Abstract
Cadmium is a non-essential toxic metal in mammals. Its toxicity is mainly caused by interactions with cellular proteins that result in protein dysfunction and the disturb normal cellular functions. Glutathione (GSH) has been reported to play a role in cadmium resistance by serving as a cofactor for MRP1/GS-X pump-mediated cadmium elimination. To further investigate the role of GSH in cadmium toxicity, we carried out comparative study using small cell lung cancer derived cell lines, SR3A, and those that were stably transfected with glutamate cysteine ligase catalytic subunit (GCLC), a rate limiting enzyme in GSH biosynthesis. These GCLC-stably transfected cell lines produced higher levels of GSH and were more resistant to cadmium toxicity than the parental cell line was. The rates of cadmium uptake were reduced in these GCLC-transfected cell lines, which were associated with down regulation of the cadmium transporter ZIP8/SLC39A8. Further analyses demonstrated that Sp1 binding site at the proximal promoter region of ZIP8 was sensitive to the GSH level, and that the expression level of transcription factor Sp1 was reduced by increased GSH levels. We also demonstrated that low concentrations of cadmium exposure downregulated ZIP8 expression with concomitant reduction of Sp1 expression. Taken together, these results demonstrate the importance of Sp1 in the regulation of ZIP8 expression. More important, our results reveal a new mechanism by which elevated GSH levels confer cadmium resistance by downregulation of ZIP8 expression through the suppression of Sp1.
Introduction
Cadmium is a toxic metal ion in humans and is considered as a type I carcinogen (Waalkes, 2003). Because of its long retention time in the human body and high mutagenicity at low concentrations, cadmium is an important environmental poison to human health. Cadmium toxicity is through interactions with proteins that subsequently cause dysfunction of protein complex and organelles (Martelli et al., 2006) Eukaryotic cells develop detoxification mechanism to defend cadmium-mediated toxicity. The cellular cadmium detoxification pathway is mostly dependent on the upregulation of metallothionein. Metallothionein consists of 61 amino acids and has multiple cysteine residues by which heavy metal ions including cadmium are immobilized and lose their toxic effects (Klaassen et al., 1999). However, studies in metallothionein knockout mouse strains showed that overall cadmium sensitivity is not determined solely by the metallothionein phenotype (Liu et al., 2001a; Liu et al., 2001b).
Another important mechanism that affects cadmium toxicity is the cadmium transport system. In eukaryotic cells, several studies have demonstrated that the cadmium efflux system is mediated by the multidrug resistance protein (MRP) family in yeast (Adle et al., 2007) and plants (Kim et al., 2007). MRP proteins belong to the ATP-binding cassette (ABC) superfamily membrane proteins, which are involved in eliminating a wide spectrum of cytotoxic agents from intracellular compartments (Borst et al., 2000). Human MRP1 is induced by cadmium exposure and its expression levels are associated with cadmium resistance (Ishikawa et al., 1996).
Despite a number of studies on the regulation of the cadmium efflux system, molecular mechanism of regulation of cadmium uptake has not been well studied. In mammals, cadmium is taken up from the gastrointestinal tract or lung through contaminated food and water or through smoking (Bridges and Zalups, 2005; Oberdorster, 1992). Studies on cadmium uptake using various organisms revealed that cadmium uptake is mainly carried out by a metal transporter with a high affinity for manganese (Martin et al., 2006). Presently several metal transporters have been identified as cadmium transporters. While in the gastrointestinal tract, the divalent metal transporter is an important mediator of cadmium in addition to other divalent metal ions including Fe2+, Cu2+, and Zn 2+ (Park et al., 2002). A recent genetic study revealed that ZIP8 plays an important role in cadmium uptake and is known to be expressed in lung rather than gastrointestinal tract (Begum et al., 2002; Dalton et al., 2005) and, a subsequent biochemical study revealed that ZIP8 has a high affinity for cadmium and manganese (He et al., 2006). In addition to these transporters, recent studies have suggested that the calcium channel in cadmium uptake, although the significance of these calcium channels in cellular defense against cadmium toxicity has not been well developed (Leslie et al., 2006).
Glutathione (GSH) is an abundant intracellular thiol-containing compounds (1 ∼ 10 mM). De novo biosynthesis of GSH is controlled by the rate-limiting enzyme, glutamate cystein ligase which catalyzes the conjugation of glutamine and cysteine. Glutamate cysteine ligase is a heterodimer consists of a catalytic subunit (GCLC) and a modifier subunit. We have previously demonstrated that overexpression of GCLC is sufficient to confer increased intracellular GSH content (Yamane et al., 1998). In the present study, we demonstrate that GCLC-mediated elevated level of GSH confer cadmium resistance through downregulation of cadmium transporter ZIP8 expression, owing to reduced expression of transcription factor Sp1 which is required for ZIP8 expression. Our results reveal a novel pathway for GSH-mediated reduction of cadmium toxicity in mammalian cells.
Materials and Methods
Cell culture and reagents
Small cell lung cancer cells (SCLC) and their doxorubicin-resistant cell line SR3A (Yamane et al., 1998) were maintained in RPMI medium supplemented with 10% fetal calf serum. We supplemented 200 μg/ml G418 for GCLC-transfected SR3A cell lines. Small cell lung cancer (SCLC) cell and HEK-293 cell lines were cultured in Dulbecco’s modified Eagle’s medium with 10% fetal bovine serum. Rabbit anti-Sp1 (PEP-2) and Sp3 (D-20) antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA) and mouse monoclonal anti-β-actin (AC-15) and hemagglutinin (HA-7) antibody were from Sigma-Aldrich (St. Louis, MO). Rabbit anti-GCLC antibody was described previously (Tatebe et al., 2002). BSO, N-acetyl cysteine (NAC), zinc chloride, manganese chloride and cadmium chloride were purchased from Sigma.
Plasmid and siRNA transfection
Synthetic siRNA targeting ZIP8 was purchased from Sigma. The siRNA sequence used in this study was 5′-GAUUUGAUCCCAAAGUCGA-3′. Control siRNA was purchased from Santa Cruz Biotechnology (Santa Cruz, CA). The Sp1 full-length or zinc finger domain (encoding amino acids 480-785) sequences were cloned in pcDNA3-HA vector. The siRNA and plasmid DNA were transfected using lipofectamine 2000 (Invitrogen (Carlsbad, CA)) following manufacture’s instructions.
Total GSH analysis
Total GSH levels were analyzed using the Colorimetric Microplate Assay for Total glutathione kit (Oxford Biomedical Research (Oxford, MI)) following manufacture’s instruction.
Sulfurhodamine B colorimetirc (SRB) and 3-(4,5-dimethylthiazol-2-yl)-2-5-disphenyltetrazoliumbromide (MTT) assays
We seeded 20,000 cells in a 96-well plate and treated them with various concentrations of cadmium for 72 hours. For the MTT assay, MTT solution (thiazolyl blue tetrazolium bromide (Sigma-Aldrich)) was added to the culture after cadmium exposure and cells were incubated for 4 hours. The medium was removed, MTT crystals were dissolved in dimethyl sulfoxide, and the conversion rate of MTT crystal was measured by absorbance at 559 nm. The SRB assay was carried out following standard protocol (Vichai and Kirtikara, 2006). Briefly, cadmium exposed cells were fixed with 10% trichloroacetic acid 30 min and were stained by 0.4% SRB solution. After washed by 1% acetic acid, SRB dye was dissolved in 10 mM Tris buffer and cell viability was measured by absorbance at 405 nm.
Cadmium uptake Analysis
Cells (2 × 105) were plated in a 6-wells plate and exposed with final concentration of 0.3 μM 109Cd (Amersham biosciences (Piscataway, NJ)) for 6 hours. After cadmium exposure, cells were washed with phosphate buffered saline (PBS) containing 0.05 mM EDTA and lysed in 0.2 N sodium hydroxide. Cadmium uptake was analyzed by measuring the radioactivity of 109Cd.
DNA plasmid constructs
ZIP8 cDNA was cloned by PCR using the following primer (forward: 5′-TTATCTCTGTCCCCCTTTGTCCTC, and the reverse primer: 5′-GCGATAAGCCTCTAAGCCTGAACT). For the luciferase assay, the promoter region of ZIP8 was cloned using forward primer (5′ CTCGAGCTCCCTTCAGGCATGAATCCTCC) and the reverse primer (5′ AAGCTTCGAAAGAACAGCAGCTCGCGACC). For deletion constructs of ZIP8 promoter, DNA fragments were amplified using the reverse primer named above and the following forward primers: F1: 5′ GCTAGCTATGTAGACAATGCAAGGG, F2: 5′ GCTAGCAAGAGGGCATGGCTGATGC, F3: 5′ GCTAGCGGGACAGGGCCCTCCTCC, F4:5′ GCTAGCTATTTGTAAAGAGCGCCGG, F5: 5′ GCTAGCAGTCTTACGTTGACACGC and F6: GCTAGCTCTCACTTCTAAGTTTGC. Amplified DNA fragments were cloned using pGEM easy vector system (Promega (Madison, WI)) and the sequence integrity was confirmed. The cloned promoter sequence was digested with NheI/HidIII and ligated into pGL3 basic vector (Promega). For mutation analysis, mutations were introduced using the QuickChange Site Directed Mutagenesis Kit (Stratagene (La Jolla, CA)) with following oligo nucleotides pairs; (MT1: 5′ CACGTGCTACGGTTTCGGGGACAGGGCCCTCCTCC and 5′ GGAGGAGGGCCCTGTCCCCGAAACCGTAGCACGTG, MT2: 5′ CACGTGCTACGGAGGCGGGGACTTTGCCCTCCTCC and 5′ GGAGGAGGGCAAAGTCCCCGCCTCCGTAGCACGTG, MT:3 5′ CACGTGCTACGGAGGCGTTTACAGGGCCCTCCTCC and 5′ GGAGGAGGGCCCTGTAAACGCCTCCGTAGCACGTG).
