Abstract
The IL-23 pathway plays a pivotal role in the development of chronic mucosal inflammation seen in the inflammatory bowel diseases. Multiple studies have now established the contribution of the interleukin 23 receptor gene (IL23R) to Crohn’s Disease (CD) risk in general and of the IL23R R381Q variant in particular. The aim of this work was to estimate the total contribution of this gene to CD risk test using a haplotype approach.
Methods
763 CD subjects and 254 controls were genotyped for single nucleotide polymorphisms in the IL23R gene using Illumina and ABI methods. Haplotypes were assigned using PHASEv2 and tested for association with CD by chi-square and permutation.
Results
Haplotypes with both increased and decreased risk for CD were observed in 2 of the 4 observed blocks (Block 2 H1: 55.4% control, 64% CD, p=0.019; H2: 64.5% control, 54.4% CD, p=0.006; Block 3 H1: 55.8% control, 64.4% CD, p=0.013; H2: 47.0% control, 36.6% CD, p=0.001). The population attributable risk for these haplotypes was substantially larger than that estimated for the IL23R R381Q variant (Block 2 H1 and block 3 H1 ~20%, compared with ~4% for Block 3 H6, containing the variant).
Discussion
These observations suggest that IL23R makes a substantial contribution to Crohn’s disease susceptibility, larger than that estimated from the population frequency of the R381Q variant. These observations also support the expectation that finding “hits” from genome wide association studies will be but an important chapter in the story of unraveling the genetic contribution to Crohn’s disease, rather than the final chapter that brings clarity to all the plot twists of a complicated story.
The IL-23 pathway plays a pivotal role in the development of chronic mucosal inflammation seen in the inflammatory bowel diseases (McGovern and Powrie 2007). Furthermore, the contribution of the interleukin 23 receptor gene (IL23R) to Crohn’s Disease (CD) risk has now been established by a genome wide association study (GWAS) (Duerr et al. 2006), confirmed in a pediatric cohort (Dubinsky et al. 2007), and confirmed the large multi-disease study from the UK (2007). The association of IL23R variants with CD has also been observed in populations ascertained from Scotland (Van Limbergen et al. 2007), New Zealand (Roberts et al. 2007), Continental Europe (Buning et al. 2003; Libioulle et al. 2007; Weersma et al. 2007), North America (Baldassano et al. 2007), Brazil (Baptista et al. 2008), and Israel (Leshinsky-Silver et al. 2007), but not Japan (Yamazaki et al. 2007). In general, most of the confirmation studies have focused on the “protective” effect of the single-nucleotide polymorphism (SNP), rs11209026, a coding SNP that alters Arginine 381 to Glutamine (R381Q). In the original report, the Glutamine allele was present in ~7% of controls and 2-3% of CD cases (Duerr et al. 2006).
When NOD2 (CARD15) was identified as the first gene for CD, the initial findings suggested that three mutations within the gene were associated with disease susceptibility (Hugot et al. 2001; Ogura et al. 2001). However, the initial observations also suggested that the CD association with these three SNPs did not account for all of the linkage signal with CD at the IBD1 locus (Hampe et al. 2002). A portion of the linkage signal, at least, could be explained by other mutations within NOD2 that were not identified in the early NOD2 association studies (Lesage et al. 2002). This initial experience with NOD2 suggests that, once a susceptibility gene has been identified for a complex genetic trait, additional association studies with other variants are necessary in order to evaluate the role of a particular gene in disease etiopathogenesis more fully.
One strategy to test for the role of other variation in a putative susceptibility gene is to examine the association of major haplotypes within the disease under study (Crawford and Nickerson 2005). The rationale for this approach is that, by testing the major haplotypes in a population, the genetic variation at a high frequency in a population may be tested prior to knowing all of the individual variants that may be present. A further advantage of the haplotype approach is that the identification of haplotypes associated with disease susceptibility identify those patients that should be given priority in a deep sequencing project aimed at uncovering variation.
