Abstract
Although allosteric regulation is the ‘second secret of life’, the molecular mechanisms that give rise to allostery currently elude understanding. In my opinion, experimental progress is hampered by a commonly used but misleading definition of allostery as protein structural changes that are elicited by the binding of a single ligand. Allostery is more strictly defined in functional terms as a comparison of how one ligand binds in the absence, versus the presence, of a second ligand. Therefore, as each of the two binding events involves two protein complexes, a study of allostery must consider four complexes and not just two. Such a comparison can distinguish allosteric from non-allosteric protein changes, the importance of which is frequently overlooked. When a study of all four complexes is not feasible, an alternative, albeit limited, strategy can identify subsets of allosteric-specific changes.
Restricting allostery to a functional definition
The post-genomic era has renewed focus on protein function and regulation. Allosteric regulation (see Glossary) is intrinsic to the control of most metabolic and signal-transduction pathways. As a result, allosteric regulation enables a defining principle of life, enabling living organisms to adapt to ever changing environmental conditions. Monod’s [1] recognition of this important biological role led to the historical description of allostery as ‘the second secret of life’, second only to the genetic code.
Classically, allosteric regulation, as applied to the study of enzymes1, has had three defining characteristics: (i) the effector is not chemically identical to the substrate, (ii) the effector elicits a change in a functional property of the protein (e.g. binding of a second ligand or altered catalytic properties), and (iii) the effector binds at a site that is topographically distinct from (i.e. does not overlapping [2]) the functional site of the protein (e.g. active site or orthosteric site). Allostery is most often associated with protein functions that respond to changes in concentrations of small molecules. However, the same principles apply when the function of a protein (or other macromolecule) is altered upon association with other proteins, DNA or membranes. In addition to currently known allosteric effectors, many unidentified allosteric proteins and/or effectors are predicted [3]. Given the central role that allostery has in biology, modulating allosteric responses holds promise for drug design. Indeed, the current pharmaceutical interest in developing allosteric drugs is being driven by the natural specificity and selectivity profiles and concentration-independent limits of allosteric regulation [4–7]. However, there is a paucity of information on precise molecular mechanisms by which proteins are allosterically regulated, a deficiency that prohibits the full potential of rational drug design.
The current challenge to understanding allosteric mechanisms is the correlation of allosteric function with relevant protein structural (conformational and/or dynamic) changes. Unfortunately, a growing number of phenomena are described as ‘allosteric’ (Box 1), which, in turn, confuses the necessary correlations between structure and function. In my (and others’) opinion, this confusion arises from the commonly used definition that allostery is any ligand-induced change in protein conformation and/or dynamics. This definition does not account for the functional characteristic of allostery (one ligand altering protein function involving a second ligand) that was introduced in the preceding paragraph. Furthermore, this misleading definition implies that mechanisms of allostery can be completely revealed by structural studies comparing only two protein complexes, one with an allosteric effector bound and one with no effector bound [8, 9]. Moreover, there is often no discrimination between whether the substrate is or is not bound in the latter complex. Structural differences between these two complexes probably do not identify all changes that are important to allosteric function, thereby providing an overly restrictive view of allostery. Moreover, assigning all structural differences between these two complexes as ‘allosteric’ does not distinguish between changes that are and are not part of an allosteric mechanism, thus yielding an under-restricted view of allostery. Given these problems, inconsistencies in the terminology used to describe allostery are also common. These discrepancies have prompted this overview of the fundamental concept of allostery and associated terminology (see the Glossary) with the intent of aiding future correlations between structure and function aimed at defining allosteric residues within a protein. Other common concepts that limit a molecular understanding of allostery are discussed at length in Supplementary Table 1.
Box 1: Comparing definitions of allostery
The current literature contains several inconsistent definitions of allostery (listed here). These definitions share features of energetic coupling and protein structural changes, but differ in other respects as detailed here:
1) Energetic coupling between two binding events [2, 11, 12]
This is the original definition and the basis of my discussion.
2) Energetic coupling between a protein structural change and a binding event [10, 15]
This definition is in common use, but, as presented in the main text, could misguide structural studies aiming to understand functional regulation. Ligand binding most often modifies protein structure. However, additional structural changes caused by binding of a second ligand are also expected to contribute to the allosteric mechanism (see Figure 1 and 2 in the main text). Therefore, structural changes elicited by binding of a single ligand cannot account for all changes necessary for functional regulation. It is this functional regulation that influences biology sufficiently to be designated ‘the second secret of life’. ‘Induced-fit’ and/or ‘conformational selection’, instead of allostery, describe single-ligand-induced structural changes, whether changes are local or long- range [27].
3) Energetic coupling between a covalent modification and a binding event [2, 28, 29]
A covalent modification cannot be considered as a ligand that undergoes binding; however, there are obvious parallels that can be drawn.
4) Energetic coupling between an amino acid side chain of the protein and a binding event [30]
Mutations of amino acid residues are not relevant to regulation in the ‘normal’ biological system. Furthermore, a mutation might influence any of the other scenarios listed here.
5) Non-additivity of mutant cycles (i.e. energetic coupling between two amino acid side chains) [31].
Although accurately applied to date [31], without additional constraints this definition includes energetic coupling events relevant to protein stability.
6) Mutual influence between two substrates binding in the same active site [32].
Because of potential direct interactions of two substrates, this scenario is excluded from allostery [2].
