Abstract
The docking and fusion of cargo-containing vesicles with target membranes of eukaryotic cells is mediated by the interaction of SNARE proteins present on both vesicle and target membranes. In many cases, the target membrane SNARE, or t-SNARE, exists as a complex of syntaxin with a member of the SNAP-25 family of palmitoylated proteins. We have identified a novel human kinase SNAK (SNARE kinase) that specifically phosphorylates the nonneuronal t-SNARE SNAP-23 in vivo. Interestingly, only SNAP-23 that is not assembled into t-SNARE complexes is phosphorylated by SNAK, and phosphorylated SNAP-23 resides exclusively in the cytosol. Coexpression with SNAK significantly enhances the stability of unassembled SNAP-23, and as a consequence, the assembly of newly synthesized SNAP-23 with syntaxin is augmented. These data demonstrate that phosphorylation of SNAP-23 by SNAK enhances the kinetics of t-SNARE assembly in vivo.
INTRODUCTION
Vesicular transport is required for many diverse processes of membrane and protein trafficking in eukaryotic cells. The interaction of integral membrane proteins on vesicles (termed v-SNAREs) with membrane-associated proteins on target organelles (termed t-SNAREs) is widely believed to impart some degree of specificity to this process (Rothman, 1994; Hay and Scheller, 1997). Data from a number of experimental systems have shown that the interaction of v-SNAREs on a donor membrane with t-SNAREs on an acceptor membrane is required for membrane fusion (Nichols et al., 1997; Weber et al., 1998), although the precise mechanism of fusion and the contribution of other proteins to the fusion process remain controversial.
In neurons and neuroendocrine cells, the t-SNAREs consist of the integral membrane protein syntaxin and the palmitoylated protein SNAP-25 (reviewed by Ferro-Novick and Jahn, 1994; Bennett and Scheller, 1994; Südhof, 1995), whereas in nonneuronal tissues, SNAP-23 functionally replaces SNAP-25 (Ravichandran et al., 1996; Sadoul et al., 1997). There is considerable evidence that both syntaxin and SNAP-23 family members are required for t-SNARE function and that t-SNAREs exist as heterodimers of syntaxin and SNAP-23 family members (Söllner et al., 1993; Brennwald et al., 1994; Rea et al., 1998; Ungermann and Wickner, 1998; Weber et al., 1998; St.-Denis et al., 1999). For example, fusion of v-SNARE–containing phospholipid vesicles with t-SNARE vesicles requires that the t-SNARE complex contain both syntaxin and SNAP-25 (Weber et al., 1998). In addition, the ability of insulin to stimulate GLUT4 translocation in adipocytes requires functional syntaxin and SNAP-23 proteins (Rea et al., 1998), and we have recently shown that these proteins exist in a complex before and after insulin stimulation (St.-Denis et al., 1999). Because the formation of the t-SNARE heterodimer is the rate-limiting step in the assembly of the yeast v-SNARE/t-SNARE fusion complex (Nicholson et al., 1998), it is likely that the initial interaction of syntaxin with SNAP-23 family members is an essential step leading to the docking and fusion of vesicles with target membranes.
Despite the importance of the SNAP-23/syntaxin t-SNARE complex for membrane traffic in eukaryotic cells, factors that regulate the assembly of this heterodimer in vivo are not known. There is evidence that the association of syntaxin with the mammalian sec1 homologue munc18 prevents the binding of syntaxin to SNAP-23 (Araki et al., 1997; Riento et al., 1998), although a recent analysis of t-SNARE binding to yeast Sec1p calls into question the role of Sec1 family members in t-SNARE assembly (Carr et al., 1999). To identify proteins regulating SNARE function in hematopoietic cells, we have screened a yeast two-hybrid B lymphocyte cDNA library with the plasma membrane t-SNARE syntaxin 4. We had used this library to isolate SNAP-23 (Ravichandran et al., 1996), a t-SNARE that binds to syntaxin and regulates such diverse processes as transcytosis (Low et al., 1998a), transferrin recycling (Leung et al., 1998), insulin-stimulated GLUT4 translocation (Rea et al., 1998), and mast cell exocytosis (Guo et al., 1998). We now report the isolation and identification of a novel serine/threonine kinase, termed SNAK (for SNARE kinase), that phosphorylates nascent SNAP-23 and significantly enhances the in vivo assembly of SNAP-23/syntaxin t-SNARE complexes.
MATERIALS AND METHODS
cDNA Cloning of SNAK
The human B lymphocyte cDNA library in the vector pACTI was screened with the use of the cytosolic domain of human syntaxin 4 as described previously (Ravichandran et al., 1996). A 1-kilobase cDNA encoding a novel kinase was isolated during this screen, and while we were screening a human placenta λgt11 cDNA library to obtain a full-length clone of this cDNA, the sequence of a full-length clone corresponding to our partial cDNA fragment was reported (Nagase et al., 1995). The coding region for this clone, termed KIAA0137/HUMKG17 (GenBank accession number D50927), was amplified by PCR with the use of human B lymphocyte cDNA and specific primers and subcloned into the mammalian expression vector pCDM8 (Invitrogen, Carlsbad, CA). The complete sequence of this cDNA, termed SNAK, has been deposited in GenBank (accession pending). The SNAK Mg2+-binding mutant was generated by site-directed mutagenesis of residues Asp-390, Phe-391, and Gly-392 to three alanines with the use of the Quick-Change mutagenesis protocol (Stratagene, La Jolla, CA). The sequence of all DNA constructs was verified by automated sequence analysis.
Northern Blot Analysis
The expression of SNAK mRNA was analyzed by Northern blot analysis. A multiple human tissue Northern blot containing 2 μg of poly(A)+ RNA from heart, brain, placenta, lung, liver, skeletal muscle, kidney, and pancreas (Clontech, Palo Alto, CA) was probed with a 460-base pair SNAK probe (encompassing amino acids 1–119) under high-stringency hybridization conditions as described previously (Valdez et al., 1999).
Antibodies
Immune sera directed against SNAK were generated by immunizing rabbits with synthetic peptides corresponding to the 17 amino-terminal amino acids of SNAK coupled to keyhole limpet hemocyanin. SNAP-23 antiserum has been described elsewhere (Low et al., 1998b), and syntaxin 4 antiserum (Mandon et al., 1996) was a gift from Dr. Mark Knepper (National Heart, Lung, and Blood Institute, National Institutes of Health, Bethesda, MD).