Luciferase assay
The luciferase assay was carried out using a dual luciferase assay kit (Promega), according to the manufacturer’s instructions. In brief, cells cultured in 24 well dishes were transfected with 0.3 μg of pGL3 vector and 2 ng of pRLII vector using lipofectamine 2000 (Invitrogen (Carlsbad, (CA)) following manufacture’s instruction. Forty-eight hours after transfection, cells were lysed with passive lysis buffer. Cell lysates were cleared by centrifuge and supernatants were used for dual luciferase assay. The promoter activity of ZIP8 was represented as a fold induction from the empty pGL3 basic vector to standardize deviation from different cell lines.
Northern blotting
Northern blotting was carried out using the standard protocol. Total RNA was isolated by TRIZOL reagent using the standard protocol (Invitrogen). Fifteen micrograms of total RNA was separated in 1% formaldehyde-denatured agarose gel and transferred to a Hybond N+ membrane (Amersham Biosciences). The DNA probe was synthesized with a random prime DNA labeling kit (Roche, Basel, Switzerland) using ZIP8, Sp1 or glyceraldehyde 3-phosphate dehydrogenase cDNA and hybridized to RNA on the membrane at 45°C overnight. The membrane was washed three times with 2x SSC with 0.1% sodium dodecyl sulfate and the blots were visualized by X-ray film or the Phosphor imaging system.
Electrophoretic mobility shift assay (EMSA)
For EMSA analysis, double-stranded oligonucleotide DNA (5′ CACGTGCTACGGAGGCGGGGACAGGGCCCTCCTCC, only upper strand sequence is shown) was annealed and radioactively labeled by T4 kinase (New England Biolabs , Ipswich, MA). The labeled DNA oligonucleotide was purified by 8% PAGE. Radiolabeled DNA probe (10,000 cpm) was incubated at ambient temperature with 3 μg of nuclear extract, 3 μg of poly dI.dC in binding buffer containing 10 mM Tris HCl, 50 mM NaCl, 1 mM DTT, 5% Glycerol, 0.1 mg/ml BSA and 1 mM MgCl2. For the super-shift assay, 1.0 μg of anti-Sp1 or anti-Sp3 antibody was added to the reaction mixture. The reaction mixture was separated by 4% PAGE. The shift bands were visualized on X-ray film.
Real-time polymerase chain reaction (PCR)
The ABI 7900 system was used for real-time PCR analysis. For real time PCR, the following oligonucleotide DNA were used: human ZIP8 (F: 5′ CCTTATGTGTGATCGAGAGCCATTC, R: GTAATTCCTGAGATCATTGTTGGGC), human β̃-actin (F: 5′ GAGGCCCAGAGCAAGAGAG, R: 5′ AGAGGCGTACAGGGATAGCA), human CacnαG1 (F: 5′ CAAAGATGCACCTCATCTGC, R: 5′ ACTCTAAGCTGCTTCTGGTC). Amplification was monitored by fluorescence of SyberGreen dye (Invitrogen) and expression level was calculated by the cycling time (Ct) value. Expression level of ZIP8 was normalized by the expression level of β-actin.
Western blotting analysis
Cells were harvested and washed with phosphate buffered saline (PBS). Cell pellets were suspended in RIPA buffer and incubated on ice for 30 min. Cell lysates were cleared by centrifuge and protein concentrations were determined by Protein Assay reagent (Biorad Laboratories, Hercules, CA). Cell lysates were boiled in SDS sample buffer, separated by SDS-PAGE and transferred to PVDF membrane. The membrane was washed with TBST and blocked with 5% non-fat milk. Target proteins were detected with rabbit anti-Sp1, rabbit anti-Sp3, mouse anti β-actin, mouse anti-HA epitope or rabbit anti-GCLC antibodies. Proteins were further labeled with HRP conjugated secondary antibody (Pierce, Rockford, IL) and visualized with X-ray film.
Chromatin immunoprecipitation (ChIP) assay
The ChIP assay was performed according to the manufacturer’s instructions (Upstate, Waltham, MA). The precipitated DNA samples were amplified using oligonucleotide primers specific to the ZIP8 promoter (F: 5′ AATGGGTCACCACGTGCTAC, R: 5′ TTAATCGGAAGCACTCGCTG). PCR products were separated on 2% Agarose gel and visualized using ethidium bromide. For quantitative analysis, ChIP samples were analyzed by real-time PCR using the same oligonucleotide primers.
Preparation of lentiviral recombinants
Lentiviruses were generated using pLKO.1 vector (Addgene (Cambridge, MA)). The shRNA sequences used for targeting Sp1 were 5′-CCAGGTGCAAACCAACAGATT-3′ and 5′-GCTGGTGGTGATGGAATA-3′ for #2 and #3, respectively. The sequences for ZIP8 shRNA 325 and 715 are 5′-AAATTCTCTGTCATCTGTCCA-3′ and 5′-AATGGTCATACCCACTTTGGA-3′, respectively.
Statistical Analysis
Statistical significance was analyzed using Student’s t-test or one-way ANOVA and following the Student—Newman—Keuls post-hoc test. P-values <0.05 were considered to be statistically significant.
Result
Increased GCLC levels are associated with cadmium resistance
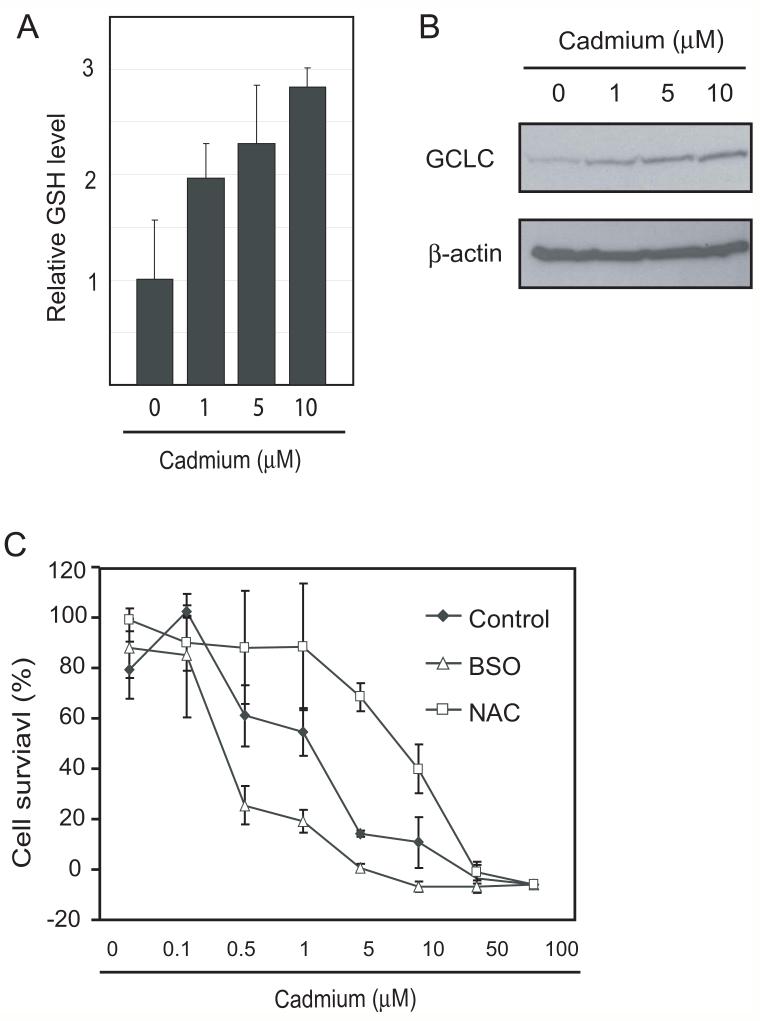
To study the role of GSH in cellular defense against cadmium toxicity, we first analyzed the effects of cadmium exposure on cellular GSH level. As shown in Fig.1A, cadmium exposure increased intercellular GSH levels in a dose-dependent manner in SCLC cells. This upregulation of GSH correlated with the expression level of GCLC (Fig. 1B).
Fig. 1. Protective role of GSH in cadmium toxicity.
A and B. SCLC cell lines were treated with 0, 1, 5 and 10 μM of cadmium for 20 hours and cellular GSH and GCLC expression levels were analyzed. C, The role of GSH in cadmium toxicity was examined by 24 hours pretreatment with 100 μM of BSO or 3 mM of NAC followed by exposure to various concentrations of cadmium for 72 hours. Cadmium toxicity was analyzed by SRB assay. Error bars represent standard deviation from three independent experiments. *p<0.05
To evaluate the role of upregulated GSH in cellular defense against cadmium, we modulated intercellular GSH level by BSO, an inhibitor for GSH biosynthesis, or by NAC. The 24 hours of treatment with 100 μM BSO effectively depleted GSH to less than 50% of control and 3 mM NAC treatment increased GSH level about three times (data not shown). The SCLC cells were pretreated with BSO or NAC for 24 hours and effect on cadmium sensitivity was determined. As shown in Fig. 1C, treatment with BSO sensitized cells to cadmium toxicity whereas with NAC significantly increased cadmium resistance.
