Abstract
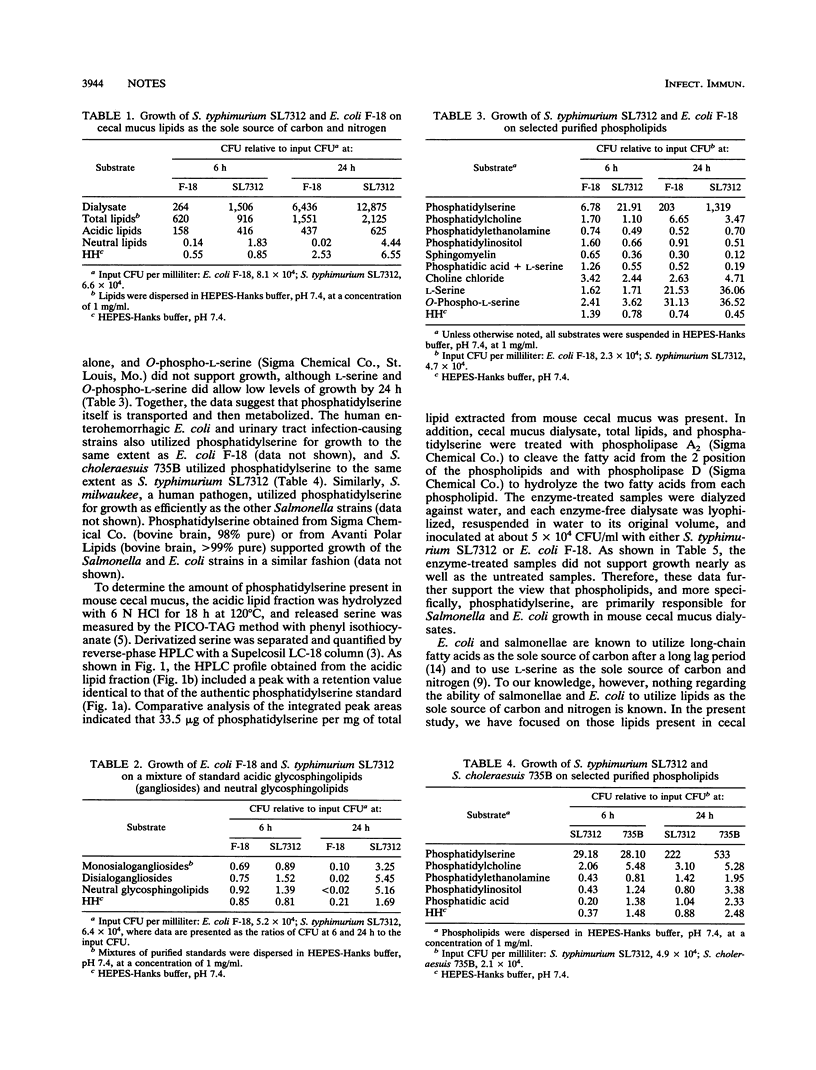
Salmonella choleraesuis (a pig pathogen), Salmonella typhimurium (a virulent strain in mice), and three strains of Escherichia coli (including a human enterohemorrhagic strain, a human urinary tract isolate, and a human fecal isolate) grew as well in vitro utilizing the lipids derived from mouse cecal mucus as the sole source of carbon and nitrogen as they did in mouse crude cecal mucus. Further analysis of the total lipid extracts of mucus dialysates showed that the acidic lipid fraction supported growth nearly as well as the total lipid fraction. Interestingly, among the many purified acidic lipids from mucus which were tested and analyzed, including several phospholipids, only phosphatidylserine was found to support the growth of all of these enteric bacteria, including Salmonella milwaukee, a human pathogen. The possible role of growth on pure phosphatidylserine in the pathogenesis of salmonellae is discussed.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ando S., Isobe M., Nagai Y. High performance preparative column chromatography of lipids using a new porous silica, Iatrobeads. I. Separation of molecular species of sphingoglycolipids. Biochim Biophys Acta. 1976 Jan 22;424(1):98–105. [PubMed] [Google Scholar]
- Beachey E. H. Bacterial adherence: adhesin-receptor interactions mediating the attachment of bacteria to mucosal surface. J Infect Dis. 1981 Mar;143(3):325–345. doi: 10.1093/infdis/143.3.325. [DOI] [PubMed] [Google Scholar]
- Bidlingmeyer B. A., Cohen S. A., Tarvin T. L. Rapid analysis of amino acids using pre-column derivatization. J Chromatogr. 1984 Dec 7;336(1):93–104. doi: 10.1016/s0378-4347(00)85133-6. [DOI] [PubMed] [Google Scholar]
- Cohen P. S., Rossoll R., Cabelli V. J., Yang S. L., Laux D. C. Relationship between the mouse colonizing ability of a human fecal Escherichia coli strain and its ability to bind a specific mouse colonic mucous gel protein. Infect Immun. 1983 Apr;40(1):62–69. doi: 10.1128/iai.40.1.62-69.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen S. A., Bidlingmeyer B. A., Tarvin T. L. PITC derivatives in amino acid analysis. Nature. 1986 Apr 24;320(6064):769–770. doi: 10.1038/320769a0. [DOI] [PubMed] [Google Scholar]
- Conway P. L., Welin A., Cohen P. S. Presence of K88-specific receptors in porcine ileal mucus is age dependent. Infect Immun. 1990 Oct;58(10):3178–3182. doi: 10.1128/iai.58.10.3178-3182.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drumm B., Roberton A. M., Sherman P. M. Inhibition of attachment of Escherichia coli RDEC-1 to intestinal microvillus membranes by rabbit ileal mucus and mucin in vitro. Infect Immun. 1988 Sep;56(9):2437–2442. doi: 10.1128/iai.56.9.2437-2442.