Abstract
UDP-glucuronosyltransferases (UGTs) are critical to the detoxification of numerous drugs, environmental pollutants, and endogenous molecules. However as yet not all of the human UGTs have been cloned and characterized. cDNA clones from the UGT2A3 gene (located on chromosome 4q13) were isolated using pooled human liver RNA. Approximately 10% of clones contained a c.1489A>G nucleotide substitution yielding proteins with a residue 497 alanine (UGT2A3.2) instead of a threonine (UGT2A3.1). The allele frequency of this polymorphism (rs13128286) was 0.13 in a European-American population as determined by direct DNA sequencing. Of 81 structurally diverse glucuronidation substrates tested, UGT2A3 expressed by a baculovirus system selectively glucuronidated bile acids – particularly hyodeoxycholic acid at the 6-hydroxy position. Apparent Km values of UGT2A3.1 and UGT2A3.2 for hyodeoxycholic acid 6-glucuronidation were 69±7 and 44±12 µM, respectively. Of 29 different extrahepatic tissues evaluated by real-time PCR, UGT2A3 mRNA was most highly expressed in small intestine (160% of liver), colon (78% of liver) and adipose tissue (91% of liver). An in silico scan of the proximal UGT2A3 promoter/5’-regulatory region identified transcription factor consensus elements consistent with tissue selective expression in liver (HNF1), and intestine (CDX2), as well as induction by rifampicin (PXR). In LS180 human intestinal cells, rifampicin increased UGT2A3 mRNA by more than 4.5-fold compared with vehicle while levels were not significantly affected by the AhR ligand β-naphthoflavone. This is the first report establishing UGT2A3 as a functional enzyme, and represents significant progress toward the goal of having a complete set of recombinant human UGTs for comparative functional analyses.
Introduction
The UDP-glucuronosyltransferase (UGT) enzymes catalyze the glucuronidation of exogenous compounds including dietary constituents, drugs, and environmental intoxicants, as well as endogenous compounds including steroids, other hormones, neurotransmitters, and bile acids, as well as their metabolites (Bock, 2003). The resulting glucuronide metabolite is generally inactive and easily excreted, thereby representing a critical final step in the termination of the physiological, pharmacological or toxicological activity of these compounds.
The UGT enzymes have been classified primarily based on gene sequence similarity with the UGT1 and UGT2 gene families accounting for all of the currently known UGTs (Mackenzie et al., 2005). Based on evaluation of the Build 35.1 draft of the human genome sequence, cDNAs for all but 2 of the human UGTs have been cloned, recombinant enzyme expressed, and shown to have glucuronidation activity for at least one or more substrates (Mackenzie et al., 2005). These exceptions include UGT2A2 and UGT2A3.
UGT2A2 is a predicted gene splice variant of UGT2A1, the UGT isoform that was first isolated from olfactory epithelium (Jedlitschky et al., 1999). The UGT2A2 splice variant is a consequence of utilization of an alternate promoter/exon 1 region downstream of the UGT2A1 promoter/exon 1 resulting in a unique UGT2A2 exon 1 that is spliced onto the shared UGT2A1 exons 2 to 6. Although tissue distribution or glucuronidation activity of this predicted enzyme has not been reported, preliminary evaluation of high throughput mouse cDNA sequences indicates that UGT2A2 is expressed in neonatal olfactory epithelium (Genbank ID: BC058786) (Strausberg et al., 2002).
UGT2A3 was originally identified as a novel UGT through screening a guinea pig (Cavia porcellus) liver cDNA library (Smith et al., 1999). Recombinant expression of this cDNA (Genbank ID: AF175221) in CHO cells resulted in an enzyme that showed relatively low levels of glucuronidation of several phenolic substrates including p-nitrophenol and 4-methylumbelliferone. Northern blot analysis also demonstrated UGT2A3 mRNA expression in guinea pig liver, intestine, and kidney. Preliminary evidence for expression of UGT2A3 in human tissues was derived from several high throughput sequencing projects that identified cDNA sequences highly similar to guinea pig UGT2A3 by screening either pooled human tissue cDNA libraries or HepG2 human hepatoma cells (Genbank ID: AK025587) (Clark et al., 2003).
A powerful method for identification of the major UGTs responsible for metabolism of existing drugs and those under development involves screening recombinant enzymes for glucuronidation of the candidate compound (Court, 2004). However this technique relies on having a complete complement of recombinant UGTs available for screening, and as yet human UGT2A2 and UGT2A3 have not yet been functionally characterized. Consequently, the primary aim of the work described here was to clone and express recombinant human UGT2A3 and verify enzymatic activity using a diverse set of substrates known to be glucuronidated by human liver. Our results demonstrate that UGT2A3 mRNA is most highly expressed in human tissues of greatest relevance to drug clearance, including liver, gastrointestinal tract, and kidneys. However, rather than glucuronidate multiple diverse substrates, as is typical of most UGTs, UGT2A3 was found to specifically glucuronidate bile acids, all of which are endogenous products of liver and/or intestinal bacterial metabolism. We also established that UGT2A3 is a polymorphic enzyme with 2 common variants (UGT2A3.1 and UGT2A3.2), which differ in apparent enzyme affinity constant (Km). Finally, we showed that UGT2A3 expression is inducible by the pregnane X receptor (PXR; NR1I2) ligand rifampicin in a human intestinal cell line. This is the first report to establish UGT2A3 as a functional human UDP-glucuronosyltransferase and represents a significant advance toward the goal of having a complete set of recombinant human UGTs for comparative functional analyses.
Materials and methods
Reagents
Unless otherwise indicated, most reagents including, UDP-glucuronic acid (UDPGA; sodium salt), 5-hydroxytryptamine hydrochloride (serotonin), 4-nitrophenol, and 4-nitrophenol glucuronide were purchased from Sigma-Aldrich (St. Louis, MO). Proprietary drugs used to screen for glucuronidation activity (Table 1) that were not available from Sigma-Aldrich were obtained from their manufacturer. Oligonucleotide primers were synthesized by the Tufts Core Facility (Tufts University, Boston, MA) using an automated DNA synthesizer (ABI 380B, PE Applied Biosystems, Foster City, CA). Recombinant UGT1A3 and UGT2B7 were purchased from BD-Gentest.
Table 1.
Substrates screened by HPLC-MS for glucuronidation by human UGT2A3 expressed in insect cells. (+) = glucuronide peak detected, (−) = no glucuronide peak detected in UGT2A3 incubations. All substrates were glucuronidated by human liver microsomes (positive control) but not by uninfected insect cells (negative control).
| Drug or metabolite | Food constituents | Bile acids | |||
|---|---|---|---|---|---|
| (−) | 3′-Azido-3′-deoxythymidine | (−) | Curcumin | (+) | Chenodeoxycholic acid |
| (−) | 5-(4-Hydroxyphenyl)-5-phenylhydantoin | (−) | Daidzein | (−) | Cholic acid |
| (−) | 5-Hydroxyrofecoxib | (−) | Eugenol | (−) | Dehydrocholic acid |
| (−) | Acetaminophen | (−) | Genistein | (+) | Deoxycholic acid |
| (−) | Amitriptyline | (−) | Lauric acid | (+) | Hyodeoxycholic acid |
| (−) | Bezafibrate | (−) | Naringenin | (−) | Lithocholic acid |
| (−) | Carbamazepine | (−) | Resveratrol | (+) | Ursodeoxycholic acid |
| (−) | Chloramphenicol | ||||
| (−) | Clofibrate | Other xenobiotics | Steroids | ||
| (−) | Clozapine | (−) | 8-Hydroxyquinoline | (−) | 17-α-ethynylestradiol |
| (−) | Codeine | (−) | 12-Hydroxydodecanoic acid | (−) | 17-α-ethynylestradiol-3-methyl ether |
| (−) | (−) Cotinine | (−) | 1-Naphthol | (−) | 17-α-methyltestosterone |
| (−) | Diclofenac | (−) | 4-Nitrophenol | (−) | 17α-Hydroxyprogesterone |
| (−) | E-4-OH-tamoxifen | (−) | 4-Methylumbelliferone | (−) | 19-Nortestosterone |
| (−) | Etoposide | (−) | Tert-butylhydroquinone | (−) | 5α-Pregnan-3α-ol-20-one |
| (−) | Ezetimibe | (−) | 5α-Androstan-3α, 17β-diol | ||
| (−) | Furosemide | (−) | Aldosterone | ||
| (−) | (S)-(+)-Ibuprofen | (−) | Corticosterone | ||
| (−) | Imipramine | (−) | Dehydroandrosterone | ||
| (−) | Ketotifen | (−) | Dihydrotestosterone | ||
| (−) | Lamotrigine | (−) | Estradiol | ||
| (−) | Lorazepam | (−) | Estriol | ||
| (−) | Lovastatin | (−) | Estrone | ||
| (−) | Morphine | (−) | Prednisone | ||
| (−) | Mycophenolic acid | (−) | Pregnenolone | ||
| (−) | Naproxen | (−) | Testosterone | ||
| (−) | (−)-Nicotine | ||||
| (−) | Oxazepam | Other endogenous substrates | |||
| (−) | Probenecid | (−) | All-trans retinoic acid | ||
| (−) | Propofol | (−) | Arachidonic acid | ||
| (−) | Propranolol | (−) | Bilirubin | ||
| (−) | Salicylic acid | (−) | Linoleic acid | ||
| (−) | Sulfaphenazole | (−) | Serotonin | ||
| (−) | Trifluoperazine | (−) | 5-Hydroxytryptophol | ||
| (−) | Trovafloxacin | (−) | 15-Hydroxyeicosatetraenoic acid | ||
| (−) | Valproic acid | ||||
| (−) | Z-4-OH-tamoxifen |
Microsomes
HLMs were prepared by differential ultracentrifugation as previously described (Court et al., 1997). Microsomal pellets were suspended in 0.1 M potassium phosphate buffer (pH 7.5) containing 20% glycerol and kept at −80°C until use. Protein concentrations of HLM samples were determined using the bicinchoninic acid protein assay (Pierce, Rockford, IL) with bovine serum albumin as the standard. The quality of the liver samples was ascertained by reference to at least 10 glucuronidation activities measured in this laboratory using the same set of livers. Livers that consistently showed low activity values (>2-fold lower for all measured activities) relative to the median activity value for the entire liver set had been excluded from study as we have previously reported (Court et al., 2004; Krishnaswamy et al., 2005).
