Abstract
Tech is a RhoA guanine nucleotide exchange factor (GEF) that is highly enriched in hippocampal and cortical neurons. To help define its function, we have conducted studies aimed at identifying partner proteins that bind to its C-terminal PDZ ligand motif. Yeast two hybrid studies using the Tech C-terminal segment as bait identified MUPP1, a protein that contains 13 PDZ domains and has been localized to the post-synaptic compartment, as a candidate partner protein for Tech. Co-transfection of Tech and MUPP1 in hEK293 cells confirmed that these full-length proteins interact in a PDZ-dependent fashion. Furthermore, we confirmed that endogenous Tech co-precipitates with MUPP1, but not PSD-95, from hippocampal and cortical extracts prepared from rat brain. In addition, immunostaining of primary cortical cultures revealed co-localization of MUPP1 and Tech puncta in the vicinity of synapses. In assessing which PDZ domains of MUPP1 mediate binding to Tech, we found that Tech can bind to either PDZ domain 10 or 13 of MUPP1 as mutation of both these domains is needed to disrupt their interaction. Taken together, these findings demonstrate that Tech binds to MUPP1 and suggest that it regulates RhoA signaling pathways in the vicinity of synapses.
Keywords: RhoA, GEF, PDZ domain, synapse, hippocampus
Rho signaling pathways play a prominent role in regulating neuronal morphology and have been implicated in several inherited forms of mental retardation (Bennarroch, 2007). Accordingly, these findings have focused attention on understanding how these pathways are regulated in neurons. Although the prototypical Rho family members are expressed ubiquitously, many RhoGEFs and RhoGAPs, such as PDZ-RhoGEF and oligophrenin-1, display substantial tissue heterogeneity (Kuner et al., 2002; Govek et al., 2004). Presumably, this heterogeneity provides a mechanism for tailoring the activity of these powerful signaling networks to the needs of cell types that differ drastically in their morphology.
To understand how Rho signaling pathways operate in neurons, it will be important to identify and characterize the components of these pathways that are expressed in these cells. To this end, we have been studying a neuronal RhoA GEF, called Tech, that is highly enriched in brain compared to a broad range of peripheral tissues (Marx et al., 2005). However, it is also expressed in spinal cord and peripheral nerve (Maystadt et al., 2007). Within the brain it is expressed selectively in neurons of the cortex and hippocampus. Furthermore, expression of constitutively active Tech constructs in cultured cortical neurons decreases dendritic complexity, an effect that is mediated by RhoA (Marx et al., 2005). Additional evidence for the importance of Tech in neuronal function has been provided by the identification of a point mutation in the human orthologue of Tech, PLEKHG5, in a pedigree with an autosomal recessive form of lower motor neuron disease (Maystadt et al., 2007).
As identification of proteins that interact with Tech may provide important clues to understanding its regulation and function, we have conducted a yeast two hybrid screen to look for partner proteins that bind to the PDZ ligand motif located at Tech’s extreme C-terminus. As described below, these studies have led to the identification of MUPP1, a scaffold protein that contains 13 PDZ domains (Ullmer et al., 1998), as a protein that interacts with Tech in vivo.
Materials and methods
1. Plasmids and antibodies
Rat Tech cDNA was previously subcloned in pRK5-myc (Marx et al., 2005). Rat MUPP1 cDNA, and MUPP1 PDZ-domain dyad constructs, which contain two neighboring PDZ domains, were previously subcloned in pCDNA3 (Becamel et al. 2001; Sitek et al. 2003). A MUPP1 construct in which PDZ domain 10 has been inactivated, MUPP1m10, was constructed in two sequential mutagenic steps using the Stratagene Quikchange II XL site-directed mutagenesis kit, using full-length wild-type MUPP1 as a template. First we introduced the following mutations, L1623P and G1624V, the two most conserved residues in the canonical GLGL sequence of PDZ domain 10. We then proceeded to make the following substitutions: G1622R and L1625R. The nucleotide sequence for the mutated residues is CGCCCGGTACGG (translates to RPVR). A mutant version of the dyad MUPP1 fragment containing domains 10 and 11, MUPP1(m10&11), was also constructed using the Stratagene Quikchange II XL kit, using MUPP1(10&11) as template. Two amino acid mutations were introduced, namely L1623P and G1624V. MUPP1 constructs with C-terminal truncations were generated by introducing an amber termination signal just upstream of the truncated domains. HA-MUPP1, HA-MUPP1Δ10 and HA-MUPP1Δ7Δ10 were gifts from R. Javier (Baylor College of Medicine, Texas; Lee et al., 2000). GST-TAPP2 was a gift from M. Deak (University of Dundee, Scotland; Kimber et al., 2002).
