Abstract
This work for the first time demonstrates that synthetic polymers enhance uptake and nuclear import of plasmid DNA (pDNA) through the activation of cellular trafficking machinery. Nonionic block copolymers of poly(ethylene oxide) and poly(propylene oxide), Pluronics, are widely used as excipients in pharmaceutics. We previously demonstrated that Pluronics increase the phosphorylation of IκB and subsequent NFκB nuclear localization as well as upregulate numerous NFκB-related genes. In this study, we show that Pluronics enhance gene transfer by pDNA/polycation complexes (“polyplexes”) in a promoter-dependent fashion. Addition of Pluronic P123 or P85 to polyethyleneimine-based polyplexes had little effect on polyplex particle size but significantly enhanced pDNA cellular uptake, nuclear translocation and gene expression in several cell lines. When added to polyplex-transfected cells after transfection, Pluronics enhanced nuclear import of pDNA containing NFκB–binding sites, but have no effect on import of pDNA without these sites. All together, our studies suggest that Pluronics rapidly activate NFκB, which binds cytosolic pDNA that possesses promoters containing NFκB binding sites and consequently increases nuclear import of pDNA through NFκB nuclear translocation.
INTRODUCTION
Nonviral gene delivery systems, such as DNA incorporated in cationic lipids, polycations, or nanoparticles, are safer, cheaper and easier to produce in comparison with viral vectors (1) Unfortunately, these systems are also much less efficient in delivering DNA and initiating gene expression than the viruses. This is due to low efficacy of the nonviral vectors in overcoming numerous barriers for gene delivery into a cell. These barriers include (i) cellular uptake by endocytosis, (ii) escape from endosomes prior to delivery to lysosomes, (iii) release to the cytoplasm, and (iv) transport to the nucleus. Nuclear delivery of the DNA is believed to be among the most challenging barriers for every type of nonviral vector (2). Its significance is evident from numerous studies showing that microinjection of plasmid DNA (pDNA) into the cytoplasm results in negligible expression compared to expression of the same pDNA injected directly in the nucleus (3-7). It is well known that nuclear import substrates, such as transcription factors, contain nuclear localization signals (NLSs) that are recognized by nuclear import factors (8). Hence, NLS-containing peptides or proteins were coupled to pDNA to facilitate its nuclear delivery (9-21). As an alternative, we posit that some safe and nontoxic pharmaceutical excipients can activate selected cell signaling pathways (22, 23) and harness cell trafficking machinery to enhance nuclear transport of pDNA without DNA modification. In particular, a nonionic block copolymer of poly(ethylene oxide) and poly(propylene oxide), Pluronic, can activate a transcription factor, NFκB in cells and enhance gene expression in vitro and in vivo without inducing a cytotoxic effect (24, 25). It is known that NFκB can bind pDNA in cytoplasm and transport it to the nucleus through nuclear transport machinery (26-28). In this study we demonstrate for the first time that Pluronic enhances both cellular uptake and nuclear transport of pDNA delivered into cells with DNA/polycation complexes (“polyplexes”) and suggest that by combining Pluronic with existing non-viral vectors the efficiency of gene delivery can be safely increased.
MATERIALS AND METHODS
Materials
Nonionic block copolymers Pluronic P123 and Pluronic P85 were kindly provided by BASF Corp (Parsippany, NJ). SP1017, a mixture of 0.25 % Pluronic L61 and 2 % Pluronic F127 was kindly provided by Supratek Pharma Inc. (Montreal, Canada). Exgen 500, a 22 kD linear PEI, was purchased from MBI Fermentas Inc. (Hanover, MD). The cyanine dimmer dye, YOYO-1, was purchased from Molecular Probes (Carlsbad, CA). Plasmid gWIZLuc (encoding luciferase, under control of CMV promoter) was purchased from Gene Therapy Systems, Inc. (San Diego, CA) and plasmid pEGFP-N1 (encoding enhanced green fluorescent protein (GFP)) was purchased from Clontech (palo Alto, CA). “PathDetect® cis-Reporting Systems” containing luciferase reporter gene with NFκB and AP-1 response elements (pNFκB-luc and pAP-1-luc) were purchased from Stratagene (La Jolla, CA). All the plasmids were expanded in DH5α E. coli and isolated using Qiagen endotoxin-free plasmid giga-prep kits according to the supplier's protocol. TRIzol reagent was purchased from Invitrogen (Carlsbad, CA). Oligo GEArray® Mouse NFκB Signaling Pathway Microarray was purchased from SuperArray Bioscience Corporation (Frederick, MD). Methyl-β-cyclodextrin (MBCD) and Sucrose was purchased from Sigma Aldrich (St Loius, MO). Bovine serum albumin was purchased from Fischer Scientific (Waltham, MA)
Synthesis of PB25080 and P85PEI conjugates
Cationic graft or block copolymers P85PEI and PB25080 were synthesized by Dr. Vinogradov at University of Nebraska Medical Center (UNMC). Briefly, in case of P85PEI (2 kDa) 20% of free terminal hydroxyl groups of Pluronics P85 were activated by 1,1′-carbonyldiimidazole (CDI) in anhydrous acetonitrile for 4 h at 20°C and then reacted with equimolar quantity of branched PEI (MW 2 kDa) in 20% ethanol for 48 h at 20°C. The conjugate was purified from free PEI by gel permeation chromatography on Sephadex G-25 using 20% ethanol as eluent (29). To obtain PB25080 PEG (MW 8 kDa) was modified by one terminal hydroxyl group with 4,4′-dimethoxytrityl (DMT) chloride to obtain DMT-PEG (30). DMT-PEG was activated with CDI in acetonitrile and reacted with ethylenediamne to obtain monoamino-PEG. Monoamino-PEG was treated with d-Biotin p-nitrophenyl ester in 5 mL of dimethylformamide containing 1% (v/v) of triethylamine for 18 h at 40 °C to obtain biotin-PEG. Biotin-PEG was treated again with CDI and then reacted with PEI (25 kDa) to obtain PB25080 (31).
Labeling of Pluronic
Ahydrous Pluronic P123 (2 mg) was activated with CDI (213 mg) in 10 ml of acetonitrile for 2 h at 37°C and reacted with ethylenediamine (313mg) in 20 ml ethanol for 12 h at room temperature. The reaction mixture was dialyzed in a 2 kD cutoff membrane against 15 % ethanol for 72 h (change ethanol twice) and lyophilized. Amino-Pluronic (0.4 mg) was dissolved in 2 ml acetonitrile, mixed with 2 ml 0.1M NaH2CO3 buffer and reacted with 25 mg of rhodamine B isothiocyanate (RITC) or fluorescein isothiocyanate (FITC) in 1 ml dimethylformamide added drop-wise over 10 min with stirring. After 2 h incubation at room temperature RITC- or FITC-labeled Pluronic was dialyzed in a 2 kD cutoff membrane in 20% ethanol for 72 h at 4°C in dark and then lyophilized. Similar procedure was used for Pluronic P85. For cell uptake studies the labeled Pluronic 123 was mixed with the unlabeled copolymer at 1:10 ratio.
