Abstract
Microbial detection requires the recognition of pathogen-associated molecular patterns (PAMPs) by pattern recognition receptors (PRRs) that are distributed on the cell surface and within the cytosol. The nucleotide-binding oligomerization domain (NOD)-like receptor (NLR) family functions as an intracellular PRR that triggers the innate immune response. The mechanism by which PAMPs enter the cytosol to interact with NLRs, particularly muropeptides derived from the bacterial proteoglycan cell wall, is poorly understood. PEPT2 is a proton-dependent transporter that mediates the active translocation of di- and tripeptides across epithelial tissues, including the lung. Using computational tools, we initially established that bacterial dipeptides, particularly γ-D-glutamyl-meso-diaminopimelic acid (γ-iE-DAP), are suitable substrates for PEPT2. We then determined in primary cultures of human upper airway epithelia and transiently transfected CHO-PEPT2 cell lines that γ-iE-DAP uptake was mediated by PEPT2 with an affinity constant of approximately 193 μM, whereas muramyl dipeptide was not transported. Exposure to γ-iE-DAP at the apical surface of differentiated, polarized cultures resulted in activation of the innate immune response in an NOD1- and RIP2-dependent manner, resulting in release of IL-6 and IL-8. Based on these findings we report that PEPT2 plays a vital role in microbial recognition by NLR proteins, particularly with regard to airborne pathogens, thereby participating in host defense in the lung.
Keywords: human, lung, bacterial, cell surface molecules, acute phase reactants
CLINICAL RELEVANCE
This research demonstrates that peptide transporters expressed in human lung epithelia contribute to the uptake of select bacterial peptides, leading to specific activation of innate immune response.
Pathogen-associated molecular patterns (PAMPs) are recognized by cell surface or cytosolic pattern recognition receptors (PRRs) (1). One family of intracellular PRRs include the nucleotide-binding oligomerization domain (NOD)-containing proteins (NODs or NOD-like receptors [NLRs]) that sense intracellular pathogen invasion and trigger signaling cascades, thereby activating the host immune response (2–4). NOD1 and NOD2 mediate intracellular recognition of bacterial proteoglycan (PGN)-derived molecules through recognition by their carboxy-terminal leucine-rich repeat region (5). This results in recruitment of the downstream kinase protein RIP2 and initiation of NF-κB–mediated transcription and elaboration of newly synthesized cytokines and chemokines (6). Biochemical and functional analyses have identified γ-D-glutamyl-meso-diaminopimelic acid (γ-iE-DAP) and muramyl dipeptide (MDP) as the minimal bacterial muropeptide epitopes recognized by NOD1 and NOD2, respectively, resulting in immune activation (5, 7).
Enzymes that are naturally present in the epithelial lining fluid (e.g., lysozyme) are believed to act in concert to degrade the bacterial cell wall as a front-line defense mechanism, thereby liberating muropeptide fragments that include both γ-iE-DAP and MDP. The dipeptides may also be derived as byproducts of bacterial PGN biosynthesis, cell growth, and division; therefore, it is plausible that these particular PAMPs reside in the airway microenvironment upon pathogen invasion. However, the mechanism(s) by which PAMPs enter the cytosol to interact with NOD1 and NOD2 and activate innate immunity is poorly understood, particularly with respect to extracellular pathogens whose cell wall derivatives cannot readily enter cells.
PEPT2 (SLC15A2) is a proton-dependent solute transporter that mediates the active translocation of peptides across epithelial tissues, including the lung (8). PEPT2 displays promiscuous ligand affinity and mediates, in addition to its natural di- and tripeptide substrates, the uptake of numerous peptidomimetic drug molecules, such as β-lactam antibiotics and antivirals. Our previous work and that of others characterized PEPT2 expression and function in humans in situ in bronchial, bronchiolar, and alveolar type II epithelial cells (9–11). We first reported the functional characterization of PEPT2-mediated transport across the apical surface of fully differentiated, polarized, primary human upper airway epithelial cultures (hUAECs, henceforth referred to as human lung epithelial cells [hLECs]). Extending upon these observations, we hypothesized that PEPT2 could also mediate bacterial dipeptide transport, thereby serving as an initiator of intracellular pathogen recognition by PRRs.
Here, we demonstrate that PEPT2 selectively transports bacterial dipeptides into primary human cultures and related human cell lines. In particular, studies were conducted in differentiated, polarized, lung epithelia grown at an air-to-liquid interface, thereby providing an opportunity to simulate microbial invasion from the airway interface. We observed that γ-iE-DAP is selectively transported across the apical interface and into lung epithelia, thereby triggering the host immune response. Moreover, our results indicate that in humans, the lung epithelium can discriminate between different pathogens by virtue of subtle structural differences that impact cellular entry of specific bacterial muropeptides by a specific membrane transporter. The role that we describe for PEPT2 in microbe recognition introduces a new mode of communication between the cells that constitute the mucosal barrier in the lung and the critical interface that they defend.
MATERIALS AND METHODS
Pharmacophore Mapping
The PEPT1 pharmacophore was generated as described previously (12). The bacterial peptides were mapped onto this pharmacophore using “Fast Fit” algorithm and analyzed. Higher “Fast fit” score indicates better fit to the centroid of the pharmacophore features. A second HIPHOP pharmacophore was developed with the three highest-affinity PEPT2 substrates from a recent study, namely, Trp-Trp-Trp, Trp-Trp and Leu-Arg-Pro (13) and this model was used to map the muropeptides.
