Abstract
Pancreatic Derived Factor (PANDER) is a novel cytokine-like protein dominantly expressed within the endocrine pancreas. Our previous study demonstrated that the PANDER promoter was both tissue-specific and glucose-responsive. Surrounding the PANDER transcriptional start site are several putative A- and E-Box elements that may bind to the various pancreatic transcriptional factors of MafA, BETA2/NeuroD, and Pancreatic Duodenal Homeobox-1 (PDX-1). To characterize the transcriptional regulatory factors involved in PANDER gene expression, we performed co-transfection reporter gene analysis and demonstrated upregulation by all three transcription factors, with the greatest individual increase stemming from PDX-1. Potential binding of PDX-1 to A-box (TAAT) regions of the PANDER promoter was demonstrated by chromatin immunoprecipitation (ChIP) and further corroborated by electrophoretic mobility shift assay (EMSA). Binding of PDX-1 to the A box regions was inhibited by mutagenized (TAGT) oligonucleotides. Site-directed mutagenesis of the three PDX-1 A-box binding motifs revealed that A box sites 2 and 3 in combination were critical for maximal gene expression and deletion resulted in a 82% reduction in promoter activity. Furthermore, deletion of A-box sites 2 and 3 completely diminished the glucose-responsiveness of the PANDER promoter. Our findings demonstrate that PANDER is a potential PDX-1 target gene and the A-box sites within the promoter region are critical for basal and glucose-stimulated PANDER expression.
Keywords: PANDER, promoter, islet, glucose, reporter gene, β-TC3
1. INTRODUCTION
Pancreatic Derived Factor (PANDER, FAM3B), is a member of a recently-discovered family of predicted cytokines, FAM3. Structurally, PANDER is comprised of 235 amino acids (including four conserved cysteine residues) whose major secondary topological feature is a series of four α-helices organized in an up-up-down-down fashion, also found in well-known cytokines such as IL-2, -3, -4, -5, -6, -7, -8, -9, -10, and –13 [1,2]. However, despite this ostensible structural homology, amino acid comparisons reveal that, in fact, PANDER bears absolutely no similarity to any known cytokines based on sequence alignment. PANDER is primarily expressed in the islets of Langerhans of the pancreas within both alpha and beta cells colocalizing with both insulin and glucagon, respectively [2]. The biological function of PANDER has yet to be determined.
Similar to insulin, the expression and secretion of PANDER, is both cell type-specific and glucose-responsive [3–5]. Glucose has been demonstrated to have a profound impact on PANDER and has been shown to (i) increase PANDER promoter activity in insulinoma cell lines and primary islets [3]; (ii) upregulate PANDER mRNA and protein expression [5]; and (iii) induce secretion of PANDER from both insulinoma cells and primary islets [4]. Therefore, in a fashion resembling insulin [6], glucagon [7], and Nkx6.1 [8], the tissue specificity of PANDER expression is controlled, at least to some extent, by its promoter region. However the specific transcriptional factors capable of interaction and regulation of the PANDER promoter have yet to be examined.
The genomic region of the PANDER promoter, especially between base pairs −338 (within the 5′-UTR) and +491, contains a number of cis-acting binding motifs that are common throughout endocrine pancreatic genes and necessary for islet-specific expression and glucose-mediated induction of gene expression [3]. In addition, our previous deletion studies in β-cell lines indicated that this region of the PANDER promoter contained the minimal activated and glucose-regulated regions. For example, these include A boxes (5′-TAAT-3′), demonstrated to bind the essential pancreatic transcription factor Pancreatic/Duodenal Homeobox-1 (PDX-1) [9], E-Boxes (5′-CANNTG-3′), which bind transcription factor β-cell E box Transactivator 2 (BETA2/NeuroD) [10], and others. Arguably, the most important of these potential transcription factors is PDX-1, a homeodomain protein present in pancreatic β-cells, and to a lesser extent in pancreatic δ cells and certain endocrine cells in the small intestine [11]. PDX-1 has been demonstrated to be essential in the development of the pancreas [11–13]. Preclusion of PDX-1 expression during embryological development, for example, results in complete agenesis of the pancreas. While the fetus is still viable, though, the neonate dies within days of birth [13–15]. The postnatal effects of PDX-1 on endocrine pancreas function have also been heavily investigated and PDX-1 is known to be a necessary regulator of the insulin gene, as well as of GLUT2 [16] and glucokinase [17]. Empirical data have shown that islets whose PDX-1 expression was attenuated showed decreased insulin secretion and overall β cell mass with subsequent onset of type 2 diabetes [18–21].
The mere presence of these putative transcription factor binding sites does not obligatorily denote protein binding due to, for example, chromatin packing that would proscribe the approach of regulatory proteins to the promoter region in question (ie. DNA methylation). However, the identification of multiple potential PDX-1, MafA, and BETA2/NeuroD binding sites within the promoter regions of PANDER and other key endocrine pancreatic genes to which these proteins have been proven to bind suggests that the PANDER promoter is also a likely regulatory target. Our current investigation evaluated the effects of the three candidate transcription factors we selected (MafA, BETA2/NeuroD, and PDX-1) to, either individually or synergistically, upregulate PANDER expression, as quantified by reporter gene analysis. These experiments showed a statistically significant greater effect of PDX-1 on this upregulation (although all three transcription factors did upregulate PANDER expression above basal level in NIH-3T3 cells). As a result, we chose to sharpen our focus on PDX-1 specifically. Our goals became, first, to corroborate PDX-1 upregulation of PANDER by site-directed mutagenesis of the PDX-1 binding sites (A boxes) in the PANDER promoter. Second, both EMSA and ChIP were utilized to demonstrate the potential PDX-1 interaction and regulation of the PANDER promoter via the A boxes. Finally, we demonstrated that the specific A box sites are critical for the glucose-responsive expression of the PANDER promoter.
2. MATERIALS and METHODS
2.1. Cell Culture, Transient Transfection, and Luciferase Assay
Transient transfections and luciferase assay were performed as previously described [3]. NIH-3T3 cells were cultured at 37°C and 95% O2/5% CO2 for one week in a T175 flask in Dulbecco’s Modified Eagle Medium supplemented with 10% heat-inactivated fetal bovine serum, 100 U/ml penicillin, 100 μg/ml streptomycin, and 2 mM L-glutamine. Five equimolar transient co-transfections of PANDER-luciferase plasmids and plasmids encoding either pcDNA3.1 (empty vector for negative control), MafA, PDX-1, or BETA2 (all pcDNA3.1 plasmids were a kind gift from Dr. Roland Stein of Vanderbilt University), as well as a condition containing all three transcription factor genes, were each prepared in triplicate at a density of 1 × 105 cells per well in a 24-well culture treated dish (Becton-Dickinson). PANDER-luciferase and transcription factor-containing constructs were co-transfected with phRL-TK plasmid (Promega) to provide a metric for transfection efficiency obtained by LipofectAMINE 2000 (Invitrogen).
