Abstract
We are studying endoplasmic reticulum–associated degradation (ERAD) with the use of a truncated variant of the type I ER transmembrane glycoprotein ribophorin I (RI). The mutant protein, RI332, containing only the N-terminal 332 amino acids of the luminal domain of RI, has been shown to interact with calnexin and to be a substrate for the ubiquitin-proteasome pathway. When RI332 was expressed in HeLa cells, it was degraded with biphasic kinetics; an initial, slow phase of ∼45 min was followed by a second phase of threefold accelerated degradation. On the other hand, the kinetics of degradation of a form of RI332 in which the single used N-glycosylation consensus site had been removed (RI332-Thr) was monophasic and rapid, implying a role of the N-linked glycan in the first proteolytic phase. RI332 degradation was enhanced when the binding of glycoproteins to calnexin was prevented. Moreover, the truncated glycoprotein interacted with calnexin preferentially during the first proteolytic phase, which strongly suggests that binding of RI332 to the lectin-like protein may result in the slow, initial phase of degradation. Additionally, mannose trimming appears to be required for efficient proteolysis of RI332. After treatment of cells with the inhibitor of N-glycosylation, tunicamycin, destruction of the truncated RI variants was severely inhibited; likewise, in cells preincubated with the calcium ionophore A23187, both RI332 and RI332-Thr were stabilized, despite the presence or absence of the N-linked glycan. On the other hand, both drugs are known to trigger the unfolded protein response (UPR), resulting in the induction of BiP and other ER-resident proteins. Indeed, only in drug-treated cells could an interaction between BiP and RI332 and RI332-Thr be detected. Induction of BiP was also evident after overexpression of murine Ire1, an ER transmembrane kinase known to play a central role in the UPR pathway; at the same time, stabilization of RI332 was observed. Together, these results suggest that binding of the substrate proteins to UPR-induced chaperones affects their half lives.
INTRODUCTION
Newly synthesized proteins of the endomembrane system and most secreted proteins of eukaryotic cells enter the secretory pathway through the translocation channel at the membrane of the endoplasmic reticulum (ER). The lumen of the ER is the site where translocated proteins assume their secondary structure and where assembly of oligomeric complexes occurs. Additionally, newly synthesized proteins undergo cotranslational and posttranslational modifications in the lumen of the ER, some of which allow transient interactions of the folding polypeptide chains with a set of ER-resident proteins (Hammond and Helenius, 1995; Leitzgen and Haas, 1998). Only after acquiring a fully folded, native conformation can proteins complete their journey through the secretory pathway (Hurtley and Helenius, 1989). The monitoring of the conformational status of newly synthesized polypeptides is confined to the lumen of the ER, which houses an efficient quality control system composed of chaperones resident in this subcellular compartment. Failure of nascent proteins to obtain their correct three-dimensional conformations usually results in their retention in the ER and consequent degradation in the cytosol mediated by the ubiquitin-proteasome pathway, a process known to date as ER-associated degradation (ERAD) (Klausner and Sitia, 1990; Brodsky and McCracken, 1997; Sommer and Wolf, 1997).
In eukaryotic cells, a wide variety of secretory and membrane proteins have one or more N-linked glycans in their exoplasmic domains that contribute not only to their conformational maturation but also to their multiple biological functions (Varki, 1993). In fact, glycans provide polar surface groups, thus enhancing the solubility and preventing the aggregation of the polypeptides, on the one hand, and enabling the nascent glycoproteins to interact with a number of ER-resident chaperones, on the other. In this regard, two lectin-like proteins of the ER, calnexin, a transmembrane protein, and its soluble homologue, calreticulin, have been demonstrated to play an important role (Helenius, 1994; Helenius et al., 1997). Although other ER chaperones involved in quality control largely rely on the recognition of polypeptide determinants on the substrate protein, the interaction between calnexin/calreticulin and glycoproteins is mediated by N-linked oligosaccharides. Thus, these two lectin-like proteins, which act in a very similar manner, represent exclusive ER components of the quality control machinery for nascent glycoproteins. Because it became evident that calnexin is able to bind to both folded and unfolded forms of N-glycosylated ribonucleases, direct recognition of nonglycosylated domains on substrate proteins by these lectin-like proteins has become less favored (Rodan et al., 1996; Zapun et al., 1997). Indeed, calnexin and calreticulin appear to function as retention devices, allowing the substrate proteins to profit from the folding environment of the ER. Polypeptides that succeed in reaching their mature conformation are released from the lectin-like proteins as soon as their folding process is completed (Helenius, 1994; Trombetta and Helenius, 1998). Glycoproteins that persist in a malfolded or incompletely assembled state are delivered to cytosolic degradation mediated by the ubiquitin-proteasome pathway. Recent evidence indicates that the efficiency of targeting of glycoproteins to proteolysis in the cytoplasm appears to be dependent on the structure of their N-linked glycans, which results from the activity of various glycosidases present in the ER (Jakob et al., 1998; Liu et al., 1999). Furthermore, a prolonged half life of a mutated form of carboxypeptidase Y was observed in yeast strains in which the gene encoding the ER-resident α1,2-mannosidase had been disrupted (Knop et al., 1996). A role of calnexin in ERAD has been postulated for different substrates of the ubiquitin-proteasome pathway, such as the cystic fibrosis transmembrane conductance regulator, major histocompatibility complex class I heavy chains, apolipoprotein B, the PiZ variant of α1-antitrypsin (α1-AT), and RI332, a mutant form of the glycoprotein ribophorin I containing only the N-terminal 332 amino acids of its luminal domain (for review, see Ivessa et al., 1999).
Another well-characterized ER chaperone is the luminal protein BiP, which apparently interacts with hydrophobic stretches exposed on the surface of newly synthesized, unfolded, misfolded, and unassembled proteins (Leitzgen and Haas, 1998). In addition, cellular stress conditions that result in the accumulation of unfolded proteins in the ER trigger the increased expression of BiP and other chaperones, a phenomenon termed “unfolded protein response” (UPR) (Shamu et al., 1994). In this pathway, the ER transmembrane kinase Ire1p serves as a proximal sensor for the presence of unfolded proteins in the lumen of the ER, but it also functions as a site-specific endoribonuclease that initiates the splicing of the mRNA coding for the UPR-specific transcription factor Hac1p, eventually resulting in the enhanced transcription of UPR-regulated genes such as the one encoding BiP (Chapman et al., 1998; Sidrauski et al., 1998). A role for BiP in ERAD has been proposed based on the observation that the half life of a complex between BiP and different immunoglobulin light chains correlates with the half life of those unassembled subunits (Knittler et al., 1995; Skowronek et al., 1998). Furthermore, genetic evidence from yeast indicates the functional involvement of BiP in the degradation pathway for a mutated form of carboxypeptidase Y (Plemper et al., 1997).
In this study, we investigated the role of N-linked glycans in the degradation pathway of a substrate for ERAD with the use of RI332, a soluble luminal variant of the type I ER transmembrane glycoprotein ribophorin I, as a reporter protein. We show that the features of degradation of RI332 are dependent on the presence of the N-linked glycan of RI332 and its structure, which affects the interaction of the protein with calnexin, suggesting a role for this lectin-like protein in the proteolytic pathway. In addition, the half life of the substrate protein also appears to be influenced by its ability to bind to the ER chaperone BiP that is induced after stress exposure of cells.
MATERIALS AND METHODS
Reagents
The mammalian expression vectors pCI-neo and pcDNA3 were purchased from Promega (Madison, WI) and Invitrogen (Groningen, The Netherlands), respectively. Protein A–Sepharose CL-4B beads, Hybond C extra nitrocellulose membrane, and the ECL kit were obtained from Amersham Pharmacia Biotech (Uppsala, Sweden). Geneticin (G418 sulfate), minimal essential medium, methionine-free RPMI 1640, other cell culture components, lipofectin, and the Plus reagent were from Life Technologies (Grand Island, NY). Trypsin from bovine pancreas, l-methionine, aprotinin, leupeptin, l-leucyl-l-leucyl-l-leucine, PMSF, ATP, chicken egg albumin, CHAPS, calcium ionophore A23187, chloroquine, and brefeldin A (BFA) were purchased from Sigma (Deisenhofen, Germany). Carbobenzoxy-l-leucyl-l-leucyl-l-leucinal (ZLLL) and carbobenzoxy-l-leucyl-l-leucyl-l-norvalinal (ZLLNva) were from Peptides International (Louisville, KY). Endoglycosidase H (endo H), castanospermine, and restriction enzymes were obtained from Boehringer Mannheim Biochemicals (Mannheim, Germany). Tunicamycin, deoxymannojirimycin, and kifunensine were purchased from Toronto Research Chemicals (Toronto, Canada). EXPRE35S35S protein labeling mix containing l-[35S]methionine and EN3HANCE were from New England Nuclear (Boston, MA). Dithio bis (succinimidyl propionate) (DSP) was from Pierce (Rockford, IL), NP40 was from Fluka (Buchs, Switzerland), digitonin was from Calbiochem (La Jolla, CA), and the Chameleon double-stranded site-directed mutagenesis kit containing the pWhitescript (pWS) cloning vector was obtained from Stratagene (La Jolla, CA). X-Omat Blue XB-1 and BioMax MR x-ray films were from Eastman Kodak (Rochester, NY).
Antibodies
The polyclonal rabbit antibody against rat liver ribophorin I (Marcantonio et al., 1984; Yu et al., 1990; Tsao et al., 1992) and the polyclonal anti-calnexin antibody (de Virgilio et al., 1998) have been described previously. A mouse monoclonal anti-BiP antibody was purchased from Stress-Gen Biotechnologies (Victoria, Canada), a mouse monoclonal anti-c-myc antibody (Ab-1) was from Oncogene Research Products (Cambridge, MA), and a goat anti-mouse polyclonal antibody conjugated with HRP was from Sigma.