Materials
Ala-γ-D-Glu-Diaminopimelic acid (γ-iE-DAP) and Ala-α-D-Glu-DAP (α-iE-DAP) were purchased from Anaspec (San Jose, CA). N-Acetylmuramyl-L-alanyl-D-isoglutamine hydrate (MDP) was from Sigma (St. Louis, MO). Human PEPT2 cDNA was a kind gift from Dr. Mathias Hediger, Harvard Medical School (Boston, MA). Anti-PEPT2 antibody was a kind gift from Dr. David Smith, University of Michigan (Ann Arbor, MI). All radiolabeled chemicals were from Moravek Biochemicals (Brea, CA).
Lung Cell Isolation and Cell Culture
Human donor lungs were collected with approval from The Ohio State University Institutional Review Board. Lung cell culture was conducted as previously described (9). and maintained in a 1:1 mixture of Dulbecco's modified Eagle's media (DMEM) and Ham's F12 media (DMEM/F12) (Invitrogen, Carlsbad, CA) supplemented with 2% Ultroser G and antibiotics at 37°C, and 5% CO2. Wild type CHO-K1 cells (CCL-61), transiently transfected PEPT2-CHO cells, and BEAS-2B cells were cultured in DMEM supplemented with 10% FBS and 250 U/ml Penicillin-G/Streptomycin (Invitrogen).
Transport Experiments
All transport experiments were performed as described previously (9). The culture inserts with hLECs were washed three times and conditioned for 2 hours with 0.2 ml apical (pH = 6.5) and 0.4 ml basal transport buffer (BL) (pH = 7.4). For construction of a Cornish-Bowden Plot, three different [3H]-Gly-Sar (1 Ci/mmol) (Moravek Biochemicals) concentrations—2.27 nM, 22.72 nM, and 44.54 nM—were selected using a 0–500 μM γ-iE-DAP concentration range. The AP chamber received 0.2 ml mix of γ-iE-DAP, [3H]-GlySar and [14C]-Mannitol (53 mCi/mmol). All experiments were the average of three repetitions. A similar strategy was used for immune activation studies. Briefly, culture inserts were washed and conditioned with phosphate-buffered saline for 20 minutes, followed by washing with sterile apical transport buffer (AP) (DMEM/F-12 w/o phenol red, 50:50 mix, pH 6.5). Three hundred microliters of BL (DMEM/F-12 w/o phenol red, 50:50 mix, pH 7.9; Gibco, Carlsbad, CA) was placed on the basolaterally and 100 μl of AP buffer apically. hLECs were treated apically with 5 μg/ml of the active bacterial dipeptides γ-ieDAP, MDP, or α-iE-DAP. For competitive inhibition studies, the apical chamber received a 10,000-fold excess of Gly-Sar (0.1 mM) before addition of muropeptides. At the end of a 4-hour incubation the hLECs were washed extensively and 200 μl of buffer was placed on each side of the chamber. A total cell supernatant (apical and basal) was collected at the end of 24 hours, snap frozen, and stored at −80°C for cytokine analysis.
Transfection and Uptake Experiments
CHO-K1 cells were seeded in 24-well culture plates. Cells were transfected at 80 to 90% confluence with human PEPT2 cDNA with Lipofectamine 2000 (Invitrogen) using a standard protocol. Forty-eight hours after transfection, the cell monolayers were used for uptake studies as described above.
Real-Time PCR
RNA was isolated with TRIzol reagent followed by reverse transcription of 2 μg of total RNA with ThermoScript RNase H− reverse transcriptase (Invitrogen), then diluted to 100 μl. Between 20 and 60 ng of cDNA was used for quantitative PCR with SYBR green I PCR master mixture and the PRISM 7700 apparatus (Applied Biosystems, Foster City, CA). The total volume of the PCR reaction was set at 20 μl and included 2 μl of cDNA template and 0.25 μM of each primer. Primer pairs intentionally spanned introns to avoid false negatives by amplification of genomic DNA. Relative copy numbers (RCN) and expression ratios of NOD1 and NOD2 were normalized to two housekeeping genes (GAPDH and CAP-1 [cAMP-accessory protein]) and calculated with the following equation: RCN = E−ΔCt × 100.
Cytokine Determination
Measurement of human IL-6 and IL-8 concentrations in cell culture supernatants was performed using the Immulite automated chemiluminometer (Siemens Medical Solutions Diagnostics, Los Angeles, CA). Manufacturer-supplied standards were used for calibration before sample analysis. Samples were measured over a range of dilutions to validate accurate quantitation.
NOD1 Overexpression
The human full-length NOD1 cDNA was a generous gift from Dr. Gabriel Nunez, University of Michigan (Ann Arbor, MI). The 2,862-bp cDNA sequence (accession: NM_006092) was inserted into a pcDNA3 vector with KpnI and XhoI restriction sites with a CMV promoter region and N-terminal flag tag. BEAS-2B cells were transiently transfected for 2 hours using Lipofectamine 2000 (Invitrogen). Twenty-four hours later, cells were subjected to muropeptide challenge and analyzed.
Small Interfering RNA Assay
The following NOD1 small interfering RNA (siRNA) used in this study was purchased from Dharmacon (Lafayette, CO). BEAS-2B cells were transfected with 5-nM ON-TARGETplus SMARTpool siRNA by using HiPerfect transfection reagent (Qiagen, Valencia, CA) according to the manufacturer's protocol. Forty-eight hours after transfection, cells were subjected to muropeptide challenge and analyzed.