Cloning of human UGT2A3
Human UGT2A3 cDNA (coding region only) was amplified and cloned from human liver total RNA by RT-PCR. Briefly, 2 ug of total RNA (pooled from 10 different human livers in order to capture the most common UGT2A3 mRNA variant) was reverse transcribed (Superscript II, Invitrogen), cDNA amplified by PCR (Platinum HiFi, Invitrogen) using primers Pri189 (TTG CAG ATC AGT GTG TGA GGG AAC TG) and Pri190 (CCC CAT CAG GTC TTT CTT GAA TTT GG), and cloned into pcDNA3.1. Isolated clones were sequenced using primers Pri189, Pri190 and Pri276 (ACA GGA AAC CAA CTA CGA TGT AAT).
UGT2A3 expression in mammalian and insect cells
Initial studies were performed in an attempt to express UGT2A3 cDNA by transient and stable transfections of the human embryonic kidney (HEK) 293 cell line. However since the resultant membrane preparations showed poor enzyme expression, we scaled up enzyme production using the Baculovirus-insect cell system which is most commonly used for commercial recombinant UGT preparations. For this the enzyme coding region was subcloned via the pENTR vector into the Baculodirect baculovirus expression system (Invitrogen). Resulting virus was amplified by 3 passages through cultured Sf9 cells, and protein subsequently expressed in 50 mL suspension cultures of Sf21 cells with cells harvested 2 days post-infection. Crude cell extracts were prepared by washing harvested cells 3 times in phosphate buffered saline, lysing cells by resuspending in pure water and sonicating briefly 3 times on ice, followed by addition of an equal volume of 100 mM potassium phosphate buffer (pH 7.5). Resultant homogenates were stored at −80°C until assay. The UGT2A3.2 variant enzyme (T497A) was generated in parallel with the wild-type enzyme. For this, a pENTR vector encoding the variant enzyme was derived by site-directed mutagenesis (Quikchange, Stratagene) of the pENTR plasmid encoding the wild-type enzyme through substitution of a guanine for an adenine at cDNA position 1489 (relative to adenine of the start codon) resulting in an alanine substituted for a threonine at amino acid position 497 (relative to the start methionine).
Immunoblotting of recombinant UGT2A3
Insect cell homogenates (50 ug protein per well) were immunoblotted using a method we have described previously (Court, 2001). Blots were probed using the RAL antibody (a generous gift of Dr Brian Burchell, The University of Dundee, Dundee, Scotland), which readily detects both UGT1 and UGT2 family isoforms (Coughtrie et al., 1988). Pooled human liver microsomes (n=54) obtained from our human liver bank (details as given previously (Hesse et al., 2004)) were used as a positive control.
Glucuronidation activity screen
Both wild-type UGT2A3.1 and variant UGT2A3.2 recombinant enzyme preparations were screened for glucuronidation activity using a large range (n = 81) of structurally diverse substrates, with molecular weights ranging from 138 (salicylic acid) to 588 (etoposide), and including drugs, drug metabolites, environmental intoxicants, food constituents, bile acids, steroids, and other endogenous hormones and neurotransmitters (Table 1). Pooled human liver microsomes and uninfected insect cells were used as positive and negative controls, respectively. Incubations (100 uL) were performed as previously described with minor modifications (Court, 2005). Aglycone substrate concentration was 100 uM and UDPGA concentration was 5 mM. Protein concentration was 0.5 mg / mL and incubation time was 6 hours. Reactions were terminated by addition of 50 uL of acetonitrile containing 5% acetic acid, vortexing, and cooling on ice. After centrifuging for 10 minutes at 14,000 g supernatants were dried in a vacuum oven at 45°C, reconstituted with 100 uL of water and analyzed by LC-MS.
LC-MS apparatus consisted of an HPLC instrument (Surveyor, Thermofinnigan, Somerset, NJ) with a 2 mm × 150 mm reverse phase column (Synergi Fusion-RP, Phenomenex), serially connected to a UV absorbance diode array detector (Surveyor) and an ion trap mass spectrometry detector with electrospray ionization source (Deca XP Plus, Thermofinnigan, Somerset, NJ). The mass detector was set to alternately monitor both positive and negative ions in full scan mode over a 120 to 2000 m/z range. Mobile phase was flowed at 0.5 mL / min and consisted of 10 mM formic acid in water (mobile A) and acetonitrile (mobile B) run as a linear gradient starting at 100% mobile A and increasing to 100% mobile B over 15 minutes followed by equilibration at 100% mobile A for 10 minutes resulting in a 25 minute total run time. Glucuronide metabolites were identified in positive control samples (HLM incubations) as chromatographic peaks with a major ion mass exactly 176 mass units greater than the aglycone substrate. The retention times of these glucuronide peaks were then used to identify possible glucuronide metabolites in recombinant UGT2A3 incubations in comparison with negative control incubations (insect cells).
Hyodeoxycholic acid glucuronidation assay
A hyodeoxycholic acid glucuronidation assay was developed based on the method described above except that the LC-MS method was adjusted to enable separation of individual glucuronide isomers (i.e. 3-hydroxy, 6-hydroxy and 24-carboxy). Briefly, an isocratic mobile phase consisting of 10 mM ammonium acetate (pH 4.5), methanol, and acetonitrile (mixed at 52:26:22, respectively) was run at 0.5 mL / min using a 2 mm × 150 mm C18 column (Synergi Hydro-RP, Phenomenex). Negative ions with m/z− of 567.4 (hyodeoxycholic acid glucuronides) and 391.6 (hyodeoxycholic acid) were measured.
Hyodeoxycholic acid glucuronide standards were not available for identification or quantitation purposes. Consequently, individual glucuronides were identified by use of recombinant UGT2B7 (primarily produces the 6-hydroxy glucuronide), UGT1A3 (primarily produces 24-carboxy glucuronide) and effects of β-glucuronidase (hydrolyzes all glucuronides) and alkalinization (hydrolyzes 24-carboxy glucuronide only) as previously reported (Gall et al., 1999). Hyodeoxycholic acid glucuronide concentrations were approximated by use of a standard curve generated using 0.5 to 10 uM hyodeoxycholic acid and results expressed as glucuronide concentration equivalents.
Preliminary studies were conducted to ensure glucuronide formation by evaluated enzyme preparations were linearly dependent on incubation time (up to 6 hours) and enzyme concentration (up to 2 mg protein / mL). The effect of hyodeoxycholic acid concentration on UGT2A3 (wild-type and variant) glucuronidation activity using expressed enzyme as source was evaluated over a concentration range of 5 to 200 µM with an incubation time of 3 hours and protein concentration of 2 mg / mL. Substrate concentrations over 200 µM were not evaluated since they appeared to result in decreased activity consistent with substrate inhibition. Activity data were then used to derive kinetic parameter estimates of apparent Km and Vmax by nonlinear regression analysis (Sigmaplot).
UGT2A3 mRNA quantitation in human tissues
Real-time PCR analysis (Applied Biosystems, USA) was used to quantitate and compare UGT2A3 mRNA expression (normalized to 18S rRNA expression) in a range of human tissues based on a method we have reported previously (Kaivosaari et al., 2007) with minor modifications as follows. Pooled total RNA was prepared from our human liver bank samples (n=47). All donors were of self-described European-American ancestry and included both males (n=36) and females (n=11), while other donor details have been reported previously (Hesse et al., 2004). Total RNA from human kidney (male Asian donor), fetal liver (63 European-Americans), small intestine (5 European-Americans), colon (3 European-Americans), trachea (pooled Male European-Americans), lung (male Asian donor), adrenal gland (62 European-Americans), bone marrow (7 European-Americans), heart (male Asian donor), placenta (7 European-Americans), prostate (47 European-Americans), salivary gland (24 European-Americans), skeletal muscle (2 European-Americans), spleen (14 European-Americans), testis (19 European-Americans), thymus (9 European-Americans), thyroid (65 European-Americans), uterus (10 European-Americans), breast carcinoma (female European-American donor), colon adenocarcinoma (female European-American donor), cerebellum (24 European-Americans), fetal brain (59 European-Americans), and whole brain (male Asian donor) were purchased from BD-Clontech (USA). Total RNA from human stomach (male European-American donor), pancreas (male European-American donor), ovary (female African American donor), breast (female European-American donor), adipose (female donor), and prostate adenocarcinoma (male donor) were from Ambion (USA). Human olfactory epithelium cDNA was a generous gift from Dr Xinxin Ding (Wadsworth Center, New York State Department of Health, Albany, NY).
Primer pair sequences were as follows: CCC CTC GAT GCT CTT AGC TGA GTG T (18S-rRNA-forward), CGC CGG TCC AAG AAT TTC ACC TCT (18S-rRNA-reverse), ATG AGG TAA CAG TAT TGA CTC ACT CAA AGC (UGT2A3-forward), and GTT GAT AAG CCT GGC AAG ACA TTC A (UGT2A3-reverse). Amplification specificity was ensured initially by sequencing of representative PCR products, and in each run by PCR product duplex melting temperature analysis. Negative controls included exclusion of cDNA template and reverse transcription enzyme. Linearity of response over the concentration measured was ensured using standard curves of PCR threshold cycle number versus concentration of template, which were derived from serial dilutions of purified PCR product. For each tissue, cDNA reactions were performed in triplicate and quantitative PCR reactions were performed in duplicate. UGT2A3 mRNA results for each tissue were normalized to 18S rRNA level expressed as a mean (±SE) percentage relative to that of the (pooled) adult liver values. Assay precision as reflected by the coefficient of variation of replicates averaged 19%, which compares well with previously published values (Karlen et al., 2007).
UGT2A3 exon 6 sequencing
Genomic DNA was isolated from our human liver bank tissues representing 48 individual European-American donors (details published previously (Court et al., 2004)) using DNAzol (Invitrogen, Carlsbad, CA). PCR reactions were performed using Platinum Hifi polymerase (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions. The complete UGT2A3 exon 6 was amplified by genomic PCR using primers Pri312 (TGT TCT TGG CTT GCA TAA CAT ATA CTA CGG) and Pri 190 (sequence given above). Thermal cycler (PTC200, MJ Research, Waltham, MA) settings included an initial denaturation step (95°C) for 10 minutes followed by 40 cycles of denaturation (95°C) for 30 sec, anneal (63°C) for 30 sec, and extend (72°C) for 1 minute with a final extension cycle at 72°C for 10 minutes. PCR products were treated with alkaline phosphatase (ExoSAPit, USB) and directly sequenced using primer Pri312.