Cell culture
Mouse fibroblasts NIH 3T3 cell line and human prostate cancer PC-3 cell line were cultured in Dulbecco's Modified Eagle's Medium (DMEM) and RPMI 1640, respectively, supplemented with 10% heat-inactivated fetal bovine serum (FBS), 100 U/mL penicillin/100 μg/mL streptomycin (complete media). All tissue material media was obtained from Gibco Life Technologies, Inc. (Grand Island, NY). Stable transfected C2C12-CMVluc cells were generated as described previously (24). Cells were seeded at a density of 50,000 cells per well in 24 well plates and allowed to adhere for 24 - 48 h prior to transfection. For confocal microscopy, cells were seeded onto untreated glass coverslips at 50,000 cells per coverslip.
Formation of polyplexes and transfection
Transfection solutions were prepared in 1.5 mL Eppendorf tube by mixing (i) 45 μL of pDNA (0.1 mg/mL) or 90 μL of YOYO-1 labeled pDNA (0.05 mg/mL), (ii) 9 μL of Pluronic P123 solution (Pluronic P85 or SP1017 when indicated), (iii) 150 μL of DMEM and (iv) polycations. For transfections, cell transport and localisation studies the polycation nitrogen to pDNA phosphate ratios (N/P ratio) were kept constant N/P = 8 for PB25080 and P85PEI and N/P = 9 for Exgen 500. For physicochemical characterization the N/P ratios were varied as indicated below. The solutions were vortexed for 30 sec immediately after addition of polycation, incubated for 5 min at room temperature, and supplemented with 1270 μL of complete media. Cells were incubated with transfection solution (250 μL per well or 500 μL per coverslip) for 2 h at 37°C (5% CO2), rinsed with phosphate buffer saline (PBS), incubated for additional 24 h in complete media and analyzed for luciferase or GFP expression as previously described (24).
Particle size
Effective hydrodynamic diameters were measured by photon correlation spectroscopy using a ‘ZetaPlus’ Zeta Potential Analyzer (Brookhaven Instrument) equipped with the Multi-Angle Option at 22°C at an angle of 90°. Prior to particle formation, all reagents were filtered through a Whatman 200 nm polysulfone filter. Data represents average of five separate 3 min analyses. All systems exhibited stable effective diameters over the measurement time.
Transmission electron microscopy (TEM)
Polyplexes were prepared in Eppendorf tube by first, mixing 100 μL of 0.1 M phosphate buffer (pH7.0), 120 μL of 0.1mg/ml gWIZLuc, and 6 μL of 5% Pluronic P123 and, second, adding polycation, vortexing for 30 seconds and dilution with phosphate buffer to 500 μL. A drop of the sample solution was allowed to settle on a Formavar precoated grid for 1 min, the excess sample was wicked away with filter paper and a drop of staining solution (1% uranyl acetate) was allowed to contact the sample for 1 min. The samples were analyzed using a Philips 410 TEM microscope.
Flow cytometry analysis
Polyplexes containing YOYO-1 labeled pDNA (32) with/without Pluronic were incubated with NIH 3T3 or PC-3 cells for 2 h at 37°C (Figure 3). In select experiments cells were pre-treated for 30 min with inhibitors of endocytosis (2 mM MBCD, or 0.25 M sucrose), and then the same inhibitors were also present during subsequent incubation with the polyplex in presence or absence of the copolymer. Cells were washed by PBS, trypsinized, and suspended in 1% FBS. The mean fluorescence intensity was analyzed using Becton Dickinson FACStarPlus flow cytometer operating under Lysis II (San Jose, CA), equipped with an argon ion laser (excitation − 488 nm; emission filter − 530 ± 30 nm bandpass). Data were acquired in linear mode and visualized in logarithmic mode. Data from 10,000 events were gated using forward and side scatter parameters to exclude debris and dead cells. In a separate study the flow cytometry analysis was performed to quantify the amount of DNA in nuclei isolated after transfection of the cells as described below.
Fig. 3.
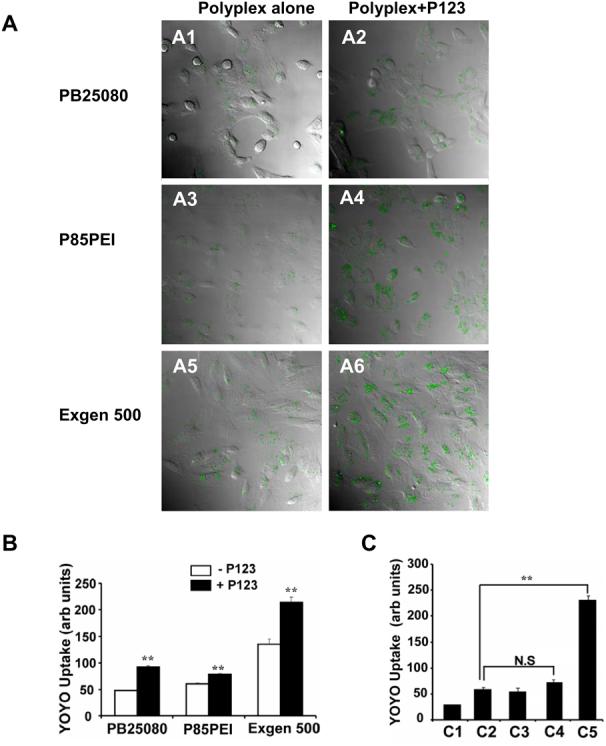
Effect of 0.03% w/v Pluronic P123 on uptake of the YOYO-1 labeled gWIZLuc pDNA in NIH 3T3 and PC-3 cells. (A) Confocal microscopy images of live cells 2 h after NIH 3T3 cells transfection with PB25080-(A1, A2), P85PEI- (A3, A4) and Exgen 500- (A5, A6) based polyplexes without (A1, A3, A5) or with (A2, A4, A6) Pluronic P123. (B) Flow cytometry under the same transfection conditions as (A) comparing groups with/without Pluronic. (C) Flow cytometry of (C1) PC-3 cells alone and PC-3 cells transfected for 2h with (C2) naked pDNA, (C3) naked pDNA with Pluronic P123, (C4) PB25080/pDNA polyplex, and (C5) PB25080/pDNA polyplex with Pluronic P123. (B,C) Statistical comparisons between (B) untreated and Pluronic P123 treated groups or (C) pDNA alone and polyplex-treated groups were made as follows ** p < 0.01, n.s., not significant (n = 3).
Intracellular trafficking
Cells were transfected with Exgen 500-based polyplex with/without RITC-labeled Pluronic P85. At the end of each time point cells were washed, fixed (using 4% parafomaldehyde), labeled with ToPro-3 iodide and examined under confocal microscope.
Nuclear translocation
Cells were transfected for 2 hr with YOYO labeled pAP-1-luc or pNFκB-luc using Exgen 500, washed twice with PBS, and then incubated in fresh medium with/without 0.03 % Pluronic P123 for additional time points. Localization of YOYO-1 labeled DNA was examined under the confocal microscope and flow cytometery was performed on nuclei isolated after transfection in these cells.
