Abstract
Quiescent nuclei from differentiated somatic cells can reacquire pluripotence, the capacity to replicate, and reinitiate a program of differentiation after transplantation into amphibian eggs. The replication of quiescent nuclei is recapitulated in extracts derived from activated Xenopus eggs; therefore, we have exploited this cell-free system to explore the mechanisms that regulate initiation of replication in nuclei from terminally differentiated Xenopus erythrocytes. We find that these nuclei lack many, if not all, pre-replication complex (pre-RC) proteins. Pre-RC proteins from the extract form a stable association with the chromatin of permeable nuclei, which replicate in this system, but not with the chromatin of intact nuclei, which do not replicate, even though these proteins cross an intact nuclear envelope. During extract incubation, the linker histones H1 and H10 are removed from erythrocyte chromatin by nucleoplasmin. We show that H1 removal facilitates the replication of permeable nuclei by increasing the frequency of initiation most likely by promoting the assembly of pre-RCs on chromatin. These data indicate that initiation in erythrocyte nuclei requires the acquisition of pre-RC proteins from egg extract and that pre-RC assembly requires the loss of nuclear envelope integrity and is facilitated by the removal of linker histone H1 from chromatin.
INTRODUCTION
During development of the vertebrate organism, a majority of cells eventually exit the cell cycle early in G1 phase and enter an “out-of-cycle” or quiescent state often referred to as G0 (Pardee, 1989). Exit from the cell cycle is reversible in certain cell types; however, in others, such as terminally differentiated frog and avian erythrocytes, it is not (Leonard et al., 1982). The mechanisms underlying this loss of proliferative capacity have not been clearly defined; however, it seems likely that many of the changes that accompany differentiation in these cells collectively contribute to this irreversible arrest. In the adult animal, major transitions in chromatin composition and structure occur during erythrocyte differentiation, and these changes have been implicated in the generation and/or maintenance of the quiescent state. In the frog, for example, histone H10 gradually accumulates during differentiation, leading to a high content of linker histones on the chromatin (Allan et al., 1981; Dimitrov and Wolffe, 1996), and it has been suggested that this high linker histone content promotes the hypercondensation and inactivation of erythrocyte chromatin (Thomas and Maclean, 1975; Allan et al., 1981; Wolffe, 1989). Indeed, this idea is supported by studies that demonstrate that the overexpression of chicken histone H5 (H10) in proliferative somatic cells results in chromatin compaction and the inhibition of DNA replication and transcription (Sun et al., 1989; Aubert et al., 1991). Importantly, the overall level of H1 in these overexpressing cells was similar to that observed in terminally differentiated erythrocytes.
Although the proliferative arrest of nucleate Xenopus laevis erythrocytes is irreversible in vivo, reactivation of DNA replication and transcription does occur when isolated erythrocyte nuclei are introduced into an activating environment such as enucleated Xenopus eggs (for review, see Gurdon, 1986). These reactivated nuclei resemble embryonic nuclei both structurally and functionally, becoming pluripotent for frog development (Gurdon and Uehlinger, 1966; Brun, 1978). The reactivation of mature erythrocyte nuclei has been recapitulated in vitro using Xenopus egg extracts (Coppock et al., 1989; Wolffe, 1989), and in this system, quiescent nuclei decondense (Leno and Laskey, 1991; Blank et al., 1992; Leno and Munshi, 1997), regain transcriptional competence (Wolffe, 1989; Dimitrov and Wolffe, 1996), and initiate DNA replication (Coppock et al., 1989; Leno and Laskey, 1991; Blank et al., 1992; Wangh et al., 1995; Leno and Munshi, 1997).
A general requirement for initiation of cellular DNA replication in eukaryotes is the coordinated assembly of a pre-replication complex (pre-RC) at origin sites. In Xenopus egg extracts, pre-RC assembly on sperm chromatin and even purified DNA occurs before nuclear envelope assembly and involves the sequential binding of origin recognition complex (ORC) proteins, Cdc6, and minichromosome maintenance (MCM) proteins to DNA (see review by Romanowski and Madine, 1996, 1997; Walter et al., 1998). S-phase–promoting factors, such as cdk2/cyclin E, subsequently accumulate within the intact nucleus and are thought to trigger initiation at sites where pre-RCs are assembled (Jackson et al., 1995; Hua et al., 1997; Walter et al., 1998). Initiation of replication is accompanied by the loss of Cdc6 (Coleman et al., 1996; Hua and Newport, 1998) and MCM proteins from chromatin (Chong et al., 1995; Kubota et al., 1995; Madine et al., 1995a,b; Romanowski et al., 1996a), whereas ORC remains bound to DNA until chromosome condensation at mitosis (Carpenter et al., 1996; Romanowski et al., 1996b; Rowles et al., 1996). It appears that reinitiation of replication is blocked during S and G2 phases by the nuclear envelope, which prevents the reassociation of MCM proteins with chromatin until passage through mitosis and entry into the next cell cycle (Madine et al., 1995b).
The molecular mechanisms regulating the reactivation of replication in quiescent nuclei have not been determined. However, several observations indicate that nuclear envelope integrity plays an important role. Indeed, nuclei isolated from contact-inhibited cultured cells (Leno and Munshi, 1994; Fang and Benbow, 1996), and avian erythrocytes (Leno and Munshi, 1997) require nuclear envelope permeabilization for initiation in egg extract. Although the molecular basis for this requirement is unknown, the recent demonstration that restoration of replication competence in nuclei from quiescent fibroblasts requires an activity from egg extract (Munshi and Leno, 1998) coupled with the observation that the level of Mcm3, an essential member of the pre-RC, is reduced in quiescent cells both in vivo (Musahl et al., 1998) and in vitro (Stoeber et al., 1998) raises the interesting possibility that permeabilization of quiescent nuclei may be required for the reassembly of pre-RCs on chromatin, much like it is in G2-phase nuclei (Madine et al., 1995b). Thus, nuclear envelope integrity could also play a role in regulating initiation within quiescent cell nuclei by modulating pre-RC assembly on chromatin.
Accompanying the reactivation of replication in erythrocyte nuclei by egg extract is the replacement of somatic H1 histones with the embryonic linker histone B4 and HMG1, another chromosomal protein found in early embryonic chromatin (Blank et al., 1992; Dimitrov and Wolffe, 1996). These transitions in chromatin composition are mediated by the molecular chaperone nucleoplasmin (NPL) and play an essential role in the reacquisition of transcriptional competence in these nuclei (Dimitrov and Wolffe, 1996). What role, if any, these changes in chromatin structure play in the reactivation of DNA replication is not clear. However, recent work showing that the assembly of somatic H1 on embryonic chromatin reduces the frequency of initiation in egg extract by limiting pre-RC assembly (Lu et al., 1998) raises the interesting possibility that efficient initiation also requires the removal of H1 from erythrocyte chromatin.
In this report we have used Xenopus egg extract to investigate the mechanisms regulating the reactivation of replication in nuclei from terminally differentiated Xenopus erythrocytes. We find that these nuclei lack essential components of the pre-RC, including XORC, XCdc6, and XMCM proteins. Pre-RC proteins from the extract form a stable association with the chromatin of permeable nuclei, which replicate in this system, but not with the chromatin of intact nuclei, which do not replicate, even though these proteins are able to cross an intact nuclear envelope. Thus, an intact nuclear envelope prevents initiation in quiescent nuclei, at least in part, by preventing the assembly of pre-RCs on chromatin. Erythrocyte nuclei contain histone H1 and H10 that are removed from the chromatin by the molecular chaperone NPL during reactivation in the extract. Immunodepletion of NPL from the extract prevents the removal of H1 from chromatin, limits pre-RC assembly, and reduces the frequency of initiation within permeable nuclei, all of which are restored by readdition of NPL to the depleted extract. Furthermore, restoring the overall H1 content on erythrocyte chromatin in control (NPL-containing) extract inhibits replication to the same extent as that observed in NPL-depleted extract. Thus, a high level of somatic H1 on erythrocyte chromatin, whether the result of NPL depletion or the addition of exogenous H1 to NPL-containing extract, inhibits replication to a similar extent. Moreover, intact G1-phase tissue culture nuclei, which contain fully assembled pre-RCs, replicate to very similar levels in mock-depleted and NPL-depleted extracts, suggesting that once pre-RC assembly is complete, removal of H1 may no longer be required for replication in the extract. Taken together, these data indicate that loss of nuclear envelope integrity and the removal of somatic linker H1 from erythrocyte chromatin are required for the acquisition of essential pre-RC proteins from the extract and the reactivation of DNA replication in this system.
MATERIALS AND METHODS
Preparation of Xenopus Egg Extract and Xenopus Erythrocyte Nuclei
Interphase extracts were prepared from activated eggs of X. laevis as previously described (Lu et al., 1997). Blood (2.7 ml), obtained from anesthetized X. laevis adults by cardiac puncture, was collected in a tube containing 0.3 ml of ice-cold anticoagulant solution (0.14 M NaCl, 0.1 M trisodium citrate, 10 mM Tris-HCl, pH 7.4) and then diluted in 30 ml of ice-cold buffer C (0.14 M NaCl, 15 mM trisodium citrate, 0.25 mM PMSF, 2.5 μg/ml leupeptin, pepstatin, and aprotinin, 10 mM Tris-HCl, pH 7.4). Erythrocytes were sedimented by centrifugation at 1500 rpm for 5 min at 0°C in a Jouan (Winchester, VA) CR4–22 swing-out centrifuge and rinsed twice in buffer C. The “buffy coat” containing nonerythroid cells was removed after each rinse. More than 99% of the cells in the final sediment were mature erythrocytes.
For permeabilization of cells with streptolysin-O (SLO), freshly isolated erythrocytes were resuspended in 1,4-piperazinediethanesulfonic acid (PIPES) buffer (50 mM KCl, 2 mM EGTA, 5 mM MgCl2, 1 mM DTT, 1 μg/ml leupeptin, pepstatin, and aprotinin, 50 mM PIPES-KOH, pH 7.0) to a final concentration of 4 × 105 cells/ml. An equal volume of PIPES buffer containing 1.6 IU/ml SLO (Murex Diagnostics, Norcross, GA) was added, and the cells were incubated on ice for 10 min with gentle inversion of the tubes every minute. The cells were then sedimented by centrifugation as described above, rinsed twice in PIPES buffer to remove unbound SLO, and finally resuspended at room temperature in PIPES buffer. The permeable cells (intact nuclei) were counted in a hemacytometer.