Western Analysis of Primary hLECs
Cell lysis buffer (Cell Signaling, Beverly, MA) with 1 mM PMSF and 1× Complete Mini protease inhibitor cocktail (Roche, Indianapolis, IN) was directly added to the apical surface of the transwell chamber to lyse cells. Equal amounts of lysate were resolved in a 10% SDS-PAGE gel (Bio-Rad, Hercules, CA) and transferred to a nitrocellulose membrane (Amersham Biosciences, Little Chalfont, UK), then subjected to Western analysis. Signal was detected with an ECL kit (Amersham Biosciences) and a Fluor-S Multi-Imager Max/Bio-quantity one (Bio-Rad). The following antibodies were used: anti-NOD1 (1:1,000; IMGENEX, San Diego, CA); anti-NOD1 (1:1,000; Dr. D. Philpott, Paris, France); anti-RIP2 (1:1,000; Cell Signaling); anti–β-actin (1:5,000; MP Biomedicals, OH); goat anti-mouse IgG-HRP (1:3,000; Cell Signaling); and goat anti-rabbit IgG-HRP (1:3,000; Zymed, San Francisco, CA).
Immunoprecipitation Experiments of hLECs
Cultured hLECs were stimulated with 250 units/ml of IFN-γ, or 100 ng/ml of TNF-α. After 24 hours, cells were recovered and lysed in Cell lysis buffer (Cell Signaling) with 1 mM PMSF and 1× Complete Mini protease inhibitor cocktail (Roche), and lysates were clarified by centrifugation (10 min at 13,200 rpm). Equal amounts of sample were pre-cleared by incubation of Recombinant Protein G (rProtein G) Agarose (Invitrogen Corporation) for 30 minutes. Immunoprecipitation of NOD1 was performed by incubating overnight at 4°C with commercially available anti-NOD1 (2 μl) antibody or pre-immune rabbit serum (2 μl) followed by a 1-hour incubation with rProtein G. Immune complexes were washed extensively with lysis buffer, boiled in Laemmli buffer, and separated by 10% SDS/PAGE. Proteins were then transferred to nitrocellulose membranes and immunoblotted with the anti-NOD1 antibody (kind gift of Dr. Philpott).
Co-Immunoprecipitation of RIP2
The BEAS-2B cell line was transfected with 1 μg flag tagged NOD1 (kind gift of Dr. Gabriel Nunez). Twenty-four hours later, cells were stimulated with 5 μg/ml iE-DAP for 0, 5, 15, and 30 min. Cells were recovered and lysed as described above. Immunoprecipitation of flag tagged NOD1 was performed by incubating overnight at 4°C with commercially available monoclonal anti-flag (1 μl) (Sigma) antibody or mouse IgG (1 μl) followed by a 1-hour incubation with anti-flag M2 beads (Sigma). Immune complexes were washed extensively with lysis buffer, boiled in Laemmli buffer, and separated by SDS/PAGE using a 10% gel. Proteins were then transferred on to nitrocellulose membranes and detected by immunoblotting with the anti-RIP2 antibody (kind gift from Dr. M. Wewers, Ohio State University, Columbus, OH).
NF-κB Activation Study
BEAS-2B cells were co-transfected with firefly and renilla luciferase constructs. Four hours after transfection, cells were washed once then submersed in 1 ml of AP buffer (pH = 6.5). A dose of 5 μg/ml of γ-iE-DAP, or α-iE-DAP, was then administered. The plate was then placed in a 37°C incubator for 2 hours. Cells were washed and then lysed in Passive Lysis Buffer (Promega, Madison, WI). Cells were scrapped on ice, collected, vortexed for 15 seconds, and then centrifuged at 13,200 rpm for 10 minutes at 4°C. Activity was then measured following the dual luciferase protocol from Promega. Luciferase values were normalized for transfection efficiency by renilla expression. NF-κB assays were performed in triplicate.
Statistical Analysis
Data are expressed as mean values ± SD. To evaluate the significance of the difference between two sample means, a two-tailed, separate-variance Student's t test was performed. Differences were considered statistically significant at P ≤ 0.05.
RESULTS
Computational Modeling Infers that γ-iE-DAP Is a Substrate for PEPT2
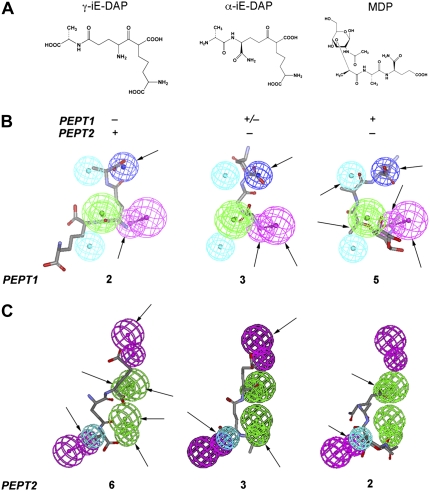
To differentiate the affinity of the muropeptides γ-iE-DAP, α-iE-DAP, and MDP for the two paralogous PEPT transporters with divergent structural requirements, we predicted their relative affinity using a previously established PEPT1 pharmacophore (12) and a novel PEPT2 pharmacophore model. We used the “Fast Fit” algorithm to make predictions based on the proximity of a substrate's functional groups to the pharmacophore centroids. Using the PEPT1 pharmacophore, we noted that γ-iE-DAP mapped to two out of seven pharmacophore groups, whereas α-iE-DAP and MDP fit three and five feature points, respectively. The new PEPT2 pharmacophore was developed with high-affinity substrates (13) and contained two hydrogen bond acceptors, two hydrogen bond donors, and one hydrophobic feature, using nine pharmacophore centroids. When used to fit the bacterial peptides, γ-iE-DAP occupied six features (Figure 1A), whereas α-iE-DAP (Figure 1C) and MDP (Figure 1D) mapped poorly to the pharmacophore feature points. Overall, computational mapping suggested a relatively high and selective affinity for γ-iE-DAP toward PEPT2, but not PEPT1, whereas MDP possessed high and selective affinity toward PEPT1, but not PEPT2.
Figure 1.