The aim of this study was to estimate the full contribution of IL23R variation to CD by examining the association of IL23R haplotypes with CD in our Caucasian population from Los Angeles. We report that haplotypic variation in IL23R accounts for a substantially larger proportion of the population attributable risk for CD than would be observed by genotyping only the IL23R R381Q variant.
MATERIALS AND METHODS
Subjects
Recruitment of subjects at the Cedars-Sinai Medical Center Inflammatory Bowel Disease center was conducted under the approval of the Cedars-Sinai Medical Center Institutional Review Board. Disease phenotype was assigned using a combination of standard endoscopic, histological, and radiographic features (Mow et al. 2004).
Selection of SNPs
SNPs were selected using “Tagger” (de Bakker 2004) and data from the International HapMap Project (2003; Barrett et al. 2005; Frazer et al. 2007). SNPs that “tagged” major Caucasian haplotypes and that were compatible with Illumina technology were genotyped in the initial phases of this study. Since we were interested in major genetic effects rather than rare alleles, the goal of “tagging” was to find a set of tagSNPs in linkage disequilibrium with all SNPs in the HapMap data with a minor allele frequency ≥5%.
Genotyping
DNA was isolated from Epstein Barr virus transformed lymphoblastoid cell lines using proteinase K digestion, organic extraction, and ethanol precipitation (Sambrook et al. 1989). Single nucleotide markers (SNPs) were genotyped following the respective manufacturer’s protocols using either: (1) the oligonucleotide ligation assay, Illumina Golden Gate technology (Shen et al. 2005) (Illumina, San Diego, CA) or (2) the 5’-extension reaction, TaqMan MGB technology (Livak 1999; Kutyavin et al. 2000) (Applied Biosystems, Foster City, CA). Consistency of SNP genotyping between the two methods was checked by genotyping 100 samples with both methods. The dbSNP accession numbers or “rs numbers” for the SNPs tested in this study are listed in Table 1.
Table 1.
Association of IL23 Haplotypes with CD
| Gene | Percent Controls with Haplotype Present | Percent CD with Haplotype Present | p empirical* | Odds Ratio | 95% Confidence Interval | Estimate of Population Attributable Risk or of Prevented Fraction | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| IL23R | n=257 | n=763 | |||||||||||
| 1) Block 1 | rs12041056 | rs1884444 | rs6671221 | ||||||||||
| H1 | G | C | A | 42.9% | 44.5% | ||||||||
| H2 | A | A | G | 43.5% | 41.9% | ||||||||
| H3 | G | A | G | 13.2% | 13.2% | ||||||||
| 2) Individual SNP between Block 1 and Block 2 | rs1569922 | ||||||||||||
| risk | C | 85.3% | 93.0% | 0.0001 | 2.3 | 1.49,3.65 | + 0.50 | ||||||
| 3) Block 2 | rs1004819 | rs790631 | |||||||||||
| risk | H1 | T | A | 55.4% | 64.0% | 0.019 | 1.43 | 1.07,1.91 | + 0.19 | ||||
| protective | H2 | G | A | 64.5% | 54.4% | 0.006 | 0.65 | 0.49,0.88 | - 0.22 | ||||
| H3 | G | C | 45.4% | 49.7% | |||||||||
| 4) Block 3 | rs7530511 | rs7528924 | rs2201841 | rs10489628 | rs11209026 | rs1343151 | |||||||