Given the considerations listed, I return to the first definition, which is also the functional basis for the original articulation of allostery [2]. As such, allostery occurs when one ligand binds to a protein differently in the absence, versus presence, of the second ligand. Thus, allostery is a subset of energy-coupled events in a protein, but not all energy-coupled events are allosteric. In addition, most of the scenarios listed here could involve long-range structural changes in the protein. Therefore, allostery can involve long-range structural changes, but not all long-range structural changes elicited by ligand binding are allosteric.
Identifying allostery: functional signature and structural correlation
As a reminder, allosteric regulation is classically defined by three characteristics: (i) the effector (X) is not chemically identical to the substrate (A), (ii) the effector elicits a change in a functional property of the protein (E) and (iii) the effector binds at a site on E that is topographically distinct from the active site. In enzymology, systems that demonstrate altered substrate affinity upon effector binding are referred to as ‘K-type’ systems, and those with altered catalysis (kcat or Vmax) are described as ‘V-type’ systems [10]. K-type systems are the most commonly studied and, consequently, are the focus of this overview.
Consider a functional view of allosteric regulation. In a K-type system, the affinity of a protein for one ligand (e.g. substrate; A) is altered when the protein has a second ligand bound (e.g. effector; X). Binding of A to an E in the absence of an X requires two protein species:
It follows that in the saturating presence of X, two additional protein complexes must be considered:
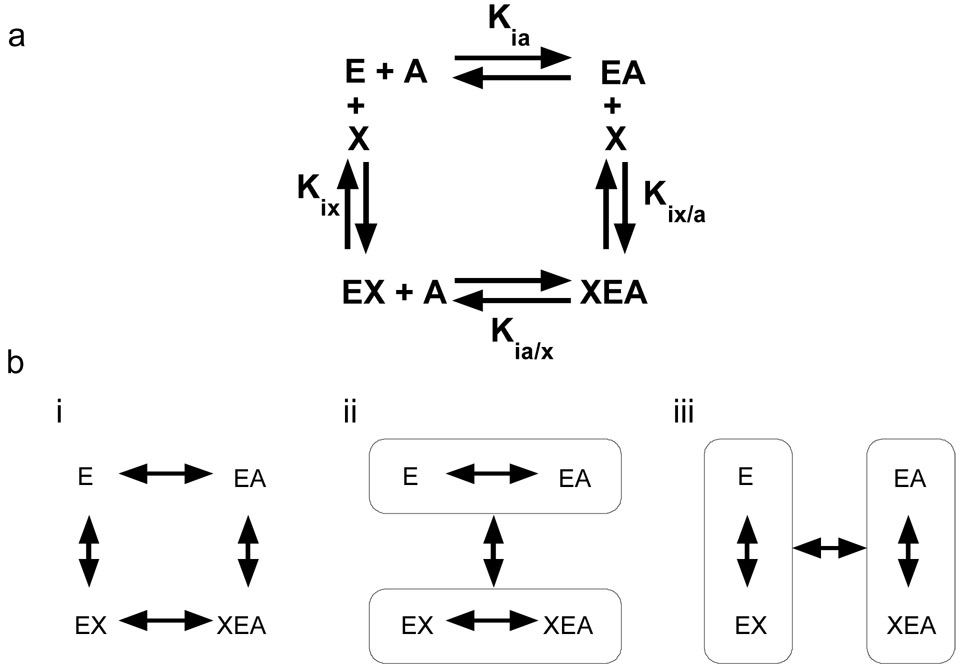
Analysis of the linked equilibrium (linkage analysis) that comprises allosteric regulation considers all four enzyme complexes in a thermodynamic energy cycle [11–18], as shown in Figure 1a. If the system is allosteric, the binding of A must change in the presence of X (Kia≠Kia/x). Therefore, the free energies of the two binding events represented by these constants are different. However, the four binding constants in Figure 1a are not independent because the free energy of the conversion of E to XEA must be independent of whether X or A binds first [11]. The difference between the free energy of binding of A to E in the presence, versus absence, of X quantifies the allostery. Because free energy values are related to binding constants through a logarithmic function, the relevant difference becomes a ratio of dissociation constants. Therefore, the relationship between dissociation constants that defines the allosteric coupling constant (Qax) is:
| (Eqn 1) |
If Qax > 1, the allosteric effector causes increased affinity of the protein for A. If Qax <1, the allosteric effector causes decreased affinity of the protein for A. If Qax =1, there is no allosteric coupling between A and X. Because Qax is a ratio of dissociation constants, the magnitude of Qax is independent of any one dissociation constant (Box 2). Simply summarized, Qax is a comparison for how one ligand binds in the absence of versus the saturating presence of the second ligand.
Figure 1.
Thermodynamic energy cycle of allostery. Allostery can be analyzed as a thermodynamic energy cycle. This analysis demonstrates that a structure/function correlation aimed at understanding the allosteric mechanism must consider four enzyme complexes. Each enzyme complex may be a single protein conformation, an equilibrium of a limited number of conformational substates, or an ensemble of conformational substates (a dynamic structure). (a) The energy cycle for an enzyme (E) which binds one substrate (A) and one allosteric effector (X). b(i) Differences between the conformation/dynamics of the enzyme complexes within circles in (ii) and (iii) are due to binding. (ii) The structural/dynamic differences that occur when A binds in the absence vs. in the saturating presence of X are allosteric effects. (iii) The structural (conformational/dynamic) differences that occur when X binds in the presence vs. in the absence of A are allosteric effects. The two presentations of allosteric effects in (ii) and (iii) are due to reciprocity.