UGT2A3 induction in LS180 human intestinal cell line
LS180 cells (ATCC) were seeded onto 6-well plates and grown to 80% confluency using Dulbecco’s Modified Eagle’s Medium (DMEM), supplemented with 10% charcoal stripped fetal bovine serum, 0.1 mM non-essential amino acids, 25 mM HEPES buffer, 100 units/ml penicillin, and 0.1 mg/ml streptomycin. Compounds assayed for induction included rifampin (10 µM), dexamethasone (1 µM) and β-napthoflavone (50 µM) dissolved in DMSO (0.1% final concentration). Cells were incubated in 2 mL of media containing these treatments (or 0.1% DMSO vehicle alone) for 24 hr, after which cells were trypsinized, washed 3 times in phosphate buffered saline and total RNA isolated. Relative UGT2A3 mRNA and 18S rRNA levels were measured by real-time PCR as described above. UGT1A1 mRNA levels were also assayed in the same samples using primers GCT TTT GTC TGG CTG TTC CCA CT (UGT1A1-forward) and TCG AAG GTC ATG TGA TCT GAA TGA GA (UGT1A1-reverse). Experiments were conducted in quadruplicate and data expressed as a mean (± SE) ratio relative to vehicle control. The effect of induction treatment relative to that of control was evaluated by Mann-Whitney U test with a p value of less than or equal to 0.05 considered statistically significant.
Results
Identification of a novel UGT expressed in human liver
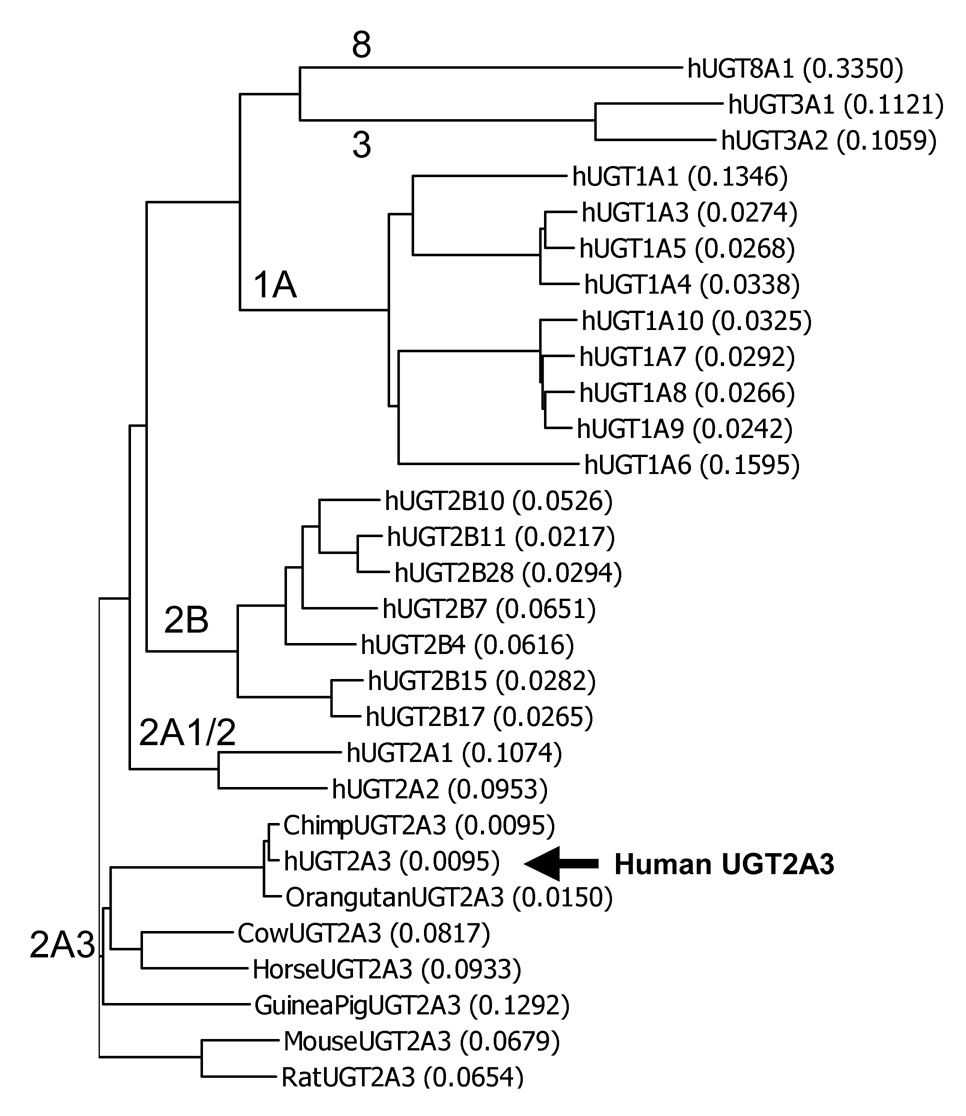
A BLAST sequence homology search (http://www.ncbi.nlm.nih.gov/BLAST/) of the entire human genome sequence was performed using the complete coding region of the guinea pig UGT2A3 cDNA sequence (Genbank ID: AF175221). A single novel UGT2A3-like gene with a predicted full length mRNA transcript (by homology to AF175221) was identified on chromosome 4q13 with the entire gene spanning 21,948 bp (including protein start to stop codons). The UGT2A3 gene was located within a large cluster of other genes encoding all known human UGT2B and UGT2A isoforms as well as a series of UGT2A and UGT2B-like pseudogenes as described previously (Mackenzie et al., 2005). The adenine nucleotide of the start codon was located at 476,626 of the Genbank reference sequence NT_077444.3 with the gene on the negative strand. The gene was predicted to contain 6 exons spliced into mature mRNA with protein coding region of 1581 bp (527 amino acids) in length. Expressed sequence tag (EST) analysis showed mRNA expression in human liver, as well as small intestine and kidney. Consequently, human liver total RNA was used to amplify and clone the entire coding region of the predicted cDNA by reverse transcription PCR using primers located in the 5’ - and 3’-untranslated regions. Phylogenetic analysis (Figure 1) of the derived novel UGT amino acid sequence and all other known human UGT isoforms indicated closest homology with the known UGT2A subfamily members (UGT2A1 and UGT2A2) and a more distant relationship with the UGT2B and UGT1A subfamily isoforms, and also with the UGT3 and UGT8 family enzymes. The naming of the novel UGT isoform as human UGT2A3 was subsequently submitted to and approved by the UGT nomenclature committee (http://som.flinders.edu.au/FUSA/ClinPharm/UGT/). The complete sequence of the cloned UGT2A3 cDNA was submitted to Genbank with accession number AY542891.
Figure 1.
Dendrogram showing amino acid sequence similarity for all known human UGTs (hUGTs) and all known UGT2A3 orthologs identified in 7 other mammalian species. Sequences were aligned by Clustal W and tree generated by Growtree as implemented in Vector NTI version 9. Each tree bifurcation identifies individual sequences and/or groups of sequences with greatest similarity. The length of each tree branch is proportional to the number of amino acid differences (expressed as a proportion of total sequence length) between a particular sequence and its theoretical ancestral sequence (shown in parentheses for each sequence). Genbank sequence files for UGT2A3 orthologs are given in the text, while those for hUGTs are provided elsewhere (Mackenzie et al., 2005).
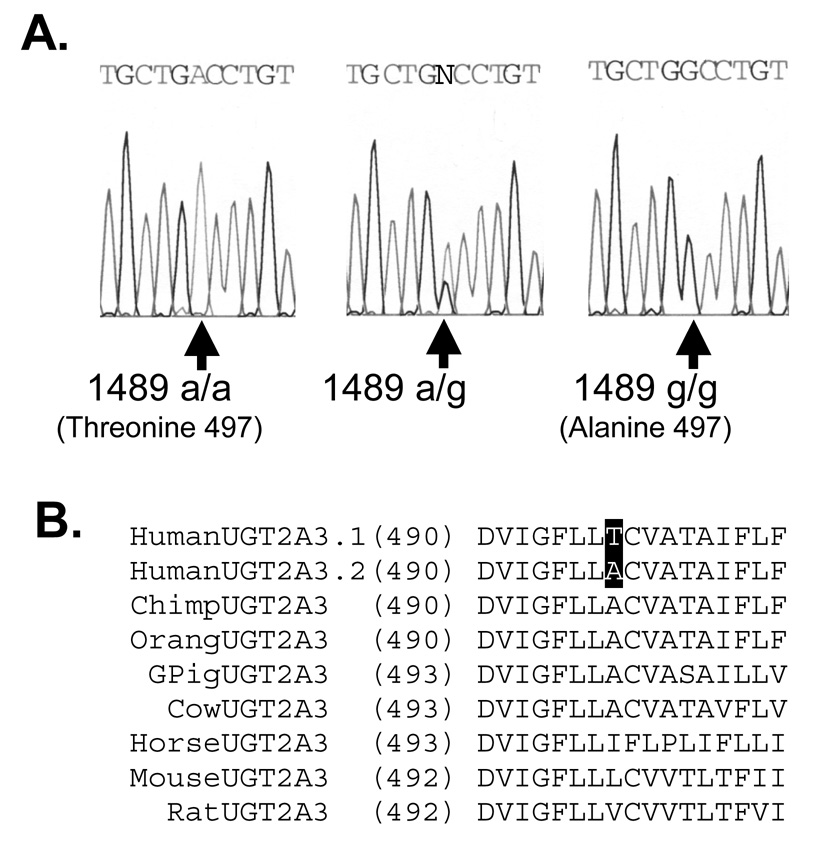
UGT2A3 gene encodes a polymorphic enzyme
Approximately 10% of amplified UGT2A3 cDNA clones that were sequenced contained an adenine to guanine nucleotide substitution at position 1489 (1489A>G relative to adenine of start codon) yielding a predicted amino acid substitution of threonine to alanine at position 497 (T497A relative to start methionine). A search of the SNP database indicated that this SNP (rs13128286) was also detected by several high throughput screening projects although had not yet been validated by direct experimentation. This also appeared to be the only coding (cSNP) identified in the UGT2A3 gene by these methods. Consequently, to confirm the presence of the 1489A>G cSNP (termed here UGT2A3*2), the entire exon 6 region of the UGT2A3 gene was sequenced using genomic DNA extracted from our human liver bank samples representing 48 individual European-American donors. As shown in Figure 2A, analysis of sequence chromatograms identified 37 individuals that were homozygous wild-type (497 T/T), 10 individuals that were heterozygous (497 T/A) and 1 individual that was homozygous variant (497 A/A) resulting in a minor allele frequency for the UGT2A3*2 polymorphism of 0.13. This distribution of genotypes was consistent with the expected Hardy-Weinberg equilibrium (X2 = 0.11; p = 0.71).