Nuclei isolation
NIH 3T3 cells were plated in 100×20 mm tissue culture plates (BD Biosciences, San Jose, CA) and grown overnight. Nuclei were isolated using Qproteome Nuclear Protien kit (Qiagen, Germantown, MD). Briefly, after transfection of YOYO-1 labeled DNA cells were washed twice with PBS, removed gently using a scraper and centrifuged (12,000 rpm, 5 min., 4°C). The pellets were lysed using the Qiagen lysis buffer supplemented with protease inhibitor solution and 0.1M dithiothreitol, centrifuged, resuspended in the Qiagen detergent solution, centrifuged again and then finally, the nuclei were resuspended in 1% bovine serum albumin in PBS.
Immunocytochemistry
Cells were pretreated with DMEM without FBS for 30 min, and then treated with 0.03% FITC-labeled Pluronic P123 for 1 h, washed and fixed with 4% paraformaldehyde. Rabbit anti-caveolin-1–Cy3 antibody (1:100) (Sigma Aldrich, St Louis, MO) and mouse anti-clathrin antibody (Affinity Bioreagents, Golden, CO) (1:10) were incubated in blocking buffer overnight at 4°C. For detection of anti-clathrin antibody specific IgG antibody conjugated to Alexa 568 (Invitrogen Inc, Carlsbad, CA) was added to cells for 1 h at 37°C. After extensive washing cells were labeled with ToPro-3 iodide, mounted with antifade reagent and examined under confocal microscope.
NFκB signaling pathway microarray
C2C12-CMVluc cells were treated with 1% Pluronic P85 for 5 min, washed twice with PBS, and incubated for additional 3 h. Cells were collected and total RNA was isolated with TRIzol reagent, then quantified and qualified by UV spectrophotometry and Gel electrophoresis, respectively. Oligo GEArray® with 3 μg of total RNA of each sample was performed following the manufacture's protocol (cDNA synthesis; cRNA synthesis, labeling and amplification; cRNA cleanup; Oligo GEArray hybridization and detection). Statistical analysis was performed using the GEArray® Expression Analysis Suite software.
Statistical analysis
Unless indicated otherwise statistical comparisons are made using Student's t-test.
RESULTS
Polyplex particle size and morphology
All polycations when mixed with the pDNA formed stable nanoparticles with effective diameters from approximately 85 nm for PB25080 to 180 nm for Exgen 500 (Table 1). Effective particle diameters were practically unchanged or changed slightly upon addition of Pluronic 123 using different orders of mixing (Table 1), N/P ratios (Figure 1A), or concentrations of the copolymer (Figure 1B). Similar results were observed with Pluronic P85 (not shown). Furthermore, the particle morphology as determined by TEM remained appear to be spherical (Figure 1C). All together, these results demonstrate that addition of Pluronic results in little change in polyplex size or morphology.
Table 1.
Effect of Pluronic P123 on particle size for different polyplex compositions and component mixing orders
| Polyplexa | Effective Diameter (nm)b | Polydispersity |
|---|---|---|
| DNA + P123 | N/A | N/A |
| DNA + PB25080 | 85 ± 3 | 0.096 |
| DNA+P123+PB25080 | 76 ± 6 | 0.229 |
| DNA+PB25080+P123 | 72 ± 5 | 0.213 |
| DNA+P85PEI | 150 ± 2 | 0.155 |
| DNA+P123+P85PEI | 120 ± 2 | 0.212 |
| DNA+Exgen 500 | 180 ± 4 | 0.164 |
| DNA+P123+Exgen 500 | 160 ± 6 | 0.241 |
Polyplexes were formed at N/P = 8 for PB25080 and P85PEI and N/P = 9 for Exgen 500.
Data are mean ± SEM (n = 5).
Fig. 1.
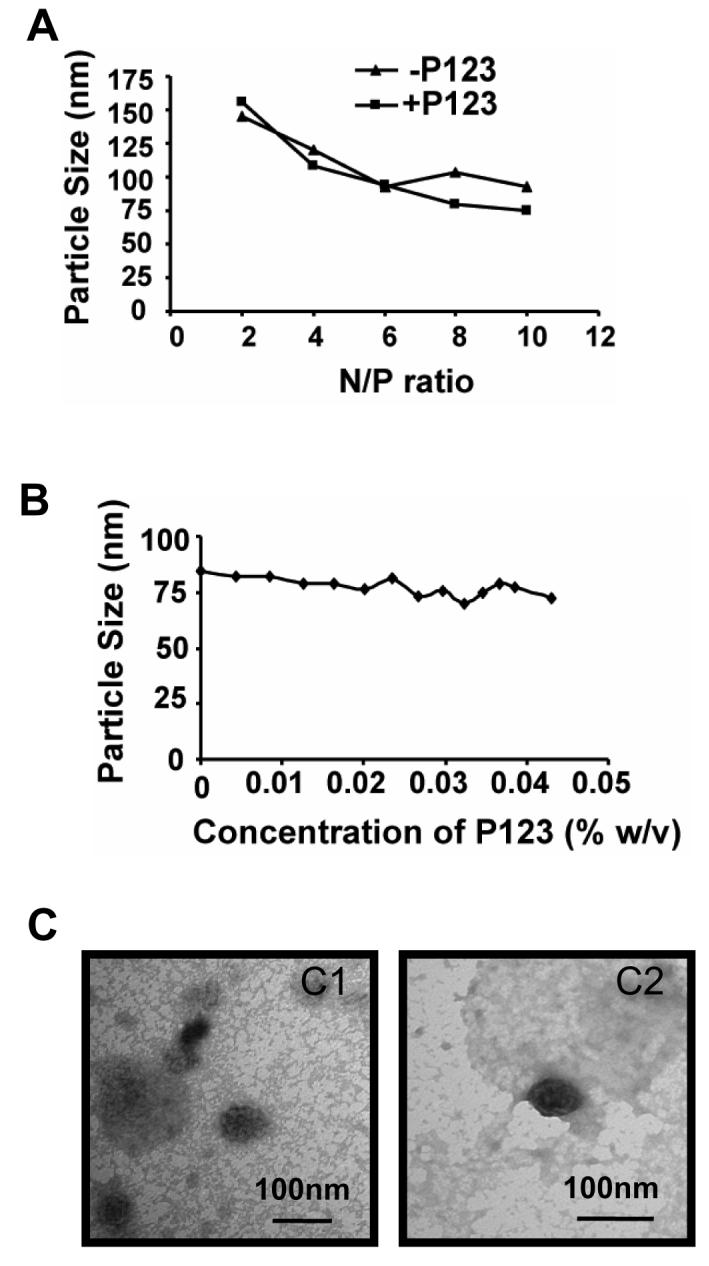
Effect of Pluronic P123 on the particle size and morphology of PB25080-based polyplex: (A-C) Particle effective diameters were measured at (A) various N/P ratios; and (B) different Pluronic concentrations. (C) TEM images of PB25080-based polyplex without (C1) and with (C2) Pluronic P123. (B,C) Polyplexes were formed at N/P = 8. (A,C) Pluronic P123 concentration was 0.03 % w/v. (A,B) All measurements were performed in phosphate buffer, pH 7.0.
