Abstract
During repair of damaged DNA, the oxidized base 8-oxoguanine (8-oxoG) is removed by 8-oxoguanine—DNA glycosylase (Ogg) in eukaryotes and most archaea, whereas in most bacteria it is removed by formamidopyrimidine—DNA glycosylase (Fpg). We report the first characterization of a bacterial Ogg, Clostridium acetobutylicum Ogg (CacOgg). Like human OGG1 and Escherichia coli Fpg (EcoFpg), CacOgg excised 8-oxoguanine. However, unlike hOGG1 and EcoFpg, CacOgg showed little preference for the base opposite the damage during base excision and removed 8-oxoguanine from single-stranded DNA. Thus, our results showed unambiguous qualitative functional differences in vitro between CacOgg and both hOGG1 and EcoFpg. CacOgg differs in sequence from the eukaryotic enzymes at two sequence positions, M132 and F179, which align with amino acids (R154 and Y203) in human OGG1 (hOGG1) found to be involved in opposite base interaction. To address the sequence basis for functional differences with respect to opposite base interactions, we prepared three CacOgg variants, M132R, F179Y, and M132R/F179Y. All three variants showed a substantial increase in specificity for 8-oxoG·C relative to 8-oxoG·A. While we were unable to definitively associate these qualitative functional differences with differences in selective pressure between eukaryotes, Clostridia, and other bacteria, our results are consistent with the idea that evolution of Ogg function is based on kinetic control of repair.
DNA is continuously damaged by reactive oxygen species (ROS)1 generated during metabolism (1, 2), and by ultraviolet light and ionizing radiation. Damaged nucleotide bases, if bypassed during replication, are prone to mispair, resulting in mutations. Oxidative DNA damage is mainly repaired by base excision repair (BER) (reviewed in ref 3). During BER, a DNA glycosylase cleaves the N-glycosylic bond of the damaged base (monofunctional activity) or cleaves the glycosylic bond and nicks the DNA strand in a lyase reaction (bifunctional activity). Other enzymes prepare the site for nucleotide replacement via DNA polymerases, and finally the site is religated.
One of the most common oxidative events is oxidation of guanine to 7,8-dihydro-8-oxoguanine (8-oxoG) (4), which can mispair during replication causing G·C to T·A transversions (5, 6). In Escherichia coli, Fpg removes 8-oxoG with bifunctional activity. A protein in eukaryotes having functional, but not structural, similarity was first cloned by van der Kemp et al. (7), from Saccharomyces cerevisiae, and named Ogg (oxoguanine glycosylase). A human homologue, hOGG1, has also been cloned and characterized (8–14), and nullizygous mice lacking Ogg1 demonstrate an increased mutation rate (15). Thus, removal of 8-oxoG from the genome is important to maintain genomic stability, and this enzymatic function is conserved across kingdoms (1). In general, 8-oxoG is removed in eubacteria by an Fpg homologue whereas eukaryotes and archaea use an Ogg homologue. Eukaryotic Ogg1 will remove 8-oxoguanine, 8-oxoadenine, 2,6-diamino-4-hydroxy-5-formamidopyrimidine (Fapy), and me-Fapy when paired opposite a cytosine but demonstrates significantly less base excision activity when paired opposite A, T, or G (7, 8, 10–14, 16, 17). Should an Ogg1 remove 8-oxoguanine that has mispaired opposite adenine during replication, transversion of G·C to T·A would be fixed; thus the opposite-base specificity of Ogg1 functions to prevent mutations. The Ogg1 lyase (strand nicking) reaction is uncoupled from the base excision reaction and is much slower (11). In addition, the Ogg1 lyase reaction shows even greater specificity for the base opposite the lesion (11). The structure of hOGG1 has been solved in both unliganded (18) and liganded forms (19, 20). These structures demonstrate, as is seen with other DNA glycosylases, that the nucleoside lesion is everted from the DNA helix into a catalytic pocket, with concomitant bending of the DNA backbone. The residues that contact the everted 8-oxoguanine and the estranged cytosine have also been determined (19). Previously characterized Ogg proteins from eubacteria [Thermotoga maritima (21)] and archaea [Methanococcus jannaschii (22) and Archaeoglobus fulgidus (23)] belong to the Ogg2 clade (24) (Figure 1A) or to a separate class of Ogg (AGOG) (25). Members of the Ogg2 clade lack the N-terminal region of Ogg1 (22, 23) and demonstrate a low sequence identity to hOGG1 (13−19%). These enzymes do not demonstrate significant opposite base specificity (21–23). The archaeal Ogg from Pyrobaculum aerophilum (PaeAGOG, neither an Ogg1 nor Ogg2) also demonstrates no opposite base specificity and will cut single-stranded DNA (25).
Figure 1.
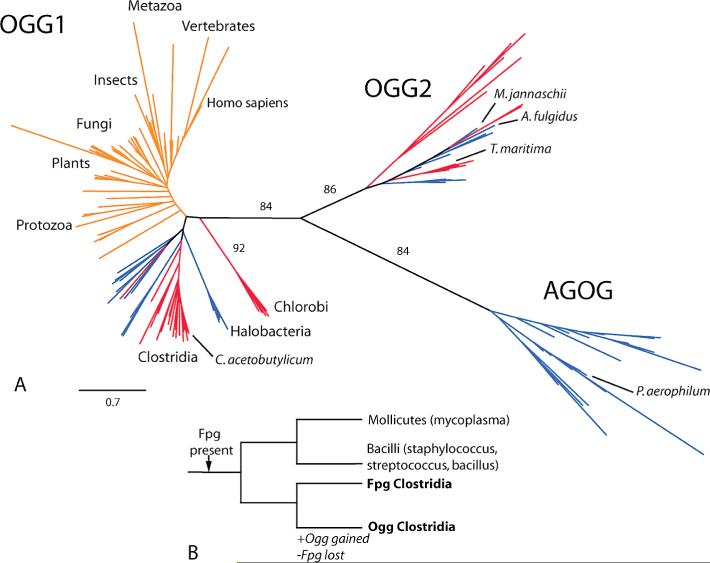
(A) Phylogenetic tree of Ogg homologues. Eukaryotes are denoted by orange lines, archaea by blue lines, and eubacteria by red lines. The edge length corresponding to 0.7 substitution per site is shown in the lower left of the figure. Significant bootstrap support is noted along relevant edges. (B) Phylogenetic tree of the Firmicutes. Transfer of Ogg to Clostridia occurred with simultaneous loss of Fpg.
Ogg1 homologues have now been identified in a significant number of bacterial species (24) (Figure 1A). The largest group of bacterial Ogg1 homologues occurs in Firmicutes, with most in the genus Clostridium, which are obligate anaerobic, Gram-positive, rod-shaped, endospore-forming bacteria. Many Clostridia colonize mammalian hosts and are also found in the soil. Clostridium acetobutylicum is a solvent-producing bacterium, originally isolated from soil, and was used industrially to produce the solvents acetone, butanol, and 2-propanol before the age of petroleum. Although the organism is anaerobic, it may encounter brief, nonlethal exposures to oxygen. The C. acetobutylicum homologue of hOGG1, CacOgg, shares the predicted helix—hairpin—helix domain and functional residues with hOGG1. However, CacOgg has a nonconserved substitution of the hOGG1 154R to M. In hOGG1, 154R has been shown (19) to directly contact the base opposite the damaged lesion, and replacement of hOGG1 154R with H abrogates opposite base specificity (19, 26).
Here we describe, for the first time, the expression, purification, and characterization of an Ogg1 cloned from eubacterial DNA. We show that CacOgg demonstrates reduced opposite base specificity and that the CacOgg variants M132R and F179Y partially restore opposite base specificity. The double variant M132R/F179Y shows increased discrimination against the opposite base A for both base removal and strand nicking but does not affect discrimination against opposite G or T during base removal. Furthermore, we demonstrate that duplex DNA is required for the CacOgg lyase reaction but not the glycosylase reaction.
MATERIALS AND METHODS
Sequence Identification, Phylogeny, and Structure Visualization
Homologous proteins were identified in the NCBI protein sequence database using BLAST (27) family tools based on conserved domain profiles, position-specific descriptions derived from an alignment of multiple sequences. NCBI Conserved Domain Database entries were used for identification of Fpg and RecA homologues [pfam06831 (28) and cd00983 (29), respectively]. Conserved domain profiles for Ogg1, Ogg2, and AGOG were constructed on the basis of sequence and structure information using CDTree (30). Sets of homologous sequences were aligned using T-Coffee (31) and MAFFT version 6.240 (32) and inspected using Pfaat (33), Jalview (34), or Seaview (35) to review both the identification and alignment processes, as well as to remove phylogenetically uninformative sites. Phylogenetic trees were constructed using the neighbor joining algorithm (36) [Phylip NEIGHBOR (37)] based on maximum likelihood-derived distances (Phylip PROTDIST). We used Phylip SEQBOOT to create 100 bootstrap replicates. The phylogenetic trees were used to assign bacterial sequences to the HhH-GPD Ogg1, Ogg2, and AGOG clades. Secondary structure predictions were obtained using PSIPRED (38). PyMOL (39) was used for structure visualization and graphics (Figure 2B).
Figure 2.