| risk | H1 | C | A | C | C | G | C | 55.8% | 64.4% | 0.013 | 1.43 | 1.07,1.91 | + 0.19 |
| protective | H2 | C | A | T | T | G | T | 47.0% | 36.6% | 0.001 | 0.65 | 0.48,0.86 | - 0.16 |
| H3 | C | A | T | T | G | C | 20.3% | 23.0% | |||||
| H4 | C | G | T | C | G | C | 19.5% | 21.2% | |||||
| H5 | T | G | T | C | G | C | 19.5% | 19.8% | |||||
| H6 | C | A | T | C | A | T | 12.0% | 8.4% | -04 | ||||
| 5) Block 4 | rs11209032 | rs1495965 | |||||||||||
| H1 | G | A | 82.4% | 76.9% | |||||||||
| H2 | A | G | 57.7% | 63.2% | |||||||||
| H3 | G | G | 19.2% | 18.1% |
Statistical Analyses
Haplotype blocks were determined using Haploview v4 (Barrett et al. 2005) using the confidence interval method (Gabriel et al. 2002). Haplotypes of subjects were inferred from individual genotyping data using PHASE v2 (Stephens et al. 2001). The association of the presence of a haplotype was tested using logistic regression. The impact of multiple testing was assessed by permuting the phenotypes with respect to the genotypes. All p-values reported have been corrected by this permutation test. Since the haplotypes were inferred from individual SNP data, we corrected for uncertainty in the haplotype assignment by including the probability of haplotype assignment as a covariate in the analysis. This correction for uncertainty usually had no impact because most of the assignments had a greater than 90% probability of assignment. However, all p-values shown have also been corrected for the uncertainty in haplotype assignment (Cordell 2006). Population attributable risk was estimated by assuming: (1) the frequency of a particular haplotype in the controls reflected the population frequency of that haplotype, and (2) the odds ratio for the association of a given haplotype reflected the relative risk of that haplotype for Crohns disease (Sugimura et al. 2003). Haplotypes are numbered in the order of frequency in controls seen in both our own controls and in publicly available data.
RESULTS
Single nucleotide polymorphisms (SNPs) in IL23R were genotyped in a CD case-control cohort and used to infer haplotype block and haplotype structure in the Caucasian sample from Los Angeles using Haploview v4. The existence of the blocks and haplotypes studied here was also confirmed using trio data from the International HapMap Project (result not shown). Using the “confidence interval method” (Gabriel et al. 2002), four haplotype blocks were observed plus one region of linkage disequilibrium (LD) defined by an individual SNP, rs1569922, between blocks 1 and 2. The SNPs forming these blocks are given in Figure 1 and Table 1.
Figure 1. The Four Haplotype Blocks of IL23R.
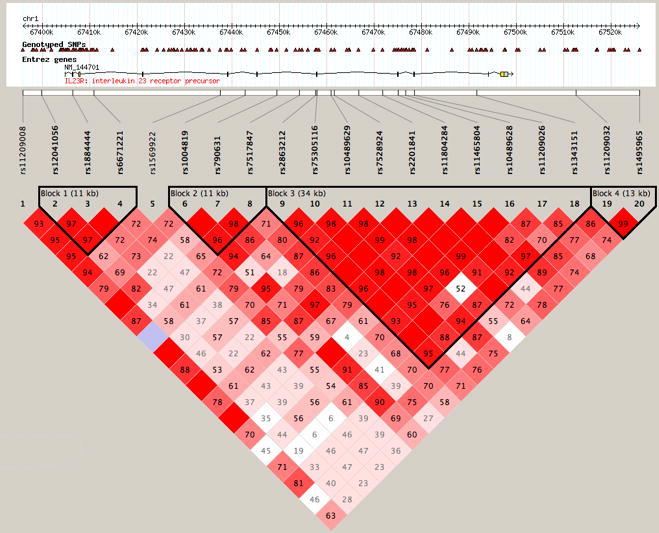
The position of the SNPs genotyped is shown in relationship to the chromosome 1 coordinates (NCBI build 36) and to the IL23R gene. Numbers indicate the extent of D’ between two SNPs. The dark lines show the block structure of IL23R as determined using the confidence interval method (Gabriel et al. 2002).
