); Macular Degeneration Center, Casey Eye Institute, Oregon Health & Science University, Portland, OR (P.J.F., M.L.K., E.S.); Department of Ophthalmology & Visual Sciences, University of Kentucky, Lexington, KY (M.E.K., W.C., J.A.); Department of Human Genetics, University of Utah School of Medicine, Salt Lake City, UT (S.W., L.J.); Institute of Genetic Medicine (P.L.T., L.D., N.K.), Department of Molecular Biology and Genetics (N.K.), Department of Oncology (G.P.), Department of Biostatistics (C.W., G.P.), and Wilmer Eye Institute (J.S.S., P.C., B.C., D.J.Z., N.K.), Johns Hopkins University, Baltimore, MD; Richard E. Hoover Rehabilitation Services for Low Vision and Blindness, Greater Baltimore Medical Center, Baltimore, MD (J.S.S.); Departments of Pathology and Ophthalmology, Keck School of Medicine of the University of Southern California, Doheny Eye Institute, Los Angeles, CA (D.R.H.); Laboratory of Statistical Genetics, Rockefeller University, 1230 York Avenue, New York, NY (S.C.H.).
1,
Lynn Jorde
Lynn Jorde, Ph.D.
1Center for Human Molecular Biology and Genetics, Sichuan Academy of Medical Sciences and Sichuan Provincial People's Hospital, Chengdu, China (Z.Y.); Department of Ophthalmology & Visual Sciences, Moran Eye Center, University of Utah School of Medicine, Salt Lake City, UT (Z.Y., C.S., D.G., Z.T., H.C., R.C., X.Y., Y. C., J.Z., X.M., V.S.H., J.H., J.B., E.P., S.P., Y.K., L.L., N.A.Z., P.S.B., A.S., B.Y., K.Z.); Program in Human Molecular Biology & Genetics, Eccles Institute of Human Genetics, University of Utah School of Medicine, Salt Lake City, UT (C.S., D.G., Z.T., H.C., R.C., X.Y., H.C., J.Z., X.M., J.H., J.B., E.P., S.P., Y.K., L.L., B.Y., K.Z.); Department of Ophthalmology, Shiley Eye Center, University of California San Diego, San Diego, CA (Z.Y., Y.C., J.Z., K.Z.); Macular Degeneration Center, Casey Eye Institute, Oregon Health & Science University, Portland, OR (P.J.F., M.L.K., E.S.); Department of Ophthalmology & Visual Sciences, University of Kentucky, Lexington, KY (M.E.K., W.C., J.A.); Department of Human Genetics, University of Utah School of Medicine, Salt Lake City, UT (S.W., L.J.); Institute of Genetic Medicine (P.L.T., L.D., N.K.), Department of Molecular Biology and Genetics (N.K.), Department of Oncology (G.P.), Department of Biostatistics (C.W., G.P.), and Wilmer Eye Institute (J.S.S., P.C., B.C., D.J.Z., N.K.), Johns Hopkins University, Baltimore, MD; Richard E. Hoover Rehabilitation Services for Low Vision and Blindness, Greater Baltimore Medical Center, Baltimore, MD (J.S.S.); Departments of Pathology and Ophthalmology, Keck School of Medicine of the University of Southern California, Doheny Eye Institute, Los Angeles, CA (D.R.H.); Laboratory of Statistical Genetics, Rockefeller University, 1230 York Avenue, New York, NY (S.C.H.).
1,
Giovanni Parmigiani
Giovanni Parmigiani, Ph.D.
1Center for Human Molecular Biology and Genetics, Sichuan Academy of Medical Sciences and Sichuan Provincial People's Hospital, Chengdu, China (Z.Y.); Department of Ophthalmology & Visual Sciences, Moran Eye Center, University of Utah School of Medicine, Salt Lake City, UT (Z.Y., C.S., D.G., Z.T., H.C., R.C., X.Y., Y. C., J.Z., X.M., V.S.H., J.H., J.B., E.P., S.P., Y.K., L.L., N.A.Z., P.S.B., A.S., B.Y., K.Z.); Program in Human Molecular Biology & Genetics, Eccles Institute of Human Genetics, University of Utah School of Medicine, Salt Lake City, UT (C.S., D.G., Z.T., H.C., R.C., X.Y., H.C., J.Z., X.M., J.H., J.B., E.P., S.P., Y.K., L.L., B.Y., K.Z.); Department of Ophthalmology, Shiley Eye Center, University of California San Diego, San Diego, CA (Z.Y., Y.C., J.Z., K.Z.); Macular Degeneration Center, Casey Eye Institute, Oregon Health & Science University, Portland, OR (P.J.F., M.L.K., E.S.); Department of Ophthalmology & Visual Sciences, University of Kentucky, Lexington, KY (M.E.K., W.C., J.A.); Department of Human Genetics, University of Utah School of Medicine, Salt Lake City, UT (S.W., L.J.); Institute of Genetic Medicine (P.L.T., L.D., N.K.), Department of Molecular Biology and Genetics (N.K.), Department of Oncology (G.P.), Department of Biostatistics (C.W., G.P.), and Wilmer Eye Institute (J.S.S., P.C., B.C., D.J.Z., N.K.), Johns Hopkins University, Baltimore, MD; Richard E. Hoover Rehabilitation Services for Low Vision and Blindness, Greater Baltimore Medical Center, Baltimore, MD (J.S.S.); Departments of Pathology and Ophthalmology, Keck School of Medicine of the University of Southern California, Doheny Eye Institute, Los Angeles, CA (D.R.H.); Laboratory of Statistical Genetics, Rockefeller University, 1230 York Avenue, New York, NY (S.C.H.).
