Abstract
DNA damage is accepted as a consequence of thymidylate deprivation induced by chemotherapeutic inhibitors of thymidylate synthase (TS), but the types of damage and signaling responses remain incompletely understood. Thymidylate deprivation increases dUTP and uracil in DNA, which is removed by Base Excision Repair (BER). Because BER requires a synthesis step, strand break intermediates presumably accumulate. Thymidylate deprivation also induces cell cycle arrest during replication. Homologous recombination (HR) is a means of repairing persistent BER intermediates and collapsed replication forks. There are also intimate links between HR and S-phase checkpoint pathways. In this study, the goals were to determine the involvement of HR-associated proteins and DNA damage signaling responses to thymidylate deprivation. When RAD51, which is a central component of HR, was depleted by siRNA cells were sensitized to raltitrexed (RTX), which specifically inhibits TS. To our knowledge, this is the first demonstration in mammalian cells that depletion of RAD51 causes sensitivity to thymidylate deprivation. Activation of DNA damage signaling responses was examined following treatment with RTX. Phosphorylation of replication protein A (RPA2 subunit) and formation of damage-induced foci were strikingly evident following IC50 doses of RTX. Induction was much more striking following RTX treatment than with hydroxyurea, which is commonly used to inhibit replication. RTX treatment also induced foci of RAD51, γ-H2AX, phospho-Chk1, and phospho-NBS1, although the extent of co-localization with RPA2 foci varied. Collectively, the results suggest that HR and S-phase checkpoint signaling processes are invoked by thymidylate deprivation and influence cellular resistance to thymidylate deprivation.
Keywords: homologous recombination, RAD51, replication protein A, thymidylate deprivation, damage foci, antifolates
1. INTRODUCTION
Folate- and nucleotide-based inhibitors of thymidylate synthetase (TS) continue to be used as main line therapies for a number of cancers. TS inhibitors are presumed to exert their therapeutic effects through DNA damage resulting from thymidylate deprivation, yet the precise types of DNA damage and DNA damage signaling responses that occur following TS inhibition remain incompletely understood. TS inhibition and thymidylate deprivation can cause DNA damage via several inter-related mechanisms. TS inhibition causes an increase in dUTP, which can become incorporated into DNA [1]. The base excision repair (BER) pathway actively removes genomic uracil [2]. BER requires a DNA resynthesis step following excision, but elevated dUTP causes reintroduction of uracil into DNA to create a ‘futile cycling’ of BER [1]. Additionally for fluoropyrimidines, the metabolite FdUTP can be incorporated into DNA, which has received attention recently [3–5]. Also, an implicit but less explored source of DNA damage caused by TS inhibitors is stalled replication forks that eventually collapse as a result of prolonged thymidylate deprivation and nucleotide pool imbalance.
There are a number of reasons to suspect the involvement of homologous recombination (HR) following TS inhibition regardless of which damage predominates. We have previously shown that the antifolate raltitrexed (RTX, Tomudex™), which is specific for TS, induces sister chromatid exchanges, which are DNA crossover events mediated by HR [6]. HR is invoked as a means of resolving stalled replication forks [7, 8]. Unresolved BER strand break intermediates appear to be processed by HR [9]. Thymidylate deprivation in S. cerevisiae has been shown to induce recombination [10], and there is evidence in murine cells that thymidylate deprivation can induce events suggestive of recombination [11, 12]. Collectively, the observations suggest that HR is likely involved in the response to thymidylate deprivation in mammalian cells.
There are extensive links and crosstalk between HR and the ATM/ATR signaling pathways that respond to DNA damage and stalled replication forks [13]. Replication Protein A (RPA) is a heterotrimeric protein that is essential for DNA replication and DNA repair processes. Its biochemical activity is to bind to and presumably protect single strand DNA generated during replication. ATM and ATR phosphorylate the 32 kDa subunit (RPA2) of RPA at multiple sites in response to DNA damage and replication stress [14]. Evidence suggests that ATR is activated in response to all types of replication stress, whereas the ATM response is specific for double strand breaks [13]. The downstream cascade includes the Chk1 and Chk2 signaling kinases among targets that number in the hundreds [15]. The MRN complex includes MRE11, RAD50, and NBS1, a complex that appears to act both upstream and downstream of ATR signaling via interactions with RPA [16, 17]. Recruitment of the MRN complex then stimulates RAD51 loading onto DNA facilitated by RPA, RAD52, and BRCA2 to initiate homology searching [8]. Chk1 has also been shown to be required for HR [18].
In this study, the activation of early DNA damage responses was examined, including phosphorylation of RPA and formation of damage foci in response to TS inhibition in HT-29 colon adenocarcinoma cells, which have been used in studies of TS inhibitors, and in HeLa cells, which have been extensively used in studies of damage foci formation. We also depleted by siRNA the RAD51 protein, which is a central component of HR. We used RTX, which is specific for TS [19]. The goals were to elucidate the DNA damage signaling responses to thymidylate deprivation and determine the involvement of HR. The results demonstrate that an ~IC50 dose of RTX induces a potent S-phase signaling response involving HR-associated proteins, which suggests that these processes likely contribute to cellular resistance to thymidylate deprivation.
2. Materials and methods
2.1. Chemicals and antibodies
Hydroxyurea (HU), Bovine albumin (BSA), thiazolyl blue tetrazolium bromide (MTT), β-glycerophosphate, β-mercaptoethanol (BME), phenylmethanesulfonyl fluoride (PMSF), sodium fluoride, sodium bicarbonate, dimethyl sulfoxide (DMSO), Giemsa Stain, and sodium othovanadate were purchased from Sigma (St. Louis, MO). Raltitrexed (RTX) was generously provided by AstraZeneca (U.K.). Anti-phospho-Histone-H2AX (Serine 139) was purchased from Upstate Biotech (Temecula, CA). Anti-RPA32 (RPA2) monoclonal antibody was purchased from Kamiya Biomedical Company (Seattle, WA). Anti-phospho-RPA2 (Seine 4/Serine 8) polyclonal antibody was purchased from Bethyl Biotech Inc. (Montgomery, TX) and anti-Rad51 polyclonal antibody was obtained from Santa Cruz Biotech (Santa Cruz, CA). Anti-phospho-pS317-Chk1 and anti-phospho-p95-NBS1 antibodies were purchased from Cell Signaling Technology (Danvers, MA).
2.2. Cell culture, drug treatment, and Western analysis
The HT-29 human colon cancer cells and HeLa human cervical cancer cells were obtained from American Type Culture Collection (Manassas, VA), and maintained at 37°C in a humidified atmosphere and 5% CO2 in DMEM (Invitrogen, Carlsbad CA) supplemented with 10% (v/v) FBS (Hyclone, Logan UT) and 1% penicillin/streptomycin (Invitrogen). A total of 0.6 × 106 cells were seeded into 60 mm dishes 24 h prior to drug treatments for 24 h. Untreated and treated cells were then harvested, whole cell extracts prepared, and western blots performed as described previously [6, 20, 21]. Equal protein loading was confirmed by β-actin (Abcam, Cambridge, MA).