GST Phosphorylation Assay
GST fusion proteins encoding full-length human SNAP-23, the cytosolic portion of rat VAMP 2, and the cytosolic portion of human syntaxin 4 were described previously (Ravichandran et al., 1996). GST-SNAK was generated by subcloning the entire coding region of SNAK into pGEX-4T (Pharmacia, Piscataway, NJ). His6-SNAP-23 was generated by subcloning the entire coding region of human SNAP-23 (Low et al., 1998b) into the EcoRI and XhoI sites of pET-28a (Novagen, Madison, WI). GST fusion proteins and His6-SNAP-23 were isolated with the use of standard protocols. In vitro phosphorylation of GST, GST-syntaxin 4, GST-VAMP 2, and GST-SNAP-23 fusion proteins was performed by immobilizing 3 μg of each fusion protein onto glutathione-Sepharose beads and incubating each sample for 10 min at 30°C with 100 ng of GST-SNAK and 1 μCi of [γ-32P]ATP in 50 μl of buffer (20 mM HEPES, pH 7.0, 25 mM NaCl, 1 mg/ml BSA, 1 mM EDTA, 3% glycerol, 7 mM MgCl2, 7 mM CaCl2, and proteinase inhibitors). After extensive washing, the samples were solubilized by boiling in SDS sample buffer and analyzed by SDS-PAGE. After staining with Coomassie blue, the gels were dried and subjected to autoradiography. In vitro phosphorylation of His6-SNAP-23 was performed by incubating His6-SNAP-23 (1.5 μg) with various amounts of GST-SNAK and 1 μCi of [γ-32P]ATP for 30 min at 30°C in 20 μl of buffer (10 mM HEPES, pH 7.0, 150 mM NaCl, 10 mM MgCl2, 1 mM CaCl2). The reaction was terminated by boiling the sample in SDS-PAGE sample buffer, and the samples were analyzed by SDS-PAGE and autoradiography.
Cell Culture, SDS-PAGE, and Immunoprecipitation
HeLa cells were transiently transfected with plasmid DNA with the use of calcium phosphate or Lipofectamine (Life Technologies, Grand Island, NY), radiolabeled with [35S]methionine or [32P]orthophosphate, lysed, and subjected to immunoprecipitation and SDS-PAGE as described (Anderson and Roche, 1998). In some experiments, cells were pulse labeled with [35S]methionine for 15 min, the cells were washed, and complete medium was added for various times before cell isolation. Cells were lysed in 1% Triton X-100 in a buffer of 10 mM Tris, pH 7.4, 150 mM NaCl with protease and phosphatase inhibitors. Nuclei were removed by low-speed centrifugation, cell lysates were precleared with control antibodies, and specific immunoprecipitation with the use of antibodies bound to protein A–agarose was performed as described (Anderson and Roche, 1998). Immunoblotting of immunoprecipitates was performed by probing the blots with anti-SNAP-23 antisera, and the results were visualized with the use of HRP-protein A and ECL. Control studies confirmed that the 32P signal was not detectable during the chemiluminescence experiments.
Subcellular Fractionation
Transfected HeLa cells were harvested by trypsinization and washed in PBS. The cells were resuspended in hypotonic buffer (Valdez et al., 1999) and were disrupted by repeated passage of cells through a 25-gauge syringe. Nuclei and unbroken cells were removed by centrifugation at 1,000 × g, and the postnuclear supernatant was subjected to centrifugation at 15,000 × g for 1 h at 4°C to isolate membranes (pellet) and cytosol (supernatant). Control experiments demonstrated that essentially identical amounts of SNAP-23 and syntaxin were present in 15,000 × g and 100,000 × g pellets. The membrane pellet and cytosolic supernatant were adjusted to a final concentration of 1% Triton X-100 in a buffer containing equal parts lysis buffer and hypotonic cell resuspension buffer before preclearing and specific immunoprecipitation, as described above.
RESULTS
Identification of SNAK
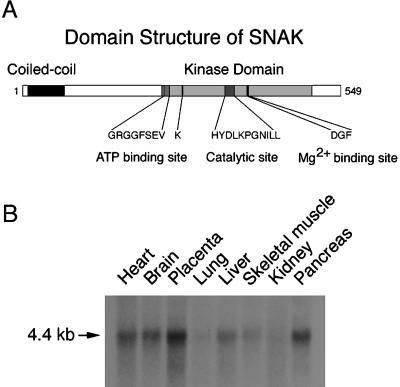
To identify proteins regulating SNARE function in hematopoietic cells, we have used the yeast two-hybrid system with the plasma membrane t-SNARE syntaxin 4 as bait. In addition to identifying SNAP-23 in this screen, we obtained a partial cDNA encoding a novel human serine/threonine kinase. Analysis of a full-length clone of this kinase revealed a 549-amino acid protein that contains a single coiled-coil domain of seven heptad repeats at the amino terminus, no hydrophobic transmembrane domain, and several motifs conserved among serine/threonine kinase proteins at the carboxy terminus (Figure 1A). Northern blot analysis revealed that mRNA for this kinase was ubiquitously expressed (Figure 1B), and Western blot analysis with a polyclonal rabbit antiserum revealed that this protein was expressed at low levels in most tissues examined (data not shown). Given our method of identifying this protein and subsequent biochemical and functional analyses, we have termed this kinase SNAK, for SNARE kinase.
Figure 1.
Identification of SNAK, a SNARE kinase. (A) Domain structure of SNAK. The presence of the coiled-coil domain within SNAK was identified by the COILS algorithm (Lupas, 1996), and the presence of the kinase domain and the specified subdomains was determined by aligning the primary sequence of SNAK with that of other serine/threonine kinases. (B) Northern blot analysis of SNAK expression. A human tissue Northern blot containing poly(A)+ RNA from heart, brain, placenta, lung, liver, skeletal muscle, kidney, and pancreas was probed with a SNAK probe under high-stringency hybridization conditions. A single 4.4-kilobase transcript was detected in all tissues examined in this and other Northern blots.