Figure 2.
Coding single nucleotide polymorphism identified in human UGT2A3 gene exon 6. A. Shown are the sequence chromatograms from representative homozygous reference (1489 a/a), heterozygote (1489 a/g), and homozygous variant (1489 g/g) DNA samples. This polymorphism is predicted to result in a threonine or alanine at amino acid position 497 resulting in either UGT2A3.1 or UGT2A3.2 protein variants, respectively. DNA was extracted and PCR amplified from human liver bank specimens. B. Alignment of all known UGT2A3 orthologs showing that the less common UGT2A3.2 protein variant is most likely the ancestral form, while there is incomplete species conservation of the alanine at this position.
The ancestral origin of this polymorphism and conservation of amino acid sequence at this position was determined by alignment of the human UGT2A3 wild-type sequence (UGT2A3.1), the human variant sequence (UGT2A3.2) with UGT2A3 sequences from all other mammalian species identified in the Genbank database (accessed 1-11-2008) including chimpanzee (Pan troglodytes; XM_526602.2), orangutan (Pongo pygmaeus; Q5RFJ3), Guinea pig (Cavia porcellus; AF175221.1), cow (Bos taurus; XM_589136.3), horse (Equus caballus; XM_001498022.1), mouse (Mus musculus; NM_028094.2) and rat (Rattus norvegicus; XM_223289.2). As shown in Figure 2B the less common human UGT2A3.2 is likely the ancestral allele since alanine was identified at this position in a closely related primate species (chimp and orangutan) as well as in several other mammalian species including Guinea pig and cow. However there was not complete conservation of alanine at this position in that horse, mouse, and rat showed substitutions with isoleucine, leucine, and valine (respectively).
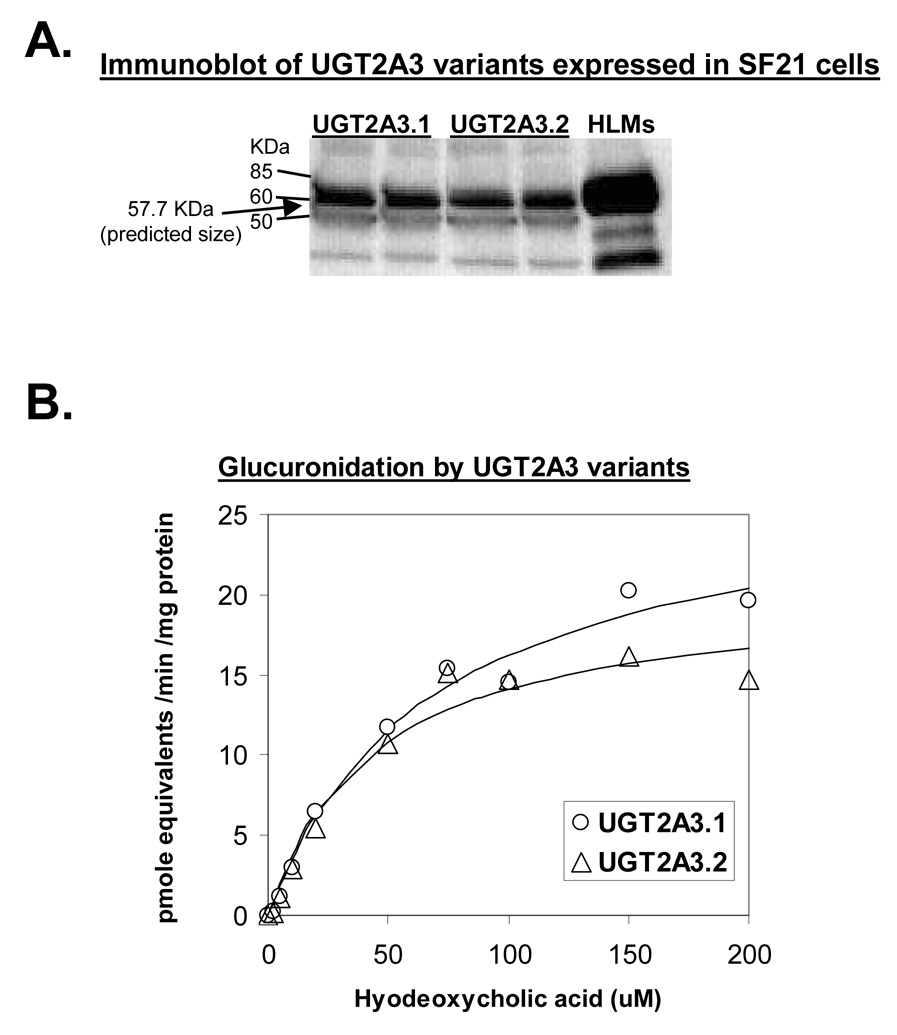
Recombinant expression of UGT2A3
Initial studies involving either transient or stable transfection of HEK293 mammalian cells failed to produce sufficient protein (either native or epitope tagged) that could be readily detected by immunoblotting. Subsequently, recombinant expression using a baculovirus-insect cell system yielded ample enzyme protein for immunoblotting and functional studies. Immunoblotting analysis using a UGT specific antibody yielded a single major band with a molecular size of approximately 56 to 58 kDa (Figure 3A). Several smaller minor bands were also observed possibly representing proteolytic cleavage products or nonspecific antibody binding. Based on the translated cDNA sequence (Vector NTI ver. 9.1), the nascent protein size of UGT2A3 was expected to be 60.3 kDa. However, cleavage of an N-terminal signal peptide sequence found in all other UGT enzymes was predicted by SignalP version 3.0 (Bendtsen et al., 2004) to occur between amino acids 23 and 24. This would yield a mature protein of 57.7 kDa in size, which agrees well with the observed immunoblotting results and is similar in size to other human UGTs.
Figure 3.
UGT2A3 variants expressed in a baculovirus-insect cell system. A. Immunoblot of homogenate of SF21 insect cells (50 ug protein / lane) that had been infected with baculovirus containing cDNAs coding for UGT2A3.1 or UGT2A3.2. Pooled human liver microsomes (5 ug) were used as positive control. Blots were probed using a sheep anti-rat UGT antibody that readily recognizes all UGT1 and UGT2 family enzymes. Arrow at 57.7 kDa indicates the predicted size of the mature UGT2A3 protein. B. Plot of hyodeoxycholic acid concentration versus glucuronide formation for UGT2A3.1 and UGT2A3.2 recombinant enzymes. Shown also are fitted curves using the Michaelis-Menten enzyme kinetic model. Derived enzyme kinetic parameters are given in Table 2. Glucuronide standards were not available and so activities are expressed as glucuronide equivalents using a hyodeoxycholic acid standard curve.
UGT2A3 selectively glucuronidates bile acids
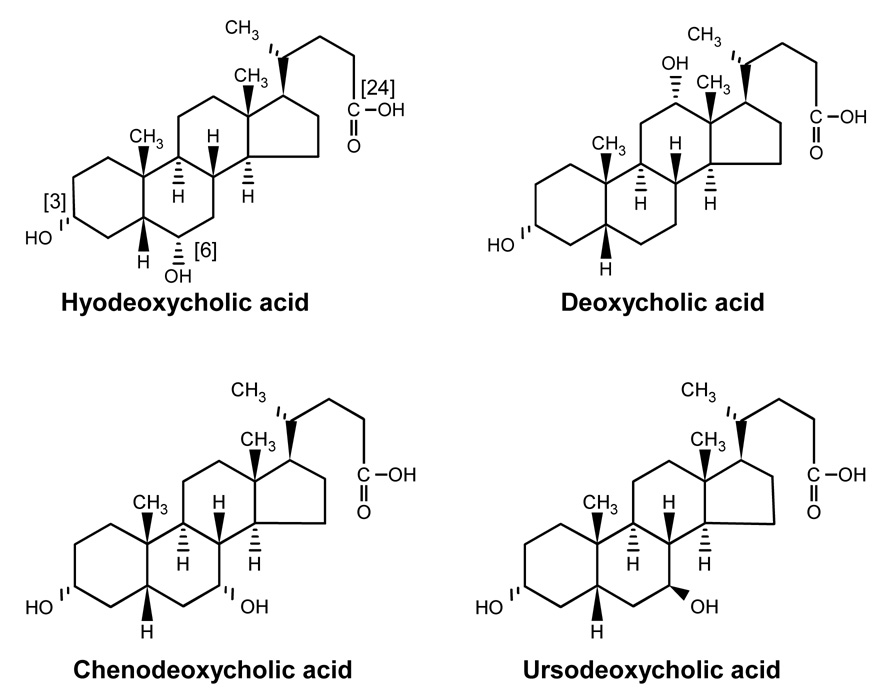
A large number of potential substrates (N=81) known to be glucuronidated by human liver were screened for glucuronidation by recombinant UGT2A3.1 and UGT2A3.2 using long duration incubations coupled with identification of metabolite peaks formed as glucuronides by HPLC-MS. The positive control was human liver microsomes, while the negative control was uninfected insect cells. As shown in Table 1, only 4 of these substrates showed evidence for glucuronidation by UGT2A3 including hyodeoxycholic acid, deoxycholic acid, chenodeoxycholic acid, and ursodeoxycholic acid, all of which are naturally occurring bile acids (see Figure 4). Hyodeoxycholic acid appeared to be the most active substrate with 5 to 20 times higher glucuronide HPLC peak heights compared with these other substrates. Consequently, hyodeoxycholic acid was chosen as a substrate for further enzyme characterization.
Figure 4.
Structures of bile acids that were glucuronidated by UGT2A3. Shown on hyodeoxycholic acid are the possible glucuronidation sites including 3-hydroxy, 6-hydroxy, and 24-carboxy.
To ensure that insect cell expressed UGT2A3 behaved similarly to mammalian cell expressed UGT2A3, microsomes of HEK293 cells that had been transiently transfected with a UGT2A3.1 pcDNA3.1 construct were also evaluated for glucuronidation of hyodeoxycholic acid, as well as two phenolic substrates that had been previously reported to be glucuronidated by guinea pig UGT2A3(Smith et al., 1999). While there was low but detectable glucuronidation activity for hyodeoxycholic acid (~2 pmoles/min/mg protein), glucuronidation of 4-nitrophenol and 4-methylumbelliferone was not detected (< 1 pmole / min / mg protein). Empty vector control transfected HEK293 cells showed no activity for any substrate evaluated.