2.3. Cytotoxicity
Viability and clonogenic assays were performed to determine toxicity. Viability was measured by MTT as described previously [20]. In brief, 5000 cells were plated on 96-well plates 24 h prior to treatment. Cells were treated with various concentrations of RTX for 24 h, and then grown in drug- free medium for 3 days. MTT was added 5 h prior to pelleting the cells, removing media and resuspending the dye in DMSO. Absorbance was measured using a plate reader at 595 nm (Bio-Tek UV808 Microplate Reader, Winooski, VT). Cell viability and IC50 values were calculated as a percentage compared to untreated controls. Colony-forming assays were performed as previously described [20]. Briefly, 1000 cells were seeded in 100 mm dishes 24 h prior to the addition of drug. Following 10 to 14 days, the cells were washed, fixed with methanol, and stained with Giemsa (Sigma, St. Louis, MO). Colonies were counted using an ACOLYTE colony counter (Symbiosis, Hingham MA).
2.4. RAD51 knockdown
The pKD-rad51-v1 and non-expressing backbone control vectors were purchased from Upstate Biotech (Temecula, CA). The pKD-rad51-V1 vector expresses a short hairpin small RNA that when intracellularly processed produces RAD51 siRNA. The vectors were mixed at a 1:2 ratio with FuGene®6 (Roche, Indianapolis IN) and transfected into HT-29 and HeLa cells by drop-wise addition according to the manufacturer’s protocol. Cells were harvested after 24, 48, 72, and 96 h for western analysis to determine the extent of knockdown. In the colony-forming assay, 72 h after transfection cells were treated for 24 h with the indicated concentration of RTX and then plated.
2.5. Indirect Immunofluorescence
Prior to treatment (24 h), 3 × 105 log phase HT-29 or HeLa cells were seeded in 12-well plates containing coverslips pretreated with collagen and UV. Cells were treated in media containing 30 nM RTX solution or an equal volume of phosphate-buffered saline (PBS) for the untreated control for 24 h. After treatment, the coverslips were washed with cold Hanks’ balanced buffer and fixed with 4% paraformadehyde at room temperature for 10 min. After washing, cells were permeablized with 0.3% triton X-100 in PBS for 15 min. Coverslips were washed and blocked with 2.5% milk in PBS at 4°C overnight. After washing, coverslips were incubated with primary antibody at 4°C overnight. Primary antibodies were used at the following ratios: anti-RAD51, 1:500; anti-RPA2, 1:500; anti-phospho-Chk1, 1:1000; anti-γH2AX 1:1000; anti-phospho-NBS1 1:500. Secondary antibody incubation with Alexa Fluor 568 anti-mouse IgG and Alexa Fluor 488 anti-rabbit IgG at ratio (1: 250) was for 1 h at room temperature (Invitrogen, Carlsbad, CA). After 10 washes with cold PBS, the coverslips were mounted by using ProlLong® gold antifade with DAPI (Invitrogen), dried completely, sealed and stored in the dark. Image data were captured with a ZEISS Axiovert 200 M fluorescence microscope with AxioVision Rel 4.5 software, and a 63X objective under oil. Adobe® Photoshop software CS was used to analyze images and NIH Image J software (http://rsb.info.nih.gov/ij/) was used to quantify data by counting foci number in at least 50 cells per sample, with a foci intensity of at least 30 intensity units. Cells were scored as positive for damage-induced foci if there were ≥10 spots per cell for HT-29 and ≥5 for HeLa. The threshold levels to score foci as positive for co-localization were a foci size of at least four pixels and 200 intensity units. Co-localization was scored as a percent of the number of spots containing at least 25% yellow compared to the total number of foci. The averaged data are from three independent experiments.
2.6. Flow cytometry for DNA content
Flow cytometry experiments were performed in similar fashion to those described previously [20, 21]. Briefly, exponentially growing cells (1.5 × 106) were seeded 24 h prior to treatment.
Cells were treated with RTX or HU at the doses listed in the text for 24 h. Cells were harvested, washed twice with cold PBS, fixed with cold 70% ethanol, and washed with 1% bovine serum albumin in phosphate buffered saline. After staining overnight (50 μg/ml propidium iodide, 0.1 mg/mL of RNase A and 1% BSA in PBS), DNA content was analyzed by using a Cytomics FC-500 Flow Cytometer with CXP software version 2.2 (Beckmen Coulter, Fullerton, CA.).
3. Results
3.1. Relationship between RTX treatment and RAD51
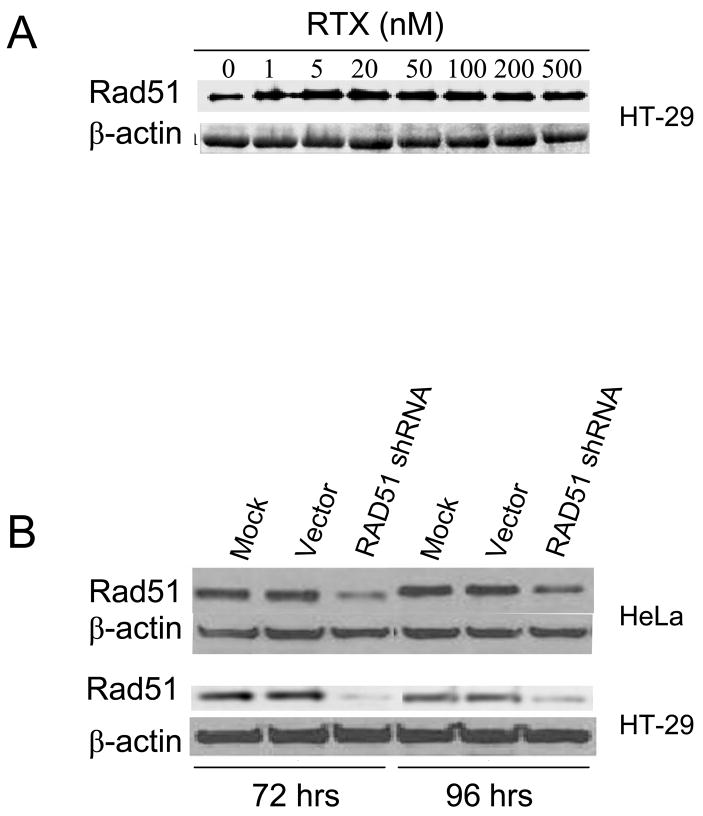