Enhancement of gene expression with Pluronic
Significant enhancement of luciferase expression was observed upon addition of Pluronic P123, Pluronic P85 or SP1017 in both NIH 3T3 and PC-3 cells transfected with various polyplexes (Figure 2A-C). Overall protein levels in the cells treated with polyplexes were not affected by adding Pluronic (not shown) although the portion of cells expressing the transgene increased (Figure 2D). A similar enhancement was observed with several other polyplex types (Superfect, linear PEI) and in a different cell line (C2C12) (24). Thus, there is compelling evidence that Pluronic block copolymers enhance transfection by different polyplexes in numerous cell types.
Fig. 2.
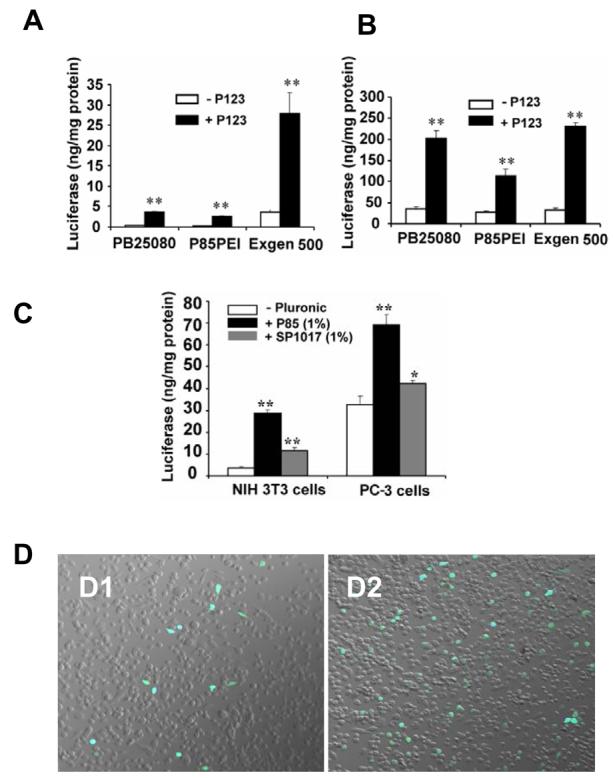
Enhancement of gene expression by Pluronic block copolymers in cells transfected with polyplexes. (A,B) Luciferase gene expression in NIH 3T3 (A) and PC-3 cells (B) transfected with different types of polyplexes containing gWIZLuc pDNA with/without 0.03% Pluronic P123 (** p < 0.01, n = 3). (C) Luciferase gene expression in NIH 3T3 and PC-3 cells transfected with Exgen 500-based polyplex with/without 1% Pluronic P85 or SP1017 (*p < 0.05, **p < 0.01, n = 3). (D) Confocal microscopy images of GFP gene expression in PC-3 cells transfected with Exgen 500-based polyplex containing pEGFP-N1 pDNA without (D1) or with (D2) Pluronic P123.
Effect of Pluronic on DNA uptake in cells
Addition of Pluronic P123 considerably enhanced the uptake of polyplex-delivered DNA in NIH 3T3 cells (Figure 3A). The magnitude of the effect varied for each polyplex type (1.3, 1.6, and 1.9-fold for P85PEI, Exgen 500 and PB25080 respectively), but the differences in the presence of Pluronic were significant (Figure 3B). An even greater enhancement of uptake of polyplex-delivered DNA with Pluronic was observed in PC-3 cells (Figure 3C). In contrast, Pluronic did not affect uptake of the naked DNA. The effects of Pluronic on cell uptake were observed as early as 30 min after exposure (Figure 4A,B). Notably, Pluronic was internalized during the same period, but exhibited little if any colocalization with the DNA (Figure 4A,B). Interestingly, Pluronic exhibited greater co-localization with caveolin-1 compared to clathrin (Figure 4C,D). This is consistent with a recent study suggesting that caveolae-mediated endocytosis is a preferred pathway for cellular uptake of Pluronic individual molecules (33). At the same time, PEI-based polyplexes enter cells through both caveolae- and clathrin-mediated endocytosis (34, 35). Inhibitors of these pathways, MBCD for caveolae and hypertonic sucrose for clathrin, each significantly decreased uptake of pDNA delivered with polyplexes without and with Pluronic (Table 2). However, the effect of sucrose was much more pronounced than that of MBCD, suggesting clathrin-mediated endocytosis is a preferred route for the polyplex. Hence, Pluronic and polyplex are likely to have different internalization pathways and the transport pathways of pDNA apparently did not change in the presence of Pluronic.
Fig. 4.
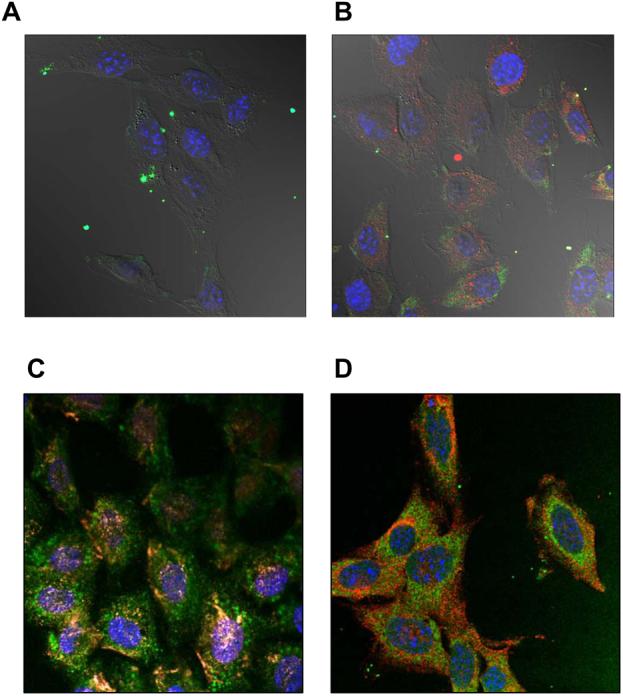
Cellular localization of fluorescently labeled DNA and/or Pluronic 123 examined by confocal microscopy on fixed NIH 3T3 cells. (A, B) Cells were transfected for 30 min with Exgen 500-based polyplex without (A) or with (B) 0.03 % Pluronic P123. Signals correspond to YOYO-1 labeled gWIZLuc pDNA (green), RITC-labeled Pluronic P123 (red) and ToPro-3 stained nuclei (blue), respectively. (C, D) Cells were exposed to 0.03 % Pluronic P123 alone for 60 min and then examined for co-localization of Pluronic P123 with caveolin-1 (C) and clathrin (D). Signals correspond to FITC-labeled Pluronic P123 (green), caveolin-1 (red), clathrin (red) and ToPro-3 stained nuclei (blue).
Table 2.
Effect of endocytosis inhibitors on uptake of YOYO-labeled gWIZLuc pDNA in NIH 3T3 cells.
| Polyplex | Mean Fluorescence (Arb units)a |
|
|---|---|---|
| −P123 | +P123 | |
| DNA + PB25080 | 100 ± 3 | 158±3 |
| DNA+ PB25080+MBCD | 75 ± 6 ** | 111 ± 7 ** |
| DNA+ PB25080+Sucrose | 50 ± 1*** | 71 ± 1 *** |
Statistical comparisons were made for inhibitor treated and non-treated groups
p < 0.01
p<0.001, n=3.