Box 2: Determining the magnitude of Qax
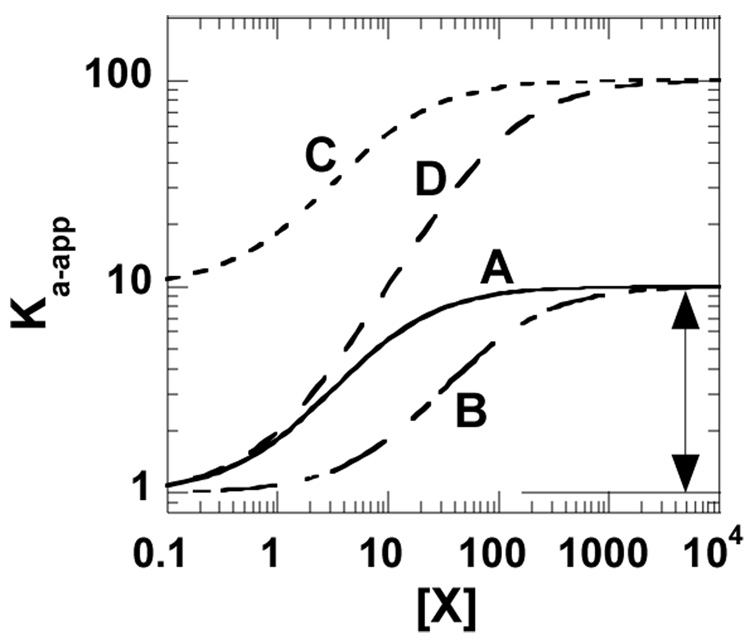
The strategy outlined in the text to distinguish allosterically relevant structural changes stresses the correlation of changes with the magnitude of the parameter Qax. Therefore, a brief review of methods for measuring Qax is warranted. One established method is to monitor the affinity (or apparent affinity, Ka-app, derived from initial velocity techniques, see [13]) of the protein for one ligand as a function of the concentration of the second ligand [12, 16]. On a log-log plot, the allosteric coupling is the difference between the upper and lower plateaus (Figure I). Although other methods for evaluating Qax are being developed for systems that use a single substrate and a single effector [33], they have not been adapted to more complex systems [13, 14].
Because the allosteric coupling is a comparison of dissociation (or affinity) constants (Equation 1 in main text), it is independent of either the substrate affinity in the absence of effector or the effector affinity in the absence of substrate. Therefore, when an allosteric system is perturbed (e.g. introduction of mutations or modification of ligand), the varied experimental conditions may alter the ligand affinities, allosteric coupling or both [19, 26]. This is graphically exemplified in Figure I, in which curves A, B, and C share a common Qax value. Curve D represents a condition with an altered value of Qax.
To correlate protein structural changes with allosteric function, it is necessary to identify protein structural (conformational and/or dynamic) changes that occur when the first ligand binds in the absence of the second ligand and that differ from changes that occur when the first ligand binds in the presence of the second ligand. A structural characterization of protein changes associated with a ligand binding event requires the comparison of protein structures for two enzyme complexes (the E/EA pair or the E/EX pair). It follows that a structural comparison aimed at understanding allostery (a comparison of two binding constants; see Equation 1) must include comparisons of protein structures for four enzyme complexes (Figure 1b). Restated, structural changes in E that occur when either A binds or X binds are due to ligand binding. Importantly, these non-allosteric changes are not limited to the ligand binding site. Allosteric changes are a consequence of both A and X being bound simultaneously to E. Advanced considerations regarding the relationship between Qax and structure are presented in Box 3.
Box 3: Advanced considerations
There are known cases of allostery for which the value of Qax=1. Here are two known underlying scenarios that can give rise to this ‘silent’ allosteric coupling. Each has a functional consequence that is masked by either compensation between entropy and enthalpy or energy compensation. Therefore, these cases are very different from long-range structural changes caused by the binding of a single ligand.
Condition 1
Because Qax is the dissociation constant for the following reaction,
Qax can be converted into free energies (ΔGax) [11, 12]. ΔGax in turn comprises ΔHax and -TΔSax components. If these two equally oppose each other, then Qax would be unity (i.e. there is no allosteric coupling). However, the protein could experience structural changes associated with each of the two components. There are multiple reported examples of this compensation between entropy and enthalpy [34–37].
Condition 2
Multiple allosteric pathways can exist in a single protein [20–23, 38– 46]. Consider a protein that contains only two communicating pathways, one of which has a Qax > 1 (enhancement of binding affinity), whereas the second has a Qax < 1 (antagonism of binding affinity). If the magnitudes of the absolute values of the two Qax parameters are equivalent, then no overall allosteric coupling will be observed. However, structural changes associated with the two pathways may be present.
Considering that any one protein might demonstrate both compensation between entropy and enthalpy and energy compensation between multiple communication pathways, conservation of allosteric mechanisms through evolution is questionable. Additionally, multiple types of energy-coupled events might involve long-range structural changes (Box 1). Any of these events could be conserved in a protein family. Therefore, proposed correlations between allosteric function and evolutionarily and/or co-evolutionarily conserved residues [47–49] require further testing (Supplemental Table 1).
