Abstract
Metal ions facilitate the folding of the hairpin ribozyme, but do not participate directly in catalysis. The metal complex cobalt (III) hexaammine supports folding and activity of the ribozyme and also mediates specific internucleotide photocrosslinks, several of which retain catalytic ability. These crosslinks imply that the active core structure organized by [Co(NH3)6]3+ is different from that organized by Mg2+ and that revealed in the crystal structure (1). Residues U+2 and C+3 of the substrate, in particular, adopt different conformations in [Co(NH3)6]3+. U+2 is bulged out of loop A and stacked on residue G36, whereas the nucleotide at position +3 is stacked on G8, a nucleobase crucial for catalysis. Cleavage kinetics performed with +2 variants and a C+3 U variant correlate with the crosslinking observations. Variants that decreased cleavage rates in magnesium up to 70-fold showed only subtle decreases or even increases in observed rates when assayed in [Co(NH3)6]3+. Here, we propose a model of the [Co(NH3)6]3+-mediated catalytic core generated by MC-SYM that is consistent with these data.
Interactions between cations and RNA molecules are critical for the biological activity of RNA, in that metal ions promote RNA folding events and RNA-catalyzed reactions, including RNA processing reactions and peptide bond formation (2). In the hairpin and hammerhead ribozymes, cations function to facilitate folding into the active conformations, but play little or no direct role in catalysis (3–6). Folding and cleavage activity of the hairpin ribozyme can be supported by high concentrations (>1 M) of monovalent ions (4), moderate concentrations (2 to 20 mM) of magnesium and some other divalent ions (7), or by low concentrations (~1 mM) of the trivalent complex [Co(NH3)6]3+..This complex serves as an analogue of hexahydrated magnesium, in that it cannot make inner-sphere binding interactions with RNA (3).
Catalysis by the hairpin ribozyme is preceded by a major conformational change, in which the two domains of the ribozyme-substrate complex come into close association with one another. This docking step is accompanied by changes in the orientation of the Watson-Crick helical elements within the complex, which can be monitored by biochemical and biophysical methods, including FRET, electrophoretic mobility, transient electric birefringence, and hydroxyl radical footprinting (8–10). Concomitantly, extensive interactions between the two major non-helical regions are formed, and result in the positioning of the likely catalytic bases, G8 and A38, at the scissile phosphodiester linkage. These latter conformational changes can be monitored by the photocrosslinking and fluorescence behavior of the affected nucleobases (11). The scope of overall conformational change can be visualized by comparing the NMR structures of the individual domains with the crystallographic structures of complexes of the ribozyme with analogs of substrates, reaction intermediates and products (1, 12–14).
Photochemical crosslinking methods have been used to examine the global conformational change that occurs upon docking (15–17), and to examine details of the conformational changes associated with docking. In most cases the results have been confirmed by high-resolution studies. Stacking of U42 upon G21 in the undocked conformation (16) was confirmed by NMR analysis (13), and the unstacking of these bases upon formation of the docked complex was monitored by loss of crosslinking (17). The docked complex did not yield spontaneous photocrosslinks, but it was found that [Co(NH3)6]3+ could induce crosslinking in this conformation between closely located (presumably stacked) bases. This process is initiated by absorption of photons by the metal ion complex at wavelengths considerably longer than those absorbed by unmodified RNA (C. Kraemer-Chant, J. Heckman, D. Lambert, and J. M. Burke, in preparation). Two active-site, [Co(NH3)6]3+-induced photocrosslinks from the docked complex linked the catalytic nucleobases (G8 and A38) (Fig. 1) to the substrate nucleobases that span the active site, A-1 and G+1, respectively (18), (P. Chan and J. M. Burke, unpublished results). This photocrosslinking behavior suggests that G8 is stacked upon A-1 and the nucleotide at position 38 (2-aminopurine) is stacked upon G+1, and these findings have been confirmed by crystallographic studies (1, 14). The relevance of these crosslinked species was also confirmed by the observation that the covalently crosslinked RNA strands, when reconstituted in a complex, retained the ability to carry out cleavage chemistry at the correct site (18), (P. Chan and J. M. Burke, unpublished results). The same [Co(NH3)6]3+ crosslinking reactions that resulted in G8 to A-1 crosslinks also yielded a G36 to U+2 crosslink. In this case the implied stacking interaction or close approach of residue G36 in Loop B and U+2 of the substrate strand was not in agreement with the crystal structure (they are separated by approximately 10–15 Å). Nevertheless, the crosslinked strands could be reconstituted to yield cleavage activity, suggesting that covalent attachment of G36 and U+2 is at least permissive for the reaction.
Figure 1.
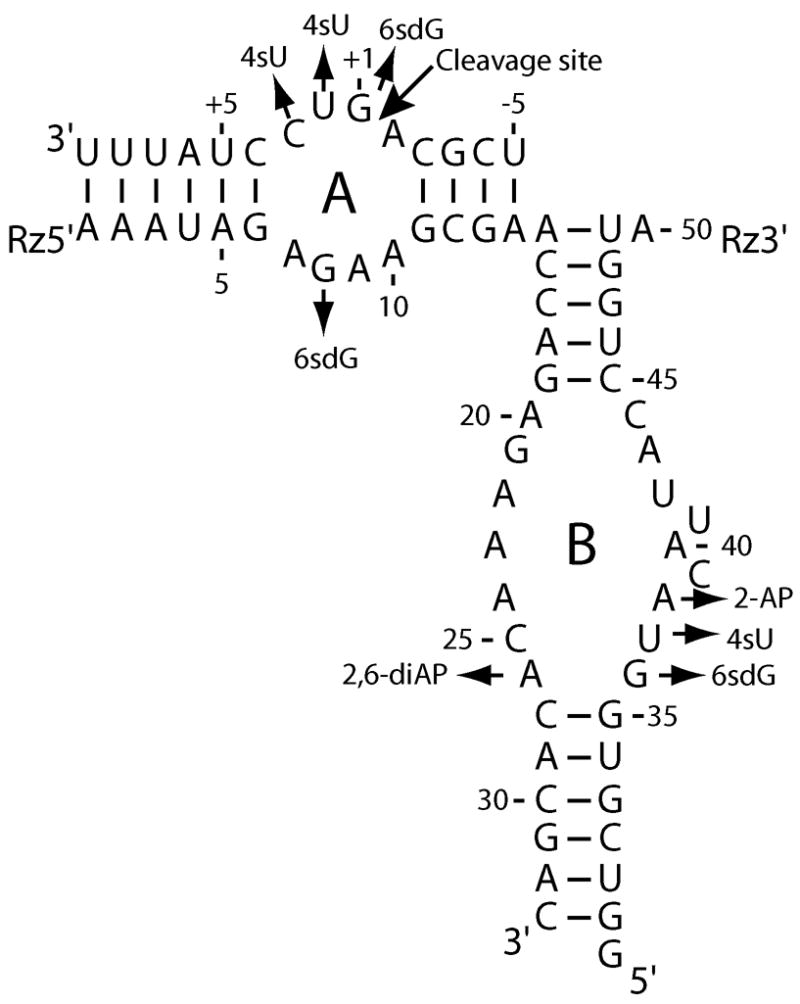
The hairpin-ribozyme substrate complex. Secondary structure of the hairpin ribozyme complex used in this study includes modifications in helices 1 and 2 in the substrate and ribozyme 5′ strand as well as a rate-enhancing U39C mutation in ribozyme 3′ strand. Domains A and B, and helices 1–4 are indicated. Cleavage site is shown by an arrow. Nucleotides at positions +1, +2, +3, 8, 26, 36, 37, 38 were individually substituted by modified nucleotides, i.e. 4-thiouridine (4sU), 6-thiodeoxyguanosine (6sdG), 2,6-diaminopurine (2,6diAP) or 2-aminopurine (2-AP) as indicated by small arrows.
