Abstract
Mechanisms regulating myofibroblastic differentiation of fibroblasts within fibroblastic foci in idiopathic pulmonary fibrosis (IPF) remain unclear. Epigenetic processes, including DNA methylation, produce heritable but potentially reversible changes in DNA or its associated proteins and are prominent in development and oncogenesis. We have shown that Thy-1 suppresses myofibroblastic differentiation of lung fibroblasts and that fibroblasts in fibroblastic foci are Thy-1(−). Epigenetic down-regulation of Thy-1 has been demonstrated in cellular transformation and clinical cancer. We hypothesized that epigenetic regulation of Thy-1 affects the lung fibroblast fibrogenic phenotype. RT-PCR, methylation-specific PCR (MSP), and bisulfite genomic sequencing were used to determine the methylation status of the Thy-1 promoter in Thy-1(+) and Thy-1(−) lung fibroblasts, and MSP–in situ hybridization (MSPISH) was performed on fibrotic tissue. Thy-1 gene expression is absent in Thy-1(−) human and rat fibroblasts despite intact Thy-1 genomic DNA. Cytosine-guanine islands in the Thy-1 gene promoter are hypermethylated in Thy-1(−), but not Thy-1(+), fibroblasts. RT-PCR and MSP demonstrate that, in IPF samples in which Thy-1 expression is absent, the Thy-1 promoter region is methylated, whereas in lung samples retaining Thy-1 expression, the promoter region is unmethylated. MSPISH confirms methylation of the Thy-1 promoter in fibroblastic foci in IPF. Treatment with DNA methyltransferase inhibitors restores Thy-1 expression in Thy-1(−) fibroblasts. Epigenetic regulation of Thy-1 is a novel and potentially reversible mechanism in fibrosis that may offer the possibility of new therapeutic options.
Keywords: pulmonary fibrosis, epigenetic processes, Thy-1 antigen, fibroblasts
CLINICAL RELEVANCE
We report reversible epigenetic suppression of Thy-1 in lung fibroblasts in vitro and evidence of Thy-1 promoter hypermethylation in fibroblastic foci in idiopathic pulmonary fibrosis (IPF), suggesting an important role for epigenetic regulation as a novel pathogenic mechanism in IPF.
The most pernicious form of pulmonary fibrosis is idiopathic pulmonary fibrosis (IPF), which is a chronic, fatal pulmonary disorder with an approximately 70% mortality rate 5 years after diagnosis (1). The pathogenesis of IPF is poorly understood. A distinct feature of IPF is the development of fibroblastic foci (FF), which represent areas of active fibrosis (2). The majority of cells within FF are myofibroblasts, which are contractile fibroblasts felt to be the main source of abnormal extracellular matrix production in IPF (3, 4). Pulmonary fibroblasts are heterogeneous; one of the most extensively characterized in vitro models is based on Thy-1 surface expression. Thy-1 (CD90) is a glycosylphophatidylinositol-linked outer membrane leaflet glycoprotein that is expressed on subsets of neural cells, lymphocytes, and fibroblasts (5). The differential expression of Thy-1 in lung fibroblasts affects multiple aspects of the fibrogenic phenotype (5, 6). Thy-1 is an important regulator of cell–cell and cell–matrix interactions and regulates intracellular signaling pathways (7, 8). Most normal human lung fibroblasts are Thy-1(+), whereas in FF the majority of myofibroblasts are Thy-1(−) (9). Studies show that Thy-1(+) and Thy-1(−) subsets have distinct functional properties. Rat lung Thy-1(−) fibroblasts can activate latent TGF-β and express higher levels of α–smooth muscle actin, desmin, myosin, MyoD, myocardin, and myogenin. Thy-1(−) fibroblasts also are more contractile and are more resistant to collagen-induced apoptosis compared with Thy-1(+) rat lung fibroblasts (10, 11).
These differences between Thy-1(+) and Thy-1(−) fibroblasts indicate that Thy-1(−) fibroblasts are more completely differentiated myofibroblasts and that the absence of Thy-1 expression correlates with a more profibrotic myofibroblast phenotype (9). Because most normal lung fibroblasts are Thy-1(+), we investigated the potential mechanisms regulating the emergence of Thy-1(−) fibroblasts in FF. Thy-1 is regulated developmentally and spatially (12) by post-transcriptional mechanisms such as occur in axonal growth in mice and rats (13) or through alterations in steady-state Thy-1 mRNA levels, such as in T-lineage cells in mouse (14). Cell-surface Thy-1 may also be regulated by protein shedding (9). In some types of cancer, such as nasopharyngeal carcinoma, Thy-1 expression can be regulated by the epigenetic mechanism of promoter region methylation (15), in which down-regulation of Thy-1 is associated with a more invasive/metastatic clinical phenotype.
Although extensive studies have been done regarding epigenetic regulation in cancer, few studies have investigated this mechanism in fibrotic diseases. In this study, we show that epigenetic regulation may also play an important role in lung fibrosis. We demonstrate that methylation of the Thy-1 gene promoter region causes loss of Thy-1 gene expression in rat and human lung fibroblasts, whereas methyltransferase inhibition can induce partial re-expression of the gene in vitro. We confirmed this finding by sequencing the promoter region with bisulfite-modified DNA in lung fibroblasts. In archived IPF lung samples, we showed that loss of Thy-1 expression correlates with hypermethylation of the promoter region of the Thy-1 gene. In situ methylation specific PCR in samples from patients with IPF confirmed that hypermethylation of the promoter region of Thy-1 occurs in FF.
MATERIALS AND METHODS
Cell Culture and Tissue Sources
Primary rat lung fibroblasts were cultured as described previously (16). Primary human lung fibroblasts (HNL-24) were cultured from uninvolved human lung tissue obtained at lung resection (49-year-old woman, lung cancer) and used before passage (P) 10; CCL-210 human lung fibroblasts (20-year-old woman, head trauma) purchased from American Type Culture Collection at unknown passage number and used at P < 8 since purchase. Both primary cell lines were sorted for surface expression of Thy-1 by flow cytometry. Human lung tissues were obtained with informed consent at biopsy, autopsy, or lung transplantation at the University of Alabama or the Instituto Nacional de Enfermedades Respiratorias; the diagnosis of IPF is based on the pathologic and clinical history findings, which fulfilled the criteria of the American Thoracic Society and European Respiratory Society (17). All patients with IPF had the pathologic diagnosis of usual interstitial pneumonia (UIP). Ages ranged from 53 to 71 years, and 60% of the patients were female.