SNAK Phosphorylates SNAP-23 In Vitro and In Vivo
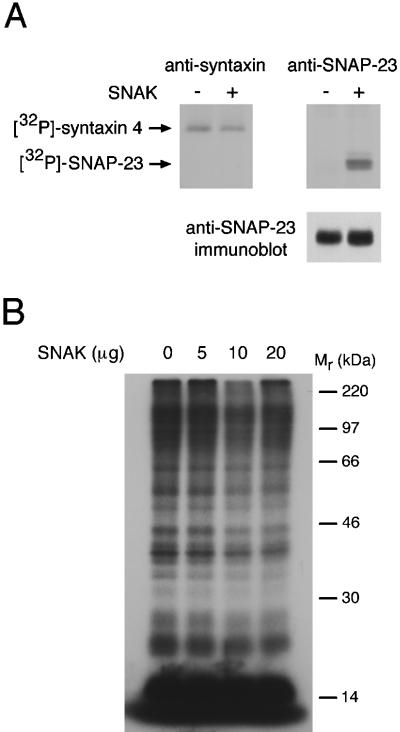
To determine if SNAK possessed t-SNARE kinase activity, we performed in vitro phosphorylation reactions with the use of GST-SNARE fusion proteins as substrates. SNAK did not phosphorylate GST and only weakly phosphorylated GST-VAMP 2 and GST-syntaxin 4 (Figure 2A). Similar results were obtained when we examined the phosphorylation of GST-syntaxin 1, GST-syntaxin 2, GST-syntaxin 3, GST-VAMP 1, and GST α-SNAP (data not shown). Surprisingly, GST-SNAP-23 was a very good substrate for SNAK and was a much better substrate than syntaxin 4, the t-SNARE used to isolate SNAK from the yeast two-hybrid screen.
Figure 2.
SNAK phosphorylates SNAP-23 in vitro. (A) GST-SNAK was incubated with immobilized GST, GST-syntaxin 4, or GST-SNAP-23 in the presence of [γ-32P]ATP at 30°C for 10 min. The reaction was terminated by extensive washing, and in vitro phosphorylation reactions were analyzed by SDS-PAGE and autoradiography. The relative mass (Mr) of 14C-labeled molecular weight markers is indicated. (B) His6-SNAP-23 (1.5 μg) was incubated with the indicated amount of GST-SNAK in the presence of [γ-32P]ATP at 30°C for 30 min. The reaction products were analyzed by SDS-PAGE and autoradiography.
Using a His6-SNAP-23 fusion protein and GST-SNAK, we confirmed that SNAP-23 is an excellent substrate for SNAK. Very little SNAK was required to observe robust phosphorylation of SNAP-23, and the phosphorylation was observed only in the presence of His6-SNAP-23 (Figure 2B). In addition to SNAP-23, SNAK itself was phosphorylated in each of these studies, demonstrating that this kinase is capable of either cis or trans autophosphorylation.
We next set out to determine if SNAK possessed SNARE kinase activity in vivo. Although most cell lines we have examined possess endogenous syntaxin 4, SNAP-23, and SNAK, the expression of these proteins is generally quite low and the detection of the metabolically radiolabeled endogenous proteins is extremely difficult. For this reason, HeLa cells were transiently transfected with t-SNARE cDNAs in the presence or absence of SNAK and the cells were radiolabeled with [32P]orthophosphate to monitor in situ phosphorylation. In agreement with in vitro phosphorylation studies (Foster et al., 1998; Risinger and Bennett, 1999), we found that syntaxin 4 was basally phosphorylated in HeLa cells and that coexpression of SNAK did not alter syntaxin 4 phosphorylation (Figure 3A). By contrast, coexpression with SNAK dramatically increased the phosphorylation of SNAP-23. Similar increases in phosphorylation were obtained with the use of the neuronal SNAP-23 homologue SNAP-25 as the substrate (data not shown). Immunoblot analysis demonstrated that coexpression with SNAK only slightly enhanced the amount of SNAP-23 protein present in the cells at steady state, a result that we attribute to enhanced SNAP-23 stabilization (see below). In addition, analysis of total 32P cell lysates did not reveal a gross increase in protein phosphorylation in cells overexpressing SNAK alone (Figure 3B), arguing in favor of a specific target for SNAK in vivo. Together, these data demonstrate that SNAP-23 is the predominant t-SNARE substrate for SNAK.
Figure 3.
SNAK phosphorylates SNAP-23 in vivo. (A) Transfected HeLa cells expressing syntaxin 4 or SNAP-23 in the absence (−) or presence (+) of SNAK were labeled with [32P]orthophosphate, lysed in Triton X-100, immunoprecipitated with the indicated antiserum, and analyzed by SDS-PAGE and fluorography. Equivalent fractions of each anti-SNAP-23 immunoprecipitate were also analyzed by immunoblotting with a SNAP-23 antiserum (lower panel). (B) HeLa cells were mock transfected or transfected with 5, 10, or 20 μg of a SNAK expression vector by calcium phosphate coprecipitation and labeled with [32P]orthophosphate for 2 h. The cells were lysed in Triton X-100, nuclei and debris were removed by centrifugation in a microfuge, and equal amounts of each lysate were analyzed by SDS-PAGE and fluorography. The relative mass (Mr) of 14C-labeled molecular weight markers is indicated.
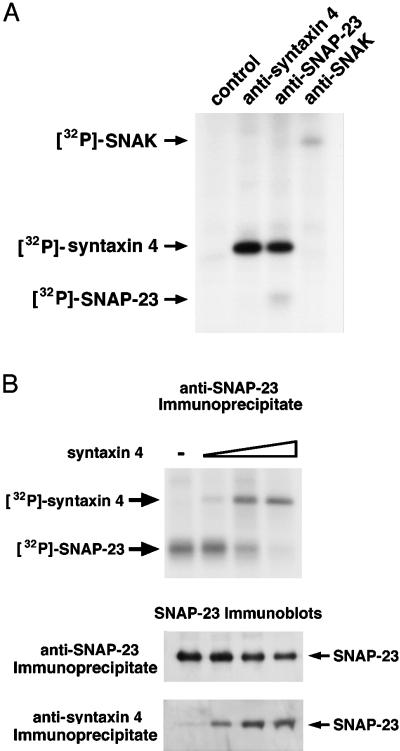
SNAP-23 in t-SNARE Complexes Is Not Phosphorylated by SNAK
Because SNAP-23 can exist either as an unassembled monomer or bound to syntaxin in a heterodimeric t-SNARE complex in vivo (Foster et al., 1998; Riento et al., 1998; St.-Denis et al., 1999), we examined the phosphorylation status of SNAP-23 in t-SNARE complexes by coexpressing syntaxin 4, SNAP-23, and SNAK in HeLa cells radiolabeled with [32P]orthophosphate. Similar amounts of [32P]syntaxin 4 were present in anti-syntaxin 4 or anti-SNAP-23 immunoprecipitates (Figure 4A), revealing that a significant fraction of the [32P]syntaxin 4 in the sample was indeed associated with SNAP-23. Interestingly, although [32P]SNAP-23 was observed in the SNAP-23 immunoprecipitate, absolutely no [32P]SNAP-23 was present in the syntaxin 4 immunoprecipitate, demonstrating that phosphorylated SNAP-23 was not present in t-SNARE complexes. In agreement with the autophosphorylation of SNAK observed in vitro, we found that SNAK was also phosphorylated in vivo (Figure 4A). Furthermore, we did not detect either syntaxin 4 or SNAP-23 in anti-SNAK immunoprecipitates, demonstrating that the interaction between SNAK and these t-SNAREs is either weak or transient.