Twenty-four hours following transfection, transfected NIH-3T3 cells were washed with Versene EDTA and subjected to lysis in 200 μl of 1X Passive Lysis Buffer (Promega). Luciferase activities of the transcription factor-containing plasmids and the phRL-TK construct were assessed consecutively via the Dual-Luciferase Reporter Assay System (Promega). Luciferase activity was measured for 10 s (with a 2-s delay) utilizing a PharMingen Monolight 3010 luminometer (Analytical Luminescence Laboratory). Normalization of the variation in transfection efficiency was determined as the ratio of transcription factor construct activity to that of the corresponding co-transfected phRL-TK.
2.2. Chromatin Immunoprecipitation
Chromatin immunoprecipitation (ChIP) was performed using the EZ-ChIP (Upstate) kit. Manufacturer’s instructions were followed accordingly unless otherwise noted. Confluent β-HC9 insulinoma cells were cultured as described above. Cells were fixed with 1% (v/v) fresh formaldehyde to cross-link DNA with associated transcription factors. After 10 min incubation at room temperature, 1 ml 10X glycine was added and gently swirled to quench the unreacted CH2O. Samples were sonicated using a Branson Sonifier 250 set to setting 5 and duty constant of 50%.
For immunoprecipitation, one of the following antibodies were each added to one of the three lysate samples: 1.0 μg of normal mouse IgG (Upstate), 1.0 μg of mouse monoclonal anti-RNA polymerase II (Upstate), and 1.0 μg of mouse monoclonal anti-PDX-1 (Upstate).
2.3. PCR of Immunoprecipitated Chromatin
Following purification of immunoprecipitated chromatin, PCR was performed on the samples. The primers used in the PCR reactions had been previously designed to amplify specific regions of the PANDER promoter, specifically bp −338/+1, +1/+100 and +200/+491. Primer sequences have been described previously [3]. Two sets of tubes were prepared for PCR. One contained water, 10x Buffer II (Invitrogen), appropriate primers, Accuprime Taq (Invitrogen), and immunoprecipitated DNA. The other set contained water, 10x Buffer II, appropriate primers, Accuprime Taq (Invitrogen), and input DNA (i.e. not immunoprecipitated). The PCR reagents were all added to thin-walled, flat-topped 0.5-ml PCR tubes (Molecular Bioproducts). The PCR reactions were run in a MJ Research PTC-200 DNA Engine thermocycler, using the following conditions: 94°C for 3 min (one cycle, for initial denaturation), 94°C for 20 s (denaturation)/59°C for 30 s (annealing)/72°C for 30 s (extension) (32 cycles), with one cycle of 72°C for 2 min for final extension. The PCR samples were analyzed on a 1% agarose/TAE gel and analyzed using a Fuji Intelligent DarkBox.
2.4. Site-Directed Mutagenesis
The three A boxes within the murine PANDER promoter were mutagenized using the QuikChange II Site-Directed Mutagenesis Kit (Stratagene). Based on previous research demonstrating successful abrogation of PDX-1 binding to the insulin promoter [22], the PANDER A box motif was similarly altered from 5′-TAAT-3′ to 5′-TAGT-3′. The three A boxes were subjected to combinatorial mutagenesis to produce all possible permutations of mutagenized and wild type PDX-1 binding sites (Figure 2A). The plasmids, designed to fit manufacturer’s specifications regarding length and melting temperature, employed the PANDER promoter sequence previously determined [3].
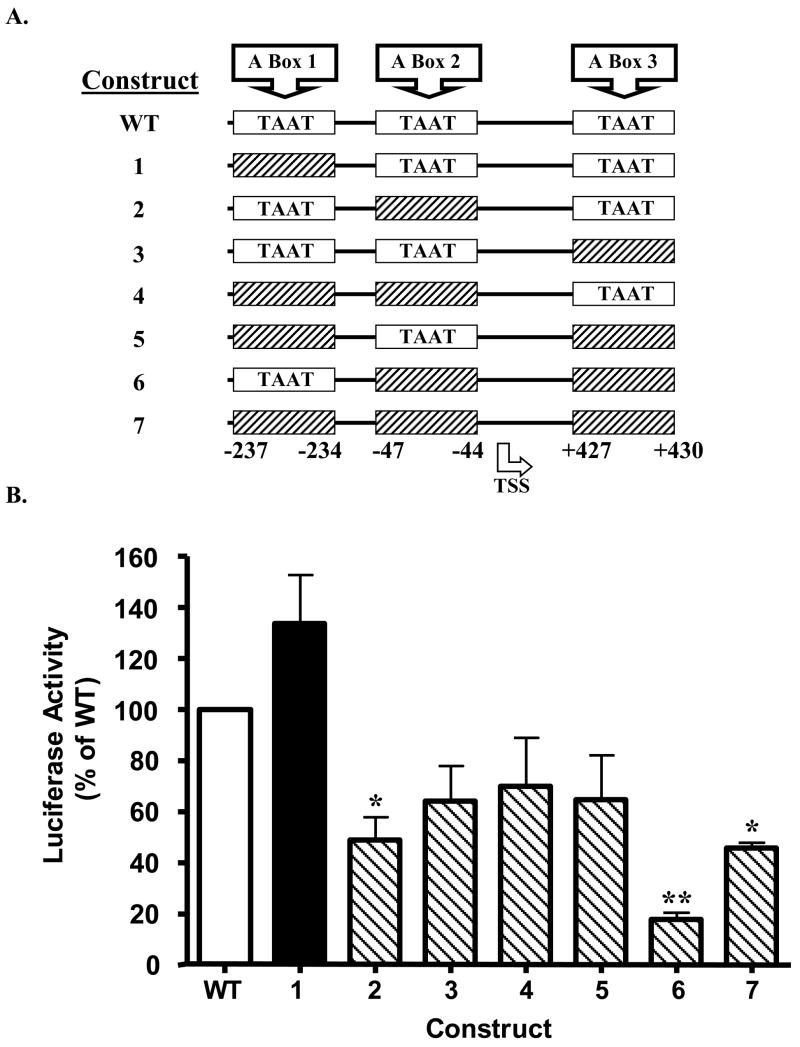
Figure 2. Effects of site-directed mutagenesis of PDX-1 binding motif on PANDER promoter activity.
(A) Structure of the mutagenized PANDER promoter luciferase constructs. A schematic diagram representing the PANDER promoter’s three A boxes, either wild type (i.e. black, 5′-TAAT-3′) or mutagenized (hatched bars, 5′-TAGT-3′). A plasmid encoding the −338 to +491 region of the PANDER gene, containing three A boxes (5′-TAAT-3v PDX-1 binding motifs) were subjected to site-directed mutagenesis to create seven mutant constructs, each containing a combination of one, two, or all three A boxes as shown above. Nucleotide locations are numbered and shown at the bottom of the diagram. TSS refers to transcriptional start site. (B) Luciferase activity of PANDER promoter luciferase constructs. Mutagenized PANDER promoter luciferase constructs (as diagramed in Figure 2A) were transiently co-transfected with Renilla luciferase reporter plasmid phRL-TK into β-HC9 cells in complete DMEM. Twenty-four hours following transfection, luciferase activity was detected by luminometry. WT denotes wild-type PANDER promoter luciferase construct that contains all 3 intact A box (TAAT) regions. Data are shown as mean ± S.E.M. percentage luciferase activity (firefly/renilla) relative to cells transfected with WT construct (open white bar). Luciferase activity above WT construct is shown with black filled bar whereas observed activity below WT construct is represented by back slashed bar. These data encompass two independent experiments with each construct transfected in triplicate. Single and double asterisk indicates P < 0.05 and 0.01 as determined by ANOVA, respectively, between mutagenized (shown according to construct number) and wild-type PANDER-luciferase plasmid (indicated by WT).