Cell Treatments
Cells were seeded in culture flasks 24 hours before treatment. Culture medium with 5 μM 5-aza-2′-deoxycytidine (AZA), a demethylation agent (Sigma, St. Louis, MO) was added to the cells. The medium was changed every 24 hours with freshly added AZA. After 3 days, cells were harvested for total RNA and DNA extraction.
Total RNA, DNA Extraction and Modification, RT-PCR, PCR, and Real-Time RT-PCR
Genomic DNA was extracted from cells and tissues using the DNeasy Tissue kit from Qiagen, and total RNA was extracted using the RNeasy mini kit (Qiagen, Germantown, MD). Bisulfate modification of DNA was performed by using a DNA modification kit (Zymo Research, Orange, CA). Total RNA (1 μg) was used for making cDNA with the Advantage RT-for-PCR kit (Clontech, Mountain View, CA). PCR primers are as previously described (10) or as listed in Table 1. Real-time RT-PCR was performed by using iQ SYBR Green Supermix with a iCycler iQ (Bio-Rad, Hercules, CA); each sample was performed in triplicate and normalized to GAPDH as previously described (10). A linear regression approach was used to estimate input mRNA in comparison to that of GAPDH without the need for assumption of equal PCR efficiencies (18).
TABLE 1.
PRIMER SETS FOR PCR AND REAL-TIME RT-PCR
| Primer Sets | |
|---|---|
| Real-time RT-PCR | |
| Rat GAPDH | F 5′-ATGCCGCCTGGAGAAACC-3′ |
| R 5′-AGAATGGGAGTTGCTGTTGAAG-3′ | |
| Rat Thy-1 | F 5′-GCCTGACCCGAGAGAAGAAGAAG-3′ |
| R 5′-TGGTGGTGAAGTTCGCTAGAGTAAG-3′ | |
| Rat DNMT1 | F 5′-AACCTACCACGCCGACATC-3′ |
| R 5′-GCCGCCCTGTGAGTAATCC-3′ | |
| Rat DNMT3a | F 5′-CCAAGTGGACCGCTACATC-3′ |
| R 5′-ACTCCTGGATATGCTTCTGTG-3′ | |
| Rat DNMT3b | F 5′-ACAGCGGAGGATGCCAAGC-3′ |
| R 5′-TCTTCCAGGTCAGGATCAGTAGTG-3′ | |
| Rat MBD1 | F 5′-AAGGGAGAGACGCAGGACATTG-3′ |
| R 5′-AGGCAGCAGATGGAGCACAC-3′ | |
| Rat Mecp2 | F 5′-GAAGAGTGGAAAGGGACTGAAGAC-3′ |
| R 5′-GCGTGATGGTGGTGATGATGG-3′ | |
| Human Thy-1 | F 5′-CCTGACCCGTGAGACAAAGAAG-3′ |
| R 5′-GCTAGTGAAGGCGGATAAGTAGAG-3′ | |
| PCR | |
| Rat Thy-1 | F 5′-GCGAGCCTCAACCTCACTG-3′ |
| R 5′-CACCTGACTCTGCCTCCTAAG-3′ | |
| Human Thy-1 | F 5′-AGAGCCTTCGTCTGGACTG-3′ |
| R 5′-TGGTTCGGGAGCGGTATG-3′ |
Sequencing of the Thy-1 Promoter
For analysis of the Thy-1 promoter region for possible aberrations such as insertion/deletion or mutation leading to loss of Thy-1 expression, we amplified the promoter regions of rat and human Thy-1 from genomic DNA extracted from the Thy-1(+)/Thy-1(−) cell lines. The primers for sequencing are rat F 5′-AAGAGGAGGCTGCAAGCTAG-3′, R 5′-TTGGAACTCGTGAGGTAGCC-3′; human F 5′-ACTAGGCAGGGATGAGCAAG-3′, R 5′-ACGAAGGCTCTGGTCCACTA-3′. The PCR products were amplified and cleaned with a PCR Purification kit (Qiagen) and directly sequenced at the nucleic acid sequencing core facility at the University of Alabama.
Bisulfite Conversion and Methylation-Specific PCR
Bisulfite converts unmethylated cytosine into uracil, whereas methylated cytosine is left unchanged. Extracted DNA samples (Qiagen DNeasy tissue kit) were bisulfite-converted using the EZ DNA Methylation-Gold kit (Zymo Research) according to the manufacturer's instructions. The methylation status of Thy-1 in the rat and human Thy-1(+) and Thy-1(–) lung fibroblasts and IPF lung tissues were investigated by methylation-specific PCR (MSP). The sequences of the Thy-1 promoter region of rat and human were retrieved from Ensembl (www.ensembl.org) with rat Thy-1 ID ENSRNOG00000006604 and human Thy-1 ID ENSG00000154096. GC-rich regions of Thy-1 promoter region were identified by Methprimer software (19) to fulfill the criteria of a CpG island in rat and human gene sequences, and appropriate primers were designed. MSP primer sets for rat and human Thy-1 promoter CpG islands in methylated (M) or unmethylated (U) states are as shown in Table 2, and their positions within the Thy-1 promoter are shown in Figure 2 (closed arrows). All PCRs were performed under the following conditions: 95°C for 5 minutes, then 38 cycles of 95°C for 30 seconds, 60°C for 30 seconds, and 72°C for 30 seconds, followed by 5 minutes of 72°C extension.
TABLE 2.
METHYLATION-SPECIFIC PCR AND BISULFITE GENOMIC SEQUENCING (BGS) PRIMER SETS
| Primer Sets | |
|---|---|
| Rat M | F 5′-AGTTTTCGTCGGGTGATTTC-3′ |
| R 5′-CAAAAACTCAATTTATTTCTAACGCT-3′ | |
| Rat U | F 5′-GAGTTTTTGTTGGGTGATTTTGT-3′ |
| R 5′-AAAAACTCAATTTATTTCTAACACT-3′ | |
| Rat BGS | F 5′-AGTTTTTGGTTTTGTTTTTTATTTAGAG-3′ |
| R 5′-ACTAACCCCACCCTAATACTATTTC-3′ | |
| Human M | F 5′-TATTTTTATATTAATGCGGGATCGT-3′ |
| R 5′-ACTAACCCACCCTAATACTATTTC-3′ | |
| Human U | F 5′-TTATTTTTATATATTAATGTGGGATTGT-3′ |
| U 5′-CCACCTAAACTAAAATCTTCCACT-3′ |
Definition of abbreviations: M = methylated; U = unmethylated.