The analysis of RNA structure is complicated by the conformational plasticity of RNA molecules, which can fold into inactive conformations, some of which are quite stable. The distribution of conformational isomers of an RNA molecule can clearly be influenced by such factors as (i) the ionic composition of a solution (2), (ii) base substitutions or other covalent modifications (19), and (iii) crystallographic packing (20, 21). Fortunately, the catalytic activity of ribozymes provides a chemical readout on the ability of the RNA to fold into an active conformation. It appears that the active conformations can themselves be stable (heavily populated) or unstable (sparsely populated). In our view, the accumulated evidence concerning the hairpin and hammerhead ribozymes suggests that the active hairpin ribozyme structure is quite stable, while the active hammerhead ribozyme structure is formed only transiently (22).
In this work, we have investigated whether the variant docked structure implied by the [Co(NH3)6]3+-induced G36 to U+2 photocrosslink actually functions in solution and generates biochemically observable consequences. We have incorporated nucleobases with increased photoreactivity (23–26) (e.g. 4-thiouridine (4sU) and 6-thiodeoxyguanosine (6sdG)) in order to compare crosslinks obtainable in Mg2+ versus [Co(NH3)6]3+. We have also tested the cleavage rates of substrates with nucleotide substitutions at the +2 position, (and ribozymes with other substitutions) comparing performance in Mg2+ and [Co(NH3)6]3+. Results suggest that these two metals support distinguishably different active structures of the hairpin ribozyme, and that the differences in structure are consistent with differential performances of mutant ribozyme complexes under the two sets of conditions. They have led us to present an alternative structure for an active hairpin ribozyme. Distribution of the RNA between the two conformations described by the crystal structure and by this crosslinking work could be influenced by the binding of the metal ions, crystallographic packing, or both.
MATERIALS AND METHODS
RNA preparation
All oligoribonucleotides were prepared by solid-phase synthesis using standard RNA phosphoramidite chemistry. Reagents were purchased from Glen Research. Following deprotection, RNA was purified by denaturing gel electrophoresis and reverse-phase HPLC, as previously described (11).
Crosslinking assays
For large-scale preparative crosslinking, the ribozyme-substrate complexes were assembled by incubating the 5′-32P-end-labeled strand (approximately 200 nM) with an excess of the other two strands (Fig. 1) (>400 nM) in reaction buffer (50 mM Tris-HCl, pH 8 or 9) for 10 min at 37 °C (high pH resulted in higher yields of most [Co(NH3)6]3+-induced photocrosslinks). The solutions were allowed to equilibrate for 2 min at room temperature. Cleavage reactions were initiated by addition of 1 mM [Co(NH3)6]3+ or 20 mM MgCl2. Reactions were immediately irradiated in 96-well plates for 3–5 min at 312 nm using a hand-held UV lamp (UVH, 6W, International Biotechnologies, Inc.). Crosslinked species were fractionated by denaturing 20% polyacrylamide gel electrophoresis and isolated as described by Pinard et al. (18).
Mapping of the crosslinked species
Crosslinking sites were mapped as described in Pinard et al. (15). Briefly, the purified, desalted crosslinked species (with one strand end-labeled) were subjected to partial hydrolysis with alkali or with T1 RNase. Control end-labeled strands were similarly digested. Samples were fractionated on denaturing 20% polyacrylamide gels and exposed to X-ray film. Resulting sequencing patterns could be read in the alkali track up to the nucleotide preceding the crosslink, because species beyond that point are part of a branched RNA that migrates more slowly.
Reconstitution of the ribozyme-substrate complex with crosslinked strands and activity assays
Crosslinked species contained two covalently joined strands out of the three used to make up the ribozyme-substrate complex (Fig. 1). The ribozyme-substrate complex was reconstituted by incubating the purified 32P-labeled crosslinked strands in standard reaction buffer (50 mM Tris-HCl pH 7.5), in the presence of 200 nM of the missing strand for 10 minutes at 37 °C. Cleavage reactions were initiated by the addition of 1 mM [Co(NH3)6]3+ and were allowed to proceed for 2 hours at 25 °C. The reactions were stopped by the addition of an equal volume of 90% formamide, 1 mM EDTA, and loaded directly onto a denaturing 20% polyacrylamide gel.
Cleavage assays of substrate variants
All reactions were performed in 50 mM Tris-HCl (pH 7.5), 12 mM MgCl2 or 1 mM [Co(NH3)6]3+ at 25 °C. Ribozyme-substrate complexes (with 5-fold molar excess of ribozyme) were preincubated at 37 °C for 10 minutes in reaction buffer. The solutions were then allowed to equilibrate at 25 °C for 5 minutes. Cleavage reactions were initiated by adding the metal ions. Aliquots of the reaction (2μl) were then taken and quenched with 18 μl of loading solution (90% formamide, 1 mM EDTA). Reaction products were separated on 20% denaturing polyacrylamide gels and quantified using a BioRad Molecular Imager FX system. Cleavage rates were determined by non-linear regression using the Origin software (Microcal Software, Inc.).
Molecular Modeling
Models of the catalytic core were generated using the constraint-satisfaction program MC-SYM (27, 28). The structural constraints used to generate the MC-SYM script were derived from data generated in this work, from previously published topographic work (11, 29), and crystallographic data (1, 14). A-form RNA was assumed for all Watson-Crick helices. Specific residues in loop A (A-1, G+1 and U+2), and loop B (G36, U37, A38 and C39) were allowed to adopt all sugar pucker conformations, and glycosyl angles were taken from crystallographic data except for U+2 (1, 14). Solutions that were similar (= 2Å rmsd) were combined by MC-SYM. Energy-minimization of the backbone of a representative model was done through molecular mechanics calculations by the molecular simulation program Sander, from the Amber 7 suite of programs (30) using the Amber 2002 force field for RNA. All 1–4 electrostatic interactions were set to a factor of 1.2, and the distance-dependent dielectric model (ε = 4Rij) for the Coulombic representation of electrostatic interactions was used. Energy minimization was performed using the steepest descent for the first 100 steps, then the conjugate gradient method was applied until the maximum derivative was less that 0.1 kcal/mol Å. Figures were prepared using MolScript (31). To obtain the scripts email J.M. Burke.
RESULTS
Folding in the presence of cobalt (III) hexaammine differs from that in magnesium
[Co(NH3)6]3+ and photoreactive nucleobases were used to generate UV-induced crosslinks within a complex of the hairpin ribozyme and its substrate. The construct used for these studies (SV5) is shown in Figure 1. It consists of three strands of RNA: substrate, ribozyme 5′ strand, and ribozyme 3′ strand. It is a structurally and kinetically well-behaved construct that eliminates some misfolding associated with the original hairpin ribozyme (10). Crosslinked species were mapped by partial alkaline hydrolysis (Table 1) and their catalytic activity was examined through cleavage assays following purification and reconstitution.
Table 1.