Figure 4.
SNAP-23 in t-SNARE complexes is not phosphorylated. (A) HeLa cells expressing syntaxin 4, SNAP-23, and SNAK were labeled with [32P]orthophosphate, lysed in Triton X-100, subjected to immunoprecipitation with the indicated antiserum, and analyzed by SDS-PAGE and fluorography. (B) HeLa cells were transfected with a constant amount of SNAP-23 and SNAK cDNA (2 μg each) and increasing amounts of syntaxin 4 cDNA (0, 0.1, 0.5, or 2 μg) with the use of Lipofectamine. The cells were then labeled with [32P]orthophosphate, lysed, immunoprecipitated with the indicated antiserum, and directly analyzed by SDS-PAGE and fluorography (upper panel). The immunoprecipitates were also probed by immunoblotting with a SNAP-23 antiserum to detect total SNAP-23 and t-SNARE–associated SNAP-23 (lower panels).
The absence of [32P]SNAP-23 in t-SNARE complexes suggests that free SNAP-23 is the preferred substrate for SNAK in vivo and led us to predict that promoting SNAP-23 assembly into t-SNARE complexes would prevent SNAP-23 phosphorylation by SNAK. To test this hypothesis directly, we transfected cells with constant amounts of SNAP-23 and SNAK together with various amounts of syntaxin and labeled the cells with [32P]orthophosphate to monitor in situ phosphorylation. In the absence of syntaxin 4, SNAP-23 was readily phosphorylated (Figure 4B), confirming that free SNAP-23 is indeed a good substrate for SNAK. Expression of increasing amounts of syntaxin 4 in the cells enhanced t-SNARE complex assembly, as indicated by a dose-dependent increase in SNAP-23 coprecipitation with the syntaxin 4 antibody and a dose-dependent increase in [32P]syntaxin 4 coprecipitation with the SNAP-23 antibody. In agreement with our hypothesis, we found that the extent of SNAP-23 phosphorylation in the cells was inversely proportional to the amount of SNAP-23 present in t-SNARE complexes, because [32P]SNAP-23 was not detected in cells expressing large amounts of syntaxin 4 (Figure 4B). These data confirm that SNAP-23 in t-SNARE complexes is not a substrate for SNAK phosphorylation in vivo.
Because free SNAP-23 is readily phosphorylated by SNAK and SNAP-23 in the assembled t-SNARE complex is not, we performed in vitro binding assays to determine if phosphorylated SNAP-23 was capable of binding to syntaxin. In agreement with many reports examining the binding of SNAP-23 to syntaxin (Ravichandran et al., 1996; Araki et al., 1997; Steegmaier et al., 1998), GST-syntaxin bound readily to nonphosphorylated His6-SNAP-23 in vitro (data not shown). However, 32P-labeled His6-SNAP-23 that was phosphorylated by SNAK did not bind specifically to the GST-syntaxin 4 fusion protein (data not shown). This result, together with our data obtained in vivo, strongly suggest that not only is SNAP-23 in a preformed t-SNARE complex not a substrate for SNAK but also that phosphorylated SNAP-23 is unable to bind to syntaxin.
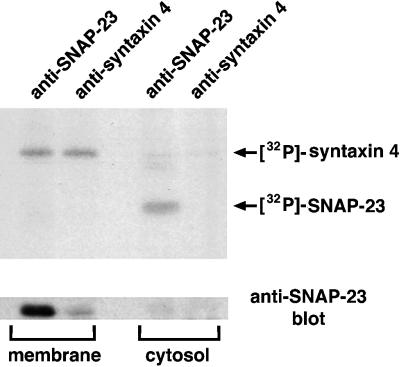
Phosphorylated SNAP-23 Resides in the Cytosol
To further analyze the status of SNAP-23 in t-SNARE complexes, we examined the intracellular location of [32P]SNAP-23 in HeLa cells expressing SNAP-23, SNAK, and subsaturating amounts of syntaxin. Like SNAP-25 (Veit et al., 1996; Gonzalo and Linder, 1998), SNAP-23 binds to syntaxins and associates with membranes only after posttranslational palmitoylation (Koticha et al., 1999; Vogel and Roche, 1999). Subcellular fractionation studies demonstrated that although SNAP-23 was clearly associated with [32P]syntaxin 4 in membrane-associated t-SNARE complexes, the SNAP-23 in these complexes was not phosphorylated (Figure 5). By contrast, despite the low level of SNAP-23 protein present in the cytosol, [32P]SNAP-23 was readily detected in the cytosolic fraction and was not assembled into syntaxin 4–containing t-SNARE complexes. (We attribute the enhanced SNAP-23 levels in the SNAP-23 immunoprecipitate compared with the syntaxin 4 immunoprecipitate to the presence of either free, nonphosphorylated SNAP-23 in the membranes or to SNAP-23 molecules associated with other syntaxin isoforms in these cells.) Together, these data unambiguously demonstrate that the phosphorylated pool of SNAP-23 resides in the cytosol and is not associated with syntaxin.
Figure 5.
Phosphorylated SNAP-23 resides in the cytosol. HeLa cells expressing syntaxin 4, SNAP-23, and SNAK were labeled for 3 h with [32P]orthophosphate. After mechanical cell disruption, the postnuclear supernatant was separated into a membrane fraction and a cytosol fraction as described in the text. Triton X-100 was added to each fraction, immunoprecipitations were performed with the use of the indicated antiserum, and equivalent portions of the immunoprecipitates from each fraction were analyzed by SDS-PAGE and fluorography (upper panel). The immunoprecipitates were also probed by immunoblotting with a SNAP-23 antiserum to detect the total amount of SNAP-23 present in each sample (lower panel).
