SUMMARY
Tetraloops are a common building block for RNA tertiary structure and most tetraloops fall into one of three well-characterized classes: GNRA, UNCG, and CUYG. Here, we present the sequence and structure of a fourth highly conserved class of tetraloop that occurs only within the ζ-ζ′ interaction of group IIC introns. This GANC tetraloop was identified, along with an unusual cognate receptor, in the crystal structure of the group IIC intron and through phylogenetic analysis of intron RNA sequence alignments. Unlike conventional tetraloop-receptor interactions, which are stabilized by extensive hydrogen bonding interactions, the GANC-receptor interaction is limited to a single base stack between the conserved adenosine of the tetraloop and a single purine of the receptor, which consists of a one to three nucleotide bulge and does not contain an A-platform. Unlike GNRA tetraloops, the GANC tetraloop forms a sharp angle relative to the adjacent helix, bending by approximately 45° towards the major groove side of the helix. These structural attributes allow GANC tetraloops to fit precisely within the group IIC intron core, thereby demonstrating that structural motifs can adapt to function in a specific niche.
Keywords: tetraloop, motif, RNA structure, group II intron, ribozyme
Tetraloops are among the most common RNA structural elements. These motifs have been divided into three main classes based on their sequence: GNRA, UNCG, and CUYG, with each sequence corresponding to a unique structure.1,2 While some sequence variability is observed,3 nearly all structured tetraloops belong to one of these three classes. Here, we present phylogenetic and structural evidence for a fourth class of conserved tetraloop, together with its cognate receptor. The GANC tetraloop is found within the ζ-ζ′ interaction of group IIC introns, and it was structurally characterized during analysis of the 3.1 Å crystal structure of the Oceanobacillus iheyensis intron4 (PDB5 ID: 3BWP). The ζ-ζ′ interaction is one of several tertiary contacts that join intron domains I and V. 6 While ζ-ζ′ is also found in the IIA and IIB introns, it consists of a canonical GNRA tetraloop and receptor in those contexts.6
Group IIC introns are smaller than the well-studied IIA and IIB introns, and they are found only in bacteria.7 DV is the most highly conserved substructure within group II introns and consists of two helices that are separated by a bulge and capped by the ζ′ tetraloop (Fig 1A). This bulge, together with a conserved triad of nucleotides in the lower stem of DV,8 is essential for catalytic activity by the intron.9 The upper helix of DV is typically five base pairs in length, but it contains only three base pairs in IIC introns.7 In addition to changes in the morphology of DV, there are numerous structural simplifications in IIC introns,7,10 and phylogenetic analysis has suggested that they represent the most primitive class of introns.11 Group IIC introns are highly catalytically reactive in vitro, even in the absence of associated proteins.10,12
Figure 1.
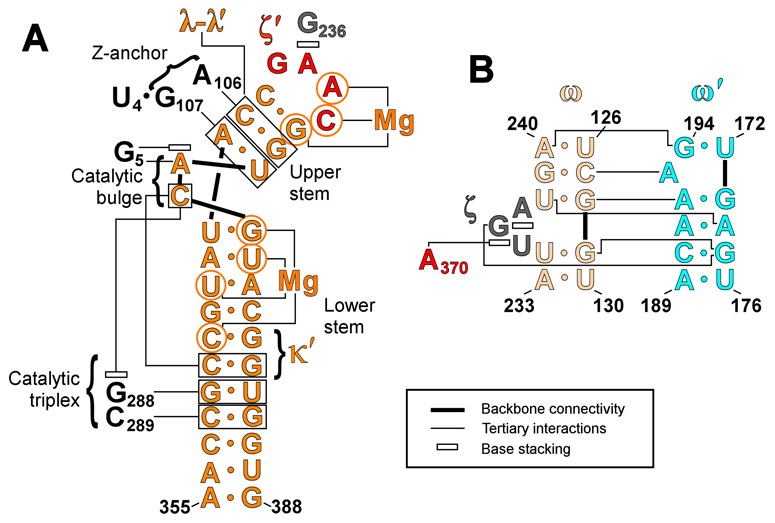
Secondary structures of domain V and the ω–ω’ region of domain I that contains the receptor. Color coding is the same as that in Table 1 and in subsequent figures. Tertiary interaction sites are indicated with labels and greek letters. (A) Secondary structure of domain V. Interactions between domain V and other intron bases are indicated. Note that κ-κ′ was not well ordered in the crystal structure; therefore, no specific interactions are shown.(B) Secondary structure of the tetraloop receptor and ω-ω′ ribose zipper.