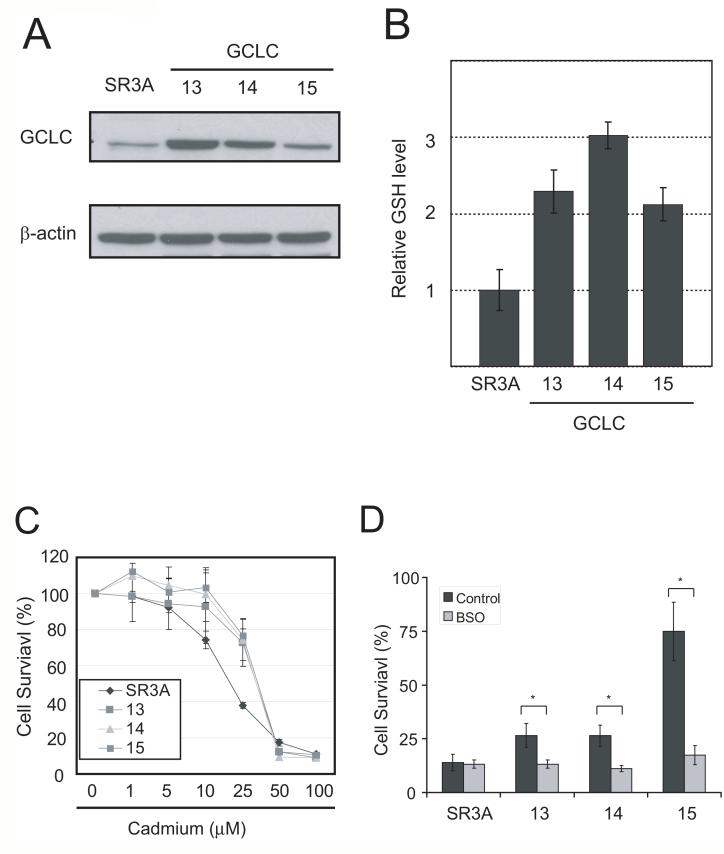
SR3A was a doxorubicin-resistant cell line established from SCLC. SR3A cells contain 1.5-fold higher GCLC mRNAs as compared with that in SCLC cells (Yamane et al., 1998). No induction of GCLC expression by cadmium at the same concentration range in SR3A cells was found. This is perhaps because the endogenous GCLC levels are already high (Fig. 2A, upper panel). To study how elevated GSH levels affect cadmium toxicity, we used GCLC-transfected cell lines SR3A-13, SR3A-14 and SR3A-15. These cells express two to four times higher GCLC and almost equally higher levels of GSH than parental cell line (Fig. 2A lower panel, and B). We found that GCLC-transfected cell lines exhibited significant resistance to cadmium (Fig. 2C). To verify the role of GSH in this increased cadmium resistance, we treated cells with BSO. As shown in Fig. 2D, BSO treatment reversed cadmium resistance in GCLC-transfected cell lines with minimum effect on parental cell line. These results indicate that GCLC-mediated upregulation of GSH level lead to enhanced resistance to cadmium toxicity.
Fig. 2. GCLC overexpression confer cadmium resistance.
A. (upper) Western blotting analysis showing no induction of GCLC expression by the treatment of cadmium at the concentrations indicated. (bottom) Western blotting analysis on GCLC levels in parental and GCLC-stably transfected SR3A cell lines. B. GSH levels in the GCLC-transfected cell lines. C. Cadmium sensitivity in GCLC stably transfected cell lines. Cells were seeded on a 96-well plate and exposed them to various concentrations of cadmium. D. Cadmium sensitivity was also examined by 72 hours of 25 μM cadmium exposure with or without 100 μM BSO. Error bar represent standard deviation from three independent experiments. *p<0.05
Decreased cadmium uptake is associated with downregulation of the ZIP8 transcript in the GCLC-transfected cell lines
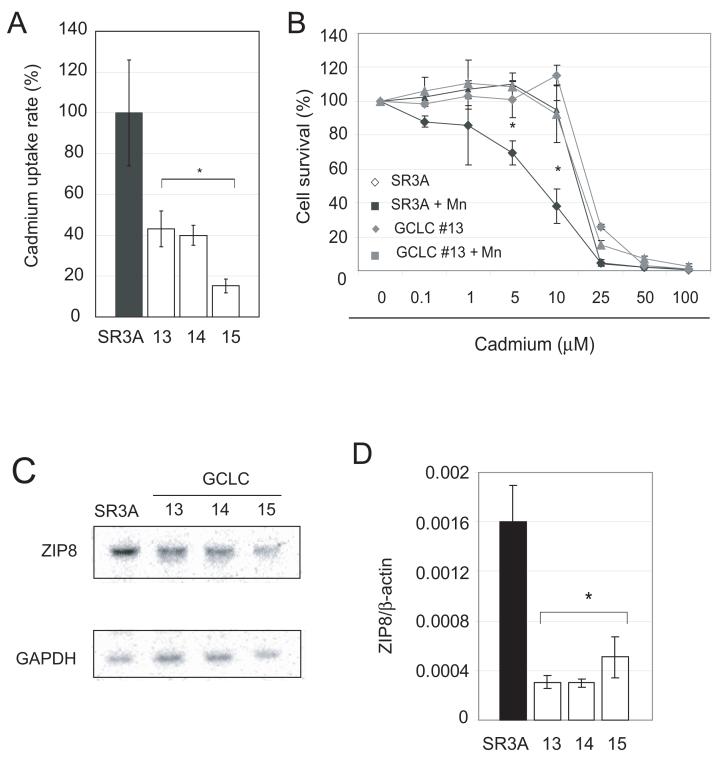
We next investigated whether the increased cadmium resistance in the GCLC-transfected cells were due to reduction in the rates of cadmium uptake. As shown in Fig. 3A, GCLC-overexpressing cells showed reduced cadmium uptake compared with the parental cells. To confirm the contribution of cadmium uptake, we used manganese, which is a known antagonist of cadmium uptake. Cells were exposed to cadmium with or without 100 μM of manganese, and cadmium sensitivity was analyzed using the MTT assay. As shown in Fig. 3B, manganese effectively protected cells against cadmium toxicity in parental SR3A but not in GCLC-transfected cell lines. The cadmium sensitivity of manganese-treated SR3A cells was similar to that of GCLC-transfected cell lines SR3A-13 (Fig. 3B) as well as clones SR3A-14 and SR3A-15 (data not shown). These data indicated that the cadmium transporter is most likely involved in the increased cadmium resistance.
Fig. 3. Downregulation of cadmium transporter ZIP8 in GCLC-overexpressing cells.
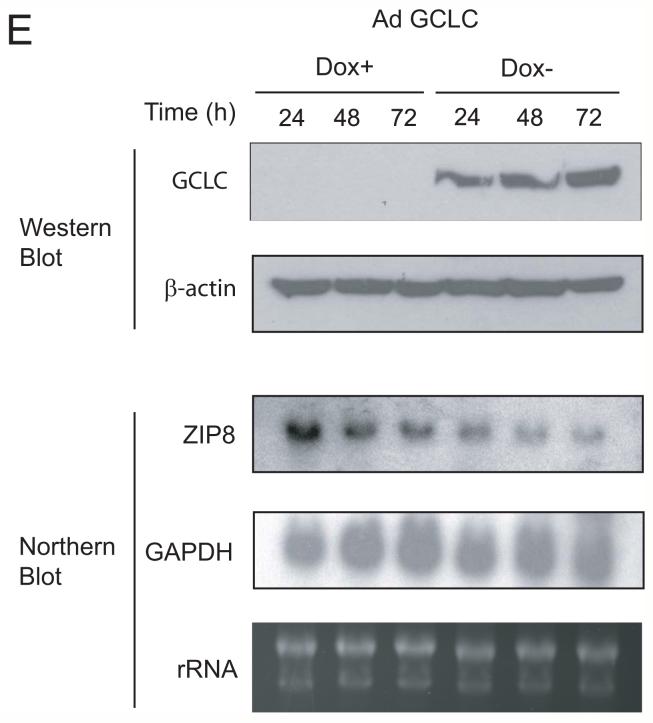
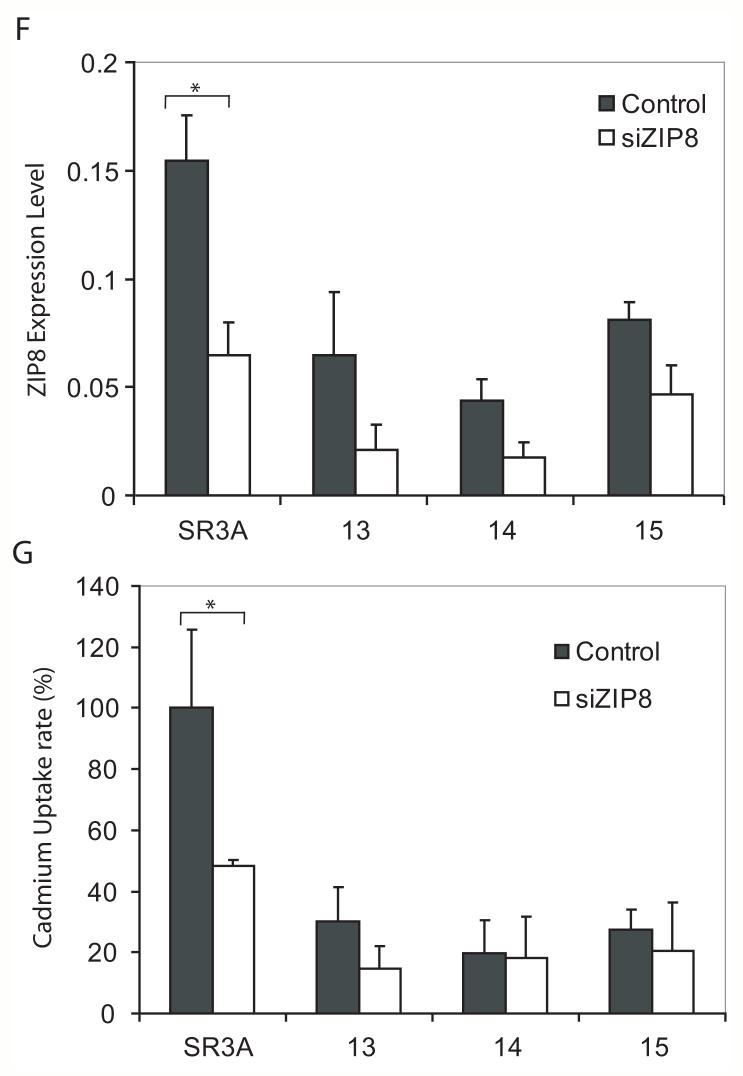
A. Effects of GCLC overexpression on cadmium uptake rate. Cells were treated with 0.3 μM 109Cd for 6 hours and cadmium uptake rates were analyzed. The cadmium uptake rate is represented as a percentage of the cadmium uptake of the parental SR3A cell line. Experiments were done triplicate and error bars represent standard deviations. B. Protective effect of manganese against cadmium toxicity. SR3A cell and SR3A-13 was exposed to various concentrations of cadmium in the presence or absence of 100 μM manganese for 72 hours. Survival cell fraction was assayed by MTT conversion. C, D. Down regulation of ZIP8 in GCLC-transfected cell lines. The expression levels of ZIP8 in SR3A and GCLC-transfected cell lines (SR3A-13, 14 and 15) were analyzed by Northern blotting and Real-time PCR. E. Effect of GCLC on expression level of ZIP8 was also evaluated by transduction with Tet-off adenovirus GCLC expression vector using SCLC cell. Cells were harvested 24, 48 and 72 hours after transduction, and the expression level of GCLC and ZIP8 was analyzed by western blotting and northern blotting, respectively. F, G. Effect of ZIP8 knock-down on cadmium uptake. Parental SR3A and GCLC-transfected cell lines were transfected with 100 nM of control or ZIP8 siRNA. Seventy-two hours after transfection, cells were harvested and subjected for real-time PCR. The transfected cells were also subjected to a cadmium uptake assay. Error bars represent standard deviation from six independent experiments. *p<0.05
Recently, ZIP8/SLC39A8 was identified as a potent cadmium transporter with a high affinity for manganese (He et al., 2006). Therefore, we determined whether ZIP8 plays a role in cadmium resistance in GCLC transfected cell lines. By northern blotting and real-time PCR analysis, we found the parental SR3A cell expressed significantly higher levels of ZIP8 transcripts than the GCLC-transfected cell lines (Figs. 3C, 3D).