Permeable nuclei were prepared using lysophosphatidylcholine (LPC). Sedimented erythrocytes were resuspended in 5 ml of buffer P (60 mM KCl, 15 mM NaCl, 340 mM sucrose, 15 mM β-mercaptoethanol, 0.5 mM spermidine, 0.15 mM spermine, 2.5 μg/ml leupeptin, pepstatin, and aprotinin, 15 mM HEPES-KOH, pH 7.5). An equal volume of buffer P containing 2 mg/ml LPC (Sigma, St. Louis, MO) was added, and the cells were incubated for 20 min at room temperature with gentle inversion of the tubes every minute. Permeabilization was stopped by adding 5 ml of ice-cold buffer P containing 3% BSA, and the resultant permeable nuclei were sedimented by centrifugation at 2750 rpm for 10 min at 0°C in a Jouan CR4–22 centrifuge. The nuclei were then rinsed in buffer P, sedimented, resuspended, and counted in a hemacytometer. Both intact and permeable nuclei were diluted with buffer to a final concentration equivalent to 1 μg of DNA/μl, assuming a DNA mass of 6.3 pg per diploid nucleus (Dawid, 1965). The permeability of plasma and nuclear membranes was determined by incubating an aliquot of SLO- or LPC-treated cells with affinity-purified TRITC-labeled immunoglobulin G (IgG) for 5 min. The percentage of nuclei excluding the labeled IgG was determined by fluorescence microscopy (Leno and Munshi, 1994).
Cell Culture and Synchronization
BALB/c 3T3 cells (CCL 163; American Type Culture Collection, Manassas, VA) were grown in Dulbecco’s modified Eagle’s medium supplemented with 10% calf serum, 100 U/ml penicillin G, 100 μg/ml streptomycin sulfate, and 0.25 μg/ml amphotericin B (all from Life Technologies, Gaithersburg, MD). Cells were synchronized in G1 phase by release from a nocodazole-induced mitotic arrest. Nocodazole was added to the culture medium at 0.04 μg/ml for 3 h; mitotic cells (mitotic index > 95%) were collected on ice, and nocodazole was washed out with two rinses of ice-cold complete growth medium. Cells were resuspended and replated for 3 h at which time ∼85% of the cells were in G1 as determined by flow cytometry. S-phase contaminants in these cultures were identified by pulsing with 100 μM bromodeoxyuridine (BrdU) for 15 min before nuclear isolation.
In Vitro DNA Replication
Freshly isolated intact or permeable erythrocyte nuclei or intact G1-phase mouse 3T3 cell nuclei (Munshi and Leno, 1998) were incubated at 3 ng of DNA/μl of extract supplemented with an energy-regenerating system (60 mM creatine phosphate, 150 μg/ml creatine phosphokinase), 100 μg/ml cycloheximide, 2 mM ATP, and either 100 μCi/ml [α-32P]dATP (800 Ci/mmol; New England Nuclear, Boston, MA) or 20 μM 5-biotin-16-dUTP (Boehringer Mannheim, Indianapolis, IN). dNTPs were added to a final concentration of 50 μM to readjust pool sizes after dilution (Cox and Leno, 1990). Mouse somatic linker histone H1c was prepared as described (Lu et al., 1997) and, where indicated, added at 5.68 μM to egg extract. An equivalent volume of water was added in control reactions. All incubations were performed at 22°C. Incorporation of [α-32P]dATP or biotinylated dUTP was determined as previously described (Lu et al., 1997, 1998). Density substitution experiments were performed essentially as described (Lu et al., 1998), except that reactions were diluted with ice-cold buffer A, and the nuclei were sedimented before DNA extraction.
Immunofluorescence Microscopy and Western Blotting
The detection of individual pre-RC proteins by immunofluorescence microscopy was carried out as described (Lu et al., 1998) with modifications as specified in each experiment. For Western blotting of chromatin-associated pre-RC proteins, erythrocyte nuclei, with or without extract incubation, were diluted with HE′ buffer (50 mM HEPES-KOH, pH 7.6, 50 mM KCl, 5 mM MgCl2, 2 mM β-mercaptoethanol, 0.5 mM spermine, 0.15 mM spermidine, 1 μg/ml leupeptin, pepstatin, and aprotinin, 0.1% Triton X-100) and sedimented through 15% sucrose in NIBS buffer (50 mM HEPES-KOH, pH 7.6, 50 mM KCl, 5 mM MgCl2, 2 mM β-mercaptoethanol, 0.5 mM spermine, 0.15 mM spermidine) at 2750 rpm for 10 min at 0°C in a Jouan CR4–22 centrifuge. Proteins from each sample were separated on 7.5% SDS-PAGE gels and transferred to a nitrocellulose membrane (Micron Separations, Westboro, MA) by electroblotting. Membranes were blocked in TTBS-M (25 mM Tris-HCl, pH 8.0, 137 mM NaCl, 2.7 mM KCl, 10% dried milk, 0.5% Tween-20) and incubated for 1 h in primary antibody. Membranes were then incubated in goat anti-rabbit HRP-conjugated secondary antibody in TTBS-M. Blots were developed with enhanced chemiluminescence using the ECL immunoblotting kit (Amersham Pharmacia Biotech, Uppsala, Sweden).
To determine the levels of NPL in egg extract by Western blotting, 1 μl of extract was diluted with 4 μl of extraction buffer (50 mM HEPES-KOH, pH 7.6, 50 mM KCl, 5 mM MgCl2, 2 mM β-mercaptoethanol) and then with 5 μl of 2× SDS-PAGE sample buffer (125 mM Tris-HCl, pH 6.8, 10% SDS, 50% glycerol, 100 mM β-mercaptoethanol). Samples were run on 15% polyacrylamide gels and blotted onto nitrocellulose membranes. Membranes were processed as described above, except that PA3C5 hybridoma culture supernatant, containing 10% dried milk, was used directly as the source of primary antibody.
Purification of NPL and Monoclonal Antibody Production and Purification
NPL was purified from Xenopus eggs essentially as described by Dingwall et al. (1982), and a molar extinction coefficient of 13,980 M/cm at 280 nm was used to determine the final concentration of protein (Pace et al., 1995). Mouse anti-NPL monoclonal antibody was derived from the hybridoma clone PA3C5 (Dilworth et al., 1987). Production and purification of antibody were as previously described (Philpott et al., 1991). An extinction coefficient of 1.35 g/cm at 280 nm (IgG) was used to determine the protein concentration (Harlow and Lane, 1988).
Immunodepletion of NPL
Immunodepletion of NPL from egg extract was performed essentially as described (Philpott and Leno, 1992). Anti-NPL monoclonal antibody PA3C5 was incubated at 2.5 μg/μl with protein A-Sepharose beads (Amersham Pharmacia Biotech) in HEPES buffer for 30 min at room temperature. The beads were rinsed three times in extraction buffer to remove unbound antibody. Egg extract was thawed, supplemented with an energy-regenerating system and 100 μg/ml cycloheximide, mixed with a half-volume of antibody-coated beads, and incubated on ice for 25 min. Beads were then sedimented to the bottom of a pipette tip (Chong et al., 1997), and the flow-through extract was subjected to a second round of immunodepletion. Mock-depleted extract was prepared using HEPES buffer without PA3C5 antibody. For reconstituted samples, NPL was added to the depleted extract to a final concentration of 500 ng/μl, the physiological concentration in egg extracts (Philpott and Leno, 1992).
Isolation of Chromatin-bound Proteins
To isolate chromatin-bound basic proteins, samples were diluted with buffer A, and the nuclei were sedimented by centrifugation at 2000 rpm for 10 min at 0°C in a Jouan CR4–22 centrifuge. Basic proteins were extracted from chromatin by addition of HCl to a final concentration of 0.5 M, lyophilized, and analyzed by SDS-PAGE as previously described (Lu et al., 1997).
Alkaline Agarose Gel Electrophoresis
Alkaline agarose gel electrophoresis was performed essentially as described (Lu et al., 1998). Samples were run on a 1% Nusieve 3:1 gel (FMC Bioproducts, Rockland, ME) in alkaline running buffer (30 mM NaOH, 1 mM EDTA), fixed in 5% trichloroacetic acid solution, dried, and subjected to autoradiography.
RESULTS
An Intact Nuclear Envelope Prevents Replication of Xenopus Erythrocyte Nuclei in Egg Extract by Preventing the Assembly of Prereplication Complexes on Chromatin
In Xenopus egg extracts, permeabilization of the nuclear envelope is required for the initiation of replication in nuclei from quiescent cultured cells (Leno and Munshi, 1994; Fang and Benbow, 1996; Munshi and Leno, 1998) and from terminally differentiated chicken erythrocytes (Leno and Munshi, 1997). The molecular basis for this requirement is not clear. Our hypothesis is that quiescent nuclei are incompetent for replication because they lack functional pre-RCs and that envelope permeabilization is required for assembly of pre-RCs by egg extract. We have used Xenopus erythrocyte nuclei to test this hypothesis to exploit the availability of antibodies that recognize Xenopus pre-RC proteins (Madine et al., 1995a,b; Romanowski et al., 1996a,b; Lu et al., 1998). However, an essential first step in our approach was to confirm that loss of nuclear envelope integrity is also a requirement for initiation in erythrocyte nuclei from Xenopus.
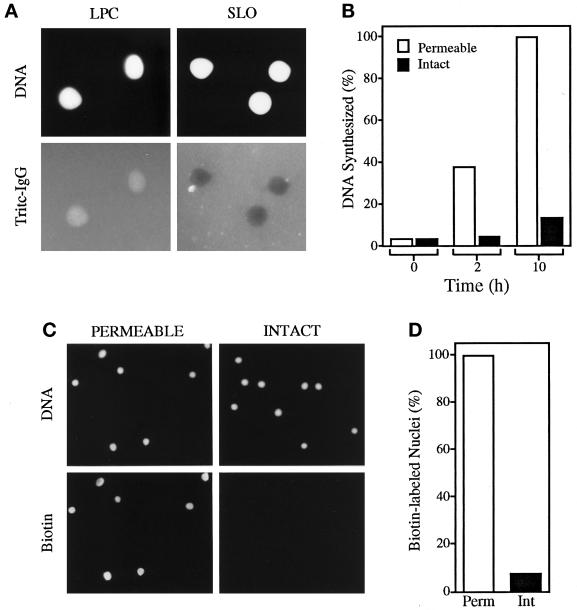
Nuclei were obtained by treating Xenopus erythrocytes with the bacterial exotoxin SLO or with LPC, and nuclear envelope integrity was determined by incubating treated cells with TRITC-labeled IgG (Leno et al., 1992; Leno and Munshi, 1997). IgG is excluded from nuclei with intact nuclear envelopes but not from nuclei with detergent-damaged (permeable) envelopes. We consistently found that >90% of SLO-prepared nuclei excluded IgG (Figure 1A, SLO), whereas >99% of the LPC-prepared nuclei did not (Figure 1A, LPC). To determine the replication competence of SLO (intact) and LPC (permeable) nuclei, we incubated each at 3 ng of DNA/μl of extract supplemented with [α-32P]dATP (Figure 1B) or biotinylated dUTP (Figure 1, C and D) for various periods as described in MATERIALS AND METHODS. As expected, permeable nuclei replicated to a much greater extent than intact nuclei in our time course experiments (Figure 1B). Furthermore, the limited replication within the intact sample was restricted to very few nuclei (<10%; Figure 1, C, INTACT, and D, Int), whereas virtually all permeable nuclei initiated replication under identical conditions (Figure 1, C, PERMEABLE and D, Perm). Density substitution experiments confirmed that incorporation of label was the result of a single round of semiconservative DNA replication (our unpublished observation). Thus, permeabilization of the nuclear envelope is required for replication of Xenopus erythrocyte nuclei by egg extract.