Sequence Conservation
The unusual tetraloop in domain V of group IIC introns is not found in any other known RNA, and it contains the consensus sequence GANC. Alignments of group IIC intron sequences reveal that the first, second, and fourth position of this tetraloop are completely conserved (Table 1). All four nucleotides are observed in the third position, although adenosine and cytidine are the most common variants (~44% of sequences in each case).
Table 1. Alignment of GANC tetraloop and cognate receptor sequences in group IIC introns.
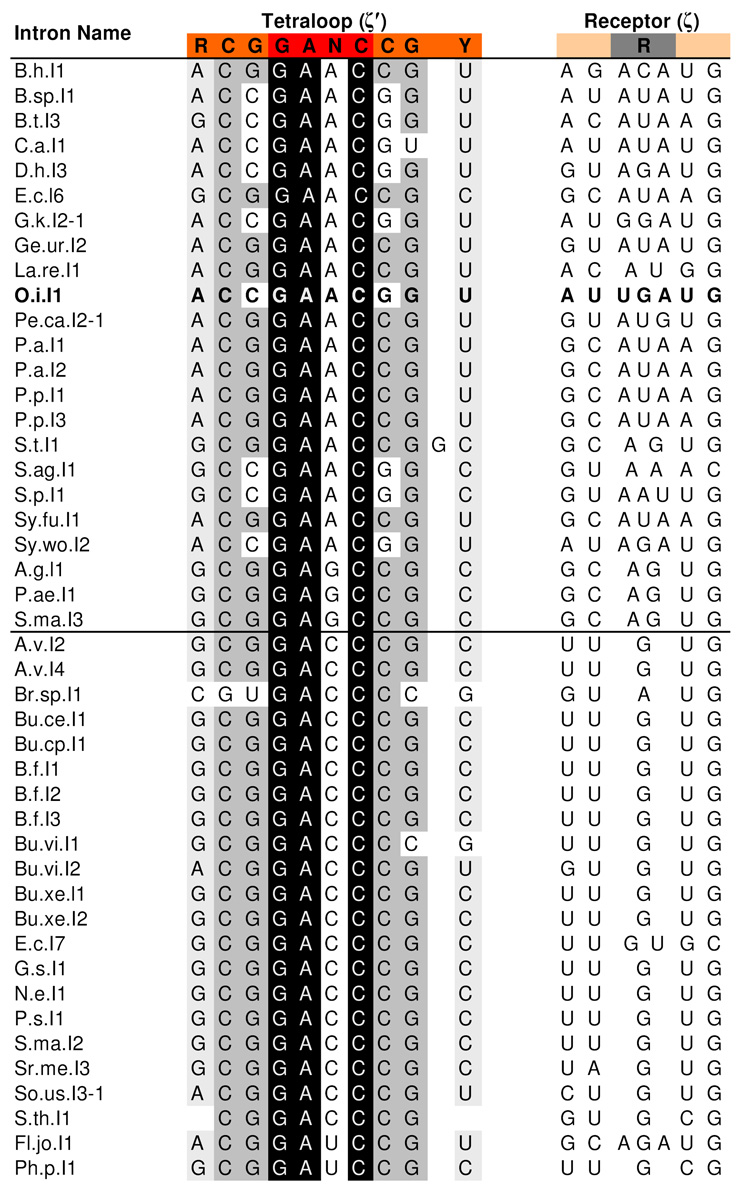 |
Footnote: The consensus sequence is shown at the top and nucleotides are color-coded as in Figure 1. Sequences of the O. iheyensis intron are indicated in bold. Conserved bases of the tetraloop are indicated with a black background for complete conservation (100% A, C, or G) a dark gray background for strong conservation (>75% C or G) and a light gray background for weaker conservation (>95% purine or pyrimidine). Conservation within the receptor is attributable to sequence constraints imposed by the ω-<′ ribose zipper and therefore highlighting was omitted. A horizontal line through the middle of the alignment indicates that there are two different subclasses of group IIC introns: The top class, which includes the O. iheyensis intron, generally has a three-base receptor and a purine (usually an adenosine) in the third position of the tetraloop. The bottom class has a single bulged G receptor and a pyrimidine (usually a cytidine) in the third position of the tetraloop. Sequences were retrieved from the database for mobile group II introns.30 All known IIC intron sequences are shown except for Cl.sp.I1 and M.sp.I1, in which ζ′ consists of a GNRA tetraloop, and D.p.I2 and Pe.th.I2, in which ζ′ consists of an AGGCC pentaloop.