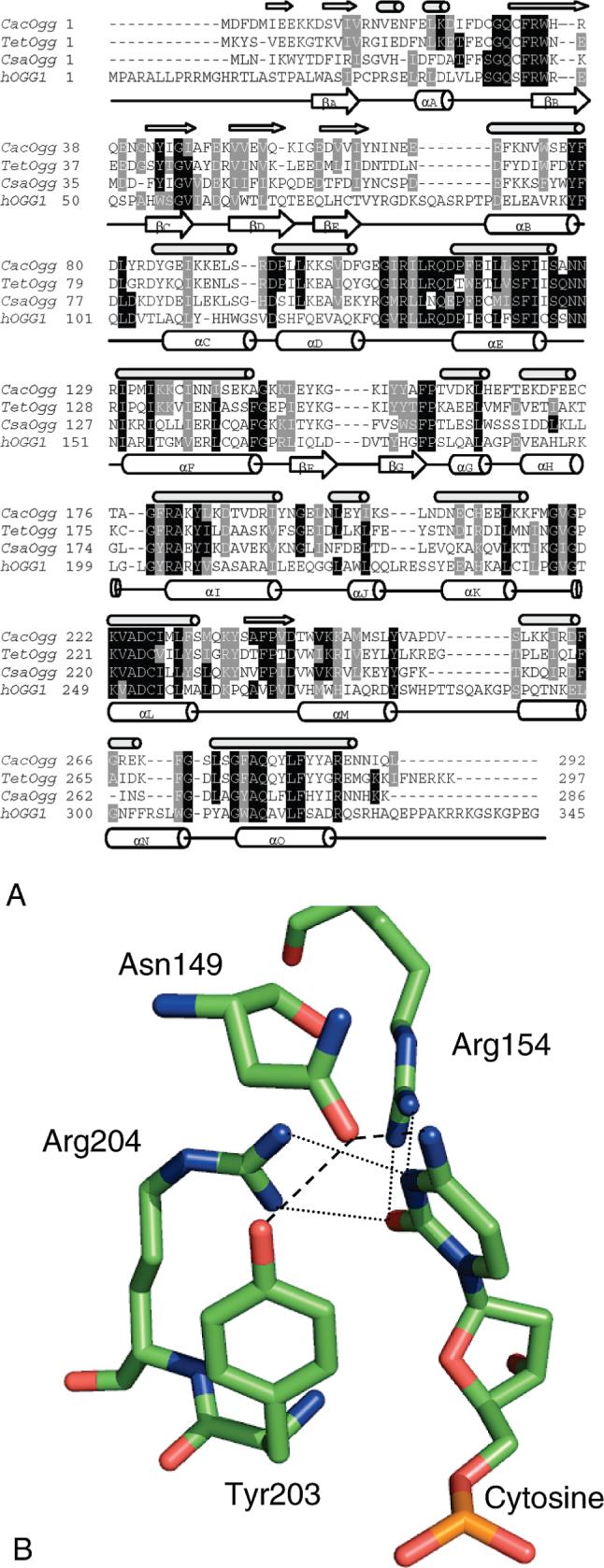
(A) Sequence alignment of CacOgg, hOGG1, and putative Oggs from Thermoanaerobacter ethanolicus (Tet) and Caldicellulosiruptor saccharolyticus (Csa). The PSIPRED prediction (36) of CacOgg secondary structure is indicated by small arrows ( β-sheets) and small cylinders (α-helices) directly above the CacOgg sequence. The hOGG1 secondary structure (18) is indicated by large arrows and cylinders drawn under the hOGG1 sequence. (B) Model of hOGG1 interactions with the base opposite the lesion as redrawn from the crystal structure (1EBM). Both dashed and dotted lines indicate H-bonding.
Cloning, Expressing, and Purifying Enzymes
Genomic DNA of C. acetobutylicum (ATCC 824) generously supplied by Dr. George N. Bennett (Rice University) was used to amplify and clone CacOgg into the pTYB2 vector. Site-directed mutants of CacOgg (CacOggM132R, CacOggF179Y, and the double variant CacOgg M132R/F179Y) were constructed using the Stratagene QuikChange XL site-directed mutagenesis kit. Wild-type and variant CacOggs were expressed in ER2566 fpg- E. coli cotransfected with a pLysS RIR vector (40). After overnight induction with IPTG at 16 °C, cell pellets were sonicated in lysis buffer (50 mM Tris-HCl, 500 mM NaCl, 1 mM EDTA, pH 8.0, containing 1 mM PMSF), clarified by centrifugation, loaded onto a chitin column according to the manufacturer's specifications [Impact system (New England Biolabs, Ipswich, MA)], and eluted using 50 mM dithiothreitol (DTT). Pooled fractions were then loaded onto a Q column using ÄKTAprimeplus (GE Healthcare Bio-Sciences Corp., Piscataway, NJ), eluted with a salt gradient, and concentrated on a Vivaspin 10000 MWCO ultraconcentrator. The concentrate was dialyzed into storage buffer (20 mM glycylglycine buffer, pH 7.6 at 4 °C, 100 mM NaCl, 1 mM DTT, and 50% glycerol) overnight at 4 °C and stored at −20 °C until use. hOGG1 and EcoFpg enzymes used in this paper were from our laboratory stocks (40). Enzyme concentration was determined by the Bio-Rad protein assay using bovine serum albumin as a standard. All enzymes were purified to apparent homogeneity (data not shown). As expected, CacOgg and variants CacOggM132R, CacOggF179Y, and CacOggM132R/F179Y migrate similarly, with a predicted MW of 34419 Da.
Substrates
A 35-mer oligonucleotide and its complementary strand were prepared by Midland Certified Reagent Co. (Midland, TX) and gel purified on a 20% urea gel. The sequence of the strand with the damage was 5′-TGTCAATAGCAAGXGGAGAAGTCAATCGTGAGTCT-3′, where X = 8-oxoguanine (8-oxoG), thymine glycol (Tg), 5-hydroxycytosine (5OHC), 5-hydroxyuracil (5OHU), 5,6-dihydrothymine (DHT), 5,6-dihydrouracil (DHU), uracil, or the normal base G. The oxidized damage guanidinohydantoin (Gh), a gift of Dr. Cynthia Burrows (University of Utah), was also used in the 14-mer sequence context 5′-GCGTCCAXGTCTAC-3′, where X = Gh. The damaged strand was 5′ end-labeled with [γ-32P]ATP using T4 polynucleotide kinase, ethanol precipitated, and mixed with unlabeled DNA at a 1:10 ratio. The oligonucleotide used in single turnover experiments was not mixed with unlabeled DNA. It was then annealed to its complementary strand with C, A, T, or G opposite the lesion in a 1:1 ratio (10 mM glycylglycine buffer, 50 mM NaCl, 1 mM EDTA, pH 8.0) by heating to 95 °C and slowly cooling to room temperature. Duplex or single-stranded oligonucleotides containing uracil were treated with EcoUDG to prepare abasic (AP) substrates.
Schiff Base Assays
The Schiff base assay (41) was used to estimate the fraction of active protein for kinetic calculations. Enzyme (100 nM) was reacted with various concentrations of substrate (25−250 nM) in the presence of the reducing agent sodium borohydride (NaBH4, 50 mM). The sodium concentration in the Schiff base reactions was adjusted to 100 mM final concentration using the CacOgg glycosylase assay buffer containing 50 mM NaCl. Reactions were electrophoresed on a 12% urea gel, and the gel bands were analyzed using a Molecular Imager FX (Bio-Rad). Active enzyme fractions of various preparations (maximum concentration of substrate trapped divided by the enzyme concentration, multiplied by 100) ranged from 45% to 100% active (data not shown).
DNA Glycosylase Reactions
Enzymes were incubated with substrates in CacOgg glycosylase assay buffer (10 mM glycylglycine buffer, pH 8.0 at 37 °C, containing 1 mM EDTA, 100 mM NaCl, 0.1 mg/mL BSA, and 5% glycerol) at 37 °C for various times as indicated in the figure legends. The reactions were terminated by addition of an equal volume of formamide and heated to 95 °C for 3 min. For quantitation of the base excision activity separately from the bifunctional enzyme activity, NaOH was added to reactions to a final concentration of 0.1 N prior to formamide addition. Reactions using AP substrates were incubated on ice following formamide addition. The substrate concentration was 5 nM and the enzyme concentration was 1 nM, unless otherwise noted in the figure legends. Reaction volumes were 10−100 μL as noted in the figure legends. Reactions were electrophoresed on a 12% urea gel, and the gel bands were analyzed using a Molecular Imager FX (Bio-Rad Laboratories, Inc., Hercules, CA).
Association of Glycosylase Transfer and Loss during Firmicute Evolution
We tested the hypothesis that the loss of Fpg and gain of Ogg by horizontal transfer in the Firmicute phylogeny are independent. Under this null hypothesis
where p(edge = e|#events = 1) is the probability of an event (loss or transfer) occurring on edge e conditional upon only one such event occurring in the phylogeny, le is the length of edge e, and L = ∑ele is the length of the tree. The probability of coincident events under the null hypothesis is
where {le} was obtained from a RecA phylogeny.
Enzyme Kinetic Analysis
The kinetic model includes glycosylase and lyase reactions as well as product binding (Scheme 1). Statistical analysis was performed using the R Language and Environment for Statistical Computing (42). In addition to the kinetic parameters, parameters were included to account for unreactive substrate as well as the small amount of added DNA present initially as either product. The differential equations associated with the reaction model were integrated using the R odesolve package (43). Point estimation was accomplished using the R stats package (42). Confidence intervals and regions were obtained using the profile likelihood method (44). Michaelis constants
where c ∈ {ES,EP}, and their confidence intervals were estimated directly, as opposed to propagating error obtained on the kinetic constants.