Construction of Expression Plasmids
An EcoRI fragment was excised from double-stranded DNA prepared from a M13mp18 phage clone containing the rat ribophorin I cDNA and was subcloned in the pWS vector (pWS-RI). cDNAs coding for RI332 or RI332-Thr were constructed on pWS-RI by site-directed mutagenesis according to the manufacturer’s instructions for the Chameleon double-stranded site-directed mutagenesis kit. In a first step, the primer 5′GATGCGGTTTGTATAACACGTCGACGATGAGCAAGTG3′ was used to produce the RI332 cDNA in which Asp-333 of the mature ribophorin I polypeptide is replaced by a stop codon. At the same time, a SalI site was introduced 4 base pairs downstream of the new stop codon, and the single KpnI site present in pWS was changed to a SrfI site. Subsequently, the BamHI-XhoI fragment containing the RI332 cDNA was excised from this construct, subcloned in the pWS vector containing the KpnI site, and subjected to a second round of site-directed mutagenesis. In this reaction, Asn-275 of ribophorin I was replaced by Thr with the use of the primer 5′CCGGGATGAAATCGGTACTGTTAGTACTAGCCACCTCC3′. During this step, a ScaI site was introduced 4 base pairs downstream of the newly created Thr codon without changing the remaining amino acid sequence. The identity of all cDNAs generated by site-directed mutagenesis was confirmed by DNA sequencing acording to the dideoxy nucleotide analogue chain-termination method (Sanger et al., 1977). The SalI-EcoRI fragments containing the RI332 and RI332-Thr cDNAs were isolated from the appropriate pWS constructs and used for the preparation of the expression plasmid vectors as follows. Because of the newly introduced SalI site in the first step of site-directed mutagenesis (see above), these fragments are essentially devoid of the complete portion of the ribophorin I cDNA downstream of the new stop codon after amino acid 332. The SalI-EcoRI RI332 and RI332-Thr cDNA fragments were subcloned in pcDNA3 with the use of its EcoRI-XhoI sites, and, in a second step, the excised EcoRI-XbaI fragments were subcloned into the mammalian expression vector pCI-neo with the use of the same sites. The latter constructs were used for expression in HeLa cells.
Cell Culture and Transfections
HeLa cells were grown at 37°C in minimal essential medium supplemented with 7% FCS, penicillin G (100 IU/ml), streptomycin sulfate (100 μg/ml), and amphotericin B (250 ng/ml). The cells were transfected with the expression constructs (the RI332 and RI332-Thr cDNAs contained in pCI-neo) by the lipofectin method according to the manufacturer’s instructions, with the use of 1 μg of DNA and 10 μl of lipofectin reagent on cells cultured in a 6-cm dish and an incubation time of 18 h. Permanent transformants of HeLa cells expressing RI332 or RI332-Thr (designated HeLa-RI332 and HeLa-RI332-Thr cells) were obtained after selection for growth in the presence of G418 (1 g/l). Single clones of highly expressing cells were selected by pulse labeling, followed by immunoprecipitation with anti-ribophorin I antibodies, and cultured in the continued presence of G418 (500 mg/l). HeLa-RI332 cells plated on a 10-cm dish were also transiently transfected with 8 μg of pcDNA3 vector containing the murine IRE1 (mIRE1) cDNA with a c-myc tag at its C terminus (a kind gift of Dr. David Ron, Skirball Institute of Biomolecular Medicine, New York, NY), with the use of lipofectin together with the Plus reagent. Cells were analyzed 48 h after transfection.
Treatment of Cells with Drugs, Cell Labeling, and Immunoprecipitations
The transfected HeLa-RI332 and HeLa-RI332-Thr cells were grown in 35-mm dishes at near confluence (5–8 × 105 cells per dish). Cells were preincubated at 37°C with castanospermine (1 mM), tunicamycin (5 μg/ml), kifunensine (2 μg/ml), deoxymannojirimycin (2 mM), or A23187 (5 μM) for 1 h or with ZLLL (50 μM), ZLLNva (40 μM), NH4Cl (50 mM), or chloroquine (0.1 mM) for 90 min in complete medium, for another 30 min in methionine- and serum-free medium, and then subjected to pulse-chase incubations in the continuous absence or presence of the drugs, as described previously (Tsao et al., 1992). For pulse-chase experiments in the presence of BFA (5 μg/ml), no preincubation was performed and the drug was included only in the starvation and pulse-chase media. Cells were lysed, and immunoprecipitations were performed under stringent conditions with the use of the polyclonal anti-ribophorin I antibody, as reported elsewhere (Tsao et al., 1992). When required, immunoprecipitates were treated with endo H, as described previously (Rosenfeld et al., 1984). Samples were analyzed by SDS-PAGE followed by fluorography. Quantitations of immunoprecipitations were performed by scanning densitometry with ImageQuant software (Molecular Dynamics, Sunnyvale, CA). For the determination of degradation kinetics, the intensities of the bands corresponding to the truncated ribophorin I variants were normalized with respect to the intensity of the endogenous intact ribophorin I band in the same lane. When short pulse periods were used, the highest intensity of labeling of the truncated ribophorins relative to that of the endogenous HeLa cell intact ribophorin I usually occurred at 5 min of chase. Therefore, the relative intensities at different chase times were expressed as percentages of the 5-min value. First-order kinetics of degradation were deduced and used for the plots shown (see also Tsao et al., 1992).
Sequential Immunoprecipitations with Anti-Ribophorin I and Anti-Calnexin Antibodies
These sequential immunoprecipitations were performed as described previously (de Virgilio et al., 1998)
Sequential Immunoprecipitation with Anti-BiP and Anti-Ribophorin I Antibodies
Immunoprecipitations with anti-BiP antibodies were performed as described previously (Knittler and Haas, 1992). Briefly, HeLa-RI332 and HeLa-RI332-Thr cells, plated in 35-mm dishes and grown at near confluence (5–8 × 105 cells per dish), were pretreated with tunicamycin or calcium ionophore A23187, as described above, and pulse labeled in the continuous absence or presence of the drugs for 30 min. Cells were lysed in 200 μl of NET buffer (150 mM NaCl, 5 mM EDTA, 50 mM Tris-Cl, pH 7.4, 0.5% NP40) or in 200 μl of ATP-NET buffer (150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 50 mM Tris-Cl, pH 7.4, 5 mM MgCl2, 0.5% NP40) and incubated on ice for 30 min. Cell lysates were centrifuged in an Eppendorf centrifuge at 13,000 rpm for 10 min. The supernatants of ATP-NET buffer lysates were incubated on ice for another 30 min in the presence of freshly prepared ATP (5 mM, pH 7.0), and the supernatants of NET buffer lysates were incubated in the absence of ATP. Immunoprecipitations were performed with anti-BiP antibodies conjugated to protein A–Sepharose beads (2 μl/800 μl total incubation volume) in the presence of 0.1 M H3BO3, 25 mM Na2B4O7, pH 8.3, 75 mM NaCl, 0.5% NP40, and 0.05% ovalbumin. Immunocomplexes were washed with wash buffer containing 0.1 M H3BO3, 25 mM Na2B4O7, pH 8.3, 1 M NaCl, and 0.5% NP40 and analyzed directly by SDS-PAGE, as described previously (Tsao et al., 1992). Alternatively, the immunocomplexes were eluted from protein A–Sepharose beads by boiling for 5 min in lysis buffer containing 25 mM Tris-Cl, pH 7.4, 95 mM NaCl, 3 mM EDTA, 2% SDS, and a mixture of protease inhibitors (1.7 μg/ml aprotinin, 1 μg/ml leupeptin, 1 μg/ml l-leucyl-l-leucyl-l-leucine, 5 mM PMSF). The eluates were subjected to a second round of immunoprecipitations in the presence of 0.6% SDS and 1% Triton X-100 with the use of anti-ribophorin I antibodies (4 μl/ml) before analysis, as described previously (Tsao et al., 1992).
Chemical Cross-Linking with Anti-BiP and Anti-Ribophorin I Antibodies
Cross-linking analysis has been described previously (Melnick et al., 1994). For the experiments performed in this study, the procedure described by Melnick et al. was modified as follows. HeLa-RI332 and HeLa-RI332-Thr cells, cultured in 35-mm dishes at near confluence (5–8 × 105 cells per dish), were preincubated and pulse labeled for 30 min in the absence or presence of tunicamycin. Cells were washed twice with wash buffer (130 mM NaCl, 20 mM Bicine, pH 8) and lysed in 300 μl of lysis buffer (150 mM NaCl, 5 mM KCl, 5 mM MgCl2, 50 mM Bicine, pH 8, 0.2% digitonin). The lysates were incubated in the absence or presence of ATP (5 mM, pH 7.0) on ice for 30 min and for another 15 min in the absence or presence of freshly prepared DSP (100 μg/ml). The excess of cross-linker was inactivated by incubation with 10 mM glycine for 15 min on ice, followed by boiling at 95°C for 2 min. SDS and Triton X-100 were added to final concentrations of 0.4 and 0.85%, respectively. Immunoprecipitations with anti-ribophorin I (4 μl/ml) or anti-BiP (4 μl/ml) antibodies were performed and analyzed by SDS-PAGE (Tsao et al., 1992).
Western Blotting with Anti-BiP and Anti-c-myc Antibodies
HeLa-RI332 and HeLa-RI332-Thr cells, grown in 10-cm dishes, were kept untreated or preincubated with tunicamycin (5 μg/ml) for up to 4 h. Cell solubilization was performed in Triton X-100–containing buffer, as described previously (Hermann et al., 1997). To detect the presence of insoluble material after the extraction with Triton X-100, the pellet remaining after ultracentrifugation was resuspended in 100 μl of lysis buffer containing 2% SDS (see above) and sonicated for 10 s to dissolve the material completely. For the analysis of BiP, the Triton X-100 extracts from each sample (20 μg of protein) and aliquots of the resuspended pellets, corresponding to the same amount of cellular material contained in the supernatants, were used. Samples were subjected to SDS-PAGE, and the proteins were transferred to a nitrocellulose membrane. The immunodetection of BiP was performed according to the instructions of the ECL kit with the use of anti-BiP antibodies at a dilution of 1:3,000 followed by goat anti-mouse immunoglobulin (Ig) G conjugated with HRP at a dilution of 1:10,000.
For the immunodetection of the c-myc–tagged mIre1, Triton X-100 extracts (30 μg of protein), anti-c-myc antibodies at a dilution of 1:100, and HRP-conjugated goat anti-mouse IgGs at a dilution of 1:5000 were used.
RESULTS
From previous studies, it is known that the truncated ribophorin I variant, RI332, is a substrate for ERAD dependent on the ubiquitin-proteasome pathway (Tsao et al., 1992; de Virgilio et al., 1998). RI332 is a glycoprotein with three potential N-glycosylation sites, only one of which, located at Asn-275 of the mature protein, is used by oligosaccharyl transferase (Kreibich, unpublished observations). The aim of this study was to define the role of N-linked glycans in the degradation of the truncated protein.