Conservation is also observed among the three base pairs in the upper stem of domain V. The closing base pair of this helix is typically G-C (78%), although the most common variation is a C-G pair (20%), as in the case of the O. iheyensis intron discussed here. Importantly, the latter variant only occurs when the third base of the tetraloop is a purine. Interestingly, this preference for G-C over C-G is the opposite of that seen for GNRA and UNCG tetraloops.13,14 The CUYG tetraloop shows the G-C preference seen here; however, this is due to a direct interaction between the second base of the tetraloop and the minor groove of the closing base pair.15 The first and second base pairs of the upper DV stem also show strong conservation, with a 98% preference for an RY and a CG pair, respectively. However, these base pairs are involved in a base quartet and triple with the z-anchor region of domain I (Fig 1A).4 Given the functional importance of these interactions, the conservation of the constituent nucleotides can be attributed to their role in stabilizing the z-anchor network, which includes the ζ-ζ’ interaction found in other classes of group II introns.16
The receptor for the GANC tetraloop consists of a bulged region in the D(i) stem of domain I, corresponding to nucleotides 235 through 237 in the O. iheyensis intron.4 This bulge ranges in size from one to three bases and always contains at least one purine (Table 1). Receptor sequences covary with the third base of the tetraloop (GANC). When this base is a purine, the receptor bulge is normally three bases, although it is occasionally only two. Additionally, an adenosine is always present in the bulge, with a guanosine also being present in 39% of the sequences. When the third base of the tetraloop is a pyrimidine, the receptor bulge is almost always a single guanosine. The sequence of the helix in this region is conserved as well; however, this helix is involved in the ω-ω′ ribose zipper (Fig 1B),4 and the conservation patterns are consistent with those seen in ribose zipper motifs.17