Scheme 1: Reaction Modela.
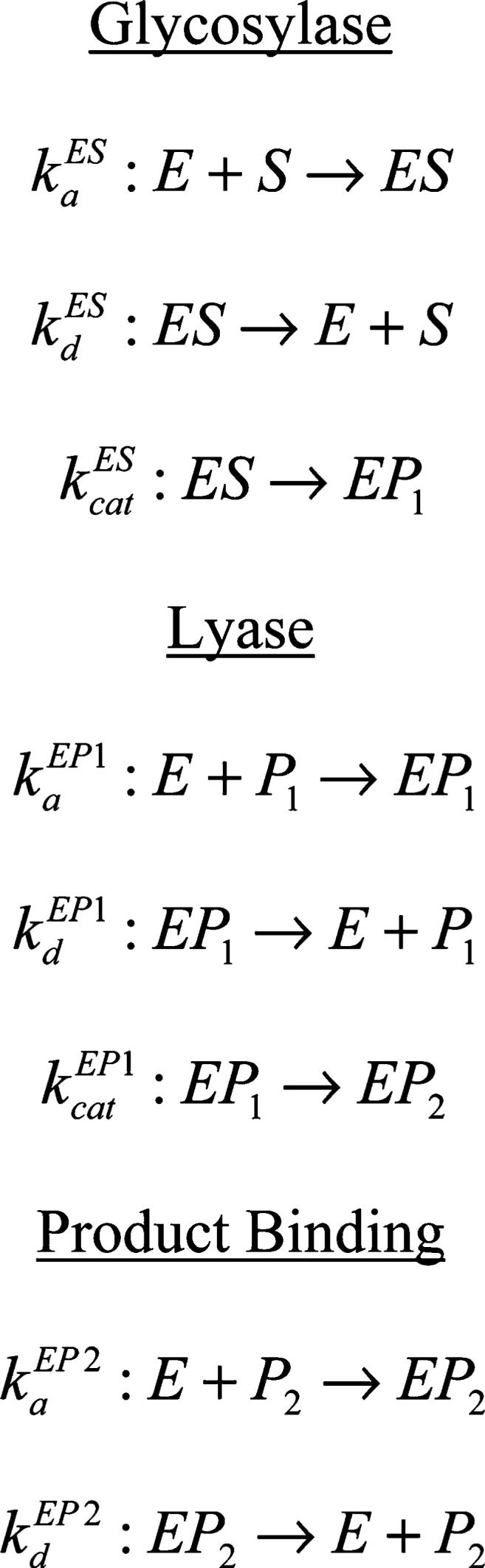
a Abbreviations: E, enzyme; S, oxidative lesion substrate; P1, product of the glycosylase reaction, the nascent AP substrate; P2, product of the lyase reaction, nicked DNA.
RESULTS
Bacterial Proteins Were Identified That Share a More Recent Common Ancestor with hOGG1 Orthologues Than with Other HhH-GPD Family Members
We found Ogg1 homologues primarily in two bacterial phyla (Figure 1A), the genus Clostridium and the genus Chlorobi. In addition, an Ogg1 homologue was found in one planctomycetes and numerous archaeal representatives. The homologues from Clostridia demonstrated about 25−31% amino acid sequence identity to hOGG1 and have the hallmarks of the Ogg1 sequence including the HhH-GPD region and the absolutely conserved catalytic residues (homologous to hOGG1 K249 and D268). Secondary structure predictions (Figure 2A) (38) suggest nearly the same pattern of β-sheet and α-helix structure as found in hOGG1 by Bruner et al. (19), a feature that distinguishes it from other HhH-GPD members, including Ogg2. Similarly, the Ogg1 clade and Ogg2 clade in the Ogg1/Ogg2 phylogeny (Figure 1A) are separated by an edge describing more than three substitutions per site, confirming that the two clades are very different in sequence. The topology of the phylogeny suggests horizontal transfer of Ogg1 prior to the divergence of eukaryotes from their common ancestor. Notably, the eubacterial and archaeal Ogg1 proteins are found primarily in anaerobes.
Gain of Ogg during firmicute evolution is associated with loss of a functional Fpg (Figure 1B). Only one clostridium, Clostridium phytofermentans, retains an Fpg sequence, which is expected to be inactive due to mutation of several key residues (data not shown). We used RecA protein sequences to estimate the topology and relative branch lengths of a firmicute phylogeny. The phylogenetic profiles of Ogg and Fpg were consistent with the existence of a single branch associated with both gain of Ogg and loss of selection pressure for maintaining a functional Fpg, which corresponds to statistical significance at p < 0.01. We note that an alternative phylogeny exists (45), but it is still consistent with our results.
C. acetobutylicum appears to have the full complement of enzymes involved in repair of 8-oxoguanine in particular [the GO system (46)] and base excision repair in general (reviewed in ref 3). Sequence comparison, using BLASTN to compare examples of known genes with the C. acetobutylicum genome, suggested C. acetobutylicum genes include, in addition to CacOgg, homologues of Udg (two examples), Nth, Mag, MutY, Nfo, Xth, Pol1, and DNA ligase. C. acetobutylicum also has seven genes coding for Nudix domain proteins, one of which is likely to be a homologue of MutT. Thus no anomalies in base excision repair systems were observed.
Comparison of Eukaryotic with Eubacterial Ogg1 Sequences Suggests Very Different Interactions with the DNA Strand Opposite the Lesion
The residues of hOGG1 that have been shown (19) to contact with the everted lesion (G42, C253, Q315, and F319) are observed in CacOgg (Figure 2A). Four of the six hOGG1 residues contacting the lesion-containing DNA strand [N150, N151, G245, K249, V250, and H270 (unless otherwise specified, positions refer to hOGG1)] are seen in CacOgg, the exceptions being R and W in place of N151 and H270, respectively. Thus, damaged base specificity and contacts to the lesion and to the lesion-containing strand are expected to be nearly identical between CacOgg and hOGG1.
However, there are substantial sequence differences between hOGG1 and CacOgg involving residues that contact the opposite base and opposite strand (Figure 2). The C. acetobutylicum homologue of hOGG1 has a nonconservative substitution of an amino acid (M for R154), which has been shown to recognize the Watson—Crick face of cytosine opposite 8-oxoG (19). Mutation of R154 in hOGG1 abrogates opposite base specificity (19, 26). CacOgg differs at a second position (F for Y203) presumed to play a substantial, although indirect, role in interactions with the opposite base. The Y203 hydrogen bonds to N149 through its hydroxyl, an interaction not available to F. N149 and thus presumably Y203 play a critical role in opposite strand interactions by contacting the base opposite the lesion (19). Y203 also inserts between bases in the opposite strand to bend the DNA (19), a function that could be replaced by F in CacOgg.
There are also differences between hOGG1 and CacOgg residues that contact the opposite strand at positions distant from the base opposite the lesion. Amino acids near the C-terminus of hOGG1 (Q287 and Q294, part of loop S286–Q294, between the αM and αN helices, Figure 2A) contact the DNA phosphate backbone and are conserved among mammalian sequences, with residue K289 being highly conserved among eukaryotes (data not shown). This contact is about 5 base pairs (one-half helical turn) 5′ to the estranged base (3′ to the lesion). In contrast, the CacOgg sequence, based on our alignment (Figure 2A), lacks the full length of the hOGG1 loop from S286 to Q294. CacOgg also lacks the C-terminal region of hOGG1 (residues 330−345), which may also contact the DNA (19, 47). It has been demonstrated that the C-terminus of hOGG1 enhances DNA binding to hOGG1 but is not required for catalysis (19, 47). The solved structures of hOGG1 do not include the C-terminus (18, 19, so the exact nature of C-terminal contacts with DNA is not known.