The MUPP1 rabbit polyclonal antibody, mup5-6, was prepared previously against a GST fusion protein containing the segment located between PDZ domains 5 and 6 (Sitek et al. 2003). The C-terminal Tech rabbit polyclonal antibody was prepared previously (Marx et al., 2005) against a peptide fragment that extended from amino acid residues −41 to −25, counting from the extreme C-terminus of the rat sequence. The N-terminal Tech rabbit polyclonal antibody was prepared against an N-terminal fragment, amino acids 1 to 74, that was fused to GST. The following antibodies were obtained for commercial sources: myc mouse monoclonal (Invitrogen), GST goat polyclonal (Amersham Pharmacia), PSD-95 mouse monoclonal (Chemicon), bassoon mouse monoclonal (Stressgen).
2. Yeast two-hybrid screen
To construct the bait, a C-terminal fragment of rat Tech extending from amino acid 721 to 1039, the extreme C-terminus, was subcloned into the SalI and NotI sites of pBDLeu as a Gal4-DNA-binding domain fusion (GBD). A yeast two-hybrid screen was carried out by co-transforming pBDLeu Tech and a rat forebrain cDNA library fused to the Gal4 activation domain into the yeast strain y190. The brain library was provided by P. Worley and T. Lanahan. Approximately 1 × 106 transformed yeast cells were incubated for 10 days at 30°C on yeast nitrogen base and agar medium supplemented with 40 mg/L adenine, 30 mg/L lysine, 20 mg/L methionine, 30 mg/L tyrosine, 20 mg/L uracil, 50 mM 3-aminotriazole but lacking histidine, leucine and tryptophan.
3. Transfection of and co-precipitation from hEK 293 cells
Human embryonic kidney (hEK) 293 cells were cultured in 10% fetal bovine serum (FBS) , 2 mM glutamine, 100 U/mL penicillin, 100 µg/mL streptomycin, high glucose Dubelcco’s modified Eagle medium (DMEM) from Invitrogen. We performed transfections of hEK 293 cells using Lipofectamine and PLUS reagents (Invitrogen). We incubated 5 µg of each plasmid with a 60 to 80% confluent culture in a 10-cm dish for 3 to 4 h in serum-free DMEM. Cells were harvested 16 to 20 h after transfection in 500 µL IP buffer (50 mM Tris HCl pH 7.5; 150 mM NaCl; 4 mM EDTA; 1% (v/v) NP40; 2X Complete mini EDTA-free protease inhibitor complex from Roche). Cell homogenates were incubated on ice for 10 min then centrifuged at 15000 g for 10 min. The supernatant was then precleared with 50 µL 50% (in IP buffer) immobilized protein A on agarose (Pierce) for 30 min with rotation. The precleared supernatant was divided in two 300-µL fractions for plus and minus antibody assays. For plus antibody immunoprecipitation, we used 1 µL myc antibody or 0.7 µL MUPP1 antibody. The supernatants were incubated with the indicated antibodies at 4°C with rotation for 1.5 h. We then added 50 µL 50% immbolized protein A agarose, and extended the incubation for an additional 1 h. The agarose beads were washed three times with IP buffer. We added Laemli sample buffer and boiled for 2 minutes. Sample proteins were separated on 8 or 10% polyacrylamide gels, and then transferred to a nitrocellulose membrane (Bio-Rad) for immunoblotting. Membranes were probed with myc antibody (1:5000), MUPP1 antibody (1:70000), or GST antibody (1:1000) in 1% nonfat dry milk, 0.05% Tween-20 in Tris-buffered saline.
4. Brain extracts and co-IP from brain
Hippocampi were rapidly dissected on ice from adult rats. Two hippocampi were Dounce homogenized in 1 mL IP buffer. Tissue homogenate was incubated on ice for 10 min, and then centrifuged at 15000 g for 10 min. Tissue extract was precleared as described above for hEK 293 cell extracts, followed by immunoprecipitation with 5 µL Tech antibody or 0.7 µL MUPP1 antibody. Immunoblotting was performed with Tech antibody (1:500), MUPP1 antibody (1:70000) or PSD-95 antibody (1:1000). For immunoprecipitation from cortical lysates, we used approximately 150 mg of tissue, a comparable mass to that of two hippocampi.
5. Neuronal culture and immunocytochemistry
Hippocampi or cerebral cortices were dissected from E17 rat embryos. Tissue was treated with papain and DNase, followed by trituration in neuronal medium: neurobasal medium (Invitrogen), 5% FBS (Hyclone), 2 mM GlutaMAX-I (Invitrogen), 100 U/mL penicillin, 100 µg/mL streptomycin. Resulting cell suspension was passed through a 70-µm filter and cells were then plated over polylysine-coated coverglasses at a density of 1100 cells/mm3. Neurons were fed glia-conditioned, 1% FBS neuronal medium supplemented with 2% B27 (Invitrogen) every 3 to 4 days.