Phosphorylation of SNAP-23 by SNAK Enhances t-SNARE Complex Assembly
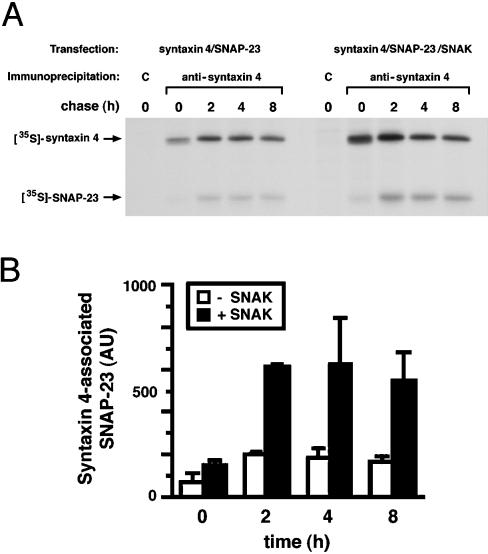
The failure to detect phosphorylated SNAP-23 in t-SNARE complexes suggested that there might be competition between phosphorylation of SNAP-23 and the assembly of SNAP-23 into t-SNARE complexes. If this were correct, overexpression of SNAK, which significantly enhances phosphorylation of SNAP-23, would inhibit t-SNARE complex assembly in vivo. Contrary to this expectation, pulse-chase biosynthesis experiments with [35S]methionine revealed that significantly more newly synthesized SNAP-23 was assembled with syntaxin 4 in cells expressing SNAK compared with cells that were not expressing SNAK (Figure 6A). Interestingly, we routinely observed a slight increase in the expression of syntaxin 4 in cells cotransfected with SNAP-23 and SNAK. We attribute this to enhanced stabilization of syntaxin in the t-SNARE complexes in cells expressing SNAK, as we have noted that, unlike free syntaxin, syntaxin bound to SNAP-25 is resistant to proteolytic degradation in vitro (data not shown). To account for this slight increase in syntaxin 4 protein, therefore, the amount of SNAP-23 associated with syntaxin 4 at each time point was normalized for the total amount of syntaxin 4 present in the sample (Figure 6B). The results of quantitative phosphorimager analysis of independent experiments confirm that, instead of inhibiting t-SNARE assembly, phosphorylation of SNAP-23 actually promotes the assembly of SNAP-23 into t-SNARE complexes.
Figure 6.
Phosphorylation by SNAK promotes SNAP-23 assembly with syntaxin. (A) HeLa cells expressing SNAP-23 and syntaxin 4 in the absence or presence of SNAK were labeled with [35S]methionine for 15 min and chased in complete medium. After incubation for various times at 37°C, the cells were harvested, lysed in Triton X-100, immunoprecipitated with the use of control (C) or anti-syntaxin 4 serum, and analyzed by SDS-PAGE and fluorography. (B) The relative amount of [35S]SNAP-23 coprecipitated with the syntaxin 4 serum at different times of chase in the absence (□) or in the presence (▪) of SNAK was determined by phosphorimager analysis. The amount of [35S]SNAP-23 present is expressed in arbitrary phosphorimager units (AU). Each value represents the results from two independent experiments with SD.
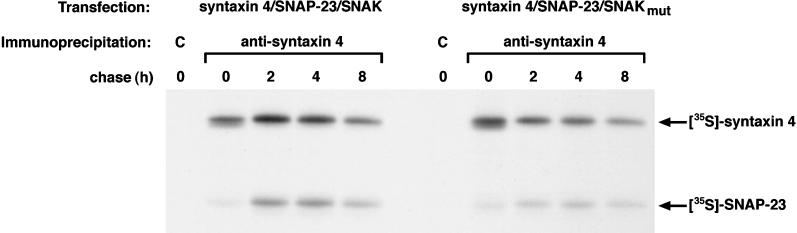
To confirm that the effect of SNAK on t-SNARE complex assembly was dependent on its kinase activity, we generated an inactive form of SNAK by mutagenesis of the Mg2+-binding site (to eliminate kinase activity). Pulse-chase biosynthesis studies performed in the presence of either wild-type or mutant SNAK revealed that kinase activity was required for SNAK-mediated enhancement of SNAP-23/syntaxin t-SNARE complex assembly (Figure 7). To demonstrate this in another way, we performed this same assembly assay in the absence or presence of the serine/threonine kinase inhibitor staurosporine. Staurosporine prevents SNAK kinase activity in vivo, and inclusion of staurosporine during the chase completely eliminated the effect of SNAK on enhanced t-SNARE complex assembly (data not shown). These data demonstrate that the stimulation of t-SNARE complex assembly by SNAK is due to its kinase activity.
Figure 7.
SNAK kinase activity is required to promote SNAP-23 assembly with syntaxin. HeLa cells expressing SNAP-23 and syntaxin 4 in the presence of wild-type SNAK (left panel) or SNAP-23 and syntaxin 4 in the presence of a catalytically inactive SNAK mutant (right panel) were labeled with [35S]methionine for 15 min and chased for various times. After cell lysis in Triton X-100, lysates were immunoprecipitated with the use of control (C) or anti-syntaxin 4 serum and analyzed by SDS-PAGE and fluorography.