Specificity of CacOgg with Respect to Damaged Nucleotides Is Similar to hOGG1
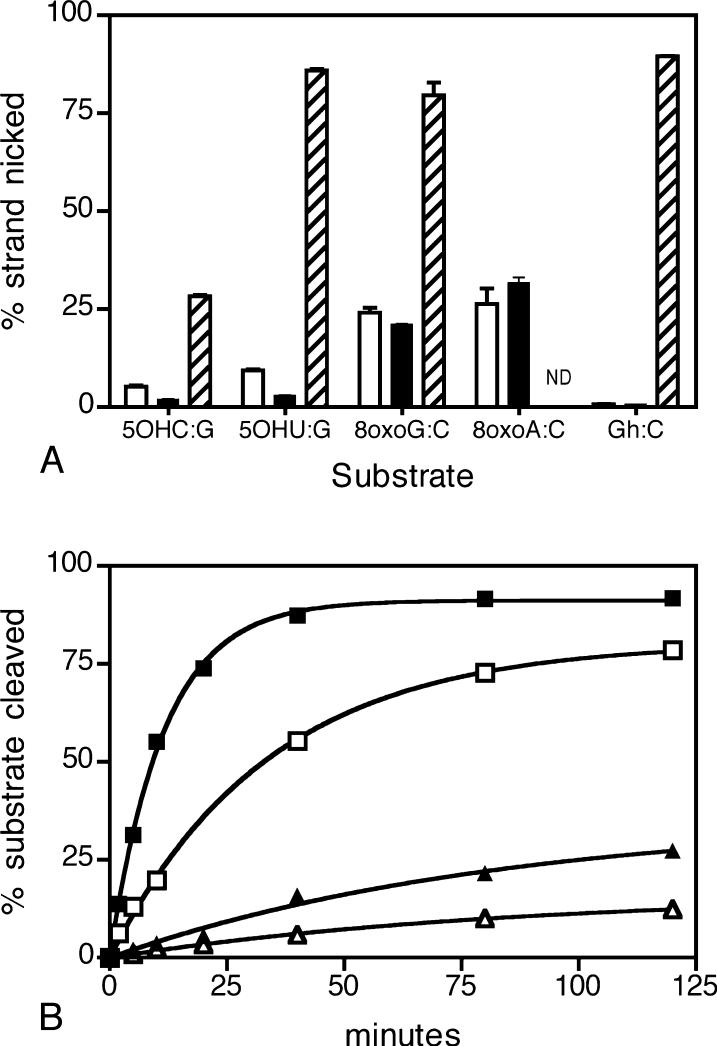
We measured CacOgg bifunctional activity on several DNA lesions using 35-mer oligonucleotides containing a single damage, mismatch, or normal DNA. Similar to hOGG1, CacOgg did not cleave oxidized pyrimidines DHT·A, DHU·G, or Tg·A, the deaminated pyrimidine U·G, the mismatch G·T, or normal DNA G·C, even at a 10-fold excess of enzyme over substrate concentration (data not shown). Like hOGG1 but unlike EcoFpg, CacOgg was unable to efficiently remove 5OHC·G or 5OHU·G (Figure 3A). CacOgg, like hOGG1, did remove 8-oxoG·C and 8-oxoA·C but not the further oxidation product of 8-oxoG, guanidinohydantoin (Gh) opposite C. EcoFpg recognizes both 8-oxoG·C and Gh·C [Figure 3A (48)]. The CacOgg glycosylase reaction was rapid relative to lyase activity (700-fold), similar to hOGG1. The time course (Figure 3B) demonstrates that, after the base excision reaction, the nascent AP oligo is released from the enzyme. More precisely, the difference between the amount of product accumulated between glycosylase and the lyase assays can exceed the amount of enzyme present, demonstrating enzyme turnover. Thus lyase activity, while much slower, does not limit glycosylase activity.
Figure 3.
(A) Bifunctional reaction substrate specificity. Bars indicate the percentage of substrate cleaved by CacOgg (no fill), hOGG1 (black), and EcoFpg (diagonals). The substrates are 5OHC·G, 5OHU·G, 8-oxoG·C, 8-oxoA·C, and Gh·C (ND = not determined). The reaction was performed under single turnover conditions (100 nM enzyme, 10 nM substrate) in a reaction volume of 10 μL, incubated at 37 °C for 30 min, and stopped by the addition of formamide as described in Materials and Methods. The results of three independent experiments are reported as the mean ± SEM. (B) The base excision and the bifunctional activity of CacOgg and hOGG1. Base excision activity is indicated by squares [CacOgg (■) and hOGG1 (□)], and bifunctional activity is indicated by triangles [CacOgg (▲) and hOGG1 (△)]. Each reaction was prepared as a pool of 160 μL, and 10 μL aliquots were removed at various times and added to NaOH (for base excision measurements) or formamide (for bifunctional measurements) as described in Materials and Methods. Substrate (20 nM) was in excess of enzyme (1 nM). A representative experiment is shown.
Comparison (Table 1) of the Km for the base excision reaction (24 nM) with the Km for the lyase reaction (1.2 nM) reveals that CacOgg binds preferentially to an AP oligonucleotide rather than an 8-oxoG oligonucleotide, which may reflect the additional work required to evert 8-oxoG or the use of binding energy during the glycosylase reaction (49).
Table 1.
Multiple Turnover Kinetics for CacOgga
| glycosylase |
lyase |
||||
|---|---|---|---|---|---|
| Km (nM) | kcat (min−1) | efficiency | Km (nM) | kcat (min−1) | efficiency |
| 24 (8, 39) | 31.2 (2.04, inf) | 1.32 (1.02, 1.74) | 1.2 (0.93, 1.6) | 0.0462 (0.045, 0.048) | 0.0372 (0.0306, 0.0558) |
Enzyme = 1 nM, substrate = 5−20 nM. Kinetics were modeled from time course data using each 8-oxoG·C and AP·C as substrates. 95% confidence intervals are listed in parentheses.
In summary, CacOgg demonstrates both specificity for the damaged base and reaction mechanism similar to hOGG1.
Opposite Strand Interactions of CacOgg Differ from hOGG1
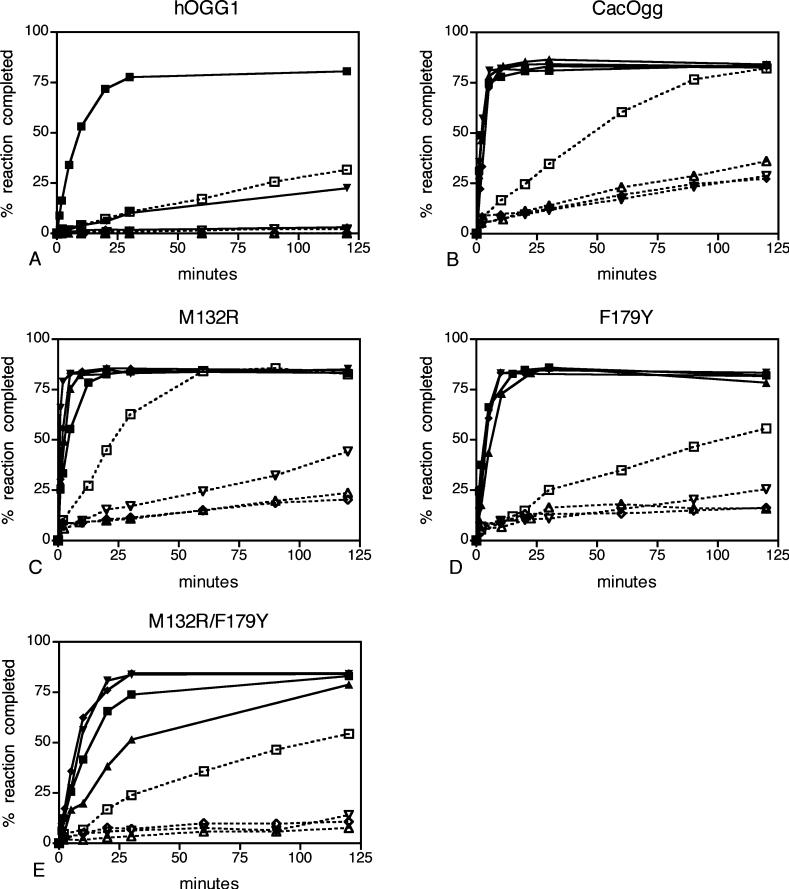
We examined the base excision and bifunctional activity of CacOgg on a duplex oligo with 8-oxoG paired opposite all four bases using multiple turnover conditions. CacOgg efficiently removed 8-oxoG opposite each of the four bases (Figure 4B, solid lines. In each panel of Figure 4, opposite base C is represented by squares, A with triangles, T with inverted triangles, and G with diamonds). As previously shown (8, 11, 14), hOGG1 removes 8-oxoG when opposite C but not when paired with G or A (Figure 4A, solid lines). When 8-oxoG is paired with T, hOGG1 will remove the lesion base but does not appear to turn over (Figure 4A, compare inverted triangles with squares).
Figure 4.
Opposite base specificity of the glycosylase and bifunctional activity in hOGG1, CacOgg, and variants. The glycosylase activity is indicated by closed symbols and solid lines, and bifunctional activity is indicated by open symbols and dashed lines. The substrate is 8-oxoG opposite C (■, □), A (▲, △), T (▼, ▽), and G (◆, ◇). (A) hOGG1. (B) CacOgg. (C) M132R. (D) F179Y. (E) M132R/F179Y. Each reaction was prepared as a 150 μL pool, and 10 μL aliquots were removed at t minutes and added to NaOH or formamide as described in Materials and Methods. Substrate (5 nM) was in excess of enzyme (CacOgg, 1 nM; hOGG1, 1.67 nM). A representative experiment is shown.
When both glycosylase and lyase activities are measured (Figure 4B, dashed lines), CacOgg turns over fastest with C opposite the lesion. Although CacOgg will remove the base and nick the strand when A, T, or G is opposite 8-oxoG, the reaction is slower. In contrast, the hOGG1 bifunctional activity opposite C is much weaker than that of CacOgg, and no bifunctional activity is observed when the opposite base is A, T, or G (Figure 4A, dashed lines). CacOgg product release is faster than the lyase reaction. Thus, unlike hOGG1, under excess substrate conditions the rate of product formation is limited by the lyase reaction, not product release. CacOgg nicks the strand of an abasic substrate·N (data not shown) at a similar rate to its bifunctional reaction on 8-oxoG·N, as is also observed for hOGG1 (50).