Haplotypes in cases and controls were assigned using PHASEv2 and tested for association with Crohn’s disease. No association was observed between CD and haplotypes in blocks 1 and 4; but CD was significantly associated with SNP rs1569922 and haplotypes in blocks 2 and 3 (Table 1). A “risk” haplotype, or a haplotype inferring increased CD risk, and a “protective” haplotype, or a haplotype inferring decreased CD risk, was observed in both blocks 2 and 3 (Block 2 H1: 55.4% control, 64% CD, p=0.019; H2: 64.5% control, 54.4% CD, p=0.006; Block 3 H1: 55.8% control, 64.4% CD, p=0.013; H2: 47.0% control, 36.6% CD, p=0.001). Furthermore, within each block, the odds ratio (OR) for CD significantly increased with the number of copies of the risk haplotype and significantly decreased with the number of copies of the protective haplotype (Figure 2a & 2b).
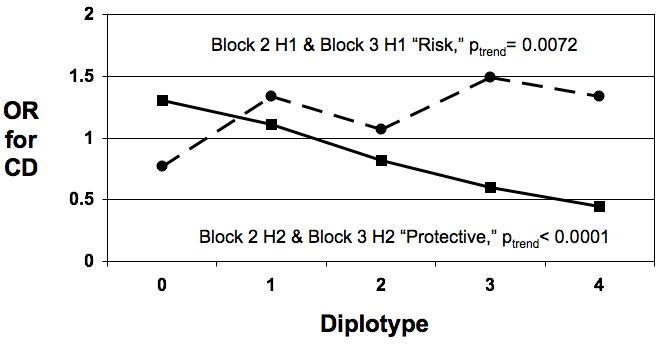
Figure 2. Odds Ratio for CD Increases With Increasing Number of Risk Haplotypes.
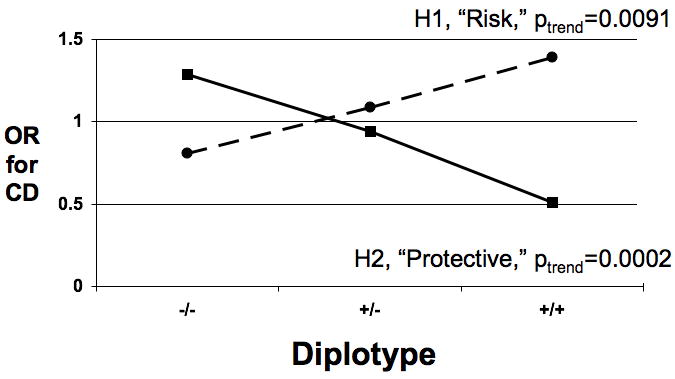
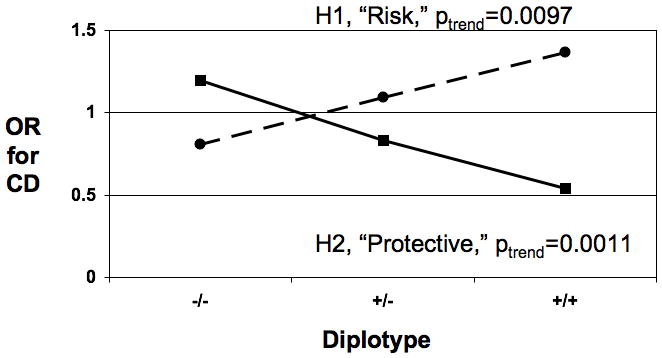
- The odds ratio (OR) for CD with increasing number of copies of risk (dotted line) or protective (solid line) haplotypes is shown in the left chart for block 2 and in the right chart for block 3.
- OR with increasing number of copies of risk (dotted line) or protective (solid line) haplotypes across blocks 2 and 3 combined.
As Figure 2c shows, when the risk haplotypes from blocks 2 and 3 were combined, the OR for CD significantly increased with the number of copies of risk haplotypes from OR=0.77 for 0 risk copies to OR=1.3 for 3 copies, p(trend)=0.0072). Concomitantly, when the protective haplotypes from blocks 2 and 3 were combined, the OR significantly decreased with the number of copies of protective haplotypes from OR=1.3 for 0 protective copies to OR=0.45 for 4 copies, p(trend) <0.0001).