Figure 1.
Permeabilization of the nuclear envelope is required for replication of Xenopus erythrocyte nuclei by egg extract. (A) Nuclei were prepared by treating erythrocytes with LPC or SLO, and nuclear envelope integrity was determined by incubating nuclei with TRITC-labeled IgG (Tritc-IgG). Total DNA (DNA) was stained with Hoechst 33258. (B) Permeable (LPC) and intact (SLO) nuclei were incubated in egg extract, supplemented with [α-32P]dATP, for various times as indicated. DNA replication is expressed as a percentage of incorporated label in the permeable sample at 10 h. The mass of DNA synthesized in this sample was 0.89 ng/μl of extract, representing ∼30% of the input DNA. (C) Permeable and intact nuclei were incubated in extract supplemented with 20 μM biotinylated dUTP for 4 h. Nuclei were isolated and stained for total DNA (DNA) and with Texas Red-streptavidin to detect biotin-dUTP incorporation into nascent DNA (Biotin). A representative field of nuclei is shown. (D) Two hundred nuclei from each sample in C were examined for Texas Red fluorescence. The percentages of biotin-labeled nuclei are shown.
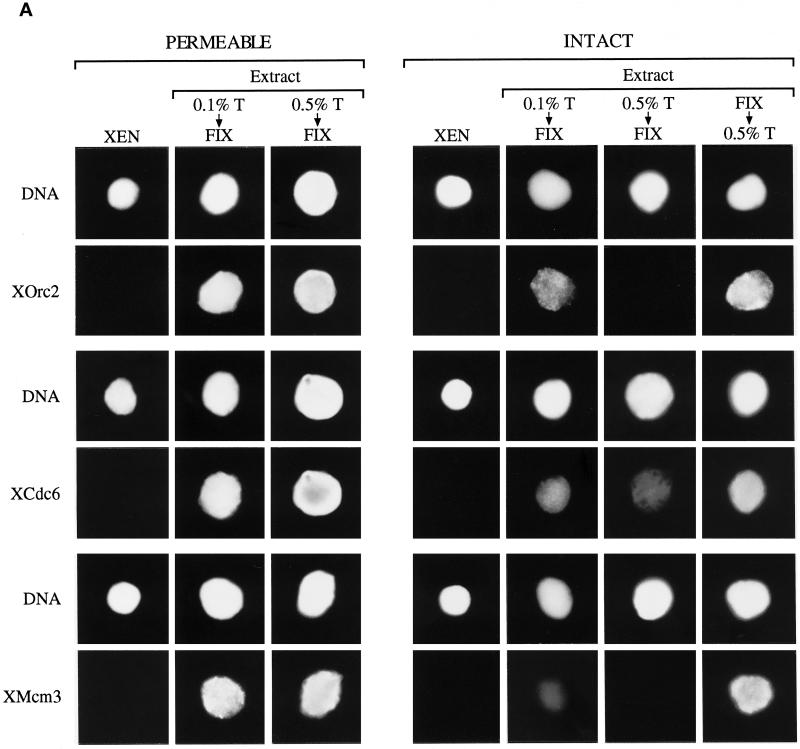
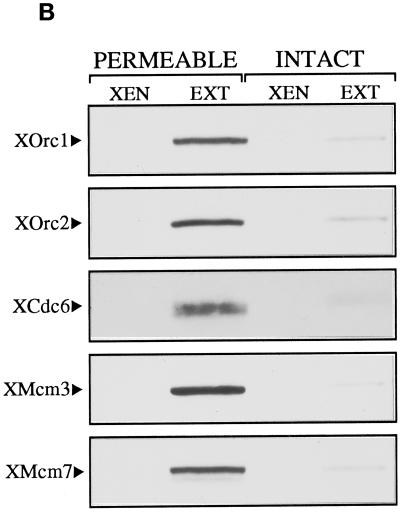
We next examined intact and permeable erythrocyte nuclei for the presence of pre-RC proteins both before and after extract incubation. Permeable nuclei were incubated in extract for 45 min, before initiation of replication (our unpublished observation), whereas intact nuclei, which do not initiate replication in the extract (Figure 1, C and D), were incubated for up to 4 h. Nuclei were sedimented onto coverslips, treated with different concentrations of Triton X-100, before or after fixation with paraformaldehyde, and probed with antibodies that recognize the Xenopus pre-RC proteins XOrc2, XCdc6, and XMcm3. The immunofluorescence results are shown in Figure 2A. Pre-RC proteins were undetectable within erythrocyte nuclei in the absence of extract incubation (XEN), irrespective of the method of nuclear isolation. However, after incubation in the extract, all three pre-RC proteins were found to stably associate with the chromatin from permeable nuclei (PERMEABLE, 0.5% T ↓ FIX) but not with the chromatin from intact nuclei (INTACT, 0.5% T ↓ FIX), even though all were able to cross an intact nuclear envelope (INTACT, FIX ↓ 0.5% T). Low levels of XOrc2 and XCdc6 were detected within intact nuclei after low-stringency detergent extraction (INTACT, 0.1% T ↓ FIX) but not after high-stringency extraction (INTACT, 0.5% T ↓ FIX). By contrast, an increase in stringency had no observable effect on the association of pre-RC proteins with chromatin from permeable nuclei (PERMEABLE, compare 0.1% T ↓ FIX with 0.5% T ↓ FIX). The differential association of pre-RC proteins with chromatin from intact and permeable nuclei was confirmed by Western blot (Figure 2B). In this case, nuclei incubated in egg extract (EXT) were treated under low-stringency conditions (0.1% Triton X-100) before analysis by PAGE. All five pre-RC proteins examined (XOrc1, XOrc2, XCdc6, XMcm3, and XMcm7) were stably associated with chromatin from permeable nuclei (PERMEABLE, EXT) but not with chromatin from intact nuclei (INTACT, EXT). Taken together, these data illustrate several important points. First, terminal differentiated erythrocyte nuclei lack essential components of the pre-RC. Second, reactivation of DNA replication by egg extract involves the assembly of pre-RC proteins on erythrocyte chromatin. Third, an intact nuclear envelope prevents initiation in these quiescent nuclei at least in part by preventing the assembly of pre-RCs on chromatin.
Figure 2.
An intact nuclear envelope prevents the assembly of pre-RCs on erythrocyte chromatin. (A) Permeable erythrocyte nuclei were incubated in egg extract for 45 min, diluted with buffer, sedimented on coverslips, and treated with either 0.1% or 0.5% Triton X-100 before fixation with paraformaldehyde. Intact nuclei were incubated in extract for 4 h, diluted, sedimented, and treated with 0.1 or 0.5% Triton X-100 before fixation or with 0.5% Triton X-100 after fixation. Incubated (Extract) and unincubated (XEN) nuclei were then probed with antibodies to the pre-RC XOrc2, XCdc6, and XMcm3. Primary antibodies were detected with fluorochrome-conjugated secondary antibody, which was visualized by fluorescence microscopy. Total DNA (DNA) was stained with Hoechst 33258. (B) Permeable and intact nuclei were incubated in extract as described in A. Incubated (EXT) and unincubated (XEN) nuclei were diluted with buffer containing 0.1% Triton X-100 and sedimented, and the chromatin proteins were separated by SDS-PAGE and transferred to nitrocellulose. Western blots were probed with antibodies to XOrc1, XOrc2, XCdc6, XMcm3, and XMcm7, incubated with enzyme-conjugated secondary antibody, and developed with enhanced chemiluminescence using the ECL immunoblotting kit.
An Intact Nuclear Envelope Reduces the Rate and Extent of H1 Removal and B4 Assembly on Erythrocyte Nuclei by Egg Extract
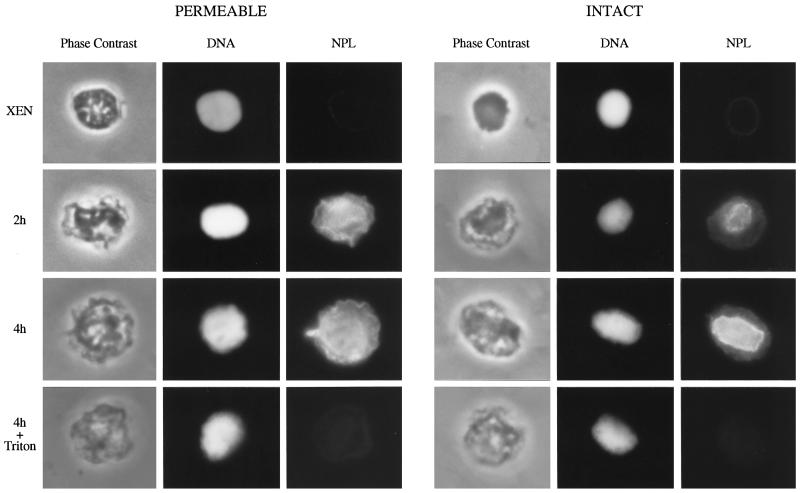
The replacement of somatic linker histones H1 and H10 with the embryonic linker histone B4 and HMG1 facilitates the acquisition of transcriptional competence in erythrocyte chromatin by egg extract (Dimitrov and Wolffe, 1996). Furthermore, the replacement of B4 with somatic H1 reduces the frequency of initiation of replication in egg extract by limiting the assembly of pre-RCs on embryonic chromatin (Lu et al., 1997, 1998). Therefore, if the removal of H1 from erythrocyte chromatin is required for pre-RC assembly, then an intact envelope could prevent this assembly by preventing the removal of H1. The selective removal of H1s from erythrocyte chromatin is mediated by the molecular chaperone NPL (Dimitrov and Wolffe, 1996). NPL accumulates within both intact and permeable erythrocyte nuclei after a 2-h incubation in egg extract (Figure 3, NPL, 2h). By 4 h (4h), no clear difference in the extent of import was observed between these nuclei. Treatment with Triton X-100 before fixation resulted in the loss of NPL from all nuclei (4h + Triton), consistent with its role as a soluble nucleoplasmic protein. Thus, NPL can cross an intact erythrocyte nuclear envelope, allowing us to determine whether H1 removal occurs within intact nuclei under conditions in which pre-RC assembly does not (Figure 2, A and B).
Figure 3.