Since both HLMs and UGT2B7 have been shown to glucosidate hyodeoxycholic acid (Mackenzie et al., 2003), we also evaluated whether UGT2A3 was capable of this activity. In contrast to HLMs, which readily formed hyodeoxycholic acid glucoside (m/z+ = 555.6) in the presence of UDP-glucose, insect cell and mammalian cell expressed UGT2A3 preparations were not capable of utilizing this alternate sugar donor (data not shown).
UGT2A3 allozymes vary in hyodeoxycholic acid glucuronidation kinetics
Enzyme kinetic parameters were determined for hyodeoxycholic acid 6-hydroxyglucuronidation by the wild-type UGT2A3.1 and variant UGT2A3.2 insect cell preparations. As shown in Figure 3B, both enzyme variants were active in glucuronidation of hyodeoxycholic acid with Km values in the low micromolar range. A comparison of the kinetic parameters derived for each enzyme variant (given in Table 2) indicated that apparent Km estimates were somewhat lower for UGT2A3.2 (44±12 µM) compared with UGT2A3.1 (69 ± 7 µM). However, this was offset by somewhat higher Vmax values such that Vmax/Km values were essentially similar.
Table 2.
Enzyme kinetic parameter estimates obtained for hyodeoxycholic acid glucuronidation by human UGT2A3 variants (UGT2A3.1 and UGT2A3.2) expressed in SF21 insect cells.
| UGT2A3.1 |
UGT2A3.2 |
|||
|---|---|---|---|---|
| Estimate | SE | Estimate | SE | |
| Vmax (pmoles/min/mg) | 27 | 2 | 20 | 2 |
| Km (µM) | 69 | 7 | 44 | 12 |
| Vmax / Km (mL/ min /gm) | 0.40 | 0.04 | 0.46 | 0.13 |
UGT2A3 glucuronidates hyodeoxycholic acid at the 6-hydroxy position
Hyodeoxycholic acid contains 3 potential sites for glucuronidation (3-hydroxy, 6-hydroxy, and 24-carboxy) and the HPLC assay we used for activity screening employed a rapid solvent gradient that would not differentiate between these glucuronide isomers. Consequently, we developed an isocratic HPLC method that enabled us to separate the glucuronides formed by HLMs as well as by human intestinal and kidney microsomes. This method was then used to determine the regioselectivity of UGT2A3 in comparison with recombinant UGT1A3 and UGT2B7. These latter UGT isoforms were evaluated since previous work (Gall et al., 1999) indicates that UGT1A3 preferentially glucuronidates at the 24-carboxy position, while UGT2B7 glucuronidates at the 6-hydroxy position, thereby enabling identification of each of these glucuronide peaks (see Methods section for details).
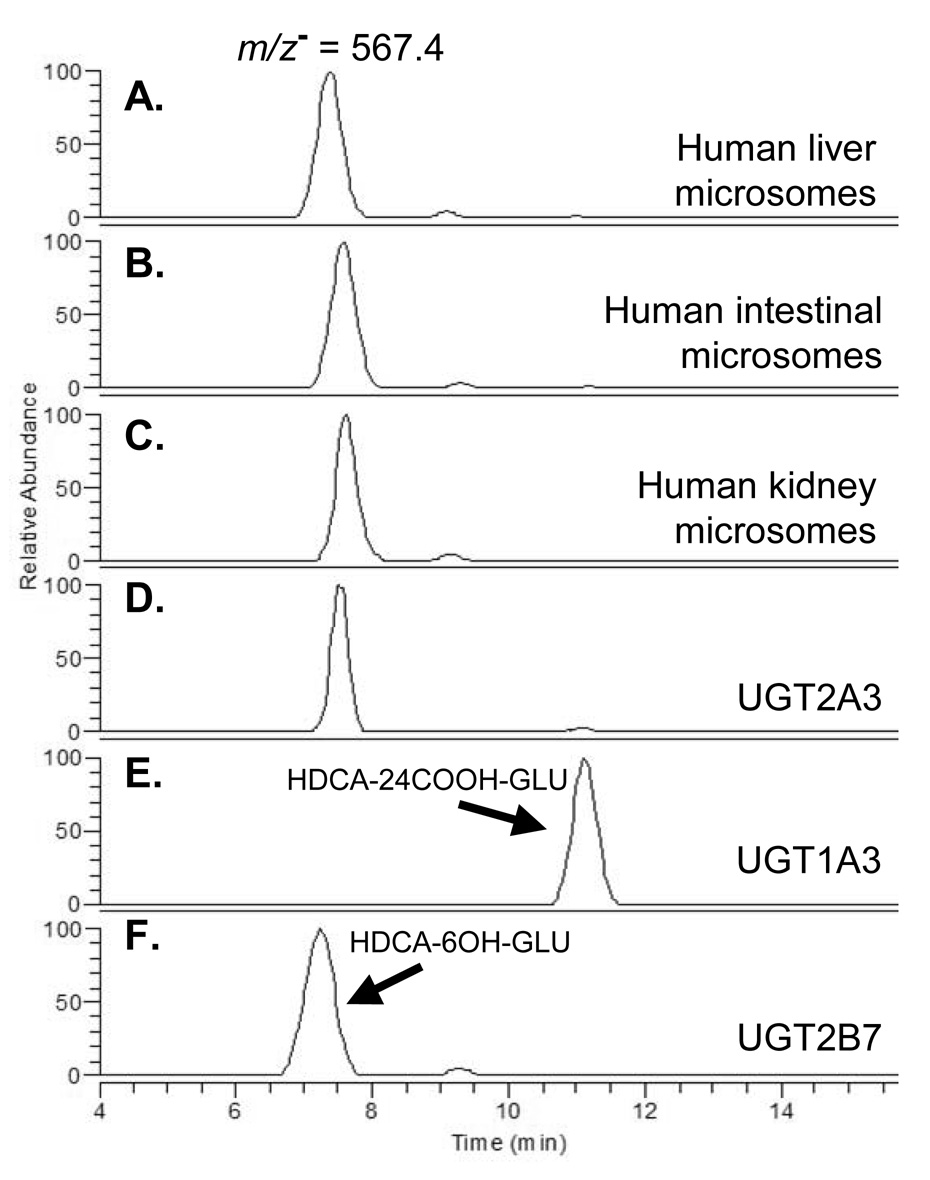
As shown in Figure 5, a major glucuronide peak most likely representing the 6-hydroxy-hyodeoxycholic acid glucuronide was identified at 7.3 minutes in the UGT2B7 incubation. This was also the major glucuronide peak formed by human liver, kidney and intestinal microsomes as well as by UGT2A3.1 and UGT2A3.2. The UGT1A3 incubation exclusively formed a glucuronide peak with a retention time of 11.2 minutes, most likely representing the 24-carboxy-hyodeoxycholic acid glucuronide. Much smaller 24-carboxy peaks (relative to the 6-hydroxy peak) were also identified in human liver and intestinal microsome incubations, as well as UGT2A3.1 and UGT2A3.2, but not in UGT2B7 or human kidney microsome incubations. As expected for alkaline sensitive acyl-glucuronides, the 24-carboxy peak (but not the 6-hydroxy peak) was lost after treatment with sodium hydroxide. The order of glucuronide peak elution from the HPLC column (6-hydroxy then 24-carboxy) is identical to that recently reported by Caron et al (Caron et al., 2006) further supporting our peak assignments. Another small glucuronide peak was identified at 9 minutes retention time in human liver, intestinal and kidney microsomes and also in UGT2B7 incubations, but not in UGT1A3, UGT2A3.1 or UGT2A3.2 incubations. By exclusion, this could represent the 3-hydroxy-hyodeoxycholic acid glucuronide, although this contention would need to be confirmed by definitive structural studies such as nuclear magnetic resonance. All three putative glucuronide peaks were absent in negative control incubations (use of boiled enzyme or exclusion of UDPGA).
Figure 5.
HPLC-MS chromatograms of hyodeoxycholic acid glucuronide positional isomers. Samples were generated using either human tissue microsomes or recombinant UGTs incubated with hyodeoxycholic acid and UDP-glucuronic acid and separated by HPLC with mass spectrometry detection monitoring the predicted negative ion of hyodeoxycholic acid glucuronide (m/z−=567.4). This method enabled complete separation of positional isomers and showed that UGT2A3, UGT2B7, liver, kidney and intestines glucuronidate hyodeoxycholic acid at the 6-hydroxy position, while UGT1A3 glucuronidates hyodeoxycholic acid at the 24-carboxy position.