Summary of photocrosslinking experiments
| Ribozyme nt | XL Agent | Target | Other XL | Metal | Activity |
|---|---|---|---|---|---|
| none | G21/U421 | Mg++ | − | ||
| A-1/G8 | Co(NH3)63+ | + | |||
| U+2/G36 | Co(NH3)63+ | + | |||
| G8 | 6-thio-dG8 | A-1 | Mg++ | − | |
| G+1 | Co(NH3)63+ | − | |||
| A26 | DiAP26 | Mg++ | N/D | ||
| Co(NH3)63+ | N/D | ||||
| G36 | 6-thio-dG36 | Mg++ | N/D | ||
| U+2 | Co(NH3)63+ | + | |||
| U37 | 4-thio-U37 | Mg++ | N/D | ||
| U+2 | Co(NH3)63+ | − | |||
| U+2/G36 | Co(NH3)63+ | + | |||
| A38 | 2-AP382 | Mg++ | N/D | ||
| G+1 | Co(NH3)63+ | + | |||
| Substrate nt | |||||
| G+1 | 6-thio-dG+1 | G6 | Mg++ | − | |
| G21/U42 | Mg++ | − | |||
| U+23/G8 | Co(NH3)63+ | − | |||
| C+3/G36 | Co(NH3)63+ | − | |||
| G21/A43 | Co(NH3)63+ | − | |||
| U+2 | 4-thio-U+2 | A9 | Mg++ | − | |
| G8 | Co(NH3)63+ | − | |||
| G36 | Co(NH3)63+ | − | |||
| C+3 | 4-thio-U+3 | A9 | Mg++ | − | |
| G8 | Co(NH3)63+ | + | |||
| RLB4 | Co(NH3)63+ | + | |||
| U+2/G36 | Co(NH3)63+ | + |
Obtained at 254 nm.
Chan, P. and Burke, J.M. unpublished results.
Impossible to determine clearly which substrate residue was mapped since deoxyG+1 does not give a band after T1 or alkali digestion. The cut off appeared to be a mixture of U+2 and C+3.
We were unable to identify unambiguously the residue involved in this crosslink although it is reproducible.
One of the [Co(NH3)6]3+-induced crosslinks described above was observed between the ribozyme and substrate, and was mapped to substrate nucleobase U+2 and ribozyme G36 (Fig. 2A). In the primary structure, U+2 is adjacent to the cleavage site nucleotide G+1, and G36 is close to the putative catalytic base A38. The three-dimensional crystallographic structure (1) shows G+1 stacked on A38, but U+2 is 10–15 Å distant from G36, with O4 of U+2 forming a single hydrogen bond with N2 of G8, one of the catalytic nucleobases. Importantly, activity assays of the U+2 • G36 crosslinked RNA showed that it retains cleavage activity whether assayed in [Co(NH3)6]3+ or in Mg2+ (Fig. 2B). Together, these results indicate that the conformation which gives rise to the U+2 • G36 crosslink is active and different from the fold observed in the crystallographic structures.
Figure 2.
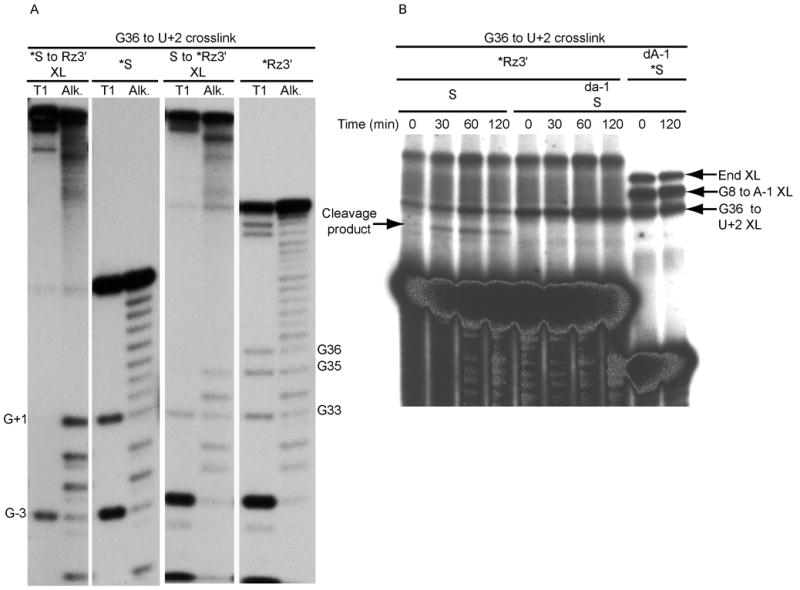
G36/U+2 cobalt hexaammine-induced crosslinks. A) Mapping of G36/U+2 cobalt hexaammine-induced crosslink. The crosslinked sites were mapped by limited alkaline hydrolysis (Alk) and partial digestion with RNase T1 (T1). The species consisting of 5′-32P end-labeled substrate crosslinked to ribozyme 3′ strand (*S to Rz3′ XL), and the end-labeled substrate control (*S) are paired on the left. The species consisting of 5′-32P end-labeled ribozyme 3′ strand crosslinked to substrate (S to *Rz3′ XL) and end-labeled ribozyme 3′ strand control (*Rz3′) are paired on the right. Sites of T1-induced cleavages are indicated by the numbered guanosine (note that the G+1 band appears in the alkali track, but is poorly cleaved by T1 RNase because of its proximity to the branched crosslink structure). B) Cleavage assay with crosslinked end-labeled ribozyme 3′ strand, crosslinked to cleavable or noncleavable (2′-deoxy A-1) substrate. Since ribozyme 3′ strand is crosslinked to residue U+2, cleavage removes the 5′ cleavage product (5nt) from the crosslinked species, generating a band with slightly increased mobility. Crosslinks were generated as described in Materials and Methods, then simply incubated to permit cleavage. Crosslinks made with end-labeled noncleavagle substrate (at right) identify the crosslinked species.
Investigating the alternative fold
To further examine the differences between this active fold and the crystallographic structure, we introduced a series of alternative nucleotides at substrate position +2. All of the bases introduced at +2 (G, A, C) were found to be able to crosslink to G36, while no crosslink was observed when an abasic linkage was present (data not shown). To ensure that the +2 • 36 crosslink was not unique to the construct used (SV5) (32, 33), we irradiated a construct with a U39 as in the wild-type sequence (34), and again observed both the U+2 • G36 and A-1 • G8 crosslinks, indicating that the fold that gives rise to these crosslinks is not unique to the SV5 construct used in the present study (data not shown). These results suggest that the ability of the nucleobase at substrate position +2 to stack on G36 in the presence of [Co(NH3)6]3+ is a general property of folding and catalysis of the hairpin ribozyme.
While [Co(NH3)6]3+ can initiate photocrosslinking through the absorbance of long-wavelength UV light (312 and 365 nm), it is also likely that it may occupy distinct binding sites and not simply compete for those occupied by Mg2+, the most relevant biological ligand (35). Additionally, [Co(NH3)6]3+ has been shown to make outer-sphere interactions with more ligands than Mg2+ does in DNA, and to promote the rearrangement of B-DNA into Z-DNA (36). Therefore, we sought to determine whether the +2 • 36 crosslink could be obtained in the absence of [Co(NH3)6]3+. Since the +2 • 36 crosslink is not obtained when unmodified RNA is irradiated in Mg2+-containing solutions, we examined the effects of introducing the photoreactive nucleotides 4-thiouridine (4sU) and 6-thiodeoxyguanosine (6sdG) at positions +2 and 36.
Upon irradiation of the complex containing 4sU at substrate position +2, a single crosslinked species was observed in Mg2+-containing solutions (Table 1). This crosslink maps from substrate 4sU+2 to ribozyme residue A9, is inactive, and is likely to result from the undocked structure of the internal loop in domain A (12, 37) (data not shown). In the presence of [Co(NH3)6]3+, two crosslinks were obtained, one from 4sU+2 to G36 (Fig. 3), and the second from 4sU+2 to ribozyme G8 (data not shown). Neither crosslinked species retained detectable catalytic activity when tested in [Co(NH3)6]3+. The presence of 4sU at +2 is not itself inhibitory for cleavage; thus, the lack of cleavage activity of the crosslink from 4sU+2 to G36 might be due to chemical rearrangement of crosslinked 4sU+2.
Figure 3.