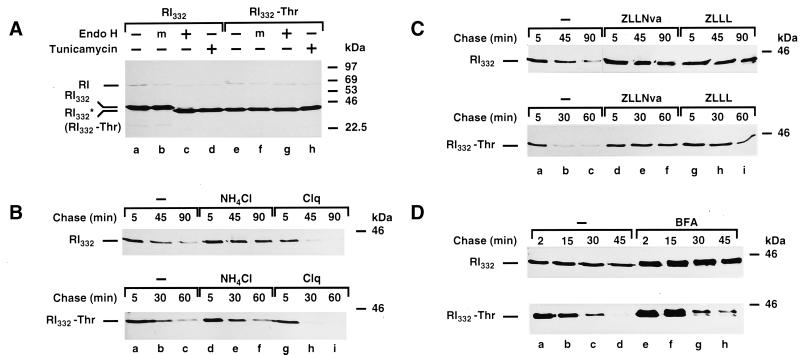
For this purpose, RI332-Thr, a variant of RI332 in which the single used N-glycosylation consensus site has been removed by site-directed mutagenesis, was constructed and expressed in HeLa cells. It was necessary to ensure that the mutant protein remained nonglycosylated. Therefore, a digestion with endo H was performed on anti-ribophorin I immunoprecipitates from lysates of HeLa cells expressing RI332 and RI332-Thr (Figure 1A). It became clear that endo H treatment resulted in a shift of the band corresponding to RI332 toward higher electrophoretic mobility (∼2 kDa; compare lanes a, b, and c), which was at the same level as RI332-Thr, digested or not with endo H (lanes e-g). A band of the same size was also observed when the immunoprecipitations were performed on lysates from cells treated with tunicamycin (lanes d and h). These observations indicate that RI332-Thr is not modified by N-glycosylation.
Figure 1.
Nonlysosomal and proteasome-dependent degradation of the glycosylated and nonglycosylated ribophorin I variants, RI332 and RI332-Thr. (A) HeLa-RI332 and HeLa-RI332-Thr cells, plated in 35-mm dishes to a density of 5–8 × 105 cells per dish, were left untreated (lanes a–c and e–g) or treated with tunicamycin (5 μg/ml) for 1 h (lanes d and h). The cells were then incubated in serum- and methionine-free medium for 30 min and metabolically labeled with the same medium containing [35S]methionine (250 μCi/ml) for 30 min in the continued absence or presence of the drug. Cells were lysed with SDS-containing buffer and processed for immunoprecipitation with a polyclonal rabbit anti-ribophorin I antibody. The immunoprecipitates were left untreated (lanes a, d, e, and h), mock treated (m; lanes b and f), or digested with endo H (lanes c and g) overnight at 37°C. Finally, all samples were analyzed by SDS-PAGE (10% gels) followed by fluorography. (B and C) HeLa-RI332 and HeLa-RI332-Thr cells were left untreated (B and C, lanes a–c) or preincubated with NH4Cl (50 mM; B, lanes d–f), chloroquine (Clq, 0.1 mM; B, lanes g–i), ZLLNva (40 μM; C, lanes d–f), or ZLLL (50 μM; C, lanes g–i). (D) Cells were incubated in serum- and methionine-free medium in the absence (lanes a–d) or presence of BFA (5 μg/ml; lanes e–h) for 30 min. (B–D) Pulse labeling was carried out in the same medium containing [35S]methionine (250 μCi/ml) for 10 min (B and C) or 5 min (D), followed by chase incubations in complete medium supplemented with unlabeled l-methionine (5 mM) in the continued absence or presence of the drugs for the times indicated. Cell lysis, immunoprecipitations, and sample analysis were performed as described for panel A. Note that because of the lower incorporation of [35S]methionine in the presence of chloroquine, the truncated ribophorins are less apparent already at 45 and 30 min of chase. RI indicates the position of the native ribophorin I, and RI332* indicates the position of nonglycosylated RI332, observed in the presence of tunicamycin.
The Glycosylated and Nonglycosylated Truncated Ribophorin I Variants Follow Similar Degradation Pathways
Pulse-chase experiments indicated that RI332-Thr is a short-lived protein, as has been shown for its glycosylated counterpart (see Figure 1, B–D, and below). We wanted to determine whether the turnover of RI332-Thr is independent of lysosomal proteases, as is the turnover of RI332 (Tsao et al., 1992). Therefore, HeLa-RI332 and HeLa-RI332-Thr cells were metabolically labeled with [35S]methionine and subjected to subsequent chase incubations in the presence or absence of two different lysosomotropic agents, NH4Cl (Figure 1B, lanes d–f) and chloroquine (Clq; lanes g–i). In both cases, degradation of RI332 and RI332-Thr was hardly affected by these treatments, suggesting that the two polypeptides are not degraded by the lysosome.
We have previously demonstrated that RI332 is degraded by the ubiquitin-proteasome pathway in a CHO cell line (de Virgilio et al., 1998). To determine if the nonglycosylated variant is also a substrate for proteasomal degradation, the effect of two proteasome inhibitors, ZLLNva (Figure 1C, lanes d–f) and ZLLL (lanes g–i), on the half life of the truncated proteins expressed in HeLa cells was assessed. As expected, RI332-Thr was indeed stabilized by these agents, indicating that its degradation is proteasome dependent.
It has already been shown that treatment of HeLa-RI332 cells with BFA does not inhibit the degradation of the truncated protein. In addition, time-dependent O-glycosylation of RI332 occurred (Ivessa et al., 1992; Tsao et al., 1992). Similarly, RI332-Thr appeared to be subjected to the same modification in response to BFA treatment (Figure 1D, lanes e–h).
Summarizing these results, both truncated ribophorin I mutants, despite the absence or presence of an N-linked glycan, at least at first glance, follow similar degradation pathways.
The N-linked Glycan Is Involved in the Initial Phase of Proteolysis of RI332
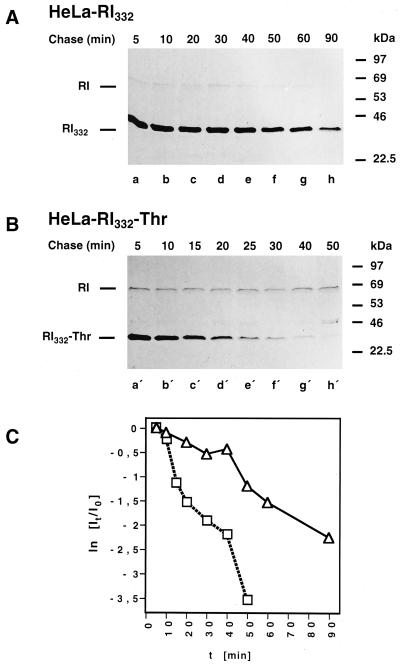
In a previous study, it was established that the degradation of RI332 follows biphasic kinetics (Tsao et al., 1992). It was of interest, therefore, to compare the degradation kinetics of RI332-Thr with that of the N-glycosylated variant by pulse-chase analysis. As shown in Figure 2, A and B, the first, slow phase of degradation, readily detected for RI332, is essentially omitted in the case of the nonglycosylated protein. A quantitation of this observation by scanning densitometry is shown in Figure 2C. This result indicates that the initial, slow phase of RI332 degradation may depend on the presence of the N-linked glycan on the protein.
Figure 2.
Kinetics of degradation of RI332 and RI332-Thr expressed in HeLa cells. HeLa-RI332 (A) and HeLa-RI332-Thr (B) cells were pulse labeled for 10 min and chased for up to 90 or 50 min, respectively. The cells were lysed, and anti-ribophorin I immunoprecipitations were performed. All samples were analyzed by SDS-PAGE followed by fluorography. (C) Quantitations of the bands corresponding to radiolabeled full-length (RI) and truncated ribophorin I variants were obtained by scanning densitometry. Semilog plots showing the degradation kinetics of RI332 (▵) and RI332-Thr (□) were established as described in MATERIALS AND METHODS. Data from three independent experiments are shown.
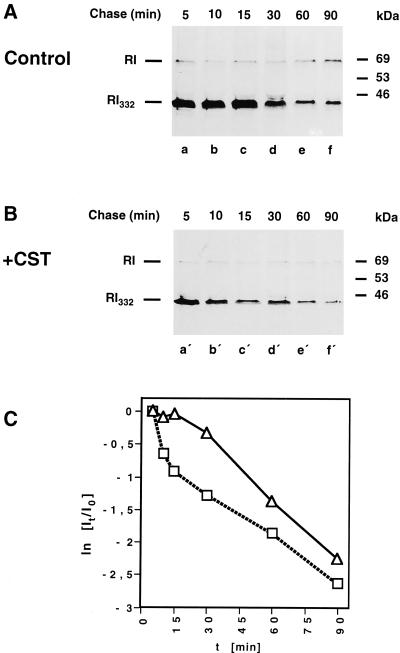
From our previous studies (de Virgilio et al., 1998), it became evident that RI332 interacts with calnexin, a lectin-like ER transmembrane protein that recognizes newly synthesized glycoproteins in their monoglucosylated form. Thus, it was conceivable that the interaction of the N-linked oligosaccharide of RI332 with calnexin may be a proximal cause of the first, slow phase of degradation of the truncated ribophorin I. To test this hypothesis, the kinetics of degradation of RI332 was established in cells preincubated with castanospermine, an inhibitor of glucosidases I and II. As a consequence of this treatment, the interaction of glycoproteins and calnexin has been demonstrated to be prevented (Helenius, 1994). As shown in Figure 3, immediate rapid proteolysis of RI332 occurred in castanospermine-treated cells, comparable to that observed for RI332-Thr (Figure 2, B and C). It is not clear why the rate of RI332 degradation in drug-treated cells, although initially very rapid, levels off with time, as was consistently observed. The kinetics of degradation of RI332-Thr was not affected by the inhibitor (our unpublished results).
Figure 3.
The initial, slow phase of RI332 degradation is omitted in the presence of castanospermine. HeLa-RI332 cells were kept untreated (A) or preincubated with castanospermine (CST; 1 mM) (B). All cell cultures were pulse labeled for 10 min and chased for up to 90 min in complete medium in the continued absence or presence of the drug. Cells were lysed, and RI332 and ribophorin I were immunoprecipitated and analyzed by SDS-PAGE and fluorography. (C) Quantitations of the bands corresponding to radiolabeled ribophorin I (RI) and RI332 to establish the degradation kinetics of RI332 in the absence (▵) or presence (□) of castanospermine were performed as described in the legend of Figure 2C and in MATERIALS AND METHODS. Data from six independent experiments are shown.
Calnexin Interacts with the Glycosylated Variant during the First, Slow Degradation Phase
Because the latter result suggests that calnexin binding may define the initial kinetics of RI332 turnover, the interaction between calnexin and the substrate protein was investigated by sequential immunoprecipitation of the two proteins from lysates of labeled cells chased for different periods of time (Figure 4). RI332 was reprecipitated from anti-calnexin immunoprecipitates obtained from HeLa-RI332 cell lysates under nonstringent conditions (see also de Virgilio et al., 1998). Some interaction of RI332 with calnexin was observed as early as after 5 min of chase (lane a), but maximal coprecipitation was evident at 15 min of chase (lane c), indicating that some time may be required for the formation of the complex. Both time points correspond to the initial, slow phase of RI332 degradation. In contrast, an interaction of RI332 with calnexin after 45 min of chase, i.e., shortly after the onset of the second, rapid phase of degradation, was detectable only on very long exposures of the x-ray films (lane e). In quantitative terms, at 45 min of chase only 5% of the RI332 molecules were recovered from the anti-calnexin immunoprecipitates compared with the 15-min time point. It should be noted, however, that at 45 min of chase nearly 80% of the RI332 that was present at the 15-min time point was still left undegraded (see Figure 2C). Thus, it appears that most of the RI332 remaining at the beginning of the second proteolytic phase had already been released from calnexin binding. Furthermore, at 15 min of chase, when maximal interaction of RI332 and calnexin was detected, no complex between these proteins was found in castanospermine-treated cells (lane d), indicating that the treatment of the cells with this drug was effective at preventing glycoprotein binding to the lectin-like protein. In addition, as may have been expected, RI332-Thr did not interact with calnexin (lanes g and h).