UGT2A3 is expressed primarily in human intestines, liver, and adipose tissue
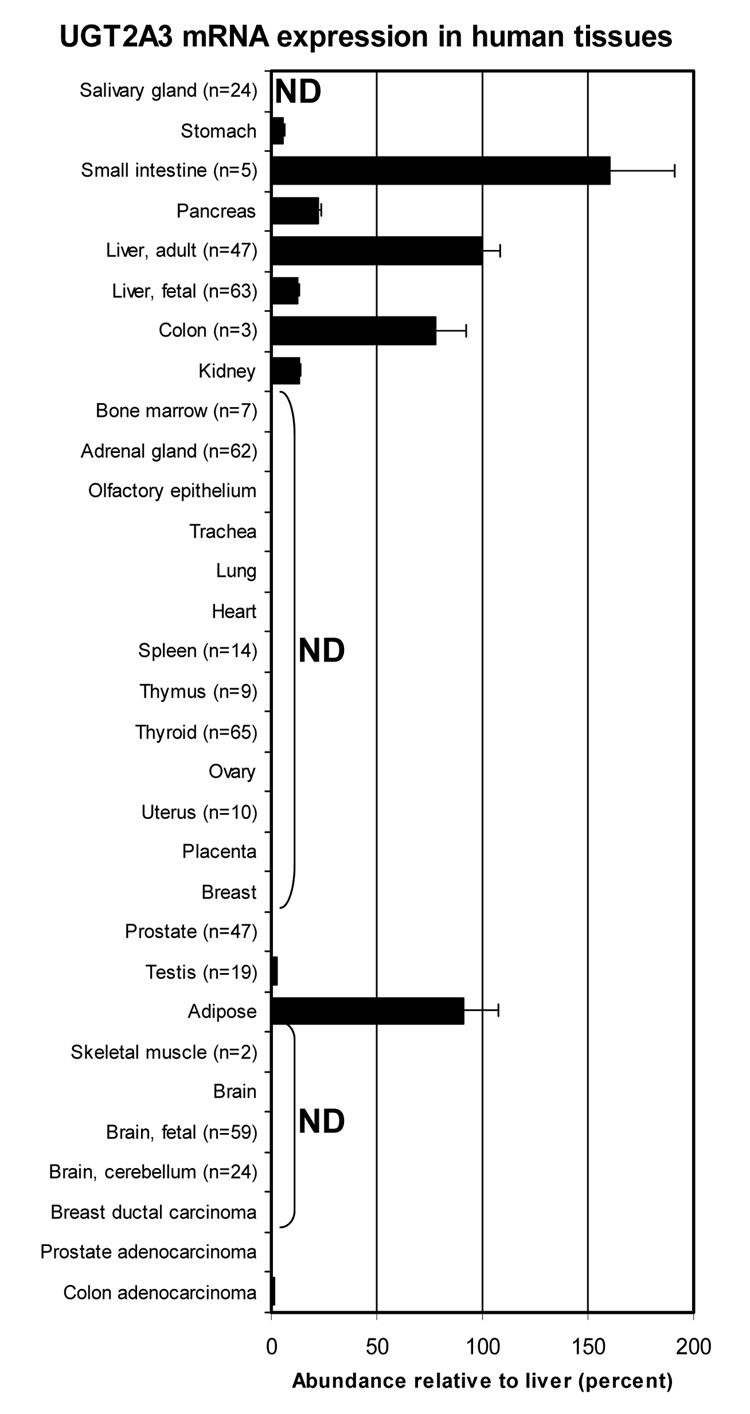
A range of human tissues were screened by quantitative real-time PCR to identify tissues expressing UGT2A3 in addition to adult human liver. As shown in Figure 6, UGT2A3 was highly expressed in the intestinal tract with greatest expression in the small intestine (60% higher than liver) followed by colon (78% that of liver). Relatively high expression was also found in adipose tissue (91% of liver). Somewhat lower levels were found in pancreas (22% of liver), stomach (6% of liver), kidney (13% of liver), fetal liver (13% of liver) and testis (3% of liver). Trace levels (<1% of liver) were found in bone marrow, adrenal, and colon adenocarcinoma, while the remaining tissues were below the level of detection.
Figure 6.
Distribution of UGT2A3 mRNA in human tissues. RNA samples from commercial sources and our human liver bank (details provided in text) were assayed for UGT2A3 mRNA content by real-time quantitative RT-PCR. Results were normalized to 18S rRNA content using standard curves generated for each molecule. ND = not detected. Bar gives average (n = 6) with standard error. A tissue without a visible bar indicates that UGT2A3 mRNA was detected but abundance was less than 1% relative to adult liver.
UGT2A3 proximal promoter contains putative HNF1α, CDX2, PXR and GR consensus elements
The UGT2A3 mRNA transcription start site was identified at −26 bp relative to the translation start site based on the mRNA sequence NM_024743.2, which contains the most 5’-untranslated mRNA sequence of all human UGT2A3 sequences deposited to date in the Genbank database. Upstream of this, the promoter contains a TATA box element position located at 70 to 73 bp upstream of the start translation codon.
Rodent UGT2A3 was recently shown to be regulated by pregnane X receptor (PXR; NR1I2) and glucocorticoid receptor (GR; NR3C1) activators in rat liver and intestine (Hartley et al., 2004). Consequently, further in silico analysis (Matinspector Version 7.4.8.2 with Matrix Family Library Version 6.3, www.genomatix.de) was used to scan the proximal UGT2A3 promoter/5’-regulatory region for the presence of putative binding sites for transcription factors previously shown to be important for tissue specific expression of UGTs in liver (hepatocyte nuclear factor 1; HNF1) and intestine (caudal-related homologue 2; CDX2), as well as for PXR and GR binding elements (Gregory et al., 2004). Using this approach, multiple HNF1 sites and one CDX2 site could be identified within 250 bp of the promoter. Interestingly, a PXR/RXR heterodimer DR4 site was found to partially overlap the CDX2 site, while a GR IR3 site was nested between the two HNF1 sites.
UGT2A3mRNA is induced by rifampicin in LS180 human intestinal cell line
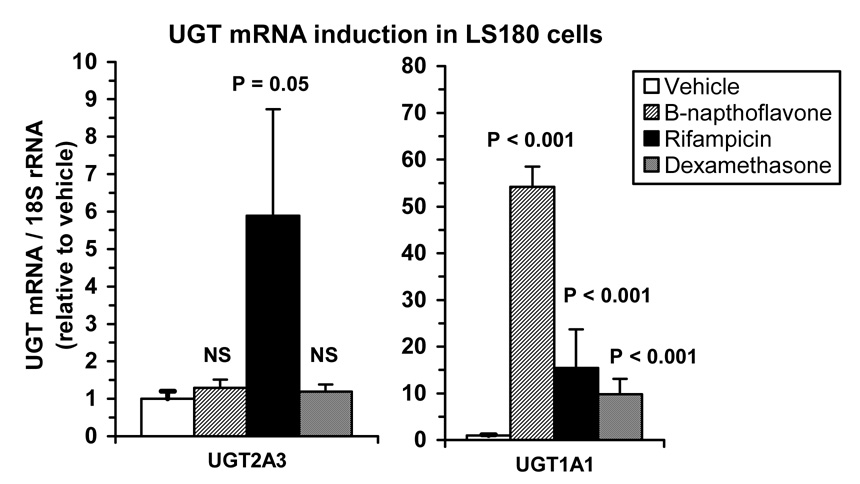
LS180 human intestinal cells were used to evaluate the potential for induction of UGT2A3 mRNA expression by a PXR ligand (rifampicin) and a GR ligand (dexamethasone) in this model system. The effect of treatment with β-napthoflavone (a model inducer of the arylhydrocarbon receptor (AhR)) was also evaluated as a negative control. As shown in Figure 7, rifampicin increased UGT2A3 mRNA by almost 6-fold (p=0.05; Mann-Whitney U test) compared with vehicle control, while levels were not significantly affected by dexamethasone or β-napthoflavone (p>0.05). By comparison, mRNA levels of UGT1A1, which is regulated by PXR, GR, and AhR (Xie et al., 2003), were increased by 54-fold (p<0.001), 15-fold (p<0.001) and 10-fold (p<0.001) by β-napthoflavone, rifampicin, and dexamethasone, respectively.
Figure 7.
Induction of UGT2A3 mRNA by the PXR ligand, rifampicin. LS180 human intestinal cells were treated with 50 µM β-napthoflavone, 10 µM rifampin, , 1 µM dexamethasone or vehicle (0.1% DMSO) and assayed for UGT2A3 and UGT1A1 mRNA content (normalized to 18s rRNA) by real-time quantitative RT-PCR. Experiments were conducted in triplicate twice (n=6) and data expressed as a mean ± SE ratio relative to vehicle control. Differences from control were evaluated by Mann-Whitney U test.
Discussion
This is the first report describing the cloning and functional characterization of human UGT2A3. In the only previous functional analysis of a UGT2A3 ortholog (i.e. from guinea pig liver), only 7 substrates were evaluated, and of those only 2 (p-nitrophenol and 4-methylumbelliferone) showed reactivity, albeit at quite low levels. However, to our surprise, we were unable to show glucuronidation by UGT2A3 of p-nitrophenol, 4-methylumbelliferone or any other small phenolic substrate. Consequently, we expanded our screen of potential substrates to include a diverse set of 81 different compounds known to be glucuronidated in human liver through a variety of functional groups including amines and carboxyls, as well as hydroxyls. Out of these substrates, only 4 compounds showed reactivity with UGT2A3, and all of these were bile acids. These included 2 of the 3 major human bile acids (deoxycholic acid and chenodeoxycholic acid), as well as several of the lower abundance human bile acids (ursodeoxycholic acid and hyodeoxycholic acid). Although we were unable to demonstrate glucuronidation activity for the 2 other bile acids we evaluated, including cholic acid (the remaining major bile acid) and lithocholic acid, limitations in assay sensitivity could have precluded detection of glucuronides of these compounds.
Bile acids are products of cholesterol metabolism by the liver, and are attributed with a number of important physiological functions, including solubilization of dietary lipids and fat soluble vitamins for absorption from the intestines, maintenance of a normal bile outflow from the liver, and provision of an elimination pathway for cholesterol (Heubi et al., 2007). Bile acids normally undergo substantial enterohepatic recycling with about 90% of that excreted through the biliary system being reabsorbed and returned to the liver. However, because of the potential for cytotoxicity, the circulating levels of bile acids are tightly controlled. Furthermore, in conditions where bile flow is limited (such as in cholestatic liver disease), the accumulation of excess toxic bile acids can result in significant liver injury. Finally, exogenous administration of relatively nontoxic bile acids has been shown beneficial for the treatment of cholestatic liver disease in animal models and in people (Lindor, 2007). Ursodeoxycholic acid (ursodiol) is currently the only approved therapy for primary biliary cirrhosis in the USA.
Although glucuronidation generally represents a relatively minor pathway in bile acid metabolism, under certain circumstances glucuronidation may be of substantial importance. For example, it has been shown that an important pathway for clearance of the cholestatic and highly hepatotoxic bile acid, lithocholic acid, involves 6α-hydroxylation to form hyodeoxycholic acid followed by glucuronidation and elimination in urine (Xie et al., 2001). Induction of this pathway by PXR ligands such as rifampin (Wietholtz et al., 1996) as well as by lithocholic acid itself (Staudinger et al., 2001) can protect against bile acid induced liver injury (Xie et al., 2001). Rifampin has also been shown effective in treating pruritis associated with cholestasis (Khurana and Singh, 2006).
The results of the present work identify hyodeoxycholic acid as the most active bile acid substrate for UGT2A3 and show that glucuronidation occurs at the 6α-hydroxy position. This is consistent with previous work using human liver microsomes that showed 6α-hydroxy substituted bile acids were the most actively glucuronidated bile acids (Radominska-Pyrek et al., 1987). Furthermore, the apparent Km for hyodeoxycholic acid glucuronidation reported by the same group for human liver microsomes (36 µM) is similar to values we determined for UGT2A3.1 and UGT2A3.2 (69 and 44 µM, respectively). The difference in Km values between UGT2A3 variants is relatively small, but may be of significance particularly at physiologically relevant substrate concentrations. Circulating levels of individual bile acids appear to be in the high nanomolar range under normal circumstances, but may reach micromolar levels with cholestasis (Burkard et al., 2005; Ye et al., 2007).