Fourteen-day old in vitro neurons were washed with phosphate buffered saline (PBS) and fixed in 8% formaldehyde in PBS for 10 min., permeabilized with 0.1% Triton X-100 and blocked with 2% (w/v) bovine serum albumin (BSA) in PBS for 1 h at room temperature. Since both Tech and MUPP1 antibodies were generated in rabbits, we used two approaches to perform double staining for these antigens. In one we decreased the titer of Tech antibody so that it could only be detected with the enhanced sensitivity provided by Tyramide amplification. In this approach, incubation with Tech antibody (1:1000) overnight at 4°C was followed by another 4°C overnight incubation with biotinylated anti-rabbit IgG (1:2000). Avidin-Biotinylated enzyme complex (Vectastain ABC from Vector) followed by Cy3-conjugated Tyramide (TSA Fluorescence Systems from Perkin Elmer) were used following the secondary antibody step. In the other approach cultures were processed for staining with Tech antibody (1:600) and fluorophore-conjugated anti-rabbit IgG. The secondary antibody step was followed by an additional blocking step with unconjugated anti-rabbit IgG (1:250; Jackson ImmunoResearch), prior to adding MUPP1 antibody (1:5000). Both methods worked well and we confirmed that omission of either primary antibody abolished staining with the corresponding fluorophore. To process cultures for triple staining for Tech, MUPP1 and Bassoon, cultures were stained first for Tech using the Tyramide approach, and then incubated overnight at 4°C with Bassoon (1:2000) and MUPP1 (1:2000) antibodies. Cells were then incubated for 1 h at room temperature with secondary antibodies: FITC-conjugated anti-mouse IgG (Vector) and Cy5-conjugated anti-rabbit IgG (Jackson ImmunoResearch). Omission of MUPP1 antibody blocked the Cy5 signal, confirming that Cy5 secondary does not detect Tech antibody under these conditions. In control experiments we confirmed that preincubation of the Tech C-terminal antibody with its antigen peptide abolished staining.
6. GST pull-down assay
BL21-Gold(DE3) bacteria (Stratagene) were transformed with GST constructs, and single colonies were grown in a Lysogenic Broth (LB) starter culture overnight. 200 mL LB were inoculated with 5 mL starter culture at 37°C with shaking for approximately 2 h until absorbance at 600 nm reached between 0.6 and 0.8. Culture was induced with 0.25 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) and grown at 30°C for 4 h. Cells were pelleted by centrifugation at 3000 × g for 15 min. Cells were resuspended in lysis buffer [50 mM Tris pH 8.0, 150 NaCl, 0.5% (v/v) NP-40, 1 × Complete EDTA-free protease inhibitor complex (Roche)]. Resuspension was incubated with lysozyme for 0.5 h, then sonicated to homogenize lysate. Lysate was centrifuged at 15000 × g for 30 min. Supernatant was collected and stored at −80°C until use.
Cleared lysates were thawed and protein concentration was determined with BCA assay (Pierce), according to manufacturer’s instructions. Glutathione-sepharose beads (Amersham-Pharmacia) were washed and resuspended in lysis buffer, to make a 50%-bead slurry. 200 µL bead slurry was incubated with 2 µg bacterial lysate for 1 h at 4°C. Beads were washed with lysis buffer.
Tech transfected hEK 293 cells were harvested 20 h after transfection in lysis buffer. Homogenates were cleared by centrifugation, as described in immunoprecipitation procedure. Cleared homogenates were precleared with unbound 100 µL glutathione-sepharose bead slurry fo 1 h at 4°C. Extracts were then incubated with 100 µL of GST protein-bound glutathione beads for 2 h at 4°C. Beads were then washed with lysis buffer. Laemli sample buffer was added to beads. Samples were boiled, and separated by polyacrylamide gel electrophoresis for analysis with Coomassie stain or immunoblotting.
Results
Interaction of recombinant Tech and MUPP1 proteins
To identify candidate proteins that interact with the type I PDZ ligand sequence motif present at the C-terminus of Tech, SEV (Songyang et al., 1997), we performed a yeast two hybrid screen of a rat brain cDNA library using a C-terminal Tech fragment as bait. We isolated three inserts that encoded C-terminal fragments of MUPP1, all of which contained PDZ domain 10. In addition, we found two clones that encoded GIPC, a PDZ domain-containing protein that has been reported previously to interact with Tech (Liu and Horowitz, 2006). Accordingly, we focused on analyzing Tech’s interaction with MUPP1. To check that the MUPP1 clones did not represent false positives, we used a MUPP1 fragment that extends from PDZ domain 10 to the end of the protein to confirm that induction of beta-galactosidase reporter activity depended on the presence of both the C-terminal Tech bait fragment and this MUPP1 prey fragment.
To assess whether Tech is able to bind to full-length MUPP1, we co-transfected hEK293 cells with a rat MUPP1 construct and a Tech construct containing an N-terminal myc tag. Both of these recombinant proteins were readily detected by immunoblotting (Figure 1; leftmost lane). Following co-transfection with myc-Tech and MUPP1 constructs, lysates were processed for immunoprecipitation with either myc or MUPP1 antibodies. As expected, immunoprecipitation with MUPP1 antibodies brought down Tech and vice versa (Figure 1; second and third lanes from left). To check whether these proteins interact in a PDZ-dependent fashion, we generated a truncated Tech construct (TechΔPDZ) missing the final three amino acids, SEV, thought to be critical for mediating interaction of this type I PDZ ligand motif with PDZ domains (Songyang et al., 1997). Although this truncated construct displayed similar levels of expression as the full-length Tech construct, it does not co-precipitate with MUPP1, confirming that their interaction is PDZ-dependent (Figure 1; three lanes labeled TechΔPDZ + MUPP1). In addition, this negative result demonstrates that the co-precipitation observed with full-length Tech and MUPP1 is not due to non-specific adherence of these constructs to the agarose beads used in these studies.