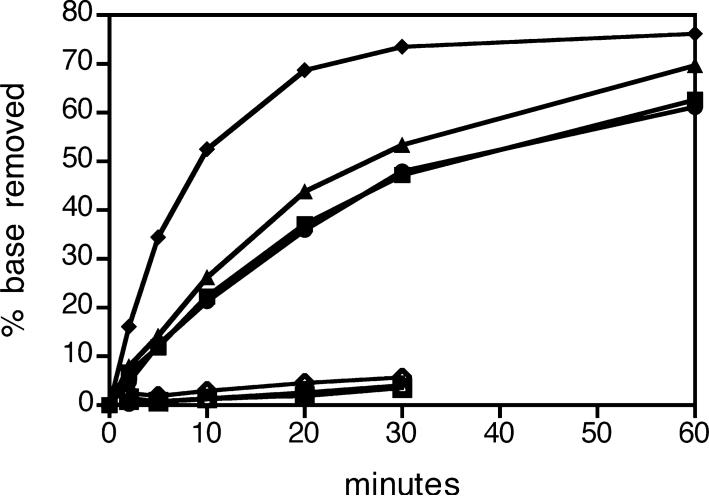
Unlike hOGG1 and EcoFpg (51), CacOgg removes 8-oxoG from a single-stranded substrate with an 8-oxoG lesion (Figure 5) and forms a Schiff base intermediate (data not shown). However, it cannot catalyze the bifunctional reaction (Figure 5). In contrast, Fpg (52), but not CacOgg, cleaves single-stranded AP substrates (data not shown).
Figure 5.
Glycosylase and bifunctional activity of CacOgg and variants on single-stranded DNA. CacOgg wild type (•), CacOggM132R (▲), CacOggF179Y (▼), and CacOgg double variant (◆) remove 8-oxoG from single-stranded DNA. Bifunctional activity is indicated by open symbols. Each reaction was prepared as a 120 μL pool, and 10 μL aliquots were removed at various times and added to NaOH as described in Materials and Methods to determine base removal or added to formamide to determine base removal plus strand nicking. Substrate (5 nM) was in excess of enzyme (1 nM). The results reported are of a typical experiment.
Site-Directed Mutation of a Single Amino Acid Changes the Opposite Base Specificity of CacOgg to More Closely Resemble That of hOGG1
As described earlier, CacOgg differs in sequence from hOGG1 at several positions that, in the latter enzyme, interact with the opposite strand. In particular, on the basis of the crystal structure of the hOGG1—substrate complex, R154 and Y203 (which align with M132 and F179, respectively, in CacOgg) appear to play an important role in interacting with C opposite 8-oxoG (Figure 2B). To test whether these residues play a role in CacOgg, we examined the opposite base specificity of three variants: M132R, F179Y, and M132R/F179Y.
Multiple turnover enzyme kinetics suggested that the M132R increased specificity for nicking opposite C relative to the other bases (Figure 4C, dashed lines). Specifically, M132R (Figure 4C) exhibited a more rapid accumulation of nicked product opposite C (open squares) relative to the CacOgg wild type (Figure 4B). While nicked product accumulation was also more rapid with T opposite the damage, reaction rates for A and G opposite the damage were slower. The F179Y (Figure 4D, dashed lines) bifunctional reaction rate was slower for all bases opposite the damage relative to the CacOgg wild type, although F179Y also exhibited a more rapid accumulation of nicked product opposite C relative to A, T, or G. While nicked product accumulation with the M132R/F179Y double variant (Figure 4E, dashed lines) was somewhat slower than with wild-type CacOgg, the lyase reaction opposite the other three bases was very slow, indicating increased preference for C on the opposite strand. All three variants nicked about an equimolar amount of substrate, or less, after 120 min when A or G was opposite the lesion, demonstrating no reaction turnover. The lyase activity was also compared, using an AP-containing oligonucleotide duplex opposite all four bases (data not shown). The results were similar to the bifunctional reaction.
CacOgg wild type, M132R, and F179Y all removed 8-oxoG rapidly under multiple turnover conditions, regardless of the opposite base (Figure 4B–D, solid lines). Surprisingly, M132R removed 8-oxoG opposite C less efficiently compared to A, T, or G, likely due to impaired product release (compare with single turnover results, below). Although the double variant removed 8-oxoG·A less efficiently than 8-oxoG·C, making it more like hOGG1 than CacOgg, it did not discriminate against 8-oxoG·T or 8-oxoG·G; thus the conversion to hOGG1-like activity was not fully achieved.
Additionally, all variants retained the ability to remove 8-oxoG from single-stranded DNA (Figure 5). M132R/F179Y, rather than losing recognition of single-stranded DNA, catalyzed lesion removal faster than wild type.
In contrast to the multiple turnover studies with limiting enzyme, in the single turnover experiment with excess enzyme the rate at which product accumulates is not heavily influenced by the rate at which product dissociates. Single turnover kinetic measurements shown in Table 2 clearly demonstrate that M132R and F179Y have increased specificity for base removal opposite C relative to A, about 17- and 12-fold, respectively. The overall efficiency of the M132R variant for 8-oxoG·C base removal was 4-fold greater than wild-type CacOgg, and for 8-oxoG·A base removal was 3.5-fold less than wild type. The efficiency of the F179Y variant on 8-oxoG·A base removal was significantly less (about 14-fold) than that of wild type. The combined effect (greater than 50-fold) of the two amino acid substitutions on discrimination (against 8-oxoG·A compared to 8-oxoG·C) was greater than seen with either individual substitution.
Table 2.
Single Turnover Kineticsa
| enzyme | substrate | glycosylase efficiency kcat/Kd (min−1 nM−1) | 95% conf intervals | lyase catalytic rate kcat (min−1) | 95% conf intervals |
|---|---|---|---|---|---|
| CacOgg | 8-oxoG·C | 0.420 | 0.378−0.456 | 0.0420 | 0.0378−0.0468 |
| CacOgg | 8-oxoG·A | 0.384 | 0.276−0.576 | 0.0252 | 0.0168−1.02E+06 |
| M132R | 8-oxoG·C | 1.800 | 1.620−2.040 | 0.0960 | 0.0900−0.1080 |
| M132R | 8-oxoG·A | 0.108 | 0.078−0.156 | 0.0342 | 0.0252−0.0444 |
| F179Y | 8-oxoG·C | 0.336 | 0.300−0.384 | 0.0234 | 0.0210−0.0258 |
| F179Y | 8-oxoG·A | 0.027 | 0.018−0.033 | 0.0078 | 0.0054−0.0090 |
| M132R/F179Y | 8-oxoG·C | 1.080 | 0.900−1.200 | 0.1140 | 0.0960−0.1380 |
| M132R/F179Y | 8-oxoG·A | 0.021 | 0.016−0.025 | 0.0072 | 0.0048−2.64E+14 |
| hOGG1 | 8-oxoG·C | 0.270 | 0.180−0.390 | 0.0234 | 0.0174−0.0582 |
The efficiency (kcat/Kd) of the glycosylase reaction and kcat of the bifunctional reaction for CacOgg and variants with the substrates 8-oxoG·C and 8-oxoG·A were obtained under single turnover conditions (0.025 nM 8-oxoG·C and 0.25, 1, or 2.5 nM enzyme for the glycosylase reaction and 2.5, 12, or 25 nM enzyme for the bifunctional reaction). Each reaction was prepared as a 70 μL pool, and 10 μL aliquots were removed at various times and added to NaOH or formamide as described in Materials and Methods.
The single turnover kinetic measurements of lyase activity (Table 2) reproduced the multiple turnover result and demonstrated that M132R increased the catalytic rate for the lyase reaction opposite C. F179Y also shows a 3-fold increase in preference for C compared to A, greater than the CacOgg wild type 1.7-fold preference for C compared to A. The ratio of lyase catalytic rate, opposite C relative to A, was substantially larger for the M132R/F179Y double variant relative to CacOgg wild type (16-fold), consistent with variant synergism.
These data are summarized graphically in Figure 6. Plotting the glycosylase efficiency versus the lyase catalytic rate (taken as the log of the values) yields a comparison of the properties of the wild-type enzyme and variants. The confidence intervals of the values are represented by the ellipses. The joint distributions of glycosylase efficiency and lyase catalytic rate with a substrate containing C on the opposite strand clearly indicate that, with high statistical significance, the M132R mutant (either alone, green, or in combination with the F179Y mutation, blue) increases both the glycosylase efficiency and the lyase catalytic rate relative to the CacOgg wild type (black).
Figure 6.
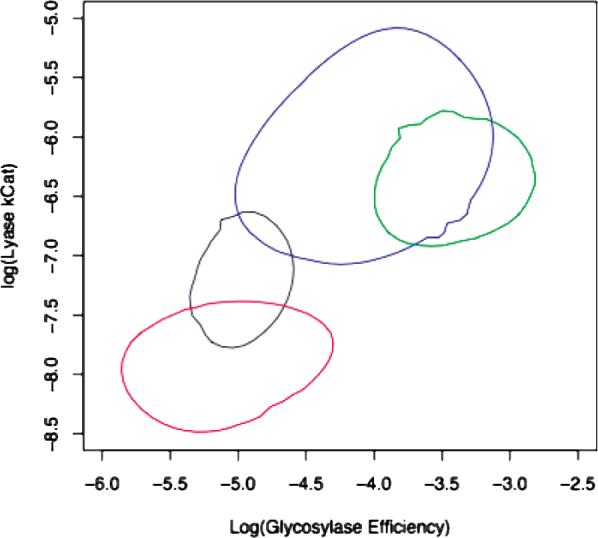
Plot of the mean (center of ellipse) and confidence intervals (edge of ellipse) of log glycosylase efficiency (nM−1 s−1) versus log lyase kcat (s−1. Key: CacOgg, black; M132 R, green; F179Y, red; M132R/F179Y, blue. The values were generated using the reaction model in Scheme 1 from data presented in Table 2.