Enhancement of nuclear transport of DNA by Pluronic
Initial confocal microscopy of cellular localization of YOYO-1 labeled DNA suggested that presence of Pluronic during transfection enhanced nuclear transport of the gWIZLuc plasmid at different time points (Figure 5). Specifically, in Pluronic treatment groups the DNA exhibited enhanced perinuclear localization 2h after transfection and increased nuclear uptake 3 h after transfection. To exclude possible effects of the copolymer on the early stage of DNA internalization the subsequent experiments first transfected cells with a polyplex without Pluronic (for 2 h) and then treated cells with Pluronic for different periods. In these experiments along with the gWIZLuc plasmid we also used two other plasmids, pNFκB-luc and pAP-1-luc, containing NFκB and AP-1, response elements respectively. Figure 6 presents the results both as quantification of the DNA in isolated nucleus as well as confocal images in live cells. The addition of Pluronic resulted in a quantitative increase in accumulation of the gWIZLuc and pNFκB-luc plasmids in isolated nuclei. The gWIZLuc is controlled by a CMV promoter, which also contains NFκB binding sites. In contrast, no increase in nuclei accumulation was observed with the pAP-1-luc, which does not contain NFκB binding sites. All together, these results suggest that Pluronic can enhance nuclear translocation of polyplex-delivered DNA in a promoter-dependent fashion.
Fig. 5.
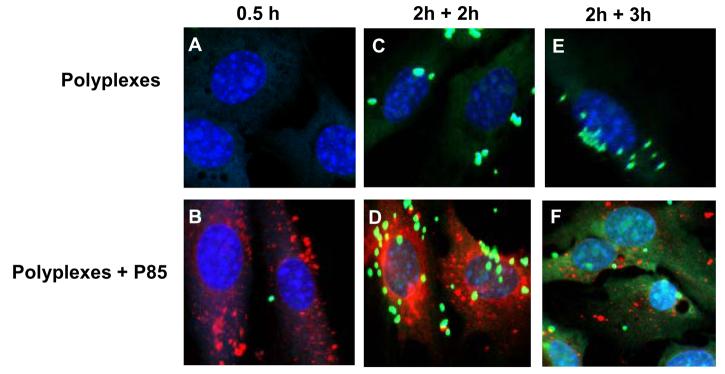
Effect of Pluronic on intracellular localization of DNA in NIH 3T3 cells. Confocal microscopy images of cells transfected with Exgen 500-based polyplexes with (B, D, F) or without (A, C, E) 1% Pluronic P85. Cells were exposed to polyplex for (A, B) 30 min or (C-F) 2 h and then incubated for additional 2 (C, D) or 3 h (E, F) in a fresh medium. Signals correspond to YOYO-labeled pDNA (green), RITC-labeled Pluronic P85 (red) and ToPro-3 stained nuclei (blue), respectively. This figure is representative of multiple areas in confocal images.
Fig. 6.
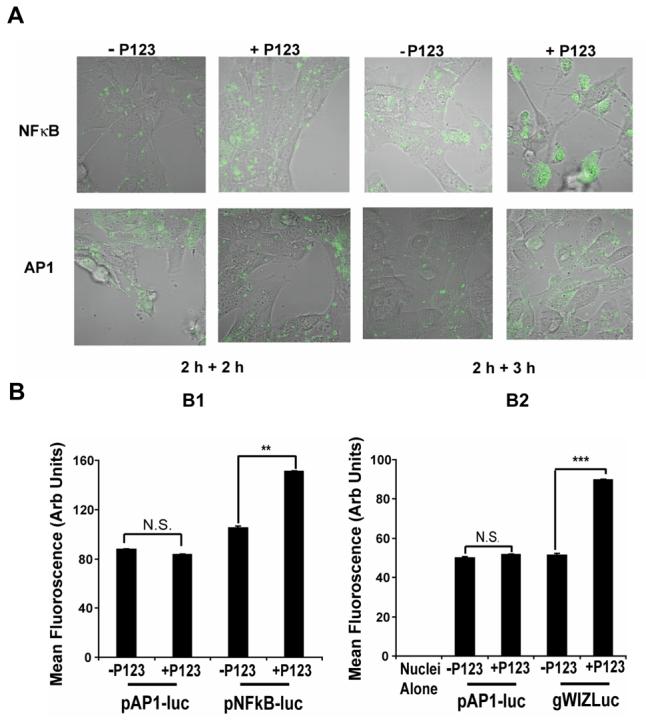
Promoter-dependent enhancement of nuclear transport of pDNA by Pluronic P123 in NIH 3T3 cells. (A) Confocal microscopy images of live cells transfected (2 h) with pNFκB-luc or pAP-1-luc using Exgen 500 polyplexes and then cultured for 2 h or 3 h with or without 0.03% Pluronic P123. Signal corresponds to YOYO-1 labeled pDNA. (B) Flow cytometery of isolated nuclei of cells transfected with pAP-1-luc (as a control B1, B2) and either pNFκB-luc (B1) or gWIZLuc (B2). Cells were transfected using the same procedure as in (A) and then cultured for 3 h with or without 0.03% Pluronic P123. (B) ** p < 0.01, *** p < 0.001, n.s., not significant (n = 3).
Activation of NFκB signaling pathway by Pluronic
Pluronic was previously shown to activate the NFκB signaling pathway by inducing phosphorylation of IκB (24). Here we examined the effect of Pluronic on NFκB-related genes using microarray (Figure 7). Pluronic P85 significantly upregulated 39 out of 113 genes related to NFκB-mediated signal transduction (supplement data, Table S-1).
Fig. 7.
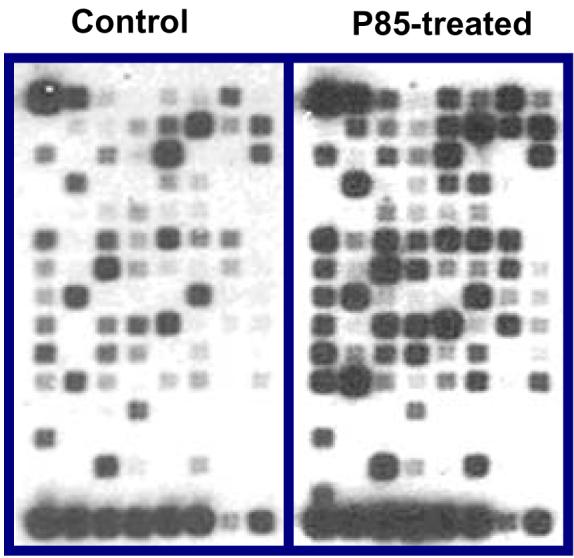
Effect of Pluronic on NFκB related genes expression in C2C12-CMVluc cells. Cells were exposed to 1 % Pluronic P85 for 5 min, washed by PBS and incubated for additional 3 h. Total RNA was isolated and assayed using Oligo GEArray® Mouse NFκB Signaling Pathway Microarray. A representative set of images shown for control and Pluronic treated groups. The triplicate averaged data presented in supplement (Table S-1).