Figure 1. Co-immunoprecipitation of recombinant Tech and MUPP1: Dependence on Tech PDZ ligand motif.
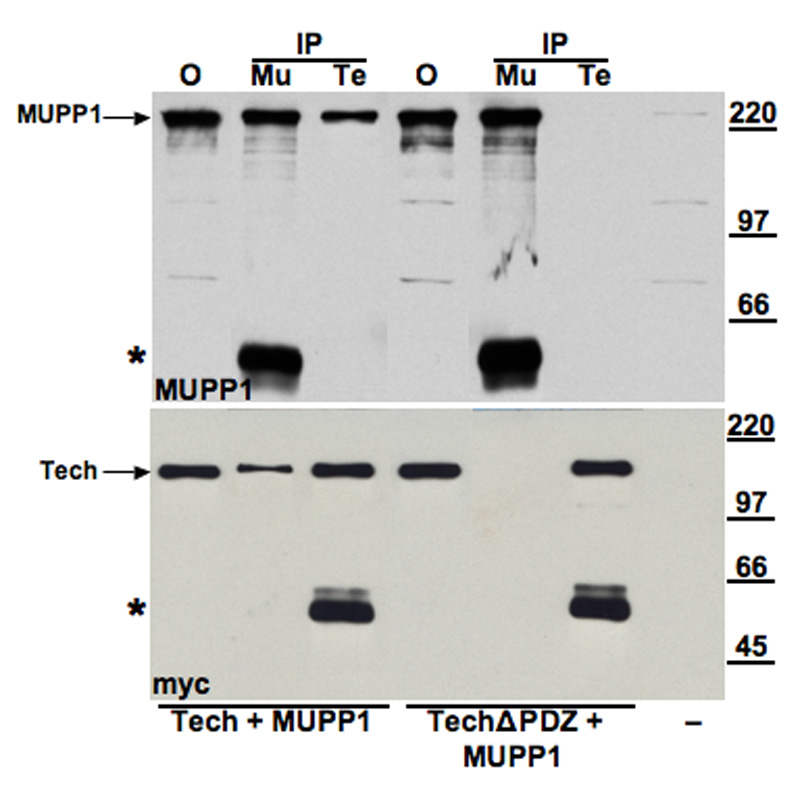
Lysates obtained from hEK 293 cells that were either untransfected (−; far right lane) , or co-transfected with MUPP1 and either myc-Tech or myc-TechΔPDZ constructs were processed for immunoprecipitation with either MUPP1 antibody (Mu) or myc antibody (Te) and analyzed by immunoblot. Upper blot was incubated with MUPP1 antibody; lower blot with myc antibody to detect Tech. Leftmost lane shows levels of recombinant MUPP1 and mycTech (arrows) in the offered sample (O). As shown in the adjacent lanes labeled IP, both MUPP1 and myc-Tech proteins are present in the pellet obtained from immunoprecipitations performed with either MUPP1 or myc antibodies. In contrast, as shown in the lanes labeled TechΔPDZ + MUPP1, co-transfection with the truncated Tech construct blocks co-immunoprecipitation of these proteins. Asterisks near the bottom of each blot mark the location of bands formed by the antibodies used for immunoprecipitation. In this and subsequent figures showing results of co-immunoprecipitation studies, the “offered” sample lane contains 2.5% of the total sample volume processed for immunoprecipitation. 25% of the immunoprecipitate was added to each of the ip lanes. Thus, bands of equal intensity in the offered and ip lanes indicate that 10% of the protein being assayed was immunoprepicipitated.
Association of endogenous Tech and MUPP1 proteins
To assess whether these proteins also bind to each other in vivo, we performed similar co-precipitation studies on extracts prepared from rat hippocampus or cortex, two regions enriched with Tech. For these studies, we used Tech antibodies directed to either the N- or C-terminal portions of Tech, both of which are able to immunoprecipitate Tech from brain lysates (Figure 2A; upper blot). We found that both antibodies also brought down MUPP1 (Figure 2A; lower blot). Although the extreme C-terminus of Tech would be expected to be inaccessible as an epitope if bound to MUPP1, these C-terminal antibodies were generated against a segment located further upstream, amino acids −25 to −41 (Marx et al., 2005). To check the specificity of these immunoprecipitation studies, we confirmed that neither of these proteins is in the immunoprecipitate if Tech antibodies are omitted from this procedure (Figure 2A and B). In addition, we checked that pre-incubation of the C-terminal antigen peptide with the C-terminal, but not the N-terminal Tech antibody, blocked precipitation of both these proteins.
Figure 2. Endogenous MUPP1 and Tech co-immunoprecipitate from rat hippocampus.