For mutagenesis of A box 1, the forward primer used was 5′-AGACCACTGTGACTCTAGTGAGCTAAAAAGTTCT-3′ and the reverse primer 5′-AGAACTTTTTAGCTCACTAGAGTCACAGTGGTCT-3′ (mutagenized sequence is bolded and underlined). For mutagenesis of A box 2, the forward primer used was 5′-CTCATTTCCCTAAATAGTCATCCATCAGTAAA-3′ and the reverse primer was 5′-TTTACTGATGGGATGACTATTTAGGGAATGAG-3′. For mutagenesis of A box 3, the forward primer used was 5′-CCCAAGGGCGGTCATAGTCTATGTGCAGCTCAA-3′ and the reverse primer was 5′-TTGAGCTGCACATAGACTATGACCGCCCTTGGGG-3′.
These primer sets were used in a PCR amplification of the wild type PANDER-luciferase construct −338/+491 [3]. The PCR reactions were run in a MJ Research PTC-200 DNA Engine thermocycler using the following conditions: 95°C for 30 sec (one cycle, for denaturation), 95°C for 30 s (denaturation)/55°C for 70 s/(annealing)/68°C for 5 min (extension) (32 cycles), and finally one cycle of 68°C for 5 min for final extension.
The PCR products resulting from each of these amplifications were treated with restriction endonuclease DpnI (Stratagene) to digest parental (non-mutated) plasmid and chromosomal DNA. The DpnI-treated plasmids were then cultured and amplified using the QIAprep Spin Miniprep Kit (Qiagen). A sample of the amplified plasmid was sequenced by the Children’s Hospital of Philadelphia Sequencing Core to verify the success of the site-directed mutagenesis procedure. Large scale preparation of the sequenced and mutagenized plasmids was performed using the EndoFree Plasmid Maxi Kit (Qiagen).
The product plasmids were then transiently transfected into cultured β-HC9 insulinoma cells. Transient transfections and luciferase assay were performed as described above for NIH-3T3 cells, modified from the literature [3]. Eight equimolar transient transfections of PANDER-luciferase plasmids, either wild type (positive control) or one of the seven site-directed mutagenesis-derived A box mutant constructs, were each prepared in triplicate at a density of 2 ×105 cells per well in a 24-well culture treated dish (Beckton-Dickinson). PANDER-luciferase constructs were co-transfected with phRL-TK plasmid (Promega) to normalize transfection efficiency obtained by Lipofectamine 2000 (Invitrogen). Three independent transfections were performed with each construct being tested in triplicate. Twenty-four hours after transfection, β-HC9 cells were washed with versene EDTA and lysed in 200 μl of Passive Lysis Buffer (Promega). Luciferase activities of the mutagenized plasmids and the phRL-TK construct were assessed and normalized as described above and previously [3].
2.5. Electrophoretic Mobility Shift Assay
Electrophoretic Mobility Shift Assay (EMSA) was performed using the LightShift Chemiluminescent EMSA kit (Pierce). Manufacturer’s instructions were followed accordingly unless otherwise noted. Oligonucleotide sequences representing the three A boxes and portions of their flanking regions were 3′-biotinylated using the Biotin 3′ End DNA Labeling Kit (Pierce). Additionally, mutagenized primer sets were also biotinylated using the Pierce kit. Manufacturers’ instructions were followed accordingly unless otherwise noted. The oligonucleotides for the EMSA were the same ones employed for site-directed mutagenesis.
Three classes of EMSA binding reactions were prepared. Each reaction class included three separate conditions, one for each of the three A boxes present in the PANDER promoter. First, as a negative control, only the chemical reagents (see following) and biotinylated wild type (i.e. 5′-TAAT-3′) oligonucleotides were included. The second class of reactions included rat PDX-1 protein (150 ng) in addition to biotinylated wild type oligonucleotides. This condition, therefore, was intended to display retarded mobility in the biotinylated DNA due to PDX-1 binding to the A box present in each oligonucleotide. The third class of EMSA binding reactions included biotinylated mutagenized oligonucleotides (i.e. 5′-TAGT-3′) and rat PDX-1 protein. Thus, as PDX-1 is unable to bind to a mutated A box sequence [23], there is no mobility shift of the biotinylated DNA expected from this preparation. Each of the nine reactions (three reaction classes, each comprising trials for the three A boxes in the PANDER promoter) also included 1X Binding Buffer (Pierce), 10% glycerol (Pierce), 1 μg μl−1 poly (dI · dC) (Pierce), 0.2 mM HEPES buffer (pH 7.9), and 150 mM KCl (Pierce). Total volume for each reaction was brought to 20 μl with ddH2O. After combining all of the reagents, the binding reactions were incubated at room temperature 20 min. Following incubation, 5 μl of 5X loading buffer (0.2M Tris · HCl [pH 6.8], 0.03% bromophenol blue, 33% glycerol [v/v], and 9.3% ddH2O [v/v]) was added to each completed binding reaction. Reaction mixtures were applied to a 5% native polyacrylamide gel and transferred to a Hybond-N+ nylon membrane (Amersham) using a current of 380 mA in a stir bar-mixed bath of chilled 0.5X TBE for 30 min. Following transfer, protein+DNA complexes were cross-linked to the nylon membrane via a Fisher BioTech Electrophoresis Systems 312 nm Variable Intensity Transilluminator FBTV-816 set to 80% intensity, with protein side down, for 15 min. Gel shifts were then detected on the membrane with a chemiluminescent nuclei acid detection module (Pierce) according to manufacturer’s protocol. Images were then visualized using the Fuji Intelligent Darkbox.
2.6. Glucose-responsive Experiments
The glucose-responsive expression of the mutagenized contructs was evaluated by transient transfection of β-TC3 cells as previously described [3].
2.7. Statistical Analysis
Data are presented as mean ± S.E.M. Statistical significance of differences between groups was analyzed by paired Student’s t test or by one-way analysis of variance (ANOVA) when more than two groups were compared.