DISCUSSION
Inefficient nuclear translocation of DNA is believed to be the Achilles' heel of gene therapy (36). Coupling NLS peptides with pDNA to facilitate its nuclear import has been a major approach to address this problem (9-21). Our work demonstrates a simpler and potentially safer approach using a well known pharmaceutical excipient, Pluronic block copolymer, to activate intrinsic nuclear translocation machinery and enhance nuclear gene transfer with polyplex-delivered pDNA. Pluronics are amphiphilic block copolymers that are widely used in pharmaceutical compositions and were evaluated successfully as intravenous agents in cancer therapy clinical trials (37). Astafieva et al. were first to report that addition of Pluronic P85 can increase the uptake and expression of pDNA delivered by poly(N-ethyl-4-vinylpyridinium)-based polyplex (38). In this work, we demonstrate for the first time that Pluronics additionally increase nuclear transport of pDNA delivered with polyplexes in a promoter-dependent manner. To demonstrate the effects of Pluronics we evaluated three representative polycations: 1) a linear PEI, Exgen 500; 2) biotin-modified graft copolymer of PEO and PEI, PB25080, and 3) a graft/block copolymer of Pluronic P85 and PEI, P85PEI. Among a variety of polycations used in polyplexes, linear PEI is, perhaps, one of the most efficacious (39, 40). Two other PEI-based polycations evaluated in this work were developed in our laboratory to enhance polyplex stability in the body by grafting hydrophilic PEO chains to PEI (PB25080) (30) or enhance membrane interactions of polyplex by grafting amphiphilic Pluronic P85 to PEI (P85PEI) (41).
We previously suggested that Pluronic can increase gene transfer by binding with polyplexes and decreasing particle size (32). Since Pluronic is not charged, this interaction involves hydrophobic binding of poly(propylene oxide) chains of Pluronic with non-polar sites in a polyplex. However, contrary to previously studied polyplex systems, which aggregated in the absence of Pluronic (32), the polycations used in this work each formed stable polyplexes with pDNA. Addition of Pluronic to these polyplexes resulted in little if any change in particle size. Furthermore, confocal microscopy suggested that Pluronic distribution within the cells was different from that of pDNA at each time point, i.e. Pluronic is not transported along with the DNA but may act independently, perhaps by affecting cell transport machinery, such as activating NFκB signaling pathway (as described later). Therefore, we propose that Pluronic acts as a biological response modifier, presumably, by binding with cellular membranes and activating cell signaling pathways that enhance nuclear gene delivery (22, 23). This is supported by the observation that nuclear transport of the pDNA was enhanced when Pluronic was added to cells after polyplex transfection (Figure 6). The time course for the nuclear entry of pDNA, 4 to 5 h after beginning of transfection, is consistent with a previous report (42). Polyplexes formed by Exgen 500 rapidly escape the endosomes, spread into the cytoplasm, and then dissociate within 4 h, followed by the nuclear uptake of pDNA (43, 44). Previously, Pitard et al. reported that co-injection of pDNA and Pluronic L64 into the cytosol increased the percentage of cells expressing a reporter gene (45), suggesting that Pluronic may enhance transport of naked pDNA from the cytosol into the nucleus.
The involvement of the NFκB signaling pathway was directly demonstrated by the promoter-dependence of Pluronic effect on pDNA nuclear uptake. The plasmids containing NFκB binding sites, gWIZLuc and pNFκB-luc exhibited increased unclear uptake with Pluronic, while the pAP-1-luc plasmid, which lacks NFκB binding site did not. Evidence that support the activation of the NFκB pathway by Pluronic was shown by phosphorylation of IκB within 5 minutes (24), increased NFκB nuclear localization (25), and upregulation of numerous NFκB-related genes. It has also been shown that NFκB can bind cytosolic DNA and transport it to the nucleus through nuclear import machinery (27, 46, 47). Furthermore, the NLS of NFκB alone can transport DNA across the plasma membrane and into the cytosol (47). Thus, the early stages of gene transfer may also be affected by the activation of NFκB, which would explain the increased cellular uptake of pDNA observed in this work. All together, we propose that Pluronics rapidly activate NFκB (24), NFκB binds pDNA that contain NFκB-binding sites, then increases nuclear import of a pDNA through the NF-κB transport into the nucleus. Furthermore, following the delivery of pDNA to the nucleus, Pluronics increase transcriptional activation of genes through the NFκB signaling pathway (24). We present evidence here that Pluronic co-localizes in cells with caveolin-1, which regulates NFκB activation (48). In addition, an extensive study describing binding of Pluronic with caveolae and disruption of caveolae-mediated endocytosis has been recently published (33). Hence, there is a possibility that Pluronic activates NFκB through the disruption of caveolae.
The safety of Pluronic formulations may be an important advantage for their use in nonviral gene delivery. Even though NFκB was activated, inflammatory responses to Pluronics are mild and no cytotoxic effects are seen at their effective doses in many cell lines (24). More importantly, clinical trials in human subjects reinforce the safety of Pluronic formulations, such as SP1017 (37), which was also used in this study. This suggests a possibility of optimizing Pluronic formulations to safely enhance gene therapy. Pluronic may be delivered simultaneously with polyplexes or administered after polyplexes. For example, after intravenous administration Pluronic block copolymers were shown to accumulate in the tumors (data in preparation), which are same sites where polyplexes may be targeted. In addition, both polyplexes and Pluronic can be administered locally (49). Furthermore, the fact that the increased nuclear transfer was observed with Pluronic added after transfection opens a possibility for consequent treatments with the polyplex and Pluronic. It is also important to note that Pluronics may be not the only polymers capable of affecting nuclear delivery of pDNA via modulation of cell signaling. Hence, it is advisable to evaluate the effects of all polymer transfection systems, especially those that produce very strong enhancements of gene expression and may have membrane-active hydrophobic moieties (50-52). It is noteworthy that most polyplex transfection systems require a considerable excess of polycation that cannot incorporate in the complex with DNA and may directly affect cell signaling machinery. Finally, this study underscores the need to match the polymeric formulation with pDNA structure to take full advantage of potential polymer effects on signal transduction.
Supplementary Material
ACKNOWLEDGEMENTS
This study was supported by Tobacco Settlement Funds awarded to University of Nebraska, and National Institutes of Health R01 CA116591 to (AVK). We would like to thank Dr. Sergey Vinogradov for assistance in synthesizing PB25080 and P85PEI. We also thank UNMC Confocal and Flow Cytometery Core facilities for technical assistance.