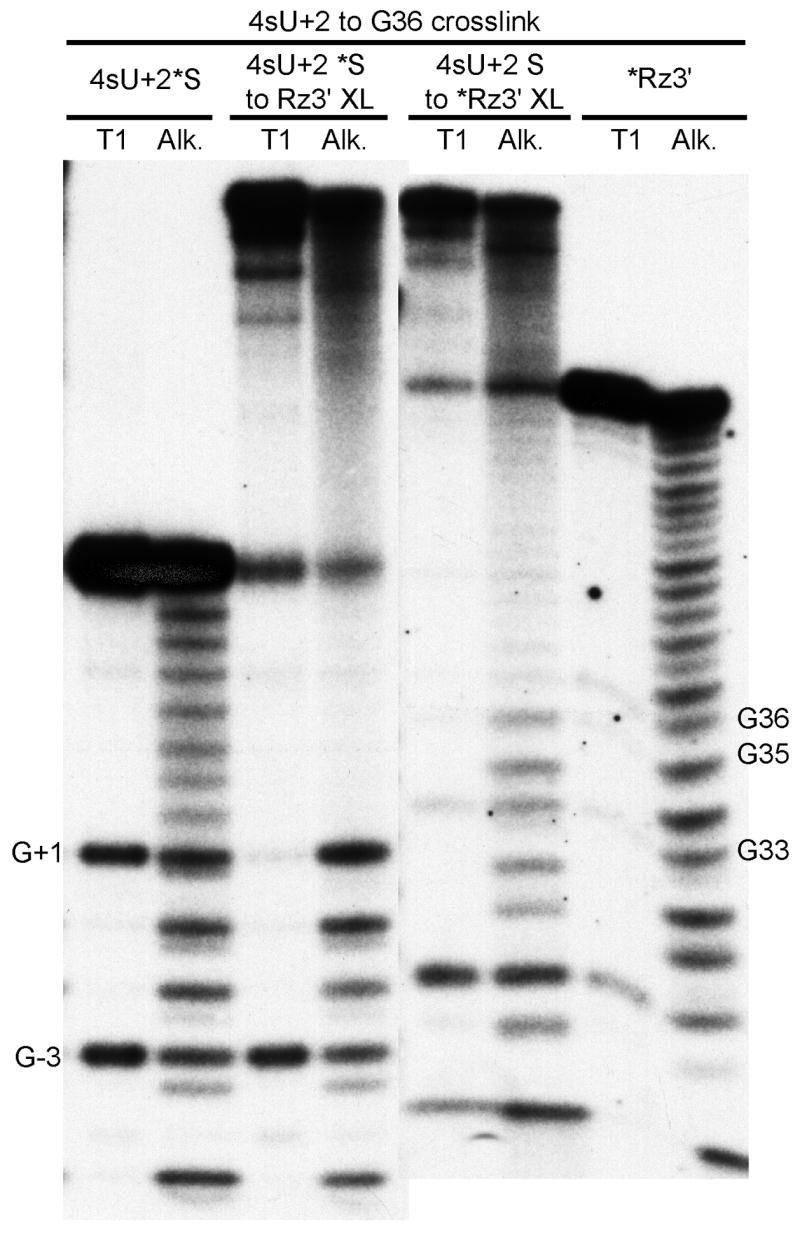
Mapping of 4sU+2/G36 crosslinked sites. The crosslinked sites were mapped by limited alkaline hydrolysis (Alk) and partial digestion with RNAse T1 (T1). The species consisting of 5′-32P end-labeled 4sU+2 substrate crosslinked to ribozyme 3′ strand (4sU+2*S to Rz3′ XL), and the end-labeled 4sU+2 substrate control (4sU+2*S) are paired on the left. The 5′-32P end-labeled ribozyme 3′ strand crosslinked to 4sU+2 substrate (4sU+2S to *Rz3′ XL), and end-labeled ribozyme 3′ strand control (*Rz3′) are paired on the right. Sites of T1-induced cleavages are indicated by the numbered guanosine.
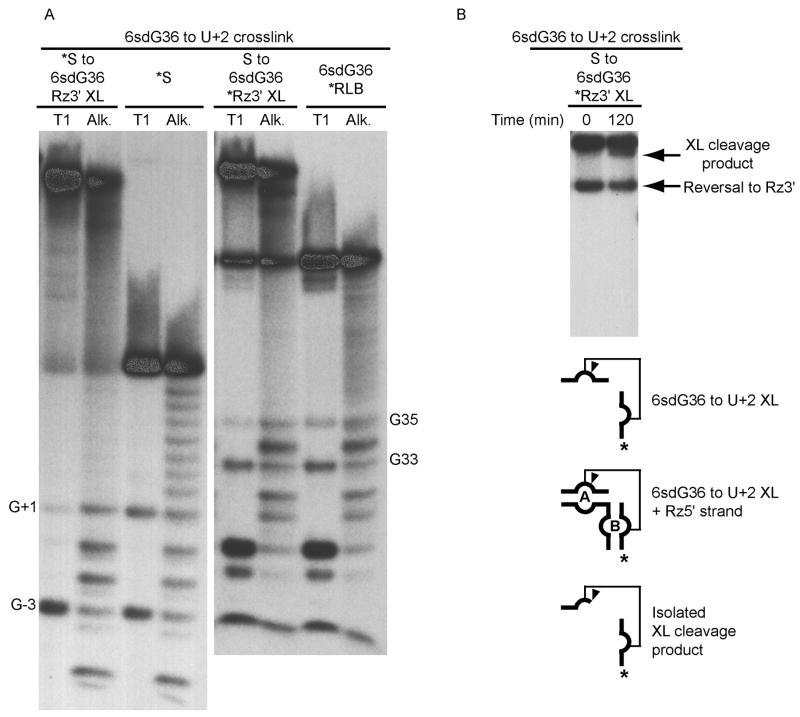
Before using ribozymes containing 6sdG36 in crosslinking experiments, we conducted a control to determine the effect of the deoxyribose substitution at position 36 on catalytic activity, and found that the dG36 ribozyme showed activity virtually identical to that of the unmodified ribozyme (data not shown). Irradiation of the ribozyme-substrate complex containing 6sdG36 in the presence of Mg2+ showed that no crosslinks were formed. However, a catalytically active 6sdG36 • U+2 crosslinked species was observed when [Co(NH3)6]3+ was present (Table 1) (Fig. 4). Together, these results suggest that [Co(NH3)6]3+ stabilizes a catalytically-active conformation of the ribozyme-substrate complex, in which U+2 has flipped out of the internal loop of domain A to form a specific stacking interaction with G36 in domain B.
Figure 4.
6sdG36 crosslink. A) Mapping of 6sdG36 crosslinked sites. The crosslinked sites were mapped by limited alkaline hydrolysis (Alk) and partial digestion with RNAse T1 (T1). The species consisting of 5′-32P end-labeled substrate crosslinked to 6sdG36 ribozyme 3′ strand, (*S to 6dG36Rz3′ XL), and the end-labeled substrate control (*S) are paired on the left. The species consisting of 5′-32P end-labeled 6sdG36 ribozyme 3′ strand crosslinked to substrate (S to 6sdG36*Rz3′ XL), and end-labeled 6sdG36 ribozyme 3′ strand control (6sdG36*Rz3′) are paired on the right. Sites of ribonuclease T1-induced cleavages are indicated by the numbered guanosine. B) Activity assay for isolated crosslinked species. Ribozyme 5′ strand (Rz5′) was added to crosslinked products labeled on ribozyme 3′ strand (*Rz3′), in the presence of 1 mM cobalt hexaammine for 2 hours. The crosslink cleavage product and the reversal to ribozyme 5′ strand are shown by arrows.