Figure 4.
RI332 interacts with calnexin only in the absence of castanospermine. HeLa-RI332 (lanes a–f) and HeLa-RI332-Thr (lanes g and h) cells were preincubated, pulse labeled for 10 min, and chased for the times indicated in the absence (lanes a, c, e, and g) or in the presence (lanes b, d, f, and h) of castanospermine (CST; 1 mM). Cells were lysed in buffer containing CHAPS (2%), and anti-calnexin immunoprecipitations were performed under nonstringent conditions in the presence of 1% of the same detergent. The second steps of the sequential immunoprecipitations were carried out under stringent conditions in the presence of SDS (0.6%) and Triton X-100 (1%). Only ribophorin I (RI) and RI332 or RI332-Thr reprecipitated under stringent conditions from anti-calnexin immunoprecipitations obtained under nonstringent conditions are shown for each chase time point. As a control, RI332 and RI332-Thr were immunoprecipitated under stringent conditions from cell lysates of HeLa-RI332 and HeLa-RI332-Thr cells metabolically labeled for 10 min (lanes i and j, respectively). All samples were analyzed by SDS-PAGE and fluorography. Exposure times: lanes a and b, 7 d; lanes c, d, and f–h, 10 d; lane e, 30 d; lanes i and j, 1 d.
Inhibition of Mannose Trimming Prolongs the Initial Step in the Degradation Kinetics of RI332
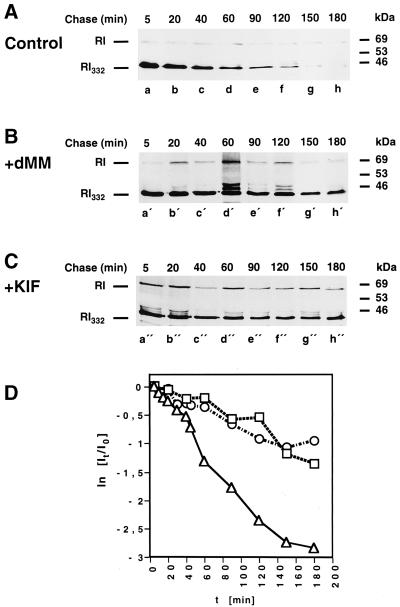
While newly synthesized glycoproteins undergo cycles of binding and release from calnexin, some mannose residues of the N-linked oligosaccharide core are removed by ER-resident mannosidases. As a result of the progressive loss of mannose residues, the glycoproteins lose their capability to interact with calnexin (Helenius, 1994). It is conceivable, therefore, that, on the contrary, inhibition of ER mannosidase activities allows for an extended interaction of newly synthesized glycoprotein substrates with calnexin, which in turn may contribute to their enforced retention in the ER.
To determine if inhibition of mannose trimming influences the half life of the truncated ribophorin I variants, the effects of two inhibitors of the ER α-mannosidase, deoxymannojirimycin (Figure 5B) and kifunensine (Figure 5C) (Liu et al., 1997, 1999), on the degradation kinetics of RI332 were observed. As expected, the half life of RI332 was prolonged to a significant extent after treatment of the cells with the inhibitors. In fact, in the presence of deoxymannojirimycin and kifunensine, 26 and 39%, respectively, of the protein synthesized during the pulse labeling remained in the last chase time point, when essentially all of the protein had already been degraded in control cells (Figure 5A). Furthermore, it became evident that the transition to the second, rapid phase of RI332 degradation kinetics was prevented by the incubations with the ER mannosidase inhibitors.
Figure 5.
RI332 is stabilized in kifunensine- or deoxymannojirimycin-treated HeLa cells. HeLa-RI332 cells were left untreated (A) or preincubated with deoxymannojirimycin (dMM; 2 mM) (B) or kifunensine (KIF; 2 μg/ml) (C). The cells were pulse labeled for 10 min and chased for up to 3 h in the continued absence or presence of the drugs. Anti-ribophorin I immunoprecipitations performed on cell lysates were analyzed by SDS-PAGE and fluorography. (D) The kinetics of RI332 degradation in the absence (▵) or in the presence of dMM (□) or KIF (○) were established as described in the legend of Figure 2 and in MATERIALS AND METHODS. Data from three independent experiments are shown.
Together, these data strongly favor the idea that the interaction with calnexin of N-linked oligosaccharides present on a glycoprotein substrate significantly contributes to the prolonged retention of the protein in the lumen of the ER before it is disposed of.
Binding of the Substrate Proteins to UPR-induced BiP Correlates with Their Stabilization
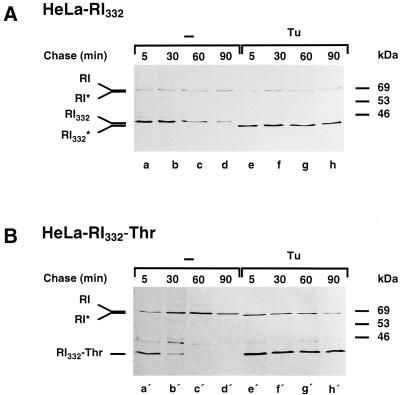
We have shown above that rapid monophasic proteolysis of RI332 in castanospermine-treated cells (Figure 3) and of RI332-Thr (Figure 2) takes place, in which case an interaction of the substrate proteins with calnexin is excluded. We reasoned that similar results concerning the kinetics of degradation of the truncated proteins should be obtained when N-glycosylation is inhibited altogether by exposure of cells to tunicamycin. To our surprise, however, nonglycosylated RI332 and RI332-Thr were dramatically stabilized after tunicamycin pretreatment of the HeLa cell transformants expressing the proteins, prolonging the average half lives of these proteins more than 10-fold (Figure 6, compare lanes a–d with e–h and a′–d′ with e′–h′).
Figure 6.
The truncated ribophorin I variants are stabilized in tunicamycin-treated HeLa cells. HeLa-RI332 (A) and HeLa-RI332-Thr (B) cells were left untreated (lanes a–d and a′–d′) or preincubated with tunicamycin (Tu, 5 μg/ml; lanes e–h and e′–h′). The cells were pulse labeled for 10 min and chased for up to 90 min in the continued absence or presence of the drug. Anti-ribophorin I immunoprecipitations performed on cell lysates were analyzed by SDS-PAGE and fluorography. RI* and RI332* indicate the positions of nonglycosylated endogenous ribophorin I and RI332, respectively, observed in the presence of tunicamycin.
Tunicamycin is known to trigger the UPR (Shamu et al., 1994), resulting in the induction of a number of ER-resident proteins, among them BiP/GRP78, GRP94, and calreticulin. We wanted to determine the extent to which the rate of synthesis of one of the major UPR-induced proteins, BiP, is increased under the experimental conditions used. Indeed, levels of metabolically labeled BiP were highly enhanced (∼15- to 20-fold) in tunicamycin-treated HeLa-RI332 and HeLa-RI332-Thr cells (Figure 7A). However, the increase of the total amount of BiP present in the drug-treated cells was much less pronounced, as determined by Western blotting (Figure 7B). Specifically, the amounts of BiP recovered from Triton X-100 extracts were elevated by only 10–20% in drug-treated cells compared with controls (Figure 7B, compare lanes a′ with b′–d′). To determine if tunicamycin incubation causes massive aggregation of luminal ER proteins, including BiP, the insoluble material remaining after detergent extraction was solubilized in an SDS-containing buffer and probed for the presence of the chaperone. Even in untreated cells, ∼30% of the total amount of BiP was detected in this Triton X-100–insoluble fraction (compare lanes a′ and e′), a figure that increased to only 40% in drug-exposed cells (compare lanes b′–d′ with f′–h′). Altogether, the amount of insoluble and thus probably aggregated BiP doubled, and the total level of BiP increased by 40% after tunicamycin treatment. These results suggest that the BiP molecules synthesized during tunicamycin action could represent a rather small fraction of the total amount of preexisting and constitutively expressed BiP in HeLa cells.
Figure 7.
The synthesis of BiP is highly induced by tunicamycin treatment in HeLa cells. (A) HeLa-RI332 and HeLa-RI332-Thr cells were left untreated (lanes a and e) or preincubated with tunicamycin (Tu; 5 μg/ml) for 1 h (lanes b and f), 2 h (lanes c and g), or 4 h (lanes d and h). Cells were pulse labeled in the continued absence or presence of the drug for 30 min. Cell lysis and immunoprecipitations with the monoclonal mouse anti-BiP antibody were performed as described in MATERIALS AND METHODS. Samples were analyzed by SDS-PAGE and fluorography. (B) HeLa-RI332 and HeLa- RI332-Thr cells, grown in 10-cm dishes, were left untreated (lanes a′ and e′) or preincubated with tunicamycin (5 μg/ml) for the times indicated (lanes b′–d′ and f′–h′). Cell extracts were prepared in Triton X-100–containing buffer, and the pellets were dissolved in an SDS-containing buffer. Cell extracts (20 μg of total protein, corresponding to approximately one-tenth of each extract; lanes a′–d′) and corresponding amounts of the dissolved pellets (lanes e′–h′) were subjected to SDS-PAGE. After transfer of the proteins to nitrocellulose, immunodetection was performed with the use of the anti-BiP antibody and the ECL kit. (C) HeLa-RI332 cells, grown in 6-cm dishes, were preincubated and pulse labeled for 30 min in the absence (lanes a" and c") or presence of tunicamycin (5 μg/ml; lanes b" and d"). Anti-BiP immunoprecipitates obtained from cell lysates were subjected to SDS-PAGE, and the proteins were transferred to a nitrocellulose membrane. The membrane was probed by Western blot analysis with the use of anti-BiP antibodies (lanes c" and d"), and after decay of the signal generated by the ECL reaction, the immunoprecipitates were analyzed by autoradiography with the use of BioMax MR x-ray film (lanes a" and b").