The population attributable risk (PAR) for a genetic risk factor estimates the percentage of CD cases that would not exist if that risk factor were not present in a given population and the prevented fraction estimates the protection due to the presence of a protective factor. While this type of analysis is widely recognized to be a simplification of the contribution of any single genetic locus to disease susceptibility, it does provide a rough estimate of the relative contribution of the effect size that different loci have on disease susceptibility. The prevented fraction was approximately 4% for the protective IL23R R381Q variant (rs11209026, located in block 3, on H6). A substantially greater reduction in CD risk was observed for the protective haplotypes reported here, a PAR of 22% for block 2 H2 and 16% for block 3 H2 in this sample (Table 1). In addition, a large PAR was observed for the risk haplotypes reported here, approximately 10% for either block 2 H1 or block 3 H1. These values are visually compared in Figure 3.
Figure 3. The Effect of the Risk and Protective Haplotypes Is Greater Than The Effect of the IL23R R381Q SNP.
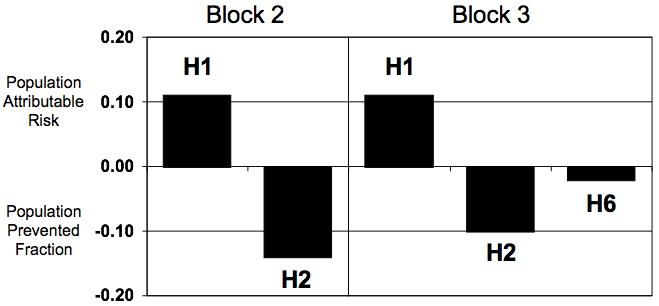
Population attributable risk (positive parts of the bar graph) or prevented fraction (negative parts of the bar graph) was estimated as outlined in Methods. H1 is the risk haplotype in each block and H2 is the protective haplotype in each block as identified in this report. H6 contains the R381Q variant.
DISCUSSION
The significant genetic associations and high population attributable risks reported here underscore the importance of IL23R genetic variation to CD susceptibility. While 10 SNPs were associated with CD in the original report of a genome wide association of ileal CD, the greatest association for an individual SNP was observed for rs11209026 (R381Q) (Duerr et al. 2006). For this SNP, the prevented fraction, or the amount of disease that would not be present if the genetic variant did not exist in the Caucasian population, is roughly 4%. In contrast, the IL23R risk and protective haplotypes identified here (Figure 1, Table 1) are at a higher frequency in our sample of the Caucasian CD population and thus substantially raise the population attributable risk and prevented fraction (Figure 3).
The limitations of this study include the fact that the haplotypes were inferred from data for individual SNPs and multiple comparisons were required for haplotype analysis. We have attempted to account for these problems by including the uncertainty in haplotype assignment as a covariate in the logistic regression analysis and by calculating empirical p-values using permutation of the phenotypes with respect to genotypes. Since the probability of correct haplotype assignment was greater than 90% for a large proportion of the subjects, this correction had only a slight effect on the observed significance of the associations reported here.
These data support the concept that the initial observation of an association between a genetic variant and a disease phenotype or quantitative trait should be followed by further genotyping and analysis of the genetic locus under study. Such follow-up should include performing haplotype association studies using haplotype tagging SNPs to “cover” the gene as well as directly sequencing the gene itself in order to identify SNPs that are potentially associated with the phenotype but are rare in the general population and so not included in analysis by “tagSNPs.” In this way, the genome wide association studies being performed currently represent the first step, rather than the goal, of the genetic dissection of a complex trait such as CD (Taylor et al. 2007).
While additive to increase CD risk, no interaction between CARD15/NOD2 mutations and IL23R risk haplotypes were observed (analysis not shown). This observation may suggest that CARD15/NOD2 and IL23R variants define two separate pathways to intestinal inflammation in humans. Extensive work with mouse models of intestinal inflammation, developed by “knocking out” many different immune-related genes (Sartor 2006), has demonstrated that there are multiple pathways to intestinal inflammation. If so, then IL23R variation may be useful to distinguish CD subtypes with different underlying pathophysiological mechanisms, and raises the possibility that therapies targeted at IL23 signalling may be more effective in the IL23-related CD subtype(s) rather than in CD as a whole.