1,
Donald J Zack
Donald J Zack, M.D., Ph.D.
1Center for Human Molecular Biology and Genetics, Sichuan Academy of Medical Sciences and Sichuan Provincial People's Hospital, Chengdu, China (Z.Y.); Department of Ophthalmology & Visual Sciences, Moran Eye Center, University of Utah School of Medicine, Salt Lake City, UT (Z.Y., C.S., D.G., Z.T., H.C., R.C., X.Y., Y. C., J.Z., X.M., V.S.H., J.H., J.B., E.P., S.P., Y.K., L.L., N.A.Z., P.S.B., A.S., B.Y., K.Z.); Program in Human Molecular Biology & Genetics, Eccles Institute of Human Genetics, University of Utah School of Medicine, Salt Lake City, UT (C.S., D.G., Z.T., H.C., R.C., X.Y., H.C., J.Z., X.M., J.H., J.B., E.P., S.P., Y.K., L.L., B.Y., K.Z.); Department of Ophthalmology, Shiley Eye Center, University of California San Diego, San Diego, CA (Z.Y., Y.C., J.Z., K.Z.); Macular Degeneration Center, Casey Eye Institute, Oregon Health & Science University, Portland, OR (P.J.F., M.L.K., E.S.); Department of Ophthalmology & Visual Sciences, University of Kentucky, Lexington, KY (M.E.K., W.C., J.A.); Department of Human Genetics, University of Utah School of Medicine, Salt Lake City, UT (S.W., L.J.); Institute of Genetic Medicine (P.L.T., L.D., N.K.), Department of Molecular Biology and Genetics (N.K.), Department of Oncology (G.P.), Department of Biostatistics (C.W., G.P.), and Wilmer Eye Institute (J.S.S., P.C., B.C., D.J.Z., N.K.), Johns Hopkins University, Baltimore, MD; Richard E. Hoover Rehabilitation Services for Low Vision and Blindness, Greater Baltimore Medical Center, Baltimore, MD (J.S.S.); Departments of Pathology and Ophthalmology, Keck School of Medicine of the University of Southern California, Doheny Eye Institute, Los Angeles, CA (D.R.H.); Laboratory of Statistical Genetics, Rockefeller University, 1230 York Avenue, New York, NY (S.C.H.).
1,
Nicholas Katsanis
Nicholas Katsanis, Ph.D.
1Center for Human Molecular Biology and Genetics, Sichuan Academy of Medical Sciences and Sichuan Provincial People's Hospital, Chengdu, China (Z.Y.); Department of Ophthalmology & Visual Sciences, Moran Eye Center, University of Utah School of Medicine, Salt Lake City, UT (Z.Y., C.S., D.G., Z.T., H.C., R.C., X.Y., Y. C., J.Z., X.M., V.S.H., J.H., J.B., E.P., S.P., Y.K., L.L., N.A.Z., P.S.B., A.S., B.Y., K.Z.); Program in Human Molecular Biology & Genetics, Eccles Institute of Human Genetics, University of Utah School of Medicine, Salt Lake City, UT (C.S., D.G., Z.T., H.C., R.C., X.Y., H.C., J.Z., X.M., J.H., J.B., E.P., S.P., Y.K., L.L., B.Y., K.Z.); Department of Ophthalmology, Shiley Eye Center, University of California San Diego, San Diego, CA (Z.Y., Y.C., J.Z., K.Z.); Macular Degeneration Center, Casey Eye Institute, Oregon Health & Science University, Portland, OR (P.J.F., M.L.K., E.S.); Department of Ophthalmology & Visual Sciences, University of Kentucky, Lexington, KY (M.E.K., W.C., J.A.); Department of Human Genetics, University of Utah School of Medicine, Salt Lake City, UT (S.W., L.J.); Institute of Genetic Medicine (P.L.T., L.D., N.K.), Department of Molecular Biology and Genetics (N.K.), Department of Oncology (G.P.), Department of Biostatistics (C.W., G.P.), and Wilmer Eye Institute (J.S.S., P.C., B.C., D.J.Z., N.K.), Johns Hopkins University, Baltimore, MD; Richard E. Hoover Rehabilitation Services for Low Vision and Blindness, Greater Baltimore Medical Center, Baltimore, MD (J.S.S.); Departments of Pathology and Ophthalmology, Keck School of Medicine of the University of Southern California, Doheny Eye Institute, Los Angeles, CA (D.R.H.); Laboratory of Statistical Genetics, Rockefeller University, 1230 York Avenue, New York, NY (S.C.H.).