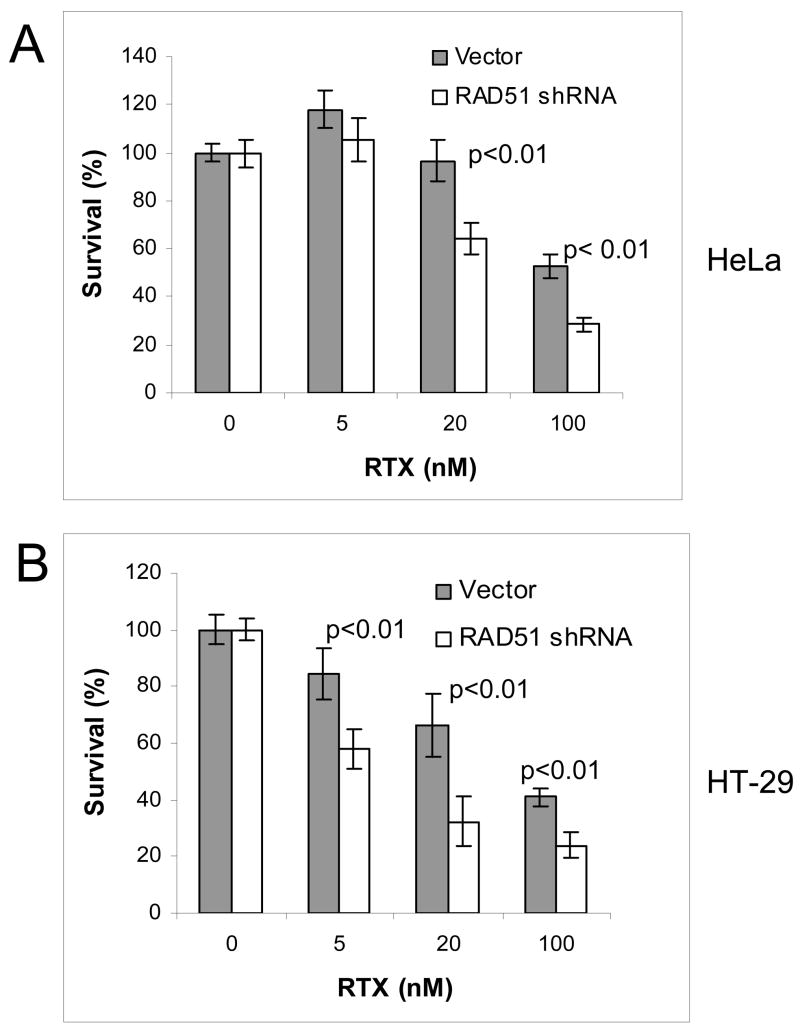
RAD51 plays a central role in strand exchange during HR. RAD51 protein in HT-29 cells was modestly induced by RTX treatment (20–60%, n=3), although the induction did not increase with higher doses (Figure 1A). In order to investigate the role of RAD51 more directly, we transiently depleted RAD51 protein by siRNA. Mice in which the Rad51 gene has been disrupted are early embryonic lethal and knockout cells fail to proliferate [22–24]. Transient antisense RNA expression was reported to reduce RAD51 levels [25], so we performed transient siRNA knockdown of RAD51. Figure 1B shows a reduction of RAD51 protein at 72 and 96 h following transfection of the RAD51 shRNA vector, while there was no change in RAD51 levels following transfection of the negative control vector. By densitometry, the shRNA vector reduced RAD51 protein levels by approximately 85% and 76% in HeLa cells and 84% and 72% in HT-29 cells at 72 and 96 h post transfection, respectively (Figure 1B). Cell viability following transfection showed that the FuGene transfection reagent alone reduced cell viability by 20% compared to mock treated (data not shown). No difference in viability was seen between cells treated with FuGene alone or FuGene plus RAD51shRNA or negative control vector, indicating that the RAD51shRNA vector did not reduce viability under these conditions. By viability assay, the IC50 for RTX in the absence of FuGene was 42 nM and 30 nM in HeLa and HT-29 cells, respectively (data not shown), which provided the dose range to utilize for colony-forming assays following transient knockdown. Colony-forming assays were performed to compare cell survival following RTX treatment in cells transfected with RAD51shRNA vector versus the negative control vector, which were performed instead of viability assays due to the duration of the experiment. Figure 2 shows that HeLa (2A) and HT-29 (2B) cells expressing the RAD51shRNA were 20–40% more sensitive to RTX than cells harboring the negative control vector (p<0.01).
Figure 1. (A) RAD51 expression following RTX treatment in HT-29 cells.
Cells were treated with increasing RTX for 24 h and analyzed for RAD51 protein. (B) RAD51 depletion by a vector expressing a small short hairpin RNA against RAD51 (RAD51 shRNA). RAD51 protein was measured by western blot. Lanes labeled “Mock” represent cells treated with FuGene transfection reagent alone and “vector” represent cells treated with control vector. By densitometry, the shRNA vector reduced RAD51 protein by approximately 85% and 76% in HeLa cells (upper panel) and 78% and 50% in HT-29 cells (lower panel) at 72 hrs and 96 hrs post transfection, respectively.
Figure 2. RAD51 depletion sensitizes HeLa (A) and HT-29 cells (B) to RTX treatment.
Cell survival was measured by colony-forming assay. Shaded bars represent cells transfected with empty control vector 72 h prior to RTX treatment. Open bars represent cells transfected with RAD51 shRNA vector 72 h prior to RTX treatment. Each bar is an average of three independent experiments (error bars are the standard deviation).
3.2. RPA is phosphorylated in response to RTX
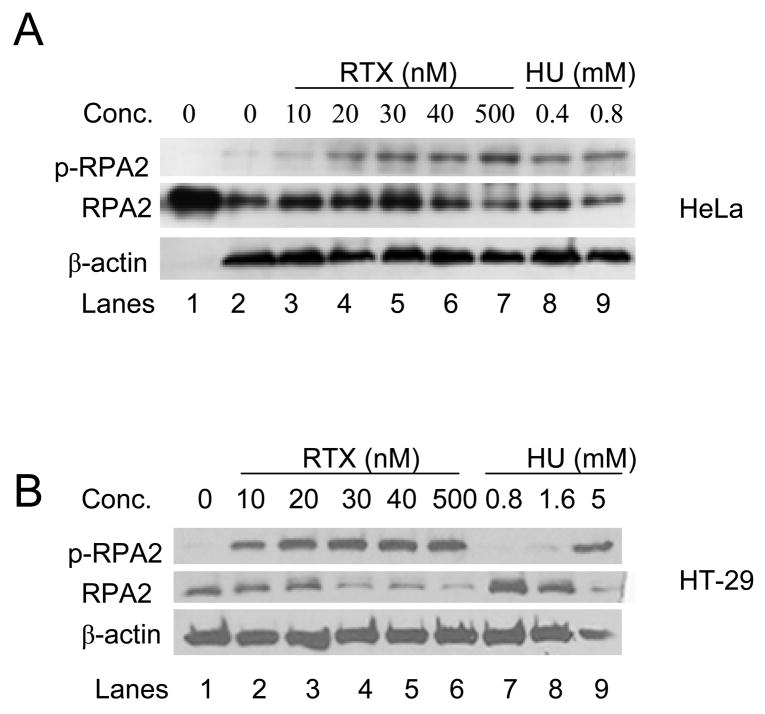
RPA is a heterotrimeric protein of which the RPA2 (32 kDa) subunit is phosphorylated at multiple sites in response to replication stress. Figure 3 shows that RTX treatment induced phosphorylation of RPA2 in HeLa (3A) and HT-29 (3B) cells from a subtoxic dose to an IC50 dose, as measured by the appearance of a band detected by an anti-phospho-RPA2 antibody. A supratoxic dose of RTX (500 nM) did not significantly increase phosphorylated RPA beyond that seen for the IC50 dose in each cell line. RPA from untreated HeLa and HT-29 cells migrated the same as human RPA protein purified from E. coli (Figure 3A and data not shown, respectively). Hydroxyurea (HU) is commonly used to induce S-phase arrest because it blocks ribonucleotide reductase, thus preventing dNTP synthesis. In agreement with a number of previous studies, HU induced RPA2 phosphorylation in HeLa cells at doses of 400 μM and 800 μM (Figure 3A, last two lanes on right). For reference, the approximate IC50 for HU in HeLa and HT-29 cells is 200 μM (data not shown). Interestingly, HU did not induce RPA2 phosphorylation in HT-29 cells until supratoxic doses of 1.6 and 5 mM were used (Figure 3B, last two lanes on right), suggesting that there is a distinct difference in S-phase checkpoint signaling induced by RTX versus HU in the HT-29 cells.