It is obvious that the >15-fold increase of metabolically labeled and immunoprecipitated BiP observed in tunicamycin-treated cells (Figure 7A) is not paralleled by the relatively small increase in the level of this chaperone as determined by Western blotting (Figure 7B). To determine if this difference is due to manifold higher amounts of BiP recovered from the immunoprecipitation in drug-treated cells, which could be a consequence of the massive induction of this chaperone, the total amounts of BiP immunoprecipitated from control and drug-treated cells were assessed by Western blotting (Figure 7C). Whereas levels of metabolically labeled and immunoprecipitated BiP increased by ∼15-fold (compare lanes b" with a"), the total amount of BiP precipitated increased by only 1.3-fold (compare lanes d" with c"). This indicates that the relative efficiency of the anti-BiP immunoprecipitation is not significantly affected by the amount of BiP molecules available; thus, as expected, the small increase of BiP immunoprecipitated in tunicamycin-treated cells corresponds roughly to the increase of total BiP present in the sample.
A possibility to explain the stabilization of RI332 and RI332-Thr in drug-treated cells could be that increased synthesis of BiP is a cause of the formation of increased amounts of complex between BiP and the truncated ribophorin I variants, or of a prolonged interaction of the chaperone with its polypeptide substrate. As a consequence, RI332 and RI332-Thr may be prevented from being degraded.
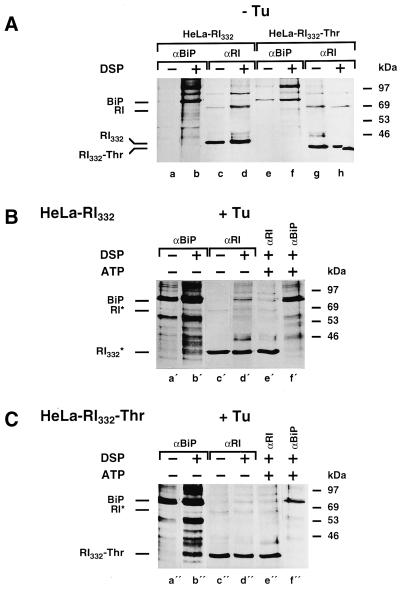
To detect the interaction of RI332 and RI332-Thr with BiP, a cross-linking experiment and immunoprecipitations with anti-BiP and anti-ribophorin I antibodies were performed on lysates from cells preincubated and metabolically labeled in the absence or presence of tunicamycin (Figure 8). When the results presented in Figure 8, A and B, as well as A and C, are compared, it is clear that nonambiguous cross-linking products between BiP and the truncated ribophorins were obtained only when the cells had been treated with tunicamycin. The cross-linked complex could be precipitated by antibodies to both components (Figure 8, B, lanes b′ and d′, and C, lanes b" and d"). Considering that the amount of BiP immunoprecipitated in tunicamycin-treated cells was increased by only ∼30% (see Figure 7C) and that the amount of truncated ribophorins was comparable in drug-treated and control cells (see Figure 6), it should be noted that the cross-links of RI332 and RI332-Thr with BiP detected only in the presence of tunicamycin cannot be explained by a difference in the extent of labeling of the truncated ribophorin I molecules.
Figure 8.
RI332 and RI332-Thr are readily cross-linked to BiP in tunicamycin-treated cells. HeLa-RI332 (A, lanes a–d, and B) and HeLa-RI332-Thr (A, lanes e–h, and C) cells were pretreated and pulse labeled for 30 min in the absence (A) or in the presence (B and C) of tunicamycin (Tu; 5 μg/ml). Cells were lysed in the presence of digitonin (0.2%) and incubated in the absence or presence of DSP (100 μg/ml) as indicated. As a control, a cross-linking experiment was carried out on a cell lysate preincubated with ATP (5 mM; B, lanes e′ and f′; C, lanes e" and f"). Ribophorin I (RI) and RI332 or RI332-Thr were immunoprecipitated from the cell lysates in the presence of SDS and Triton X-100 (A, lanes c, d, g, and h; B, lanes c′–e′; C, lanes c"–e"). Anti-BiP immunoprecipitations were performed under the same conditions (A, lanes a, b, e, and f; B, lanes a′, b′, and f′; C, lanes a", b", and f"). All samples were analyzed by SDS-PAGE followed by fluorography. Exposure times: A, 7 d; B, lanes a′, b′, and f′, 2 d; lanes c′–e′, 7 d; C, lanes a", b", and f", 1 d; lanes c"–e", 7 d.
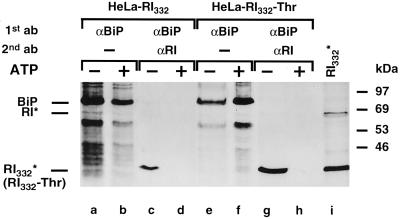
To further substantiate these results on the interaction of the truncated ribophorin I variants with BiP, a sequential immunoprecipitation experiment was performed (Figure 9). It is apparent that in the presence of tunicamycin RI332 and RI332-Thr were recovered from anti-BiP immunoprecipitates (lanes c and g), whereas no interaction of these proteins was observed in the absence of the drug (Tsao et al., 1992; our unpublished results). To verify the specificity of the coimmunoprecipitation, a control experiment was performed in the presence of ATP, which is known to mediate the release of bound proteins from BiP (Wei et al., 1995). Indeed, an association of RI332 and RI332-Thr with BiP was not detectable in the presence of ATP (lanes d and h). Similar results were also obtained in the cross-linking experiment described in Figure 8.
Figure 9.
RI332 and RI332-Thr are coimmunoprecipitated with BiP in tunicamycin-treated HeLa cells. HeLa-RI332 and HeLa-RI332-Thr cells were preincubated with tunicamycin (5 μg/ml) and pulse labeled for 30 min in the presence of the drug. Cell lysis, a subsequent 30-min incubation in the absence (lanes a, c, e, and g) or presence (lanes b, d, f, and h) of ATP (5 mM), and anti-BiP immunoprecipitations were performed as described in MATERIALS AND METHODS. For some samples, the anti-BiP immunoprecipitates were eluted from the protein A–Sepharose beads with a buffer containing SDS (2%) and used for reprecipitations under stringent conditions (lanes c, d, g, and h). One sample was subjected to anti-ribophorin I immunoprecipitation without prior anti-BiP precipitation (lane i). All samples were analyzed by SDS-PAGE and fluorography. Exposure times: lanes a, b, e, f, and i, 20 h; lanes c, d, g, and h, 14 d.
To exclude the possibility that stabilization of both RI332 and RI332-Thr after tunicamycin treatment occurred because of the requirement for functional glycoproteins in the degradation machinery, we monitored the behavior of the two ribophorin I variants in the presence of the calcium ionophore A23187, which is also known to trigger the UPR (Shamu et al., 1994). Again, not only were RI332 and RI332-Thr stabilized in the presence of the drug (Figure 10A), but the levels of newly synthesized BiP increased during A23187 treatment in both cell lines (Figure 10B), even though in both cases the effects of A23187 were less pronounced compared with those of tunicamycin (Figures 6 and 7A). Thus, in the presence of the calcium ionophore, the half life of RI332 was extended approximately fourfold and that of RI332-Thr was extended approximately twofold. The nonglycosylated variant has been reproducibly found to be less stabilized by the drug treatment than its glycosylated counterpart; it may be hypothesized that the initial retention by a calnexin-mediated mechanism of the latter variant may contribute to this difference. Moreover, in contrast to the situation observed with tunicamycin, the amounts of metabolically labeled BiP in A23187-treated cells increased only by a factor of three to five. With the use of sequential immunoprecipitation, RI332 and RI332-Thr were detected in association with BiP in the absence of ATP only when the cells were pulse labeled in the presence of the calcium ionophore (Figure 10C, lanes b" and e"); no interaction was detectable when the coimmunoprecipitations were performed with untreated samples (lanes a" and d") or in the presence of ATP (lanes c" and f").
Figure 10.
RI332 and RI332-Thr are stabilized after treatment of cells with A23187 and are coimmunoprecipitated with BiP only in the presence of the drug. (A) HeLa-RI332 and HeLa-RI332-Thr cells were left untreated (lanes a–d) or preincubated with A23187 (5 μM) (lanes e–h). The cells were pulse labeled for 10 min and chased for up to 2 h in the continued absence or presence of the drug. Anti-ribophorin I immunoprecipitations performed on cell lysates were analyzed by SDS-PAGE and fluorography. (B) HeLa-RI332 (lanes a′–c′) and HeLa-RI332-Thr (lanes d′–f′) cells were left untreated (lanes a′ and d′) or preincubated with A23187 for 30 min (lanes b′ and e′) or 90 min (lanes c′ and f′). Cells were pulse labeled in the continued absence or presence of the drug for 30 min. Cell lysis and immunoprecipitations with the monoclonal anti-BiP antibody were performed as described in MATERIALS AND METHODS. (C) HeLa-RI332 (lanes a"–c") and HeLa-RI332-Thr (lanes d"–f") cells were preincubated in the absence (lanes a" and d") or in the presence of A23187 (lanes b", c", e", and f") and pulse labeled for 30 min in the continuous absence or presence of the drug. Cell lysis, a 30-min incubation in the absence (lanes a", b", d", and e") or presence (lanes c" and f") of ATP (5 mM), and anti-BiP immunoprecipitations were performed as described in MATERIALS AND METHODS. The anti-BiP immunoprecipitates were eluted from the protein A–Sepharose beads with a buffer containing SDS (2%) and used for a second round of anti-ribophorin I immunoprecipitations under stringent conditions. All samples were analyzed by SDS-PAGE and fluorography. Exposure times: A and B, 2 d; C, 42 d.
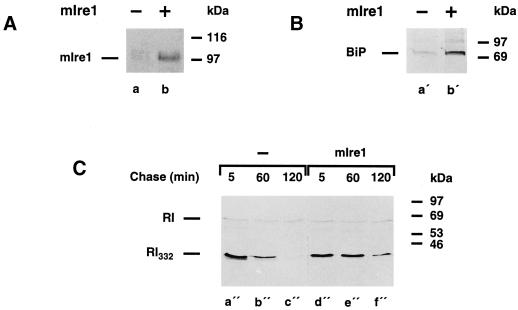
In recent years, it has been established that the UPR in yeast is mediated by the ER transmembrane protein Ire1p/Ern1p, a proximal sensing device for unfolded proteins in the lumen of the ER. Ire1p constitutes a bifunctional serine/threonine protein kinase and a site-specific endoribonuclease, both of which activities are required for the transduction of the UPR signal to the nucleus (Shamu et al., 1994; Chapman et al., 1998; Sidrauski et al., 1998). Mammalian homologues of the yeast IRE1 have been cloned, and their gene products have been characterized (Tirasophon et al., 1998; Wang et al., 1998). Thus, overexpression of mIre1 in mammalian cells leads to the activation of UPR-regulated genes, such as BiP, and of the transcription factor CHOP (Wang et al., 1998). Therefore, based on the results presented above, one would expect that the UPR induced independently of drug treatments by overexpression of mIre1, which is known to cause increased synthesis of BiP, should also result in the stabilization of RI332. For this purpose, HeLa-RI332 cells were transiently transfected with an expression vector containing the cDNA encoding mIre1 with a c-myc tag at its C terminus. The efficiency of transfection was determined to be ∼30% by indirect immunofluorescence with the use of an anti-c-myc antibody (our unpublished results). The expression of the transfected gene was also detected by Western blotting (Figure 11A). As expected, increased amounts of metabolically labeled BiP were recovered in the transfected cells (Figure 11B); quantitation showed this increase to be approximately fourfold. We indeed observed that the half life of RI332 was extended by a factor of three in these cells (Figure 11C). Taking into account the efficiency of transfection, these values clearly represent underestimates.