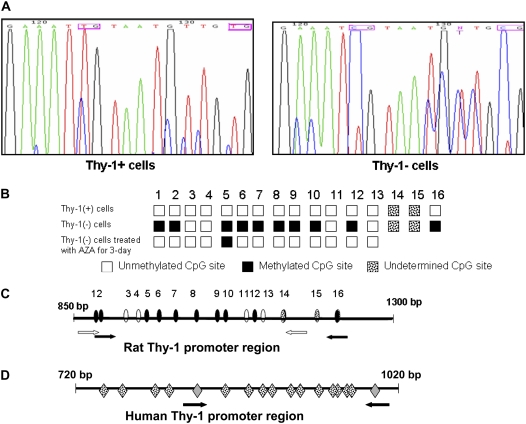
Figure 2.
Methylation status of Thy-1 promoter in Thy-1(−) and Thy-1(+) rat lung fibroblasts. (A) Bisulfite genomic sequencing (BGS) of Thy-1 promoter region using bisulfite modified DNA. In this representative portion of the promoter sequence, two CpG sites are methylated in Thy-1(−) rat fibroblasts, as indicated by T to C shift (pink boxes) (see Materials and Methods). (B) Schematic illustration of the rat Thy-1 promoter region subjected to methylation analysis, based on sequence data from Thy-1(+) cells and Thy-1(−) cells +/− treatment with AZA for 3 days. White boxes: unmethylated CpG; black boxes: methylated CpG; spotted boxes: undetermined sequence. MSP primers located at number 1 and 2 CpG islands of Forward primer and number 16 CpG of the Reverse primer. (C) The CpG island of the rat Thy-1 promoter region as determined by BGS and MSP; white arrows indicate the positions of the rat BGS primers, and black arrows indicate positions of the MSP primers. Ovals indicate CpG sites (white are unmethylated; black are methylated) as determined by BGS. Spotted ovals indicate additional CpG sites of unknown methylation status, which are within the amplified MSP product but not within region sequenced for BGS; the last one of these (number 16) is positioned within the reverse MSP primer. (D) The CpG island of human Thy-1 promoter region; mRNA starts at 1017 bp. Arrows indicate the positions of the MSP primers. Gray diamonds are CpG sites within the MSP primer sequences. Spotted diamonds indicate CpG islands (methylation status unknown.
Bisulfite Genomic Sequencing
Bisulfite-converted DNA samples were subjected to PCR with primers that did not discriminate between methylated and unmethylated sequences, and the cleaned PCR products were subjected to sequencing directly. The bisulfite genomic sequencing primers for rat are as shown in Table 2, and their positions within the Thy-1 promoter are shown in Figure 2C (open arrows). The PCR reaction conditions are described above.
Immunofluorescence
Immunofluorescence microscopy was performed as previously described (10).
Immunohistochemistry and MSP In Situ Hybridization
Immunohistochemistry (IHC) was performed as previously described (9, 20). The sections were prepared from paraffin-embedded tissues from patients with IPF. The MSP in situ hybridization (MSP-ISH) protocol was as published (20). Briefly, the paraffin-embedded samples from patients with IPF were digested with pepsin (2 mg/ml in 0.1 M HCl) for 30 minutes, washed in water for 1 minute, and air dried. The samples were incubated in 0.2 M NaOH for 10 minutes at 37°C then placed in 3 M sodium bisulfite solution and incubated at 55°C for 15 hours followed by incubation in 0.3 M NaOH for 5 minutes.
The PCR in situ hybridization protocol was followed as in Ref. 21. After 3 minutes at 94°C for denaturation, 35 cycles were performed at 94°C for 1 minute and 55°C for 1.5 minutes. One section of each slide was analyzed with 1-μM primers specific for methylated Thy-1 sequence, another section was analyzed with unmethylated primers, and third section was analyzed with no primers as control. After amplification, ISH was performed using a methylated-specific internally digoxigenin-labeled probe (1 μg/ml) diluted with Hybrisol VII (Oncor, Gaithersburg, MD). This corresponded to the M product generated by methylation-specific PCR (MSP) as described above. The amplicon and the probe were co-denatured at 95°C for 5 minutes, hybridized at 37°C for 2 hours, and washed in 1× SSC (0.15 M sodium chloride/0.015 M sodium citrate [pH7]) with 2% BSA for 10 minutes at 52°C, incubated with the anti-digoxigenin alkaline phosphatase conjugate (1:200; Roche Molecular Biochemicals, Basel, Switzerland), and exposed to the chromogen nitroblue tetrazolium and 5-bromo-4-chloro-3-indolyl phosphate (Enzo Diagnostics, Farmingdale, NY) at 37°C. The final counter stain, nuclear fast red, stains the negative cells pink in contrast to the blue signal. To determine the relative degree of Thy-1 expression or promoter hypermethylation detected by IHC, ISH, or MSP-ISH, a semiquantitative scoring system was used. For each sample, 10 high-power (400×) fields (HPFs) were examined by an experienced pathologist (GJN) who was blinded to the final diagnosis. Samples were scored as follows: 0 = no positive cells, 1 = 1 to 10 positive cells/HPF, 2 = 11 to 20 positive cells/HPF, and 3 = more than 20 cells/HPF.
Statistical Analysis
Comparisons involving three or more groups were analyzed using one-way ANOVA (Newman-Keuls method for multiple comparisons). A P value of 0.05 was used to determine statistical significance.