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutnick D., Calvo J. M., Klopotowski T., Ames B. N. Compounds which serve as the sole source of carbon or nitrogen for Salmonella typhimurium LT-2. J Bacteriol. 1969 Oct;100(1):215–219. doi: 10.1128/jb.100.1.215-219.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoskins L. C., Agustines M., McKee W. B., Boulding E. T., Kriaris M., Niedermeyer G. Mucin degradation in human colon ecosystems. Isolation and properties of fecal strains that degrade ABH blood group antigens and oligosaccharides from mucin glycoproteins. J Clin Invest. 1985 Mar;75(3):944–953. doi: 10.1172/JCI111795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick B. A., Stocker B. A., Laux D. C., Cohen P. S. Roles of motility, chemotaxis, and penetration through and growth in intestinal mucus in the ability of an avirulent strain of Salmonella typhimurium to colonize the large intestine of streptomycin-treated mice. Infect Immun. 1988 Sep;56(9):2209–2217. doi: 10.1128/iai.56.9.2209-2217.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouricout M. A., Julien R. A. Pilus-mediated binding of bovine enterotoxigenic Escherichia coli to calf small intestinal mucins. Infect Immun. 1987 May;55(5):1216–1223. doi: 10.1128/iai.55.5.1216-1223.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sajjan S. U., Forstner J. F. Characteristics of binding of Escherichia coli serotype O157:H7 strain CL-49 to purified intestinal mucin. Infect Immun. 1990 Apr;58(4):860–867. doi: 10.1128/iai.58.4.860-867.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slomiany A., Yano S., Slomiany B. L., Glass G. B. Lipid composition of the gastric mucous barrier in the rat. J Biol Chem. 1978 Jun 10;253(11):3785–3791. [PubMed] [Google Scholar]
- Stanley R. A., Ram S. P., Wilkinson R. K., Roberton A. M. Degradation of pig gastric and colonic mucins by bacteria isolated from the pig colon. Appl Environ Microbiol. 1986 May;51(5):1104–1109. doi: 10.1128/aem.51.5.1104-1109.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svennerholm L., Fredman P. A procedure for the quantitative isolation of brain gangliosides. Biochim Biophys Acta. 1980 Jan 18;617(1):97–109. doi: 10.1016/0005-2760(80)90227-1. [DOI] [PubMed] [Google Scholar]
- Wadolkowski E. A., Laux D. C., Cohen P. S. Colonization of the streptomycin-treated mouse large intestine by a human fecal Escherichia coli strain: role of growth in mucus. Infect Immun. 1988 May;56(5):1030–1035. doi: 10.1128/iai.56.5.1030-1035.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witas H., Sarosiek J., Aono M., Murty V. L., Slomiany A., Slomiany B. L. Lipids associated with rat small-intestinal mucus glycoprotein. Carbohydr Res. 1983 Aug 16;120:67–76. doi: 10.1016/0008-6215(83)88007-0. [DOI] [PubMed] [Google Scholar]