Acknowledgments
Supported by NIDDK Grant PO1DK06763, the Feintech Chair in Immunobiology (SRT) and the Board of Governor’s Chair in Medical Genetics (JIR) at Cedars-Sinai Medical Center. Genotyping was also supported in part by M01-RR000425 to the Cedars-Sinai GCRC genotyping core and by DK062413 (KDT).
Abbreviations
- CD
Crohn’s disease
- GWAS
genome wide association study
- IBD
inflammatory bowel disease
- IL23
interleukin 23, alpha subunit p19 (dbGENE id 51561)
- IL23R
interleukin 23 receptor (dbGENE id 149233)
- OR
odds ratio
- PAR
population attributable risk
- SNP
single nucleotide polymorphism
Footnotes
Found at www.ibdjournal.com
References
- The International HapMap Project. Nature. 2003;426:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldassano RN, Bradfield JP, Monos DS, Kim CE, Glessner JT, Casalunovo T, Frackelton EC, Otieno FG, Kanterakis S, Shaner JL, Smith RM, Eckert AW, Robinson LJ, Onyiah CC, Abrams DJ, Chiavacci RM, Skraban R, Devoto M, Grant SF, Hakonarson H. Association of variants of the interleukin-23 receptor gene with susceptibility to pediatric Crohn’s disease. Clin Gastroenterol Hepatol. 2007;5:972–976. doi: 10.1016/j.cgh.2007.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baptista ML, Amarante H, Picheth G, Sdepanian VL, Peterson N, Babasukumar U, Lima HC, Kugathasan S. CARD15 and IL23R influences Crohn’s disease susceptibility but not disease phenotype in a Brazilian population. Inflamm Bowel Dis. 2008 doi: 10.1002/ibd.20372. [DOI] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Buning C, Genschel J, Weltrich R, Lochs H, Schmidt H. The interleukin-25 gene located in the inflammatory bowel disease (IBD) 4 region: no association with inflammatory bowel disease. Eur J Immunogenet. 2003;30:329–333. doi: 10.1046/j.1365-2370.2003.00411.x. [DOI] [PubMed] [Google Scholar]
- Cordell HJ. Estimation and testing of genotype and haplotype effects in case-control studies: comparison of weighted regression and multiple imputation procedures. Genet Epidemiol. 2006;30:259–275. doi: 10.1002/gepi.20142. [DOI] [PubMed] [Google Scholar]
- Crawford DC, Nickerson DA. Definition and clinical importance of haplotypes. Annu Rev Med. 2005;56:303–320. doi: 10.1146/annurev.med.56.082103.104540. [DOI] [PubMed] [Google Scholar]
- de Bakker P. Tagger. 2004 http://www.broad.mit.edu/mpg/tagger/
- Dubinsky MC, Wang D, Picornell Y, Wrobel I, Katzir L, Quiros A, Dutridge D, Wahbeh G, Silber G, Bahar R, Mengesha E, Targan SR, Taylor KD, Rotter JI. IL-23 receptor (IL-23R) gene protects against pediatric Crohn’s disease. Inflamm Bowel Dis. 2007;13:511–515. doi: 10.1002/ibd.20126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duerr RH, Taylor KD, Brant SR, Rioux JD, Silverberg MS, Daly MJ, Steinhart AH, Abraham C, Regueiro M, Griffiths A, Dassopoulos T, Bitton A, Yang H, Targan S, Datta LW, Kistner EO, Schumm LP, Lee AT, Gregersen PK, Barmada MM, Rotter JI, Nicolae DL, Cho JH. A genome-wide association study identifies IL23R as an inflammatory bowel disease gene. Science. 