1,
Jayakrishna Ambati
Jayakrishna Ambati, M.D.
1Center for Human Molecular Biology and Genetics, Sichuan Academy of Medical Sciences and Sichuan Provincial People's Hospital, Chengdu, China (Z.Y.); Department of Ophthalmology & Visual Sciences, Moran Eye Center, University of Utah School of Medicine, Salt Lake City, UT (Z.Y., C.S., D.G., Z.T., H.C., R.C., X.Y., Y. C., J.Z., X.M., V.S.H., J.H., J.B., E.P., S.P., Y.K., L.L., N.A.Z., P.S.B., A.S., B.Y., K.Z.); Program in Human Molecular Biology & Genetics, Eccles Institute of Human Genetics, University of Utah School of Medicine, Salt Lake City, UT (C.S., D.G., Z.T., H.C., R.C., X.Y., H.C., J.Z., X.M., J.H., J.B., E.P., S.P., Y.K., L.L., B.Y., K.Z.); Department of Ophthalmology, Shiley Eye Center, University of California San Diego, San Diego, CA (Z.Y., Y.C., J.Z., K.Z.); Macular Degeneration Center, Casey Eye Institute, Oregon Health & Science University, Portland, OR (P.J.F., M.L.K., E.S.); Department of Ophthalmology & Visual Sciences, University of Kentucky, Lexington, KY (M.E.K., W.C., J.A.); Department of Human Genetics, University of Utah School of Medicine, Salt Lake City, UT (S.W., L.J.); Institute of Genetic Medicine (P.L.T., L.D., N.K.), Department of Molecular Biology and Genetics (N.K.), Department of Oncology (G.P.), Department of Biostatistics (C.W., G.P.), and Wilmer Eye Institute (J.S.S., P.C., B.C., D.J.Z., N.K.), Johns Hopkins University, Baltimore, MD; Richard E. Hoover Rehabilitation Services for Low Vision and Blindness, Greater Baltimore Medical Center, Baltimore, MD (J.S.S.); Departments of Pathology and Ophthalmology, Keck School of Medicine of the University of Southern California, Doheny Eye Institute, Los Angeles, CA (D.R.H.); Laboratory of Statistical Genetics, Rockefeller University, 1230 York Avenue, New York, NY (S.C.H.).
1,
Kang Zhang
Kang Zhang, M.D., Ph.D
1Center for Human Molecular Biology and Genetics, Sichuan Academy of Medical Sciences and Sichuan Provincial People's Hospital, Chengdu, China (Z.Y.); Department of Ophthalmology & Visual Sciences, Moran Eye Center, University of Utah School of Medicine, Salt Lake City, UT (Z.Y., C.S., D.G., Z.T., H.C., R.C., X.Y., Y. C., J.Z., X.M., V.S.H., J.H., J.B., E.P., S.P., Y.K., L.L., N.A.Z., P.S.B., A.S., B.Y., K.Z.); Program in Human Molecular Biology & Genetics, Eccles Institute of Human Genetics, University of Utah School of Medicine, Salt Lake City, UT (C.S., D.G., Z.T., H.C., R.C., X.Y., H.C., J.Z., X.M., J.H., J.B., E.P., S.P., Y.K., L.L., B.Y., K.Z.); Department of Ophthalmology, Shiley Eye Center, University of California San Diego, San Diego, CA (Z.Y., Y.C., J.Z., K.Z.); Macular Degeneration Center, Casey Eye Institute, Oregon Health & Science University, Portland, OR (P.J.F., M.L.K., E.S.); Department of Ophthalmology & Visual Sciences, University of Kentucky, Lexington, KY (M.E.K., W.C., J.A.); Department of Human Genetics, University of Utah School of Medicine, Salt Lake City, UT (S.W., L.J.); Institute of Genetic Medicine (P.L.T., L.D., N.K.), Department of Molecular Biology and Genetics (N.K.), Department of Oncology (G.P.), Department of Biostatistics (C.W., G.P.), and Wilmer Eye Institute (J.S.S., P.C., B.C., D.J.Z., N.K.), Johns Hopkins University, Baltimore, MD; Richard E. Hoover Rehabilitation Services for Low Vision and Blindness, Greater Baltimore Medical Center, Baltimore, MD (J.S.S.); Departments of Pathology and Ophthalmology, Keck School of Medicine of the University of Southern California, Doheny Eye Institute, Los Angeles, CA (D.R.H.); Laboratory of Statistical Genetics, Rockefeller University, 1230 York Avenue, New York, NY (S.C.H.).
1