In order to determine if other nucleotides were involved in this conformational change, we introduced photoreactive nucleobases at adjacent sites in the substrate (6sdG at position +1 and 4sU at position +3) and examined crosslinking and catalytic activity. The substitution of 6sdG at position +1 resulted in a significant decrease in catalytic activity, which is a primary consequence of the 2′-deoxy substitution (note that the corresponding ribonucleoside phosphoramidite was not available). Interestingly, this loss of activity was exhibited in the presence of Mg2+, but not when the reactions were conducted in [Co(NH3)6]3+. Control experiments showed that dG+1 had the same effect on cleavage activity.
In the presence of Mg2+, a crosslink from 6sdG+1 was observed to the ribozyme at G6. In addition, the previously characterized G21 • U42 crosslink (16) was observed in domain B. This latter spontaneous crosslink has been shown to occur only in the undocked version of Loop B (17), and together with the activity assay, suggests that the 2′-OH of substrate G+1 is important for folding into an active structure in solutions containing Mg2+. A hydrogen bond between the 2′-OH of G+1 and the exocyclic amino group of G36 was previously shown to contribute to the stability of the docked, active complex (38).
When a complex containing 6sdG+1 substrate was irradiated in the presence of [Co(NH3)6]3+, no crosslinks were obtained that originated from the modified nucleobase. Instead, we found multiple inactive crosslinks, including U+2 • G8, C+3 • G36, and G21 • A43 (Table 1). These results were somewhat surprising. Since 6sdG+1 does not inhibit cleavage activity in [Co(NH3)6]3+, it might be expected to permit formation of crosslinks, at least those normally seen in the active complex. It may be that the photochemical reaction of 6sdG+1, positioned close to G8 and G36 in the 3-dimensional structure (Fig. 6), interferes with formation of the normal [Co(NH3)6]3+-induced crosslinks.
Figure 6.
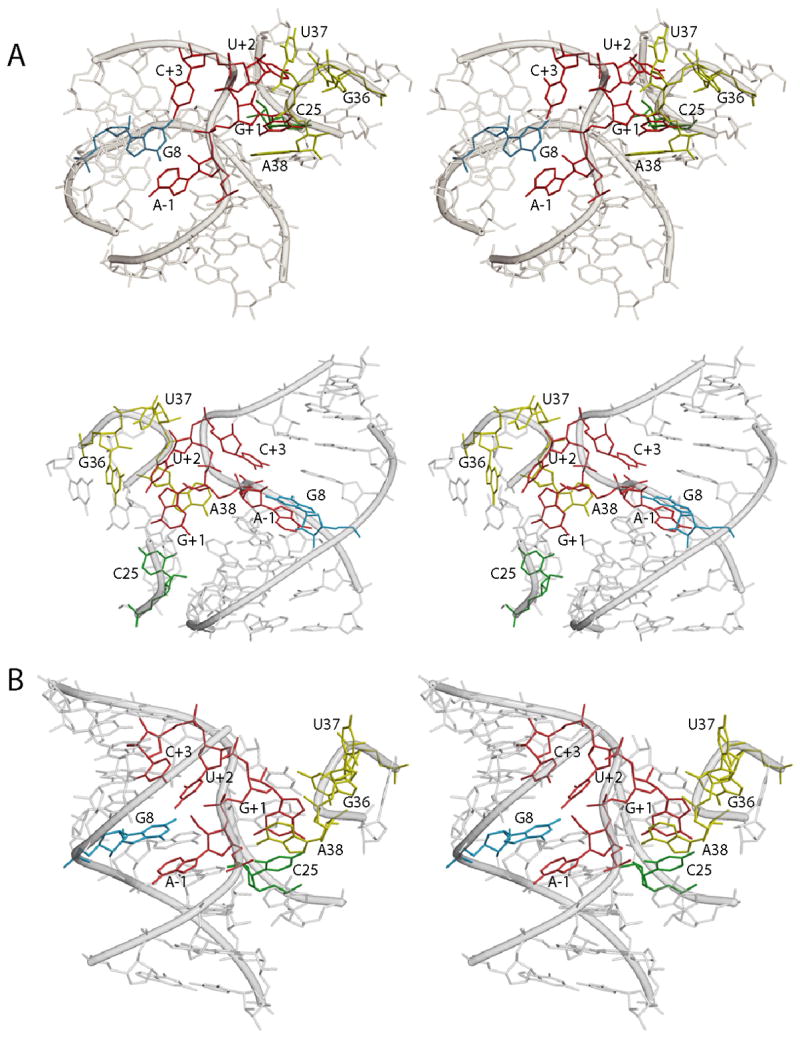
Models of the hairpin ribozyme active site. A) Two different stereo views of the MC-SYM-generated model based on biochemical and photochemical experiments. Residues A-1, G+1, U+2 and C+3 in the substrate strand are shown in red. G+1 which is stacking on A38 (yellow) and forming a Watson-Crick base pair with C25 (green) in the ribozyme 5′ strand. U+2 is bulged out of domain A and is in the vicinity of G36 and U37 (both in yellow) as suggested by crosslinking experiments. Residue C+3 forms a hydrogen bond with A7 and partially stack on G8 (blue). The latter nucleobase, in the substrate-binding strand, is stacked on A-1 and its Watson-Crick face is adjacent to the scissile bond. Remaining part of substrate and ribozyme strands are depicted in transparent gray. B) Stereo view of the crystal structure of the four-way junction hairpin ribozyme catalytic core.
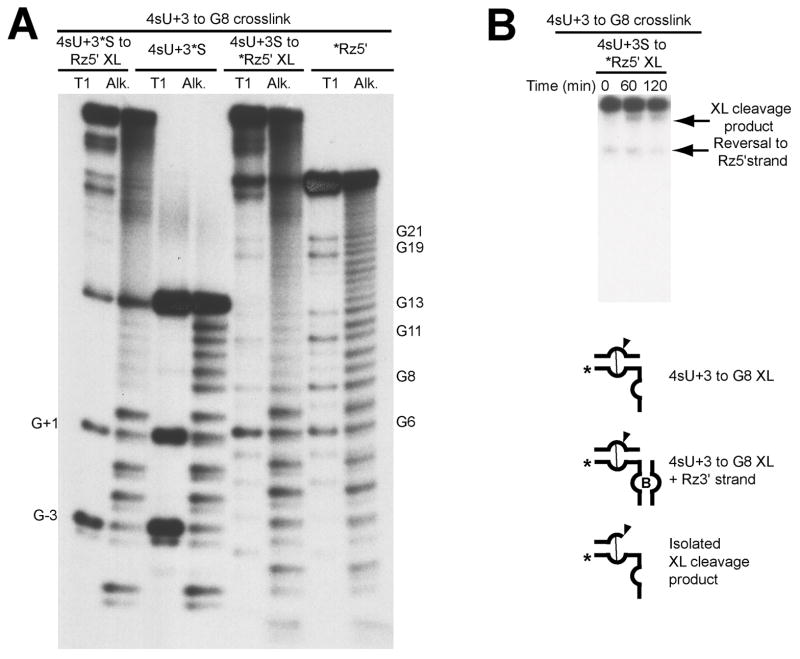
Replacement of C+3 with 4sU has different effects on catalysis in the presence of Mg2+ or [Co(NH3)6]3+ (see below; Table 2). When irradiated in solutions containing Mg2+, we obtained an inactive intra-domain crosslink to A9 (Table 1). In [Co(NH3)6]3+, we isolated three crosslinks, all of which were active. These are the U+2 • G36 crosslink described above and an active 4sU+3 • G8 intra-domain crosslink. The third crosslink links 4sU+3 to a site or sites on the ribozyme 3′ strand of the complex that could not be mapped in an unambiguous manner, indicating multiple crosslinks not resolved by preparative electrophoresis.
Table 2.