Focus on the ternary complex
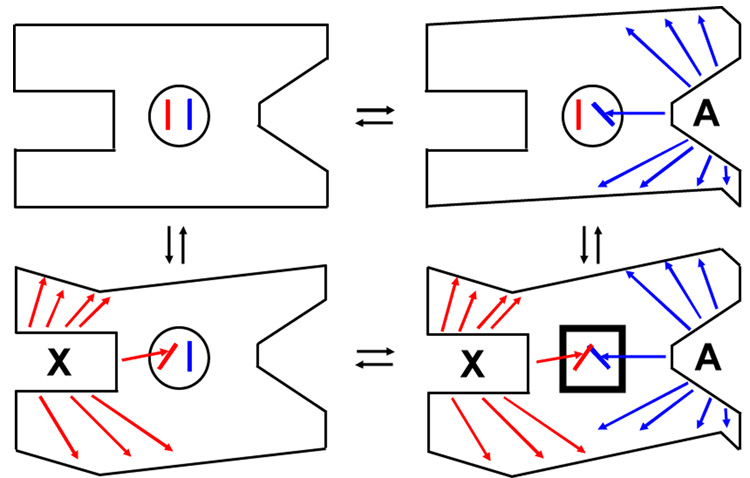
Based on the discussion here, allostery is only realized in the ternary complex. To illustrate this concept, consider the oversimplified enzyme in Figure 2. When A binds to the active site of E in the absence of X, structural (conformational and/or dynamic) changes occur. Structural changes that are not important to allostery are indicated by the change in the perimeter of the protein. The allosterically relevant change is represented by a shift in the lever in the central circle. However, this lever shift alone is not the complete allosteric mechanism. When X binds to the allosteric site of E (the two left cartoons), the protein again experiences allosterically relevant and allosterically irrelevant structural changes. Again, the allosterically relevant change is indicated by the shift in the lever in the central circle, but alone is not the complete allosteric mechanism. Allostery can only be realized when both X and A simultaneously bind to the enzyme (lower right protein). In this oversimplified model, the allosteric response would result from a steric clash of the two levers and is indicated by the conversion of the central circle to a square.
Figure 2.
A schematic of the four protein complexes of Figure 1. These simplified illustrations demonstrate how some ligand-elicited changes in the protein structure will be relevant to allostery, but others will not. They also show why allostery is only realized in the ternary complex. Ligand dependent structural changes are indicated by arrows and change in the exterior boarder of the protein. Structural changes associated with A binding are blue and those associated with X binding are red. The region with a crucial allosteric role is in the middle of the protein. The heavy square in XEA highlights allosteric changes resulting from the representative steric clash of levers.
Several caveats should be underscored when considering Figure 2. Real proteins have many potential forms of energetically unfavorable and favorable interactions, beyond a simple steric clash as illustrated. Changes in any of these interactions might contribute to the allosteric mechanism. In addition, multiple communication pathways are likely to contribute to the total allosteric response [19–24], instead of only one as illustrated. Depending on the contribution from altered protein dynamics, there may be no obvious connectivity in the conformational changes involved in any one communication pathway [25]. To introduce the final caveat, consider only a single molecular change in the protein that is required for allosteric function (e.g. movement of an amino acid side-chain). The relevant changes might occur between the complexes on the right side, but not between the complexes on the left side and vice versa. Similarly, the relevant changes could occur between the complexes on the top, but not between the complexes on the bottom and vice versa. In other words, a comparison of all four enzyme complexes identifies allosteric changes introduced in producing the XEA complex that are in addition to changes collectively introduced in producing the EA and EX complexes.
Because allostery may only be realized in the ternary complex, previous reviews of the thermodynamic analysis of allosteric systems have emphasized that the ternary complex cannot be ignored [11, 12]. However, the necessity of monitoring all four protein complexes will, at times, present a technical challenge due to difficulties in obtaining structural information of all four complexes. For example, when addressing inhibition, obtaining a homogeneous sample of the ternary XEA complex could be complicated by the mutual antagonism between the binding of A and X. A second technically challenging example is the difficulty encountered in obtaining structural information for enzyme complexes that are undergoing turnover.
An alternative strategy
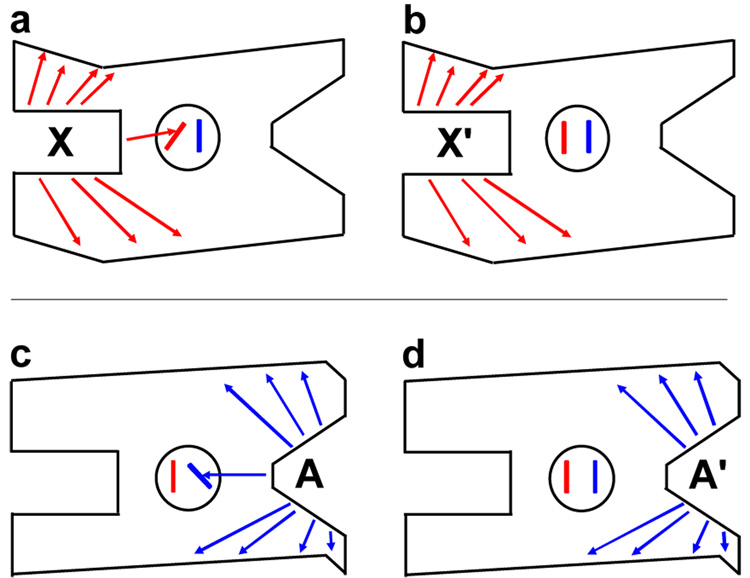
In the event that the ternary complex cannot be obtained, the desired comparison of all four enzyme complexes (Figure 1 and Figure 2) will not be possible. Therefore, we have identified an alternative strategy to identify allosteric selective protein changes. This strategy is based on our finding, in addition to those reported by others, that only specific protein-ligand interactions contribute to eliciting an allosteric response (similar to hot spots in protein-protein interactions) [26]. Knowledge of these allosterically relevant interactions can direct the identification and/or design of a non-allosteric analog, a ligand that binds competitively with the allosteric ligand but does not elicit an allosteric effect on the binding of the second ligand. Comparing the EX complex with a complex between E and a non-allosteric analog (X’) can identify allosteric-specific changes in protein properties (Figure 3a,b). A similar comparison between an EA complex and an EA’ complex (where A’ is the non-allosteric analog of A) will identify additional changes in the protein that are important to the allosteric mechanism (Figure 3c,d). However, these comparisons are not expected to identify all allosterically relevant changes in the protein structure because additional changes in protein properties are expected when the XEA complex is formed.