3. RESULTS
3.1. Effects of β-cell specific transcription factors on PANDER expression
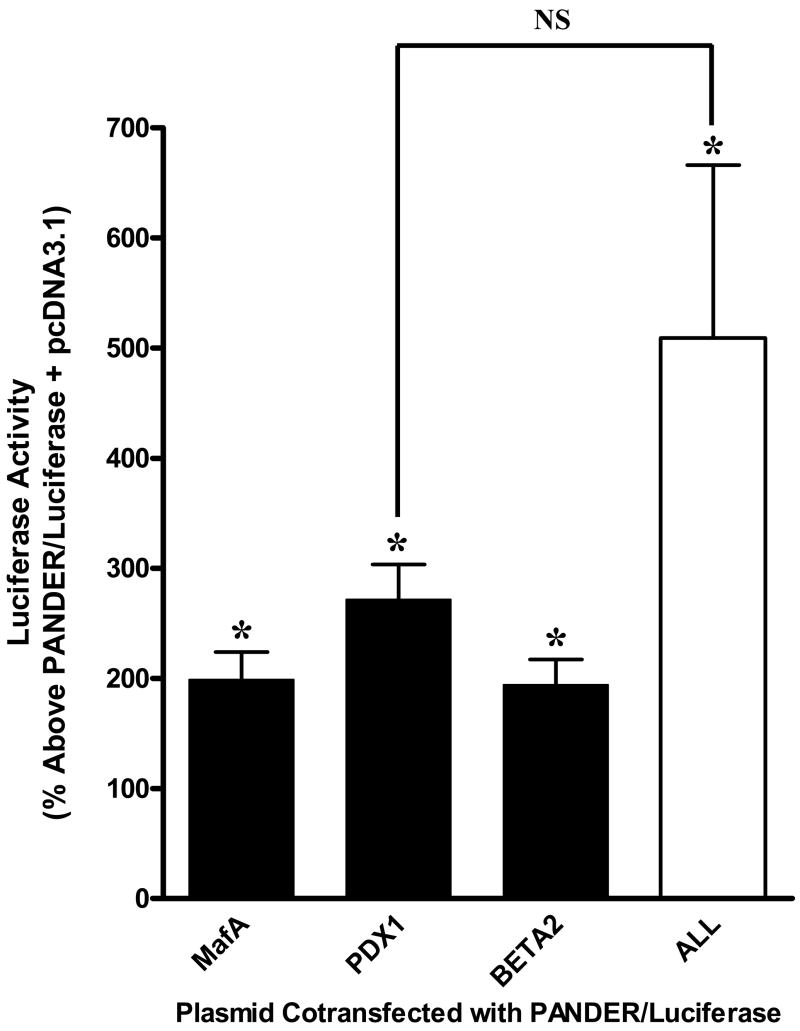
To investigate the role of the pancreatic transcription factors consisting of MafA, PDX-1, and BETA2/NeuroD on PANDER expression, we transiently co-transfected NIH-3T3 cells with a PANDER-luciferase construct (−338/+491 of the PANDER promoter) along with a plasmid encoding one of the aforementioned transcription factors (Figure 1). NIH-3T3 cells are not permissive for PANDER expression and therefore were utilized for these experiments. An analysis of the results of 3 separate experiments showed that, based on relative PANDER promoter expression above the PANDER-luciferase construct co-transfected with the empty vector of pcDNA3.1, PDX-1 increased PANDER expression by 272 ± 32%, while MafA and BETA2/NeuroD showed a smaller upregulation of PANDER expression, with increases of 199 ± 25% and 194 ± 23%, respectively. All 3 transcriptional factors significantly increased PANDER driven promoter activity (P < 0.05, paired t-test). The combination of all three transcription factors demonstrated an increase in PANDER promoter expression of 509 ± 157%. However this was not statistically greater than the increase on promoter activity by PDX-1 alone (P > 0.05, ANOVA). In contrast, PANDER promoter driven luciferase expression observed with the MafA or BETA2/NeuroD co-transfection conditions was significantly different than that found with all three transcription factors (P < 0.05, ANOVA). As a result of the greater effect of PDX-1 on PANDER expression relative to MafA and BETA2/NeuroD, we decided to further characterize the impact of PDX-1 on the PANDER promoter.
Figure 1. Effects of β-cell specific transcription factors on PANDER expression.
Plasmids encoding the transcription factors MafA, PDX-1, BETA2/NeuroD, as well as one condition using all three, were co-transfected with the PANDER and phRL-TK luciferase reporter plasmids into NIH-3T3 cells. Twenty-four hours post-transfection, luciferase activity was measured and normalized to luciferase expression of the PANDER-luciferase plasmid co-transfected with pcDNA3.1 (empty vector). Data presented is from three experiments with each construct tested in triplicate, as % above ± S.E.M PANDER-luciferase plasmid co-transfected with pcDNA3.1. The asterisk indicates a P < 0.05 as determined by paired t-test with respect to luciferase activity from the PANDER-luciferase plasmid plus pcDNA3.1 empty vector. NS denotes not significant (P > 0.05) between PDX-1 and All (MafA, PDX-1, and BETA2/NeuroD co-transfected together) luciferase activity as determined by One-Way ANOVA.
3.2. Impact of site-directed mutagenesis of PDX-1 binding motif on PANDER promoter activity
As mentioned previously, the PANDER promoter contains three A box regions surrounding the transcriptional start site (−237 to −234, −47 to −44, and +427 to +430). Their consensus TAAT sequence is a well-characterized binding site for homeodomain proteins such as PDX-1. Therefore, we sought to evaluate the contribution of the specific A-box sites in regulating PANDER promoter activity. Mutagenesis was performed on individual and multiple PDX-1 binding sites within the PANDER-luciferase construct containing the −338 to +491 genomic region by altering the TAAT core sequence located internal to the A box site to TAGT (Figure 2A). The mutagenized PANDER promoter constructs were then transfected into the murine β-HC9 insulinoma cell line. Mutagenesis of individual A box 2 reduced promoter activity by 52% (P < 0.05, ANOVA) as compared to the wild-type (WT) PANDER-luciferase plasmid with all three intact A box sites (Figure 2B, Construct 2). Combined ablation of A boxes 2 and 3 (Construct 6) had the strongest impact on PANDER promoter activity by significantly reducing luciferase expression by 82% (P < 0.01, ANOVA). Interestingly, mutagenesis of A box 2 significantly decreased promoter activity as greatly as the disruption of all 3 putative A boxes (Construct 2 vs. Construct 7, P < 0.05, ANOVA). In addition, mutagenesis of all 3 A boxes reduced promoter activity by 55%, yet mutation of A boxes 2 and 3 had the greatest reduction in expression. Taken together, these results demonstrate that A box sites 2 and 3 are important for PANDER promoter activity.