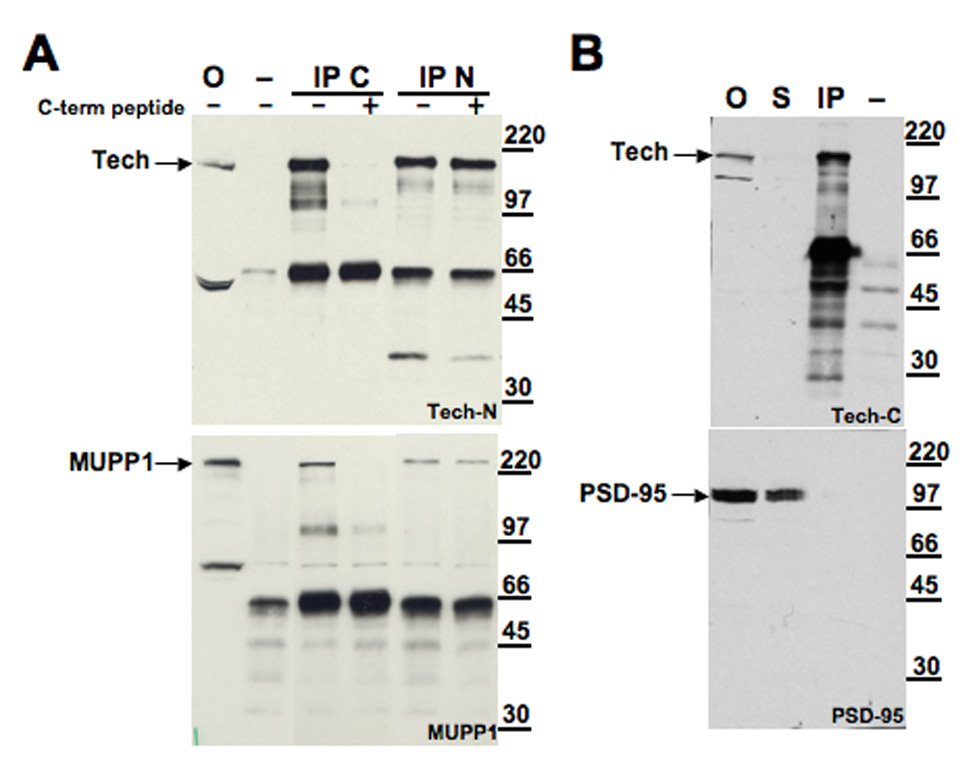
Immunoblots shown in his figure were performed on samples obtained from rat hippocampal extracts. (A) In this panel, the lanes show, from left to right, offered sample (O), minus antibody control (−) and IP using either C or N terminal Tech antibodies (IP C and IP N, respectively). Antibodies were preincubated with C-terminal antigen peptide as indicated above the blot. Blots were probed with N-terminal Tech (top) or MUPP1 (bottom) antibodies. (B) Immunoblots presented in this panel show, from left to right, offered sample (O), supernatant from immunoprecipitation (S), IP with C terminal Tech antibody (IP) and minus antibody control (−). Blots were probed with either C-terminal Tech (Tech-C; top) or PSD-95 (bottom) antibodies.
To check the specificity of the Tech-MUPP1 interaction, we also examined whether Tech would also co-precipitate with PSD-95, a multiple PDZ domain containing protein localized to synaptic zones (Kim and Sheng, 2004) that recognizes a “class I” PDZ motif like that found at the C-terminus of Tech. We found that under similar conditions, PSD-95 did not co-precipitate with Tech, even though Tech protein is markedly depleted from the supernatant (Figure 2B).
Previous immunohistochemical studies of Tech in brain sections have demonstrated that it is expressed in pyramidal cell bodies in both the hippocampus and cortex with staining extending into dendrites (Marx et al., 2005). Similarly, MUPP1 staining has been shown to be present in neuronal dendrites in these areas as well (Sitek et al., 2003). However, MUPP1 is more broadly expressed in brain and peripheral tissues than Tech. As MUPP1 has been localized to post-synaptic densities and our findings, presented above, indicate that Tech binds to MUPP1 in brain, we conducted immunohistochemical studies to assess whether Tech and MUPP1 co-localize at synaptic sites. Immunostaining of low density cortical cultures revealed Tech-positive puncta that extend from the cell body into proximal and distal dendritic processes (Figure 3B). To assess whether Tech and MUPP1 puncta co-localize, we focused on distal dendritic processes as isolated puncta were easier to visualize in this compartment. To estimate the percentage of MUPP1 puncta that were also stained for Tech, we selected over 20 MUPP1 puncta in each of 6 high power fields, while unaware of whether they were also stained for Tech. Each of these MUPP1-positive puncta was then scored as Tech positive or negative. Using this approach, we found that 65 +/− 6% (mean +/− sem, n=6) of MUPP1-positive puncta were also Tech-positive (Figure 3C–E).
Figure 3. Co-localization of Tech and MUPP1 puncta at synaptic sites.