Figure 3. RPA2 is phosphorylated in response to TS inhibition.
HeLa (A) and HT-29 (B) cells were treated with increasing RTX or HU for 24 h, then analyzed for phosphorylated RPA2 (p-RPA2) with an anti-phospho-ser4/ser8 RPA2 antibody. RPA2 was detected with an anti-RPA antibody. Human RPA2 purified from E. coli served as a control (left-most lane in the upper panel). β-actin served as the loading control.
3.3. Co-localization of proteins involved in DNA damage signaling
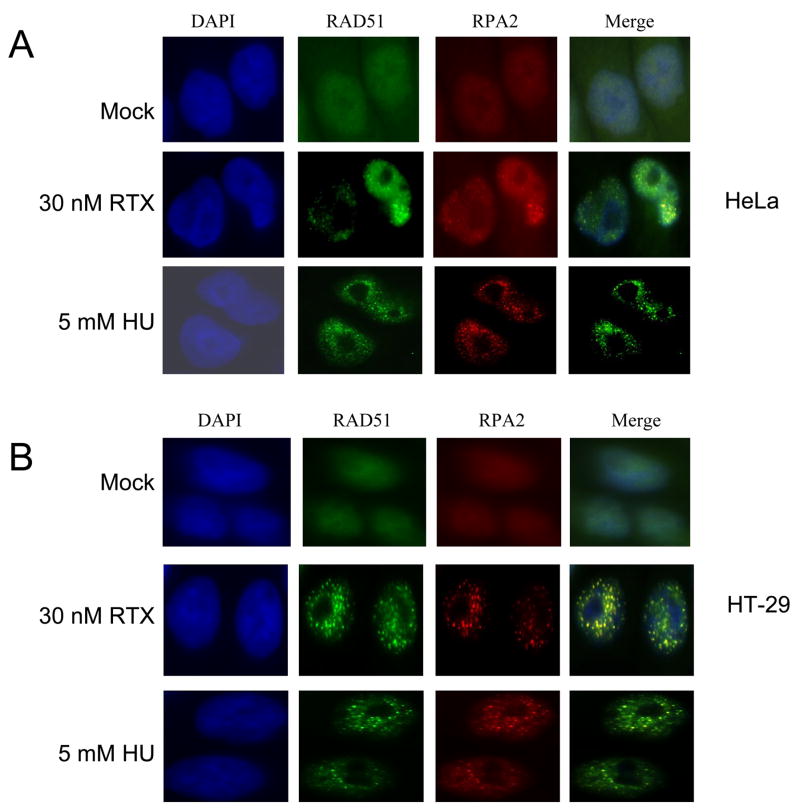
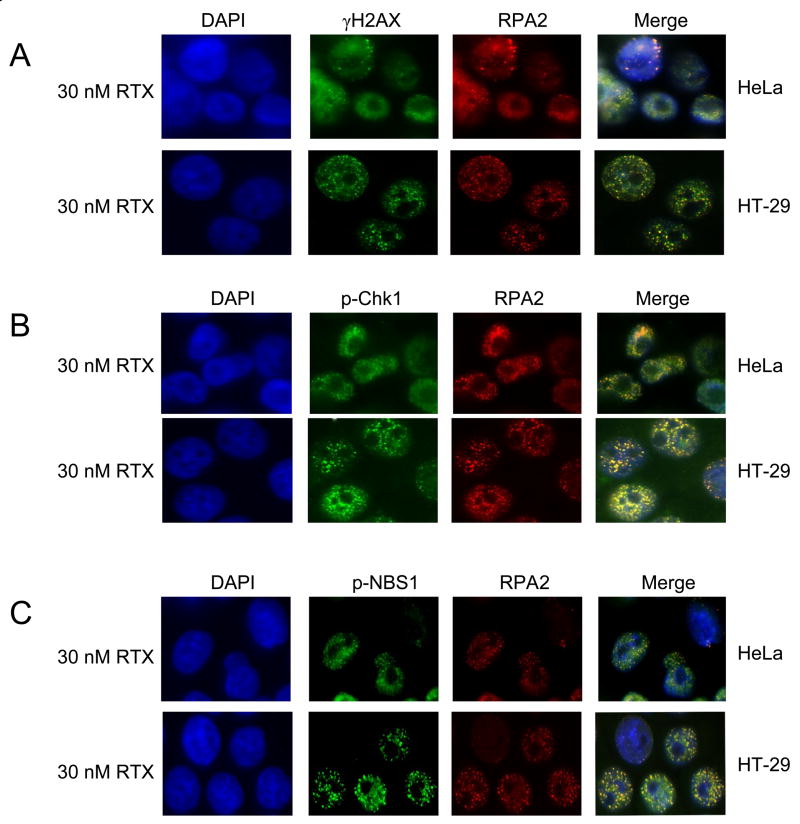
Focal points of localization for multiple proteins detected by immunofluorescence are well known phenomena following DNA damage. RPA focalization was examined in comparison to RAD51 and other key DNA damage signaling proteins. At a dose of 30 nM RTX, RPA foci were readily apparent in HeLa cells (Figure 4A). Quantification is shown in Figure 6A, in which >95% of the cells were scored as positive with ≥5 foci per cell (Materials and Methods). Foci were even more striking in HT-29 cells treated with 30 nM RTX (Figure 4B). Even at a mildly toxic dose of 5 nM RTX, RPA foci were seen in ~80% of HT-29 cells (data not shown). Figure 4 also shows that RAD51 foci were induced by RTX treatment in both cell types, implicating the initiation of HR. Figure 6B shows that the extent of co-localization for RPA and RAD51 was 56% for HeLa and 75% for HT-29 following RTX treatment. RPA and RAD51 foci were induced in both cell types treated with 5 mM HU, which is within a dose range commonly reported in studies of HU-induced damage foci and induced RPA phosphorylation (Figure 3), yet was well beyond the IC50 of ~200 μM for each cell line. We also observed phospho-RPA foci in response to RTX in the HeLa and HT-29 cells (Supplementary Figure 1). Phospho-RPA foci induced by HU were evident in the HeLa but not HT-29 cells, in agreement with the results shown in Figure 3B.
Figure 4. Focus formation and co-localization of RAD51 and RPA2 following RTX or HU treatment in HeLa (A) and HT-29 (B) cells.
Cells were mock-treated (top panels), treated for 24 h with 30 nM RTX (middle panels) or 5 mM HU (bottom panels) and examined by immunofluorescence (Materials and Methods). The columns are representative DAPI-stained (blue), RAD51 (green), RPA2 (red), and merged images of cells.