3.3. ChIP analysis of PDX-1 interaction with the PANDER promoter region
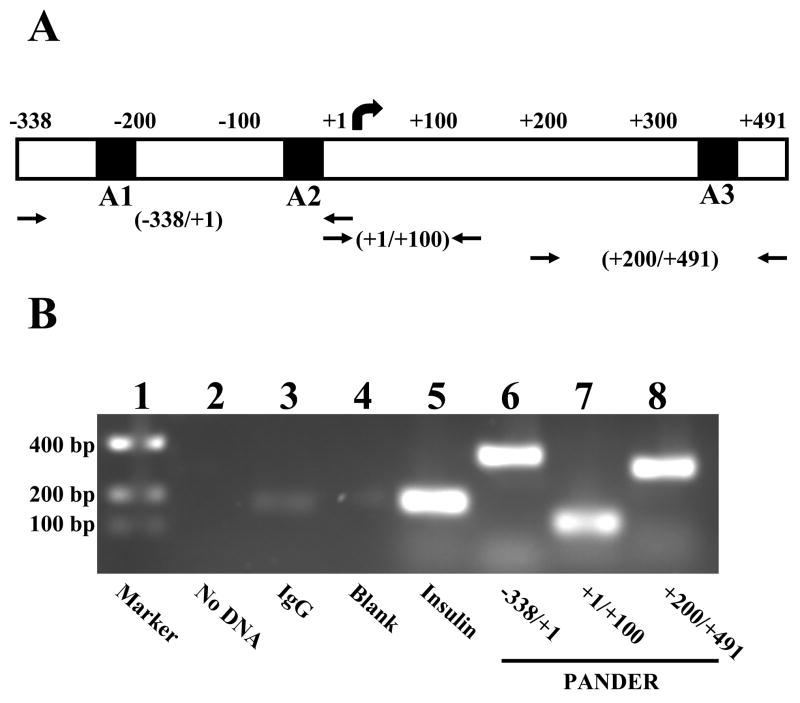
To functionally demonstrate that PDX-1 binds to the PANDER promoter, we initially performed chromatin immunoprecipitation (ChIP). We screened various regions of the previously sequenced PANDER promoter based on the number of A boxes (i.e. TAAT binding motifs) contained therein. This was based on the previously reported binding of PDX-1 to A boxes within the promoters of other genes expressed in pancreatic β-cells, most notably insulin and glucokinase. Three pairs of primer sets were selected that encompass the regions of the PANDER promoter that contain or surround the A boxes (i.e. TAAT binding motifs), namely −338/+1, +1/+100, and +200/+491 (Figure 3A). Following PDX-1 immunoprecipitation of cross-linked β-HC9 DNA with subsequent PCR amplification using PANDER promoter specific primers, our results showed robust amplification at about the expected molecular weights for the first and third primer pairs (specifically, about 338 and 291 bp, respectively), in keeping with the presence of two A boxes in the −338/+1 region and one A box in the +200/+491 region (Figure 3B, Lanes 6 and 8). A slightly weaker band was detected at approximately 100 bp for the DNA amplified with the +1/+100 primer sets (Figure 3B, Lane 7). This weaker band reflects the absence of A boxes within the amplified region itself. However, because of the stochastic nature of the chromatin shearing by sonication, the presence of a weaker band denotes the variable extent of PDX-1 binding to promoter regions closely flanking the region to be amplified by PCR. The result of the electrophoresis of input DNA corroborates these findings (data not shown). As a positive control, owing to its own, well-established binding of PDX-1 to the insulin promoter, a pair of insulin promoter primers were used to amplify the PDX-1 immunoprecipitated DNA from β-HC9 cells. As expected, PCR amplification using the insulin primers resulted in a product of expected length (about 200 bp) indicating the ChIP was successful in detecting this expected interaction between PDX-1 and the insulin promoter (Figure 3B, Lane 5). Nonetheless, ChIP analysis demonstrated that PDX-1 binds to at least one location within the PANDER promoter region.
Figure 3. ChIP analysis of PDX-1 interaction with the PANDER promoter region.
Cross-linked DNA isolated from murine insulinoma β-HC9 cells were subjected to ChIP analysis using either anti-PDX1 antibody, murine IgG antibody (negative control), or murine insulin antibody (positive control) followed by PCR amplification using primers specific for the PANDER promoter that either surround or encompass A box regions. (A) Schematic diagram of the PANDER promoter region indicating the approximate location of the A box sites (TAAT) and primers utilized for ChIP PCR amplification. A1, A2, and A3 refer to A-box sites. Arrows representing forward and reverse primers designate primer locations. (B) Representative PCR results following ChIP purification on a 2% ethidium bromide stained agarose gel. Lane 1, marker; Lane 2, no input DNA; Lane 3, mouse IgG immunoprecipitation (IP); Lane 4, blank; Lane 5, insulin IP; Lanes 6, 7, 8, PDX1 IP with PANDER specific amplification at various regions.
3.4. Binding of PDX-1 to the PANDER promoter as demonstrated by EMSA
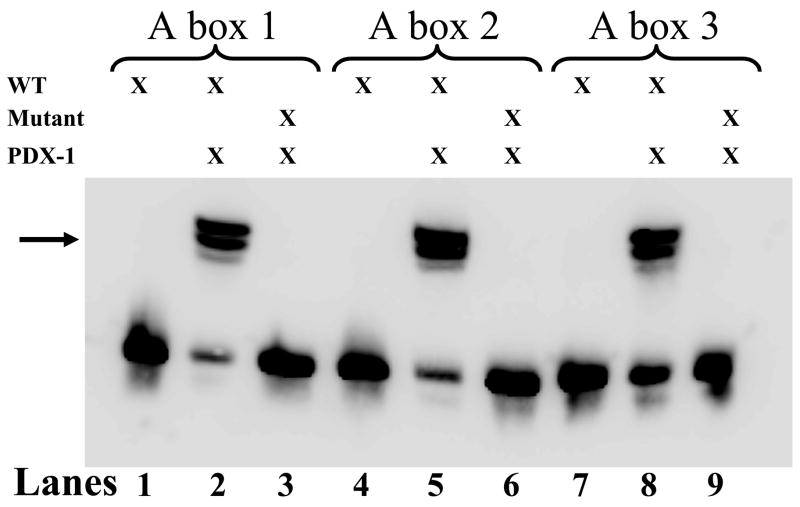
The above results were validated further by EMSA assays that examined the DNA binding specificity of the A boxes utilizing 3′-biotinylated oligonucleotides containing either the intact (wild type, 5′-TAAT-3′) A boxes with surrounding sequence or site-directed mutagenesis-derived A boxes (5′-TAGT-3′ with purified rat PDX-1 protein (Figure 4). All three A boxes demonstrate robust binding when incubated with PDX-1 (Figure 4, Lanes 2, 5, and 8). Concordantly, a marked decrease in binding was observed when the mutant A box-containing oligonucleotides were substituted for wild type oligonucleotides, indicating the binding specificity of the A box region of the PANDER promoter (Lanes 3, 6, and 9). The presence of a doublet in the shifted complex may be attributed to either proteolysis or dimerization of PDX-1. These results, in addition to the ChIP analysis, provide additional evidence for potential PDX-1 binding with the A boxes located in the PANDER promoter.
Figure 4. Binding of PDX-1 to the PANDER promoter as demonstrated by EMSA.
Chemiluminescence of mobility-shifted DNA/protein complexes. 3′-Biotinylated oligonucleotides (above, DNA) imitating either the wild type (5′-TAAT-3′) A boxes (WT) and flanking regions within the PANDER promoter, or the site-directed mutagenesis-derived (5′-TAAT-3′) A boxes (MUT), were incubated with rat PDX-1 protein (PDX-1). The reaction mixtures were then electrophoresed in a non-denaturing gel and transferred and cross-linked to a nylon membrane, incubated in Streptavidin-Horseradish Peroxidase Conjugate (Pierce), and the resulting chemiluminescence was detected photographically. Evaluation of A boxes 1 (Lanes 1–3), 2 (Lanes 4–6), and 3 (Lanes 7–9) are indicated above the membrane. Lanes 1, 4, and 7 contain biotinylated WT oligonucleotides only; Lanes 2, 5, and 8 contain biotinylated WT oligonucleotides with rat PDX-1 protein (PDX-1), and Lanes 3, 6, and 9 contain MUT oligonucleotides with rat PDX-1 protein. Arrow indicates shifted oligonucleotide/protein complex.