Footnotes
SUPPORTING INFORMATION AVAILABLE
This information is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Schaffer DV, Lauffenburger DA. Targeted synthetic gene delivery vectors. Curr Opin Mol Ther. 2000;2:155–61. [PubMed] [Google Scholar]
- 2.Wiethoff CM, Middaugh CR. Barriers to nonviral gene delivery. J Pharm Sci. 2003;92:203–17. doi: 10.1002/jps.10286. [DOI] [PubMed] [Google Scholar]
- 3.Capecchi MR. High efficiency transformation by direct microinjection of DNA into cultured mammalian cells. Cell. 1980;22:479–88. doi: 10.1016/0092-8674(80)90358-x. [DOI] [PubMed] [Google Scholar]
- 4.Graessmann M, Menne J, Liebler M, Graeber I, Graessmann A. Helper activity for gene expression, a novel function of the SV40 enhancer. Nucleic Acids Res. 1989;17:6603–12. doi: 10.1093/nar/17.16.6603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mirzayans R, Aubin RA, Paterson MC. Differential expression and stability of foreign genes introduced into human fibroblasts by nuclear versus cytoplasmic microinjection. Mutat Res. 1992;281:115–22. doi: 10.1016/0165-7992(92)90045-j. [DOI] [PubMed] [Google Scholar]
- 6.Thorburn AM, Alberts AS. Efficient expression of miniprep plasmid DNA after needle micro-injection into somatic cells. Biotechniques. 1993;14:356–8. [PubMed] [Google Scholar]
- 7.Dean DA, Dean BS, Muller S, Smith LC. Sequence requirements for plasmid nuclear import. Exp Cell Res. 1999;253:713–22. doi: 10.1006/excr.1999.4716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Friedrich B, Quensel C, Sommer T, Hartmann E, Kohler M. Nuclear localization signal and protein context both mediate importin alpha specificity of nuclear import substrates. Mol Cell Biol. 2006;26:8697–709. doi: 10.1128/MCB.00708-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aronsohn AI, Hughes JA. Nuclear localization signal peptides enhance cationic liposome-mediated gene therapy. J Drug Target. 1998;5:163–9. doi: 10.3109/10611869808995871. [DOI] [PubMed] [Google Scholar]
- 10.Chan CK, Jans DA. Enhancement of polylysine-mediated transferrinfection by nuclear localization sequences: polylysine does not function as a nuclear localization sequence. Hum Gene Ther. 1999;10:1695–702. doi: 10.1089/10430349950017699. [DOI] [PubMed] [Google Scholar]
- 11.Kaneda Y, Iwai K, Uchida T. Increased expression of DNA cointroduced with nuclear protein in adult rat liver. Science. 1989;243:375–8. doi: 10.1126/science.2911748. [DOI] [PubMed] [Google Scholar]
- 12.Hagstrom JE, Ludtke JJ, Bassik MC, Sebestyen MG, Adam SA, Wolff JA. Nuclear import of DNA in digitonin-permeabilized cells. J Cell Sci. 1997;110(Pt 18):2323–31. doi: 10.1242/jcs.110.18.2323. [DOI] [PubMed] [Google Scholar]
- 13.Ciolina C, Byk G, Blanche F, Thuillier V, Scherman D, Wils P. Coupling of nuclear localization signals to plasmid DNA and specific interaction of the conjugates with importin alpha. Bioconjug Chem. 1999;10:49–55. doi: 10.1021/bc980061a. [DOI] [PubMed] [Google Scholar]
- 14.van der Aa MA, Koning GA, d'Oliveira C, Oosting RS, Wilschut KJ, Hennink WE, Crommelin DJ. An NLS peptide covalently linked to linear DNA does not enhance transfection efficiency of cationic polymer based gene delivery systems. J Gene Med. 2005;7:208–17. doi: 10.1002/jgm.643. [DOI] [PubMed] [Google Scholar]
- 15.Zanta MA, Belguise-Valladier P, Behr JP. Gene delivery: a single nuclear localization signal peptide is sufficient to carry DNA to the cell nucleus. Proc Natl Acad Sci U S A. 1999;96:91–6. doi: 10.1073/pnas.96.1.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Branden LJ, Mohamed AJ, Smith CI. A peptide nucleic acid-nuclear localization signal fusion that mediates nuclear transport of DNA. Nat Biotechnol. 1999;17:784–7. doi: 10.1038/11726. [DOI] [PubMed] [Google Scholar]
- 17.Branden LJ, Christensson B, Smith CI. In vivo nuclear delivery of oligonucleotides via hybridizing bifunctional peptides. Gene Ther. 2001;8:84–7. doi: 10.1038/sj.gt.3301345. [DOI] [PubMed] [Google Scholar]
- 18.Liang KW, Hoffman EP, Huang L. Targeted delivery of plasmid DNA to myogenic cells via transferrin-conjugated peptide nucleic acid. Mol Ther. 2000;1:236–43. doi: 10.1006/mthe.2000.0043. [DOI] [PubMed] [Google Scholar]
- 19.Morris MC, Chaloin L, Choob M, Archdeacon J, Heitz F, Divita G. Combination of a new generation of PNAs with a peptide-based carrier enables efficient targeting of cell cycle progression. Gene Ther. 2004;11:757–64. doi: 10.1038/sj.gt.3302235. [DOI] [PubMed] [Google Scholar]
- 20.Dean DA, Strong DD, Zimmer WE. Nuclear entry of nonviral vectors. Gene Ther. 2005;12:881–90. doi: 10.1038/sj.gt.3302534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cartier R, Reszka R. Utilization of synthetic peptides containing nuclear localization signals for nonviral gene transfer systems. Gene Ther. 2002;9:157–67. doi: 10.1038/sj.gt.3301635. [DOI] [PubMed] [Google Scholar]
- 22.Kabanov AV, Batrakova EV, Sriadibhatla S, Yang Z, Kelly DL, Alakov VY. Polymer genomics: shifting the gene and drug delivery paradigms. J Control Release. 2005;101:259–71. doi: 10.1016/j.jconrel.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 23.Kabanov AV. Polymer genomics: an insight into pharmacology and toxicology of nanomedicines. Adv Drug Deliv Rev. 2006;58:1597–621. doi: 10.1016/j.addr.2006.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sriadibhatla S, Yang Z, Gebhart C, Alakhov VY, Kabanov A. Transcriptional activation of gene expression by pluronic block copolymers in stably and transiently transfected cells. Mol Ther. 2006;13:804–13. doi: 10.1016/j.ymthe.2005.07.701. [DOI] [PubMed] [Google Scholar]
- 25.Yang Z, Zhu J, Sriadibhatla S, Gebhart C, Alakhov V, Kabanov A. Promoter- and strain-selective enhancement of gene expression in a mouse skeletal muscle by a polymer excipient Pluronic P85. Journal of Controlled Release. 2005;108:496–512. doi: 10.1016/j.jconrel.2005.08.015. [DOI] [PubMed] [Google Scholar]
- 26.Muller CW, Rey FA, Sodeoka M, Verdine GL, Harrison SC. Structure of the NF-kappa B p50 homodimer bound to DNA. Nature. 1995;373:311–7. doi: 10.1038/373311a0. [DOI] [PubMed] [Google Scholar]
- 27.Mesika A, Grigoreva I, Zohar M, Reich Z. A regulated, NFkappaB-assisted import of plasmid DNA into mammalian cell nuclei. Mol Ther. 2001;3:653–7. doi: 10.1006/mthe.2001.0312. [DOI] [PubMed] [Google Scholar]