Tetraloop Structure
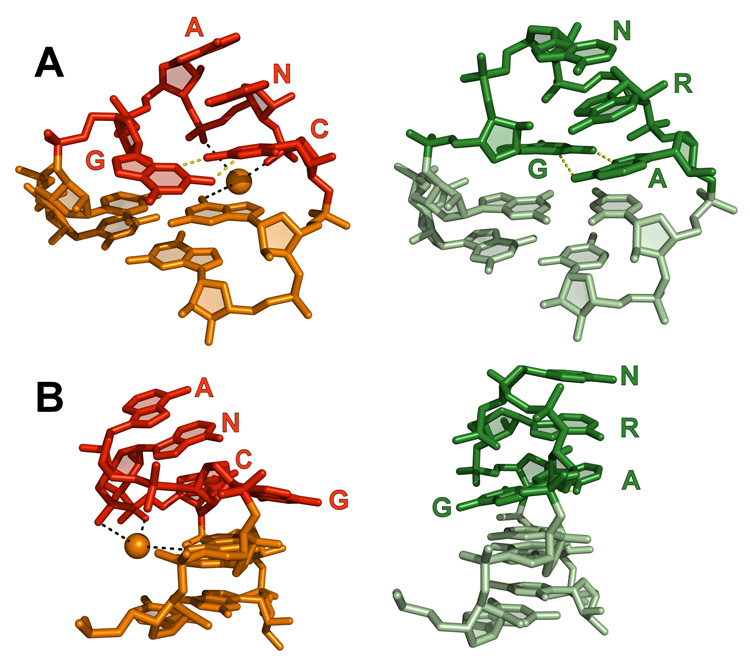
The backbone of the tetraloop adopts a U-turn-like conformation18,19 between the first and second nucleotides (Fig 2A). The second through fourth nucleotides of the tetraloop (GANC) stack on each other, and the first and fourth bases form a GC trans sugar-edge/Watson-Crick base pair (using the nomenclature of ref. 20 to describe base pairing). The structure initially appears similar to a GNRA tetraloop (Fig. 3), but there are a number of critical differences between these two tetraloop forms. Many of these differences are caused by the C2′ endo sugar pucker of the leading guanosine of the GANC, as compared to the more common C3′ endo sugar pucker for the leading nucleotide of the GNRA tetraloop. Sugar puckers in the IIC intron structure were assigned using the phosphate-glycosidic bond perpendicular distance,21,22 which measures the distance from the glycosidic bond to the 3′ phosphate. A distance of less than 3 Å corresponds to a C2′ endo sugar pucker. This distance is 1.5 Å for the leading guanosine, clearly indicating a C2′ endo pucker. This pucker switch inverts the base plane of the guanosine and moves the base by approximately 4 Å, shifting it to the minor groove side of the helix. The unusual sugar pucker also drastically alters the angle of the tetraloop relative to the helix, causing the tetraloop to bend roughly 45° towards the major groove side (Fig. 3B).
Figure 2.
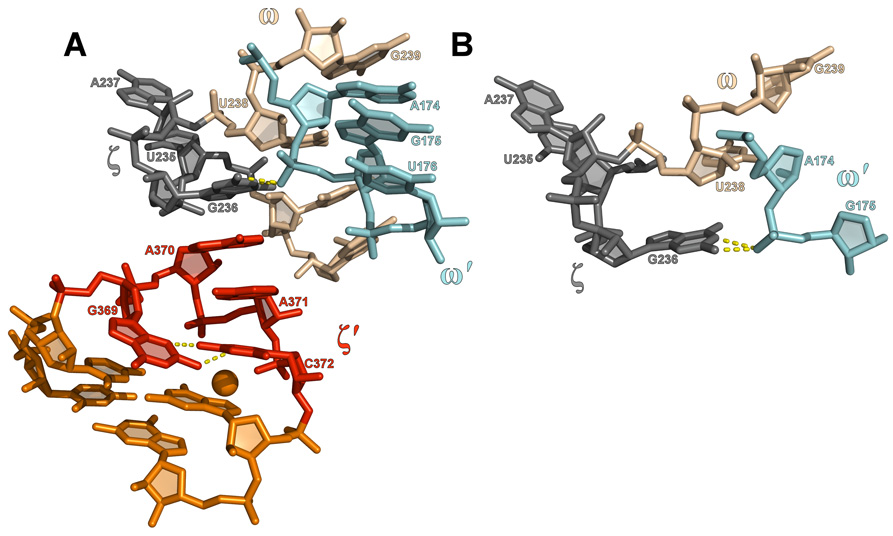
Structure of the GANC tetraloop and receptor. (A) A view of ζ′, the GANC tetraloop, docked into ζ, its cognate receptor (gray). The receptor consists of three bases bulged from ω, which is part of the ω-ω′ ribose zipper. The interaction between the tetraloop and receptor consists only of a base stack between A370 and G236. (B) Interactions within the receptor. Hydrogen bonds between G236 and a phosphoryl oxygen of G175 are shown in yellow.