Figure 11.
mIre1 overexpression induces BiP and prolongs the half life of RI332. (A) Triton X-100 extracts prepared from HeLa-RI332 cells (lane a) and from HeLa-RI332 cells transiently overexpressing mIre1 (lane b) were used to detect the c-myc–tagged version of mIre1 by Western blot analysis. (B) HeLa-RI332 cells (lane a′) and HeLa-RI332 cells transiently overexpressing mIre1 (lane b′) were pulse labeled for 30 min. Anti-BiP immunoprecipitations were performed on lysates prepared from these cells and analyzed by SDS-PAGE followed by fluorography. (C) HeLa-RI332 cells (lanes a"–c") and HeLa-RI332 cells transiently overexpressing mIre1 (lanes d"–f") were pulse labeled for 10 min and chased for up to 120 min. Anti-ribophorin I immunoprecipitations were performed on cell lysates and analyzed by SDS-PAGE and fluorography.
In summary, the latter results suggest that increased synthesis of UPR-induced proteins, such as BiP, in the presence of tunicamycin or calcium ionophore A23187, or attributable to the overexpression of mIre1, may result in the enhanced or prolonged interaction of these chaperones with the truncated ribophorin I variants. In this scenario, the UPR may ultimately be related to the stability of a protein substrate for ERAD.
DISCUSSION
In this paper, we provide insights into the role of N-linked glycans in the process of ERAD. In particular, we have compared the features of the turnover of RI332, which contains a single N-linked oligosaccharide, and those of its nonglycosylated variant, RI332-Thr. As already demonstrated, the degradation of RI332 is mediated by the ubiquitin-proteasome pathway in CHO cell transformants (de Virgilio et al., 1998). Here we have shown that the breakdown of RI332 and RI332-Thr expressed in HeLa cells is also proteasome dependent. As noted previously, RI332 is a substrate protein that is degraded with biphasic kinetics in HeLa cell transformants (Tsao et al., 1992). It was striking to observe here that the degradation kinetics of RI332-Thr was monophasic, in that the first, slow phase of degradation that occurs with RI332 was essentially absent. As a result, the overall degradation of RI332-Thr proceeded faster than that of the glycosylated form.
A role of calnexin in ERAD has been postulated by others and by us (for review, see Ivessa et al., 1999). We assumed that the slow phase of RI332 degradation was due to an initial binding of the glycoprotein to calnexin that could not occur with the nonglycosylated variant. Consistently, essentially lag-free, immediate rapid degradation of RI332 was observed in the presence of castanospermine, which impairs the interaction of glycoproteins with calnexin. On the other hand, inhibition of mannose trimming by deoxymannojirimycin and kifunensine, which leads to a prolonged interaction of glycoprotein substrates with calnexin (Liu et al., 1997, 1999), resulted in a slower rate of RI332 breakdown without a detectable transition into the second, rapid degradation phase. Direct evidence for binding of RI332 to calnexin was provided by sequential immunoprecipitation experiments. We observed calnexin-bound RI332 predominantly during the first proteolytic phase. These results allow for the conclusion that the first phase of RI332 degradation may be related to the binding of the N-linked oligosaccharide present on RI332 to the lectin-like protein calnexin. Thus, as reported previously for other glycoprotein substrates (Helenius, 1994; Helenius et al., 1997), calnexin not only appears to function in retaining newly synthesized and unfolded glycoproteins in the lumen of the ER but also seems to prevent the bound substrates from being rapidly exposed to the degradation machinery. This notion is supported by the lower rate of degradation observed during the first phase of RI332 breakdown. Possibly, binding of calnexin (or calreticulin) to glycoprotein substrates promotes their interaction with other components of the folding machinery. These include UDP-glucose:glycoprotein glucosyl transferase, which, apart from its enzymatic function, has chaperone-like activity (Helenius, 1994), as well as the thiol oxidoreductase ERp57 (also named ER60) that was shown to interact with certain polypeptides in a glycan-dependent manner (Oliver et al., 1997; Lindquist et al., 1998; Van der Wal et al., 1998). In vitro experiments have provided evidence for calnexin or its soluble homologue, calreticulin, to support ERp57-mediated folding of monoglucosylated RNase B (Zapun et al., 1998). Interestingly, although the lectin-like properties of calnexin and calreticulin appear to be essentially identical, differences in their binding to distinct substrates have been reported (Van Leeuwen and Kearse, 1996; Zhang et al., 1997). A role of calreticulin in the degradation of RI332 needs to be established. Furthermore, the elucidation of interaction partners of RI332 that determine the half life of the protein during the second, rapid proteolytic phase remains to be addressed in future work.
Consistent with our results, the prevention of binding of newly synthesized glycoproteins to calnexin by castanospermine has been shown to accelerate the proteasomal degradation of different substrates for ERAD, such as the T cell antigen receptor α-subunit, the α-subunit of the nicotine acetylcholine receptor, and apolipoprotein B (Kearse et al., 1994; Chen et al., 1998; Keller et al., 1998). Our data indicate that the interaction of calnexin with a glycoprotein substrate soon after its synthesis correlates with slow, initial turnover. For several glycoproteins that are known to bind calnexin, it has been noted that a lag phase precedes their degradation in the cytosol. This has been clearly demonstrated for the H2 subunit of the asialoglycoprotein receptor, α1-AT, the T cell antigen receptor β-subunit, and the CD3 δ-subunit (Amara et al., 1989; Wileman et al., 1990; Ciccarelli et al., 1993; Wu et al., 1994). Considering these observations, the first, slow phase of RI332 degradation could be compared with the lag phase detected for these other proteins.
If the N-glycan–dependent interaction with calnexin protects the glycoprotein substrate from degradation, inhibition of N-glycosylation by tunicamycin should lead to rapid degradation of the substrate. This was indeed reported for the major cell surface glycoprotein of chick embryo fibroblasts and IgM subunits (Olden et al., 1978; Kubo and Pelanne, 1983); in particular, lag-free and thus accelerated degradation of α1-AT was observed under these conditions (Ciccarelli et al., 1993). Recently, it was also noted that the kappa Ig light chain, a nonglycosylated substrate for ERAD (Knittler and Haas, 1992; Skowronek et al., 1998), is readily degraded in the presence of tunicamycin (Haas, personal communication). It was surprising, therefore, to find a dramatic stabilization of unglycosylated RI332 and RI332-Thr in tunicamycin-treated cells, whereas mere removal of the N-glycosylation site of RI332 caused an increased turnover of the protein. To determine whether ERAD is generally affected by tunicamycin treatment in our HeLa cell transformants, we transiently coexpressed the kappa Ig light chain in HeLa-RI332 cells. We observed that, in contrast to RI332, the degradation of the kappa chain was not inhibited in the presence of tunicamycin. This indicates that under these conditions the machinery involved in ERAD may still be efficient, yet selective for certain substrate proteins. Notably, the stabilization of the truncated ribophorin I variants in tunicamycin-treated cells is not simply due to a dramatic accumulation of unfolded proteins in the ER lumen in general, which could saturate the degradation system and thus delay retrotranslocation of substrate proteins to the cytosol and their proteolysis.
Taking into account that both RI332 and its nonglycosylated counterpart are stabilized not only in tunicamycin-treated cells but also in the presence of the calcium ionophore A23187, it seems unlikely that the stabilization of the two truncated ribophorin I variants is due to the requirement for N-glycosylation of one or more components of the targeting and degradation machinery. On the other hand, as shown previously (Shamu et al., 1994) and also as demonstrated for the cells used in this study, BiP synthesis is considerably induced in tunicamycin- and A23187-treated cells. It was striking, therefore, to find that the degradation of the truncated ribophorin I variants was inhibited by a variety of agents known to elicit the UPR. Thus, stabilization of RI332 was also observed in cells treated with thapsigargin (Ivessa et al., 1995), an inhibitor of the ER Ca2+-ATPase and a potent inducer of the UPR (Price et al., 1992; Li et al., 1993). Similarly, we noted that glucose starvation, a treatment that causes inhibition of N-linked glycosylation and induces the UPR independently of pharmacological agents (Chang et al., 1987), resulted in an increased synthesis of BiP and stabilization of RI332 and RI332-Thr (our unpublished results). BiP induction is also triggered, even more specifically, by overexpression of the ER transmembrane kinase and site-specific endoribonuclease mIre1 (Tirasophon et al., 1998; Wang et al., 1998), a mammalian homologue of yeast Ire1p/Ern1p that functions to sense a perturbed environment in the ER lumen and transmit a signal to downstream effectors, eventually leading to the enhanced transcription of UPR-inducible genes (Shamu et al., 1994; Chapman et al., 1998; Sidrauski et al., 1998). In agreement with the observations from cells treated with tunicamycin or A23187, the half life of RI332 was prolonged in cells overexpressing mIre1.
It appears, however, that the degree to which BiP synthesis is increased correlates with the half life of the truncated ribophorin I variants. Thus, BiP was highly induced in tunicamycin-treated cells, and nonglycosylated RI332 and RI332-Thr were essentially completely stabilized under these conditions. On the other hand, in cells exposed to A23187, BiP induction was less prominent and paralleled by a less pronounced stabilization of the substrate proteins. Although in HeLa-RI332 cells transfected with the mIre1 expression plasmid the induction of BiP and the degree of stabilization of the truncated protein were not comparable to those observed in drug-treated cells, it has to be considered that less than one-third of the cell population used for the experiments actually expressed the protein kinase/endoribonuclease. Therefore, it is likely that in the cotransfected cells BiP levels are increased to a more significant extent than is apparent, which in turn might coincide with a more accentuated extension of the half life of RI332.
BiP was shown to be induced after exposure of cells to BFA for an extended period of time by a transcriptional or a posttranscriptional mechanism, depending on the cell line used (Liu et al., 1992; Price et al., 1992). Consistently, the induction of BiP elicited by BFA was less pronounced than that observed after treatment of cells with the calcium ionophore A23187. In the time frame of our experiments, which included only short incubation times with BFA, such an induction was not discernible (our unpublished results). Therefore, it was perhaps not surprising to find that the truncated ribophorin I variants were not significantly stabilized in our BFA-treated HeLa cell transformants (Tsao et al., 1992; Ivessa et al., 1992; this work).