NPL accumulates within intact and permeable erythrocyte nuclei incubated in egg extract. Intact (INTACT) and permeable (PERMEABLE) erythrocyte nuclei were incubated in egg extract for 2 h (2h) and 4 h (4h) and subsequently isolated, fixed, and labeled with anti-NPL monoclonal antibdy PA3C5 and with fluorescein-conjugated goat anti-mouse secondary antibody. Total DNA was stained with Hoechst 33258 (DNA). A typical nucleus from each sample shows the intranuclear accumulation of NPL (NPL). In parallel 4-h samples, nuclei were treated with Triton X-100 before fixation (4h + Triton), resulting in the complete loss of protein, consistent with NPL’s role as a soluble nucleoplasmic protein.
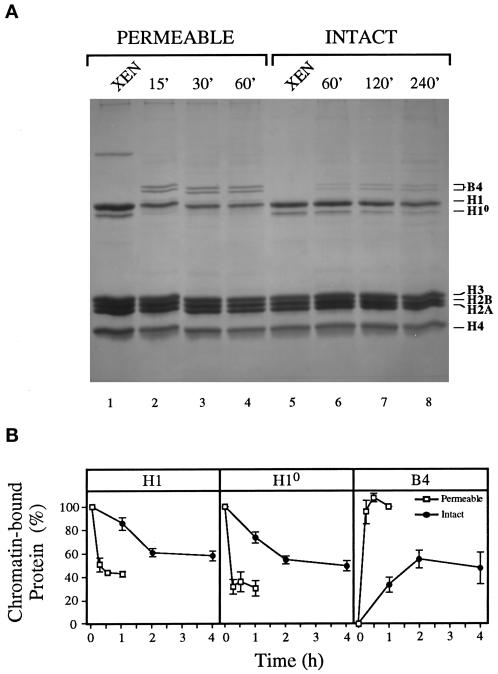
Intact and permeable nuclei were incubated in the extract for various times and isolated, and the acid-soluble nuclear proteins were separated by SDS-PAGE and stained with Coomassie blue (Figure 4A). The levels of H1, H10, and B4 proteins were quantitated by densitometry, normalized with the core histones in each sample, and a mean value was derived from three independent experiments in which three different egg extracts were used (Figure 4B). The replacement of H1s with B4 on erythrocyte chromatin within permeable nuclei reached a plateau by 15 min, whereas replacement within intact nuclei required >2 h to reach a plateau. Furthermore, the replacement within intact nuclei was less extensive than that observed within permeable nuclei for each of the linker histones, i.e., ∼40 versus ∼60% for loss of H1, ∼50 versus ∼70% for loss of H10, and ∼50 versus 100% for acquisition of B4. The amounts of H1 and H10 on unincubated XEN and the amount of B4 assembled on the chromatin of permeable nuclei after 60 min were designated as 100%. Virtually identical protein profiles were obtained when intact or permeable nuclei were treated with detergent after extract incubation but before acid extraction (our unpublished observation), demonstrating the stable association of these linker histones with chromatin. Thus, these data demonstrate that an intact nuclear envelope reduces both the rate and extent of H1 removal and B4 assembly in erythrocyte nuclei by egg extract. The protein observed directly above H1 in the permeable XEN sample (Figure 4A, XEN, PERMEABLE, lane 1) is most likely BSA, which is used to stop membrane permeabilization by LPC. It is important to note, however, that no experimental differences were observed between permeable nuclei with or without this protein.
Figure 4.
An intact nuclear envelope reduces the remodeling of erythrocyte chromatin by egg extract. (A) Permeable and intact erythrocyte nuclei were incubated in extract for various times as indicated. Incubated and unincubated (XEN) nuclei were then diluted and sedimented, and the chromatin-associated proteins were extracted with acid, resolved by SDS-PAGE, and visualized with Coomassie blue. The positions of the embryonic linker histone B4, somatic linker histones H1 and H10, and core histones H3, H2B, H2A, and H4 are indicated. (B) The chromatin-bound H1, H10, and B4 proteins were quantitated by densitometry and normalized with the core histones in each sample shown in A. Shown are the mean percentages of each chromatin-bound protein, along with the SEM, from three separate experiments in which three different extracts were used. The amounts of H1 and H10 on unincubated XEN and the amount of B4 assembled on the chromatin of permeable nuclei after 60 min were designated as 100% bound protein.
The Removal of Somatic H1 from Erythrocyte Chromatin Facilitates DNA Replication in Egg Extract
The extent to which H1s are removed from chromatin within intact erythrocyte nuclei (Figure 4) could account for the failure of pre-RC proteins to stably bind chromatin (Figure 2) and initiate DNA replication (Figure 1) if, for example, it were below a critical threshold required for the assembly of functional pre-RCs. If this idea is correct, then limiting the removal of H1s from chromatin within permeable nuclei should have the same effect. Immunodepletion of NPL from the extract prevents the removal of H1 from chromatin, whereas addition of purified NPL back to a depleted extract restores this activity (Dimitrov and Wolffe, 1996). Therefore, we used NPL-depleted egg extracts to determine the replication competence of permeable erythrocyte nuclei in the absence of H1 removal.
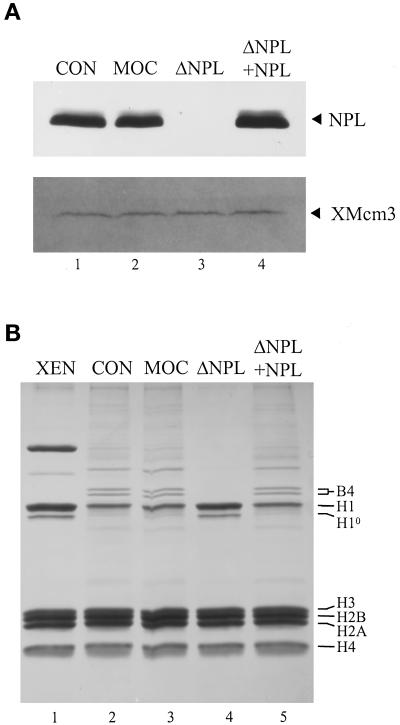
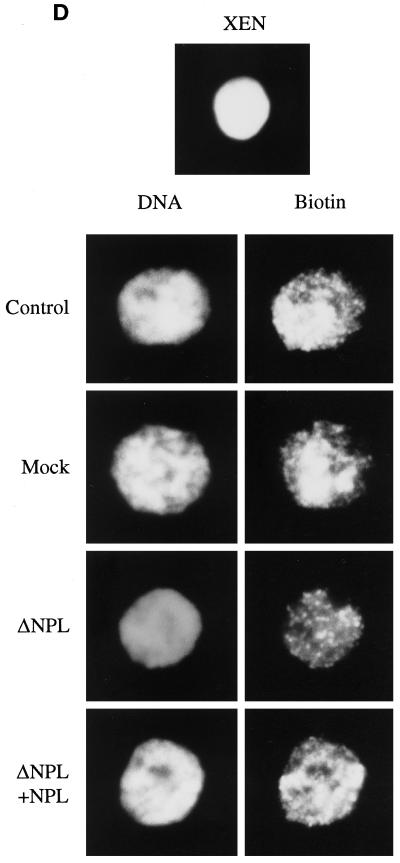
The monoclonal antibody PA3C5 was used to deplete virtually all of the NPL from our extracts (Figure 5A, compare lane 3 with lanes 1 and 2), consistent with earlier reports (Philpott et al., 1991; Philpott and Leno, 1992). The Western blot shown in Figure 5A was also probed with anti-XMcm3 to demonstrate equal loading among the samples and to show that at least one pre-RC protein is not codepleted with NPL (Figure 5A, ΔNPL, lane 3). Incubation of permeable erythrocyte nuclei in depleted extract prevented the loss of H1s from chromatin as well as the assembly of B4 (Figure 5B, compare lanes 2 and 3 with lane 4). Addition of purified NPL back to depleted extracts restored the levels of H1s to control and mock depletion levels (Figure 5B, compare lane 5 with lanes 2 and 3). Permeable erythrocyte nuclei were then incubated in undepleted “control” extract (CON), mock-depleted extract (MOC), NPL-depleted extract (ΔNPL), and depleted extract reconstituted with NPL to the physiological concentration (ΔNPL+NPL) and assayed for DNA replication by the incorporation of [α-32P]dATP (Figure 6A). Shown are the mean values from three separate experiments in which three different extracts were used. In the absence of NPL, replication was reduced nearly 50% relative to the control sample (Figure 6A, compare ΔNPL with CON). Furthermore, replication was restored to control levels in depleted extract reconstituted with NPL (compare ΔNPL+NPL with CON). Replication was not inhibited in mock-depleted extract (MOC). Interestingly, >95% of nuclei initiate replication in NPL-depleted extract as judged by the incorporation of biotin-dUTP into nascent DNA (Figure 6D). However, the intensity of streptavidin fluorescence within these nuclei was markedly reduced relative to nuclei incubated in control extract, mock-depleted extract, and depleted extract reconstituted with NPL, consistent with the data shown in Figure 6A.
Figure 5.
NPL mediates the removal of H1s from erythrocyte chromatin in egg extract. (A) NPL-depleted (ΔNPL) and mock-depleted (MOC) extracts were prepared as described in MATERIALS AND METHODS. Purified NPL was added to depleted extract at a final concentration of 500 ng/μl (ΔNPL+NPL), the concentration of NPL in our extracts. Control extract (CON) was stored on ice during the depletion procedure. Proteins from 1 μl of each extract were separated by SDS-PAGE and transferred to nitrocellulose. Shown is a Western blot probed with the anti-NPL monoclonal antibody PA3C5 and with anti-XMcm3 polyclonal antibody. (B) Permeable erythrocyte nuclei were incubated in control (CON), mock-depleted (MOC), NPL-depleted (ΔNPL), or depleted extract reconstituted with NPL (ΔNPL+NPL) for 15 min. Incubated and unincubated (XEN) chromatin samples were sedimented and rinsed, and the chromatin-bound basic proteins were extracted with acid, resolved by SDS-PAGE, and visualized with Coomassie blue. The positions of the embryonic linker histone B4, somatic linker histones H1 and H10, and core histones H3, H2B, H2A, and H4 are indicated.
Figure 6.