DISCUSSION
CacOgg Exhibits Damaged Base Specificity Similar to hOGG1 but Novel Opposite Base Specificity
In bacteria, the GO system (46) protects organisms from the consequences of 8-oxoguanine-mediated transversions. Such transversions can proceed in either direction (G·C to T·A or vice versa) via two intermediates, Watson—Crick 8-oxoG·C and Hoogsteen 8-oxoG·A pairs. MutM (Fpg) or Ogg, which remove 8-oxoG opposite C, limits G·C → T·A transversions. MutT, which removes 8-oxodGTP from the nucleotide pool, limits T·A → G·C transversions. 8-oxoG·A, however, may result either from G·C or from T·A when the aforementioned protective mechanisms fail so the cellular phenotype with respect to base excision repair of 8-oxoG·A should reflect their relative efficiencies. Selection for MutY activity, present in eukaryotes and bacteria (including C. acetobutylicum), suggests that 8-oxoG·A pairs arise predominantly from G·A.
The ability of CacOgg to excise 8-oxoG opposite A is puzzling because it suggests that the fate of 8-oxoG·A pairs in the cell is indeterminate. It seems most likely that CacOgg and CacMutY are so effective that CacOgg-mediated 8-oxoG·C → T·A transversions, although possible, are rare, consistent with the hypothesis that 8-oxoguanine is primarily formed during transient aerobic exposure in anaerobes. The result might then be either no selective pressure for CacOgg opposite base specificity or altered selective pressures that impact opposite base specificity indirectly. For example, selection pressure for faster product release may be manifest by fewer interactions with the opposite strand and therefore reduced opposite base specificity.
A Single Amino Acid Substitution Alters Opposite Base Specificity of CacOgg
hOGG1 uses unique recognition of estranged C as a mechanism to exclude A, T, and G from the opposite base binding pocket (19). Kuznetsov et al. (53) find that discrimination of opposite bases occurs during an equilibrium binding step, when hOGG1 amino acid residues plug the space of the everted lesion effectively only when C is the estranged base. However, the opposite base specificity of CacOgg is very different from hOGG1. By examination of the hOGG1 structure and comparison to the CacOgg sequence, the reason for the minimal opposite base specificity during the CacOgg base excision reaction seems to lie in substitution of the amino acids in the opposite base binding pocket. First, the hydrogen-bonding contacts with the estranged nucleotide demonstrated by hOGG1 R154 are not possible with CacOgg M132. Substitution of the CacOgg M132 with R increased the opposite base specificity of the base removal reaction, although it was still able to remove 8-oxoG opposite A, T, or G rapidly. This variant enzyme also demonstrated an increased rate of strand nicking opposite C. M132R did not release the nascent AP site as readily when opposite C, which may have contributed to the faster lyase rate opposite C observed in this variant if more glycosylase—lyase reaction coupling occurred.
Our results address the possibility that, although the role of the planar ring of Y203 in hOGG1 is conserved by an F substitution, the role that the Y203 hydroxyl group has in stabilizing N149, which does contact the opposite base, may be more important in determining opposite base specificity. Replacing the CacOgg F179 with Y also increased opposite base specificity during base removal and strand nicking, although it was still able to remove the lesion easily regardless of the opposite base. The CacOgg F179Y variant demonstrated a decreased rate of strand nicking regardless of the opposite base. When both M132 and F179 were changed to analogous hOGG1 residues (the double variant), greater opposite base specificity during base removal was observed, as defined by greater discrimination against opposite A. Surprisingly, the double variant did not prevent base removal when G or T was opposite the lesion, suggesting that there are important aspects of the hOGG1 enzyme that contribute to opposite base discrimination but have yet to be identified.
The opposite base recognition by hOGG1 may also be affected by the opposite strand contacts of the loop S286–Q294 (5′ to the estranged base, 3′ to the lesion), which probably do not exist in CacOgg. Studies altering the amino acids of this loop in hOGG1 may be in order to determine the effect of the loop on opposite base specificity.
Wild-Type CacOgg Demonstrates Opposite Base Specificity during the Lyase Reaction
Although CacOgg shows no opposite base discrimination during the glycosylase reaction, it prefers C opposite during the lyase reaction. hOGG1 also has more stringent opposite base requirements during the lyase reaction than the base excision reaction. Bjoras et al. (18) demonstrate that upon hOGG1 binding of 8-oxoG a conformational change occurs, forcing the hydrogen bonds between N149 and K249 to be disrupted and allowing N149 and R204 to swing into contact with the estranged cytosine. At this point, hOGG1 has four contacts with the 8-oxoG base, an additional six contacts with the lesion-containing strand, and four contacts with the opposite base. In addition, hOGG1 may be contacting the opposite strand with the loop described above. The scenario expected for CacOgg is similar, except there may be only two contacts with the opposite base and no loop contact, since it is not present in CacOgg. When the AP substrate is bound, hOGG1 and CacOgg would not be expected to have the four contacts with 8-oxoG base; thus there would be a greater contribution from the contacts with the opposite base. Without 8-oxoG to force the N149 position shift observed by Bjoras et al. (18), the role of the opposite base in inducing conformational change to allow catalysis to occur may be greater.
This proposal is supported by our observations with single-stranded substrate catalysis. Because CacOgg cannot complete the bifunctional reaction on single-stranded DNA, the enzyme appears to require an opposite strand before the lyase reaction can occur (Figure 5). The opposite strand contacts may help to hold the apurinic site in the catalytic pocket as well as draw N149 out of the catalytic site. In contrast, when 8-oxoG is present in a single-stranded substrate, removal of the base by CacOgg, and the variants, can occur. In this case, insertion of the 8-oxoG base into the active site is sufficient to induce the conformational change for the glycosylase reaction, but without 8-oxoG to hold the site open or the opposite strand to induce an open site, the lyase reaction on single-stranded AP-containing DNA cannot occur.
Origin of Ogg1 in Prokaryotes
The phylogenetic analysis of Eisen and Hanawalt (54) suggests that an ancestor common to extant bacteria possessed Fpg but not Ogg1, while an ancestor of eukaryotes contained Ogg1. By our analysis horizontal transfer accounts for the presence of Ogg1 among eukaryotes and Clostridia, as well as other bacteria indicated in Figure 1A. Identification of the direction of the transfer would require rooting the phylogeny. The replacement of Fpg by Ogg among bacteria is consistent with the possibility that Ogg conferred better fitness for the environmental niche that certain members of Clostridia were occupying. However, associated loss of genes from bacteria and gain of genes from their environment that support comparable function are no doubt sufficiently frequent that the observed replacements are also consistent with drift. The apparent association of the gain of Ogg1 with an anaerobic niche is similarly consistent with selection for replacement of Fpg by Ogg1.
We show that CacOgg and EcoFpg have different substrate specificities (CacOgg will not remove Gh from duplex DNA but will remove 8-oxoG from single-stranded DNA. EcoFpg will not remove 8-oxoA from duplex DNA, nor can it remove 8-oxoG from single-stranded DNA, but it will nick single-stranded DNA at an abasic site) and different mechanisms of actions (uncoupled base excision and lyase for CacOgg versus coupled for EcoFpg). It is not clear whether these differences led to the replacement of Fpg with Ogg1 in Clostridia. The bacteria with Ogg1 are anaerobes, and several are thermophiles or green sulfur bacteria. It is interesting to speculate whether Fpg was lost before or after divergence to these phenotypes. The bacteria harboring Ogg1 have known associations with eukaryotes during evolution: the phototropic green sulfur bacteria are believed to be forerunners of plant chloroplasts, and several Clostridia are commensal with vertebrates.
Biological Significance of CacOgg Recognition of Single-Stranded DNA
It is interesting that CacOgg can effectively remove 8-oxoG from single-stranded DNA sequences. The homologous hOGG1 does not remove lesions from single-stranded DNA (50); however, the archaeal enzyme with functional homology, PaeAGOG, does (25). Lesion removal from single-stranded DNA has not been examined in Ogg2 enzymes (21–23). While the double variant becomes more hOGG1-like in its interactions with duplex 8-oxoG DNA, it can still remove 8-oxoG from single-stranded DNA, unlike hOGG1, supporting the idea that the residues lining the opposite base pocket are not the only contributors to opposite base specificity.
However, the relevance of the ability of these enzymes to remove lesions from single-stranded DNA is not apparent. The base excision reaction of CacOgg on single-stranded DNA is 14-fold slower than when removing lesions from double-stranded DNA, and DNA is rarely found in single-stranded form in the cell. It is likely that the ability to remove lesions from single-stranded DNA is a consequence of other functionalities of the enzyme and has not been selected against during evolution because the activity is so low that it is not of biological importance.
Kinetic Comparison of CacOgg and hOGG1
The glycosylase activity of CacOgg is much faster than that of hOGG1. Similar to hOGG1, the CacOgg lyase reaction is not coupled to and is slower than the glycosylase reaction. hOGG1 and CacOgg kinetics can be modeled with the same scheme, which predicts that the enzymes act as monofunctional glycosylases when the concentration of 8-oxoG exceeds that of the available AP site. Although enzyme binding to an AP site is tighter than binding to the 8-oxoG lesion, the off-rate of the enzyme from the AP site is of sufficient magnitude to expect that, when an excess of 8-oxoG is present, the enzyme releases nascent AP and rebinds to an 8-oxoG lesion. This perspective contradicts evidence that hOGG1 remains bound in complex with AP substrate after base removal (55). The kinetic scheme we propose is an attempt to reconcile the observation that both hOGG1 and CacOgg remove multiple lesions before significant strand nicking is observed, with the observed tighter binding of either Ogg with AP versus 8-oxoG. C. acetobutylicum has genes homologous to endonuclease IV (nfo) and exonuclease III (xth), which would be expected to rapidly repair most AP sites, similar to human. In the case of hOGG1, the enzyme may hand off the nascent AP DNA to the next enzyme in a cascade for further repair (56). Thus, in vivo, the lyase activity of either CacOgg or hOGG1 likely occurs only in the absence of significant amounts of 8-oxoG lesion.