RESULTS
Thy-1 Genomic DNA Is Intact in Human IPF Tissues and Thy-1(−) Fibroblasts; Thy-1 Expression in IPF Tissues and Cultured Fibroblasts Corresponds to the Methylation Status of the Thy-1 Promoter
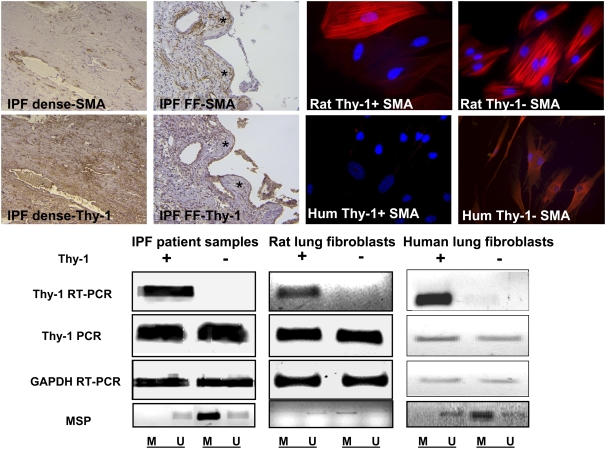
We previously demonstrated that Thy-1 is absent in the FF of IPF and in Thy-1(−) lung fibroblasts and that the absence of Thy-1 correlates to myofibroblastic differentiation (9, 10). In areas of dense fibrosis where myofibroblasts are scant, Thy-1 is expressed in fibroblasts, whereas in fibroblastic foci, Thy-1 is absent from myofibroblasts and is present in epithelium (Figure 1, IHC panels at upper left). To test the possibility that mutation of the Thy-1 gene sequence leads to the reduced or absent Thy-1 expression in these situations, we sequenced the promoter region of Thy-1 in rat lung fibroblasts. There is no difference in the gene sequence in Thy-1+ and Thy-1(−) cells (data not shown). PCR for genomic DNA shows that the Thy-1 gene is intact in the Thy-1(−) cells and tissues as shown in DNA gels in Figure 1, whereas RT-PCR demonstrates Thy-1 mRNA only in Thy-1(+) rat and human lung fibroblasts and in human samples expressing Thy-1. The results exclude the possibility that the absence of Thy-1 expression in the cells or tissues is due to genetic deletion or mutation. To investigate the mechanism of the absence of expression of Thy-1, we analyzed whether promoter region hypermethylation could be responsible for the down-regulation of Thy-1 expression. MSP on DNA extracted from archived specimens indicated that the samples in which Thy-1 expression was absent demonstrated strong methylation at the promoter region of Thy-1, whereas the ones in which Thy-1 expression is present are unmethylated (Figure 1, lower left gel images). Because of the heterogeneous nature of fibrosis in IPF, fibroblastic foci are often admixed with areas of relatively normal lung tissue and areas of dense inactive fibrosis in biopsy samples. Accordingly, some of the samples contained methylated PCR and unmethylated PCR product, indicating heterogeneity within tissues (faint U band in the Thy-1(−) sample; Figure 1, lower left). Our results in cultured fibroblasts show that the promoter region of Thy-1 in Thy-1(−) rat lung fibroblasts is predominantly methylated (M band in Figure 1; lower middle panel), whereas in Thy-1(+) cells it is predominantly unmethylated (U band), as is the case in human Thy-1(+) and Thy-1(−) lung fibroblasts (Figure 1, lower right panels).
Figure 1.
Upper left: The four micrographs demonstrating immunohistochemistry staining (original magnification: ×200) for α-smooth muscle actin (SMA) and Thy-1 on serial sections from patients with idiopathic pulmonary fibrosis (IPF), demonstrating areas of dense fibrosis (IPF dense) or fibroblastic foci (IPF FF). FF are indicated by asterisks. Upper right: Four fluorescence micrographs demonstrating rat or human Thy-1(+) or Thy-1(−) lung fibroblasts stained with antibody to SMA conjugated to Cy3 (original magnification: ×400). Nuclei are counterstained with Hoechst reagent. PCR and RT-PCR of Thy-1 from Thy-1(+) and Thy-1(−) rat and human lung fibroblasts and representative samples from patients with IPF (positive or negative for Thy-1 expression) are shown. Lower panel: DNA gels stained with ethidium bromide, demonstrating results of RT-PCR analysis of Thy-1 expression, PCR amplification of Thy-1 genomic DNA, RT-PCR for glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as a control, and methylation-specific PCR (MSP) using primers described in Table 2 and depicted in Figures 2C and 2D. MSP products are shown in the lowermost panels. M and U refer to primers specific for methylated and unmethylated promoter fragments, respectively.
Specific CpG Sites within CpG Islands in the Thy-1 Promoter Region Are Hypermethylated in Thy-1(−) Fibroblasts
To confirm our findings, we performed direct bisulfite sequencing on rat lung fibroblasts. Bisulfite chemically converts unmethylated cytosine to uracil but has no effect on methylated cytosine (Figure 2A) (22). Our findings demonstrate that 9 out of 13 CpG sites in the Thy-1 promoter region we sequenced are hypermethylated in Thy-1(−) fibroblasts (Figure 2B). Two of the sites shown are partially methylated (>50%)( number 1 and 10 CpG sites in Figure 2B). The CpG sites in the rat and human promoter, their methylation status if known, and the primers used for MSP and bisulfite sequencing are depicted in Figures 2C and 2D.
Immunohistochemistry and In Situ MSP
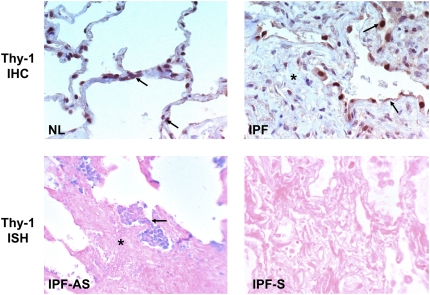
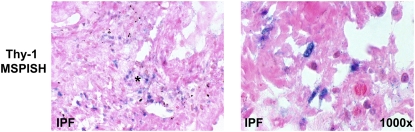
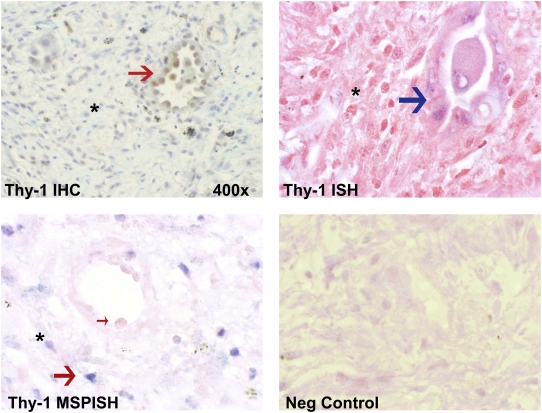
To more clearly explore Thy-1 methylation status relative to Thy-1 expression specifically within FF in patients with IPF, we performed IHC, ISH, and MSP-ISH (21). The latter technique can demonstrate the methylation status of the promoter region of Thy-1 within specific histologic regions and can be compared with Thy-1 mRNA expression by ISH and to Thy-1 protein expression by IHC within adjacent sections. IHC in normal lung demonstrates Thy-1 expression in elongated cells within alveolar septae corresponding to the location of interstitial fibroblasts, consistent with our previous studies (Figure 3, upper left). In IPF, Thy-1 is absent from mesenchymal cells within fibrotic areas but is expressed in the epithelium overlying fibrotic foci (Figure 3, upper right). ISH confirms the absence of Thy-1 expression in mesenchymal cells and its presence in the epithelial cells overlying FF (Figure 3, middle left). The MSP-ISH findings indicate that within mesenchymal cells in FF, the absence of Thy-1 expression by IHC and ISH correlates to the presence of hypermethylation in the Thy-1 promoter (blue nuclei in spindle-shaped cells; Figure 3, lower panels). Six cases of IPF and three normal control lungs were examined for Thy-1 expression and Thy-1 promoter hypermethylation in situ by IHC, ISH, and MSP-ISH. Within FF, Thy-1 scoring by IHC and ISH was 0 for all samples, whereas in regenerating epithelium overlying FF, the Thy-1 scoring by IHC and ISH was 1 to 2+. Normal lung tissue had Thy-1 scores of 1 to 2+ in alveolar septae. By MSP-ISH, Thy-1 promoter methylation was 2 to 3+ in FF but was 0 in regenerating epithelium and in normal alveoli. Figure 4 demonstrates IHC, ISH, and MSP-ISH in the same area in serial sections from one patient with IPF, again demonstrating Thy-1 protein and mRNA in epithelium overlying fibroblastic foci (Figure 4, red and blue arrows, upper panels) but no Thy-1 in fibroblasts within FF. MSP-ISH shows Thy-1 promoter hypermethylation in fibroblastic cells (Figure 4, large red arrow, lower left) but not in overlying epithelium (Figure 4, small arrow).