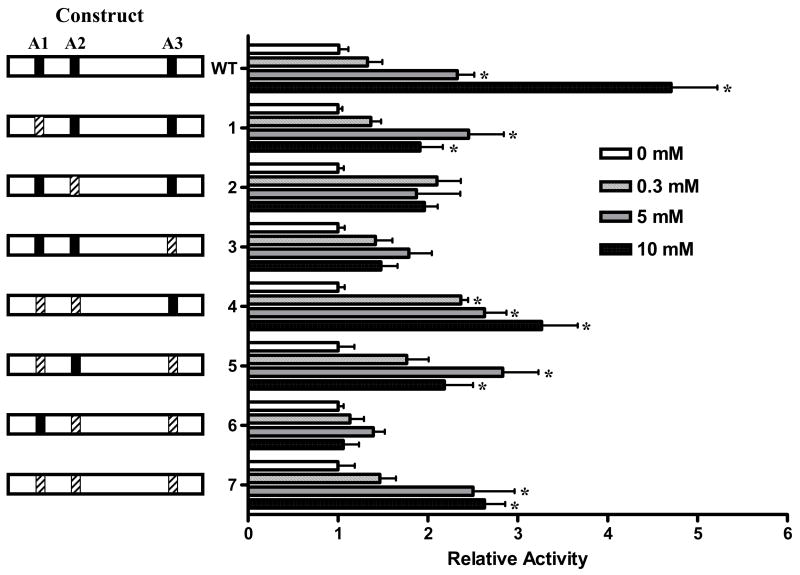
3.5. Mutagenesis of A boxes impairs glucose-responsiveness of the PANDER promoter
PDX-1 is a critical and essential factor involved in glucose modulation of the insulin promoter and endogenous insulin mRNA levels [24–26]. In addition, our previous results and those of others have demonstrated that glucose increases PANDER promoter activity, mRNA, and protein secretion in both insulinoma cell lines and primary islets [3–5]. The potential presence of multiple PDX-1 binding sites (A boxes) in conjunction with the glucose-responsive nature of PANDER implies that PDX-1 may be pivotal in the glucose regulation of PANDER expression. Therefore, we sought to determine if disruption of the PDX-1 binding sites impact the glucose-responsive expression of the PANDER promoter. This potential interaction was explored using the series of A box mutagenized PANDER/luciferase constructs for transfection into β-TC3 cells under increasing glucose conditions (Figure 5). PANDER/luciferase constructs containing mutagenized A box sites of 2 and 3, alone or in combination, resulted in significantly diminished glucose-responsiveness (Figure 5, constructs 2, 3, and 6). In contrast, alteration of A box 1, even in the presence of mutagenized A box sites 2 and 3, was still significantly glucose-responsive (constructs 1, 4, 5, and 7) (P > 0.05, ANOVA). As expected, the wild-type PANDER promoter was glucose-responsive with maximal induction at 10 mM (P < 0.05, ANOVA). Mutagenesis of A boxes 2 and 3 in combination resulted in a complete abrogation of glucose-induced PANDER promoter activity (Figure 5, construct 6). This result was also concordant with previously demonstrating that A box sites 2 and 3 are also critical for basal expression as shown in Figure 2. Therefore, the A box regions appear critical for glucose-responsive PANDER expression with particular importance on A boxes 2 and 3.
Figure 5. Mutagenesis of A boxes impairs glucose-responsiveness of the PANDER promoter.
(A) Mutagenized PANDER/luciferase plasmids are shown on the left side of the above figure and denoted by construct number as described previously (Figure 2A). A boxes are designated as A1, A2, and A3. Intact A boxes (5′-TAAT-3′) are represented by a black bar whereas mutagenized sites (5′-TAGT-3′) are shown by hatched bars. PANDER/luciferase constructs were transiently co-transfected with Renilla luciferase reporter plasmid phRL-TK into βTC3 cells in glucose-free DMEM. Four hours following transfection, the medium was replaced with DMEM containing incremental concentrations of D-glucose (i.e., 0 mM (white bar), 0.3 mM (vertical hatched bar), 5 mM (gray bar), 10 mM (black bar)). Twenty-four hours post-transfection, luciferase activity was detected by luminometry. Luciferase activity is shown as fold above respective construct at 0 mM glucose and normalized to renilla expression of the phRL-TK plasmid. All data are shown as mean ± S.E. from three independent experiments with each construct evaluated in triplicate. Asterik indicates a P value < 0.05 for the comparison of the PANDER/luciferase construct with that of the activity at 0 mM glucose according to ANOVA.
4. DISCUSSION
Promoters regulating genes involved in metabolic homeostasis share many similar characteristics in terms of genomic organization and functional expression. Metabolic regulating genes of insulin [27] and glucagon [7] have tissue-restricted (primarily islet) expression due to the complex interaction between highly conserved sequence binding motifs located within the promoter region and various islet-specific and ubiquitous transcriptional factors. These various trans-acting factors such as PDX-1, MafA, and Beta2/Neuro D serve a multi-purpose role in governing both the tissue-specificity and physiologically (nutrient or hormonal) induced expression via interaction with conserved binding motifs located within the 5′ flanking region of gene. Of these various elements, some of the most conserved in terms of β-cell specific expression are the A and E boxes [28] serving as binding domains for PDX-1 [6] and BETA2/NeuroD, respectively [29].
Insulin serves an essential role in metabolic homeostasis and is tightly regulated at multiple levels from gene transcription to insulin production and subsequent release. As previously characterized, the insulin promoter contains both A and E boxes and these regions are highly conserved and are pivotal determinants of β-cell specific and glucose-responsive expression [6,27]. PANDER and insulin share many similar characteristics that include (i) islet-specific expression (unlike insulin, PANDER is also found in alpha cells)[1,2]; (ii) localization within secretory granules of pancreatic β-cells [30]; (iii) glucose-induced promoter activity [3,5]; (iv) similar promoter organization containing both A and E box elements within close proximity of the transcriptional start site [3]; (v) glucose-induced mRNA expression [5]; (vi) glucose-induced protein expression [4,5]; and (vii) glucose-induced protein secretion in insulinoma cell lines or primary islets [4,5]. This report has now supported an additional similarity by demonstrating that a major transactivator of the insulin gene, PDX-1, also interacts with and regulates PANDER. Our current results demonstrate that PDX-1, MafA, and BETA2/NeuroD can upregulate PANDER promoter activity in a non-islet cell line with PDX-1 having the strongest stimulatory impact. Site-directed mutagenesis of the putative A boxes revealed that sites 2 and 3 were critical for PANDER promoter activity. Functional binding of PDX-1 to these A box sites was demonstrated via both EMSA and ChIP analysis. Furthermore, these A box regions were necessary for the glucose-responsive expression of PANDER.