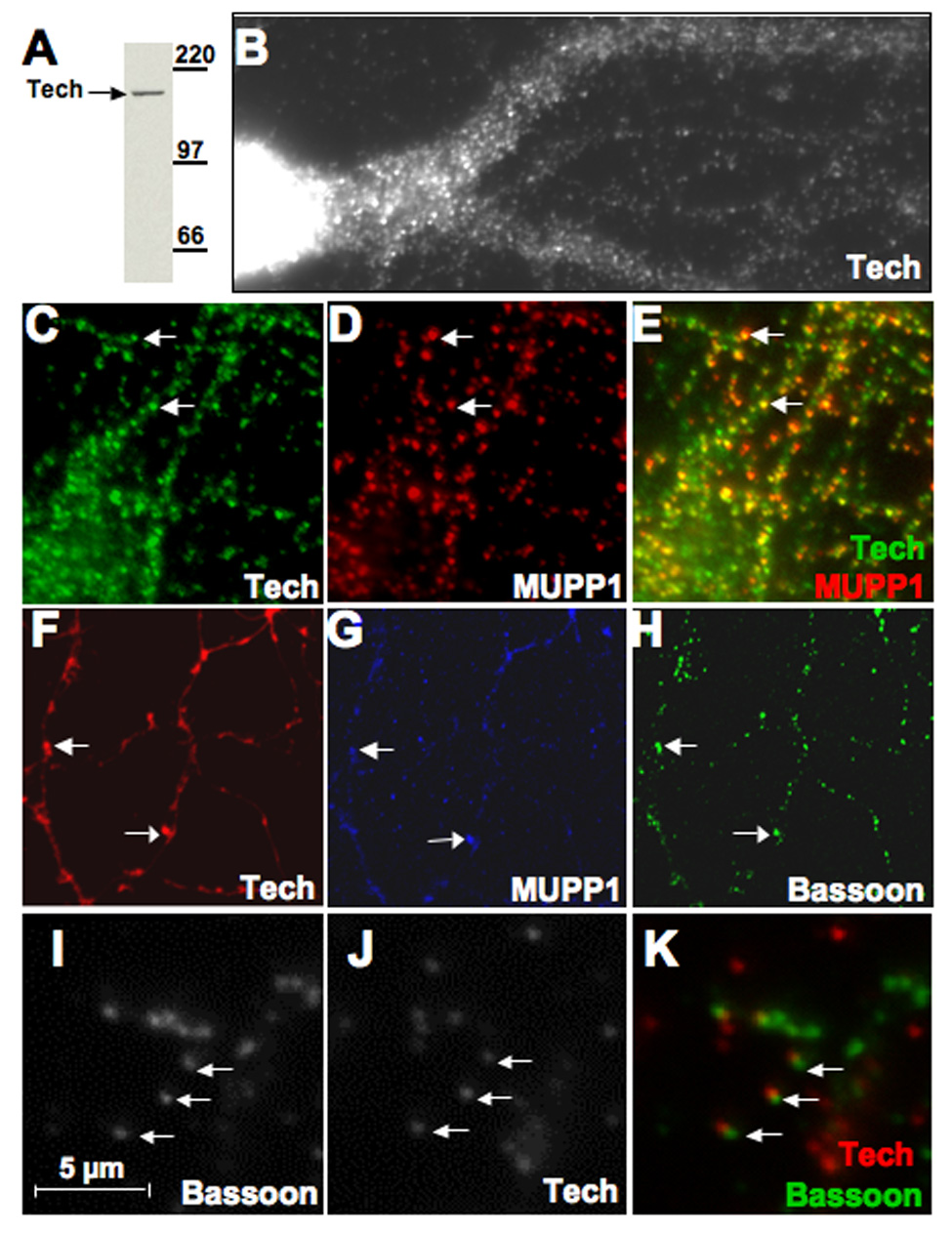
(A) Immunoblot shows specificity of C-terminal Tech antibody on culture extract. (B)Tech immunostaining in the cell body and proximal dendrites of a cultured hippocampal neuron (14 DIV) shows highly punctate pattern. (C–E) Double staining for Tech and MUPP1 in distal dendritic processes reveals many examples (arrows) of co-localization in isolated puncta. (F–H) Images of fluorescent staining shown in these panels were taken of the same field of cultured hippocampal neurons (14 DIV) processed for triple staining with antibodies against (F) Tech, (G) MUPP1 and (H) Bassoon, a presynaptic marker. Arrows point to two examples of Tech and MUPP1 co-localization juxtaposed to Bassoon puncta. (I–K) Higher power image of double immunostaining for Bassoon and Tech shows that Tech and bassoon puncta are offset from each other (arrows).
To assess whether Tech and MUPP1 co-localize at synaptic sites, we processed cultures for triple staining with antibodies to Tech, MUPP1 and bassoon, a pre-synaptic marker (Garner et al., 2000). In these studies, we found that puncta positive for both and Tech and MUPP1 are closely juxtaposed to puncta labeled with bassoon (Figure 3F–H). Higher power images of cultures double-stained for Tech and bassoon showed that these puncta are slightly offset, consistent with a post-synaptic localization of Tech (Figure 3 I–K). Using the scoring approach employed above to estimate co-localization of bassoon and Tech, we found that 45 +/− 4% (mean +/− sem, n=11 high power fields) of bassoon puncta located in distal dendrities are also Tech positive. Taken together, these findings indicate that Tech is located in the post-synaptic compartment and is associated with MUPP1.