To validate these results, we utilized tetracycline (tet)-off GCLC expression system, adenoviral vector AdE1.tTA. GCLC (Savaraj et al., 2005). SCLC cells were transduced with AdE1.tTA. GCLC in the presence (+) or absence (-) of tet for 24, 48, or 72 hrs. Levels of GCLC expression were increased approximately 11- and 30-fold in cells grown in the absence of tet for 24 and 48 hr, respectively, as compared with those treated with tet. Comparable levels of elevated GSH were found in the GCLC-overexpressing samples (Savaraj et al., 2005). Levels of ZIP8 mRNA were reduced in the GCLC-overexpressing cells (Fig. 3E) as compared correspondingly to the cells cultured under tet(+) conditions. These results, taken together, strongly suggest that the expression of ZIP8 levels was negatively regulated by intracellular GSH contents.
To confirm the contribution of ZIP8 in SR3A cell lines, we transfected ZIP8 siRNA and analyzed the expression of ZIP8 mRNA levels and cadmium uptake rates. As shown in Fig. 3F, transfection of ZIP8 siRNA significantly down regulated ZIP8 expression to about 50 % of control levels in SR3A cells and their GCLC-transfected variants. A significant reduction in cadmium uptake and slightly decreased those of GCLC transfected cell lines were observed (Fig. 3G). These differences in the effect of ZIP8 siRNA on cadmium uptake may be due to the already low levels of expression of ZIP8 in GCLC-transfected cells and/or to the background levels contributed by other metal transporters.
Analysis of ZIP8 promoter activity
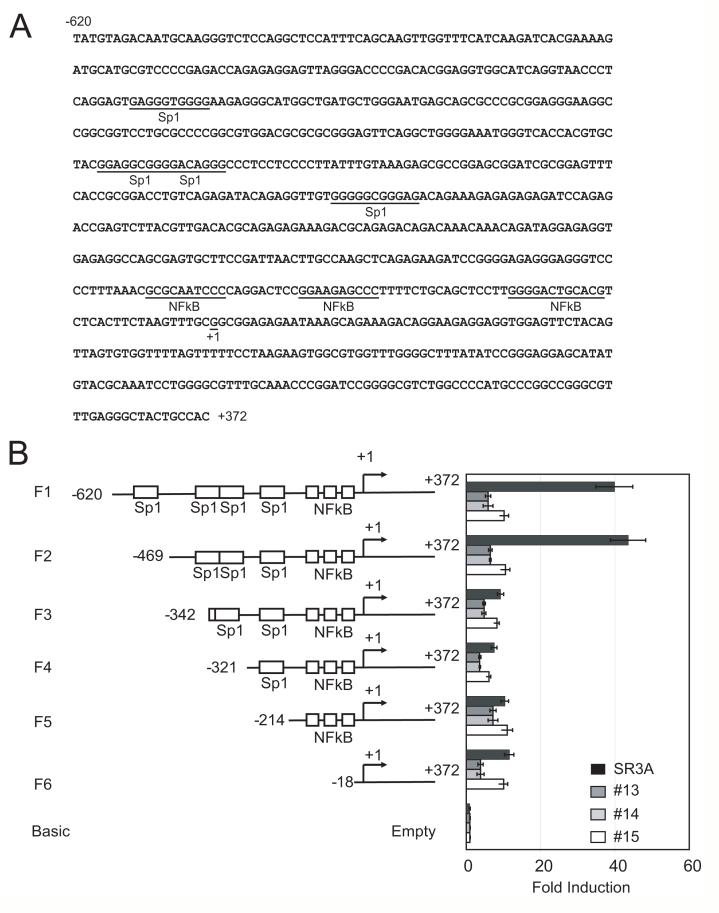
Next, we analyzed ZIP8 promoter activity in order to identify the mechanism that downregulates ZIP8 expression level in GCLC-transfected cell lines. Sequences upstream of the ZIP8 gene were analyzed by the computational method to identify potential promoter region and transcriptional start sites (Fig. 4A). We did not observe the canonical TATA-box, but several GC-box and several putative nuclear factor κB (NF-κB) binding sites were located in this region. Based on this information a 620 bp fragment that included the transcription start site from ZIP8 promoter region was amplified by PCR from human genomic DNA. The resulting fragment was cloned into the pGL3 basic vector for the reporter expression assay. The reporter construct was transfected into SR3A cells, and GCLC-transfected cell lines and promoter activity was analyzed. GCLC-transfected cell lines had a drastic reduction in luciferase activity compared with that in the parental cell line. These results suggested that transcription factor(s) interacting with cis-acting elements within the -620 nt of the ZIP8 promoter may be downregulated in GCLC-transfected cells.
Fig. 4. Analysis of ZIP8 proximal promoter activity.
A. ZIP8 promoter sequence. Transcription factor binding sites and the transcription initiation site (+1) are shown. B. ZIP8 promoter deletion and their promoter activity. Parental SR3A and -13, -14 and -15 cell lines cultured in 24 well plates were transfected with pGL3 vector coding various deletion of ZIP8 promoter (-620/+372, -469/+372, -342/+372, -321/+372 and 214/+372) or empty pGL3 vector and pRLII vector. Seventy-two hours after transfection, the promoter activity of each deletion was analyzed. Promoter activities of each construct were represented by fold induction from that of the empty vector. Error bars represent standard deviation from three independent experiments. *p<0.05
To determine the locations of the cis-acting elements, we generated several ZIP8 promoter deletion constructs. As shown in Fig. 4B, deletion mapping of ZIP8 promoter activity revealed that removing sequence between -469 nt (construct F2) and -342 (Construct F3) resulted in a drastic reduction of reporter activity in SR3A cells. These results suggested that sequences within -469 nt and -342 nt harbor a cis-element that supports basal expression of ZIP8 and that trans-acting factor that interacts with this element may be downregulated in GCLC-transfected cells.
Regulation of ZIP8 promoter by Sp1
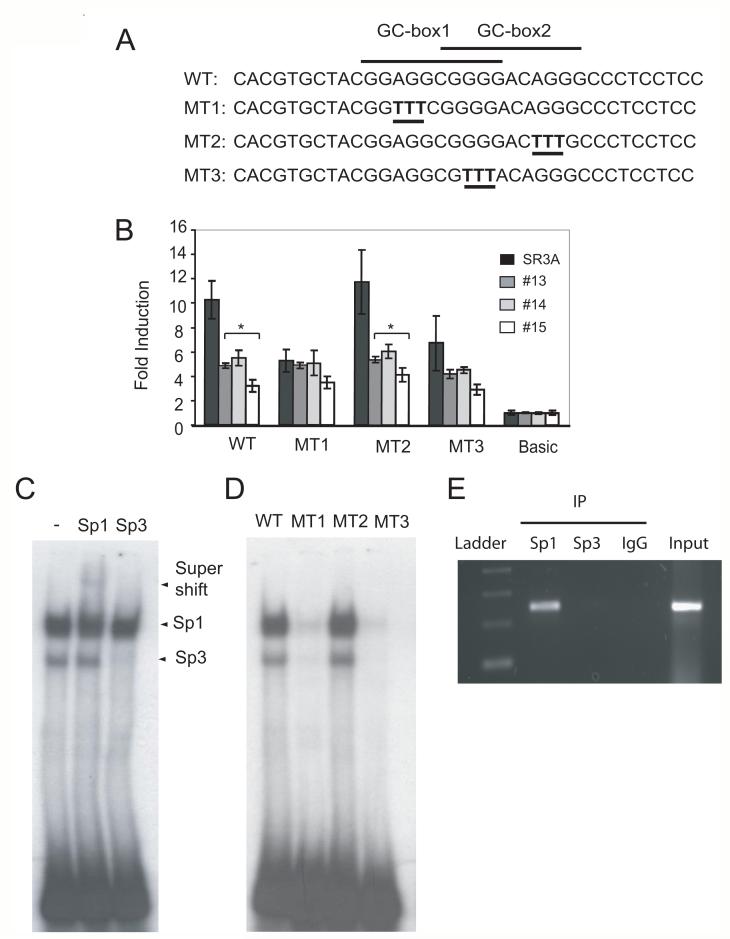
Examining nucleotide sequence between -469 nt and -342 nt, we found two overlapping GC-boxes. To investigate whether these GC-boxes are responsible for the basal transcription activity of the ZIP8 promoter, we generated three mutants (MT1, MT2, and MT3) containing altered nucleotides in these GC-boxes (Fig. 5A) in the F2 reporter construct. These mutant reporter constructs were transfected into SR3A and its GCLC-transfected variants. MT2 did not show reduction in the reporter activity, whereas MT1 and MT3 did (Fig.5B). These results demonstrated that GC-box 1 is involved in the GSH-mediated transcriptional regulation of ZIP8.