Pharmacophore modeling predicts interaction between γ-iE-DAP and PEPT2. (A) Chemical structures of Ala-γ-D-Glu-diaminopimelic acid (γ-iE-DAP), Ala-α-D-Glu-diaminopimelic acid (α-iE-DAP), and N-acetylmuramyl-L-Ala-D-isoglutamine (MDP) and their predicted interaction with the small oligopeptide transporters PEPT1 and PEPT2. Individual muropeptides mapped to (B) the PEPT1 and (C) PEPT2 pharmacophores. The number of pharmacophore feature points occupied by each molecule is listed below each figure, and arrows indicate the location of positive interactions. The color coded spheres represent pharmacophore feature points: purple, hydrogen bond donor; green, hydrogen bond acceptor; and cyan, hydrophobic.
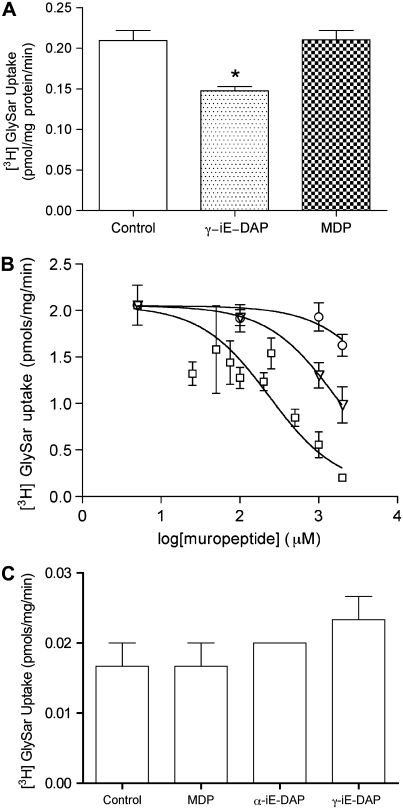
γ-iE-DAP Competitively Inhibits PEPT2-Mediated Substrate Translocation
To evaluate the potential affinity of γ-iE-DAP and MDP for PEPT2, we undertook conventional inhibition studies using [3H]-GlySar, a prototypical PEPT2 substrate, in cultured primary hLECs. We consistently observed that γ-iE-DAP but not MDP competitively inhibited the transport of [3H]-GlySar (Figure 2A). We then quantified the affinity of γ-iE-DAP for PEPT2 using three different concentrations of [3H]-GlySar and serial dilutions of [γ-iE-DAP], thereby confirming competitive uptake in hLECs (data not shown). When the data were transformed using the Cornish-Bowden method (14), we obtained a Ki of approximately 193 μM for γ-iE-DAP, within the range expected for PEPT2.
Figure 2.
PEPT2-mediated uptake of γ-iE-DAP. (A) Inhibition by muropeptides of transepithelial [3H]-GlySar transport across hLECs monolayers. [3H]-GlySar was tested at 45.44 nM concentration and γ-iE-DAP and MDP both were tested at 454.4 μM concentrations (n = 2, all determinations are ± SD). (B) Dose–reponse curves for [3H]-GlySar uptake in the presence of γ-iE-DAP (squares), α-iE-DAP (triangles), and MDP (circles) in PEPT2-transfected CHO-K1 cells. CHO-K1 cells were seeded in a 24-well culture plate and transfected with human PEPT2 cDNA using Lipofectamine 2000. At 48 hours after transfection, cell monolayers were assayed for 10 minutes with 100 nM [3H]-Gly-Sar in the presence of increasing muropeptide concentrations up to their solubility limit (0–2,000 μM). Data were normalized for total protein content (n = 3, all determinations are ± SD). (C) Assessment of [3H]-GlySar uptake in wild-type CHO-K1 cells in the absence (Control) or presence of a 10,000-fold excess of muropeptides (n = 3 ± SD).
To further validate these findings, we transiently expressed wild-type PEPT2 in CHO-K1 cells, an adherent cell line not endogenously expressing PEPT2. [3H]-GlySar uptake was again monitored in the presence of increasing concentrations of γ-iE-DAP, α-iE-DAP, and MDP. From these data we calculated the transporter inhibition constant for γ-iE-DAP to be 236.2 μM, also characteristic of a high-affinity, low-capacity transporter substrate (Figure 2B). Parallel experiments with MDP did not demonstrate competitive inhibition (IC50 > 10 mM), consistent with the above findings. Interestingly, the enantiomer α-iE-DAP also inhibited the [3H]-GlySar uptake in PEPT2-transfected CHO-K1 cells, albeit to a lesser extent (IC50 ∼ 1.63 mM), indicating that both stereoisomers are recognized by PEPT2.
Control experiments with wild-type CHO-K1 cells were performed to rule out the occurrence of an active GlySar flux mediated by other potentially present peptide transport systems. As shown in Figure 2C, [3H]-GlySar uptake in wt CHO-K1 cells is minimal and cannot be competitively suppressed in the presence of muropeptides. This suggests that GlySar uptake into CHO-K1 cells is governed by a passive diffusion process alone.
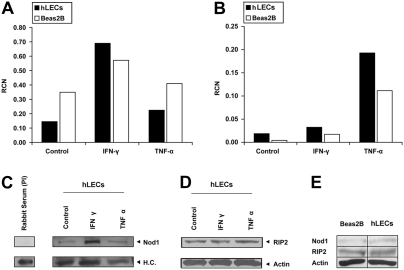
NOD1 and RIP2 Are Constitutively Expressed in Human Lung Epithelia
Previous investigations involving NLR immune activation have shown that muropeptide recognition by NLRs activates nuclear factor kappa B (NF-κB)–mediated transcription, thereby resulting in cytokine release (2). However, to our knowledge, lung epithelia have never been carefully examined. Thus, we surveyed hLEC and BEAS-2B cultures to determine the presence of key molecules involved in NOD-mediated signaling. As shown, NOD1 and NOD2 mRNA were constitutively expressed in both cell types. Also, consistent with previous reports (15), NOD1 expression was specifically induced by IFN-γ (Figure 3A), whereas NOD2 was induced by TNF-α (16) (Figure 3B). In agreement with mRNA expression, NOD1 protein was also increased after IFN-γ exposure (Figure 3C), whereas RIP2, an essential kinase involved in NLR-mediated signaling, was constitutively present and unaffected by cytokine exposure (Figure 3D). We further demonstrate by direct comparison that both hLECs and BEAS-2B cells constitutively express NOD1 and RIP2 (Figure 3E).