Figure 3.
A comparison between GANC (red) and GNRA (green) tetraloops. GNRA tetraloop coordinates derive from the structure of the P4–P6 domain of the group I intron31 (PDB5 ID: 1HR2), which is superimposable with almost all other known GNRA tetraloops.28 (A) A front view of the tetraloops, shown in identical orientations. Note that the first nucleotide of the GANC tetraloop extends into the minor groove, while the first nucleotide of the GNRA tetraloop is pushed towards the major groove of the adjacent stem. A Mg2+ ion is observed in the major groove, interacting with the final two bases of the GANC tetraloop and the closing base pair. The closing base pair observed here is the opposite that of the consensus base pair, so it is unknown if this Mg2+ is a conserved feature of the GANC tetraloop. (B) A side view of the tetraloops. The GANC tetraloop bends approximately 45° towards the major groove side of the adjacent helix, while the GNRA tetraloop exhibits no such bending. Also note that the Watson-Crick edges of the second through fourth nucleotides of the GNRA tetraloop push outward, toward the minor groove, allowing this tetraloop to readily insert into a receptor helix. In the GANC tetraloop, these nucleotides are stacked on the closing base pair and their Watson-Crick edges are not similarly accessible.
There is also a subtle, but critically important difference in base stacking between the GNRA and GANC tetraloops. In the GNRA tetraloop, the first base of the tetraloop stacks on the closing base pair, while the second through fourth bases (GNRA) are forced towards the minor groove side of the helix. This makes the Watson-Crick edges of these bases easily accessible and allows them to insert into the helix of a tetraloop receptor, as is seen in the canonical GNRA-receptor interaction.23 In the GANC tetraloop, the opposite occurs. The first base is pushed towards the helix minor groove, while the second through fourth bases (GANC) stack on the closing base pair. Thus, steric constraints between the closing base pair and the receptor helix prevent the GANC tetraloop from inserting into a receptor helix.
Another interesting aspect of the tetraloop structure is that a Mg2+ binding site is formed by the tetraloop and the closing base pair (Fig 3A, 3B). In the O. iheyensis intron discussed here, a Mg2+ ion forms an inner-sphere contact with the phosphoryl oxygen of the final base of the tetraloop (GANC) and is also bound by two outer-sphere ligands consisting of the phosphoryl oxygen of the third base of the tetraloop (GANC) and the O6 of the guanosine in the closing base pair. However, this structure contains a C-G closing base pair, whereas the consensus sequence is G-C. With a G-C base pair, the guanosine O6 would be shifted by approximately 1.8 Å; thus, it is unlikely that this Mg2+ binding site would be the same in cases where the sequence contains the consensus closing base pair.
To assess the uniqueness of the tetraloop structure, we performed an (η,θ) search24 using the AMIGOS II software.25 The η and θ values for the four nucleotides of the tetraloop are (57,158), (102,230), (167,241), and (156,238). The sequential string of these coordinates (the “worm signature” for the motif24) was used to search the RNA05 26 and Wadley et al.25 datasets, but no matches were found, indicating that this tetraloop is structurally unique amongst solved RNA structures. It is unknown if the tetraloop maintains this particular structure when it is in the free form, undocked to its cognate receptor. Further structural work will be required to characterize the unbound conformation of this tetraloop.
Receptor Structure
The receptor consists of a one to three base bulge that is located within the ω helix of the ω-ω′ ribose zipper (Fig. 1B). In the O. iheyensis intron, the sequence is UGA. The guanosine of the receptor flips away from the flanking uridine and adenosine residues, which then stack on each other in a position approximately 9 Å away from the guanosine moiety (Fig. 2A). The extruded guanosine stacks on the second base of the tetraloop (GANC), thereby serving as the major recognition determinant for theζ–ζ’ interaction. The guanosine configuration appears to be reinforced by a bifurcated hydrogen bond to the phosphoryl oxygen of G175, which is part of the ω′helix (Fig. 2B). Both the N1 and N2 atoms of the guanosine are within hydrogen bonding distance (2.8 Å and 2.6 Å, respectively) of the OP1 atom of G175.