Figure 3.
Thy-1 expression by immunohistochemistry (IHC) (upper panels) and in situ hybridization (ISH) (middle panels) in normal (NL) and IPF lung samples. Arrows indicate Thy-1 expression in elongated alveolar septal cells in NL and in abnormal epithelium overlying fibroblastic foci (asterisk) in IPF. AS and S (middle panels) refer to antisense and sense (negative control) probes for ISH, respectively. Lower panels: Thy-1 promoter hypermethylation in IPF demonstrated by MSP-in situ hybridization (MSP-ISH). Blue nuclear signal indicates hypermethylation signal in elongated cells within fibroblastic foci (asterisk). (Original magnification: ×400, except where indicated.)
Figure 4.
IHC, ISH, and MSP-ISH on serial sections from the same patient. Upper left: IHC shows Thy-1 protein in epithelial cells (red arrow) overlying fibroblastic focus (asterisk). Upper right: ISH shows Thy-1 mRNA expressed in epithelial cells (blue arrow). Lower left: MSP-ISH shows Thy-1 promoter methylation in fibroblastic cells within fibroblastic foci (large red arrow) but absent from epithelial cells (small arrow). (Original magnification: ×1,000, except where indicated.)
Thy-1 Is Re-expressed in Thy-1(−) Cells after Demethylation Treatment
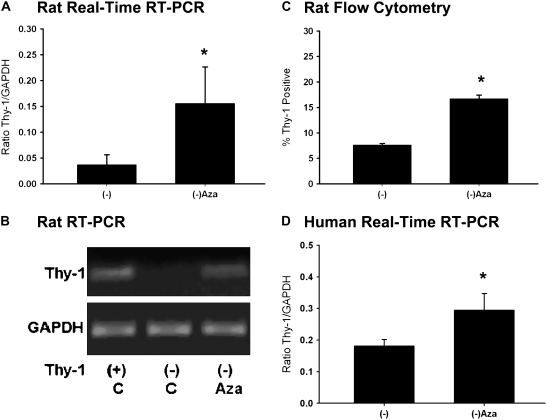
To determine whether interfering with epigenetic mechanisms could induce re-expression of Thy-1 in vitro, we treated Thy-1(−) cells with AZA, which can inhibit de novo and maintenance methylation (23). AZA treatment leads to the re-expression of Thy-1 mRNA in Thy-1(−) rat lung fibroblasts (Figures 5A and 5B). We confirmed the demethylation effect of AZA by bisulfite sequencing after treating the cells with AZA (see Figure 2B), which indicated that only one CpG site remains methylated, compared with nine methylated CpG sites without treatment. The expression of Thy-1 protein on the cell surface also increased after treatment with AZA, as indicated by flow cytometry using CD90 (Thy-1) antibody (Figure 5C). Human Thy-1(−) lung fibroblasts showed results similar to the Thy-1(−) rat lung fibroblasts (Figure 5D). The mostly Thy-1(−)HNL-24 human lung fibroblasts demonstrated promoter hypermethylation by MSP; after treatment with AZA, the cells had increased Thy-1 expression compared with untreated cells (Figure 5D). These results suggested that the decreased expression of Thy-1 in Thy-1(−) cells was related to reversible epigenetic mechanisms, including DNA promoter region methylation.
Figure 5.
Increased Thy-1 expression in Thy-1(−) cells after demethylation treatment. (A) Histogram of real-time RT-PCR results for Thy-1 expression in rat lung fibroblasts by standard curve method; 5 μM AZA treated Thy-1(−) rat lung fibroblasts (3 d) compared with control (DMSO). (B) Representative RT-PCR results. (C) Thy-1 expression on cell surface by flow cytometry in Thy-1(−) rat lung fibroblasts and Thy-1(−) cells treated with AZA for 3 days. Cells were stained with FITC-conjugated anti-CD90 (Thy-1) antibody. (D) Histogram of real-time RT-PCR results for Thy-1 expression in human lung fibroblasts; 5 μM AZA treated Thy-1(−) human lung fibroblasts (3 days) compared with control (−). Experiments were performed in triplicate, and data are expressed as the ratio of Thy-1 to GAPDH by standard curve method. *P < 0.05 versus Thy-1(−) by ANOVA.
mRNA Expression Levels of DNA Methyltransferases and Methyl Binding Proteins Do Not Correlate with Thy-1 Promoter Region Methylation
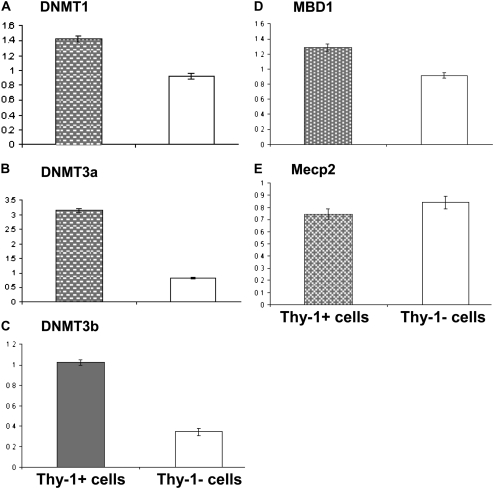
The underlying mechanisms for the Thy-1 promoter region demethylation or methylation in Thy-1(+) or Thy-1(−) cells, respectively, are unclear. We compared the mRNA expression levels of DNA methyltransferases (DNMTs) and methyl-CpG–binding domain proteins (MBD) to investigate the possible mechanism for the observed methylation differences. By real-time RT-PCR normalized to GAPDH, DNMT expression is unexpectedly higher in Thy-1(+) cells than the Thy-1(−) cells (Figure 6). The expression of two principal MBD does not differ significantly between the subsets. This suggests the methylation pattern of Thy-1 promoter region cannot be attributed to the differences in expression of methyltransferases and/or methyl-binding proteins at the mRNA level.