Figure 3.
Lung epithelial expression of nucleotide-binding oligomerization domain (NOD)-related factors. Total RNA and protein were obtained from primary human upper airway epithelial cells (hLECs) after overnight culture with or without 250 units/ml of IFN-γ, or 100 ng/ml of TNF-α. (A) NOD1/CARD4 and (B) NOD2/CARD15 mRNA was quantified by real-time PCR and expressed as relative copy number (RCN). GAPDH and CAP were used as internal controls to standardize and quantify mRNA expression. (C) Whole cell lysates were immunoprecipitated with affinity-purified anti-Nod1/Card4 antibody (IMGENEX), and then immunoblotted with an anti-Nod1/Card4 antibody (provided by Dr. Dana Philpott). Pre-immune rabbit serum was used as a negative control. (D) Western analysis of RIP2. Samples were loaded with 40 μg/well protein from the same lysate, and then the membrane was blotted with an anti-RIP2 antibody. β-Actin served as control to determine equal loading. (E) An immunoblot demonstrating constitutive levels of NOD1 and RIP2 proteins in both BEAS-2B and hLEC lung epithelial cells; 80 μg/well protein was loaded for each sample.
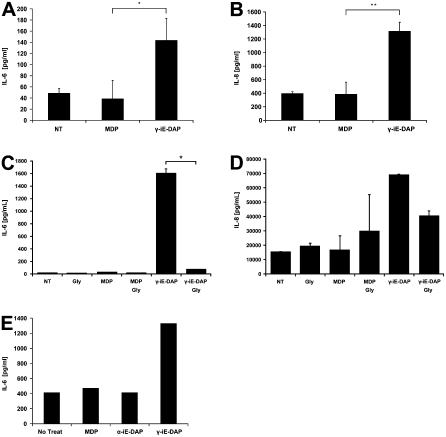
γ-iE-DAP-Mediated Immune Activation Is Inhibited by GlySar, the Prototypical PEPT2 Substrate
The apical surface of polarized hLEC cultures were exposed to γ-iE-DAP or MDP and supernatants evaluated for cytokine release. A significant increase in IL-6 and IL-8 release was observed after exposure to γ-iE-DAP, but not MDP (Figures 4A and 4B). In addition, γ-iE-DAP–mediated IL-6 and IL-8 release was significantly inhibited upon co-incubation with the nonimmunoreactive dipeptide GlySar, a dipeptide ligand with a known high affinity for PEPT2 (Figures 4C and 4D). Likewise, overexpression of PEPT2 augmented the response of lung epithelia to γ-iE-DAP, when compared with nontransfected cultures, resulting in increased IL-6 release (Figure 4E). The fact that the γ-iE-DAP–induced immune response was inhibited by co-incubation with GlySar suggests that the cellular entry of both compounds is mediated by a common membrane translocation factor. These findings expand upon the above studies demonstrating that PEPT2-mediated GlySar uptake was inhibited by γ-iE-DAP; which taken together, provides evidence that GlySar and γ-iE-DAP mutually inhibit each other's cellular uptake. Collectively, this indicates that PEPT2 plays an auxilliary role in activation of the NLR-mediated innate immune response by transporting γ-iE-DAP.
Figure 4.
γ-iE-DAP activates innate immunity in lung epithelia. Primary hLECs were cultured with the bacteria muropeptides MDP or γ-iE-DAP for 24 hours, and then cell culture supernatants were analyzed for IL-6 and IL-8 release. As shown, γ-iE-DAP induced the release of (A) IL-6 and (B) IL-8, whereas MDP did not. Next, we repeated the same experiment but in the presence or absence of the competitive PEPT2 transport inhibitor Gly-Sar for 4 hours. The constituents were then removed and cells were cultured in normal medium for an additional 20 hours. Supernatants were collected and again analyzed for (C) IL-6 or (D) IL-8. As shown, GlySar significantly reduced the amount of both IL-6 and IL-8 release only in cultures exposed to γ-iE-DAP, whereas MDP- or GlySar-treated cultures were unaffected. Data are expressed as mean values ± σ of n = 3 (P ≤ 0.05). To verify these findings, we overexpressed human PEPT2 in BEAS-2B cultures and then evaluated IL-6 after exposure to MDP, γ-iE-DAP, or the nonimmunoreactive enantiomer α-iE-DAP. (E) Compared with PEPT2 transfected untreated cultures, only the cultures exposed to γ-iE-DAP exhibited an increase in IL-6 release.