Immunodepletion of NPL from egg extract inhibits initiation but not elongation in erythrocyte nuclei. (A) Permeable erythrocyte nuclei were incubated in control (CON), mock-depleted (MOC), NPL-depleted (ΔNPL), or depleted extract reconstituted with NPL (ΔNPL+NPL) containing [α-32P]-dATP for 8 h. The samples were processed as described in MATERIALS AND METHODS. DNA replication is expressed as a percentage of the control sample that was designated as 100%. The data shown are mean values ± SE from three separate experiments in which three different extracts were used. (B) Permeable erythrocyte nuclei were incubated for 8 h in NPL-depleted extract supplemented with BrdUTP and [α-32P]dATP. DNA was purified from each sample and centrifuged to equilibrium in a cesium chloride gradient. The refractive index of every fifth fraction was determined. The radioactivity in each fraction was measured by liquid scintillation, and the counts per minute (cpm) are shown. The expected densities of heavy/light DNA (HL, 1.75 g/ml) and heavy/heavy DNA (HH, 1.79 g/ml) are indicated. (C) Permeable erythrocyte nuclei were incubated for 8 h in mock-depleted extract (MOC), NPL-depleted extract (ΔNPL), or mock-depleted extract supplemented with aphidicolin at 20 μg/ml (MOC+ APH), each containing [α-32P]dATP. DNA was precipitated and separated on a 1% agarose gel under alkaline denaturing conditions. An autoradiogram of the nascent DNA is shown. (D) Permeable erythrocyte nuclei were incubated in control (CON), mock-depleted (MOC), NPL-depleted (ΔNPL), or depleted extract reconstituted with NPL (ΔNPL+NPL) containing 20 μM biotinylated dUTP for 8 h. Bulk DNA was stained with Hoechst 33258 (DNA), and nascent DNA was labeled with fluorescein-conjugated streptavidin (Biotin). An unincubated nucleus is also shown (XEN). The samples were processed as described in MATERIALS AND METHODS.
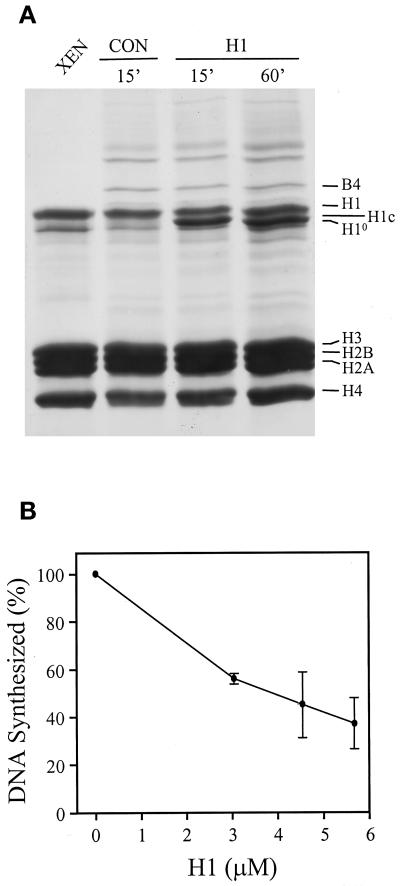
The immunodepletion data shown in Figures 5 and 6A demonstrate two important points. First, NPL is required for the removal of somatic H1 from erythrocyte chromatin and for the deposition of the cleavage stage linker histone B4. Second, NPL facilitates the replication of erythrocyte nuclei in egg extract. To investigate whether NPL facilitates replication by removing somatic H1 from chromatin, we added increasing concentrations of purified H1 to NPL-containing (control) extract and assayed the extent of inhibiton of replication at each concentration (Figure 7). We found that addition of H1 to a final concentration of 5.68 μM restored the overall linker histone content on chromatin to the level observed in unincubated erythrocyte nuclei (Figure 7A, compare H1 with XEN). Chromatin-bound H1 levels were determined by densitometry and normalized to the core histones in each sample. Purified somatic H1 (mouse H1c) migrates between Xenopus H1 and H10 in our SDS-PAGE gels. The association of H1 with erythrocyte chromatin inhibited replication in control extract in a dose-dependent manner (Figure 7B). Thus, a high level of somatic H1 on erythrocyte chromatin, whether the result of NPL depletion (Figure 5B) or the addition of exogenous H1 to NPL-containing (control) extract (Figure 7A), results in the inhibition of replication (compare Figure 6A, ΔNPL, with Figure 7B, 5.68 μM H1). Taken together, these data argue that NPL facilitates replication in egg extract by removing somatic H1 from erythrocyte chromatin.
Figure 7.
Assembly of somatic H1 on erythrocyte chromatin inhibits replication in extracts containing NPL. (A) Permeable erythrocyte nuclei were incubated (140 ng of DNA/μl of extract) in extract without (CON) or with histone H1c (H1; 5.68 μM) for 15 or 60 min as indicated. The chromatin-associated proteins from unincubated nuclei (XEN) and from incubated nuclei were isolated and analyzed as described in Figure 4. The positions of the embryonic linker histone B4, somatic linker histones H1 and H10, somatic mouse linker histone H1c, and core histones, H3, H2B, H2A, and H4 are indicated. Protein levels were quantitated by densitometry and normalized to the core histones in each sample. (B) Permeable erythrocyte nuclei were incubated for 8 h in extract without (0 μM H1) or with increasing concentrations of H1c (3.41, 4.54, and 5.68 μM H1). The samples were processed as described in MATERIALS AND METHODS. DNA replication is expressed as a percentage of the control sample that was designated as 100%.
Histone H1 on Erythrocyte Chromatin Reduces Pre-RC Assembly and the Frequency of Initiation in Egg Extract
The data presented so far indicate that removal of somatic H1s from chromatin facilitates the replication of erythrocyte nuclei in egg extract (Figures 4, 5, 6, A and D, and 7). In theory, H1 could inhibit replication in at least two ways: first, by reducing the number of active replication forks, i.e., the frequency of initiation; or second, by preventing fork movement, i.e., elongation. To distinguish between these possibilities, we first incubated permeable nuclei in NPL-depleted extract supplemented with BrdUTP and [α-32P]dATP for 8 h and separated the nascent DNA by centrifugation to equilibrium in a cesium chloride gradient. A typical density substitution profile is shown in Figure 6B. A single peak of radioactivity was detected at a density of ∼1.75 g/ml (heavy/light DNA, HL), demonstrating that erythrocyte nuclei undergo a single round of semiconservative DNA replication in NPL-depleted extract and ruling out extensive DNA repair. These data also argue against partial strand synthesis, which would resolve at densities between hemisubstituted (HL) and unsubstituted DNA (Krude et al., 1997; Mahbubani et al., 1997).
The relative size of nascent DNA strands produced in NPL-depleted extracts was determined by alkaline agarose gel electrophoresis (Figure 6C). Virtually all nascent DNA was found in a high-molecular-weight form (ΔNPL) indistinguishable from that observed in mock-depleted extract (MOC). In contrast, a range of lower-molecular-weight forms was observed when replication forks were arrested with aphidicolin shortly after initiation in mock-depleted extract (MOC+APH). Results essentially identical to those described in Figure 6C were obtained when the concentration of DNA was increased 10-fold, i.e., from 3 to 30 ng/μl of extract, demonstrating that even at high DNA concentration, replication elongation occurs in NPL-depleted extract (our unpublished observations). Taken together, these data demonstrate that elongation does occur in the presence of histone H1 and suggest that the inhibition of replication we observe in NPL-depleted extract is due to a reduction in the frequency of initiation.
The assembly of somatic histone H1 on embryonic chromatin has been shown to reduce the frequency of initiation in egg extract by limiting the assembly of pre-RCs on DNA (Lu et al., 1998). To determine whether pre-RC assembly on erythrocyte chromatin is limited in NPL-depleted extract, a Western blot, containing the chromatin-bound proteins from permeable erythrocyte nuclei incubated for 45 min in control (CON), mock-depleted (MOC), NPL-depleted (ΔNPL), and depleted extract reconstituted with NPL (ΔNPL+NPL), was probed with antibodies to several pre-RC proteins (Figure 8A). In each case, the level of the pre-RC protein in the NPL-depleted extract was reduced relative to that observed in control and mock-depleted samples. Protein levels were restored to control levels when NPL was added back to the depleted extract. Thus, NPL facilitates the assembly of pre-RCs on erythrocyte chromatin in egg extract. Given the essential role of pre-RCs in the establishment of replication competence, the data presented here raise the interesting possibility that removal of H1 from erythrocyte chromatin by NPL facilitates pre-RC assembly on DNA, thereby increasing the frequency of initiation and the overall extent of replication in egg extract.
Figure 8.
NPL facilitates pre-RC assembly on erythrocyte chromatin in egg extract. Permeable erythrocyte nuclei were incubated in control (CON), mock-depleted (MOC), NPL-depleted (ΔNPL), or depleted extract reconstituted with NPL (ΔNPL+NPL) for 45 min. Initiation events occurred within virtually all nuclei in both mock-depleted and NPL-depleted extracts by 60 min, as judged by the incorporation of biotin-dUTP into nascent DNA (our unpublished observation). Each sample was diluted with buffer containing 0.1% Triton X-100 and sedimented, and the chromatin proteins were separated by SDS-PAGE and transferred to nitro-cellulose. Western blots were probed with antibodies to XOrc1, XOrc2, XCdc6, XMcm3, and XMcm7, incubated with enzyme-conjugated secondary antibody, and developed with enhanced chemiluminescence using the ECL immunoblotting kit. (B) Intact G1-phase 3T3 nuclei were incubated in mock-depleted (MOC) or NPL-depleted (ΔNPL) extract for 6 h. The samples were pro-cessed as de-scribed in MATERIALS AND METHODS. DNA replication in the NPL-depleted sample represents a mean value derived from eight separate experiments in which two different extracts were used and is expressed as a percentage of the mock-depleted sample that was designated as 100%. The SEM for the depleted sample is shown.
Alternatively, NPL could facilitate replication of permeable erythrocyte nuclei by some other mechanism, such as a direct effect on replication proteins themselves. Our demonstration that the maintenance of somatic linker histone content on erythrocyte chromatin inhibits replication, even in the presence of NPL, argues against this possibility (Figure 7). Furthermore, intact G1-phase tissue culture nuclei, which contain fully assembled pre-RCs, replicate to similar levels in mock-depleted (MOC) and NPL-depleted (ΔNPL) extracts (Figure 8B), indicating that once pre-RC assembly is complete, NPL’s role in facilitating replication is reduced. Therefore, these results strongly support the notion that NPL increases the frequency of initiation in the extract by promoting pre-RC assembly on chromatin.
DISCUSSION
We have used Xenopus egg extract to investigate the roles of chromatin structure and nuclear envelope integrity in the reactivation of DNA replication in nuclei from terminally differentiated Xenopus erythrocytes. We find that erythrocyte nuclei lack essential components of the pre-RC, including XOrc1, XOrc2, XCdc6, XMcm3, and XMcm7 (Figure 2). These proteins stably associate with the chromatin of permeable nuclei, which initiate replication in the extract, but not with the chromatin of intact nuclei, which do not initiate under identical conditions. The failure of pre-RC proteins to bind chromatin from intact nuclei is not due to restricted nuclear access, however, because XOrc2, XCdc6, and XMcm3 are all able to cross an intact nuclear envelope (Figure 2). Therefore, given that ORC, Cdc6, and MCM proteins are all essential for DNA replication in eukaryotic cells (reviewed by Stillman, 1996), these data argue that an intact nuclear envelope prevents initiation in erythrocyte nuclei at least in part by preventing the assembly of pre-RCs on chromatin.