As the sequence of the receptor is variable (see above), it is difficult to determine how conserved the structural features really are. However, the structure indicates that interaction with the tetraloop is limited to a base stack and that the only strictly required feature of the receptor is a bulged base, which is consistent with the sequence alignment (Table 1). In addition, the ω-ω′ ribose zipper is well conserved across IIC introns, and it is therefore possible that the interactions between the receptor purine and ω′ are conserved, even among bulges of differing sizes. An (η,θ) search24,25 revealed that the conformation of the receptor studied here is not found in any previously solved structures and is dissimilar from other, more-common bulged nucleotide motifs.
Tetraloop-Receptor Interactions
In IIA and IIB introns, the ζ-ζ′ interaction is a canonical GNRA-receptor interaction.6 In these intron classes, there are two additional base pairs between the DV bulge and the ζ′ tetraloop. These base pairs move the tetraloop closer to the receptor and rotate it by approximately 65°, which positions the GNRA tetraloop and receptor for a canonical interaction. In the context of the O. iheyensis IIC intron, a GNRA tetraloop would not be able to dock into a cognate receptor at ζ. Instead, the tetraloop would sterically clash with residues in ω′ and EBS1, potentially disrupting exon binding.
As discussed above, the interaction between the tetraloop and receptor is highly unusual in that it consists of only a base stack between the third base of the tetraloop (GANC) and a bulged purine (Fig 2A). As such, the receptor may not be capable of detecting sequence variations within the tetraloop, unlike canonical receptors that recognize variants of the GNRA tetraloop sequence.27,28 Nonetheless, the tetraloop and receptor display close covariation: The receptor purine is typically an adenosine when the third tetraloop base is a purine, and typically a guanosine when the third tetraloop base is a pyrimidine (Table 1). No hydrogen bonding is observed between the tetraloop and receptor, and the receptor does not form the A-platform motif seen in GNRA receptors.29 Finally, the last three bases of the tetraloop stack on the closing base pair, thereby adopting a configuration that prevents insertion into the receptor helix in the manner previously observed for GNRA tetraloops.
The limited nature of the interaction between the GANC tetraloop and receptor suggests that, in group IIC introns, ζ-ζ′ may contribute to the proper orientation of D5 in the core, but it is unlikely to provide strong energetic stabilization for domain V binding. However, the crystal structure reveals an elaborate network of other interactions that help dock and stabilize the entire length of DV within the core (Fig 1A). These include the major-groove triplex with J2/3, the triple and quartet with the z-anchor (which includes the λ–λ′ interaction), the Mg2+ coordination site in the interior of the lower helix, the κ-κ′ interaction, and a number of active site interactions involving the catalytic metal ions.4 While the specific configurations for some of these interactions may differ from those in IIA and IIB introns, this structure demonstrates that DV can attain a catalytically relevant conformation in the absence of an extensive interaction between ζ and ζ′.
The apparent weakness of the GANC-tetraloop receptor interaction and the complexity of its local context involving a semi-conserved ω-ω′ ribose zipper may explain why the GANC tetraloop is not more prevalent in other RNA molecules. This motif appears to be specifically adapted for the unique structural constraints of IIC introns, which possess a shorter DV that functions in tandem with the ω-ω′ motif. The fact that the GANC-tetraloop receptor interaction is unique to IIC introns challenges the notion that all RNA structure is constructed from a limited toolbox of motifs. In this case, it appears that a sufficiently complex context has given rise to a unique solution. While such contexts may be rare, this tetraloop demonstrates that they do exist and that evolution has crafted a motif that has adapted to its specific niche.