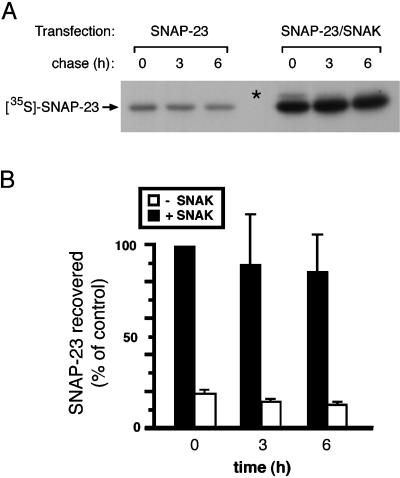
SNAK Enhances the Stability of Newly Synthesized SNAP-23
Despite the fact that we consistently observed that phosphorylated SNAP-23 was not present in t-SNARE complexes, we also found that SNAK-mediated phosphorylation of SNAP-23 actually promoted t-SNARE assembly. Because the assembly of the binary t-SNARE complex follows second-order kinetics and is proportional to the concentration of either syntaxin or SNAP-23 (Nicholson et al., 1998), we examined the possibility that coexpression with SNAK altered the expression or stability of SNAP-23. Therefore, we examined the biosynthesis of newly synthesized SNAP-23 in [35S]methionine-labeled HeLa cells expressing SNAP-23 alone or SNAP-23 together with SNAK. We were surprised to find that coexpression with SNAK resulted in a remarkable increase in the amount of newly synthesized SNAP-23 present in the cell extracts (Figure 8A). Phosphorylated SNAP-23 is clearly present in pulse-radiolabeled cells (Figure 8A, asterisk); however, phospho-SNAP-23 is dephosphorylated during the subsequent chase. The ability of SNAK to enhance the expression of SNAP-23 appears to be specific for its substrate SNAP-23, because SNAK does not lead to a similar increase in the expression of newly synthesized syntaxin 4 (see Figure 3A), syntaxin 1, VAMP 2, or other non-SNARE proteins we have examined (data not shown). Quantitative analysis of independent experiments revealed significantly more newly synthesized SNAP-23 both after the short pulse radiolabeling and after each chase point (Figure 8B). In addition, extrapolation of the kinetics of SNAP-23 degradation revealed that the half-life of SNAP-23 increased from only 6 h in the absence of SNAK to >15 h in the presence of SNAK. These data strongly suggest that coexpression with SNAK enhances t-SNARE assembly by increasing the pool of newly synthesized SNAP-23 molecules available for binding to syntaxin in vivo.
Figure 8.
SNAK enhances the expression of newly synthesized SNAP-23. (A) HeLa cells expressing SNAP-23 in the absence or presence of SNAK were labeled with [35S]methionine for 15 min and chased in complete medium. After incubation for various times at 37°C, the cells were harvested, lysed in Triton X-100, immunoprecipitated with anti-SNAP-23 serum, and analyzed by SDS-PAGE and fluorography. The mobility of phosphorylated SNAP-23 (asterisk) was determined by analysis of 32P-labeled SNAP-23 and [35S]methionine-labeled SNAP-23 on the same SDS-PAGE gel. Note that in the presence of SNAK, phospho-SNAP-23 is observed after the pulse radiolabeling with [35S]methionine. (B) The relative amount of [35S]SNAP-23 precipitated with the SNAP-23 serum in the absence (□) or in the presence (▪) of SNAK was determined by phosphorimager analysis. The recovery of [35S]SNAP-23 for each data point was expressed as a fraction of the amount of SNAP-23 recovered from cells coexpressing SNAP-23 and SNAK after the pulse radiolabeling (time 0). Each value represents the results from two independent experiments with SD.
DISCUSSION
In this paper, we have identified a novel serine/threonine kinase that interacts with t-SNAREs and specifically phosphorylates SNAP-23 both in vitro and in vivo. Furthermore, we have demonstrated that phosphorylation of SNAP-23 by SNAK enhances SNAP-23/syntaxin t-SNARE complex assembly in vivo. Phosphorylation by SNAK is the first reported type of a posttranslational modification of SNAP-23 that enhances its association with syntaxin. Because the formation of the t-SNARE heterodimer is widely believed to precede the formation of the v-SNARE/t-SNARE fusion complex (Nicholson et al., 1998), phosphorylation of SNAP-23 by SNAK is likely to be of central importance for vesicle docking and fusion.
Remarkably, we found that only unassembled SNAP-23 was phosphorylated, whereas SNAP-23 associated with syntaxin in t-SNARE complexes was not. It is likely that SNAP-23 is dephosphorylated before syntaxin binding in vivo, because we found that nonphosphorylated SNAP-23 bound to syntaxin very well in vitro but phosphorylated SNAP-23 did not. Furthermore, if dephosphorylation occurred after syntaxin binding, we might expect to observe a small population of t-SNARE complexes possessing phospho-SNAP-23, which we did not. Therefore, our kinetic analyses argue that SNAP-23 phosphorylation precedes syntaxin binding, a process that is generally believed to occur on intracellular membranes and not in the cytosol. It is unlikely that phosphorylation is essential for membrane anchoring and palmitoylation of SNAP-23, because a SNAP-23 palmitoylation mutant is efficiently phosphorylated by SNAK yet resides almost exclusively in the cytosol (Cabaniols, unpublished observations).
Studies of SNAP-25 trafficking in PC12 cells and SNAP-23 trafficking in HeLa cells demonstrated that SNAP-23 family members exist in the cytosol for ∼20 min before they begin to accumulate on intracellular membranes and become acylated by membrane-bound palmitoyltransferases (Gonzalo and Linder, 1998; our unpublished observations). Pulse-chase and subcellular fractionation studies performed in our laboratory have confirmed that whereas newly synthesized SNAP-23 in t-SNARE complexes is predominantly membrane associated, newly synthesized free SNAP-23 resides in the cytosol. Because only cytosolic SNAP-23 is phosphorylated, it is likely that newly synthesized SNAP-23 is phosphorylated after translation but before membrane anchoring and palmitoylation.
Our failure to detect phosphorylated SNAP-23 in t-SNARE complexes initially seemed at odds with our observation that phosphorylation of SNAP-23 by SNAK enhanced t-SNARE assembly. However, we subsequently found that SNAK dramatically increased the expression and stability of newly synthesized SNAP-23 in cells. Because the formation of the t-SNARE complex has been proposed to follow simple second-order kinetics (Nicholson et al., 1998), the net effect of increasing the size of the SNAP-23 pool would be to increase the probability that newly synthesized SNAP-23 would bind to syntaxin. We should emphasize that this effect of SNAK is seen only on the newly synthesized, cytosolic pool of SNAP-23, because SNAK does not dramatically influence the steady-state level of SNAP-23 (which resides primarily on membranes). However, we did observe a slight increase in the absolute amount of SNAP-23 present in cells expressing SNAK, a result we now attribute to enhanced SNAP-23 stabilization in the membrane-bound t-SNARE complex. Therefore, these data are consistent with a mechanism of action whereby SNAK promotes the formation of binary t-SNARE complexes by increasing the pool of newly synthesized SNAP-23 available for binding to syntaxin.