NOD1 Is Required for γ-iE-DAP–Mediated Innate Immune Activation
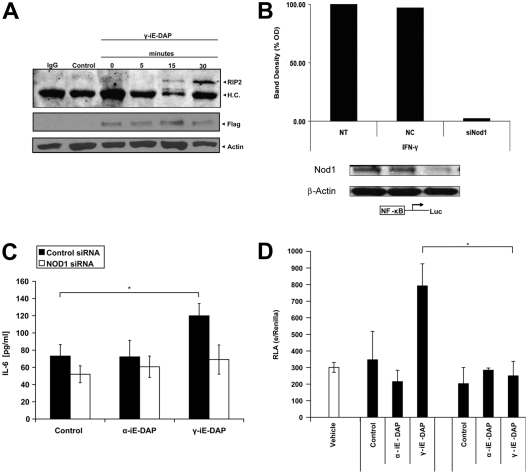
To determine whether NOD1 is required for immune activation after transport of γ-iE-DAP by PEPT2, we overexpressed FLAG-tagged NOD1 and exposed cultures to γ-iE-DAP and immunoprecipitated NOD1 to evaluate interaction with RIP2, its immediate downstream binding partner. The formation of a complex between NOD1 and RIP2 is evident between 15 and 30 minutes of dipeptide exposure (Figure 5A). Next, we determined whether NOD1 is required for PEPT2-mediated immune activation in response to γ-iE-DAP. Hence, we first evaluated the ability of NOD1-specific siRNAs to suppress endogenous protein levels. Knowing that NOD1 is at the lower threshold of detection, we evaluated cultures under IFN-γ–stimulated conditions to more thoroughly evaluate the ability of siRNA to knock down its target. As shown, only NOD1-targeted siRNA treatment resulted in a significant reduction in detectable protein levels, whereas the si-control had no effect (Figure 5B). The effect of NOD1 suppression was then evaluated to determine if it altered the epithelial response to γ-iE-DAP. As shown, siRNA-mediated knockdown of NOD1 expression inhibited the epithelial response to the muropeptide as measured by a significant decrease in IL-6 release (Figure 5C). In comparison, the non-siRNA or si-control treated samples retained their ability to respond. As an additional control, we evaluated the ability of α-iE-DAP, the enantiomer of γ-iE-DAP that does not interact with NOD1, to induce IL-6 release. As expected, α-iE-DAP did not increase IL-6 concentrations in the culture supernatants regardless of treatment conditions (Figure 5C). Further, we observed that innate immune activation with the non–NOD1-dependent stimulus, TNF-α, resulted in equivalent amounts of IL-6 release in all treatment groups, indicating that siRNA treatment alone did not negatively impact innate immune activation in BEAS2B cultures (data not shown). Finally, having observed that γ-iE-DAP induces formation of a NOD1-RIP2 complex in conjunction with cytokine release, we evaluated the ability of the muropeptide to induce NF-κB–mediated transcription. We show that γ-iE-DAP but not α-iE-DAP resulted in NF-κB–mediated transcriptional activation in BEAS-2B cultures transfected with a NF-κB promotor/luciferase reporter plasmid (Figure 5D). Furthermore, suppression of NOD1 by siRNA inhibited γ-iE-DAP–mediated luciferase expression down to baseline values. Taken together, this demonstrates that γ-iE-DAP is recognized by NOD1 in lung epithelia after PEPT2-mediated uptake, thereby initiating cell signaling events that culminate in NF-κB–mediated innate immune activation.
Figure 5.
γ-iE-DAP stimulates interaction between NOD1 and RIP2, leading to NF-κB activation. (A) Immunoprecipitation of human NOD1 was performed in BEAS-2B whole cell extracts after transfection with flag-tagged NOD1 and treatment with γ-iE-DAP (5 μg/ml). (A) Samples were resolved and then immunoblotted with an anti-RIP2 antibody. Western analysis was also performed with a monoclonal anti-flag antibody (Sigma) and β-actin (lower panels), demonstrating equal amounts of NOD1, specifically in the NOD1-transfected cultures (lanes 3–6) and equal loading in all samples. Negative controls included immunoprecipitation with a mouse IgG isotype control antibody (lane 1) or empty vector transfected cells (lane 2). (B) siRNA Suppression of NOD1 expression: since constitutive levels of NOD1 are barely detectable, we evaluated siRNA efficacy in BEAS-2B exposed to IFN-γ. Compared with the scrambled control siRNA, siRNA targeted to NOD1 resulted in a significant reduction in IFN-γ–induced protein levels. (C) Next we evaluated the ability of α-iE-DAP and γ-iE-DAP to induce NF-κB–mediated transcription in baseline, siControl, and siNOD1-treated cultures. First, BEAS-2B cultures were transfected with siRNAs, followed by transfection with an NF-κB/luciferase promotor plasmid. At 4 hours after transfection, cells were administered γ-iE-DAP or α-iE-DAP (5 μg/ml each) for 2 hours. Lysates were than analyzed for firefly luciferase activity and normalized to renilla expression. We observed that only γ-iE-DAP treatment resulted in luciferase expression, and this was significantly decreased in cultures treated with siRNA against NOD1 (data are expressed as mean values ± σ of n = 3; P ≤ 0.01). (D) To further validate these findings, BEAS-2B cultures were transiently transfected with siRNA against NOD1 or a scrambled control and then exposed to γ-iE-DAP or α-iE-DAP. Muropeptides were removed after 4 hours and then cells were cultured in normal medium for an additional 20 hours. As shown, only γ-iE-DAP treatment resulted in a significant increase in IL-6 release that was inhibited back down to baseline levels in cells receiving siRNA directed against NOD1 (data are expressed as mean values ± σ of n = 3; P ≤ 0.05).