Fig. 5. Analysis of Sp1 binding to the ZIP8 promoter.
A. Scheme of mutation on GC-box. B. Effects of mutation on ZIP8 promoter activity. Mutations were introduced into a luciferase construct bearing ZIP8 promoter sequence (F2: -469 to +372) and transfected into SR3A and 13, 14 and 15 cell lines. Seventy-two hours after transfection, promoter activity was analyzed. C. EMSA. A radio-labeled oligonucleotide probe was incubated with SR3A nuclear extract and the DNA protein complex was separated by 4% PAGE. DNA/protein complex were identified by incubation with anti-Sp1 (lane 2) or anti-Sp3 (lane 3) antibody. D. EMSA of mutant oligonucleotides. SR3A nuclear extract was incubated with WT (lane 1), MT1 (lane 2), MT2 (lane 3) or MT3 (lane 4) radioacively labeled DNA probes. Either or both of overlapping GC-boxes were introduced. E. ChIP analysis. SR3A cells were cross-linked by 1% formaldehyde. Cells were lysed, and genomic DNA was sheared by sonication. The DNA protein complex was precipitated with anti-Sp1, or anti-Sp3 antibody and control IgG. DNA was purified. ted by PCR or quantitatively analyzed by real-time PCR. Error bars represent standard deviation from three independent experiments. *p<0.05
Next, we performed an electrophoretic EMSA analysis to determine whether any transcription factors can bind to this GC-box sequence. Because two GC-box sites overlapped, we synthesized oligonucleotides that contain both GC-box sequences. A double-stranded DNA oligonucleotide probe was radioactively labeled with 32P, incubated with nuclear extract from SR3A cells, and the mixture were run on 4% polyacryamide gel electrophoresis. We found two specific protein-DNA complexes. The slow mobility complex could be partly reduced by anti-Sp1 antibody and yielded a supershift band whereas the fast mobility complex could be completely diminished by anti-Sp3 antibody, indicating that Sp1 and Sp3 can recognize this GC-box in vitro (Fig. 5C). In order to distinguish two overlapping GC-box site, we labeled WT, MT1, MT2 and MT3 oligonucleotides and performed EMSA. The DNA protein complex was only observed in WT and MT2 (Fig. 5D). Together with the results in Fig. 5C, these results indicate that only GC-box1 is required for the transcriptional activity of ZIP8 expression.
To elucidate whether Sp1, Sp3, or both, participated in the transcription regulation of ZIP8 expression, we performed a chromatin ChiP assay using Sp1 and Sp3 antibodies to precipitate the chromatin fragments engaged with Sp1 and Sp3 transcription factors. Fig. 5E shows that only Sp1, but not Sp3, bound to ZIP8 promoter in vivo. These results demonstrated Sp1 is important for the expression of ZIP8.
Sp1 was down-regulated in GCLC-transfected cell lines
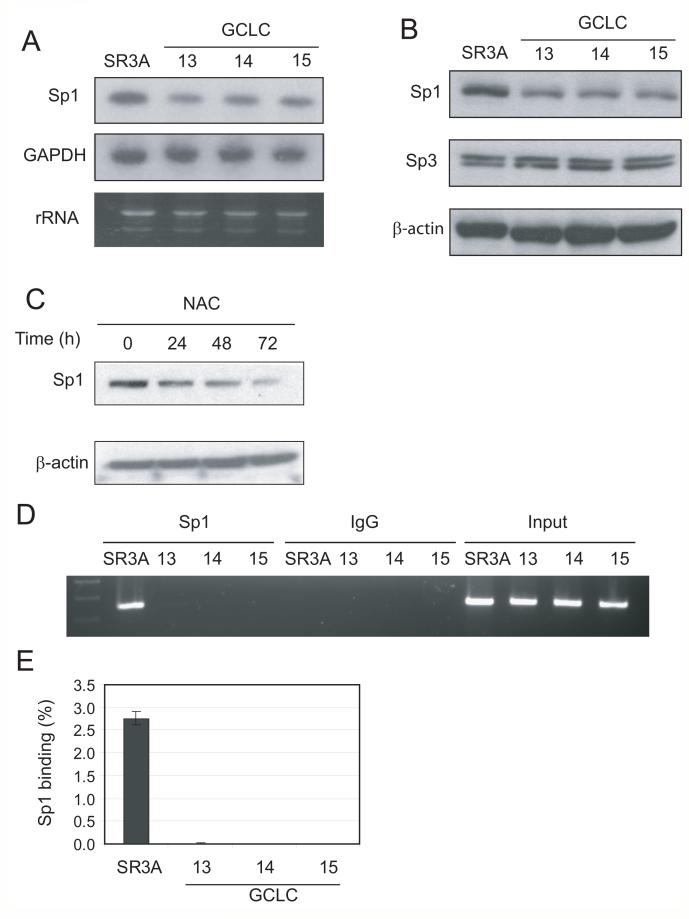
To investigate the mechanism underlying the reduced promoter activity of ZIP8 in GCLC-transfected cell lines, we measured the Sp1 levels in these cells. Levels of Sp1 mRNA (Fig. 6A) and protein (Fig. 6B) were reduced in the GCLC-transfected cell lines compared with those in un-transfected SR3A cells. Moreover, levels of Sp1 were reduced in cultured cells by continuous treatment with NAC (Fig. 6C). These results demonstrated that elevated expression of GSH downregulated Sp1 expression.
Fig. 6. Downregulation of protein and DNA binding levels of Sp1 in GCLC-transfected cell lines.
A, B. Exponentially grown SR3A and GCLC-transfected cell were harvested and Sp1 expression level was analyzed by western blotting or Northern Blotting. C. SCLC cells were treated with 5 mM NAC for 24, 48 and 72 hours and Sp1 level was analyzed by western blotting. D, E. Sp1 DNA binding on ZIP8 promoter was analyzed by ChIP assay. ChIP samples were analyzed by agarose electrophoresis and real-time PCR. Error bars represent standard deviation from three independent experiments.
Sp1 function is known to be regulated by posttranslational modification, and its protein level does not always reflect its in vivo DNA binding ability. Thus, we performed a ChIP assay using parental and GCLC-transfected cell lines to analyze the capacity of Sp1 DNA binding in vivo. A representative result of ChIP assay, shown in Fig. 6D, clearly demonstrates that Sp1 binding to ZIP8 promoter in parental SR3A cell lines but was almost undetectable in GCLC-transfected cell lines. The result of a quantitative analysis using real-time PCR result indicates that Sp1 in GCLC-transfected cell lines occupies less than 0.1% of ZIP8 promoter than that of parental cell lines (Fig. 6E). These results indicate that GCLC-overexpression reduces both Sp1 protein level and ZIP8 promoter activity.
Regulation of endogenous ZIP8 by Sp1
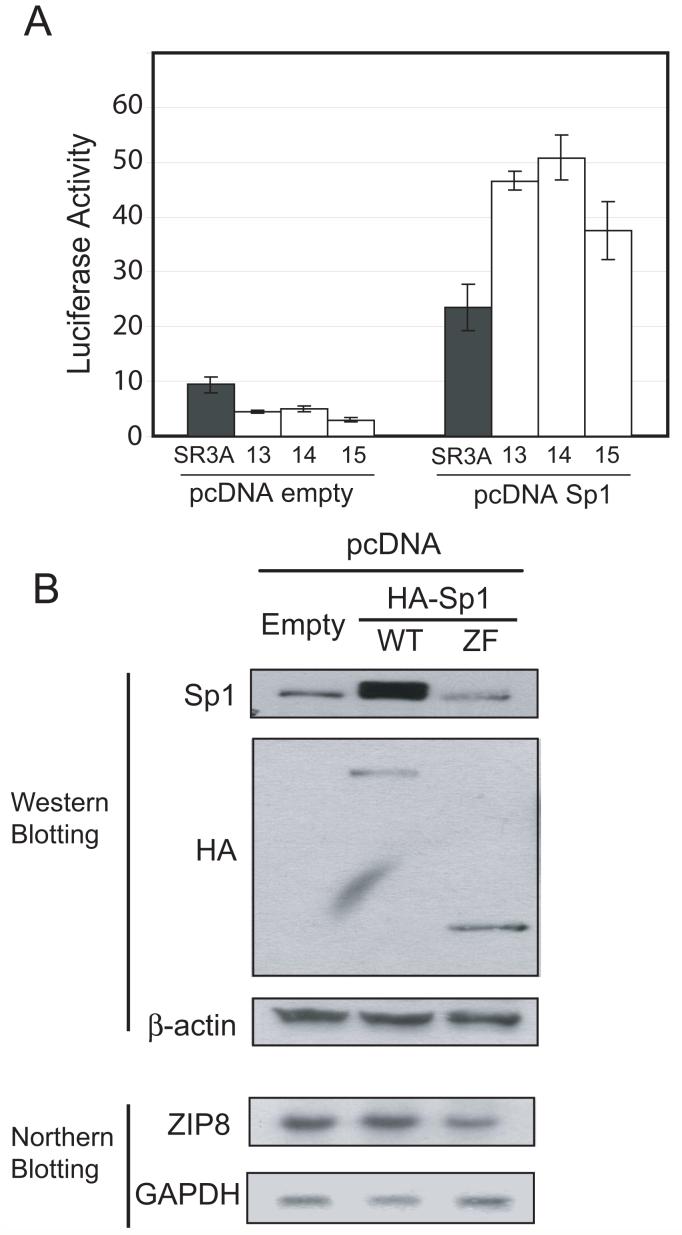
To further evaluate the role of Sp1 in ZIP8 promoter, we cotransfected Sp1 expression vector and measured ZIP8 promoter activity. As shown in Fig. 7A, expression of Sp1 upregulated ZIP8 promoter activity up to 10-fold in all four cell lines. Importantly, the luciferase activity of GCLC-transfected cell lines even exceeded that of the parental cell line when cells were cotransfected with Sp1 expression vector. These results demonstrated that expression of Sp1 enhanced the promoter activity of ZIP8.