It is noteworthy that BiP is known to exist in various differentially modified forms that are distinguished by their phosphorylation and ADP ribosylation status and that allow for the recruitment of functionally active molecules during stress exposure elicited by glucose starvation, calcium-mobilizing agents such as A23187, and tunicamycin (Freiden et al., 1992; Staddon et al., 1992; Ledford and Leno, 1994). We clearly show an increased interaction of the two ribophorin I variants with stress-induced BiP. Indeed, it has been demonstrated that the half-lives of proteins, such as specific Ig light chains, correlate with the half-lives of the complexes formed between the proteins and BiP (Knittler et al., 1995; Skowronek et al., 1998). In addition, in a CHO cell line selected for increased BiP expression comparable to that seen during the UPR, enhanced binding of the chaperone to certain secretory proteins, such as von Willebrand factor, a mutant form of factor VIII, and thyroglobulin, and consequent delayed export of these proteins from the ER have been observed (Dorner et al., 1992; Muresan and Arvan, 1998). Thus, it is conceivable that the increased interaction of the truncated ribophorin I variants with BiP or other UPR-induced proteins may result in their retention in the lumen of the ER for an extended period of time and, therefore, in their reduced accessibility to cytosolic degradation.
In summary, our results demonstrate that the half life of a substrate protein for ERAD can be modulated by features of the protein itself, such as the presence of an N-linked glycan, and its trimming status, which defines the ability of the protein to interact with ER-resident lectin-like proteins and chaperones. In the context of ERAD, this interaction causes a delay in the delivery of the substrate protein to the cytosolic degradation machinery, simultaneously increasing the probability of productive folding. On the other hand, under stress conditions degradation may also be prevented by the unusual enhanced association with stress-induced proteins in the lumen of the ER, such as BiP. This might be a consequence of an increased number of functional stress protein molecules available for the formation of a complex with the substrate protein. Such an interaction may limit the exposure of a determinant(s) on the surface of the protein required for its targeting to degradation, as suggested by Schmitz et al. (1995), resulting in the prolonged retention and stability of the substrate protein in the ER.
ACKNOWLEDGMENTS
We are deeply indebted to Dr. Ingrid G. Haas (Biochemiezentrum, Universität Heidelberg, Germany) for providing the immunoglobulin kappa light chain cDNA construct, for her help with the anti-BiP coimmunoprecipitations, and for stimulating discussions. We are very grateful to Dr. David Ron (Skirball Institute of Biomolecular Medicine, New York, NY) for his kind gift of the c-myc epitope-tagged mIre1 expression construct. We thank Dr. I.G. Haas and Dr. Myriam Ermonval (Institut Pasteur, Paris, France) for critical reading of the manuscript. Mr. Paul Breit is acknowledged for his expert photography. This work was supported by research grants from the Austrian Science Foundation (P-12562-MOB to N.E.I.) and the American Cancer Society (CB-84312 to G.K.).
Abbreviations used:
- α1-AT
α1-antitrypsin
- BFA
brefeldin A
- BiP
immunoglobulin heavy chain binding protein
- DSP
dithio bis (succinimidyl propionate)
- endo H
endoglycosidase H
- ER
endoplasmic reticulum
- ERAD
endoplasmic reticulum–associated degradation
- Ig
immunoglobulin
- mIRE1
murine IRE1
- pWS
cloning vector pWhitescript
- RI332
a truncated form of ribophorin I containing its 332 N-terminal amino acids
- RI332-Thr
nonglycosylated variant of RI332
- UPR
unfolded protein response
- ZLLL
carbobenzoxy-l-leucyl-l-leucyl-l-leucinal
- ZLLNva
carbobenzoxy-l-leucyl-l-leucyl-l-norvalinal
REFERENCES
- Amara JF, Lederkremer G, Lodish HF. Intracellular degradation of unassembled asialoglycoprotein receptor subunits: a preGolgi, nonlysosomal endoproteolytic cleavage. J Cell Biol. 1989;109:3315–3324. doi: 10.1083/jcb.109.6.3315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky JL, McCracken AA. ER-associated and proteasome-mediated protein degradation: how two topologically restricted events come together. Trends Cell Biol. 1997;7:151–156. doi: 10.1016/S0962-8924(97)01020-9. [DOI] [PubMed] [Google Scholar]
- Chang SC, Wooden SK, Nakaki T, Kim YK, Lin AY, Kung L, Attenello JW, Lee AS. Rat gene encoding the 78-kDa glucose-regulated protein GRP78: its regulatory sequences and the effect of protein glycosylation on its expression. Proc Natl Acad Sci USA. 1987;84:680–684. doi: 10.1073/pnas.84.3.680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman R, Sidrauski C, Walter P. Intracellular signaling from the endoplasmic reticulum to the nucleus. Annu Rev Cell Dev Biol. 1998;14:459–485. doi: 10.1146/annurev.cellbio.14.1.459. [DOI] [PubMed] [Google Scholar]
- Chen Y, Caherec F, Chuck SL. Calnexin and other factors that alter translocation affect the rapid binding of ubiquitin to apoB in the Sec61 complex. J Biol Chem. 1998;273:11887–11894. doi: 10.1074/jbc.273.19.11887. [DOI] [PubMed] [Google Scholar]
- Ciccarelli E, Alonso MA, Cresteil D, Bollen A, Jacobs P, Alvarez F. Intracellular retention and degradation of human mutant variant of alpha 1-antitrypsin in stably transfected Chinese hamster ovary cell lines. Eur J Biochem. 1993;213:271–276. doi: 10.1111/j.1432-1033.1993.tb17759.x. [DOI] [PubMed] [Google Scholar]
- de Virgilio M, Weninger H, Ivessa NE. Ubiquitination is required for retro-translocation of a short-lived luminal endoplasmic reticulum glycoprotein to the cytosol for degradation by the proteasome. J Biol Chem. 1998;273:9734–9743. doi: 10.1074/jbc.273.16.9734. [DOI] [PubMed] [Google Scholar]
- Dorner AJ, Wasley LC, Kaufman RJ. Overexpression of GRP78 mitigates stress induction of glucose regulated proteins and blocks secretion of selective proteins in Chinese hamster ovary cells. EMBO J. 1992;11:1563–1571. doi: 10.1002/j.1460-2075.1992.tb05201.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freiden PJ, Gaut JR, Hendershot LM. Interconversion of three differentially modified and assembled forms of BiP. EMBO J. 1992;11:63–70. doi: 10.1002/j.1460-2075.1992.tb05028.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond C, Helenius A. Quality control in the secretory pathway. Curr Opin Cell Biol. 1995;7:523–529. doi: 10.1016/0955-0674(95)80009-3. [DOI] [PubMed] [Google Scholar]
- Helenius A. How N-linked oligosaccharides affect glyco-protein folding in the endoplasmic reticulum. Mol Biol Cell. 1994;5:253–265. doi: 10.1091/mbc.5.3.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helenius A, Trombetta ES, Hebert DN, Simons JF. Calnexin, calreticulin and the folding of glycoproteins. Trends Cell Biol. 1997;7:193–200. doi: 10.1016/S0962-8924(97)01032-5. [DOI] [PubMed] [Google Scholar]
- Hermann M, Seif F, Schneider WJ, Ivessa NE. Estrogen dependence of synthesis and secretion of apolipoprotein B-containing lipoproteins in the chicken hepatoma cell line, LMH-2A. J Lipid Res. 1997;38:1308–1317. [PubMed] [Google Scholar]
- Hurtley SM, Helenius A. Protein oligomerization in the endoplasmic reticulum. Annu Rev Cell Biol. 1989;5:277–307. doi: 10.1146/annurev.cb.05.110189.001425. [DOI] [PubMed] [Google Scholar]
- Ivessa NE, De Lemos-Chiarandini C, Gravotta D, Sabatini DD, Kreibich G. The brefeldin A-induced retrograde transport from the Golgi apparatus to the endoplasmic reticulum depends on calcium sequestered to intracellular stores. J Biol Chem. 1995;270:25960–25967. doi: 10.1074/jbc.270.43.25960. [DOI] [PubMed] [Google Scholar]
- Ivessa NE, De Lemos-Chiarandini C, Tsao YS, Takatsuki A, Adesnik M, Sabatini DD, Kreibich G. O-Glycosylation of intact and truncated ribophorins in brefeldin A-treated cells: newly synthesized intact ribophorins are only transiently accessible to the relocated glycosyltransferases. J Cell Biol. 1992;117:949–958. doi: 10.1083/jcb.117.5.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivessa NE, Kitzmüller C, de Virgilio M. ER-associated protein degradation inside and outside of the endoplasmic reticulum. Protoplasma. 1999;207:16–23. [Google Scholar]
- Jakob CA, Burda P, Roth J, Aebi M. Degradation of misfolded endoplasmic reticulum glycoproteins in Saccharomyces cerevisiae is determined by a specific oligosaccharide structure. J Cell Biol. 1998;142:1223–1233. doi: 10.1083/jcb.142.5.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse KP, Williams DB, Singer A. Persistence of glucose residues on core oligosaccharides prevents association of TCR alpha and TCR beta proteins with calnexin and results speci-fically in accelerated degradation of nascent TCR alpha proteins within the endoplasmic reticulum. EMBO J. 1994;13:3678–3686. doi: 10.1002/j.1460-2075.1994.tb06677.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller SH, Lindstrom J, Taylor P. Inhibition of glucose trimming with castanospermine reduces calnexin association and promotes proteasome degradation of the alpha-subunit of the nicotinic acetylcholine receptor. J Biol Chem. 1998;273:17064–17072. doi: 10.1074/jbc.273.27.17064. [DOI] [PubMed] [Google Scholar]
- Klausner RD, Sitia R. Protein degradation in the endoplasmic reticulum. Cell. 1990;62:611–614. doi: 10.1016/0092-8674(90)90104-m. [DOI] [PubMed] [Google Scholar]