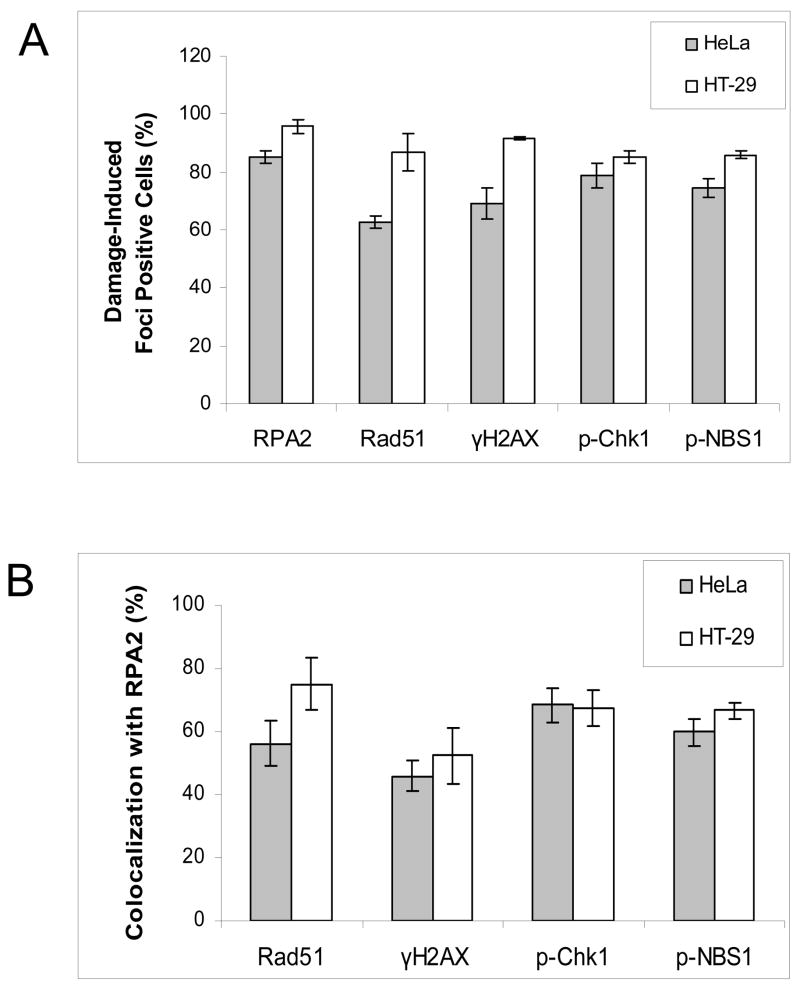
Figure 6. Quantification of damage-induced foci formation and co-localization with RPA2 induced by 30 nM RTX treatment in HeLa (shaded bars) and HT-29 (open bars) cells.
A. Cells were scored as positive for damage-induced foci as a percentage of total cells (Materials and Methods). B. Co-localization with RPA2 foci was scored as the percentage of foci containing at least 25% yellow (Materials and Methods). Each bar is an average of three independent experiments (error bars are the standard deviation).
Phosphorylation of the H2AX histone protein is a presumptive marker of DNA double strand break induction, replicational stress, and is seen following HU treatment [26]. We previously observed by western blot that RTX treatment induced γ-H2AX [21]. RTX-induced γ-H2AX foci were extensively induced, similar to that seen for RPA and RAD51 (Figure 5A). Interestingly, γ-H2AX foci overlap with RPA foci to a lesser extent than RAD51, by ~ 50% (Figure 6B). A lack of co-localization between RPA and γ-H2AX has been noted [27]. Chk1 is known to be a downstream target phosphorylated by ATM/ATR and an important regulator of S-phase progression. Distinct p-Chk1 foci were induced following RTX treatment in HeLa and HT-29 cells (Figure 5B). As part of the MRN complex, NBS1 is known to be a key target of phosphorylation during S-phase checkpoint signaling. Foci containing p-NBS1 were easily detectable following RTX treatment in HT-29 and HeLa cells (Figure 5C). The extent of co-localization for p-Chk1 and p-NBS1 with RPA foci were similar (Figure 5B). Collectively, the results demonstrate that RTX-induced thymidylate deprivation induces a robust focalization of a number of DNA damage signaling proteins at doses at or below the IC50 for each cell line.
Figure 5. Focus formation and co-localization of γ-H2AX (A), phosphorylated-Chk1 (B), phosphorylated-NBS1 (C), and RPA2 following RTX treatment.
HeLa (top panels) and HT-29 cells (bottom panels) were treated with 30 nM RTX for 24 h and examined by immunofluorescence (Materials and Methods). The columns are representative DAPI-stained (blue), RPA2 (red), and merged images of cells. Green-fluorescent images are γ-H2AX (A), p-Chk1 (B), p-NBS1 (C).
3.4. Cell cycle response to RTX and HU
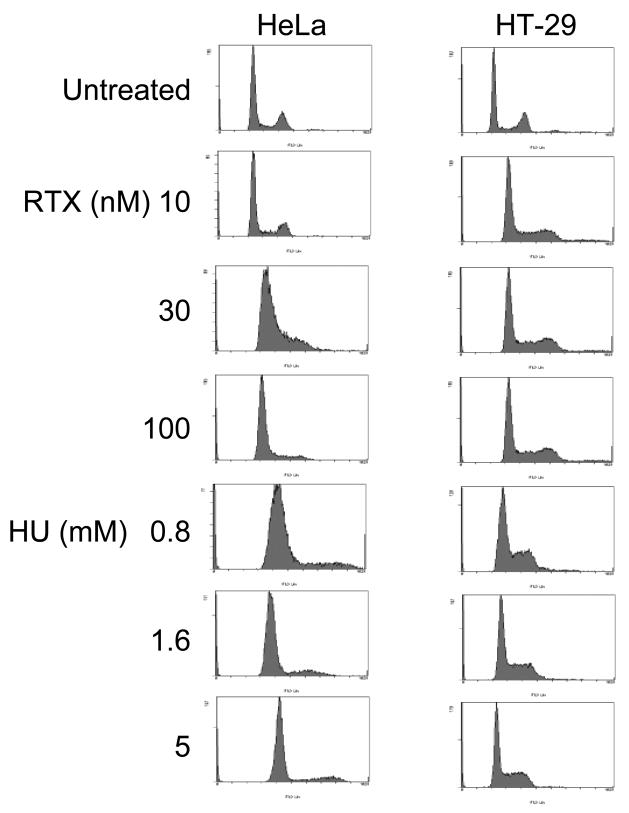
We have previously shown that RTX induces a potent S-phase arrest in MEFs and HEK293 cells [20, 21]. Cell cycle responses following RTX or HU exposure for 24 h were measured to confirm that the damage-induced foci were occurring during S-phase (Figure 7). From top to bottom, the first four left-hand panels in Figure 7 show that RTX induced an S-phase arrest in HT-29 cells, 60–71% of cells in S-phase compared to 13% for the untreated control. Even a dose of 10 nM (3-fold below the IC50 of 30 nM) induced a potent arrest. Sub G1 (apoptotic) cells do not yet appear at 24 h treatment times, in agreement with our previous studies on RTX in other cell types indicating that apoptosis begins 12–24 h after a 24 h treatment [6, 20, 21]. Treatment of HT-29 cells with increasing doses of HU produced an S-phase arrest similar to that seen for RTX, although the distribution of cells in late S-phase and G2/M appeared to decrease. The right hand top four panels show that RTX induced an S-phase arrest in HeLa cells at 30 and 100 nM, with 77 and 86 % of the cells in S-phase compared to 12% for the untreated control (IC50 of 42 nM in HeLa). The response of HeLa cells to HU was similar in that 79–86% of the cells appeared to be arrested in S-phase. Collectively, the results confirm that the RTX- and HU-induced RPA phosphorylation and foci formation of S-phase checkpoint signaling proteins occurred during S-phase arrest at the doses used.