Fig. 7. Effect of Sp1 overexpression on ZIP8 expression.
A. SR3A and GCLC-transfected cell lines were transfected with the ZIP8 pGL3 vector, with or without Sp1 expression vector. Luciferase activity was analyzed 72 hours after transfection. B. HEK-293 cell were transfected with empty vector or expression vectors encoding full-length Sp1 or the zinc finger motif of Sp1. Total protein an RNA were extracted 48 hours after transfection and endogenous and exogenous Sp1 expression was confirmed by western blotting and ZIP8 expression level was analyzed by northern blotting. Error bars represent standard deviation from three independent experiments. *p<0.05
Next we determined the role of Sp1 in endogenous ZIP8 promoter activity. HEK293 cells were transfected with expression vector encoding full-length Sp1 and a deletion mutant encoding the ZF domain. The results showed that full length Sp1 did not significantly upregulate endogenous ZIP8 expression, whereas the zinc finger domain of Sp1 downregulated ZIP8 expression level to 60% of control (Fig.7B). These results indicates that overexpression of the zinc finger domain has a dominant negative effect as reported previously (Lee et al., 2006). This dominant negative effect indicates that Sp1 DNA binding is needed for ZIP8 expression. However since Sp1 overexpression did not upregulate endogenous ZIP8 expression level, endogenous Sp1 levels in these cells may have been already high enough that additional Sp1 could not have stimulatory activity.
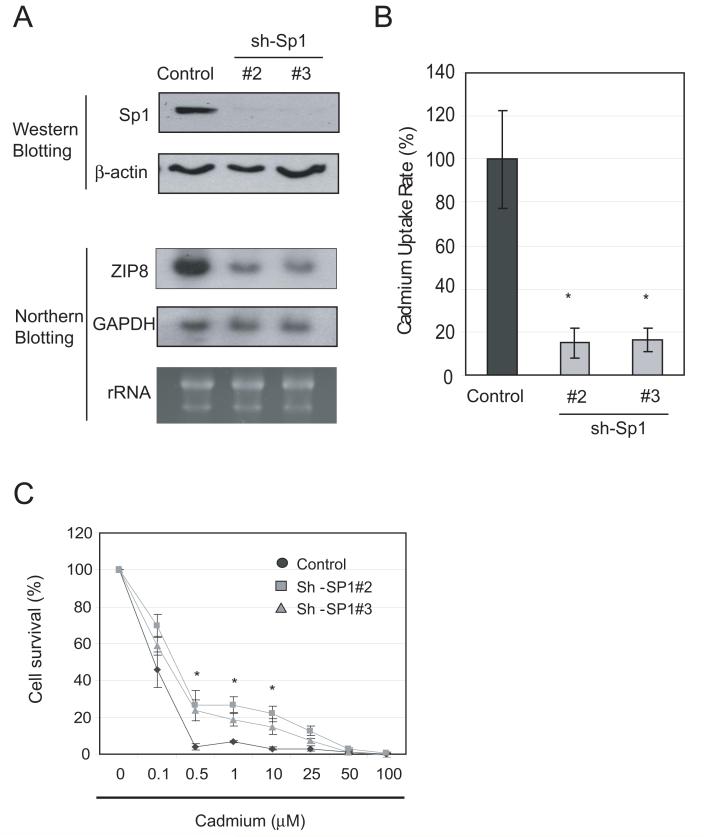
To further support the role of Sp1 in the regulation of ZIP8, we generated two lentivirus vectors encoding Sp1 shRNAs. HEK293 cells were transduced with lentivirus and Sp1 levels were analyzed by western blotting. Figure 8A shows that both shRNAs dramatically downregulated Sp1 mRNA and protein levels as analyzed by northern blotting and western blotting, respectively. Downregulation of ZIP8 by Sp1 knock-down was associated with reduced rates of cadmium uptake (Fig. 8B) and slightly enhanced cell resistance to cadmium treatment (Fig. 8C).
Fig. 8. Effect of Sp1 knockdown on cadmium toxicity.
A. Effects of Sp1 knockdown on ZIP8 expression. HEK-293 cells were transduced with lentivirus bearing control or sh-Sp1. Seventy-two hours after transduction, cells were harvested, and protein and total RNA were extracted. Expression levels of Sp1 and ZIP8 were analyzed by western blotting or northern blotting, respectively. B, C. Effects of Sp1 knockdown on cadmium sensitivity. HEK-293 cells transduced with control or sh-Sp1 vectors (#2 or #3) were subjected to cadmium uptake and MTT assays. Cadmium uptake rate was calculated and normalized to the control level. For the cadmium sensitivity assay, cells were transduced with lentivirus bearing scrambled or sh-Sp1 sequences. Seventy-two hours after transduction, cells were treated with various concentrations of cadmium for 72 hours and MTT activity was assayed. Error bars represent standard deviation from three independent experiments. *p < 0.05
Taken together, these results strongly support the important roles of Sp1 in the regulation of ZIP8 expression and that downregulation of Sp1 levels result in down-regulation of ZIP8 expression and cadmium resistance in GCLC-transfected cell lines.
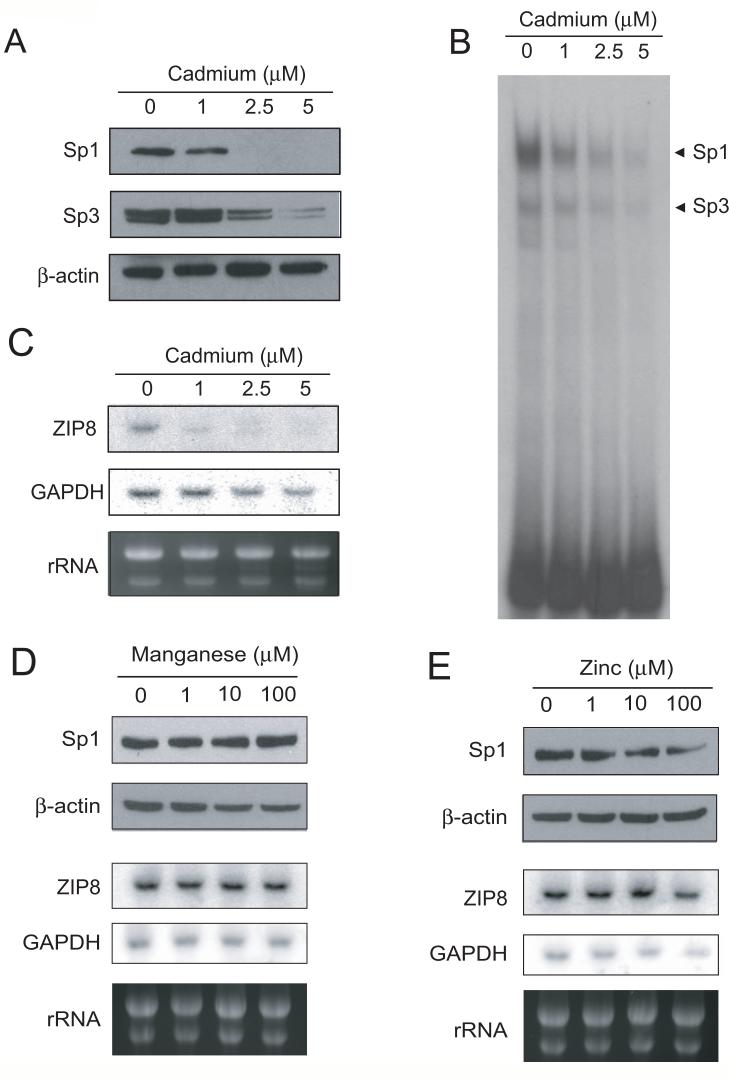
Down-regulation of Sp1 and ZIP8 by cadmium exposure
Sp1 contains three ZF at the C-terminus. Each ZF is composed of two cysteine and two histidine residues that are coordinated by zinc in a tetrahedral conformation. Cadmium, like other heavy metal ions can destabilize the folding of ZF by displacement of Zn, formation of mixed complexes, or incomplete coordination of metals (Hartwig, 2001) resulting in loss of DNA binding activity. These observations suggest that expression of ZIP8 may be downregulated by cadmium exposure through inhibition of Sp1. In order to test this hypothesis, we treated SR3A cells with various concentrations of cadmium and expression levels of Sp1 and Sp3 were determined by the western blotting. Fig. 9A shows that the expression levels of Sp1 were reduced in the cells treated with cadmium. Levels of Sp3 were also reduced, but to a lesser extent than that of Sp1. These results were also confirmed by EMSA. Sp1 DNA binding was also more sensitive to cadmium than was Sp3 binding as analyzed by EMSA (Fig. 9B). We found that expression levels of ZIP8 mRNA were also reduced in the cadmium-treated cells at 1 μM of cadmium (Fig. 9C). Importantly, Sp1 DNA binding ability reflects a decrease in ZIP8 level better than Sp1 protein level. These results suggest inactivation of Sp1 DNA binding by cadmium have role in ZIP8 downregulation. Importantly, because the IC50 of cadmium in SR3A cell line is approximately 14 μM, these results indicated that cadmium can downregulate ZIP8 expression at non-lethal concentrations.