ACKNOWLEDGEMENTS
K.S.K thanks NIH training grant T15 LM07056 for support. N.T. is a Postdoctoral Associate and A.M.P. an Investigator with the Howard Hughes Medical Institute. This work was supported by the Howard Hughes Medical Institute (HHMI) and NIH grant GM50313 to A.M.P.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Woese CR, Winker S, Gutell RR. Architecture of ribosomal RNA: constraints on the sequence of "tetra-loops". Proc Natl Acad Sci U S A. 1990;87:8467–8471. doi: 10.1073/pnas.87.21.8467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moore PB. Structural motifs in RNA. Annu Rev Biochem. 1999;68:287–300. doi: 10.1146/annurev.biochem.68.1.287. [DOI] [PubMed] [Google Scholar]
- 3.Ihle Y, Ohlenschlager O, Hafner S, Duchardt E, Zacharias M, Seitz S, Zell R, Ramachandran R, Gorlach M. A novel cGUUAg tetraloop structure with a conserved yYNMGg-type backbone conformation from cloverleaf 1 of bovine enterovirus 1 RNA. Nucleic Acids Res. 2005;33:2003–2011. doi: 10.1093/nar/gki501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Toor N, Keating KS, Taylor SD, Pyle AM. Cystal structure of a self-spliced group II intron. Science. 2008;320:77–82. doi: 10.1126/science.1153803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Costa M, Michel F. Frequent use of the same tertiary motif by self-folding RNAs. Embo J. 1995;14:1276–1285. doi: 10.1002/j.1460-2075.1995.tb07111.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Toor N, Hausner G, Zimmerly S. Coevolution of group II intron RNA structures with their intron-encoded reverse transcriptases. RNA. 2001;7:1142–1152. doi: 10.1017/s1355838201010251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boulanger SC, Belcher SM, Schmidt U, Dib-Hajj SD, Schmidt T, Perlman PS. Studies of point mutants define three essential paired nucleotides in the domain 5 substructure of a group II intron. Mol Cell Biol. 1995;15:4479–4488. doi: 10.1128/mcb.15.8.4479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schmidt U, Podar M, Stahl U, Perlman PS. Mutations of the two-nucleotide bulge of D5 of a group II intron block splicing in vitro and in vivo: phenotypes and suppressor mutations. RNA. 1996;2:1161–1172. [PMC free article] [PubMed] [Google Scholar]
- 10.Toor N, Robart AR, Christianson J, Zimmerly S. Self-splicing of a group IIC intron: 5′ exon recognition and alternative 5′ splicing events implicate the stem-loop motif of a transcriptional terminator. Nucleic Acids Res. 2006;34:6461–6471. doi: 10.1093/nar/gkl820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rest JS, Mindell DP. Retroids in archaea: phylogeny and lateral origins. Mol Biol Evol. 2003;20:1134–1142. doi: 10.1093/molbev/msg135. [DOI] [PubMed] [Google Scholar]
- 12.Granlund M, Michel F, Norgren M. Mutually exclusive distribution of IS1548 and GBSi1, an active group II intron identified in human isolates of group B streptococci. J Bacteriol. 2001;183:2560–2569. doi: 10.1128/JB.183.8.2560-2569.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Antao VP, Lai SY, Tinoco I., Jr A thermodynamic study of unusually stable RNA and DNA hairpins. Nucleic Acids Res. 1991;19:5901–5905. doi: 10.1093/nar/19.21.5901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moody EM, Feerrar JC, Bevilacqua PC. Evidence that folding of an RNA tetraloop hairpin is less cooperative than its DNA counterpart. Biochemistry. 2004;43:7992–7998. doi: 10.1021/bi049350e. [DOI] [PubMed] [Google Scholar]
- 15.Jucker FM, Pardi A. Solution structure of the CUUG hairpin loop: a novel RNA tetraloop motif. Biochemistry. 1995;34:14416–14427. doi: 10.1021/bi00044a019. [DOI] [PubMed] [Google Scholar]