Figure 6.
Expression of DNA methyltransferases (DNMTs) and methyl-CpG-binding domain proteins (MBD) by real-time RT-PCR in Thy-1(+) and Thy-1(−) rat lung fibroblasts. (A) DNMT1. (B) DNMT3a. (C) DNMT3b. (D) MBD1. (E) Mecp2. Experiments were performed in triplicate, and data are expressed as ratio of target to GAPDH by standard curve method. Primers and RT-PCR conditions were as described in Materials and Methods.
DISCUSSION
Molecular mechanisms for the emergence and/or persistence of myofibroblasts within the fibroblastic foci of IPF remain unclear. Our previous findings in vitro and in the bleomycin model and within IPF tissues indicate that the absence of Thy-1 expression in lung fibroblasts correlates with a more profibrotic myofibroblast phenotype, as shown here in Figure 1 (9, 11, 24). The mechanisms by which Thy-1 expression is regulated in fibroblasts are unclear. We previously demonstrated reversible shedding of Thy-1 associated with exposure of fibroblasts to inflammatory cytokines. However, inflammation is not a consistent feature of fibrotic disorders such as IPF (2). Loss of expression can result from genetic mutations, such as deletions or insertions, or from epigenetic modifications including promoter hypermethylation.
Hypermethylation of DNA at cytosine residues in CpG islands led to heritable gene silencing via the formation of a repressive chromatin structure (25). Epigenetic events affect gene expression without affecting the gene sequence itself and are believed to be a primary mechanism causing tumor suppressor genes to be inactive and causing certain cancers (26). Aberrant promoter methylation is a common mechanism for the transcriptional inactivation of certain genes in many types of cancer, including tumor suppressor genes, cell regulatory genes, and apoptosis-related genes. For example, the tumor suppressor gene p16 is hypermethylated in many tumor types, including colorectal, lung. and breast cancers (27).
The absence of Thy-1 expression in a rat T-cell lymphoma cell line has been shown to be associated with DNA hypermethylation in the promoter region and is reversible upon exposure to 5-azadeoxycytidine (28). Thy-1 has been proposed as a tumor suppressor gene in nasopharyngeal carcinoma (15) and is down-regulated through promoter region methylation, associated with a more invasive/metastatic clinical phenotype. In this study, we have demonstrated that the absence of Thy-1 expression in Thy-1(−) primary lung fibroblasts and in IPF samples can be attributed in part to epigenetic regulation through Thy-1 promoter hypermethylation (Figures 1–4). Demethylation in the Thy-1(−) cells restores Thy-1 gene expression (Figure 5). Human and rat lung fibroblasts demonstrate similar results.
DNA methylation is regulated in part by DNMT enzymes. This family includes DNMT1, which has maintenance DNA methylase activity; and DNMT3a and DNMT3b, which are de novo methyltransferases (22). Over-expression of DNMTs is one possible mechanism of gene hypermethylation (29). Methyl-CpG-binding proteins are a group of proteins thought to inhibit the binding of transcription factors to gene promoters and thus are proposed as another mechanism of transcription inhibition by hypermethylation (30). Although there are reports that DNMTs are overexpressed (31, 32), other studies demonstrate no association between the mRNA expression levels of DNMTs and methyl-CpG–binding proteins and the methylation status of tumor-suppressed genes (33, 34). In this study, we found neither DNMT1, -3a, -3b, nor MBD1 or Mecp2 methyl-binding proteins to be overexpressed in Thy-1(−) cells. In fact, Thy-1(+) had higher levels of mRNA for the methyltransferases. This was unexpected because DNMT expression, especially DNMT3a and -3b, is usually associated with hypermethylation (29). However, the regulation of methylation by DNMTs is complex and involves multiple molecular interactions, such that expression of a particular DNMT at the mRNA level may not correlate with hypermethylating activity for a particular gene promoter (35). Previous studies in colorectal cancer (33) and hepatocellular carcinoma (36) failed to show any significant correlation between methylation status and the DNMT mRNA expression levels. Also, because we analyzed serially passaged cell populations from established cultures, it is possible that transiently up-regulated DNMTs and/or MBD in a previous generation determined the aberrant gene methylation status, which is heritable in subsequent cell generations, and that to maintain the pattern, relatively low enzyme expression would be sufficient. Nevertheless, the demethylation of all but one CpG site in the Thy-1 promoter (Figure 2B) and the reversibility of Thy-1 expression with AZA strongly suggest that methyltransferase activity is involved in maintaining hypermethylation in Thy-1(−) fibroblasts.
Other well described epigenetic mechanisms involving chromatin modifications, such as histone deacetylation, can result in gene silencing (37). Recent studies have shown that methylation and histone modifications are related (38). More and more studies have shown that methylation of gene promoters is only one of many layers of repressive mechanisms (39–41). We also investigated whether histone deacetylation contributes to the silencing of Thy-1 gene expression: We treated the Thy-1(−) cells with trichostatin A for 72 hours, which also resulted in re-expression of Thy-1 (data not shown; studies ongoing), suggesting that promoter region methylation and histone deacetylation contribute to the silencing of Thy-1 in Thy-1(−) lung fibroblasts. Other factors, such as transcription factors, may also be affected by AZA or trichostatin A and could promote re-expression of Thy-1. The direct bisulfite sequencing of the promoter region of Thy-1 after treatment with AZA indicates that in Thy-1(−) cells, promoter region demethylation is associated with the re-expression of Thy-1 (Figures 2B and 5). By removal of methyl groups, certain genes can undergo significant changes in structure characterized by partial re-acetylation of histone H3 and H4 and re-methylation of H3(K4), but in other genes, the removal of DNA methylation does not result in appreciable histone reacetylation (40). Further investigation of the role of histone acetylation in Thy-1(−) cells is ongoing.
The FF within the IPF specimens showed down-regulation or loss of Thy-1 expression by IHC and ISH, whereas the normal samples showed normal expression of Thy-1 (Figures 3 and 4). This is consistent with our pervious findings (9). The in situ MSP findings indicate that the loss of Thy-1 expression within the FF in patients with IPF may be caused by promoter region hypermethylation of Thy-1 (Figures 3 and 4). It is possible to speculate that patients in whom Thy-1 expression in fibroblasts is lost by epigenetic regulation may be more prone to develop IPF. Alternately (or in addition), acute inflammation could result in Thy-1 protein shedding associated with additional migration/accumulation of Thy-1(−) myofibroblasts, with persistence of latent TGF-β activation, excessive matrix accumulation, and resistance to apoptosis (9–11). The demonstration of Thy-1 expression in epithelial cells overlying fibroblastic foci (Figures 1, 3, and 4) has not been previously reported. Although the significance of epithelial Thy-1 expression is unclear, it could be suggestive of epithelial-mesenchymal transition because Thy-1 is expressed on normal lung fibroblasts or could indicate emergence of a progenitor cell phenotype because Thy-1 is expressed in most stem cells (7, 8).