The rates of hyodeoxycholic acid glucuronidation we determined for UGT2A3 (about 20 pmoles / min / mg protein) are relatively low compared with values reported for other UGT isoforms such as UGT2B7 (Mackenzie et al., 2003) and UGT2B4 (Fournel-Gigleux et al., 1989) that have recombinant enzyme activities as high as 2 nmole / min / mg protein. Consequently, it is possible that UGT2A3 only contributes minimally to hyodeoxycholic acid glucuronidation in human tissues. It is also possible that our recombinant UGT2A3 was not fully active. Although we had originally intended to express UGT2A3 by stably transfecting mammalian cells, the growth of transfected cells was completely suppressed, and we instead used a baculovirus-insect cell expression system. This latter system has been widely used for UGTs (particularly for commercial purposes) and is capable of generating large amounts of protein, but may be limited in providing sufficient and perhaps mammalian-specific post-translational protein processing (such as glycosylation and phosphorylation). Further optimization of recombinant enzyme expression, perhaps through utilization of an inducible or viral-based mammalian cell expression system may be needed to achieve optimal UGT2A3 specific activities and enable direct comparisons between different UGT isoforms. Although we screened over 80 different substrates it is also quite possible that we have as yet to identify the most highly active substrates for UGT2A3. Now that recombinant human UGT2A3 is available for study, other substrates for this isoform are likely to be identified provided that it is included in glucuronidation activity screens.
UGT2A3 mRNA was most highly expressed in human liver, gastrointestinal tract and (to a lesser extent) the kidneys. Such a distribution is consistent with that previously reported for bile acid glucuronidation by human tissues (Marschall et al., 1987), but also identifies tissues known to be important for the metabolic clearance of drugs and xenobiotics by glucuronidation. This tissue distribution is essentially identical to that previously reported for guinea pig UGT2A3 (Smith et al., 1999) and more recently for mouse UGT2a3 (Buckley and Klaassen, 2007). Such similarities between species probably reflect the orthology of the UGT2A3 genes, with minimal gene sequence divergence occurring following speciation, as evidenced by the dendrogram shown in Figure 1. No UGT2A3 mRNA expression was detected in human olfactory epithelium, which has been identified previously as a primary site of expression of the only other characterized UGT2A subfamily member, UGT2A1 (Zhang et al., 2005). A novel finding was the relatively high expression of UGT2A3 mRNA in human adipose tissue. Although adipose tissue is known to express some of the UGTs involved in sex steroid metabolism (Tchernof et al., 1999), none of the sex steroids (or any other related glucocorticoid or mineralocorticoid steroid) showed reactivity with UGT2A3. Interestingly, the bile acid activated farnesoid X receptor (FXR; NR1H4) has recently been identified in adipose tissue and shown to have a role in adipocyte differentiation (Rizzo et al., 2006). Adipose tissue is also a site for accumulation of lipophilic drugs and environmental intoxicants.
Consistent with a previous microarray study that showed enhancement of UGT2A3 expression in rat liver and intestine following treatment with dexamethasone and a novel PXR ligand (Hartley et al., 2004), we identified both GR and PXR consensus elements in the proximal human UGT2A3 enhancer. We also observed consensus sites for HNF1-alpha and CDX2, which have been previously shown to be important for tissue-specific expression of UGTs in liver and intestine, respectively (Gregory et al., 2004). However, establishing whether any of these putative sites are functional will require further study including evaluation of transcription factor binding and promoter-reporter construct analyses. As a first step we showed that UGT2A3 mRNA was expressed in the LS180 human intestinal cell line and was enhanced by treatment with the PXR agonist, rifampin, but not by the GR agonist dexamethasone, or the AhR agonist β-napthoflavone. LS180 cells were used since previous work in our laboratory indicated that this cell line expresses intestinal UGTs and responds to PXR, GR, and AhR ligands as evidenced by the strong induction of UGT1A1 by more than 10-fold. The lack of induction of UGT2A3 by β-napthoflavone is consistent with an absence of identifiable AhR binding elements in the UGT2A3 regulatory region up to −2,000 bp by in silico analysis. The lack of effect of dexamethasone on UGT2A3 mRNA in the LS180 intestinal cell line is consistent with the finding by Hartley et al (2004) of UGT2A3 induction in rat liver but not intestine. It could be speculated that the response of the UGT2A3 gene to dexamethasone may require the liver enriched HNF1 and reflects the close approximation of putative GR and HNF1 sites in the UGT2A3 proximal promoter.
In conclusion, our results establish human UGT2A3 as a species-conserved, but polymorphic UDP-glucuronosyltransferase with a highly selective substrate preference and localized to tissues with a high capacity for glucuronidation of drugs, xenobiotics and endogenous substrates. This study represents a significant first step in our understanding of a hitherto uncharacterized human UDP-glucuronosyltransferase and brings us closer to the goal of having a complete set of recombinant human UGTs for comparative functional analyses.
Acknowledgements
We would like to thank Qin Hao for providing excellent technical assistance and Dr Xinxin Ding (Wadsworth Center, New York State Department of Health, Albany, NY) for providing cDNA from human nasal olfactory epithelium.
Nonstandard abbreviations
- UGT
UDP-glucuronosyltransferase
- UDPGA
UDP-glucuronic acid
- HLMs
human liver microsomes
- SNP
single nucleotide polymorphism
Footnotes
This work was funded by Pfizer Global Research and Development, USA. Other support included grants R01GM061834 and R21GM074369 from the National Institute of General Medical Sciences, National Institutes of Health, Bethesda, Maryland, USA, and the Academy of Finland, project no. 210933. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of General Medical Sciences or the National Institutes of Health.
Reprint requests should be sent to Michael H. Court, BVSc, PhD, Comparative and Molecular Pharmacogenomics Laboratory, Department of Pharmacology and Experimental Therapeutics, Tufts University, 136 Harrison Avenue, Boston, MA 02111, USA. Telephone: 617-636-2741; Fax: 617-636-6738; Email: michael.court@tufts.edu.
References
- Bendtsen JD, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: SignalP 3.0. J Mol Biol. 2004;340(4):783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- Bock KW. Vertebrate UDP-glucuronosyltransferases: functional and evolutionary aspects. Biochem Pharmacol. 2003;66(5):691–696. doi: 10.1016/s0006-2952(03)00296-x. [DOI] [PubMed] [Google Scholar]
- Buckley DB, Klaassen CD. Tissue- and gender-specific mRNA expression of UDP-glucuronosyltransferases (UGTs) in mice. Drug Metab Dispos. 2007;35(1):121–127. doi: 10.1124/dmd.106.012070. [DOI] [PubMed] [Google Scholar]
- Burkard I, Von Eckardstein A, Rentsch KM. Differentiated quantification of human bile acids in serum by high-performance liquid chromatography-tandem mass spectrometry. J Chromatogr B Analyt Technol Biomed Life Sci. 2005;826(1–2):147–159. doi: 10.1016/j.jchromb.2005.08.016. [DOI] [PubMed] [Google Scholar]
- Caron P, Trottier J, Verreault M, Belanger J, Kaeding J, Barbier O. Enzymatic production of bile Acid glucuronides used as analytical standards for liquid chromatography-mass spectrometry analyses. Mol Pharm. 2006;3(3):293–302. doi: 10.1021/mp060021l. [DOI] [PubMed] [Google Scholar]
- Clark HF, Gurney AL, Abaya E, Baker K, Baldwin D, Brush J, Chen J, Chow B, Chui C, Crowley C, Currell B, Deuel B, Dowd P, Eaton D, Foster J, Grimaldi C, Gu Q, Hass PE, Heldens S, Huang A, Kim HS, Klimowski L, Jin Y, Johnson S, Lee S, Lewis L, Liao D, Mark M, Robbie E, Sanchez C, Schoenfeld J, Seshagiri S, Simmons L, Singh J, Smith V, Stinson J, Vagts A, Vandlen R, Watanabe C, Wieand C, Woods K, Xie MH, Yansura D, Yi S, Yu G, Yuan J, Zhang M, Zhang Z, Goddard A, Wood WI, Godowski P, Gray A. The secreted protein discovery initiative (SPDI), a large-scale effort to identify novel human secreted and transmembrane proteins: a bioinformatics assessment. Genome Res. 2003;13(10):2265–2270. doi: 10.1101/gr.1293003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coughtrie MW, Burchell B, Leakey JE, Hume R. The inadequacy of perinatal glucuronidation: immunoblot analysis of the developmental expression of individual UDP-glucuronosyltransferase isoenzymes in rat and human liver microsomes. Mol Pharmacol. 1988;34(6):729–735. [PubMed] [Google Scholar]
- Court MH. Acetaminophen UDP-glucuronosyltransferase in ferrets: species and gender differences, and sequence analysis of ferret UGT1A6. J Vet Pharmacol Ther. 2001;24(6):415–422. doi: 10.1046/j.1365-2885.2001.00366.x. [DOI] [PubMed] [Google Scholar]