Figure 3.
The comparison between the a) EX and b) EX’ complexes and between c) EA and d) EA’ complexes. As presented in the text, this comparison can be used to identify structural changes relevant to allostery. This alternative strategy has been developed due to common technical challenges associated with studying the ternary complex. X’ is a non-allosteric analog that binds competitively with the X ligand. A’ is a non-allosteric analog that binds competitively with the A ligand. See Figure 2 for other details.
Future studies will need to determine if allosteric-specific changes identified by the strategy in Figure 3 are sufficient for allosteric function, or if allostery only results when these allosteric-specific changes are in addition to the changes elicited by binding of X’ and/or A’ (i.e., if the two sets of structural changes are additive to result in allosteric function). The structural resolution required to distinguish differences between the EX and EX’ complexes and between the EA and EA’ complexes is also yet to be determined.
Concluding remarks and future perspectives
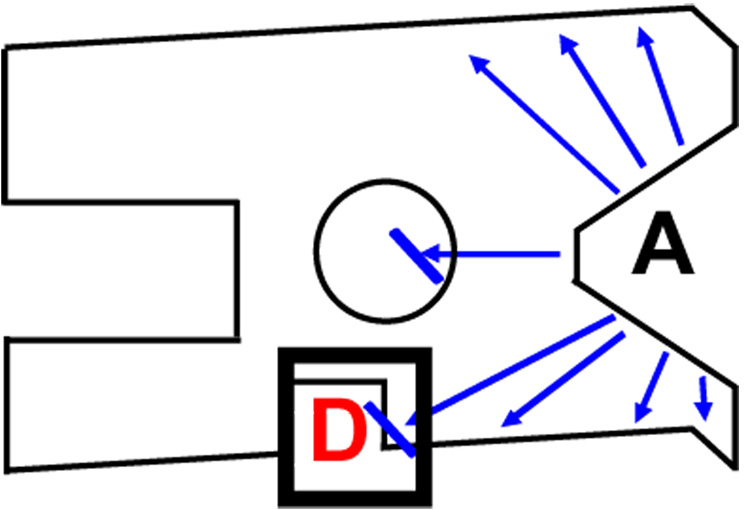
The fundamental definition of allostery, as used here, relates to the way one ligand binds to a protein in the absence of, versus the presence of, the second ligand. This simple statement should direct all structural (both conformational and dynamic) studies aimed at describing molecular mechanisms of allosteric regulation. In addition, this simple functional description of allostery can be useful for allosteric drug design by targeting; (i) known allosteric effector-binding sites, (ii) amino acid residues that participate in the native allosteric mechanism (i.e. allosteric residues) or (iii) sites on the protein that are not involved in the native allosteric mechanism but are structurally altered as a consequence of substrate binding (Figure 4). The linked equilibrium analysis and structural comparisons that can be used to describe and quantify functional allostery give rise many contradictions to common assumptions (implicit and explicit) associated with allostery; these contradictions are extensively considered in Supplementary Table 1. Therefore, the adoption of a functional based definition of allostery can have a considerable impact on future efforts to understand and utilize this ‘second secret of life’.
Figure 4.
A schematic of an allosteric drug (D) that alters substrate (A) binding using a pathway other than that used by the native effector. This schematic illustrates how an allosteric drug might use an allosteric mechanism that is different from that used by a native allosteric effector. It also demonstrates how rational drug design can target any region of the protein that is modified by the binding of A. This contrasts the example in Figure 2 (replacing X with D) which shows that allosteric drugs may target regions of the protein that are not directly modified by the binding of A. The interactions important to the allosteric function of the drug are contained in the bolded square at the bottom of the schematic. See Figure 2 for other details.