Speculatively, the frequency of oxidative lesions can be expected to be different in humans compared to C. acetobutylicum. Aerobic respiration generates a constant number of oxidative lesions, probably within a relatively narrow range of rates. In an anaerobic organism, the organism may experience a much lower background rate of oxidative lesions, with occasional high concentrations of oxidative lesions upon infrequent exposure to oxygen. The most rapid repair is likely to occur when CacOgg acts as a monofunctional glycosylase, with nascent AP sites repaired by Nfo and Xth. The uncoupled base removal and AP lyase reactions of the Ogg enzyme may have been selected for during the loss of Fpg from this subgroup of bacteria.
ACKNOWLEDGMENT
The authors thank Dr. Cynthia Burrows (University of Utah) for the gift of Gh, Dr. George N. Bennett (Rice University) for the gift of C. acetobutylicum genomic DNA, Dr. Sylvie Doublie, Dr. Scott Kathe, and Jeffrey Blaisdell for helpful discussions, and Wendy Cooper and Alicia Holmes for purifying the enzymes used in this work.
Footnotes
This work was supported by NIH Grants RO1 CA33657 and PO1 CA098993, awarded by the National Cancer Institute. The computations were performed using resources of the Vermont Advanced Computing Center [supported by NASA (NNX 06AC88G)], the Vermont Cancer Center (see below for grant), and the Vermont Genetics Network [Grant P20 RR16462 from the INBRE Program of the National Center for Research Resources (NCRR), a component of the National Institues of Health (NIH)]. The automated DNA sequencing and phophorimaging was performed in the Vermont Cancer Center DNA Analysis Facility and was supported in part by Grant P30CA22435 from the NCI. The views expressed are those of the author and do not necessarily represent the views of the NCRR, NIH, NASA, or NCI.
Abbreviations: Ogg, 8-oxoguanine—DNA glycosylase; Fpg, formamidopyrimidine—DNA glycosylase; BER, base excision repair; 8-oxoG, 7,8-dihydro-8-oxoguanine; Fapy, 2,6-diamino-4-hydroxy-5-formamidopyrimidine; DTT, dithiothreitol; Tg, thymine glycol; 5OHC, 5-hydroxycytosine; 5OHU, 5-hydroxyuracil; DHT, 5,6-dihydrothymine; DHU, 5,6-dihydrouracil; Gh, guanidinohydantoin.
REFERENCES
- 1.Cathcart R, Schwiers E, Saul RL, Ames BN. Thymine glycol and thymidine glycol in human and rat urine: a possible assay for oxidative DNA damage. Proc. Natl. Acad. Sci. U.S.A. 1984;81:5633–5637. doi: 10.1073/pnas.81.18.5633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fraga CG, Shigenaga MK, Park JW, Degan P, Ames BN. Oxidative damage to DNA during aging: 8-hydroxy-2′-deoxyguanosine in rat organ DNA and urine. Proc. Natl. Acad. Sci. U.S.A. 1990;87:4533–4537. doi: 10.1073/pnas.87.12.4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wallace SS. AP endonucleases and DNA glycosylases that recognize oxidative DNA damage. Environ. Mol. Mutagen. 1988;12:431–477. doi: 10.1002/em.2860120411. [DOI] [PubMed] [Google Scholar]
- 4.Dizdaroglu M, Nackerdien Z, Chao BC, Gajewski E, Rao G. Chemical nature of in vivo DNA base damage in hydrogen peroxide-treated mammalian cells. Arch. Biochem. Biophys. 1991;285:388–390. doi: 10.1016/0003-9861(91)90378-v. [DOI] [PubMed] [Google Scholar]
- 5.Kuchino Y, Mori F, Kasai H, Inoue H, Iwai S, Miura K, Ohtsuka E, Nishimura S. Misreading of DNA templates containing 8-hydroxydeoxyguanosine at the modified base and at adjacent residues. Nature. 1987;327:77–79. doi: 10.1038/327077a0. [DOI] [PubMed] [Google Scholar]
- 6.Wood ML, Dizdaroglu M, Gajewski E, Essigmann JM. Mechanistic studies of ionizing radiation and oxidative mutagenesis: genetic effects of a single 8-hydroxyguanine (7-hydro-8-oxoguanine) residue inserted at a unique site in a viral genome. Biochemistry. 1990;29:7024–7032. doi: 10.1021/bi00482a011. [DOI] [PubMed] [Google Scholar]
- 7.van der Kemp PA, Thomas D, Barbey R, de Oliveira R, Boiteux S. Cloning and expression in Escherichia coli of the OGG1 gene of Saccharomyces cerevisiae, which codes for a DNA glycosylase that excises 7,8-dihydro-8-oxoguanine and 2,6-diamino-4-hydroxy-5-N-methylformamidopyrimidine. Proc. Natl. Acad. Sci. U.S.A. 1996;93:5197–5202. doi: 10.1073/pnas.93.11.5197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aburatani H, Hippo Y, Ishida T, Takashima R, Matsuba C, Kodama T, Takao M, Yasui A, Yamamoto K, Asano M. Cloning and characterization of mammalian 8-hydroxyguanine-specific DNA glycosylase/apurinic, apyrimidinic lyase, a functional mutM homologue. Cancer Res. 1997;57:2151–2156. [PubMed] [Google Scholar]
- 9.Arai K, Morishita K, Shinmura K, Kohno T, Kim SR, Nohmi T, Taniwaki M, Ohwada S, Yokota J. Cloning of a human homolog of the yeast OGG1 gene that is involved in the repair of oxidative DNA damage. Oncogene. 1997;14:2857–2861. doi: 10.1038/sj.onc.1201139. [DOI] [PubMed] [Google Scholar]
- 10.Rosenquist TA, Zharkov DO, Grollman AP. Cloning and characterization of a mammalian 8-oxoguanine DNA glycosylase. Proc. Natl. Acad. Sci. U.S.A. 1997;94:7429–7434. doi: 10.1073/pnas.94.14.7429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bjoras M, Luna L, Johnsen B, Hoff E, Haug T, Rognes T, Seeberg E. Opposite base-dependent reactions of a human base excision repair enzyme on DNA containing 7,8-dihydro-8-oxoguanine and abasic sites. EMBO J. 1997;16:6314–6322. doi: 10.1093/emboj/16.20.6314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roldan-Arjona T, Wei YF, Carter KC, Klungland A, Anselmino C, Wang RP, Augustus M, Lindahl T. Molecular cloning and functional expression of a human cDNA encoding the antimutator enzyme 8-hydroxyguanine-DNA glycosylase. Proc. Natl. Acad. Sci. U.S.A. 1997;94:8016–8020. doi: 10.1073/pnas.94.15.8016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nagashima M, Sasaki A, Morishita K, Takenoshita S, Nagamachi Y, Kasai H, Yokota J. Presence of human cellular protein(s) that specifically binds and cleaves 8-hydroxyguanine containing DNA. Mutat. Res. 1997;383:49–59. doi: 10.1016/s0921-8777(96)00045-6. [DOI] [PubMed] [Google Scholar]
- 14.Radicella JP, Dherin C, Desmaze C, Fox MS, Boiteux S. Cloning and characterization of hOGG1, a human homolog of the OGG1 gene of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 1997;94:8010–8015. doi: 10.1073/pnas.94.15.8010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klungland A, Rosewell I, Hollenbach S, Larsen E, Daly G, Epe B, Seeberg E, Lindahl T, Barnes DE. Accumulation of premutagenic DNA lesions in mice defective in removal of oxidative base damage. Proc. Natl. Acad. Sci. U.S.A. 1999;96:13300–13305. doi: 10.1073/pnas.96.23.13300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nash HM, Bruner SD, Scharer OD, Kawate T, Addona TA, Spooner E, Lane WS, Verdine GL. Cloning of a yeast 8-oxoguanine DNA glycosylase reveals the existence of a base-excision DNA-repair protein superfamily. Curr. Biol. 1996;6:968–980. doi: 10.1016/s0960-9822(02)00641-3. [DOI] [PubMed] [Google Scholar]
- 17.Lu R, Nash HM, Verdine GL. A mammalian DNA repair enzyme that excises oxidatively damaged guanines maps to a locus frequently lost in lung cancer. Curr. Biol. 1997;7:397–407. doi: 10.1016/s0960-9822(06)00187-4. [DOI] [PubMed] [Google Scholar]
- 18.Bjoras M, Seeberg E, Luna L, Pearl LH, Barrett TE. Reciprocal “flipping” underlies substrate recognition and catalytic activation by the human 8-oxo-guanine DNA glycosylase. J. Mol. Biol. 2002;317:171–177. doi: 10.1006/jmbi.2002.5400. [DOI] [PubMed] [Google Scholar]
- 19.Bruner SD, Norman DP, Verdine GL. Structural basis for recognition and repair of the endogenous mutagen 8-oxoguanine in DNA. Nature. 2000;403:859–866. doi: 10.1038/35002510. [DOI] [PubMed] [Google Scholar]
- 20.Radom CT, Banerjee A, Verdine GL. Structural characterization of human 8-oxoguanine DNA glycosylase variants bearing active site mutations. J. Biol. Chem. 2007;282:9182–9194. doi: 10.1074/jbc.M608989200. [DOI] [PubMed] [Google Scholar]