2006;314:1461–1463. doi: 10.1126/science.1135245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazer KA, Ballinger DG, Cox DR, Hinds DA, Stuve LL, Gibbs RA, Belmont JW, et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, Higgins J, DeFelice M, Lochner A, Faggart M, Liu-Cordero SN, Rotimi C, Adeyemo A, Cooper R, Ward R, Lander ES, Daly MJ, Altshuler D. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- Hampe J, Grebe J, Nikolaus S, Solberg C, Croucher PJ, Mascheretti S, Jahnsen J, Moum B, Klump B, Krawczak M, Mirza MM, Foelsch UR, Vatn M, Schreiber S. Association of NOD2 (CARD 15) genotype with clinical course of Crohn’s disease: a cohort study. Lancet. 2002;359:1661–1665. doi: 10.1016/S0140-6736(02)08590-2. [DOI] [PubMed] [Google Scholar]
- Hugot JP, Chamaillard M, Zouali H, Lesage S, Cezard JP, Belaiche J, Almer S, Tysk C, O’Morain CA, Gassull M, Binder V, Finkel Y, Cortot A, Modigliani R, Laurent-Puig P, Gower-Rousseau C, Macry J, Colombel JF, Sahbatou M, Thomas G. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn’s disease. Nature. 2001;411:599–603. doi: 10.1038/35079107. [DOI] [PubMed] [Google Scholar]
- Kutyavin IV, Afonina IA, Mills A, Gorn VV, Lukhtanov EA, Belousov ES, Singer MJ, Walburger DK, Lokhov SG, Gall AA, Dempcy R, Reed MW, Meyer RB, Hedgpeth J. 3’-minor groove binder-DNA probes increase sequence specificity at PCR extension temperatures. Nucleic Acids Res. 2000;28:655–661. doi: 10.1093/nar/28.2.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesage S, Zouali H, Cezard JP, Colombel JF, Belaiche J, Almer S, Tysk C, O’Morain C, Gassull M, Binder V, Finkel Y, Modigliani R, Gower-Rousseau C, Macry J, Merlin F, Chamaillard M, Jannot AS, Thomas G, Hugot JP. CARD15/NOD2 mutational analysis and genotype-phenotype correlation in 612 patients with inflammatory bowel disease. Am J Hum Genet. 2002;70:845–857. doi: 10.1086/339432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leshinsky-Silver E, Karban A, Dalal I, Eliakim R, Shirin H, Tzofi T, Boaz M, Levine A. Evaluation of the interleukin-23 receptor gene coding variant R381Q in pediatric and adult Crohn disease. J Pediatr Gastroenterol Nutr. 2007;45:405–408. doi: 10.1097/MPG.0b013e318141a1de. [DOI] [PubMed] [Google Scholar]
- Libioulle C, Louis E, Hansoul S, Sandor C, Farnir F, Franchimont D, Vermeire S, Dewit O, de Vos M, Dixon A, Demarche B, Gut I, Heath S, Foglio M, Liang L, Laukens D, Mni M, Zelenika D, Van Gossum A, Rutgeerts P, Belaiche J, Lathrop M, Georges M. Novel Crohn disease locus identified by genome-wide association maps to a gene desert on 5p13.1 and modulates expression of PTGER4. PLoS Genet. 2007;3:e58. doi: 10.1371/journal.pgen.0030058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ. Allelic discrimination using fluorogenic probes and the 5’ nuclease assay. Genet Anal. 1999;14:143–149. doi: 10.1016/s1050-3862(98)00019-9. [DOI] [PubMed] [Google Scholar]
- McGovern D, Powrie F. The IL23 axis plays a key role in the pathogenesis of IBD. Gut. 2007;56:1333–1336. doi: 10.1136/gut.2006.115402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mow WS, Vasiliauskas EA, Lin YC, Fleshner PR, Papadakis KA, Taylor KD, Landers CJ, Abreu-Martin MT, Rotter JI, Yang H, Targan SR. Association of antibody responses to microbial antigens and complications of small bowel Crohn’s disease. Gastroenterology. 2004;126:414–424. doi: 10.1053/j.gastro.2003.11.015. [DOI] [PubMed] [Google Scholar]