Taken together with our prior studies, the findings reported here suggest that Thy-1 may function as a “fibrosis suppressor” gene. The reversibility of epigenetic suppression of Thy-1 suggests that it may be possible to restore Thy-1 expression in fibroblasts, altering the profibrotic phenotype of the cells and possibly the clinical outcome in patients with IPF. This is the first report of an epigenetic mechanism causing the silencing of a specific gene in IPF. Transcriptional silencing by CpG island methylation is a common mechanism of silencing of tumor suppressors in cancer (26). There are reports that in liver fibrosis, loss of expression of certain genes is associated with promoter region hypermethylation (42, 43). More experiments are needed to unravel how DNA methylation together with other repression mechanisms, such as histone deacetylation, may generate distinct patterns of gene silencing in IPF. These studies could give us novel insights into the causes of pulmonary fibrosis and open the possibility of novel therapeutic approaches to this fatal disease.
This work was supported in part by The Rud Polhill Senior Faculty Award from the Children's Center for Research and Innovation, R01 HL082818 from the National Heart, Lung and Blood Institute (J.S.H.), Universidad Nacional Autónoma de México Grant SDI.PTID.05.6 (M.S. and A.P.), and Research Facilities Improvement Program Grant No. C06RR 15,490 from the National Center for Research Resources (A.L.).
Originally Published in Press as DOI: 10.1165/rcmb.2007-0322OC on June 12, 2008
Conflict of Interest Statement: None of the authors has a financial relationship with a commercial entity that has an interest in the subject of this manuscript.
References
- 1.Strieter RM, Gomperts BN, Keane MP. The role of CXC chemokines in pulmonary fibrosis. J Clin Invest 2007;117:549–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pardo A, Selman M. Idiopathic pulmonary fibrosis: new insights in its pathogenesis. Int J Biochem Cell Biol 2002;34:1534–1538. [DOI] [PubMed] [Google Scholar]
- 3.Kuhn C, Boldt J, King TE, Crouch E, Vartio T, McDonald JA. An immunohistochemical study of architectural remodeling and connective tissue synthesis in pulmonary fibrosis. Am Rev Respir Dis 1989;140:1693–1703. [DOI] [PubMed] [Google Scholar]
- 4.Phan SH. Fibroblast phenotypes in pulmonary fibrosis. Am J Respir Cell Mol Biol 2003;29(3, Suppl)S87–S92. [PubMed] [Google Scholar]
- 5.Rege TA, Hagood JS. Thy-1 as a regulator of cell-cell and cell-matrix interactions in axon regeneration, apoptosis, adhesion, migration, cancer, and fibrosis. FASEB J 2006;20:1045–1054. [DOI] [PubMed] [Google Scholar]
- 6.Hagood JS, Miller PJ, Lasky JA, Tousson A, Guo B, Fuller GM, McIntosh JC. Differential expression of platelet-derived growth factor-alpha receptor by Thy-1(−) and Thy-1(+) lung fibroblasts. Am J Physiol 1999;277:L218–L224. [DOI] [PubMed] [Google Scholar]
- 7.Haeryfar SM, Hoskin DW. Thy-1: more than a mouse pan-T cell marker. J Immunol 2004;173:3581–3588. [DOI] [PubMed] [Google Scholar]
- 8.Rege TA, Hagood JS. 2006. Thy-1, a versatile modulator of signaling affecting cellular adhesion, proliferation, survival, and cytokine/growth factor responses. Biochim Biophys Acta 2006;1763:991–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hagood JS, Prabhakaran P, Kumbla P, Salazar L, MacEwen MW, Barker TH, Ortiz LA, Schoeb T, Siegal GP, Alexander CB, et al. Loss of fibroblast Thy-1 expression correlates with lung fibrogenesis. Am J Pathol 2005;167:365–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sanders YY, Kumbla P, Hagood JS. Enhanced myofibroblastic differentiation and survival in Thy-1(-) lung fibroblasts. Am J Respir Cell Mol Biol 2007;36:226–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou Y, Hagood JS, Murphy-Ullrich JE. Thy-1 expression regulates the ability of rat lung fibroblasts to activate transforming growth factor-{beta} in response to fibrogenic stimuli. Am J Pathol 2004;165:659–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Spanopoulou E, Giguere V, Grosveld F. Transcriptional unit of the murine Thy-1 gene: different distribution of transcription initiation sites in brain. Mol Cell Biol 1988;8:3847–3856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xue GP, Calvert RA, Morris RJ. Expression of the neuronal surface glycoprotein Thy-1 is under post-transcriptional control, and is spatially regulated, in the developing olfactory system. Development 1990;109:851–864. [DOI] [PubMed] [Google Scholar]
- 14.Wajeman-Chao SA, Lancaster SA, Graf LH Jr, Chambers DA. Mechanism of catecholamine-mediated destabilization of messenger RNA encoding Thy-1 protein in T-lineage cells. J Immunol 1998;161:4825–4833. [PubMed] [Google Scholar]
- 15.Lung HL, Bangarusamy DK, Xie D, Cheung AK, Cheng Y, Kumaran MK, Miller L, Liu ET, Guan XY, Sham JS, et al. THY1 is a candidate tumour suppressor gene with decreased expression in metastatic nasopharyngeal carcinoma. Oncogene 2005;24:6525–6532. [DOI] [PubMed] [Google Scholar]
- 16.McIntosh JC, Hagood JS, Richardson TL, Simecka JW. Thy1 (+) and (−) lung fibrosis subpopulations in LEW and F344 rats. Eur Respir J 1994;7:2131–2138. [DOI] [PubMed] [Google Scholar]
- 17.American Thoracic Society. Idiopathic pulmonary fibrosis: diagnosis and treatment. International consensus statement. American Thoracic Society (ATS), and the European Respiratory Society (ERS). Am J Respir Crit Care Med 2000;161:646–664. [DOI] [PubMed] [Google Scholar]
- 18.Ramakers C, Ruijter JM, Deprez RH, Moorman AF. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci Lett 2003;339:62–66. [DOI] [PubMed] [Google Scholar]
- 19.Li LC, Dahiya R. MethPrimer: designing primers for methylation PCRs. Bioinformatics 2002;18:1427–1431. [DOI] [PubMed] [Google Scholar]
- 20.Nuovo GJ. Methylation-specific PCR in situ hybridization. In T. O. Tollefsbol, editor. Epigenetics protocols. Totowa, NJ: Humana Press; 2004. pp. 261–271.