Figure 7. Cell cycle arrest in response to RTX and HU.
HeLa (left-hand panels) and HT-29 cells (right-hand panels) untreated and treated with RTX or HU for 24 h at the doses listed and examined by flow cytometry (Materials and Methods). The top row is untreated cells. The second, third, and fourth rows are cells treated with increasing concentrations of RTX. The fifth through seventh rows are cells treated with HU.
4. Discussion
To our knowledge, this study is the first to directly demonstrate that depletion of RAD51 in mammalian cells leads to increased sensitivity to chemotherapy-induced thymidylate deprivation. Other studies have noted increases in RAD51 expression in tumor cells and associations with radioresistance, recently reviewed in [28]. Immunofluorescence confirmed that RAD51 foci accumulate as a result of RTX treatment, suggesting that thymidylate deprivation induces HR events. DNA double strand breaks (DSBs) have been reported following treatment with antifolate TS inhibitors including RTX [29, 30], although it has not been directly demonstrated whether BER futile cycling and/or stalled replication forks were the source of DSBs. We previously noted by western blot that RTX treatment induced γ-H2AX in HEK293 cells independent of the activity of the UNG DNA glycosylase [21]. Immunofluorescence in the HeLa and HT-29 cells confirmed that γ-H2AX focalization occurs following RTX treatment. Hyperphosphorylated RPA and γ-H2AX foci formation have been observed following treatment with agents that alkylate DNA in the minor groove or induce DSBs, although co-localization of the two was not extensive [27]. Phosphorylation of H2AX occurs in response to replication stress induced by HU or aphidicolin [31]. However, γ-H2AX foci following HU treatment are typically the result of supratoxic concentrations (≥2 mM), and it is not definitive whether stalled replication forks cause γ-H2AX, or forks must collapse into a DSB to induce γ-H2AX. In contrast, RTX strikingly induces γ-H2AX foci at or below IC50 doses. The results suggest that thymidylate deprivation induces a more potent DNA damage signaling response as opposed to deoxynucleotide deprivation induced by HU.
Molecular markers of S-phase signaling events were examined because there appear to be intimate links between HR and S-checkpoint signaling proteins. The phosphorylation of RPA2 is thought to be one of the earliest responses to a replication fork that has stalled after encountering damage [32]. The data show that RPA2 is phosphorylated and forms damage-induced foci in response to RTX. ATR is thought to be the kinase responsible for RPA2 phsophorylation in response to replication stress, although Chk1 is also capable [33]. Deficiency in Chk1 (or inhibition with UCN-01) leads to increased initiation of replication, ATR activation, and strand breaks [34]. Chk1 appears to regulate HR via interactions with and phosphorylation of RAD51 [18]. It has also been shown that TS inhibitors induce Chk1 phosphorylation [35], and Chk1 deficiency sensitizes cells to 5-FU [36, 37]. The data in Figure 5 demonstrates that p-Chk1 forms foci following RTX treatment. Focalization of p-NBS1 was examined because of reported interactions between RPA and the MRN complex, of which NBS1 is part [16, 17]. The data supports the notion that phosphorylation and focalization of NBS1 occurs as part of a broader S-phase checkpoint in response to multiple types of DNA damage [38]. To our knowledge, the data provide the first demonstration that thymidylate deprivation induces foci of RAD51, RPA2, γ-H2AX, p-Chk1, and p-NBS1. The striking induction of these foci at or below IC50 doses of RTX in two different cell lines suggests that processes associated with HR initiation respond to the DNA damage caused by thymidylate deprivation and thus are relevant to cell death caused by chemotherapeutic TS inhibitors. Data from flow cytometry confirmed that RTX- and HU-induced S-phase arrest in HeLa and HT-29 cells at the doses used to induce RPA2 phosphorylation and co-localization of S-phase checkpoint signaling proteins.
Analyses in E. coli and S. cerevisiae have suggested that thymidylate deprivation invokes HR mechanisms, which are protective [10, 39–43]. In mammalian systems, evidence for the involvement of HR is largely lacking [44], aside from induction of sister chromatid exchanges by the antifolates methotrexate[45] and RTX [6], and suggestive evidence in a murine system utilizing a heterologous human TS gene [12]. In a separate but related study, Waldman and coworkers directly investigated recombination in human cells treated with RTX using a model system to study intrachromosomal homologous recombination and provide what we believe is the first definitive evidence that TS inhibition can stimulate accurate recombination events in human cells [46]. We believe that the evidence presented here and in the related study strongly implicate HR mechanisms as part of the cellular response to thymidylate deprivation, which is perhaps not surprising given that HR is a means of resolving collapsed replication forks and persistent BER strand break intermediates. A number of studies examining BER proficient and deficient mammalian cells have not provided consistent evidence that BER futile cycling occurs during thymidylate deprivation [3, 6, 20, 21, 47–49]. Considering the myriad cell types including cancer cells and murine embryonic fibroblasts used in the above mentioned studies, the results presented here suggest that defects in DNA damage signaling checkpoints and HR status may be an underlying modulator of the contribution of BER. For example, among commonly used colorectal cancer cell lines, HCT 116 and LoVo cells are defective for MRE11 and thus do not form MRE11/RAD50/NBS1 foci formation in response to ionizing radiation, while HT-29 cells can form MRE11/RAD50/NBS1 foci [50]. There is evidence in C. elegans that the CLK-2 kinase plays a role in response to BER intermediates generated by uracil DNA glycosylase [51]. Future studies elucidating the links among BER, DNA damage signaling circuitry and HR will help determine the precise contribution of these processes to toxicity induced by TS inhibitors.
Supplementary Material
Acknowledgments
The authors gratefully thank Drs. Douglas Pittman and Sondra Berger, Department of Pharmaceutical and Biomedical Sciences, for helpful discussions. Dr. Deanna Smith, Department of Biological Sciences, and her associates are gratefully acknowledged for assistance with obtaining immunofluorescence images. AstraZeneca is acknowledged for a gift of RTX. This research was supported by a grant from the NIH (1 R01 CA100450).