| Glossary | |
|---|---|
| Allosteric coupling | for a K-type effect, this is the ratio of the affinity of the protein for one ligand in the absence, versus presence, of a second protein-bound ligand. The magnitude of this ratio can be varied by chemically modifying the allosteric effector, mutating or covalently modifying the protein and/or changing temperature, pH or other solution conditions. |
| Allosteric drug | drugs that modify the function of a protein upon binding to the protein at a site distinct from the functional site. |
| Allosteric effector | also referred to as allosteric ligands or allosteric modulators; ligands that elicits an allosteric response upon binding to a protein. |
| Allosteric mechanism | allosteric pathways, communication pathways, allosteric communications, pathways of interaction; series of changes that (i) occur within a protein upon binding of one ligand and (ii) result in allosteric responses involving a second ligand. Multiple allosteric mechanisms can contribute to the total observed allosteric coupling. |
| Allosteric regulation (Allostery) | also termed ‘allostery’; a general term that does not distinguish function (allosteric response), magnitude (allosteric coupling), mechanism (allosteric mechanism) or physical components that act in the mechanism (allosteric residues). |
| Allosteric residues | the subset of protein residues that participate in the allosteric mechanism. Some of these residues must be in the binding sites for each of the two ligands involved in the allosteric response; other residues may be located in other regions of the protein. |
| Allosteric response | Also known as allosteric effects; the effect that binding of one ligand to a protein has on the affinity of the protein and/or catalysis of a second ligand. The two ligand-binding sites of the protein that are of interest are distinct from each other. The only difference between classic allostery (i.e. heterotropic response, heterotropic allostery or heterotropic cooperativity) and classic cooperativity (i.e. homotropic response, homotropic allostery or homotropic cooperativity) is the chemical relationship of the two ligands. Therefore, it is common to consider allostery and cooperativity as subclasses of the same phenomenon. The subclass is which the two ligands of interest are not chemically identical (classic allostery) is distinguished by the term ‘heterotropic’. The subclass in which the two ligands of interest are chemically identical (classic cooperativity) is distinguished by the term ‘homotropic’. Both homotropic and heterotropic responses can give rise to either K-type or V-type effects, as defined elsewhere. |
| Allosteric site | The binding site on the protein to which the allosteric effector binds. |
| K-type system | A system that demonstrates an allosteric response in which binding of one ligand to a protein modifies the affinity of the protein for a second ligand binding. |
| Non-allosteric analog | A ligand that does not elicit an allosteric response when it binds to the same site on the protein as the allosteric effector. |
| Reciprocity | The principle that binding of X must impact the ΔG for A binding to E to the same magnitude that the binding of A impacts the ΔG for X binding to E. |
| Structure | Used here to include both conformation and dynamic descriptions of a protein; a description of a protein ensemble at any one moment in time, which includes the average conformation and details of individual molecules (conformational substates). There can be a hierarchy of substates. The collection of all substates can be used to describe the dynamic motions that a protein molecule will sample given sufficient time. With the formalism in Figure 1 (in the main text), no assumptions about protein structures have been made. Each enzyme complex can be a single protein conformation, equilibrium of a limited number of conformational substates, or an ensemble of conformational substates. |
| V-type system | A system that demonstrates an allosteric response in which binding of an allosteric effector to an enzyme alters the catalysis (kcat or Vmax) of the enzyme. Although not the focus here, some V-type allosteric mechanisms might be analogous to the K-type allostery involving changes in ligand affinity. Such mechanisms would depend on the catalytic rate-limiting step and/or one of the two relevant binding events involving the transition-state ligand. |
Supplementary Material
Box 2, Figure I. Model Data.
This model data demonstrates potential changes that could result from modifying the allosteric effector, mutating or covalently modifying the protein, and/or changing temperature, pH, or other solution conditions. Curve A is the reference line. The allosteric coupling (Qax) for curve A is represented by the double headed arrow. Although compared to A, B has a 10-fold decrease in effector affinity in the absence of substrate and C has a 10-fold decrease in substrate affinity in the absence of effector, A, B, and C have equivalent allosteric coupling. D has a 10 fold change in allosteric coupling as compared to A.
Acknowledgements
Ideas presented here have been developed though conversations with many of my colleagues, for which I am greatly appreciative. Work in the laboratory of the author is supported by NIH grant DK78076.
Footnotes
Abbreviations: E, free enzyme; A, substrate/agonist; X, allosteric effector; AE, substrate-enzyme complex; EX, enzyme-effector complex; AEX, ternary substrate-enzyme-effector complex.
Throughout this work, we will use substrate (A) and effector (X) to distinguish the two ligands that bind to an allosteric enzyme (E). However, allostery is not restricted to enzymes: when considering a binding protein and/or receptors the term “substrate” can be substituted with “agonist” or “ligand“ and “active site” can be substituted with “orthosteric site” or “binding site”.