- Court MH. In vitro identification of UDP-glucuronosyltransferases (UGTs) involved in drug metabolism. In: Yan Z, Caldwell GW, editors. Optimization in drug discovery: In vitro methods. Totowa: Humana press, Inc.; 2004. pp. 185–202. [Google Scholar]
- Court MH. Isoform-selective probe substrates for in vitro studies of human UDP-glucuronosyltransferases. Methods Enzymol. 2005;400:104–116. doi: 10.1016/S0076-6879(05)00007-8. [DOI] [PubMed] [Google Scholar]
- Court MH, Hao Q, Krishnaswamy S, Bekaii-Saab T, Al-Rohaimi A, Von Moltke LL, Greenblatt DJ. UDP-glucuronosyltransferase (UGT) 2B15 pharmacogenetics: UGT2B15 D85Y genotype and gender are major determinants of oxazepam glucuronidation by human liver. J Pharmacol Exp Ther. 2004;310(2):656–665. doi: 10.1124/jpet.104.067660. [DOI] [PubMed] [Google Scholar]
- Court MH, von Moltke LL, Shader RI, Greenblatt DJ. Biotransformation of chlorzoxazone by hepatic microsomes from humans and ten other mammalian species. Biopharm Drug Dispos. 1997;18(3):213–226. doi: 10.1002/(sici)1099-081x(199704)18:3<213::aid-bdd15>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- Fournel-Gigleux S, Jackson MR, Wooster R, Burchell B. Expression of a human liver cDNA encoding a UDP-glucuronosyltransferase catalysing the glucuronidation of hyodeoxycholic acid in cell culture. FEBS Lett. 1989;243(2):119–122. doi: 10.1016/0014-5793(89)80111-5. [DOI] [PubMed] [Google Scholar]
- Gall WE, Zawada G, Mojarrabi B, Tephly TR, Green MD, Coffman BL, Mackenzie PI, Radominska-Pandya A. Differential glucuronidation of bile acids, androgens and estrogens by human UGT1A3 and 2B7. J Steroid Biochem Mol Biol. 1999;70(1–3):101–108. doi: 10.1016/s0960-0760(99)00088-6. [DOI] [PubMed] [Google Scholar]
- Gregory PA, Lewinsky RH, Gardner-Stephen DA, Mackenzie PI. Coordinate regulation of the human UDP-glucuronosyltransferase 1A8, 1A9, and 1A10 genes by hepatocyte nuclear factor 1alpha and the caudal-related homeodomain protein 2. Mol Pharmacol. 2004;65(4):953–963. doi: 10.1124/mol.65.4.953. [DOI] [PubMed] [Google Scholar]
- Hartley DP, Dai X, He YD, Carlini EJ, Wang B, Huskey SE, Ulrich RG, Rushmore TH, Evers R, Evans DC. Activators of the rat pregnane X receptor differentially modulate hepatic and intestinal gene expression. Mol Pharmacol. 2004;65(5):1159–1171. doi: 10.1124/mol.65.5.1159. [DOI] [PubMed] [Google Scholar]
- Hesse LM, He P, Krishnaswamy S, Hao Q, Hogan K, von Moltke LL, Greenblatt DJ, Court MH. Pharmacogenetic determinants of interindividual variability in bupropion hydroxylation by cytochrome P450 2B6 in human liver microsomes. Pharmacogenetics. 2004;14(4):225–238. doi: 10.1097/00008571-200404000-00002. [DOI] [PubMed] [Google Scholar]
- Heubi JE, Setchell KD, Bove KE. Inborn errors of bile acid metabolism. Semin Liver Dis. 2007;27(3):282–294. doi: 10.1055/s-2007-985073. [DOI] [PubMed] [Google Scholar]
- Jedlitschky G, Cassidy AJ, Sales M, Pratt N, Burchell B. Cloning and characterization of a novel human olfactory UDP- glucuronosyltransferase. Biochem J. 1999;340(Pt 3):837–843. [PMC free article] [PubMed] [Google Scholar]
- Kaivosaari S, Toivonen P, Hesse LM, Koskinen M, Court MH, Finel M. Nicotine glucuronidation and the human UDP-glucuronosyltransferase UGT2B10. Mol Pharmacol. 2007 doi: 10.1124/mol.107.037093. [DOI] [PubMed] [Google Scholar]
- Karlen Y, McNair A, Perseguers S, Mazza C, Mermod N. Statistical significance of quantitative PCR. BMC Bioinformatics. 2007;8:131. doi: 10.1186/1471-2105-8-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khurana S, Singh P. Rifampin is safe for treatment of pruritus due to chronic cholestasis: a meta-analysis of prospective randomized-controlled trials. Liver Int. 2006;26(8):943–948. doi: 10.1111/j.1478-3231.2006.01326.x. [DOI] [PubMed] [Google Scholar]
- Krishnaswamy S, Hao Q, Al-Rohaimi A, Hesse LM, von Moltke LL, Greenblatt DJ, Court MH. UDP glucuronosyltransferase (UGT) 1A6 pharmacogenetics: I. Identification of polymorphisms in the 5′-regulatory and exon 1 regions, and association with human liver UGT1A6 gene expression and glucuronidation. J Pharmacol Exp Ther. 2005;313(3):1331–1339. doi: 10.1124/jpet.104.081950. [DOI] [PubMed] [Google Scholar]
- Lindor K. Ursodeoxycholic acid for the treatment of primary biliary cirrhosis. N Engl J Med. 2007;357(17):1524–1529. doi: 10.1056/NEJMct074694. [DOI] [PubMed] [Google Scholar]
- Mackenzie P, Little JM, Radominska-Pandya A. Glucosidation of hyodeoxycholic acid by UDP-glucuronosyltransferase 2B7. Biochem Pharmacol. 2003;65(3):417–421. doi: 10.1016/s0006-2952(02)01522-8. [DOI] [PubMed] [Google Scholar]
- Mackenzie PI, Bock KW, Burchell B, Guillemette C, Ikushiro S, Iyanagi T, Miners JO, Owens IS, Nebert DW. Nomenclature update for the mammalian UDP glycosyltransferase (UGT) gene superfamily. Pharmacogenet Genomics. 2005;15(10):677–685. doi: 10.1097/01.fpc.0000173483.13689.56. [DOI] [PubMed] [Google Scholar]
- Marschall HU, Matern H, Egestad B, Matern S, Sjovall S. 6 alpha-glucuronidation of hyodeoxycholic acid by human liver, kidney and small bowel microsomes. Biochim Biophys Acta. 1987;921(2):392–397. doi: 10.1016/0005-2760(87)90041-5. [DOI] [PubMed] [Google Scholar]
- Radominska-Pyrek A, Zimniak P, Irshaid YM, Lester R, Tephly TR, St Pyrek J. Glucuronidation of 6 alpha-hydroxy bile acids by human liver microsomes. J Clin Invest. 1987;80(1):234–241. doi: 10.1172/JCI113053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzo G, Disante M, Mencarelli A, Renga B, Gioiello A, Pellicciari R, Fiorucci S. The farnesoid X receptor promotes adipocyte differentiation and regulates adipose cell function in vivo. Mol Pharmacol. 2006;70(4):1164–1173. doi: 10.1124/mol.106.023820. [DOI] [PubMed] [Google Scholar]
- Smith SA, Nagalla SR, Andrews DP, Olsen GD. Morphine regulation of a novel uridine diphosphate glucuronosyl- transferase in guinea pig pups following in utero exposure. Mol Genet Metab. 1999;68(1):68–77. doi: 10.1006/mgme.1999.2892. [DOI] [PubMed] [Google Scholar]
- Staudinger JL, Goodwin B, Jones SA, Hawkins-Brown D, MacKenzie KI, LaTour A, Liu Y, Klaassen CD, Brown KK, Reinhard J, Willson TM, Koller BH, Kliewer SA. The nuclear receptor PXR is a lithocholic acid sensor that protects against liver toxicity. Proc Natl Acad Sci U S A. 2001;98(6):3369–3374. doi: 10.1073/pnas.051551698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strausberg RL, Feingold EA, Grouse LH, Derge JG, Klausner RD, Collins FS, Wagner L, Shenmen CM, Schuler GD, Altschul SF, Zeeberg B, Buetow KH, Schaefer CF, Bhat NK, Hopkins RF, Jordan H, Moore T, Max SI, Wang J, Hsieh F, Diatchenko L, Marusina K, Farmer AA, Rubin GM, Hong L, Stapleton M, Soares MB, Bonaldo MF, Casavant TL, Scheetz TE, Brownstein MJ, Usdin TB, Toshiyuki S, Carninci P, Prange C, Raha SS, Loquellano NA, Peters GJ, Abramson RD, Mullahy SJ, Bosak SA, McEwan PJ, McKernan KJ, Malek JA, Gunaratne PH, Richards S, Worley KC, Hale S, Garcia AM, Gay LJ, Hulyk SW, Villalon DK, Muzny DM, Sodergren EJ, Lu X, Gibbs RA, Fahey J, Helton E, Ketteman M, Madan A, Rodrigues S, Sanchez A, Whiting M, Madan A, Young AC, Shevchenko Y, Bouffard GG, Blakesley RW, Touchman JW, Green ED, Dickson MC, Rodriguez AC, Grimwood J, Schmutz J, Myers RM, Butterfield YS, Krzywinski MI, Skalska U, Smailus DE, Schnerch A, Schein JE, Jones SJ, Marra MA. Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences. Proc Natl Acad Sci U S A. 2002;99(26):16899–16903. doi: 10.1073/pnas.242603899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tchernof A, Levesque E, Beaulieu M, Couture P, Despres JP, Hum DW, Belanger A. Expression of the androgen metabolizing enzyme UGT2B15 in adipose tissue and relative expression measurement using a competitive RT-PCR method [In Process Citation] Clin Endocrinol (Oxf) 1999;50(5):637–642. doi: 10.1046/j.1365-2265.1999.00709.x. [DOI] [PubMed] [Google Scholar]
- Wietholtz H, Marschall HU, Sjovall J, Matern S. Stimulation of bile acid 6 alpha-hydroxylation by rifampin. J Hepatol. 1996;24(6):713–718. doi: 10.1016/s0168-8278(96)80268-6. [DOI] [PubMed] [Google Scholar]
- Xie W, Radominska-Pandya A, Shi Y, Simon CM, Nelson MC, Ong ES, Waxman DJ, Evans RM. An essential role for nuclear receptors SXR/PXR in detoxification of cholestatic bile acids. Proc Natl Acad Sci U S A. 2001;98(6):3375–3380. doi: 10.1073/pnas.051014398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie W, Yeuh MF, Radominska-Pandya A, Saini SP, Negishi Y, Bottroff BS, Cabrera GY, Tukey RH, Evans RM. Control of steroid, heme, and carcinogen metabolism by nuclear pregnane X receptor and constitutive androstane receptor. Proc Natl Acad Sci U S A. 2003;100(7):4150–4155. doi: 10.1073/pnas.0438010100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye L, Liu S, Wang M, Shao Y, Ding M. High-performance liquid chromatography-tandem mass spectrometry for the analysis of bile acid profiles in serum of women with intrahepatic cholestasis of pregnancy. J Chromatogr B Analyt Technol Biomed Life Sci. 2007;860(1):10–17. doi: 10.1016/j.jchromb.2007.09.031. [DOI] [PubMed] [Google Scholar]
- Zhang X, Zhang QY, Liu D, Su T, Weng Y, Ling G, Chen Y, Gu J, Schilling B, Ding X. Expression of cytochrome p450 and other biotransformation genes in fetal and adult human nasal mucosa. Drug Metab Dispos. 2005;33(10):1423–1428. doi: 10.1124/dmd.105.005769. [DOI] [PubMed] [Google Scholar]