- Ogura Y, Bonen DK, Inohara N, Nicolae DL, Chen FF, Ramos R, Britton H, Moran T, Karaliuskas R, Duerr RH, Achkar JP, Brant SR, Bayless TM, Kirschner BS, Hanauer SB, Nunez G, Cho JH. A frameshift mutation in NOD2 associated with susceptibility to Crohn’s disease. Nature. 2001;411:603–606. doi: 10.1038/35079114. [DOI] [PubMed] [Google Scholar]
- Roberts RL, Gearry RB, Hollis-Moffatt JE, Miller AL, Reid J, Abkevich V, Timms KM, Gutin A, Lanchbury JS, Merriman TR, Barclay ML, Kennedy MA. IL23R R381Q and ATG16L1 T300A are strongly associated with Crohn’s disease in a study of New Zealand Caucasians with inflammatory bowel disease. Am J Gastroenterol. 2007;102:2754–2761. doi: 10.1111/j.1572-0241.2007.01525.x. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch E, Maniatis T. Molecular Cloning. Cold Spring Harbor Laboratory; New York: 1989. [Google Scholar]
- Sartor RB. Mechanisms of disease: pathogenesis of Crohn’s disease and ulcerative colitis. Nat Clin Pract Gastroenterol Hepatol. 2006;3:390–407. doi: 10.1038/ncpgasthep0528. [DOI] [PubMed] [Google Scholar]
- Shen R, Fan JB, Campbell D, Chang W, Chen J, Doucet D, Yeakley J, Bibikova M, Wickham Garcia E, McBride C, Steemers F, Garcia F, Kermani BG, Gunderson K, Oliphant A. High-throughput SNP genotyping on universal bead arrays. Mutat Res. 2005;573:70–82. doi: 10.1016/j.mrfmmm.2004.07.022. [DOI] [PubMed] [Google Scholar]
- Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet. 2001;68:978–989. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimura K, Taylor KD, Lin YC, Hang T, Wang D, Tang YM, Fischel-Ghodsian N, Targan SR, Rotter JI, Yang H. A novel NOD2/CARD15 haplotype conferring risk for Crohn disease in Ashkenazi Jews. Am J Hum Genet. 2003;72:509–518. doi: 10.1086/367848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor KD, Norris JM, Rotter JI. Genome-wide association: which do you want first: the good news, the bad news, or the good news? Diabetes. 2007;56:2844–2848. doi: 10.2337/db07-1324. [DOI] [PubMed] [Google Scholar]
- Van Limbergen J, Russell RK, Nimmo ER, Drummond HE, Smith L, Davies G, Anderson NH, Gillett PM, McGrogan P, Hassan K, Weaver L, Bisset WM, Mahdi G, Wilson DC, Satsangi J. IL23R Arg381Gln is associated with childhood onset inflammatory bowel disease in Scotland. Gut. 2007;56:1173–1174. doi: 10.1136/gut.2007.122069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weersma RK, Zhernakova A, Nolte IM, Lefebvre C, Rioux JD, Mulder F, van Dullemen HM, Kleibeuker JH, Wijmenga C, Dijkstra G. ATG16L1 and IL23R Are Associated With Inflammatory Bowel Diseases but Not With Celiac Disease in The Netherlands. Am J Gastroenterol. 2007 doi: 10.1111/j.1572-0241.2007.01660.x. [DOI] [PubMed] [Google Scholar]
- Yamazaki K, Onouchi Y, Takazoe M, Kubo M, Nakamura Y, Hata A. Association analysis of genetic variants in IL23R, ATG16L1 and 5p13.1 loci with Crohn’s disease in Japanese patients. J Hum Genet. 2007;52:575–583. doi: 10.1007/s10038-007-0156-z. [DOI] [PubMed] [Google Scholar]