- 21.Im EK, Hong CH, Back JH, Han YS, Chung JH. Functional identification of an 8-oxoguanine specific endonuclease from Thermotoga maritima. J. Biochem. Mol. Biol. 2005;38:676–682. doi: 10.5483/bmbrep.2005.38.6.676. [DOI] [PubMed] [Google Scholar]
- 22.Gogos A, Clarke ND. Characterization of an 8-oxoguanine DNA glycosylase from Methanococcus jannaschii. J. Biol. Chem. 1999;274:30447–30450. doi: 10.1074/jbc.274.43.30447. [DOI] [PubMed] [Google Scholar]
- 23.Chung JH, Suh MJ, Park YI, Tainer JA, Han YS. Repair activities of 8-oxoguanine DNA glycosylase from Archaeoglobus fulgidus, a hyperthermophilic archaeon. Mutat. Res. 2001;486:99–111. doi: 10.1016/s0921-8777(01)00081-7. [DOI] [PubMed] [Google Scholar]
- 24.Denver DR, Swenson SL, Lynch M. An evolutionary analysis of the helix-hairpin-helix superfamily of DNA repair glycosylases. Mol. Biol. Evol. 2003;20:1603–1611. doi: 10.1093/molbev/msg177. [DOI] [PubMed] [Google Scholar]
- 25.Sartori AA, Lingaraju GM, Hunziker P, Winkler FK, Jiricny J. Pa-AGOG, the founding member of a new family of archaeal 8-oxoguanine DNA-glycosylases. Nucleic Acids Res. 2004;32:6531–6539. doi: 10.1093/nar/gkh995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Audebert M, Radicella JP, Dizdaroglu M. Effect of single mutations in the OGG1 gene found in human tumors on the substrate specificity of the Ogg1 protein. Nucleic Acids Res. 2000;28:2672–2678. doi: 10.1093/nar/28.14.2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 28.Bateman A, Birney E, Cerruti L, Durbin R, Etwiller L, Eddy SR, Griffiths-Jones S, Howe KL, Marshall M, Sonnhammer EL. The Pfam protein families database. Nucleic Acids Res. 2002;30:276–280. doi: 10.1093/nar/30.1.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marchler-Bauer A, Anderson JB, Cherukuri PF, DeWeese-Scott C, Geer LY, Gwadz M, He S, Hurwitz DI, Jackson JD, Ke Z, Lanczycki CJ, Liebert CA, Liu C, Lu F, Marchler GH, Mullokandov M, Shoemaker BA, Simonyan V, Song JS, Thiessen PA, Yamashita RA, Yin JJ, Zhang D, Bryant SH. CDD: a conserved domain database for protein classification. Nucleic Acids Res. 2005;33:D192–D196. doi: 10.1093/nar/gki069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marchler-Bauer A, Anderson JB, Derbyshire MK, DeWeese-Scott C, Gonzales NR, Gwadz M, Hao L, He S, Hurwitz DI, Jackson JD, Ke Z, Krylov D, Lanczycki CJ, Liebert CA, Liu C, Lu F, Lu S, Marchler GH, Mullokandov M, Song JS, Thanki N, Yamashita RA, Yin JJ, Zhang D, Bryant SH. CDD: a conserved domain database for interactive domain family analysis. Nucleic Acids Res. 2007;35:D237–D240. doi: 10.1093/nar/gkl951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Notredame C, Higgins DG, Heringa J. T-Coffee: a novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- 32.Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–66. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Caffrey DR, Dana PH, Mathur V, Ocano M, Hong EJ, Wang YE, Somaroo S, Caffrey BE, Potluri S, Huang ES. PFAAT version 2.0: a tool for editing, annotating, and analyzing multiple sequence alignments. BMC Bioinf. 2007;8:381. doi: 10.1186/1471-2105-8-381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Clamp M, Cuff J, Searle SM, Barton GJ. The Jalview Java alignment editor. Bioinformatics. 2004;20:426–427. doi: 10.1093/bioinformatics/btg430. [DOI] [PubMed] [Google Scholar]
- 35.Galtier N, Gouy M, Gautier C. SEAVIEW and PHYLO_WIN: two graphic tools for sequence alignment and molecular phylogeny. Comput. Appl. Biosci. 1996;12:543–548. doi: 10.1093/bioinformatics/12.6.543. [DOI] [PubMed] [Google Scholar]
- 36.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 37.Felsenstein J. Using the quantitative genetic threshold model for inferences between and within species. Philos. Trans. R. Soc. London, Ser. B. 2005;360:1427–1434. doi: 10.1098/rstb.2005.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McGuffin LJ, Bryson K, Jones DT. The PSIPRED protein structure prediction server. Bioinformatics. 2000;16:404–405. doi: 10.1093/bioinformatics/16.4.404. [DOI] [PubMed] [Google Scholar]
- 39.DeLano WL. The PyMOL molecular graphics system. DeLano Scientific; Palo Alto, CA: 2002. [Google Scholar]
- 40.Bandaru V, Blaisdell JO, Wallace SS. Oxidative DNA glycosylases: recipes from cloning to characterization. Methods Enzymol. 2006;408:15–33. doi: 10.1016/S0076-6879(06)08002-5. [DOI] [PubMed] [Google Scholar]
- 41.Dodson ML, Schrock RD, III, Lloyd RS. Evidence for an imino intermediate in the T4 endonuclease V reaction. Biochemistry. 1993;32:8284–8290. doi: 10.1021/bi00083a032. [DOI] [PubMed] [Google Scholar]
- 42.R Development Core Team . R Foundation for Statistical Computing. Vienna; Austria: 2007. [Google Scholar]
- 43.Setzer RW. odesolve: Solvers for Ordinary Differential Equations. R package version 5.0−18. 2007 [Google Scholar]
- 44.Cox DR. Principles of Statistical Inference. Cambridge University Press; Cambridge: 2006. [Google Scholar]
- 45.Wolf M, Muller T, Dandekar T, Pollack JD. Phylogeny of Firmicutes with special reference to Mycoplasma (Mollicutes) as inferred from phosphoglycerate kinase amino acid sequence data. Int. J. Syst. Evol. Microbiol. 2004;54:871–875. doi: 10.1099/ijs.0.02868-0. [DOI] [PubMed] [Google Scholar]
- 46.Michaels ML, Miller JH. The GO system protects organisms from the mutagenic effect of the spontaneous lesion 8-hydroxyguanine (7,8-dihydro-8-oxoguanine). J. Bacteriol. 1992;174:6321–6325. doi: 10.1128/jb.174.20.6321-6325.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hill JW, Evans MK. Dimerization and opposite base-dependent catalytic impairment of polymorphic S326C OGG1 glycosylase. Nucleic Acids Res. 2006;34:1620–1632. doi: 10.1093/nar/gkl060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Leipold MD, Workman H, Muller JG, Burrows CJ, David SS. Recognition and removal of oxidized guanines in duplex DNA by the base excision repair enzymes hOGG1, yOGG1, and yOGG2. Biochemistry. 2003;42:11373–11381. doi: 10.1021/bi034951b. [DOI] [PubMed] [Google Scholar]
- 49.Fersht A. In: Enzyme structure and mechanism. 2nd ed. Freeman WH, editor. New York: 1985. [Google Scholar]
- 50.Zharkov DO, Rosenquist TA, Gerchman SE, Grollman AP. Substrate specificity and reaction mechanism of murine 8-oxoguanine-DNA glycosylase. J. Biol. Chem. 2000;275:28607–28617. doi: 10.1074/jbc.M002441200. [DOI] [PubMed] [Google Scholar]
- 51.Boiteux S, O'Connor TR, Lederer F, Gouyette A, Laval J. Homogeneous Escherichia coli FPG protein. A DNA glycosylase which excises imidazole ring-opened purines and nicks DNA at apurinic/apyrimidinic sites. J. Biol. Chem. 1990;265:3916–3922. [PubMed] [Google Scholar]
- 52.Neto JB, Gentil A, Cabral RE, Sarasin A. Mutation spectrum of heat-induced abasic sites on a single-stranded shuttle vector replicated in mammalian cells. J. Biol. Chem. 1992;267:19718–19723. [PubMed] [Google Scholar]
- 53.Kuznetsov NA, Koval VV, Nevinsky GA, Douglas KT, Zharkov DO, Fedorova OS. Kinetic conformational analysis of human 8-oxoguanine-DNA glycosylase. J. Biol. Chem. 2007;282:1029–1038. doi: 10.1074/jbc.M605788200. [DOI] [PubMed] [Google Scholar]
- 54.Eisen JA, Hanawalt PC. A phylogenomic study of DNA repair genes, proteins, and processes. Mutat. Res. 1999;435:171–213. doi: 10.1016/s0921-8777(99)00050-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fromme JC, Bruner SD, Yang W, Karplus M, Verdine GL. Product-assisted catalysis in base-excision DNA repair. Nat. Struct. Biol. 2003;10:204–211. doi: 10.1038/nsb902. [DOI] [PubMed] [Google Scholar]
- 56.Sidorenko VS, Nevinsky GA, Zharkov DO. Mechanism of interaction between human 8-oxoguanine-DNA glycosylase and AP endonuclease. DNA Repair (Amsterdam) 2007;6:317–328. doi: 10.1016/j.dnarep.2006.10.022. [DOI] [PubMed] [Google Scholar]