Tech binds to several MUPP1 PDZ domains
Analysis of the interaction of other proteins with MUPP1 has revealed that they interact preferentially with one or two of its PDZ domains. For example, the 5-HT2C receptor binds selectively to PDZ domain 10 (Becamel et al., 2001). In contrast, TAPP1 and 2, bind to domains 10 and 13 (Kimber et al., 2002). Accordingly, information on the selectivity of MUPP1 partner proteins for individual PDZ domains may be helpful in determining the variety of signaling complexes that can form on this scaffold. To identify which of the MUPP1 PDZ domains mediate binding to Tech, we monitored the interaction of Tech with a series of MUPP1 fragments composed of two adjacent PDZ domains. Following co-transfection of Tech with each of these fragments in hEK293 cells, lysates were processed for immunoprecipitation studies. We found that two of these MUPP1 fragments, PDZ 9&10 and PDZ 10&11, co-precipitated with Tech, whereas none of the others tested did (Figure 4A). These findings suggest that PDZ domain 10 might mediate binding of MUPP1 to Tech and fit with the initial yeast two hybrid studies which yielded C-terminal MUPP1 fragments that included PDZ domain 10. However, these studies must be interpreted with caution as several of the PDZ domain dyad constructs did not express well and so could not be evaluated properly in this assay. Nevertheless, these findings suggest that PDZ domain 10 is sufficient to mediate the Tech-MUPP1 interaction.
Figure 4. MUPP1 PDZ10 binds to Tech.
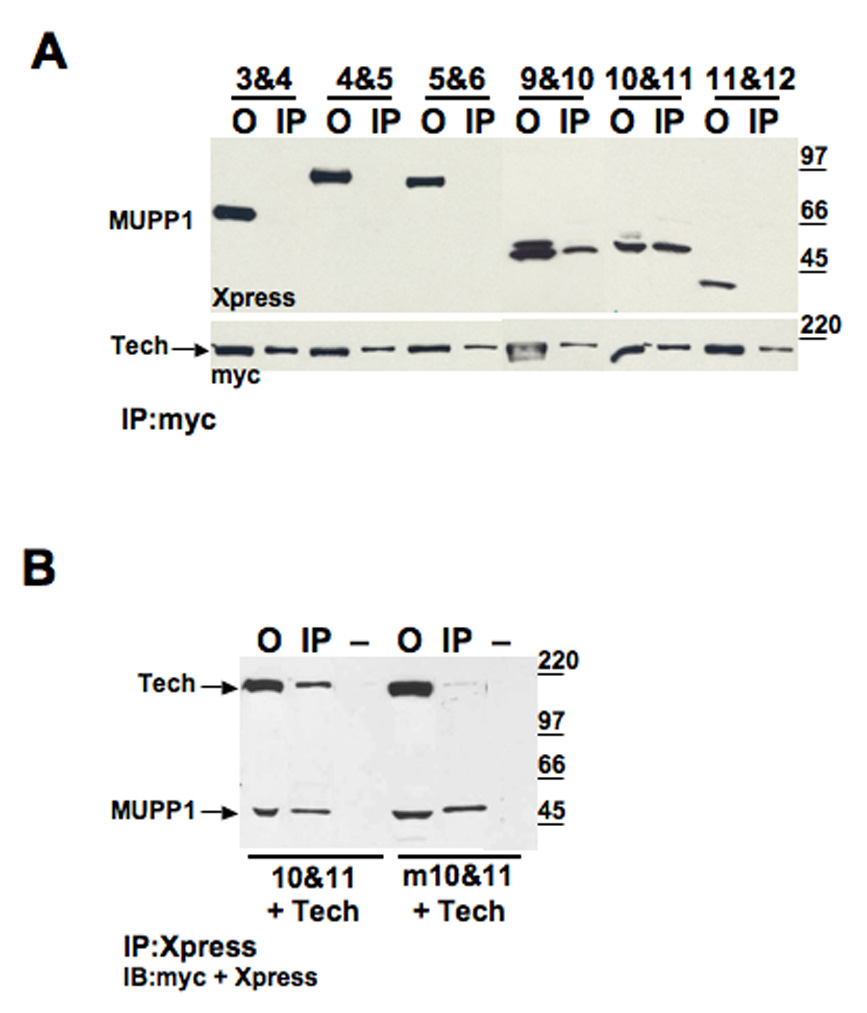
(A) HEK 293 cells were transfected with myc-tagged Tech and one of the Xpress-tagged MUPP1 fragments containing two contiguous PDZ domains. Offered sample (O) and pellet obtained by immunoprecipitation with anti-myc (IP) are shown for each of the tested constructs. Immunoblots were probed with anti-Xpress (top) to detect MUPP1 constructs, and anti-myc (bottom) to detect Tech. (B) HEK 293 cells were co-transfected with myc-tagged Tech and either MUPP1 10&11 or MUPP1 m10&11, which has a two-residue mutation in the conserved GLGL sequence of PDZ domain 10 (mutated to GPVL). From left to right, immunoblot shows offered sample (O), immunoprecipitation with anti-Xpress (IP) and minus antibody control (−) for both co-transfections. The same blot was probed with Xpress and myc antibodies.