DISCUSSION
The adult human airway is a sterile compartment that is remarkable considering its vast surface (150 m2), which encounters up to 15,000 liters of air movement each day, containing potentially hundreds of microorganisms (17). To maintain sterility, the mucociliary transport system serves as a front-line defense system that efficiently secures and removes microorganisms independent of pathogen recognition (18). Despite this, invasive organisms pose an imminent threat. In this setting, other components of the innate immune system that specifically recognize pathogens and coordinate a cellular response must come into play. NOD1 is one example of a PRR that is essential for the recognition of γ-iE-DAP, a specific muropeptide derived primarily from the cell wall of gram-negative bacteria (19). The basis of this investigation was to determine a potential mechanism that allows γ-iE-DAP, a molecule that cannot readily traverse the plasma membrane due to its charged nature at physiologic pH, to enter the cytosol, thereby associating with NOD1 and activating the host response. We tested the hypothesis that human PEPT2, a biologically relevant small peptide transporter, recognizes and transports γ-iE-DAP, thereby bridging access to NOD1 and triggering activation of the innate immune response. Here, we established that γ-iE-DAP has affinity for PEPT2 and that translocation of the muropeptide across the apical surface of lung epithelia leads to NOD1-dependent activation of innate immune defense. However, the molecular events that transpire after PEPT2-mediated translocation and presentation of γ-iE-DAP to NOD1 for immune defense activation will require further investigation.
We also demonstrate that PEPT2-mediated transport is selective, since human lung epithelia did not respond to MDP, a muropeptide derived from the cell wall of both gram-negative and gram-positive bacteria. Initially, this was not expected, since we had observed that NOD2, the intracellular PRR responsible for MDP recognition, was constitutively present in lung epithelia. Previous work has shown that MDP is a substrate for PEPT1, a paralogous member of the SLC15A proton oligopeptide transporter family (20). Interestingly, PEPT2 is abundantly expressed throughout the respiratory tract by epithelia and PEPT1 is not (10), whereas the opposite holds true of the gut epithelium. Taken with our findings, we contend that MDP was not immunoreactive due to a lack of cell uptake by lung epithelia. This is further supported by Masumoto and colleagues reporting that intestinal epithelia, cells that naturally express NOD1 but not PEPT2, are unresponsive to γ-iE-DAP (21). However, when the muropeptide was acylated to enhance membrane permeability, cells became responsive. Taken together, an explanation to account for our findings is that the structural specificity of PEPT2, expressed by lung epithelia, precludes recognition and cellular intake of MDP. If this is indeed the case, then it suggests that the respiratory tract, compared with other mucosa such as the gut, is designed to coordinate an offensive strike against gram-negative invaders while ignoring others or, more likely, that it relies on other cellular machinery to facilitate pathogen recognition. It also raises the possibility that selective evolutionary pressure in host defense mechanisms may have led to tissue-specific expression of paralogous membrane proteins.
Additional evidence for muropeptide selectivity is provided by our in silico data. Overall PEPT transporters are remarkably similar, yet they are distinct in function through subtle differences in substrate specificity. In particular, both transporters share steric, electrostatic, hydrophobic, and hydrogen-bonding features, yet the magnitude and localization of these characteristics are spatially distinct (13). We used a simple common-feature pharmacophore model for each transporter to qualitatively map the muropeptides to each transporter. Interestingly, the PEPT1 pharmacophore mapped all three molecules to differing extents, whereas the PEPT2 model was more selective and could only map γ-iE-DAP to all features; selectivity is highlighted by the fact that MDP misses a hydrogen bond donor feature and α-iE-DAP lacks the hydrophobic feature (Figure 1). Although our pharmacophores were generated from a small set of well-characterized substrates for each transporter, it appears that they both capture relevant features necessary for affinity. Thus, our in silico data support the observation that γ-iE-DAP, but not MDP, is recognized by PEPT2, and confirms the findings by Vavricka and colleagues (20) that MDP, but not γ-iE-DAP, is a substrate for PEPT1. It is also noteworthy that substrate recognition, uptake, and transfer by PEPT2 requires co-transport with H+, provided by an acidic extracellular pH (4.5–6.5) that maintains the requisite hydrogen gradient. This is interesting when taking into account that the pH of the respiratory epithelial lining fluid is maintained around 6.5 and likely further acidifies under inflammatory conditions. Therefore, the functional capacity of PEPT2 to recognize and transport bacterial muropeptides may be very well suited for this microenvironment and further enhanced during localized infection.
In summary, we report the selective uptake of γ-iE-DAP by the H+/oligopeptide co-transporter, PEPT2, in human airway epithelial cells. γ-iE-DAP uptake is associated with recognition by NOD1, activation of the NF-κB pro-inflammatory pathway and production of IL-6 and IL-8. Importantly, virtually all bacteria contain a proteoglycan layer within their cell wall, but the amount, location, and specific composition of muropeptides is quite variable. Therefore, the potential for additional interactions between different muropeptides and PEPT2, or other related transporters, likely exists. Given the biological diversity of these systems, we predict that computational modeling can be used as a platform to screen and predict novel host–pathogen interactions, possibly facilitating screening of larger muropeptide libraries. Finally, we propose that identification of this novel auxilliary role for peptide transporters in pathogen recognition may provide opportunities to develop novel therapeutic strategies to counter infection or biological agents targeted at the lung (22).
Acknowledgments
The authors are indebted to Dr. Mark Wewers and Dr. Mikhail Gavrilin for their intellectual support and technical advice. Special thanks to Lifeline of Ohio Tissue Procurement Agency, Dr. Dana Philpott for anti-NOD1 antibody, and Dr. Gabriel Nunez for the Flag-tagged NOD1 plasmid.
This work was supported by National Institutes of Health grants R01 HL86981 (D.L.K.) and R01 DK61425 (P.W.S.).
Originally Published in Press as DOI: 10.1165/rcmb.2008-0059OC on May 12, 2008
Conflict of Interest Statement: None of the authors has a financial relationship with a commercial entity that has an interest in the subject of this manuscript.