Abbreviations
- HR
homologous recombination
- DSBs
DNA double strand breaks
- TS
thymidylate synthase
- RTX
raltitrexed (Tomudex™)
- BER
base excision repair
- RPA
replication protein A
- RPA2
32kDa subunit of replication protein A
- γ-H2AX
phsophorylated histone protein 2AX
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Aherne GW, Brown S. The role of uracil misincroporation in thymineless death. In: Jackman AL, editor. Anticancer Drug Development Guide: Antifolate Drugs in Cancer Therapy. Totowa, NJ: Humana Press Inc; 1999. pp. 409–21. [Google Scholar]
- 2.Krokan HE, Drablos F, Slupphaug G. Uracil in DNA--occurrence, consequences and repair. Oncogene. 2002;21:8935–48. doi: 10.1038/sj.onc.1205996. [DOI] [PubMed] [Google Scholar]
- 3.An Q, Robins P, Lindahl T, Barnes DE. 5-Fluorouracil incorporated into DNA is excised by the smug1 DNA glycosylase to reduce drug cytotoxicity. Cancer Res. 2007;67:940–5. doi: 10.1158/0008-5472.CAN-06-2960. [DOI] [PubMed] [Google Scholar]
- 4.Fischer F, Baerenfaller K, Jiricny J. 5-Fluorouracil is efficiently removed from DNA by the base excision and mismatch repair systems. Gastroenterology. 2007;133:1858–68. doi: 10.1053/j.gastro.2007.09.003. [DOI] [PubMed] [Google Scholar]
- 5.Meyers M, Wagner MW, Mazurek A, Schmutte C, Fishel R, Boothman DA. DNA mismatch repair-dependent response to fluoropyrimidine-generated damage. J Biol Chem. 2005;280:5516–26. doi: 10.1074/jbc.M412105200. [DOI] [PubMed] [Google Scholar]
- 6.Li L, Connor EE, Berger SH, Wyatt MD. Determination of apoptosis, uracil incorporation, DNA strand breaks, and sister chromatid exchanges under conditions of thymidylate deprivation in a model of BER deficiency. Biochem Pharmacol. 2005;70:1458–68. doi: 10.1016/j.bcp.2005.08.016. [DOI] [PubMed] [Google Scholar]
- 7.Nagaraju G, Scully R. Minding the gap: the underground functions of BRCA1 and BRCA2 at stalled replication forks. DNA Repair (Amst) 2007;6:1018–31. doi: 10.1016/j.dnarep.2007.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thorslund T, West SC. BRCA2: a universal recombinase regulator. Oncogene. 2007;26:7720–30. doi: 10.1038/sj.onc.1210870. [DOI] [PubMed] [Google Scholar]
- 9.Wyatt MD, Pittman DL. Methylating agents and DNA repair responses: methylated bases and sources of strand breaks. Chem Res Toxicol. 2006;19:1580–94. doi: 10.1021/tx060164e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kunz BA, Barclay BJ, Game JC, Little JG, Haynes RH. Induction of mitotic recombination in yeast by starvation for thymine nucleotides. Proc Natl Acad Sci U S A. 1980;77:6057–61. doi: 10.1073/pnas.77.10.6057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mishina Y, Ayusawa D, Seno T, Koyama H. Thymidylate stress induces homologous recombination activity in mammalian cells. Mutat Res. 1991;246:215–20. doi: 10.1016/0027-5107(91)90124-7. [DOI] [PubMed] [Google Scholar]
- 12.Ayusawa D, Koyama H, Shimizu K, Kaneda S, Takeishi K, Seno T. Induction, by thymidylate stress, of genetic recombination as evidenced by deletion of a transferred genetic marker in mouse FM3A cells. Mol Cell Biol. 1986;6:3463–9. doi: 10.1128/mcb.6.10.3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kastan MB, Bartek J. Cell-cycle checkpoints and cancer. Nature. 2004;432:316–23. doi: 10.1038/nature03097. [DOI] [PubMed] [Google Scholar]
- 14.Binz SK, Sheehan AM, Wold MS. Replication protein A phosphorylation and the cellular response to DNA damage. DNA Repair (Amst) 2004;3:1015–24. doi: 10.1016/j.dnarep.2004.03.028. [DOI] [PubMed] [Google Scholar]
- 15.Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER, 3rd, Hurov KE, Luo J, et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–6. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- 16.Olson E, Nievera CJ, Lee AY, Chen L, Wu X. The MRE11-RAD50-NBS1 complex acts both upstream and downstream of ataxia telangiectasia mutated and Rad3-related protein (ATR) to regulate the S-phase checkpoint following UV treatment. J Biol Chem. 2007;282:22939–52. doi: 10.1074/jbc.M702162200. [DOI] [PubMed] [Google Scholar]
- 17.Olson E, Nievera CJ, Liu E, Lee AY, Chen L, Wu X. The MRE11 complex mediates the S-phase checkpoint through an interaction with replication protein A. Mol Cell Biol. 2007;27:6053–67. doi: 10.1128/MCB.00532-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sorensen CS, Hansen LT, Dziegielewski J, Syljuasen RG, Lundin C, Bartek J, et al. The cell-cycle checkpoint kinase Chk1 is required for mammalian homologous recombination repair. Nat Cell Biol. 2005;7:195–201. doi: 10.1038/ncb1212. [DOI] [PubMed] [Google Scholar]
- 19.Takemura Y, Jackman AL. Folate-based thymidylate synthase inhibitors in cancer chemotherapy. Anticancer Drugs. 1997;8:3–16. doi: 10.1097/00001813-199701000-00001. [DOI] [PubMed] [Google Scholar]
- 20.Li L, Berger SH, Wyatt MD. Involvement of base excision repair in response to therapy targeted at thymidylate synthase. Mol Cancer Ther. 2004;3:747–53. [PubMed] [Google Scholar]
- 21.Luo Y, Walla M, Wyatt MD. Uracil incorporation into genomic DNA does not predict toxicity caused by chemotherapeutic inhibition of thymidylate synthase. DNA Repair (Amst) 2008;8:162–9. doi: 10.1016/j.dnarep.2007.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lim DS, Hasty P. A mutation in mouse rad51 results in an early embryonic lethal that is suppressed by a mutation in p53. Mol Cell Biol. 1996;16:7133–43. doi: 10.1128/mcb.16.12.7133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sonoda E, Sasaki MS, Buerstedde JM, Bezzubova O, Shinohara A, Ogawa H, et al. Rad51-deficient vertebrate cells accumulate chromosomal breaks prior to cell death. EMBO J. 1998;17:598–608. doi: 10.1093/emboj/17.2.598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tsuzuki T, Fujii Y, Sakumi K, Tominaga Y, Nakao K, Sekiguchi M, et al. Targeted disruption of the Rad51 gene leads to lethality in embryonic mice. Proc Natl Acad Sci U S A. 1996;93:6236–40. doi: 10.1073/pnas.93.13.6236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Slupianek A, Hoser G, Majsterek I, Bronisz A, Malecki M, Blasiak J, et al. Fusion tyrosine kinases induce drug resistance by stimulation of homology-dependent recombination repair, prolongation of G(2)/M phase, and protection from apoptosis. Mol Cell Biol. 2002;22:4189–201. doi: 10.1128/MCB.22.12.4189-4201.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ward IM, Chen J. Histone H2AX is phosphorylated in an ATR-dependent manner in response to replicational stress. J Biol Chem. 2001;276:47759–62. doi: 10.1074/jbc.C100569200. [DOI] [PubMed] [Google Scholar]