Cleavage rates for +2 and +3 variants
| Mutations | Ions | k(obs) min−1 | k(obs)/WT |
|---|---|---|---|
| WT (U+2, C+3) | Mg++ | 0.120 | 1.0 |
| C+2 | Mg++ | 0.002 | 0.017 |
| A+2 | Mg++ | 0.009 | 0.075 |
| G+2 | Mg++ | 0.003 | 0.025 |
| Abasic | Mg++ | 0.015 | 0.125 |
| U+3 | Mg++ | 0.0131 | 0.108 |
| WT (U+2, C+3) | Co(NH3)63+ | 0.073 | |
| C+2 | Co(NH3)63+ | 0.029 | 0.40 |
| A+2 | Co(NH3)63+ | 0.023 | 0.32 |
| G+2 | Co(NH3)63+ | 0.025 | 0.34 |
| Abasic | Co(NH3)63+ | 0.090 | 1.23 |
| U+3 | Co(NH3)63+ | 0.123 | 1.68 |
Average of 2 experiments. Other experiments were performed in triplicate and values are averages.
Three other complexes containing photoactive modifications were examined. Replacement of the catalytic nucleobase G8 with 6sdG in the presence of Mg2+ showed a crosslink to substrate A-1, consistent with previous crosslinking results (11, 18) and with the crystal structures (1, 14). While the G8 • A-1 crosslink in the unmodified RNA retained catalytic activity (11, 18), the crosslink involving 6sdG was inactive, suggesting a different photoadduct structure. In the presence of [Co(NH3)6]3+ an inactive 6sdG8 • G+1 crosslink was obtained. This result is more consistent with the arrangement of G8 and G+1 observed in the structure of loop A obtained by NMR (12) and might indicate that the sulfur substitution interferes with [Co(NH3)6]3+ binding or photochemistry. Position 37 was tested because of its proximity to G36. Replacement of ribozyme U37 with 4sU revealed no crosslinks when Mg2+-containing solutions were irradiated. However, in the presence of [Co(NH3)6]3+, both the active U+2 • G36 and an inactive U+2 • 4sU37 crosslink were identified (Table 1). We also generated a complex containing 2,6-diaminopurine at position 26. Unfortunately, this modification was severely inhibitory and no crosslinks were obtained.
[Co(NH3)6]3+ suppresses the inhibition of cleavage activity by variants at substrate position +2
We measured cleavage rates of substrate +2 variants (G, A, C and abasic) in solutions containing Mg2+ (39) and [Co(NH3)6]3+, and observed that the inhibitory effects of +2 variants in Mg2+ solutions are partially or completely suppressed by [Co(NH3)6]3+. In the presence of magnesium ions, cleavage rates of U+2 variants are reduced 8- to 70-fold, while the reductions are 2- to 4-fold in the presence of [Co(NH3)6]3+. The C+3U variant showed behavior that was even more dramatic, in that the cleavage rate relative to the wild type C+3 was reduced by a factor of 10 in Mg2+, but was actually enhanced in the presence of [Co(NH3)6]3+. These observations are consistent with differences in structural roles for residues +2 and +3 in [Co(NH3)6]3+ compared to Mg2+. Table 2 summarizes all rates according to mutation and ionic conditions.
Model of the conformation stabilized by [Co(NH3)6]3+
Using active [Co(NH3)6]3+-induced crosslinks as topological constraints, we modeled the three-dimensional structure of the hairpin ribozyme catalytic core, using the constraint satisfaction program MC-SYM (27, 28). The resulting model (Fig. 6) retains many of the features of the crystallographic structures, including the active site features that are known to be essential for catalysis; specifically the G+1 • C25 base pair, stacking of catalytic nucleobase G8 on substrate A-1, stacking of catalytic A38 on G+1, and the A10 • C25, G11 • A24 ribose zipper.
The primary differences between this model and the crystallographic structures are twofold. First, U+2 is flipped out of the active site internal loop and is stacked on G36, disrupting the crystallographic hydrogen bond of U+2 to G8. Second, C+3 is partially stacked on G8, while retaining the crystallographic hydrogen bond of C+3 with A7. We are uncertain if a 4sU at position +3 can base pair with A7 and be partly responsible for the stacking inferred. Flipping U+2 out of domain A and into contact with domain B can be accomplished without a large rearrangement of the structure. Upon docking, G+1 must move out of domain A to make its essential base pair with C25, and under certain conditions, U+2 may move out with it. This modeling exercise demonstrates that differences in structure and interactions at the active site can be accommodated without a global rearrangement of the structure of the ribozyme-substrate complex.
DISCUSSION
We have used an experimental approach combining photochemical crosslinking, nucleotide substitution, activity analysis, and structural modeling to examine nucleobase stacking at and around the active site of the hairpin ribozyme-substrate complex. Our results point to a previously unidentified interaction between the two domains of the complex, in which substrate U+2 is stacked on ribozyme G36. In contrast, the published crystallographic structures (1, 14) do not show the stacking of U+2 on G36. Indeed, crystallographic studies showed that U+2 is retained within the domain A internal loop that contains the cleavage site and forms a single hydrogen bond with exocyclic amino group of G8, one of the catalytic nucleobases. This latter group also shows an inferred crystallographic hydrogen bond with the pro-Rp oxygen of the scissile bond, which appears unaffected by the conformational change (1).
U+2 is a phylogenetically conserved nucleotide within the hairpin ribozyme (40). Previous work has shown that U+2 variants are cleaved, but at substantially reduced rates (14- to 70-fold slower) in magnesium-containing solutions (39) (Table 2). The U+2 abasic variant displayed particularly interesting behavior, in that cleavage proceeded while the docked conformation of the ribozyme-substrate complex could not be detected, leading to the conclusion that cleavage could proceed via formation of a transient docked complex, as we have proposed for the hammerhead ribozyme (22). In [Co(NH3)6]3+, these U+2 variants are cleaved at rates only 2- to 3-fold slower than U+2, instead of the large reductions seen in Mg2+ (Table 2). The abasic +2 and C+3/U variants actually cleave faster than the wild-type in [Co(NH3)6]3+.
Here, we have presented photochemical crosslinking data showing that complexes containing U+2 and variants can form a docked structure in the presence of [Co(NH3)6]3+ which is characterized by the inter-domain stacking of the nucleobase at position +2 on G36. These crosslinked species retain catalytic activity, indicating that the N+2 • G36 stack is compatible with catalytic function. Together, these results provide a structural rationale for the observation that U+2 is part of a [Co(NH3)6]3+-dependent structural rearrangement of the hairpin ribozyme-substrate complex. Our results are supported by NMR analysis of this internal loop in an isolated (undocked) domain A, which indicates that U+2 is highly mobile, and is found to be flipped out into solvent (12). Furthermore, [Co(NH3)6]3+ has been shown to promote the rearrangement of B-DNA into Z-DNA by binding to additional ligands in the DNA (36), selectively stabilizing the Z-DNA conformation much more effectively than Mg2+.