Although we have not determined at the molecular level the mechanism by which phosphorylation enhances the expression and stability of SNAP-23, there are a number of possibilities to explain this phenomenon. For example, it is conceivable that phosphorylation of SNAP-23 by SNAK leads to a conformational change in the SNAP-23 molecule. Conformational alterations of protein structure are a common consequence of phosphorylation, and in this case such a change might stabilize SNAP-23 from cytosolic degradation. Alternatively, phosphorylation of SNAP-23 could result in the recruitment of a molecular chaperone that stabilizes SNAP-23 and assists in t-SNARE assembly. However, this would have to be a relatively weak interaction, because we have not observed additional proteins in our anti-syntaxin or anti-SNAP-23 immunoprecipitates.
There have been two recent reports examining the effect of phosphorylation of the neuronal SNAP-23 homologue SNAP-25 on SNARE complex assembly. Risinger and Bennett (1999) found that both Ca2+- and calmodulin-dependent protein kinase II and cyclic AMP–dependent protein kinase phosphorylated recombinant SNAP-25 but did not phosphorylate recombinant SNAP-23 in vitro. However, despite robust phosphorylation of SNAP-25, there was no noticeable effect of SNAP-25 phosphorylation on either binary (SNAP-25/syntaxin) or ternary (SNAP-25/syntaxin/VAMP) SNARE complex assembly. By contrast, Shimazaki et al. (1996) found that PKC treatment of PC12 cell extracts diminished syntaxin binding to SNAP-25, arguing in favor of an inhibitory role for SNAP-25 phosphorylation. Unlike the results of these in vitro phosphorylation studies, the effects of SNAK described in the present study are unique in that they reveal a stimulation of t-SNARE assembly upon SNAP-23 phosphorylation in vivo. Interestingly, the tryptic phosphopeptides generated by SNAK- or phorbol ester–mediated phosphorylation of SNAP-25 are different (our unpublished observations), indicating phosphorylation of distinct residues by different kinases. Together, these data strongly support the idea that phosphorylation of distinct regions of SNAP-23 family members can have either stimulatory or inhibitory consequences for t-SNARE assembly and implicate the regulation of t-SNARE assembly as an essential step in preparing target membranes for vesicle docking and fusion.
ACKNOWLEDGMENTS
We thank David Winkler for generation of oligonucleotides, peptides, and automated sequence analysis; Dr. Mark Knepper for syntaxin 4 antisera; and Drs. Al Singer, Dinah Singer, and Katherine Roche for critical reading of the manuscript.
REFERENCES
- Anderson HA, Roche PA. Phosphorylation regulates the delivery of MHC class II invariant chain complexes to antigen processing compartments. J Immunol. 1998;160:4850–4858. [PubMed] [Google Scholar]
- Araki S, Tamori Y, Kawanishi M, Shinoda H, Masugi J, Mori H, Niki T, Okazawa H, Kubota T, Kasuga M. Inhibition of the binding of SNAP-23 to syntaxin 4 by Munc18c. Biochem Biophys Res Commun. 1997;234:257–262. doi: 10.1006/bbrc.1997.6560. [DOI] [PubMed] [Google Scholar]
- Bennett MK, Scheller RH. A molecular description of synaptic vesicle membrane trafficking. Annu Rev Biochem. 1994;63:63–100. doi: 10.1146/annurev.bi.63.070194.000431. [DOI] [PubMed] [Google Scholar]
- Brennwald P, Kearns B, Champion K, Keranen S, Bankaitis V, Novick P. Sec9 is a SNAP-25-like component of a yeast SNARE complex that may be the effector of Sec4 function in exocytosis. Cell. 1994;79:245–258. doi: 10.1016/0092-8674(94)90194-5. [DOI] [PubMed] [Google Scholar]
- Carr CM, Grote E, Munson M, Hughson FM, Novick PJ. Sec1p binds to SNARE complexes and concentrates at sites of secretion. J Cell Biol. 1999;146:333–344. doi: 10.1083/jcb.146.2.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferro-Novick S, Jahn R. Vesicle fusion from yeast to man. Nature. 1994;370:191–193. doi: 10.1038/370191a0. [DOI] [PubMed] [Google Scholar]
- Foster LJ, Yeung B, Mohtashami M, Ross K, Trimble WS, Klip A. Binary interactions of the SNARE proteins syntaxin-4, SNAP23, and VAMP-2 and their regulation by phosphorylation. Biochemistry. 1998;37:11089–11096. doi: 10.1021/bi980253t. [DOI] [PubMed] [Google Scholar]
- Gonzalo S, Linder ME. SNAP-25 palmitoylation and plasma membrane targeting require a functional secretory pathway. Mol Biol Cell. 1998;9:585–597. doi: 10.1091/mbc.9.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z, Turner C, Castle D. Relocation of the t-SNARE SNAP-23 from lamellipodia-like cell surface projections regulates compound exocytosis in mast cells. Cell. 1998;94:537–548. doi: 10.1016/s0092-8674(00)81594-9. [DOI] [PubMed] [Google Scholar]
- Hay JC, Scheller RH. SNAREs and NSF in targeted membrane fusion. Curr Opin Cell Biol. 1997;9:505–512. doi: 10.1016/s0955-0674(97)80026-9. [DOI] [PubMed] [Google Scholar]
- Koticha DK, Huddleston SJ, Witkin JW, Baldini G. Role of the cysteine-rich domain of the t-SNARE component, SYNDET, in membrane binding and subcellular localization. J Biol Chem. 1999;274:9053–9060. doi: 10.1074/jbc.274.13.9053. [DOI] [PubMed] [Google Scholar]
- Leung SM, Chen D, DasGupta BR, Whiteheart SW, Apodaca G. SNAP-23 requirement for transferrin recycling in streptolysin O-permeabilized Madin-Darby canine kidney cells. J Biol Chem. 1998;273:17732–17741. doi: 10.1074/jbc.273.28.17732. [DOI] [PubMed] [Google Scholar]