Fig. 9. Effect of metal ions on ZIP8 expression.
SR3A cells were treated with various concentrations of manganese, zinc and cadmium for 20 hours. Cells were then harvested, and RNA and protein were extracted. Sp1 protein levels were analyzed by western blotting (A) and Sp1 DNA binding were analyzed by EMSA (B). The effect of cadmium exposure on ZIP8 expression was analyzed by Northern Blotting (C). The effect of manganese (D) and zinc (E) on Sp1 and ZIP8 levels were also examined by western blotting and northern blotting, respectively.
It has been shown that ZIP8 have high affinity for manganese, in addition to support transport activity of zinc, suggesting that these metal ions are also physiological substrates for ZIP8. However, treatment with 1 to 100 μM of MnCl2 (Fig. 9D) or ZnCl2 (Fig. 9E) for 20 hours did not result in significant changes in Sp1 protein and mRNA. These results suggest that the expression of ZIP8 is not regulated by these metal ions.
Discussion
The roles of GSH in regulating cadmium cytotoxicity have been studied in various model organisms and results showed that treatment with GSH derivative or its precursor NAC can suppress cadmium toxicity (Kaplan et al., 2008). Previous studies have suggested that elevated GSH enhanced cadmium efflux mediated by multidrug resistance protein (MRP) family from in yeast (Adle et al., 2007) and plant (Kim et al., 2007). In mammalian celsl, cadmium exposure upregulate both GCLC and MRP/GS-X-pump expression (Ishikawa et al., 1996). The present communication demonstrated that elevated GSH levels can downregulate cadmium uptake through downregulation of the cadmium transporter ZIP8. These results, collectively, indicate that GSH plays dual roles in regulating the cellular cadmium contents that are responsible for the acquired cadmium resistance.
Recent studies suggest that the calcium channel is involved in cadmium uptake (Leslie et al., 2006). However, siRNA based ZIP8 knockdown significantly lowered cadmium uptake rate in SR3A cell. In fact, in SCLC and SR3A cell lines, ZIP8 was expressed more than 10 times higher than that of CacnG1α, one of calcium channels proposed to be involved in cadmium transport (data not shown). This difference may reflect their different tissue distribution and because ZIP8 express highly in lung (Begum et al., 2002), ZIP8 may have potent role in olfactory cadmium intoxication.
ZIP8 was first identified as lipopolysaccharide-induced gene in monocytes and suggested regulation by NF-κB pathway (Begum et al., 2002). In fact, our promoter sequence analysis identified three putative NFκB binding sequences. However, our promoter analysis did not show any effect of deletion of these putative transcription factor binding sites. It is important to note that some studies demonstrated that Sp1 is involved in many lipopolysaccharide-induced gene expression (Hirasawa et al., 2006; Lee et al., 2005; Oh et al., 2008).
In this study, we have demonstrated that elevation of intercellular GSH levels either by overexpression of GCLC or NAC treatment downregulate Sp1 level. Although the precise mechanisms by which elevated GSH levels downregulate Sp1 expression are not known; however, several possibilities can be offered. Some studies reported an association of Sp1 with intercellular GSH or ROS levels. Oxidative stress activates MAPK signaling pathway and upregulate Sp1 protein as well as its target gene expression (Dasari et al., 2006; Schafer et al., 2003). In addition, activation of MAPK is prevented by elevated GSH level and in turn inhibit expression of Sp1 target genes (Vayalil et al., 2007). Sp1 contains three ZF domains which are important for DNA binding. Each ZF is composed of two cysteine and two histidine residues that are coordinated by zinc in a tetrahedral conformation. The structural fold of ZF, particularly the cysteine residues, is very sensitive to redox status and its stability is affected by intracellular thiol/disulfate pools such as GSH. Indeed, it has been reported that the transcription activity of Sp1 is regulated by intracellular redox conditions in either positive or negative manners, perhaps depending upon the contexts of different promoters or cell type (Dasari et al., 2006; Schafer et al., 2003; Webster et al., 2001). In fact, our results in ChIP assay showed that Sp1 DNA binding was more robustly affected in GCLC-transfected cell liens than that of Sp1 expression level. These reports complement our finding that elevated GSH level suppress ZIP8 expression through downregulation of Sp1.
The discovery that Sp1 is a transcription regulator for metal transporter ZIP8 supports the important roles of ZF-containing transcription factors in the regulation of metal ion homeostasis (Rutherford and Bird, 2004). In Sacchromyces cerevisiae, regulation of high-affinity copper uptake systems encoded by Ctr1 and Ctr3 is controlled by the ZF-containing transcription factor, Mac1. Likewise, expression of three Zn uptake systems encoded by the ZRT1, ZRT2, and FET4 genes is regulated by ZF-containing transcription factor Zap1. Because Cu and Zn are essential micronutrients but otherwise toxic at high concentrations, cells employ Mac1 and Zap1 to upregulate the expression of their respective transporters to increase transports of these metal ions under Cu- and Zn-limits conditions. Whereas cadmium is toxic, cells utilize downregulation of Sp1 to reduce expression of ZIP8 to achieve resistance to cadmium toxicity.
In addition to cadmium, ZIP8 is also known to transport zinc and manganese. Zinc was first reported to be a substrate of ZIP8 and its homeostasis is tightly regulated by its absorption and excretion (Cousins et al., 2006). One of the important pathways for maintaining zinc homeostasis is upregulation of the transcription factor MTF-1. MTF-1 itself can sense intercellular zinc concentration and upregulate genes such as metallothionein (Laity and Andrews, 2007). However, we could not find the MTF1 binding sequence on the proximal region of ZIP8 promoter, and zinc exposure did not change ZIP8 expression level. Based on its high affinity for manganese, ZIP8 is proposed as a Mn2+/HCO3- symporter (He et al., 2006). Like zinc ion, manganese is an essential metal in human. Cells have a sophisticated mechanism to maintain its homeostasis and disturbance of homeostasis is known to be related to neurotoxicity (Roth, 2006). However, in our study, high concentrations of manganese had no effect on ZIP8 expression. These results demonstrated that transcriptional regulation of ZIP8 does not seem to be involved in manganese or zinc homeostasis and suggest that other transporters may be the major players in this regard.
In summary, our results demonstrate that both GSH and cadmium exposure can suppress ZIP8 expression through the downregulation of Sp1, thus revealing a new pathway of GSH-mediated enhancement of cadmium resistance. These results bear important implications that it is possible to use antioxidants in combating cadmium toxicity.
Supplementary Material
Acknowledgments
This study was supported in part by the grants CA79085 and CA72404 from the National Cancer Institute.