Our structural model (Fig. 6A) shows U+2 having been flipped out of the domain A internal loop, and stacking on G36. In this model, catalytic base G8 is now stacked between substrate bases A-1 and the A7-C+3 base pair. In the crystal structure (1), which contains Ca2+ as its metal ion, (Fig. 6B), U+2 closely approaches G8, making an inferred hydrogen bond from the O4 of U+2 to the exocyclic amino group of G8. Variants at +2 are cleaved very slowly in Mg2+, and their (15- to 70-fold) slower cleavage rates seem to correlate with their inability to make this interaction. In [Co(NH3)6]3+, the U+2 variants are only 2- 3-fold slower in cleavage than U+2, with no distinct differences between C, G or A (Table 2). Indeed, the abasic +2 variant cleaves faster than U+2, suggesting that the N+2 has a different role in the reaction in [Co(NH3)6]3+, and that it is not making the hydrogen bond interaction with G8. The model also shows a slight change in position of C+3, required by the alteration in the backbone at U+2. The change is reflected in the ability to observe an active crosslink from a 4sU+3 variant to G8 in [Co(NH3)6]3+ but not in Mg2+(Table 1), suggesting a close approach or stack between the two residues. It also correlates with the observation that C+3/U is a detrimental variant if measured for cleavage in Mg2+ (~10-fold slower), but is a favorable variant (1.7 times faster) in [Co(NH3)6]3+. The changes in the backbone around U+2 and C+3 do not alter the conformation of the cleavable linkage between A-1 and G+1, and this may be the reason that the putative alternative fold, even though it has biochemical implications for variants, allows the ribozyme to remain active.
Overall, there appear to be three ways in which our discovery of the U+2 • G36 stack can be reconciled with the crystallographic structures. First, the structure containing the U+2 • G36 stack might represent an important folding intermediate, leading to the formation of an active structure represented by the crystal structures. A second formal possibility is that the crystal structure could be a folding intermediate on the pathway to the U+2 • G36 stacked structure. Third, the ribozyme could have significant plasticity around its active site, so that both structures are active, and the degree to which they are populated is influenced by the specific ionic conditions and RNA constructs that were used. With respect to this third possibility, the results of single-molecule spectroscopy show significant kinetic heterogeneity in the folding and reactions of the hairpin ribozyme (41, 42). Although the structural differences between these populations are not yet known, the behavior of U+2 that we have identified in this work could well contribute to some of this heterogeneity.
Cobalt hexaammine is not a biologically relevant metal complex, but its ability to support folding and activity of many RNAs, plus its ability to initiate photocrosslinking with unmodified RNA and long-wavelength UV light, makes it a useful tool for the study of RNA structure. In this study, it has provided a rare opportunity to investigate the detailed differences in structure and biochemistry induced by folding the same small ribozyme molecule with two different metal ions.
Figure 5.
4sU+3 crosslink. A) Mapping of 4sU+3 crosslinked sites. The crosslinked sites were mapped by limited akaline hydrolysis (Alk) and partial digestion with RNAse T1 (T1). The species consisting of 5′-32P end-labeled 4sU+3 substrate crosslinked to ribozyme 5′ strand (4sU+3*S to Rz5′ XL), and the end-labeled 4sU+3 substrate control (4sU+3*S) are paired on the left. The species consisting of 5′-32P end-labeled ribozyme 5′ strand crosslinked to 4sU+3 substrate (4sU+3S to *Rz5′ XL), and end-labeled ribozyme 5′ strand control (*Rz5′) are paired on the right. Sites of T1-induced cleavages are indicated by the numbered guanosine. B) Activity assay for isolated crosslinked species. Ribozyme 3′ strand (Rz3′) was added to crosslinked products,labeled on ribozyme 5′ strand as indicated, in the presence of 1 mM cobalt hexaammine for up to 2 hours. The crosslink cleavage product and the reversal to ribozyme 5′ strand are shown by arrows.
Acknowledgments
We would like to thank Kenneth Hampel and Michael Fay for helpful discussions and insights, and Gulnar Pothiawala and Michaela Essam-Agbesi for preliminary work on cleavage assays of +2 variants. Additionally, we thank Michael Fay for synthesis of RNA and computer technical support, as well as Anne MacLeod for manuscript preparation. We acknowledge the Vermont Cancer Center DNA Analysis Facility (Grant P3OCA22435 from the National Cancer Institute) for use of their phosphorimager.
ABBREVIATIONS AND TEXTUAL FOOTNOTES
- 4sU
4-thiouridine
- 6sdG
6-thiodeoxyguanosine
- EDTA
ethylenediaminetetraacetic acid
- FRET
Florescence Resonance Energy Transfer
- HPLC
High Performance Liquid Chromatography
- NMR
Nuclear Magnetic Resonance
- Tris
Tris(hydroxymethyl)aminomethane
Footnotes
This work is supported by grant AI44186 to J.M.B. from the National Institutes of Health.
References
- 1.Rupert PB, Ferre-D’Amare AR. Crystal structure of a hairpin ribozyme-inhibitor complex with implications for catalysis. Nature. 2001;410:780–786. doi: 10.1038/35071009. [DOI] [PubMed] [Google Scholar]
- 2.Pyle M. Metal ions in the structure and function of RNA. J Biol Inorg Chem. 2002;7:679–690. doi: 10.1007/s00775-002-0387-6. [DOI] [PubMed] [Google Scholar]
- 3.Young KJ, Gill F, Grasby JA. Metal ions play a passive role in the hairpin ribozyme catalysed reaction. Nucleic Acids Res. 1997;25:3760–3766. doi: 10.1093/nar/25.19.3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Murray JB, Seyhan AA, Walter NG, Burke JM, Scott WG. The hammerhead, hairpin and VS ribozymes are catalytically proficient in monovalent cations alone. Chem Biol. 1998;5:587–595. doi: 10.1016/s1074-5521(98)90116-8. [DOI] [PubMed] [Google Scholar]
- 5.Curtis EA, Bartel DP. The hammerhead cleavage reaction in monovalent cations. RNA. 2001;7:546–552. doi: 10.1017/s1355838201002357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.O’Rear JL, Wang S, Feig AL, Beigelman L, Uhlenbeck OC, Herschlag D. Comparison of the hammerhead cleavage reactions stimulated by monovalent and divalent cations. RNA. 2001;7:537–545. doi: 10.1017/s1355838201002461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chowrira BM, Berzal-Herranz A, Burke JM. Ionic requirements for RNA binding, cleavage, and ligation by the hairpin ribozyme. Biochemistry. 1993;32:1088–1095. doi: 10.1021/bi00055a014. [DOI] [PubMed] [Google Scholar]
- 8.Walter NG, Burke JM, Millar DP. Stability of hairpin ribozyme tertiary structure is governed by the interdomain junction. Nat Struct Biol. 1999;6:544–549. doi: 10.1038/9316. [DOI] [PubMed] [Google Scholar]
- 9.Walter NG, Hampel KJ, Brown KM, Burke JM. Tertiary structure formation in the hairpin ribozyme monitored by fluorescence resonance energy transfer. EMBO J. 1998;17:2378–2391. doi: 10.1093/emboj/17.8.2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hampel KJ, Walter NG, Burke JM. The solvent-protected core of the hairpin ribozyme-substrate complex. Biochemistry. 1998;37:14672–14682. doi: 10.1021/bi981083n. [DOI] [PubMed] [Google Scholar]