The absence of XCdc6 and XMcm3 from the chromatin of terminally differentiated erythrocytes is consistent with the observations that these proteins are virtually undetectable in cultured mammalian cells induced to exit the cell cycle by serum deprivation (Williams et al., 1997; Musahl et al., 1998; Yan et al., 1998). Our data are also consistent with studies in yeast that show a dramatic reduction in Mcm3, and the disappearance of the prereplicative footprint from origin DNA, after induction of a G0-like state (Diffley et al., 1994; Young and Tye, 1997). However, the presence of a postreplicative footprint in G0 yeast cells (Diffley et al., 1994) along with relatively high levels of Orc2 in quiescent mammalian cells (Musahl et al., 1998; Stoeber et al., 1998) indicate that ORC proteins remain associated with chromatin during reversible growth arrest. In contrast, XOrc1 and XOrc2 are undetectable within terminally differentiated erythrocyte nuclei (Figure 2), which, in vivo, are permanently withdrawn from the cell cycle (Leonard et al., 1982). Conceivably, the chromatin-bound ORC that remains during reversible arrest could serve to target other pre-RC proteins back to their original sites, thereby ensuring the preservation of origin specificity (Gilbert et al., 1995; Lawlis et al., 1996; Stoeber et al., 1998) and the replication timing program (Fangman and Brewer, 1992; Jackson and Pombo, 1998) during S-phase reentry. In the absence of cell cycle reentry, however, marking origins of replication would appear to be unnecessary.
The absence of pre-RC proteins from erythrocyte chromatin could be explained in two ways. First, these proteins may fail to assemble on erythrocyte chromatin during the final cell cycle of the differentiation program. This idea is intriguing in light of the fact that terminal differentiation of avian erythroid progenitors proceeds in precise synchrony, suggesting that the decision to enter G0 may be programmed in advance by a “master switch” rather than in response to environmental conditions in early G1 phase (Dolznig et al., 1995). Second, pre-RCs may be disassembled during exit from the cell cycle, as has been suggested for quiescent cultured cells (Leno and Munshi, 1994; Wu and Gilbert, 1997). Additional work is required to distinguish between these two possibilities.
Permeabilization of the nuclear envelope appears to be a general requirement for initiation of replication in nuclei from post–S-phase cells (Blow and Laskey, 1988; Leno et al., 1992; Coverley et al., 1993; Madine et al., 1995b) and quiescent cells (Leno and Munshi, 1994, 1997; Fang and Benbow, 1996; Munshi and Leno, 1998; this paper, Figure 1) by egg extract. The results presented here demonstrate that an intact nuclear envelope prevents replication of erythrocyte nuclei, at least in part, by preventing the assembly of functional pre-RCs on chromatin (Figures 1 and 2), the same general mechanism by which an intact envelope prevents rereplication within G2-phase nuclei (Madine et al., 1995b; Romanowski et al., 1996b; Hua et al., 1997). However, the specific requirements for generating functional pre-RCs within erythrocyte nuclei and G2-phase nuclei are different. In the latter case, XCdc6 and XMCM proteins must rebind to replicated DNA, which retains XORC, and it is the loading of XMCM proteins on chromatin that is prevented by an intact nuclear envelope (Madine et al., 1995b; Romanowski et al., 1996b; Hua et al., 1997). By contrast, many, if not all, pre-RC proteins must be assembled on erythrocyte nuclei, including XOrc1 and XOrc2 (Figure 2), all of which are prevented from stably binding to chromatin by an intact envelope. The failure of XCdc6 and XMCM proteins to bind erythrocyte chromatin in the absence of XORC is not surprising given the stepwise assembly of the pre-RC by egg extract (reviewed by Romanowski and Madine, 1996, 1997). However, it is surprising that assembly of XORC requires envelope permeabilization (Figure 2).
There are at least four general ways in which an intact nuclear envelope could prevent XORC assembly on erythrocyte chromatin. First, differentiation-specific changes in the nuclear envelope, such as a reduction in the density of nuclear pore complexes or changes in the capacity for nuclear protein import (Feldherr and Akin, 1990, 1991, 1993), could prevent the localization of ORC proteins within intact nuclei. Our data demonstrating that XOrc2 accumulates within intact nuclei (Figure 2) argue against this idea. However, we cannot rule out the possibility that other XORC proteins fail to cross an intact envelope, thereby preventing complete XORC assembly. Second, unique features of chromatin or nuclear structure (Thomas and Maclean, 1975; Brun, 1978; Wolffe, 1989; Chen et al., 1996), which are preserved within intact erythrocyte nuclei, could prevent the stable binding of nucleoplasmic XORC proteins to DNA. Indeed, our results demonstrate that an intact envelope reduces both the rate and the extent of removal of H1s from erythrocyte chromatin (Figure 4); however, this reduction cannot adequately explain the absence of XORC from chromatin, because functional pre-RCs form on the chromatin from permeable nuclei even in the absence of H1 removal (Figures 5 and 6), although to a much lesser extent (Figure 8). Yet, other features of chromatin or nuclear structure, which have not been investigated here, could prevent the assembly of XORC on erythrocyte DNA. Third, an intact envelope could exclude a factor that is required for XORC binding to DNA similar to the hypothetical loading factor for MCM proteins (Madine et al., 1995b). Fourth, an intact nuclear envelope could prevent XORC binding by concentrating, within the nucleus, an inhibitor of pre-RC assembly. This inhibitor could be a cyclin-dependent kinase, given that addition of moderate levels of cyclin A to egg extract prevents the binding of XORC to chromatin, possibly by reducing its affinity for DNA (Hua and Newport, 1998). If correct, the inhibitory concentration of kinase, or other factor, must be reached before XORC is assembled. This would not happen with permeable nuclei, because pre-RCs form on erythrocyte chromatin before an intact nuclear envelope is reassembled by the extract (our unpublished observations). Interestingly, we have found that H1 kinase activity is much higher within intact nuclei than within permeable nuclei after a 45-min incubation in the extract (our unpublished data). The identity of this kinase and its possible effects on XORC assembly are currently under investigation.
The hypercondensed erythrocyte chromatin (Thomas and Maclean, 1975; Brun, 1978; Wolffe, 1989) is decondensed and remodeled into embryonic-like chromatin by the replacement of somatic linker histones H1 and H10 with the cleavage-stage linker histone B4 and HMG1 from egg extract (Dimitrov and Wolffe, 1996; this paper, Figure 4). These transitions in chromatin composition and structure play a dominant and essential role in the reacquisition of transcriptional competence in erythrocyte nuclei (Dimitrov and Wolffe, 1996). The data shown here indicate that these transitions in composition and structure of erythrocyte chromatin also facilitate the initiation of replication (Figure 6) apparently by promoting the assembly of pre-RCs on DNA (Figure 8A). The molecular mechanism(s) by which such transitions in chromatin structure promote pre-RC assembly is unknown. The association of XORC with replication origins may involve the reorganization and displacement of nucleosomes surrounding the origin DNA, which appears to be the case in yeast (Bell and Stillman, 1992; Diffley and Cocker, 1992). Accordingly, the replacement of somatic H1s with embryonic B4 would induce a global reorganization of inactive chromatin, thereby unmasking potential XORC binding sites on the DNA. Once unmasked, other regulatory factors could facilitate a more localized reorganization at specific sites (Gavin et al., 1998), eventually leading to XORC binding. A component of this global reorganization may be an increase in nucleosome mobility, such as that which accompanies the replacement of somatic H1s with B4 (Nightingale et al., 1996; Ura et al., 1996), generating a more dynamic and extended chromatin structure (Dimitrov et al., 1994; Nightingale et al., 1996; Ura et al., 1996) in which potential binding sites become accessible to ORC.
Another protein that plays an important role in the compaction of erythrocyte chromatin is the mature erythrocyte nuclear termination stage-specific protein (MENT) (Grigoryev et al., 1992). MENT is a non-histone protein that is associated with condensing chromatin in terminally differentiating avian erythrocytes (Grigoryev and Woodcock, 1993). This heterochromatin protein has recently been shown to mediate higher-order chromatin folding in avian granulocytes (Grigoryev et al., 1999) presumably by acting as a glue within and between nucleosome chains (Grigoryev and Woodcock, 1998). It seems likely that the remodeling of erythrocyte chromatin in egg extract would also involve the removal of MENT from compact chromatin, leaving a more decondensed and open conformation, which could also contribute to the availability of potential ORC binding sites.
NPL mediates the decondensation and remodeling of sperm chromatin in egg extract (Philpott et al., 1991; Philpott and Leno, 1992; Leno et al., 1996). Interestingly, NPL also brings about the decondensation of nuclei from cultured somatic cells, although, in this case, limited decondensation does occur even in NPL-depleted extract (Philpott et al., 1991). We also observed residual decondensation of erythrocyte nuclei in NPL-depleted extract (our unpublished data), and it is possible that this level of decondensation allows for limited pre-RC assembly (Figure 8A) and DNA replication (Figure 6). The factor or factors responsible for this NPL-independent decondensation of erythrocyte chromatin have not been identified; however, it is intriguing to speculate that this process may be mediated by a protein complex similar to the chromatin accessibility complex (CHRAC), isolated from Drosophila embryo extracts, that increases nucleosome mobility even in the presence of histone H1 (Varga-Weisz et al., 1995, 1997). This complex has been implicated in the stimulation of SV40 replication by facilitating the binding of large T-antigen to origin DNA (Alexiadis et al., 1998).
In conclusion, the data presented here indicate that loss of nuclear envelope integrity and the removal of somatic linker H1 from erythrocyte chromatin are required for the acquisition of essential pre-RC proteins from the extract and the reactivation of DNA replication in this system. Furthermore, these data also suggest that reactivation is an ordered process, requiring, first, envelope permeabilization to remove a “block” to pre-RC assembly and, second, the removal of H1 to regulate the frequency of pre-RC assembly and initiation of replication. Further analysis is required for a more precise definition of these mechanisms and to determine the role(s) they may play in the establishment and/or maintenance of the quiescent state in eukaryotic cells.
ACKNOWLEDGMENTS
We thank Ron Laskey for providing antibodies to pre-RC proteins, Steve Dilworth for the anti-NPL hybridoma cell line PA3C5, Don Sittman for purified H1c, Jennifer Johns for technical assistance, and Asmita Kumar for helpful discussions throughout this study. This work was supported by the National Science Foundation grant MCB-9506280 (to G.H.L.).