- 16.Boudvillain M, de Lencastre A, Pyle AM. A tertiary interaction that links active-site domains to the 5′ splice site of a group II intron. Nature. 2000;406:315–318. doi: 10.1038/35018589. [DOI] [PubMed] [Google Scholar]
- 17.Tamura M, Holbrook SR. Sequence and structural conservation in RNA ribose zippers. J Mol Biol. 2002;320:455–474. doi: 10.1016/s0022-2836(02)00515-6. [DOI] [PubMed] [Google Scholar]
- 18.Jucker FM, Pardi A. GNRA tetraloops make a U-turn. RNA. 1995;1:219–222. [PMC free article] [PubMed] [Google Scholar]
- 19.Quigley GJ, Rich A. Structural domains of transfer RNA molecules. Science. 1976;194:796–806. doi: 10.1126/science.790568. [DOI] [PubMed] [Google Scholar]
- 20.Leontis NB, Westhof E. Geometric nomenclature and classification of RNA base pairs. RNA. 2001;7:499–512. doi: 10.1017/s1355838201002515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Murray LW. RNA backbone rotamers and chiropraxis. Duke University; 2007. [Google Scholar]
- 22.Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB, 3rd, Snoeyink J, Richardson JS, Richardson DC. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–W383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cate JH, Gooding AR, Podell E, Zhou K, Golden BL, Kundrot CE, Cech TR, Doudna JA. Crystal structure of a group I ribozyme domain: principles of RNA packing. Science. 1996;273:1678–1685. doi: 10.1126/science.273.5282.1678. [DOI] [PubMed] [Google Scholar]
- 24.Duarte CM, Wadley LM, Pyle AM. RNA structure comparison, motif search and discovery using a reduced representation of RNA conformational space. Nucleic Acids Res. 2003;31:4755–4761. doi: 10.1093/nar/gkg682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wadley LM, Keating KS, Duarte CM, Pyle AM. Evaluating and learning from RNA pseudotorsional space: quantitative validation of a reduced representation for RNA structure. J Mol Biol. 2007;372:942–957. doi: 10.1016/j.jmb.2007.06.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Richardson JS, Schneider B, Murray LW, Kapral GJ, Immormino RM, Headd JJ, Richardson DC, Ham D, Hershkovits E, Williams LD, Keating KS, Pyle AM, Micallef D, Westbrook J, Berman HM. RNA backbone: consensus all-angle conformers and modular string nomenclature (an RNA Ontology Consortium contribution) RNA. 2008;14:465–481. doi: 10.1261/rna.657708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pley HW, Flaherty KM, McKay DB. Model for an RNA tertiary interaction from the structure of an intermolecular complex between a GAAA tetraloop and an RNA helix. Nature. 1994;372:111–113. doi: 10.1038/372111a0. [DOI] [PubMed] [Google Scholar]
- 28.Correll CC, Swinger K. Common and distinctive features of GNRA tetraloops based on a GUAA tetraloop structure at 1.4 A resolution. RNA. 2003;9:355–363. doi: 10.1261/rna.2147803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cate JH, Gooding AR, Podell E, Zhou K, Golden BL, Szewczak AA, Kundrot CE, Cech TR, Doudna JA. RNA tertiary structure mediation by adenosine platforms. Science. 1996;273:1696–1699. doi: 10.1126/science.273.5282.1696. [DOI] [PubMed] [Google Scholar]
- 30.Dai L, Toor N, Olson R, Keeping A, Zimmerly S. Database for mobile group II introns. Nucleic Acids Res. 2003;31:424–426. doi: 10.1093/nar/gkg049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Juneau K, Podell E, Harrington DJ, Cech TR. Structural basis of the enhanced stability of a mutant ribozyme domain and a detailed view of RNA--solvent interactions. Structure. 2001;9:221–231. doi: 10.1016/s0969-2126(01)00579-2. [DOI] [PubMed] [Google Scholar]