Abbreviations
- GCLC
Glutamate-cystein ligase catalytic subunit
- GSH
Gluthathione
- NAC
N-acetyl cystein
- BSO
L-buthionine-[S,R]-sulfoximine
- MTT
3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- SRB assay
sulfurhodamine B colorimetric assay
- Cd
cadmium
- Mn
Manganese
- Zn
Zinc
- ChIP
chromatin immunoprecipitation
- BSA
bovine serum albumin
- DTT
dithiothreitol
- ZF
zinc finger
- SCLC
Small cell lung cancer
References
- Adle DJ, Sinani D, Kim H, Lee J. A cadmium-transporting P1B-type ATPase in yeast Saccharomyces cerevisiae. J Biol Chem. 2007;282(2):947–955. doi: 10.1074/jbc.M609535200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begum NA, Kobayashi M, Moriwaki Y, Matsumoto M, Toyoshima K, Seya T. Mycobacterium bovis BCG cell wall and lipopolysaccharide induce a novel gene, BIGM103, encoding a 7-TM protein: identification of a new protein family having Zn-transporter and Zn-metalloprotease signatures. Genomics. 2002;80(6):630–645. doi: 10.1006/geno.2002.7000. [DOI] [PubMed] [Google Scholar]
- Borst P, Evers R, Kool M, Wijnholds J. A family of drug transporters: the multidrug resistance-associated proteins. J Natl Cancer Inst. 2000;92(16):1295–1302. doi: 10.1093/jnci/92.16.1295. [DOI] [PubMed] [Google Scholar]
- Bridges CC, Zalups RK. Molecular and ionic mimicry and the transport of toxic metals. Toxicol Appl Pharmacol. 2005;204(3):274–308. doi: 10.1016/j.taap.2004.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cousins RJ, Liuzzi JP, Lichten LA. Mammalian zinc transport, trafficking, and signals. The Journal of biological chemistry. 2006;281(34):24085–24089. doi: 10.1074/jbc.R600011200. [DOI] [PubMed] [Google Scholar]
- Dalton TP, He L, Wang B, Miller ML, Jin L, Stringer KF, Chang X, Baxter CS, Nebert DW. Identification of mouse SLC39A8 as the transporter responsible for cadmium-induced toxicity in the testis. Proc Natl Acad Sci USA. 2005;102(9):3401–3406. doi: 10.1073/pnas.0406085102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasari A, Bartholomew JN, Volonte D, Galbiati F. Oxidative stress induces premature senescence by stimulating caveolin-1 gene transcription through p38 mitogen-activated protein kinase/Sp1-mediated activation of two GC-rich promoter elements. Cancer Res. 2006;66(22):10805–10814. doi: 10.1158/0008-5472.CAN-06-1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwig A. Zinc finger proteins as potential targets for toxic metal ions: differential effects on structure and function. Antioxid Redox Signal. 2001;3(4):625–634. doi: 10.1089/15230860152542970. [DOI] [PubMed] [Google Scholar]
- He L, Girijashanker K, Dalton TP, Reed J, Li H, Soleimani M, Nebert DW. ZIP8, member of the solute-carrier-39 (SLC39) metal-transporter family: characterization of transporter properties. Mol Pharmacol. 2006;70(1):171–180. doi: 10.1124/mol.106.024521. [DOI] [PubMed] [Google Scholar]
- Hirasawa N, Torigoe M, Kano K, Ohuchi K. Involvement of Sp1 in lipopolysaccharide-induced expression of HDC mRNA in RAW 264 cells. Biochem Biophys Res Commun. 2006;349(2):833–837. doi: 10.1016/j.bbrc.2006.08.104. [DOI] [PubMed] [Google Scholar]
- Ishikawa T, Bao JJ, Yamane Y, Akimaru K, Frindrich K, Wright CD, Kuo MT. Coordinated induction of MRP/GS-X pump and gamma-glutamylcysteine synthetase by heavy metals in human leukemia cells. The Journal of biological chemistry. 1996;271(25):14981–14988. doi: 10.1074/jbc.271.25.14981. [DOI] [PubMed] [Google Scholar]
- Kaplan M, Atakan IH, Aydogdu N, Aktoz T, Ozpuyan F, Seren G, Tokuc B, Inci O. Influence of N-acetylcysteine on renal toxicity of cadmium in rats. Pediatr Nephrol. 2008;23(2):233–241. doi: 10.1007/s00467-007-0696-7. [DOI] [PubMed] [Google Scholar]
- Kim DY, Bovet L, Maeshima M, Martinoia E, Lee Y. The ABC transporter AtPDR8 is a cadmium extrusion pump conferring heavy metal resistance. Plant J. 2007;50(2):207–218. doi: 10.1111/j.1365-313X.2007.03044.x. [DOI] [PubMed] [Google Scholar]
- Klaassen CD, Liu J, Choudhuri S. Metallothionein: an intracellular protein to protect against cadmium toxicity. Annu Rev Pharmacol Toxicol. 1999;39:267–294. doi: 10.1146/annurev.pharmtox.39.1.267. [DOI] [PubMed] [Google Scholar]
- Laity JH, Andrews GK. Understanding the mechanisms of zinc-sensing by metal-response element binding transcription factor-1 (MTF-1) Arch Biochem Biophys. 2007;463(2):201–210. doi: 10.1016/j.abb.2007.03.019. [DOI] [PubMed] [Google Scholar]
- Lee J, Kosaras B, Aleyasin H, Han JA, Park DS, Ratan RR, Kowall NW, Ferrante RJ, Lee SW, Ryu H. Role of cyclooxygenase-2 induction by transcription factor Sp1 and Sp3 in neuronal oxidative and DNA damage response. Faseb J. 2006;20(13):2375–2377. doi: 10.1096/fj.06-5957fje. [DOI] [PubMed] [Google Scholar]
- Lee KW, Lee Y, Kwon HJ, Kim DS. Sp1-associated activation of macrophage inflammatory protein-2 promoter by CpG-oligodeoxynucleotide and lipopolysaccharide. Cell Mol Life Sci. 2005;62(2):188–198. doi: 10.1007/s00018-004-4399-y. [DOI] [PubMed] [Google Scholar]
- Leslie EM, Liu J, Klaassen CD, Waalkes MP. Acquired cadmium resistance in metallothionein-I/II(-/-) knockout cells: role of the T-type calcium channel Cacnalpha1G in cadmium uptake. Mol Pharmacol. 2006;69(2):629–639. doi: 10.1124/mol.105.014241. [DOI] [PubMed] [Google Scholar]
- Liu J, Corton C, Dix DJ, Liu Y, Waalkes MP, Klaassen CD. Genetic background but not metallothionein phenotype dictates sensitivity to cadmium-induced testicular injury in mice. Toxicol Appl Pharmacol. 2001a;176(1):1–9. doi: 10.1006/taap.2001.9262. [DOI] [PubMed] [Google Scholar]
- Liu Y, Liu J, Klaassen CD. Metallothionein-null and wild-type mice show similar cadmium absorption and tissue distribution following oral cadmium administration. Toxicol Appl Pharmacol. 2001b;175(3):253–259. doi: 10.1006/taap.2001.9244. [DOI] [PubMed] [Google Scholar]
- Martelli A, Rousselet E, Dycke C, Bouron A, Moulis JM. Cadmium toxicity in animal cells by interference with essential metals. Biochimie. 2006;88(11):1807–1814. doi: 10.1016/j.biochi.2006.05.013. [DOI] [PubMed] [Google Scholar]
- Martin P, Fareh M, Poggi MC, Boulukos KE, Pognonec P. Manganese is highly effective in protecting cells from cadmium intoxication. Biochem Biophys Res Commun. 2006;351(1):294–299. doi: 10.1016/j.bbrc.2006.10.035. [DOI] [PubMed] [Google Scholar]
- Oberdorster G. Pulmonary deposition, clearance and effects of inhaled soluble and insoluble cadmium compounds. IARC Sci Publ. 1992;(118):189–204. [PubMed] [Google Scholar]
- Oh YT, Lee JY, Yoon H, Lee EH, Baik HH, Kim SS, Ha J, Yoon KS, Choe W, Kang I. Lipopolysaccharide induces hypoxia-inducible factor-1 alpha mRNA expression and activation via NADPH oxidase and Sp1-dependent pathway in BV2 murine microglial cells. Neurosci Lett. 2008;431(2):155–160. doi: 10.1016/j.neulet.2007.11.033. [DOI] [PubMed] [Google Scholar]
- Park JD, Cherrington NJ, Klaassen CD. Intestinal absorption of cadmium is associated with divalent metal transporter 1 in rats. Toxicol Sci. 2002;68(2):288–294. doi: 10.1093/toxsci/68.2.288. [DOI] [PubMed] [Google Scholar]
- Roth JA. Homeostatic and toxic mechanisms regulating manganese uptake, retention, and elimination. Biol Res. 2006;39(1):45–57. doi: 10.4067/s0716-97602006000100006. [DOI] [PubMed] [Google Scholar]
- Rutherford JC, Bird AJ. Metal-responsive transcription factors that regulate iron, zinc, and copper homeostasis in eukaryotic cells. Eukaryot Cell. 2004;3(1):1–13. doi: 10.1128/EC.3.1.1-13.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savaraj N, Wei Y, Unate H, Liu PM, Wu CJ, Wangpaichitr M, Xia D, Xu HJ, Hu SX, Tien Kuo M. Redox regulation of matrix metalloproteinase gene family in small cell lung cancer cells. Free Radic Res. 2005;39(4):373–381. doi: 10.1080/10715760400029694. [DOI] [PubMed] [Google Scholar]
- Schafer G, Cramer T, Suske G, Kemmner W, Wiedenmann B, Hocker M. Oxidative stress regulates vascular endothelial growth factor-A gene transcription through Sp1- and Sp3-dependent activation of two proximal GC-rich promoter elements. The Journal of biological chemistry. 2003;278(10):8190–8198. doi: 10.1074/jbc.M211999200. [DOI] [PubMed] [Google Scholar]
- Tatebe S, Unate H, Sinicrope FA, Sakatani T, Sugamura K, Makino M, Ito H, Savaraj N, Kaibara N, Kuo MT. Expression of heavy subunit of gamma-glutamylcysteine synthetase (gamma-GCSh) in human colorectal carcinoma. Int J Cancer. 2002;97(1):21–27. doi: 10.1002/ijc.1574. [DOI] [PubMed] [Google Scholar]
- Vayalil PK, Iles KE, Choi J, Yi AK, Postlethwait EM, Liu RM. Glutathione suppresses TGF-beta-induced PAI-1 expression by inhibiting p38 and JNK MAPK and the binding of AP-1, SP-1, and Smad to the PAI-1 promoter. Am J Physiol Lung Cell Mol Physiol. 2007;293(5):L1281–1292. doi: 10.1152/ajplung.00128.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vichai V, Kirtikara K. Sulforhodamine B colorimetric assay for cytotoxicity screening. Nat Protoc. 2006;1(3):1112–1116. doi: 10.1038/nprot.2006.179. [DOI] [PubMed] [Google Scholar]
- Waalkes MP. Cadmium carcinogenesis. Mutat Res. 2003;533(12):107–120. doi: 10.1016/j.mrfmmm.2003.07.011. [DOI] [PubMed] [Google Scholar]
- Webster KA, Prentice H, Bishopric NH. Oxidation of zinc finger transcription factors: physiological consequences. Antioxid Redox Signal. 2001;3(4):535–548. doi: 10.1089/15230860152542916. [DOI] [PubMed] [Google Scholar]
- Yamane Y, Furuichi M, Song R, Van NT, Mulcahy RT, Ishikawa T, Kuo MT. Expression of multidrug resistance protein/GS-X pump and gamma-glutamylcysteine synthetase genes is regulated by oxidative stress. The Journal of biological chemistry. 1998;273(47):31075–31085. doi: 10.1074/jbc.273.47.31075. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.