References
- 1.Hoffmann JA, Kafatos FC, Janeway CA, Ezekowitz RA. Phylogenetic perspectives in innate immunity. Science 1999;284:1313–1318. [DOI] [PubMed] [Google Scholar]
- 2.Inohara C, McDonald C, Nunez G. NOD-LRR proteins: role in host-microbial interactions and inflammatory disease. Annu Rev Biochem 2005;74:355–383. [DOI] [PubMed] [Google Scholar]
- 3.Inohara N, Koseki T, del Peso L, Hu Y, Yee C, Chen S, Carrio R, Merino J, Liu D, Ni J, et al. Nod1, an Apaf-1-like activator of caspase-9 and nuclear factor-κB. J Biol Chem 1999;274:14560–14567. [DOI] [PubMed] [Google Scholar]
- 4.Inohara N, Ogura Y, Chen FF, Muto A, Nunez G. Human Nod1 confers responsiveness to bacterial lipopolysaccharides. J Biol Chem 2001;276:2551–2554. [DOI] [PubMed] [Google Scholar]
- 5.Tanabe T, Chamaillard M, Ogura Y, Zhu L, Qiu S, Masumoto J, Ghosh P, Moran A, Predergast MM, Tromp G, et al. Regulatory regions and critical residues of NOD2 involved in muramyl dipeptide recognition. EMBO J 2004;23:1587–1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bonizzi G, Karin M. The two NF-κB activation pathways and their role in innate and adaptive immunity. Trends Immunol 2004;25:280–288. [DOI] [PubMed] [Google Scholar]
- 7.Chamaillard M, Hashimoto M, Horie Y, Masumoto J, Qiu S, Saab L, Ogura Y, Kawasaki A, Fukase K, Kusumoto S, et al. An essential role for nod1 in host recognition of bacterial peptidoglycan containing diaminopimelic acid. Nat Immunol 2003;4:702–707. [DOI] [PubMed] [Google Scholar]
- 8.Freeman TC, Bentsen BS, Thwaites DT, Simmons NL. H+/di-tripeptide transporter (PepT1) expression in the rabbit intestine. Pflugers Arch 1995;430:394–400. [DOI] [PubMed] [Google Scholar]
- 9.Bahadduri PM, D'Souza VM, Pinsonneault JK, Sadee W, Bao S, Knoell DL, Swaan PW. Functional characterization of the peptide transporter PEPT2 in primary cultures of human upper airway epithelium. Am J Respir Cell Mol Biol 2005;32:319–325. [DOI] [PubMed] [Google Scholar]
- 10.Groneberg DA, Eynott PR, Doring F, Dinh QT, Oates T, Barnes PJ, Chung KF, Daniel H, Fischer A. Distribution and function of the peptide transporter PEPT2 in normal and cystic fibrosis human lung. Thorax 2002;57:55–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Groneberg DA, Nickolaus M, Springer J, Doring F, Daniel H, Fischer A. Localization of the peptide transporter PEPT2 in the lung: Implications for pulmonary oligopeptide uptake. Am J Pathol 2001;158:707–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ekins S, Johnston JS, Bahadduri P, D'Souza VM, Ray A, Chang C, Swaan PW. In vitro and pharmacophore-based discovery of novel hPEPT1 inhibitors. Pharm Res 2005;22:512–517. [DOI] [PubMed] [Google Scholar]
- 13.Biegel A, Gebauer S, Brandsch M, Neubert K, Thondorf I. Structural requirements for the substrates of the H+/peptide cotransporter PEPT2 determined by three-dimensional quantitative structure-activity relationship analysis. J Med Chem 2006;49:4286–4296. [DOI] [PubMed] [Google Scholar]
- 14.Cornish-Bowden A. A simple graphical method for determining the inhibition constants of mixed, uncompetitive and non-competitive inhibitors. Biochem J 1974;137:143–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hisamatsu T, Suzuki M, Podolsky DK. Interferon-gamma augments CARD4/NOD1 gene and protein expression through interferon regulatory factor-1 in intestinal epithelial cells. J Biol Chem 2003;278:32962–32968. [DOI] [PubMed] [Google Scholar]
- 16.Gutierrez O, Pipaon C, Inohara N, Fontalba A, Ogura Y, Prosper F, Nunez G, Fernandez-Luna JL. Induction of NOD2 in myelomonocytic and intestinal epithelial cells via nuclear factor-kappa b activation. J Biol Chem 2002;277:41701–41705. [DOI] [PubMed] [Google Scholar]
- 17.McCormack FX, Whitsett JA. The pulmonary collectins, SP-A and SP-D, orchestrate innate immunity in the lung. J Clin Invest 2002;109:707–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Widdicombe J. Relationships among the composition of mucus, epithelial lining liquid, and adhesion of microorganisms. Am J Respir Crit Care Med 1995;151:2088–2092. (discussion 2092–2083). [DOI] [PubMed] [Google Scholar]
- 19.McDonald C, Inohara N, Nunez G. Peptidoglycan signaling in innate immunity and inflammatory disease. J Biol Chem 2005;280:20177–20180. [DOI] [PubMed] [Google Scholar]
- 20.Vavricka SR, Musch MW, Chang JE, Nakagawa Y, Phanvijhitsiri K, Waypa TS, Merlin D, Schneewind O, Chang EB. HPepT1 transports muramyl dipeptide, activating NF-kappab and stimulating IL-8 secretion in human colonic Caco2/bbe cells. Gastroenterology 2004;127:1401–1409. [DOI] [PubMed] [Google Scholar]
- 21.Masumoto J, Yang K, Varambally S, Hasegawa M, Tomlins SA, Qiu S, Fujimoto Y, Kawasaki A, Foster SJ, Horie Y, et al. Nod1 acts as an intracellular receptor to stimulate chemokine production and neutrophil recruitment in vivo. J Exp Med 2006;203:203–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paddle BM. Therapy and prophylaxis of inhaled biological toxins. J Appl Toxicol 2003;23:139–170. [DOI] [PubMed] [Google Scholar]