Therefore, in intact nuclei, the pattern of PDX-1 binding should be able to be modified on a continuous and glucose-responsive basis and thus express PANDER in a tightly regulated fashion. We and others have previously demonstrated that PANDER expression is glucose responsive [3,5]. Moreover, we have now shown that A box-mediated activation of PANDER is likewise glucose responsive. It is already well established that PDX-1 activation of insulin expression is glucose responsive [31,32]. The close concordance between the regulation of PANDER and insulin genes suggest shared upstream signaling pathways. For example, glucokinase expression, like that of insulin, is regulated by PDX-1 [17], BETA2/NeuroD [33], and p300 [34]. It is eminently sensible, therefore, that similar upstream signaling pathways would result in the same transcription factors converging at each promoter. Indeed, insulin and glucokinase expression have been shown to be positively regulated via incretin activation of the phosphatidylinositol-3-kinase [35] and protein kinase A [36] signal transduction pathways in β-cells. Wang et al. recently demonstrated that glucose induced PANDER gene expression in pancreatic β-cells is via the Ca2+-PKA-ERK1/2-CREB, Ca2+-PKC-CREB, and PI3K-, ROS- related signaling pathways [5]. Furthermore, the glucose-induced insulin response is dependent upon ERK1/2 phosphorylation of a subset of transcription factors that include PDX-1 and BETA2 [37]. Likewise, the similarity in regulation of PANDER and insulin genes corroborates the shared upstream signals, in turn implying similar secretory stimuli. In this regard, we hypothesize that the physiological role of PANDER is involved with glucose homeostasis. However, animal models (ie. transgenic or knockout) are necessary to truly determine the biological role of PANDER in-vivo.
Acknowledgments
The authors would like to thank Roland Stein and Eva Henderson for supplying the pcDNA3.1 MafA, BETA2/NeuroD, PDX-1, and pcDNA3.1 plasmids. The authors also thank Lijun Yang and William L. Donelan for supplying the rat PDX-1 protein and providing technical expertise. This work was supported by grant K01-DK070744 to B.R.B. from the NIDDK, National Institutes of Health, and the Juvenile Diabetes Research Foundation.
Abbreviations used
- BETA2
BETA2 (B-cell E box transactivator 2)/NeuroD
- ChIP
chromatin immunoprecipitation
- DMEM
Dulbecco’s modified Eagle’s medium
- MafA
musculoaponeurotic fibrosarcoma A
- PANDER
PANcreatic DERived factor
- PDX-1
pancreatic/duodenal homeobox-1
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Zhu Y, Xu G, Patel A, McLaughlin MM, Silverman C, Knecht K, Sweitzer S, Li X, McDonnell P, Mirabile R, Zimmerman D, Boyce R, Tierney LA, Hu E, Livi GP, Wolf B, bdel-Meguid SS, Rose GD, Aurora R, Hensley P, Briggs M, Young PR. Cloning, expression, and initial characterization of a novel cytokine-like gene family. Genomics. 2002;80:144. doi: 10.1006/geno.2002.6816. [DOI] [PubMed] [Google Scholar]
- 2.Cao X, Gao Z, Robert CE, Greene S, Xu G, Xu W, Bell E, Campbell D, Zhu Y, Young R, Trucco M, Markmann JF, Naji A, Wolf BA. Pancreatic-derived factor (FAM3B), a novel islet cytokine, induces apoptosis of insulin-secreting beta-cells. Diabetes. 2003;52:2296. doi: 10.2337/diabetes.52.9.2296. [DOI] [PubMed] [Google Scholar]
- 3.Burkhardt BR, Yang MC, Robert CE, Greene SR, McFadden KK, Yang J, Wu J, Gao Z, Wolf BA. Tissue-specific and glucose-responsive expression of the pancreatic derived factor (PANDER) promoter. Biochim Biophys Acta. 2005;1730:215. doi: 10.1016/j.bbaexp.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 4.Yang J, Robert CE, Burkhardt BR, Young RA, Wu J, Gao Z, Wolf BA. Mechanisms of glucose-induced secretion of pancreatic-derived factor (PANDER or FAM3B) in pancreatic beta-cells. Diabetes. 2005;54:3217. doi: 10.2337/diabetes.54.11.3217. [DOI] [PubMed] [Google Scholar]
- 5.Wang O, Cai K, Pang S, Wang T, Qi D, Zhu Q, Ni Z, Le Y. Mechanisms of glucose-induced expression of pancreatic-derived factor in pancreatic {beta}-cells. Endocrinology. 2007 doi: 10.1210/en.2007-0106. [DOI] [PubMed] [Google Scholar]
- 6.German M, Ashcroft S, Docherty K, Edlund H, Edlund T, Goodison S, Imura H, Kennedy G, Madsen O, Melloul D. The insulin gene promoter. A simplified nomenclature. Diabetes. 1995;44:1002. doi: 10.2337/diab.44.8.1002. [DOI] [PubMed] [Google Scholar]
- 7.Drucker DJ, Philippe J, Jepeal L, Habener JF. Glucagon gene 5′-flanking sequences promote islet cell-specific gene transcription. J Biol Chem. 1987;262:15659. [PubMed] [Google Scholar]
- 8.Sander M, Sussel L, Conners J, Scheel D, Kalamaras J, Dela CF, Schwitzgebel V, Hayes-Jordan A, German M. Homeobox gene Nkx6.1 lies downstream of Nkx2.2 in the major pathway of beta-cell formation in the pancreas. Development. 2000;127:5533. doi: 10.1242/dev.127.24.5533. [DOI] [PubMed] [Google Scholar]
- 9.Peshavaria M, Henderson E, Sharma A, Wright CV, Stein R. Functional characterization of the transactivation properties of the PDX-1 homeodomain protein. Mol Cell Biol. 1997;17:3987. doi: 10.1128/mcb.17.7.3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Naya FJ, Stellrecht CM, Tsai MJ. Tissue-specific regulation of the insulin gene by a novel basic helix-loop-helix transcription factor. Genes Dev. 1995;9:1009. doi: 10.1101/gad.9.8.1009. [DOI] [PubMed] [Google Scholar]
- 11.Wang H, Maechler P, Ritz-Laser B, Hagenfeldt KA, Ishihara H, Philippe J, Wollheim CB. Pdx1 level defines pancreatic gene expression pattern and cell lineage differentiation. J Biol Chem. 2001;276:25279. doi: 10.1074/jbc.M101233200. [DOI] [PubMed] [Google Scholar]
- 12.Ahlgren U, Jonsson J, Jonsson L, Simu K, Edlund H. beta-cell-specific inactivation of the mouse Ipf1/Pdx1 gene results in loss of the beta-cell phenotype and maturity onset diabetes. Genes Dev. 1998;12:1763. doi: 10.1101/gad.12.12.1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Habener JF, Kemp DM, Thomas MK. Minireview: transcriptional regulation in pancreatic development. Endocrinology. 2005;146:1025. doi: 10.1210/en.2004-1576. [DOI] [PubMed] [Google Scholar]
- 14.Offield MF, Jetton TL, Labosky PA, Ray M, Stein RW, Magnuson MA, Hogan BL, Wright CV. PDX-1 is required for pancreatic outgrowth and differentiation of the rostral duodenum. Development. 1996;122:983. doi: 10.1242/dev.122.3.983. [DOI] [PubMed] [Google Scholar]