- Low SH, Chapin SJ, Wimmer C, Whiteheart SW, Komuves LG, Mostov KE, Weimbs T. The SNARE machinery is involved in apical plasma membrane trafficking in MDCK cells. J Cell Biol. 1998a;141:1503–1513. doi: 10.1083/jcb.141.7.1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low SH, Roche PA, Anderson HA, van Ijzendoorn SCD, Zhang M, Mostov KE, Weimbs T. Targeting of SNAP-23 and SNAP-25 in polarized epithelial cells. J Biol Chem. 1998b;273:3422–3430. doi: 10.1074/jbc.273.6.3422. [DOI] [PubMed] [Google Scholar]
- Lupas A. Prediction and analysis of coiled-coil structures. Methods Enzymol. 1996;266:513–525. doi: 10.1016/s0076-6879(96)66032-7. [DOI] [PubMed] [Google Scholar]
- Mandon B, Chou CL, Nielsen S, Knepper MA. Syntaxin-4 is localized to the apical plasma membrane of rat renal collecting duct cells: possible role in aquaporin-2 trafficking. J Clin Invest. 1996;98:906–913. doi: 10.1172/JCI118873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagase T, Seki N, Tanaka A, Ishikawa K, Nomura N. Prediction of the coding sequences of unidentified human genes. IV. The coding sequences of 40 new genes (KIAA0121–KIAA0160) deduced by analysis of cDNA clones from human cell line KG-1. DNA Res. 1995;2:167–174. doi: 10.1093/dnares/2.4.167. [DOI] [PubMed] [Google Scholar]
- Nichols BJ, Ungermann C, Pelham HR, Wickner WT, Haas A. Homotypic vacuolar fusion mediated by t- and v-SNAREs. Nature. 1997;387:199–202. doi: 10.1038/387199a0. [DOI] [PubMed] [Google Scholar]
- Nicholson KL, Munson M, Miller RB, Filip TJ, Fairman R, Hughson FM. Regulation of SNARE complex assembly by an N-terminal domain of the t- SNARE Sso1p. Nat Struct Biol. 1998;5:793–802. doi: 10.1038/1834. [DOI] [PubMed] [Google Scholar]
- Ravichandran V, Chawla A, Roche PA. Identification of a novel syntaxin- and synaptobrevin/VAMP-binding protein, SNAP-23, expressed in nonneuronal tissues. J Biol Chem. 1996;271:13300–13303. doi: 10.1074/jbc.271.23.13300. [DOI] [PubMed] [Google Scholar]
- Rea S, Martin LB, McIntosh S, Macaulay SL, Ramsdale T, Baldini G, James DE. Syndet, an adipocyte target SNARE involved in the insulin-induced translocation of GLUT4 to the cell surface. J Biol Chem. 1998;273:18784–18792. doi: 10.1074/jbc.273.30.18784. [DOI] [PubMed] [Google Scholar]
- Riento K, Galli T, Jansson S, Ehnholm C, Lehtonen E, Olkkonen VM. Interaction of Munc-18-2 with syntaxin 3 controls the association of apical SNAREs in epithelial cells. J Cell Sci. 1998;111:2681–2688. doi: 10.1242/jcs.111.17.2681. [DOI] [PubMed] [Google Scholar]
- Risinger C, Bennett MK. Differential phosphorylation of syntaxin and synaptosome-associated protein of 25 kDa (SNAP-25) isoforms. J Neurochem. 1999;72:614–624. doi: 10.1046/j.1471-4159.1999.0720614.x. [DOI] [PubMed] [Google Scholar]
- Rothman JE. Mechanisms of intracellular protein transport. Nature. 1994;372:55–63. doi: 10.1038/372055a0. [DOI] [PubMed] [Google Scholar]
- Sadoul K, Berger A, Niemann H, Weller U, Roche PA, Klip A, Trimble WS, Regazzi R, Catsicas S, Halban PA. SNAP-23 is not cleaved by botulinum neurotoxin E and can replace SNAP-25 in the process of insulin secretion. J Biol Chem. 1997;272:33023–33027. doi: 10.1074/jbc.272.52.33023. [DOI] [PubMed] [Google Scholar]
- Shimazaki Y, Nishiki T, Omori A, Sekiguchi M, Kamata Y, Kozaki S, Takahashi M. Phosphorylation of 25-kDa synaptosome-associated protein: possible involvement in protein kinase C-mediated regulation of neurotransmitter release. J Biol Chem. 1996;271:14548–14553. doi: 10.1074/jbc.271.24.14548. [DOI] [PubMed] [Google Scholar]
- St.-Denis JF, Cabaniols JP, Cushman SW, Roche PA. SNAP-23 participates in SNARE complex assembly in rat adipose cells. Biochem J. 1999;338:709–715. [PMC free article] [PubMed] [Google Scholar]
- Steegmaier M, Yang B, Yoo JS, Huang B, Shen M, Yu S, Luo Y, Scheller RH. Three novel proteins of the syntaxin/SNAP-25 family. J Biol Chem. 1998;273:34171–34179. doi: 10.1074/jbc.273.51.34171. [DOI] [PubMed] [Google Scholar]
- Söllner T, Bennett MK, Whiteheart SW, Scheller RH, Rothman JE. A protein assembly-disassembly pathway in vitro that may correspond to sequential steps of synaptic vesicle docking, activation, and fusion. Cell. 1993;75:409–418. doi: 10.1016/0092-8674(93)90376-2. [DOI] [PubMed] [Google Scholar]
- Südhof TC. The synaptic vesicle cycle: a cascade of protein-protein interactions. Nature. 1995;375:645–653. doi: 10.1038/375645a0. [DOI] [PubMed] [Google Scholar]
- Ungermann C, Wickner W. Vam7p, a vacuolar SNAP-25 homolog, is required for SNARE complex integrity and vacuole docking and fusion. EMBO J. 1998;17:3269–3276. doi: 10.1093/emboj/17.12.3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdez AC, Cabaniols JP, Brown MJ, Roche PA. Syntaxin 11 is associated with SNAP-23 on late endosomes and the trans-Golgi network. J Cell Sci. 1999;112:845–854. doi: 10.1242/jcs.112.6.845. [DOI] [PubMed] [Google Scholar]
- Veit M, Söllner TH, Rothman JE. Multiple palmitoylation of synaptotagmin and the t-SNARE SNAP-25. FEBS Lett. 1996;385:119–123. doi: 10.1016/0014-5793(96)00362-6. [DOI] [PubMed] [Google Scholar]
- Vogel K, Roche PA. SNAP-23 and SNAP-25 are palmitoylated in vivo. Biochem Biophys Res Commun. 1999;258:407–410. doi: 10.1006/bbrc.1999.0652. [DOI] [PubMed] [Google Scholar]
- Weber T, Zemelman BV, McNew JA, Westermann B, Gmachl M, Parlati F, Sollner TH, Rothman JE. SNAREpins: minimal machinery for membrane fusion. Cell. 1998;92:759–772. doi: 10.1016/s0092-8674(00)81404-x. [DOI] [PubMed] [Google Scholar]