- Knittler MR, Dirks S, Haas IG. Molecular chaperones involved in protein degradation in the endoplasmic reticulum: quantitative interaction of the heat shock cognate protein BiP with partially folded immunoglobulin light chains that are degraded in the endoplasmic reticulum. Proc Natl Acad Sci USA. 1995;92:1764–1768. doi: 10.1073/pnas.92.5.1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knittler MR, Haas IG. Interaction of BiP with newly synthesized immunoglobulin light chain molecules: cycles of sequential binding and release. EMBO J. 1992;11:1573–1581. doi: 10.1002/j.1460-2075.1992.tb05202.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop M, Hausner N, Wolf DH. N-Glycosylation affects endoplasmic reticulum degradation of a mutated derivative of carboxypeptidase yscY in yeast. Yeast. 1996;12:1229–1238. doi: 10.1002/(sici)1097-0061(19960930)12:12<1229::aid-yea15>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- Kubo RT, Pelanne ML. Tunicamycin inhibits the expression of membrane IgM in the human lymphoblastoid cell line Daudi. Mol Immunol. 1983;20:67–76. doi: 10.1016/0161-5890(83)90106-2. [DOI] [PubMed] [Google Scholar]
- Ledford BE, Leno GH. ADP-ribosylation of the molecular chaperone GRP78/BiP. Mol Cell Biochem. 1994;138:141–148. doi: 10.1007/BF00928456. [DOI] [PubMed] [Google Scholar]
- Leitzgen K, Haas IG. Protein maturation in the endoplasmic reticulum. Chemtracts Biochem Mol Biol. 1998;11:423–445. [Google Scholar]
- Li WW, Alexandre S, Cao X, Lee AS. Transactivation of the grp78 promoter by Ca2+ depletion: a comparative analysis with A23187 and the endoplasmic reticulum Ca(2+)-ATPase inhibitor thapsigargin. J Biol Chem. 1993;268:12003–12009. [PubMed] [Google Scholar]
- Lindquist JA, Jensen ON, Mann M, Hammerling GJ. ER-60, a chaperone with thiol-dependent reductase activity involved in MHC class I assembly. EMBO J. 1998;17:2186–2195. doi: 10.1093/emboj/17.8.2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu ES, Ou J-h, Lee AS. Brefeldin A as a regulator of grp78 gene expression in mammalian cells. J Biol Chem. 1992;267:7128–7133. [PubMed] [Google Scholar]
- Liu Y, Choudhury P, Cabral CM, Sifers RN. Intracellular disposal of incompletely folded human α1-antitrypsin involves release from calnexin and posttranslational trimming of asparagine-linked oligosaccharides. J Biol Chem. 1997;272:7946–7951. doi: 10.1074/jbc.272.12.7946. [DOI] [PubMed] [Google Scholar]
- Liu Y, Choudhury P, Cabral CM, Sifers RN. Oligosaccharide modification in the early secretory pathway directs the selection of a misfolded glycoprotein for degradation by the proteasome. J Biol Chem. 1999;274:5861–5867. doi: 10.1074/jbc.274.9.5861. [DOI] [PubMed] [Google Scholar]
- Marcantonio EE, Amar-Costesec A, Kreibich G. Segregation of the polypeptide translocation apparatus to regions of the endoplasmic reticulum containing ribophorins and ribosomes. II. Rat liver microsomal subfractions contain equimolar amounts of ribophorins and ribosomes. J Cell Biol. 1984;99:2254–2259. doi: 10.1083/jcb.99.6.2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melnick J, Dul JL, Argon Y. Sequential interaction of the chaperones BiP and GRP94 with immunoglobulin chains in the endoplasmic reticulum. Nature. 1994;370:373–375. doi: 10.1038/370373a0. [DOI] [PubMed] [Google Scholar]
- Muresan Z, Arvan P. Enhanced binding to the molecular chaperone BiP slows thyroglobulin export from the endoplasmic reticulum. Mol Endocrinol. 1998;12:458–468. doi: 10.1210/mend.12.3.0069. [DOI] [PubMed] [Google Scholar]
- Olden K, Pratt RM, Yamada KM. Role of carbohydrates in protein secretion and turnover: effects of tunicamycin on the major cell surface glycoprotein of chick embryo fibroblasts. Cell. 1978;13:461–473. doi: 10.1016/0092-8674(78)90320-3. [DOI] [PubMed] [Google Scholar]
- Oliver JD, van der Wal FJ, Bulleid NJ, High S. Interaction of the thiol-dependent reductase ERp57 with nascent glycoproteins. Science. 1997;275:86–88. doi: 10.1126/science.275.5296.86. [DOI] [PubMed] [Google Scholar]
- Plemper RK, Böhmler S, Bordallo J, Sommer T, Wolf DH. Mutant analysis links the translocon and BiP to retrograde protein transport for ER degradation. Nature. 1997;388:891–895. doi: 10.1038/42276. [DOI] [PubMed] [Google Scholar]
- Price BD, Mannheim-Rodman LA, Calderwood SK. Brefeldin A, thapsigargin, and AIF4- stimulate the accumulation of GRP78 mRNA in a cycloheximide dependent manner, while induction by hypoxia is independent of protein synthesis. J Cell Physiol. 1992;152:545–552. doi: 10.1002/jcp.1041520314. [DOI] [PubMed] [Google Scholar]
- Rodan AR, Simons JF, Trombetta ES, Helenius A. N-linked oligosaccharides are necessary and sufficient for association of glycosylated forms of bovine RNase with calnexin and calreticulin. EMBO J. 1996;15:6921–6930. [PMC free article] [PubMed] [Google Scholar]
- Rosenfeld MG, Marcantonio EE, Hakimi J, Ort VM, Atkinson PH, Sabatini DD, Kreibich G. Biosynthesis and processing of ribophorins in the endoplasmic reticulum. J Cell Biol. 1984;99:1076–1082. doi: 10.1083/jcb.99.3.1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz A, Maintz M, Kehle T, Herzog V. In vivo iodination of a misfolded proinsulin reveals colocalized signals for BiP binding and for degradation in the ER. EMBO J. 1995;14:1091–1098. doi: 10.1002/j.1460-2075.1995.tb07092.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamu CE, Cox JS, Walter P. The unfolded-protein-response pathway in yeast. Trends Cell Biol. 1994;4:56–60. doi: 10.1016/0962-8924(94)90011-6. [DOI] [PubMed] [Google Scholar]
- Sidrauski C, Chapman R, Walter P. The unfolded protein response: an intracellular signaling pathway with many surprising features. Trends Cell Biol. 1998;8:245–249. doi: 10.1016/s0962-8924(98)01267-7. [DOI] [PubMed] [Google Scholar]
- Skowronek MH, Hendershot LM, Haas IG. The variable domain of nonassembled Ig light chains determines both their half-life and binding to the chaperone BiP. Proc Natl Acad Sci USA. 1998;95:1574–1578. doi: 10.1073/pnas.95.4.1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer T, Wolf DH. Endoplasmic reticulum degradation: reverse protein flow of no return. FASEB J. 1997;11:1227–1233. doi: 10.1096/fasebj.11.14.9409541. [DOI] [PubMed] [Google Scholar]
- Staddon JM, Bouzyk MM, Rozengurt E. Interconversion of GRP78/BiP: a novel event in the action of Pasteurella multocida toxin, bombesin, and platelet-derived growth factor. J Biol Chem. 1992;267:25239–25245. [PubMed] [Google Scholar]
- Tirasophon W, Welihinda AA, Kaufman RJ. A stress response pathway from the endoplasmic reticulum to the nucleus requires a novel bifunctional protein kinase/endoribonuclease (Ire1p) in mammalian cells. Genes Dev. 1998;12:1812–1824. doi: 10.1101/gad.12.12.1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trombetta ES, Helenius A. Lectins as chaperones in glycoprotein folding. Curr Opin Struct Biol. 1998;8:587–592. doi: 10.1016/s0959-440x(98)80148-6. [DOI] [PubMed] [Google Scholar]
- Tsao YS, Ivessa NE, Adesnik M, Sabatini DD, Kreibich G. Carboxy terminally truncated forms of ribophorin I are degraded in preGolgi compartments by a calcium-dependent process. J Cell Biol. 1992;116:57–67. doi: 10.1083/jcb.116.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Wal FJ, Oliver JD, High S. The transient association of ERp57 with N-glycosylated proteins is regulated by glucose trimming. Eur J Biochem. 1998;256:51–59. doi: 10.1046/j.1432-1327.1998.2560051.x. [DOI] [PubMed] [Google Scholar]
- Van Leeuwen JEM, Kearse KP. The related molecular chaperones calnexin and calreticulin differentially associate with nascent T cell antigen receptor proteins within the endoplasmic reticulum. J Biol Chem. 1996;271:25345–25349. doi: 10.1074/jbc.271.41.25345. [DOI] [PubMed] [Google Scholar]
- Varki A. Biological roles of oligosaccharides: all of the theories are correct. Glycobiology. 1993;3:97–130. doi: 10.1093/glycob/3.2.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XZ, Harding HP, Zhang Y, Jolicoeur EM, Kuroda M, Ron D. Cloning of mammalian Ire1 reveals diversity in the ER stress responses. EMBO J. 1998;17:5708–5717. doi: 10.1093/emboj/17.19.5708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei J, Gaut JR, Hendershot LM. In vitro dissociation of BiP-peptide complex requires a conformational change in BiP after ATP binding but does not require ATP hydrolysis. J Biol Chem. 1995;270:26677–26682. doi: 10.1074/jbc.270.44.26677. [DOI] [PubMed] [Google Scholar]
- Wileman T, Pettey C, Terhorst C. Recognition for degradation in the endoplasmic reticulum and lysosomes prevents the transport of single TCR beta and CD3 delta subunits of the T-cell antigen receptor to the surface of cells. Int Immunol. 1990;2:743–754. doi: 10.1093/intimm/2.8.743. [DOI] [PubMed] [Google Scholar]
- Wu Y, Whitman I, Molmenti E, Moore K, Hippenmeyer P, Perlmutter DH. A lag in intracellular degradation of mutant alpha 1-antitrypsin correlates with the liver disease phenotype in homozygous PiZZ alpha 1-antitrypsin deficiency. Proc Natl Acad Sci USA. 1994;91:9014–9018. doi: 10.1073/pnas.91.19.9014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu YH, Sabatini DD, Kreibich G. Antiribophorin antibodies inhibit the targeting to the ER membrane of ribosomes containing nascent secretory polypeptides. J Cell Biol. 1990;111:1335–1342. doi: 10.1083/jcb.111.4.1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zapun A, Darby NJ, Tessier DC, Michalak M, Bergeron JJ, Thomas DY. Enhanced catalysis of ribonuclease B folding by the interaction of calnexin or calreticulin with ERp57. J Biol Chem. 1998;273:6009–6012. doi: 10.1074/jbc.273.11.6009. [DOI] [PubMed] [Google Scholar]
- Zapun A, Petrescu SM, Rudd PM, Dwek RA, Thomas DY, Bergeron JJ. Conformation-independent binding of monoglucosylated ribonuclease B to calnexin. Cell. 1997;88:29–38. doi: 10.1016/s0092-8674(00)81855-3. [DOI] [PubMed] [Google Scholar]
- Zhang JX, Braakman I, Matlack KE, Helenius A. Quality control in the secretory pathway: the role of calreticulin, calnexin and BiP in the retention of glycoproteins with C-terminal truncations. Mol Biol Cell. 1997;8:1943–1954. doi: 10.1091/mbc.8.10.1943. [DOI] [PMC free article] [PubMed] [Google Scholar]