- 21.Nuovo GJ, Plaia TW, Belinsky SA, Baylin SB, Herman JG. In situ detection of the hypermethylation-induced inactivation of the p16 gene as an early event in oncogenesis. Proc Natl Acad Sci USA 1999;96:12754–12759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Singal R, Ginder GD. DNA methylation. Blood 1999;93:4059–4070. [PubMed] [Google Scholar]
- 23.Jones PA. Altering gene expression with 5-azacytidine. Cell 1985;40:485–486. [DOI] [PubMed] [Google Scholar]
- 24.Rege TA, Pallero MA, Gomez C, Grenett HE, Murphy-Ullrich JE, Hagood JS. Thy-1, via its GPI anchor, modulates Src family kinase and focal adhesion kinase phosphorylation and subcellular localization, and fibroblast migration, in response to thrombospondin-1/hep I. Exp Cell Res 2006;312:3752–3767. [DOI] [PubMed] [Google Scholar]
- 25.Kass SU, Pruss D, Wolffe AP. How does DNA methylation repress transcription? Trends Genet 1997;13:444–449. [DOI] [PubMed] [Google Scholar]
- 26.Feinberg AP, Ohlsson R, Henikoff S. The epigenetic progenitor origin of human cancer. Nat Rev Genet 2006;7:21–33. [DOI] [PubMed] [Google Scholar]
- 27.Esteller M, Corn PG, Baylin SB, Herman JG. A gene hypermethylation profile of human cancer. Cancer Res 2001;61:3225–3229. [PubMed] [Google Scholar]
- 28.Sneller MC, Gunter KC. DNA methylation alters chromatin structure and regulates Thy-1 expression in EL-4 T cells. J Immunol 1987;138:3505–3512. [PubMed] [Google Scholar]
- 29.Mizuno S, Chijiwa T, Okamura T, Akashi K, Fukumaki Y, Niho Y, Sasaki H. Expression of DNA methyltransferases DNMT1, 3A, and 3B in normal hematopoiesis and in acute and chronic myelogenous leukemia. Blood 2001;97:1172–1179. [DOI] [PubMed] [Google Scholar]
- 30.Fujita N, Takebayashi S, Okumura K, Kudo S, Chiba T, Saya H, Nakao M. Methylation-mediated transcriptional silencing in euchromatin by methyl-CpG binding protein MBD1 isoforms. Mol Cell Biol 1999;19:6415–6426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Belinsky SA, Nikula KJ, Baylin SB, Issa JP. Increased cytosine DNA-methyltransferase activity is target-cell-specific and an early event in lung cancer. Proc Natl Acad Sci USA 1996;93:4045–4050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Issa JP, Vertino PM, Wu J, Sazawal S, Celano P, Nelkin BD, Hamilton SR, Baylin SB. Increased cytosine DNA-methyltransferase activity during colon cancer progression. J Natl Cancer Inst 1993;85:1235–1240. [DOI] [PubMed] [Google Scholar]
- 33.Eads CA, Danenberg KD, Kawakami K, Saltz LB, Danenberg PV, Laird PW. CpG island hypermethylation in human colorectal tumors is not associated with DNA methyltransferase overexpression. Cancer Res 1999;59:2302–2306. [PubMed] [Google Scholar]
- 34.Sato M, Horio Y, Sekido Y, Minna JD, Shimokata K, Hasegawa Y. The expression of DNA methyltransferases and methyl-CpG-binding proteins is not associated with the methylation status of p14(ARF), p16(INK4a) and RASSF1A in human lung cancer cell lines. Oncogene 2002;21:4822–4829. [DOI] [PubMed] [Google Scholar]
- 35.Brenner C, Fuks F. DNA methyltransferases: facts, clues, mysteries. Curr Top Microbiol Immunol 2006;301:45–66. [DOI] [PubMed] [Google Scholar]
- 36.Saito Y, Kanai Y, Sakamoto M, Saito H, Ishii H, Hirohashi S. Expression of mRNA for DNA methyltransferases and methyl-CpG-binding proteins and DNA methylation status on CpG islands and pericentromeric satellite regions during human hepatocarcinogenesis. Hepatology 2001;33:561–568. [DOI] [PubMed] [Google Scholar]
- 37.Strahl BD, Allis CD. The language of covalent histone modifications. Nature 2000;403:41–45. [DOI] [PubMed] [Google Scholar]
- 38.Stancheva I. Caught in conspiracy: cooperation between DNA methylation and histone H3K9 methylation in the establishment and maintenance of heterochromatin. Biochem Cell Biol 2005;83:385–395. [DOI] [PubMed] [Google Scholar]
- 39.Gregory RI, Randall TE, Johnson CA, Khosla S, Hatada I, O'Neill LP, Turner BM, Feil R. DNA methylation is linked to deacetylation of histone H3, but not H4, on the imprinted genes Snrpn and U2af1-rs1. Mol Cell Biol 2001;21:5426–5436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lande-Diner L, Zhang J, Ben-Porath I, Amariglio N, Keshet I, Hecht M, Azuara V, Fisher AG, Rechavi G, Cedar H. Role of DNA methylation in stable gene repression. J Biol Chem 2007;282:12194–12200. [DOI] [PubMed] [Google Scholar]
- 41.Lunyak VV, Burgess R, Prefontaine GG, Nelson C, Sze SH, Chenoweth J, Schwartz P, Pevzner PA, Glass C, Mandel G, et al. Corepressor-dependent silencing of chromosomal regions encoding neuronal genes. Science 2002;298:1747–1752. [DOI] [PubMed] [Google Scholar]
- 42.Kanai Y, Ushijima S, Tsuda H, Sakamoto M, Hirohashi S. Aberrant DNA methylation precedes loss of heterozygosity on chromosome 16 in chronic hepatitis and liver cirrhosis. Cancer Lett 2000;148:73–80. [DOI] [PubMed] [Google Scholar]
- 43.Okochi O, Hibi K, Sakai M, Inoue S, Takeda S, Kaneko T, Nakao A. Methylation-mediated silencing of SOCS-1 gene in hepatocellular carcinoma derived from cirrhosis. Clin Cancer Res 2003;9:5295–5298. [PubMed] [Google Scholar]