- 11.Pinard R, Hampel KJ, Heckman JE, Lambert D, Chan PA, Major F, Burke JM. Functional involvement of G8 in the hairpin ribozyme cleavage mechanism. EMBO J. 2001;20:6434–6442. doi: 10.1093/emboj/20.22.6434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cai Z, Tinoco I., Jr Solution structure of loop A from the hairpin ribozyme from tobacco ringspot virus satellite. Biochemistry. 1996;35:6026–6036. doi: 10.1021/bi952985g. [DOI] [PubMed] [Google Scholar]
- 13.Butcher SE, Allain FH, Feigon J. Solution structure of the loop B domain from the hairpin ribozyme. Nat Struct Biol. 1999;6:212–216. doi: 10.1038/6651. [DOI] [PubMed] [Google Scholar]
- 14.Rupert PB, Massey AP, Sigurdsson ST, Ferre-D’Amare AR. Transition state stabilization by a catalytic RNA. Science. 2002;298:1421–1424. doi: 10.1126/science.1076093. [DOI] [PubMed] [Google Scholar]
- 15.Pinard R, Heckman JE, Burke JM. Alignment of the two domains of the hairpin ribozyme-substrate complex defined by interdomain photoaffinity crosslinking. J Mol Biol. 1999;287:239–251. doi: 10.1006/jmbi.1999.2626. [DOI] [PubMed] [Google Scholar]
- 16.Butcher SE, Burke JM. A photo-cross-linkable tertiary structure motif found in functionally distinct RNA molecules is essential for catalytic function of the hairpin ribozyme. Biochemistry. 1994;33:992–999. doi: 10.1021/bi00170a018. [DOI] [PubMed] [Google Scholar]
- 17.Hampel KJ, Burke JM. A conformational change in the "loop E-like" motif of the hairpin ribozyme is coincidental with domain docking and is essential for catalysis. Biochemistry. 2001;40:3723–3729. doi: 10.1021/bi0028385. [DOI] [PubMed] [Google Scholar]
- 18.Pinard R, Lambert D, Heckman JE, Esteban JA, Gundlach CWt, Hampel KJ, Glick GD, Walter NG, Major F, Burke JM. The hairpin ribozyme substrate binding-domain: a highly constrained D- shaped conformation. J Mol Biol. 2001;307:51–65. doi: 10.1006/jmbi.2000.4472. [DOI] [PubMed] [Google Scholar]
- 19.Diamond JM, Turner DH, Mathews DH. Thermodynamics of three-way multibranch loops in RNA. Biochemistry. 2001;40:6971–6981. doi: 10.1021/bi0029548. [DOI] [PubMed] [Google Scholar]
- 20.Hoogstraten CG, Wank JR, Pardi A. Active site dynamics in the lead-dependent ribozyme. Biochemistry. 2000;39:9951–9958. doi: 10.1021/bi0007627. [DOI] [PubMed] [Google Scholar]
- 21.Wedekind JE, McKay DB. Crystal structure of the leadzyme at 1.8 A resolution: metal ion binding and the implications for catalytic mechanism and allo site ion regulation. Biochemistry. 2003;42:9554–9563. doi: 10.1021/bi0300783. [DOI] [PubMed] [Google Scholar]
- 22.Heckman JE, Lambert D, Burke JM. Photocrosslinking detects a compact, active structure of the hammerhead ribozyme. Biochemistry. 2005;44:4148–4156. doi: 10.1021/bi047858b. [DOI] [PubMed] [Google Scholar]
- 23.Wang L, Ruffner DE. An ultraviolet crosslink in the hammerhead ribozyme dependent on 2- thiocytidine or 4-thiouridine substitution. Nucleic Acids Res. 1997;25:4355–4361. doi: 10.1093/nar/25.21.4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bravo C, Woisard A, Fourrey JL, Laugaa P, Favre A. A Y form of hammerhead ribozyme trapped by photo-cross-links retains full cleavage activity. Biochimie. 1999;81:201–212. doi: 10.1016/s0300-9084(99)80053-4. [DOI] [PubMed] [Google Scholar]
- 25.Hiley SL, Sood VD, Fan J, Collins RA. 4-thio-U cross-linking identifies the active site of the VS ribozyme. EMBO J. 2002;21:4691–4698. doi: 10.1093/emboj/cdf462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ouellet J, Perreault JP. Cross-linking experiments reveal the presence of novel structural features between a hepatitis delta virus ribozyme and its substrate. RNA. 2004;10:1059–1072. doi: 10.1261/rna.7230604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Major F, Turcotte M, Gautheret D, Lapalme G, Fillion E, Cedergren R. The combination of symbolic and numerical computation for three-dimensional modeling of RNA. Science. 1991;253:1255–1260. doi: 10.1126/science.1716375. [DOI] [PubMed] [Google Scholar]
- 28.Major F, Griffey R. Computational methods for RNA structure determination. Curr Opin Struct Biol. 2001;11:282–286. doi: 10.1016/s0959-440x(00)00203-7. [DOI] [PubMed] [Google Scholar]
- 29.Pinard R, Lambert D, Walter NG, Heckman JE, Major F, Burke JM. Structural basis for the guanosine requirement of the hairpin ribozyme. Biochemistry. 1999;38:16035–16039. doi: 10.1021/bi992024s. [DOI] [PubMed] [Google Scholar]
- 30.Pearlman D, Case D, Caldwell J, Ross W, Cheatman T. Computer Physics Communications. University of California; San Francisco: 1995. pp. 1–41. [Google Scholar]
- 31.Kraulis PJ. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]
- 32.Butcher SE, Heckman JE, Burke JM. Reconstitution of hairpin ribozyme activity following separation of functional domains. J Biol Chem. 1995;270:29648–29651. doi: 10.1074/jbc.270.50.29648. [DOI] [PubMed] [Google Scholar]
- 33.Esteban JA, Banerjee AR, Burke JM. Kinetic mechanism of the hairpin ribozyme. Identification and characterization of two nonexchangeable conformations. J Biol Chem. 1997;272:13629–13639. doi: 10.1074/jbc.272.21.13629. [DOI] [PubMed] [Google Scholar]
- 34.Feldstein PA, Bruening G. Catalytically active geometry in the reversible circularization of 'mini-monomer' RNAs derived from the complementary strand of tobacco ringspot virus satellite RNA. Nucleic Acids Res. 1993;21:1991–1998. doi: 10.1093/nar/21.8.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Juneau K, Podell E, Harrington DJ, Cech TR. Structural basis of the enhanced stability of a mutant ribozyme domain and a detailed view of RNA--solvent interactions. Structure (Camb) 2001;9:221–231. doi: 10.1016/s0969-2126(01)00579-2. [DOI] [PubMed] [Google Scholar]
- 36.Gessner RV, Quigley GJ, Wang AH, van der Marel GA, van Boom JH, Rich A. Structural basis for stabilization of Z-DNA by cobalt hexaammine and magnesium cations. Biochemistry. 1985;24:237–240. doi: 10.1021/bi00323a001. [DOI] [PubMed] [Google Scholar]
- 37.Komatsu Y, Kumagai I, Ohtsuka E. Investigation of the recognition of an important uridine in an internal loop of a hairpin ribozyme prepared using post-synthetically modified oligonucleotides. Nucleic Acids Res. 1999;27:4314–4323. doi: 10.1093/nar/27.22.4314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Klostermeier D, Millar DP. Energetics of hydrogen bond networks in RNA: hydrogen bonds surrounding G+1 and U42 are the major determinants for the tertiary structure stability of the hairpin ribozyme. Biochemistry. 2002;41:14095–14102. doi: 10.1021/bi025551b. [DOI] [PubMed] [Google Scholar]
- 39.Walter NG, Chan PA, Hampel KJ, Millar DP, Burke JM. A base change in the catalytic core of the hairpin ribozyme perturbs function but not domain docking. Biochemistry. 2001;40:2580–2587. doi: 10.1021/bi001609f. [DOI] [PubMed] [Google Scholar]
- 40.Burke JM. Hairpin ribozyme: current status and future prospects. Biochem Soc Trans. 1996;24:608–615. doi: 10.1042/bst0240608. [DOI] [PubMed] [Google Scholar]
- 41.Zhuang X, Kim H, Pereira MJ, Babcock HP, Walter NG, Chu S. Correlating structural dynamics and function in single ribozyme molecules. Science. 2002;296:1473–1476. doi: 10.1126/science.1069013. [DOI] [PubMed] [Google Scholar]
- 42.Okumus B, Wilson TJ, Lilley DM, Ha T. Vesicle encapsulation studies reveal that single molecule ribozyme heterogeneities are intrinsic. Biophys J. 2004;87:2798–2806. doi: 10.1529/biophysj.104.045971. [DOI] [PMC free article] [PubMed] [Google Scholar]