Reference list
- 1.Monod J. Chance and Necessity: Essay on the Natural Philosophy of Modern Biology. Penguin Books Ltd; 1977. [Google Scholar]
- 2.Monod J, et al. Allosteric proteins and cellular control systems. J Mol Biol. 1963;6:306–329. doi: 10.1016/s0022-2836(63)80091-1. [DOI] [PubMed] [Google Scholar]
- 3.Lindsley JE, Rutter J. Whence cometh the allosterome? Proc Natl Acad Sci U S A. 2006;103:10533–10535. doi: 10.1073/pnas.0604452103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Groebe DR. Screening for positive allosteric modulators of biological targets. Drug discovery today. 2006;11:632–639. doi: 10.1016/j.drudis.2006.05.010. [DOI] [PubMed] [Google Scholar]
- 5.May LT, et al. Allosteric modulation of G protein-coupled receptors. Annual review of pharmacology and toxicology. 2007;47:1–51. doi: 10.1146/annurev.pharmtox.47.120505.105159. [DOI] [PubMed] [Google Scholar]
- 6.Treadway JL, et al. Glycogen phosphorylase inhibitors for treatment of type 2 diabetes mellitus. Expert opinion on investigational drugs. 2001;10:439–454. doi: 10.1517/13543784.10.3.439. [DOI] [PubMed] [Google Scholar]
- 7.Hardy JA, Wells JA. Searching for new allosteric sites in enzymes. Curr Opin Struct Biol. 2004;14:706–715. doi: 10.1016/j.sbi.2004.10.009. [DOI] [PubMed] [Google Scholar]
- 8.Schirmer T, Evans PR. Structural basis of the allosteric behaviour of phosphofructokinase. Nature. 1990;343:140–145. doi: 10.1038/343140a0. [DOI] [PubMed] [Google Scholar]
- 9.Kimmel JL, Reinhart GD. Reevaluation of the accepted allosteric mechanism of phosphofructokinase from Bacillus stearothermophilus. Proc Natl Acad Sci U S A. 2000;97:3844–3849. doi: 10.1073/pnas.050588097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Monod J, et al. On the Nature of Allosteric Transitions: a Plausible Model. J Mol Biol. 1965;12:88–118. doi: 10.1016/s0022-2836(65)80285-6. [DOI] [PubMed] [Google Scholar]
- 11.Weber G. Ligand binding and internal equilibria in proteins. Biochemistry. 1972;11:864–878. doi: 10.1021/bi00755a028. [DOI] [PubMed] [Google Scholar]
- 12.Reinhart GD. Quantitative analysis and interpretation of allosteric behavior. Methods Enzymol. 2004;380:187–203. doi: 10.1016/S0076-6879(04)80009-0. [DOI] [PubMed] [Google Scholar]
- 13.Reinhart GD. The determination of thermodynamic allosteric parameters of an enzyme undergoing steady-state turnover. Arch Biochem Biophys. 1983;224:389–401. doi: 10.1016/0003-9861(83)90225-4. [DOI] [PubMed] [Google Scholar]
- 14.Reinhart GD. Linked-function origins of cooperativity in a symmetrical dimer. Biophys Chem. 1988;30:159–172. doi: 10.1016/0301-4622(88)85013-0. [DOI] [PubMed] [Google Scholar]
- 15.Wyman J, Gill SJ. Binding and Linkage Functional Chemistry of Biological Macromolecules. University Science Books; 1990. [Google Scholar]
- 16.Di Cera E. Thermodynamic Theory of Site-Specific Binding Processes in Biological Macromolecules. Cambridge University Press; 1995. [Google Scholar]
- 17.Di Cera E, editor. Linkage Thermodynamics of Macromolecular Interactions. Academic Press; 1998. [Google Scholar]
- 18.Frieden C. Treatment of Enzyme Kinetic Data. I. the Effect of Modifiers on the Kinetic Parameters of Single Substrate Enzymers. J Biol Chem. 1964;239:3522–3531. [PubMed] [Google Scholar]
- 19.Fenton AW, et al. Identification of substrate contact residues important for the allosteric regulation of phosphofructokinase from Eschericia coli. Biochemistry. 2003;42:6453–6459. doi: 10.1021/bi034273t. [DOI] [PubMed] [Google Scholar]
- 20.Fenton AW, Reinhart GD. Mechanism of substrate inhibition in Escherichia coli phosphofructokinase. Biochemistry. 2003;42:12676–12681. doi: 10.1021/bi0349221. [DOI] [PubMed] [Google Scholar]
- 21.Fenton AW, et al. Disentangling the web of allosteric communication in a homotetramer: heterotropic activation in phosphofructokinase from Escherichia coli. Biochemistry. 2004;43:14104–14110. doi: 10.1021/bi048569q. [DOI] [PubMed] [Google Scholar]
- 22.Fenton AW, Reinhart GD. Isolation of a single activating allosteric interaction in phosphofructokinase from Escherichia coli. Biochemistry. 2002;41:13410–13416. doi: 10.1021/bi026450g. [DOI] [PubMed] [Google Scholar]
- 23.Ackers GK. Deciphering the molecular code of hemoglobin allostery. Adv Protein Chem. 1998;51:185–253. doi: 10.1016/s0065-3233(08)60653-1. [DOI] [PubMed] [Google Scholar]
- 24.Ackers GK, Holt JM. Asymmetric cooperativity in a symmetric tetramer: human hemoglobin. J Biol Chem. 2006;281:11441–11443. doi: 10.1074/jbc.R500019200. [DOI] [PubMed] [Google Scholar]
- 25.Hilser VJ, Thompson EB. Intrinsic disorder as a mechanism to optimize allosteric coupling in proteins. Proc Natl Acad Sci U S A. 2007;104:8311–8315. doi: 10.1073/pnas.0700329104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Williams R, et al. Differentiating a Ligand's Chemical Requirements for Allosteric Interactions from Those for Protein Binding. Phenylalanine Inhibition of Pyruvate Kinase(,) Biochemistry. 2006;45:5421–5429. doi: 10.1021/bi0524262. [DOI] [PubMed] [Google Scholar]
- 27.Koshland DE. Application of a Theory of Enzyme Specificity to Protein Synthesis. Proc Natl Acad Sci U S A. 1958;44:98–104. doi: 10.1073/pnas.44.2.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johnson LN, Lewis RJ. Structural basis for control by phosphorylation. Chem Rev. 2001;101:2209–2242. doi: 10.1021/cr000225s. [DOI] [PubMed] [Google Scholar]