- 27.Liu JS, Kuo SR, Melendy T. DNA damage-induced RPA focalization is independent of gamma-H2AX and RPA hyper-phosphorylation. J Cell Biochem. 2006;99:1452–62. doi: 10.1002/jcb.21066. [DOI] [PubMed] [Google Scholar]
- 28.Klein HL. The consequences of Rad51 overexpression for normal and tumor cells. DNA Repair (Amst) 2008;7:686–93. doi: 10.1016/j.dnarep.2007.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matsui SI, Arredondo MA, Wrzosek C, Rustum YM. DNA damage and p53 induction do not cause ZD1694-induced cell cycle arrest in human colon carcinoma cells. Cancer Res. 1996;56:4715–23. [PubMed] [Google Scholar]
- 30.Parsels LA, Parsels JD, Wagner LM, Loney TL, Radany EH, Maybaum J. Mechanism and pharmacological specificity of dUTPase-mediated protection from DNA damage and cytotoxicity in human tumor cells. Cancer Chemother Pharmacol. 1998;42:357–62. doi: 10.1007/s002800050829. [DOI] [PubMed] [Google Scholar]
- 31.Liu JS, Kuo SR, Melendy T. Comparison of checkpoint responses triggered by DNA polymerase inhibition versus DNA damaging agents. Mutat Res. 2003;532:215–26. doi: 10.1016/j.mrfmmm.2003.08.018. [DOI] [PubMed] [Google Scholar]
- 32.Dery U, Masson JY. Twists and turns in the function of DNA damage signaling and repair proteins by post-translational modifications. DNA Repair (Amst) 2007;6:561–77. doi: 10.1016/j.dnarep.2006.12.009. [DOI] [PubMed] [Google Scholar]
- 33.Liu JS, Kuo SR, Melendy T. Phosphorylation of replication protein A by S-phase checkpoint kinases. DNA Repair (Amst) 2006;5:369–80. doi: 10.1016/j.dnarep.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 34.Syljuasen RG, Sorensen CS, Hansen LT, Fugger K, Lundin C, Johansson F, et al. Inhibition of human Chk1 causes increased initiation of DNA replication, phosphorylation of ATR targets, and DNA breakage. Mol Cell Biol. 2005;25:3553–62. doi: 10.1128/MCB.25.9.3553-3562.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Parsels LA, Parsels JD, Tai DC, Coughlin DJ, Maybaum J. 5-fluoro-2′-deoxyuridine-induced cdc25A accumulation correlates with premature mitotic entry and clonogenic death in human colon cancer cells. Cancer Res. 2004;64:6588–94. doi: 10.1158/0008-5472.CAN-03-3040. [DOI] [PubMed] [Google Scholar]
- 36.Robinson HM, Jones R, Walker M, Zachos G, Brown R, Cassidy J, et al. Chk1-dependent slowing of S-phase progression protects DT40 B-lymphoma cells against killing by the nucleoside analogue 5-fluorouracil. Oncogene. 2006;25:5359–69. doi: 10.1038/sj.onc.1209532. [DOI] [PubMed] [Google Scholar]
- 37.Xiao Z, Xue J, Sowin TJ, Zhang H. Differential roles of checkpoint kinase 1, checkpoint kinase 2, and mitogen-activated protein kinase-activated protein kinase 2 in mediating DNA damage-induced cell cycle arrest: implications for cancer therapy. Mol Cancer Ther. 2006;5:1935–43. doi: 10.1158/1535-7163.MCT-06-0077. [DOI] [PubMed] [Google Scholar]
- 38.Robison JG, Lu L, Dixon K, Bissler JJ. DNA lesion-specific co-localization of the MRE11/RAD50/NBS1 (MRN) complex and replication protein A (RPA) to repair foci. J Biol Chem. 2005;280:12927–34. doi: 10.1074/jbc.M414391200. [DOI] [PubMed] [Google Scholar]
- 39.Kunz BA, Haynes RH. DNA repair and the genetic effects of thymidylate stress in yeast. Mutat Res. 1982;93:353–75. [Google Scholar]
- 40.Kunz BA, Taylor GR, Haynes RH. Induction of intrachromosomal recombination in yeast by inhibition of thymidylate biosynthesis. Genetics. 1986;114:375–92. doi: 10.1093/genetics/114.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nakayama H, Nakayama K, Nakayama R, Irino N, Nakayama Y, Hanawalt PC. Isolation and Genetic-Characterization of a Thymineless Death-Resistant Mutant of Escherichia-Coli-K12 - Identification of a New Mutation (Recq1) That Blocks the Recf Recombination Pathway. Mol Gen Genet. 1984;195:474–80. doi: 10.1007/BF00341449. [DOI] [PubMed] [Google Scholar]
- 42.Nakayama K, Shiota S, Nakayama H. Thymineless Death in Escherichia-Coli Mutants Deficient in the Recf Recombination Pathway. Can J Microbiol. 1988;34:905–7. doi: 10.1139/m88-157. [DOI] [PubMed] [Google Scholar]
- 43.Nakayama K, Kusano K, Irino N, Nakayama H. Thymine Starvation-Induced Structural-Changes in Escherichia-Coli DNA - Detection by Pulsed-Field Gel-Electrophoresis and Evidence for Involvement of Homologous Recombination. J Mol Biol. 1994;243:611–20. doi: 10.1016/0022-2836(94)90036-1. [DOI] [PubMed] [Google Scholar]
- 44.Berger SH, Pittman DL, Wyatt MD. Uracil in DNA: consequences for carcinogenesis and chemotherapy. Biochem Pharmacol. 2008 doi: 10.1016/j.bcp.2008.05.019. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Banerjee A, Benedict WF. Production of sister chromatid exchanges by various cancer chemotherapeutic agents. Cancer Res. 1979;39:797–9. [PubMed] [Google Scholar]
- 46.Waldman BC, Wang Y, Kilaru K, Yang Z, Bhasin A, Wyatt MD, et al. Induction of Intrachromosomal Homologous Recombination in Human Cells by Raltitrexed, an Inhibitor of Thymidylate Synthase. DNA Repair (Amst) 2008 doi: 10.1016/j.dnarep.2008.06.006. In Press. DOI:j.dnarep.2008.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Andersen S, Heine T, Sneve R, König I, Krokan HE, Epe B, et al. Incorporation of dUMP into DNA is a major source of spontaneous DNA damage, while excision of uracil is not required for cytotoxicity of fluoropyrimidines in mouse embryonic fibroblasts. Carcinogenesis. 2005;26:547–55. doi: 10.1093/carcin/bgh347. [DOI] [PubMed] [Google Scholar]
- 48.Fischer JA, Muller-Weeks S, Caradonna SJ. Fluorodeoxyuridine Modulates Cellular Expression of the DNA Base Excision Repair Enzyme Uracil-DNA Glycosylase. Cancer Res. 2006;66:8829–37. doi: 10.1158/0008-5472.CAN-06-0540. [DOI] [PubMed] [Google Scholar]
- 49.Welsh SJ, Hobbs S, Aherne GW. Expression of uracil DNA glycosylase (UDG) does not affect cellular sensitivity to thymidylate synthase (TS) inhibition. Eur J Cancer. 2003;39:378–87. doi: 10.1016/s0959-8049(02)00610-x. [DOI] [PubMed] [Google Scholar]
- 50.Giannini G, Ristori E, Cerignoli F, Rinaldi C, Zani M, Viel A, et al. Human MRE11 is inactivated in mismatch repair-deficient cancers. EMBO Rep. 2002;3:248–54. doi: 10.1093/embo-reports/kvf044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dengg M, Garcia-Muse T, Gill SG, Ashcroft N, Boulton SJ, Nilsen H. Abrogation of the CLK-2 checkpoint leads to tolerance to base-excision repair intermediates. EMBO Rep. 2006;7:1046–51. doi: 10.1038/sj.embor.7400782. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.