- 15.Jonsson J, Carlsson L, Edlund T, Edlund H. Insulin-promoter-factor 1 is required for pancreas development in mice. Nature. 1994;371:606. doi: 10.1038/371606a0. [DOI] [PubMed] [Google Scholar]
- 16.Waeber G, Thompson N, Nicod P, Bonny C. Transcriptional activation of the GLUT2 gene by the IPF-1/STF-1/IDX-1 homeobox factor. Mol Endocrinol. 1996;10:1327. doi: 10.1210/mend.10.11.8923459. [DOI] [PubMed] [Google Scholar]
- 17.Watada H, Kajimoto Y, Miyagawa J, Hanafusa T, Hamaguchi K, Matsuoka T, Yamamoto K, Matsuzawa Y, Kawamori R, Yamasaki Y. PDX-1 induces insulin and glucokinase gene expressions in alphaTC1 clone 6 cells in the presence of betacellulin. Diabetes. 1996;45:1826. doi: 10.2337/diab.45.12.1826. [DOI] [PubMed] [Google Scholar]
- 18.Holland AM, Gonez LJ, Naselli G, Macdonald RJ, Harrison LC. Conditional expression demonstrates the role of the homeodomain transcription factor Pdx1 in maintenance and regeneration of beta-cells in the adult pancreas. Diabetes. 2005;54:2586. doi: 10.2337/diabetes.54.9.2586. [DOI] [PubMed] [Google Scholar]
- 19.Lottmann H, Vanselow J, Hessabi B, Walther R. The Tet-On system in transgenic mice: inhibition of the mouse pdx-1 gene activity by antisense RNA expression in pancreatic beta-cells. J Mol Med. 2001;79:321. doi: 10.1007/s001090100229. [DOI] [PubMed] [Google Scholar]
- 20.Thomas MK, Devon ON, Lee JH, Peter A, Schlosser DA, Tenser MS, Habener JF. Development of diabetes mellitus in aging transgenic mice following suppression of pancreatic homeoprotein IDX-1. J Clin Invest. 2001;108:319. doi: 10.1172/JCI12029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Iype T, Francis J, Garmey JC, Schisler JC, Nesher R, Weir GC, Becker TC, Newgard CB, Griffen SC, Mirmira RG. Mechanism of insulin gene regulation by the pancreatic transcription factor Pdx-1: application of pre-mRNA analysis and chromatin immunoprecipitation to assess formation of functional transcriptional complexes. J Biol Chem. 2005;280:16798. doi: 10.1074/jbc.M414381200. [DOI] [PubMed] [Google Scholar]
- 22.Chakrabarti SK, James JC, Mirmira RG. Quantitative assessment of gene targeting in vitro and in vivo by the pancreatic transcription factor, Pdx1. Importance of chromatin structure in directing promoter binding. J Biol Chem. 2002;277:13286. doi: 10.1074/jbc.M111857200. [DOI] [PubMed] [Google Scholar]
- 23.Wang Z, Fang R, Olds LC, Sibley E. Transcriptional regulation of the lactase-phlorizin hydrolase promoter by PDX-1. Am J Physiol Gastrointest Liver Physiol. 2004;287:G555–G561. doi: 10.1152/ajpgi.00011.2004. [DOI] [PubMed] [Google Scholar]
- 24.Melloul D, Ben-Neriah Y, Cerasi E. Glucose modulates the binding of an islet-specific factor to a conserved sequence within the rat I and the human insulin promoters. Proc Natl Acad Sci U S A. 1993;90:3865. doi: 10.1073/pnas.90.9.3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Macfarlane WM, Read ML, Gilligan M, Bujalska I, Docherty K. Glucose modulates the binding activity of the beta-cell transcription factor IUF1 in a phosphorylation-dependent manner. Biochem J. 1994;303(Pt 2):625. doi: 10.1042/bj3030625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Petersen HV, Serup P, Leonard J, Michelsen BK, Madsen OD. Transcriptional regulation of the human insulin gene is dependent on the homeodomain protein STF1/IPF1 acting through the CT boxes. Proc Natl Acad Sci U S A. 1994;91:10465. doi: 10.1073/pnas.91.22.10465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Melloul D, Marshak S, Cerasi E. Regulation of insulin gene transcription. Diabetologia. 2002;45:309. doi: 10.1007/s00125-001-0728-y. [DOI] [PubMed] [Google Scholar]
- 28.Hay CW, Docherty K. Comparative analysis of insulin gene promoters: implications for diabetes research. Diabetes. 2006;55:3201. doi: 10.2337/db06-0788. [DOI] [PubMed] [Google Scholar]
- 29.Massari ME, Murre C. Helix-loop-helix proteins: regulators of transcription in eucaryotic organisms. Mol Cell Biol. 2000;20:429. doi: 10.1128/mcb.20.2.429-440.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu W, Gao Z, Wu J, Wolf BA. Interferon-gamma-induced regulation of the pancreatic derived cytokine FAM3B in islets and insulin-secreting betaTC3 cells. Mol Cell Endocrinol. 2005;240:74. doi: 10.1016/j.mce.2005.05.010. [DOI] [PubMed] [Google Scholar]
- 31.Stanojevic V, Habener JF, Thomas MK. Pancreas duodenum homeobox-1 transcriptional activation requires interactions with p300. Endocrinology. 2004;145:2918. doi: 10.1210/en.2003-1188. [DOI] [PubMed] [Google Scholar]
- 32.Mosley AL, Corbett JA, Ozcan S. Glucose regulation of insulin gene expression requires the recruitment of p300 by the beta-cell-specific transcription factor Pdx-1. Mol Endocrinol. 2004;18:2279. doi: 10.1210/me.2003-0463. [DOI] [PubMed] [Google Scholar]
- 33.Moates JM, Nanda S, Cissell MA, Tsai MJ, Stein R. BETA2 activates transcription from the upstream glucokinase gene promoter in islet beta-cells and gut endocrine cells. Diabetes. 2003;52:403. doi: 10.2337/diabetes.52.2.403. [DOI] [PubMed] [Google Scholar]
- 34.Roth U, Curth K, Unterman TG, Kietzmann T. The transcription factors HIF-1 and HNF-4 and the coactivator p300 are involved in insulin-regulated glucokinase gene expression via the phosphatidylinositol 3-kinase/protein kinase B pathway. J Biol Chem. 2004;279:2623. doi: 10.1074/jbc.M308391200. [DOI] [PubMed] [Google Scholar]
- 35.Buteau J, Roduit R, Susini S, Prentki M. Glucagon-like peptide-1 promotes DNA synthesis, activates phosphatidylinositol 3-kinase and increases transcription factor pancreatic and duodenal homeobox gene 1 (PDX-1) DNA binding activity in beta (INS-1)-cells. Diabetologia. 1999;42:856. doi: 10.1007/s001250051238. [DOI] [PubMed] [Google Scholar]
- 36.Wang X, Cahill CM, Pineyro MA, Zhou J, Doyle ME, Egan JM. Glucagon-like peptide-1 regulates the beta cell transcription factor, PDX-1, in insulinoma cells. Endocrinology. 1999;140:4904. doi: 10.1210/endo.140.10.7158. [DOI] [PubMed] [Google Scholar]
- 37.Khoo S, Griffen SC, Xia Y, Baer RJ, German MS, Cobb MH. Regulation of insulin gene transcription by ERK1 and ERK2 in pancreatic beta cells. J Biol Chem. 2003;278:32969. doi: 10.1074/jbc.M301198200. [DOI] [PubMed] [Google Scholar]