- 29.Kuriyan J, Eisenberg D. The origin of protein interactions and allostery in colocalization. Nature. 2007;450:983–990. doi: 10.1038/nature06524. [DOI] [PubMed] [Google Scholar]
- 30.Marvin JS, Hellinga HW. Manipulation of ligand binding affinity by exploitation of conformational coupling. Nat Struct Biol. 2001;8:795–798. doi: 10.1038/nsb0901-795. [DOI] [PubMed] [Google Scholar]
- 31.Alexiev U, et al. Evidence for long range allosteric interactions between the extracellular and cytoplasmic parts of bacteriorhodopsin from the mutant R82A and its second site revertant R82A/G231C. J Biol Chem. 2000;275:13431–13440. doi: 10.1074/jbc.275.18.13431. [DOI] [PubMed] [Google Scholar]
- 32.Masterson LR, et al. Allosteric cooperativity in protein kinase A. Proc Natl Acad Sci U S A. 2008;105:506–511. doi: 10.1073/pnas.0709214104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Velazquez-Campoy A, et al. Exact analysis of heterotropic interactions in proteins: Characterization of cooperative ligand binding by isothermal titration calorimetry. Biophys J. 2006;91:1887–1904. doi: 10.1529/biophysj.106.086561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fisher HF, Tally J. Isoergonic cooperativity: a novel form of allostery. Methods Enzymol. 1998;295:331–349. doi: 10.1016/s0076-6879(98)95047-9. [DOI] [PubMed] [Google Scholar]
- 35.Fisher HF, Tally J. Isoergonic cooperativity in glutamate dehydrogenase complexes: a new form of allostery. Biochemistry. 1997;36:10807–10810. doi: 10.1021/bi9708388. [DOI] [PubMed] [Google Scholar]
- 36.Tlapak-Simmons VL, Reinhart GD. Obfuscation of allosteric structure-function relationships by enthalpy-entropy compensation. Biophys J. 1998;75:1010–1015. doi: 10.1016/S0006-3495(98)77589-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Braxton BL, et al. Temperature-induced inversion of allosteric phenomena. J Biol Chem. 1994;269:47–50. [PubMed] [Google Scholar]
- 38.Ortigosa AD, et al. Disentangling the web of allosteric communication in a homotetramer: heterotropic inhibition of phosphofructokinase from Bacillus stearothermophilus. Biochemistry. 2004;43:577–586. doi: 10.1021/bi035077p. [DOI] [PubMed] [Google Scholar]
- 39.Kimmel JL, Reinhart GD. Isolation of an individual allosteric interaction in tetrameric phosphofructokinase from Bacillus stearothermophilus. Biochemistry. 2001;40:11623–11629. doi: 10.1021/bi010844a. [DOI] [PubMed] [Google Scholar]
- 40.Nelson SW, et al. Hybrid tetramers of porcine liver fructose-1,6-bisphosphatase reveal multiple pathways of allosteric inhibition. J Biol Chem. 2002;277:15539–15545. doi: 10.1074/jbc.M112304200. [DOI] [PubMed] [Google Scholar]
- 41.Grant GA, et al. Quantitative relationships of site to site interaction in Escherichia coli D-3-phosphoglycerate dehydrogenase revealed by asymmetric hybrid tetramers. J Biol Chem. 2004;279:13452–13460. doi: 10.1074/jbc.M313593200. [DOI] [PubMed] [Google Scholar]
- 42.Faga LA, et al. Basic interdomain boundary residues in calmodulin decrease calcium affinity of sites I and II by stabilizing helix-helix interactions. Proteins. 2003;50:381–391. doi: 10.1002/prot.10281. [DOI] [PubMed] [Google Scholar]
- 43.Jaren OR, et al. Calcium-induced conformational switching of Paramecium calmodulin provides evidence for domain coupling. Biochemistry. 2002;41:14158–14166. doi: 10.1021/bi026340+. [DOI] [PubMed] [Google Scholar]
- 44.VanScyoc WS, et al. Calcium binding to calmodulin mutants monitored by domain-specific intrinsic phenylalanine and tyrosine fluorescence. Biophys J. 2002;83:2767–2780. doi: 10.1016/S0006-3495(02)75286-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sorensen BR, et al. An interdomain linker increases the thermostability and decreases the calcium affinity of the calmodulin N-domain. Biochemistry. 2002;41:15–20. doi: 10.1021/bi011718+. [DOI] [PubMed] [Google Scholar]
- 46.Sun H, et al. Mutation of Tyr138 disrupts the structural coupling between the opposing domains in vertebrate calmodulin. Biochemistry. 2001;40:9605–9617. doi: 10.1021/bi0104266. [DOI] [PubMed] [Google Scholar]
- 47.Suel GM, et al. Evolutionarily conserved networks of residues mediate allosteric communication in proteins. Nat Struct Biol. 2003;10:59–69. doi: 10.1038/nsb881. [DOI] [PubMed] [Google Scholar]
- 48.Lockless SW, Ranganathan R. Evolutionarily conserved pathways of energetic connectivity in protein families. Science. 1999;286:295–299. doi: 10.1126/science.286.5438.295. [DOI] [PubMed] [Google Scholar]
- 49.Pendergrass DC, et al. Mining for allosteric information: Natural mutations and positional sequence conservation in pyruvate kinase. IUBMB Life. 2006;58:31–38. doi: 10.1080/15216540500531705. [DOI] [PubMed] [Google Scholar]
- 50.Subramanian S, et al. Thermodynamics of heterotropic interactions. The glutamate dehydrogenase . NADPH . glutamate complex. J Biol Chem. 1978;253:8369–8374. [PubMed] [Google Scholar]
- 51.Frauenfelder H, et al. Conformational substates in proteins. Annu Rev Biophys Biophys Chem. 1988;17:451–479. doi: 10.1146/annurev.bb.17.060188.002315. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.