Abbreviations used:
- BrdU
bromodeoxyuridine
- IgG
immunoglobulin G
- LPC
lysophosphatidylcholine
- MCM
minichromosome maintenance
- MENT
mature erythrocyte nuclear termination
- NPL
nucleoplasmin
- ORC
origin recognition complex
- PIPES
1,4-piperazinediethanesulfonic acid
- pre-RC
pre-replication complex
- SLO
streptolysin-O
REFERENCES
- Alexiadis V, Varga-Weisz PD, Bonte E, Becker PB, Gruss C. In vitro chromatin remodelling by chromatin accessibility complex (CHRAC) at the SV40 origin of DNA replication. EMBO J. 1998;17:3428–3438. doi: 10.1093/emboj/17.12.3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allan J, Cowling GJ, Harborne N, Cattini P, Craigie R, Gould H. Regulation of the higher-order structure of chromatin by histones H1 and H5. J Cell Biol. 1981;90:279–288. doi: 10.1083/jcb.90.2.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubert D, Garcia M, Benchaibi M, Poncet D, Chebloune Y, Verdier G, Nigon V, Samarut J, Mura CV. Inhibition of proliferation of primary avian fibroblasts through expression of histone H5 depends on the degree of phosphorylation of the protein. J Cell Biol. 1991;113:497–506. doi: 10.1083/jcb.113.3.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell SP, Stillman B. ATP-dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex. Nature. 1992;357:128–134. doi: 10.1038/357128a0. [DOI] [PubMed] [Google Scholar]
- Blank T, Trendelenburg M, Kleinschmidt JA. Reactivation of DNA replication in erythrocyte nuclei by Xenopus egg extract involves energy-dependent chromatin decondensation and changes in histone phosphorylation. Exp Cell Res. 1992;202:224–232. doi: 10.1016/0014-4827(92)90069-k. [DOI] [PubMed] [Google Scholar]
- Blow JJ, Laskey RA. A role for the nuclear envelope in controlling DNA replication within the cell cycle. Nature. 1988;332:546–548. doi: 10.1038/332546a0. [DOI] [PubMed] [Google Scholar]
- Brun RB. Developmental capacities of Xenopus eggs, provided with erythrocyte or erythroblast nuclei from adults. Dev Biol. 1978;65:271–284. doi: 10.1016/0012-1606(78)90027-1. [DOI] [PubMed] [Google Scholar]
- Carpenter PB, Mueller PR, Dunphy WG. Role for a Xenopus Orc2-related protein in controlling DNA replication. Nature. 1996;379:357–360. doi: 10.1038/379357a0. [DOI] [PubMed] [Google Scholar]
- Chen HY, Sun J, Hendzel MJ, Rattner JB, Davie JR. Changes in the nuclear matrix of chicken erythrocytes that accompany maturation. Biochem J. 1996;320:257–265. doi: 10.1042/bj3200257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong JPJ, Mahbubani HM, Khoo C, Blow JJ. Purification of an MCM-containing complex as a component of the DNA replication licensing system. Nature. 1995;375:418–421. doi: 10.1038/375418a0. [DOI] [PubMed] [Google Scholar]
- Chong JPJ, Thommes P, Rowles A, Mahbubani HM, Blow JJ. Characterization of the Xenopus replication licensing system. Methods Enzymol. 1997;283:549–564. doi: 10.1016/s0076-6879(97)83043-1. [DOI] [PubMed] [Google Scholar]
- Coleman TR, Carpenter PB, Dunphy WG. The Xenopus Cdc6 protein is essential for the initiation of a single round of DNA replication in cell-free extracts. Cell. 1996;87:53–63. doi: 10.1016/s0092-8674(00)81322-7. [DOI] [PubMed] [Google Scholar]
- Coppock DL, Lue RA, Wangh LJ. Replication of Xenopus erythrocyte nuclei in a homologous egg extract requires prior proteolytic treatment. Dev Biol. 1989;131:102–110. doi: 10.1016/s0012-1606(89)80041-7. [DOI] [PubMed] [Google Scholar]
- Coverley D, Downes CS, Romanowski P, Laskey RA. Reversible effects of nuclear membrane permeabilization: evidence for a positive licensing factor. J Cell Biol. 1993;122:985–992. doi: 10.1083/jcb.122.5.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox LS, Leno GH. Extracts from eggs and oocytes of Xenopus laevis differ in their capacities for nuclear assembly and DNA replication. J Cell Sci. 1990;97:177–184. doi: 10.1242/jcs.97.1.177. [DOI] [PubMed] [Google Scholar]
- Dawid IB. DNA in amphibian eggs. J Mol Biol. 1965;12:581–599. doi: 10.1016/s0022-2836(65)80313-8. [DOI] [PubMed] [Google Scholar]
- Diffley JFX, Cocker JH. Protein-DNA interactions at a yeast replication origin. Nature. 1992;357:169–172. doi: 10.1038/357169a0. [DOI] [PubMed] [Google Scholar]
- Diffley JFX, Cocker JH, Dowell SJ, Rowley A. Two steps in the assembly of complexes at yeast replication origins in vivo. Cell. 1994;78:303–316. doi: 10.1016/0092-8674(94)90299-2. [DOI] [PubMed] [Google Scholar]
- Dilworth SM, Black SJ, Laskey RA. Two complexes that contain histones are required for nucleosome assembly in vitro: role of nucleoplasmin and N1 in Xenopus egg extracts. Cell. 1987;51:1009–1018. doi: 10.1016/0092-8674(87)90587-3. [DOI] [PubMed] [Google Scholar]
- Dimitrov S, Dasso MC, Wolffe AP. Remodeling sperm chromatin in Xenopus laevis egg extracts: the role of core histone phosphorylation and linker histone B4 in chromatin assembly. J Cell Biol. 1994;126:591–601. doi: 10.1083/jcb.126.3.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrov S, Wolffe AP. Remodeling somatic nuclei in Xenopus laevis egg extracts: molecular mechanisms for the selective release of histones H1 and H10 from chromatin and the acquisition of transcriptional competence. EMBO J. 1996;15:5897–5906. [PMC free article] [PubMed] [Google Scholar]
- Dingwall C, Sharnick SV, Laskey RA. A polypeptide domain that specifies migration of nucleoplasmin into the nucleus. Cell. 1982;30:449–458. doi: 10.1016/0092-8674(82)90242-2. [DOI] [PubMed] [Google Scholar]
- Dolznig H, Bartunek P, Nasmyth K, Mullner EW, Beug H. Terminal differentiation of normal chicken erythroid progenitors: shortening of G1 correlates with loss of d-cyclin/cdk4 expression and altered cell size control. Cell Growth & Differ. 1995;6:1341–1352. [PubMed] [Google Scholar]
- Fang J, Benbow RM. Nuclear proteins of quiescent Xenopus laevis cells inhibit DNA replication in intact and permeabilized nuclei. J Cell Biol. 1996;133:955–969. doi: 10.1083/jcb.133.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fangman WL, Brewer BJ. A question of time: replication origins of eukaryotic chromosomes. Cell. 1992;71:363–366. doi: 10.1016/0092-8674(92)90505-7. [DOI] [PubMed] [Google Scholar]
- Feldherr CM, Akin D. The permeability of the nuclear envelope in dividing and nondividing cell cultures. J Cell Biol. 1990;111:1–8. doi: 10.1083/jcb.111.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldherr CM, Akin D. Signal-mediated nuclear transport in proliferating and growth-arrested BALB/c 3T3 cells. J Cell Biol. 1991;115:933–939. doi: 10.1083/jcb.115.4.933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldherr CM, Akin D. Regulation of nuclear transport in proliferating and quiescent cells. Exp Cell Res. 1993;205:179–186. doi: 10.1006/excr.1993.1073. [DOI] [PubMed] [Google Scholar]
- Gavin IM, Usachenko SI, Bavykin SG. Nucleosome structural transition during chromatin unfolding is caused by conformational changes in nucleosomal DNA. J Biol Chem. 1998;273:2429–2434. doi: 10.1074/jbc.273.4.2429. [DOI] [PubMed] [Google Scholar]
- Gilbert DM, Miyazawa H, DePamphilis ML. Site-specific initiation of DNA replication in Xenopus egg extract requires nuclear envelope integrity. Mol Cell Biol. 1995;15:2942–2954. doi: 10.1128/mcb.15.6.2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigoryev SA, Bednar J, Woodcock CL. MENT, a heterochromatin protein that mediates higher-order chromatin folding, is a new serpin family member. J Biol Chem. 1999;274:5626–5636. doi: 10.1074/jbc.274.9.5626. [DOI] [PubMed] [Google Scholar]
- Grigoryev SA, Solovieva VO, Spirin KS, Krasheninnikov IA. A novel nonhistone protein (MENT) promotes nuclear collapse at the terminal stage of avian erythropoiesis. Exp Cell Res. 1992;198:268–275. doi: 10.1016/0014-4827(92)90379-m. [DOI] [PubMed] [Google Scholar]
- Grigoryev SA, Woodcock CL. Stage-specific expression and localization of MENT, a nuclear protein associated with chromatin condensation in terminally differentiating avian erythroid cells. Exp Cell Res. 1993;206:335–343. doi: 10.1006/excr.1993.1154. [DOI] [PubMed] [Google Scholar]
- Grigoryev SA, Woodcock CL. Chromatin structure in granulocytes: a link between tight compaction and accumulation of a heterochromatin-associated protein (MENT) J Biol Chem. 1998;273:3082–3089. doi: 10.1074/jbc.273.5.3082. [DOI] [PubMed] [Google Scholar]
- Gurdon JB. Nuclear transplantation in eggs and oocytes. J Cell Sci. 1986;4:287–318. doi: 10.1242/jcs.1986.supplement_4.17. [DOI] [PubMed] [Google Scholar]
- Gurdon JB, Uehlinger V. “Fertile” intestine nuclei. Nature. 1966;210:1240–1241. doi: 10.1038/2101240a0. [DOI] [PubMed] [Google Scholar]
- Harlow E, Lane D. Antibodies: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1988. [Google Scholar]
- Hua XH, Newport J. Identification of a preinitiation step in DNA replication that is independent of origin recognition complex and cdc6, but dependent on cdk2. J Cell Biol. 1998;140:271–281. doi: 10.1083/jcb.140.2.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua XH, Yan H, Newport J. A role for Cdk2 kinase in negatively regulating DNA replication during S phase of the cell cycle. J Cell Biol. 1997;137:183–192. doi: 10.1083/jcb.137.1.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson DA, Pombo A. Replicon clusters are stable units of chromosome structure: evidence that nuclear organization contributes to the efficient activation and propagation of S phase in human cells. J Cell Biol. 1998;140:1285–1295. doi: 10.1083/jcb.140.6.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson PK, Chevalier S, Philippe M, Kirschner MW. Early events in DNA replication require cyclin E and are blocked by p21CiP1. J Cell Biol. 1995;130:755–769. doi: 10.1083/jcb.130.4.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krude T, Jackman M, Pines J, Laskey RA. Cyclin/Cdk-dependent initiation of DNA replication in a human cell-free system. Cell. 1997;88:109–119. doi: 10.1016/s0092-8674(00)81863-2. [DOI] [PubMed] [Google Scholar]