- 28.Mesika A, Kiss V, Brumfeld V, Ghosh G, Reich Z. Enhanced intracellular mobility and nuclear accumulation of DNA plasmids associated with a karyophilic protein. Hum Gene Ther. 2005;16:200–8. doi: 10.1089/hum.2005.16.200. [DOI] [PubMed] [Google Scholar]
- 29.Belenkov AI, Alakhov VY, Kabanov AV, Vinogradov SV, Panasci LC, Monia BP, Chow TY. Polyethyleneimine grafted with pluronic P85 enhances Ku86 antisense delivery and the ionizing radiation treatment efficacy in vivo. Gene Ther. 2004;11:1665–72. doi: 10.1038/sj.gt.3302355. [DOI] [PubMed] [Google Scholar]
- 30.Vinogradov SV, Bronich TK, Kabanov AV. Self-assembly of polyamine-poly(ethylene glycol) copolymers with phosphorothioate oligonucleotides. Bioconjug Chem. 1998;9:805–12. doi: 10.1021/bc980048q. [DOI] [PubMed] [Google Scholar]
- 31.Vinogradov S, Batrakova E, Li S, Kabanov A. Polyion complex micelles with protein-modified corona for receptor-mediated delivery of oligonucleotides into cells. Bioconjug Chem. 1999;10:851–60. doi: 10.1021/bc990037c. [DOI] [PubMed] [Google Scholar]
- 32.Gebhart CL, Sriadibhatla S, Vinogradov S, Lemieux P, Alakhov V, Kabanov AV. Design and formulation of polyplexes based on pluronic-polyethyleneimine conjugates for gene transfer. Bioconjug Chem. 2002;13:937–44. doi: 10.1021/bc025504w. [DOI] [PubMed] [Google Scholar]
- 33.Sahay G, Batrakova EV, Kabanov AV. Different internalization pathways of polymeric micelles and unimers and their effects on vesicular transport. Bioconjugate Chemistry (submitted) 2008 doi: 10.1021/bc8002315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rejman J, Bragonzi A, Conese M. Role of clathrin- and caveolae-mediated endocytosis in gene transfer mediated by lipo- and polyplexes. Mol Ther. 2005;12:468–74. doi: 10.1016/j.ymthe.2005.03.038. [DOI] [PubMed] [Google Scholar]
- 35.Rejman J, Conese M, Hoekstra D. Gene transfer by means of lipo- and polyplexes: role of clathrin and caveolae-mediated endocytosis. J Liposome Res. 2006;16:237–47. doi: 10.1080/08982100600848819. [DOI] [PubMed] [Google Scholar]
- 36.Verma IM, Somia N. Gene therapy -- promises, problems and prospects. Nature. 1997;389:239–42. doi: 10.1038/38410. [DOI] [PubMed] [Google Scholar]
- 37.Danson S, Ferry D, Alakhov V, Margison J, Kerr D, Jowle D, Brampton M, Halbert G, Ranson M. Phase I dose escalation and pharmacokinetic study of pluronic polymer-bound doxorubicin (SP1049C) in patients with advanced cancer. Br J Cancer. 2004;90:2085–91. doi: 10.1038/sj.bjc.6601856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Astafieva I, Maksimova I, Lukanidin E, Alakhov V, Kabanov A. Enhancement of the polycation-mediated DNA uptake and cell transfection with Pluronic P85 block copolymer. FEBS Lett. 1996;389:278–80. doi: 10.1016/0014-5793(96)00607-2. [DOI] [PubMed] [Google Scholar]
- 39.Godbey WT, Wu KK, Mikos AG. Poly(ethylenimine) and its role in gene delivery. J Control Release. 1999;60:149–60. doi: 10.1016/s0168-3659(99)00090-5. [DOI] [PubMed] [Google Scholar]
- 40.Kichler A, Chillon M, Leborgne C, Danos O, Frisch B. Intranasal gene delivery with a polyethylenimine-PEG conjugate. J Control Release. 2002;81:379–88. doi: 10.1016/s0168-3659(02)00080-9. [DOI] [PubMed] [Google Scholar]
- 41.Vinogradov SV, Batrakova EV, Li S, Kabanov AV. Mixed polymer micelles of amphiphilic and cationic copolymers for delivery of antisense oligonucleotides. J Drug Target. 2004;12:517–26. doi: 10.1080/10611860400011927. [DOI] [PubMed] [Google Scholar]
- 42.Godbey WT, Wu KK, Mikos AG. Tracking the intracellular path of poly(ethylenimine)/DNA complexes for gene delivery. Proc Natl Acad Sci U S A. 1999;96:5177–81. doi: 10.1073/pnas.96.9.5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brunner S, Furtbauer E, Sauer T, Kursa M, Wagner E. Overcoming the nuclear barrier: cell cycle independent nonviral gene transfer with linear polyethylenimine or electroporation. Mol Ther. 2002;5:80–6. doi: 10.1006/mthe.2001.0509. [DOI] [PubMed] [Google Scholar]
- 44.Itaka K, Harada A, Yamasaki Y, Nakamura K, Kawaguchi H, Kataoka K. In situ single cell observation by fluorescence resonance energy transfer reveals fast intra-cytoplasmic delivery and easy release of plasmid DNA complexed with linear polyethylenimine. J Gene Med. 2004;6:76–84. doi: 10.1002/jgm.470. [DOI] [PubMed] [Google Scholar]
- 45.Pitard B, Pollard H, Agbulut O, Lambert O, Vilquin J-T, Cherel Y, Abadie J, Samuel J-L, Rigaud J-L, Menoret S, Anegon I, Escande D. A Nonionic Amphiphile Agent Promotes Gene Delivery In Vivo to Skeletal and Cardiac Muscles. Human Gene Therapy. 2002;13:1767–1775. doi: 10.1089/104303402760293592. [DOI] [PubMed] [Google Scholar]
- 46.Ragin AD, Morgan RA, Chmielewski J. Cellular import mediated by nuclear localization signal Peptide sequences. Chem Biol. 2002;9:943–8. doi: 10.1016/s1074-5521(02)00189-8. [DOI] [PubMed] [Google Scholar]
- 47.Vacik J, Dean BS, Zimmer WE, Dean DA. Cell-specific nuclear import of plasmid DNA. Gene Ther. 1999;6:1006–14. doi: 10.1038/sj.gt.3300924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Garrean S, Gao XP, Brovkovych V, Shimizu J, Zhao YY, Vogel SM, Malik AB. Caveolin-1 regulates NF-kappaB activation and lung inflammatory response to sepsis induced by lipopolysaccharide. J Immunol. 2006;177:4853–60. doi: 10.4049/jimmunol.177.7.4853. [DOI] [PubMed] [Google Scholar]
- 49.Nguyen HK, Lemieux P, Vinogradov SV, Gebhart CL, Guerin N, Paradis G, Bronich TK, Alakhov VY, Kabanov AV. Evaluation of polyether-polyethyleneimine graft copolymers as gene transfer agents. Gene Ther. 2000;7:126–38. doi: 10.1038/sj.gt.3301052. [DOI] [PubMed] [Google Scholar]
- 50.Akinc A, Anderson DG, Lynn DM, Langer R. Synthesis of poly(beta-amino ester)s optimized for highly effective gene delivery. Bioconjug Chem. 2003;14:979–88. doi: 10.1021/bc034067y. [DOI] [PubMed] [Google Scholar]
- 51.Anderson DG, Peng W, Akinc A, Hossain N, Kohn A, Padera R, Langer R, Sawicki JA. A polymer library approach to suicide gene therapy for cancer. Proc Natl Acad Sci U S A. 2004;101:16028–33. doi: 10.1073/pnas.0407218101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Anderson DG, Akinc A, Hossain N, Langer R. Structure/property studies of polymeric gene delivery using a library of poly(beta-amino esters) Mol Ther. 2005;11:426–34. doi: 10.1016/j.ymthe.2004.11.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.