- Kubota Y, Mimura S, Nishimoto S, Takisawa H, Nojima H. Identification of the yeast MCM3-related protein as a component of Xenopus DNA replication licensing factor. Cell. 1995;81:601–609. doi: 10.1016/0092-8674(95)90081-0. [DOI] [PubMed] [Google Scholar]
- Lawlis SJ, Keezer SM, Wu J, Gilbert DM. Chromosome architecture can dictate site-specific initiation of DNA replication in Xenopus egg extracts. J Cell Biol. 1996;135:1207–1218. doi: 10.1083/jcb.135.5.1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leno GH, Downes CS, Laskey RA. The nuclear membrane prevents replication of human G2 nuclei but not G1 nuclei in Xenopus egg extract. Cell. 1992;69:151–158. doi: 10.1016/0092-8674(92)90126-w. [DOI] [PubMed] [Google Scholar]
- Leno GH, Laskey RA. The nuclear membrane determines the timing of DNA replication in Xenopus egg extracts. J Cell Biol. 1991;112:557–566. doi: 10.1083/jcb.112.4.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leno GH, Mills AD, Philpott A, Laskey RA. Hyperphosphorylation of nucleoplasmin facilitates Xenopus sperm decondensation at fertilization. J Biol Chem. 1996;271:7253–7256. doi: 10.1074/jbc.271.13.7253. [DOI] [PubMed] [Google Scholar]
- Leno GH, Munshi R. Initiation of DNA replication in nuclei from quiescent cells requires permeabilization of the nuclear membrane. J Cell Biol. 1994;127:5–14. doi: 10.1083/jcb.127.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leno GH, Munshi R. Reactivation of DNA replication in nuclei from terminally differentiated cells: nuclear membrane permeabilization is required for initiation in Xenopus egg extract. Exp Cell Res. 1997;232:412–419. doi: 10.1006/excr.1997.3520. [DOI] [PubMed] [Google Scholar]
- Leonard RA, Hoffner NJ, DiBerardino MA. Induction of DNA synthesis in amphibian erythroid nuclei in Rana eggs following conditioning in meiotic oocytes. Dev Biol. 1982;92:343–355. doi: 10.1016/0012-1606(82)90180-4. [DOI] [PubMed] [Google Scholar]
- Lu ZH, Sittman DB, Brown DT, Munshi R, Leno GH. Histone H1 modulates DNA replication through multiple pathways in Xenopus egg extract. J Cell Sci. 1997;110:2745–2758. doi: 10.1242/jcs.110.21.2745. [DOI] [PubMed] [Google Scholar]
- Lu ZH, Sittman DB, Romanowski P, Leno GH. Histone H1 reduces the frequency of initiation in Xenopus egg extract by limiting the assembly of prereplication complexes on sperm chromatin. Mol Biol Cell. 1998;9:1163–1176. doi: 10.1091/mbc.9.5.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madine MA, Khoo C, Mills AD, Laskey RA. MCM3 complex required for cell cycle regulation of DNA replication in vertebrate cells. Nature. 1995a;375:421–424. doi: 10.1038/375421a0. [DOI] [PubMed] [Google Scholar]
- Madine MA, Khoo C, Mills AD, Musahl C, Laskey RA. The nuclear envelope prevents reinitiation of replication by regulating the binding of MCM3 to chromatin in Xenopus egg extracts. Curr Biol. 1995b;5:1270–1279. doi: 10.1016/s0960-9822(95)00253-3. [DOI] [PubMed] [Google Scholar]
- Mahbubani HM, Chong JPJ, Chevalier S, Thommes P, Blow JJ. Cell cycle regulation of the replication licensing system: involvement of a Cdk-dependent inhibitor. J Cell Biol. 1997;136:125–135. doi: 10.1083/jcb.136.1.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munshi R, Leno GH. Replication of nuclei from cycling and quiescent mammalian cells in 6-DMAP-treated Xenopus egg extract. Exp Cell Res. 1998;240:321–332. doi: 10.1006/excr.1998.4019. [DOI] [PubMed] [Google Scholar]
- Musahl C, Holthoff HP, Lesch R, Knippers R. Stability of the replicative Mcm3 protein in proliferating and differentiating human cells. Exp Cell Res. 1998;241:260–264. doi: 10.1006/excr.1998.4041. [DOI] [PubMed] [Google Scholar]
- Nightingale K, Dimitrov S, Reeves R, Wolffe AP. Evidence for a shared structural role for HMG1 and linker histones B4 and H1 in organizing chromatin. EMBO J. 1996;15:548–561. [PMC free article] [PubMed] [Google Scholar]
- Pace CN, Vajdos F, Fee L, Grimsley G, Gray T. How to measure and predict the molar absorption coefficient of a protein. Protein Sci. 1995;4:2411–2423. doi: 10.1002/pro.5560041120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardee AB. G1 events and regulation of cell proliferation. Science. 1989;246:603–608. doi: 10.1126/science.2683075. [DOI] [PubMed] [Google Scholar]
- Philpott A, Leno GH. Nucleoplasmin remodels sperm chromatin in Xenopus egg extracts. Cell. 1992;69:759–767. doi: 10.1016/0092-8674(92)90288-n. [DOI] [PubMed] [Google Scholar]
- Philpott A, Leno GH, Laskey RA. Sperm decondensation in Xenopus egg cytoplasm is mediated by nucleoplasmin. Cell. 1991;65:569–578. doi: 10.1016/0092-8674(91)90089-h. [DOI] [PubMed] [Google Scholar]
- Romanowski P, Madine MA. Mechanisms restricting DNA replication to once per cell cycle: MCMs, prereplicative complexes and kinases. Trends Cell Biol. 1996;6:184–188. doi: 10.1016/0962-8924(96)10015-5. [DOI] [PubMed] [Google Scholar]
- Romanowski P, Madine MA. Mechanisms restricting DNA replication to once per cell cycle: the role of Cdc6p and ORC. Trends Cell Biol. 1997;7:9–10. doi: 10.1016/S0962-8924(97)30077-4. [DOI] [PubMed] [Google Scholar]
- Romanowski P, Madine MA, Laskey RA. XMCM7, a novel member of the Xenopus MCM family, interacts with XMCM3 and colocalizes with it throughout replication. Proc Natl Acad Sci USA. 1996a;93:10189–10194. doi: 10.1073/pnas.93.19.10189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanowski P, Madine MA, Rowles A, Blow JJ, Laskey RA. The Xenopus origin recognition complex is essential for DNA replication and MCM binding to chromatin. Curr Biol. 1996b;6:1416–1425. doi: 10.1016/s0960-9822(96)00746-4. [DOI] [PubMed] [Google Scholar]
- Rowles A, Chong JPJ, Brown L, Howell M, Evan GI, Blow JJ. Interaction between the origin recognition complex and the replication licensing system in Xenopus. Cell. 1996;87:287–296. doi: 10.1016/s0092-8674(00)81346-x. [DOI] [PubMed] [Google Scholar]
- Stillman B. Cell cycle control of DNA replication. Science. 1996;274:1659–1664. doi: 10.1126/science.274.5293.1659. [DOI] [PubMed] [Google Scholar]
- Stoeber K, Mills AD, Kubota Y, Krude T, Romonowski P, Marheineke K, Laskey RA, Williams GH. Cdc6 protein causes premature entry into S phase in a mammalian cell-free system. EMBO J. 1998;17:7219–7229. doi: 10.1093/emboj/17.24.7219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Wiaderkiewicz R, Ruiz-Carrillo A. Histone H5 in the control of DNA synthesis and cell proliferation. Science. 1989;245:68–71. doi: 10.1126/science.2740916. [DOI] [PubMed] [Google Scholar]
- Thomas N, Maclean N. The erythroid cells of anemic Xenopus laevis. I. Studies on cellular morphology and protein and nucleic acid synthesis during differentiation. J Cell Sci. 1975;19:509–520. doi: 10.1242/jcs.19.3.509. [DOI] [PubMed] [Google Scholar]
- Ura K, Nightingale K, Wolffe AP. Differential association of HMG1 and linker histones B4 and H1 with dinucleosomal DNA: structural transitions and transcriptional repression. EMBO J. 1996;15:4959–4969. [PMC free article] [PubMed] [Google Scholar]
- Varga-Weisz PD, Blank TA, Becker PB. Energy-dependent chromatin accessibility and nucleosome mobility in a cell-free system. EMBO J. 1995;14:2209–2216. doi: 10.1002/j.1460-2075.1995.tb07215.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga-Weisz PD, Wilm M, Bonte E, Dumas K, Mann M, Becker PB. Chromatin-remodeling factor CHRAC contains the ATPases ISWI and topoisomerase II. Nature. 1997;388:598–602. doi: 10.1038/41587. [DOI] [PubMed] [Google Scholar]
- Walter J, Sun L, Newport J. Regulated chromosomal DNA replication in the absence of a nucleus. Mol Cell. 1998;1:519–529. doi: 10.1016/s1097-2765(00)80052-0. [DOI] [PubMed] [Google Scholar]
- Wangh LJ, DeGrace D, Sanchez JA, Gold A, Yeghiazarians Y, Wiedemann K, Daniels S. Efficient reactivation of Xenopus erythrocyte nuclei in Xenopus egg extracts. J Cell Sci. 1995;108:2187–2196. doi: 10.1242/jcs.108.6.2187. [DOI] [PubMed] [Google Scholar]
- Williams RS, Shohet RV, Stillman B. A human protein related to yeast Cdc6p. Proc Natl Acad Sci USA. 1997;94:142–147. doi: 10.1073/pnas.94.1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolffe AP. Transcriptional activation of Xenopus class III genes in chromatin isolated from sperm and somatic nuclei. Nucleic Acids Res. 1989;17:767–781. doi: 10.1093/nar/17.2.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J-R, Gilbert DM. The replication origin decision point is a mitogen-independent, 2-aminopurine sensitive, G1-phase step that precedes restriction point control. Mol Cell Biol. 1997;17:4312–4321. doi: 10.1128/mcb.17.8.4312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Z, DeGregori J, Shohet R, Leone G, Stillman B, Nevins JR, Williams RS. Cdc6 is regulated by E2F and is essential for DNA replication in mammalian cells. Proc Natl Acad Sci USA. 1998;95:3603–3608. doi: 10.1073/pnas.95.7.3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young MR, Tye BK. Mcm2 and Mcm3 are constitutive nuclear proteins that exhibit distinct isoforms and bind chromatin during specific cell cycle stages of Saccharomyces cerevisiae. Mol Biol Cell. 1997;8:1587–1601. doi: 10.1091/mbc.8.8.1587. [DOI] [PMC free article] [PubMed] [Google Scholar]