Characterization of PDZ domains has led to the identification of a highly conserved sequence that is critical for mediating their interaction with PDZ ligands, GLGΦ, where Φ is a bulky hydrophobic amino acid, F/I/L/M/V (Doyle et al., 1996; Morais Cabral et al., 1996). To check that Tech’s interaction with PDZ domain 10 is mediated by this classical PDZ interaction, we confirmed that mutation of two of these residues, L1623P and G1624V, in domain 10, greatly decreased the ability of the PDZ 10&11 construct to interact with Tech (Figure 4B).
To check directly whether the PDZ 10 domain when expressed alone is able to interact with Tech and to test the possible involvement of some of the other PDZ domains that did not express well in hEK293 cells, we tested the ability of several recombinant GST-MUPP1 PDZ domain constructs generated in bacteria to interact with recombinant Tech generated in 293 cells. As expected, we found that PDZ domain 10 binds to Tech. However, we also found that a dyad construct containing PDZ domains 12 and 13 binds to Tech as well. In contrast, a construct containing PDZ domains 8 and 9 does not bind to Tech (Figure 5). In parallel experiments using the TechΔPDZ construct, we confirmed that interactions of Tech with the domain 10 and domain 12&13 constructs are dependent on Tech’s PDZ ligand motif. These findings suggest that Tech can bind to both domain 10 and either domain 12 or 13. Since in previous experiments, a construct containing domains 11&12 was inactive (Figure 4A), we infer that this interaction is likely mediated by domain 13 not 12. Previous studies analyzing the interaction of a pair of phosphoinositide binding proteins, TAPP1 and 2, with MUPP1 demonstrated that these proteins are able to bind to PDZ domains 10 and 13 (Kimber et al., 2002). Thus, our results obtained using MUPP1 fragments suggest that Tech may share this binding profile.
Figure 5. GST-PDZ domain pull-downs.
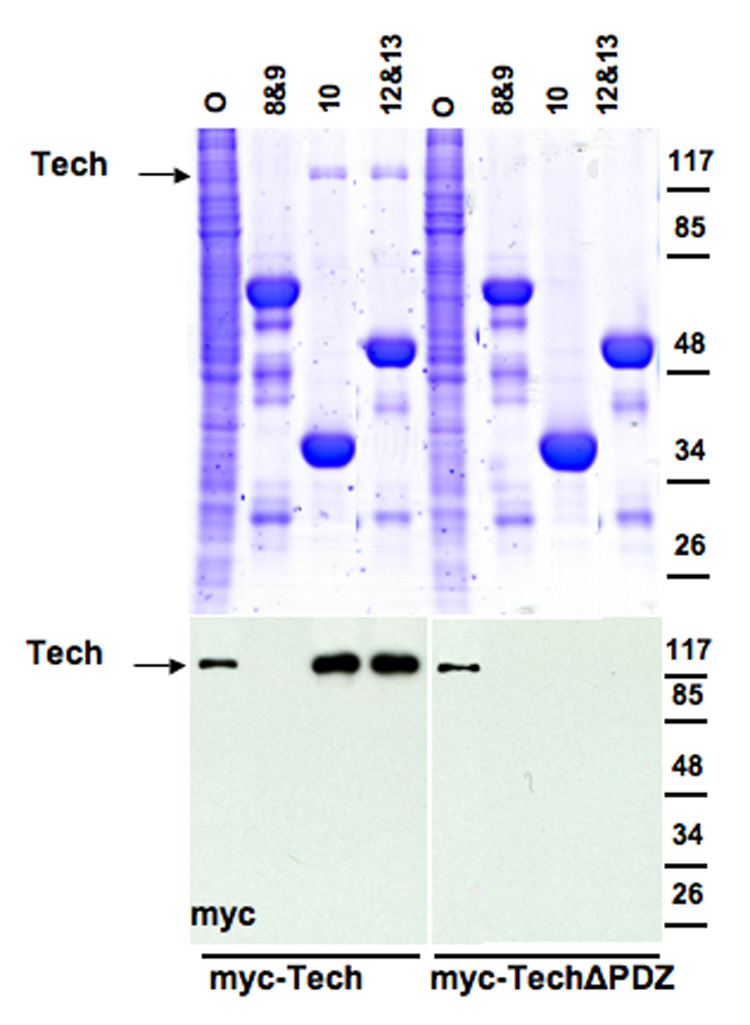
GST-fused PDZ domain constructs were expressed in bacteria. Bacterial lysates were bound to glutathione beads, which were then used to pull down either recombinant Tech or TechΔPDZ expressed in hEK 293 cells. Gels were loaded with offered sample from hEK 293 lysate (O), and with the pellets from GST pull downs, from beads bound to GST-PDZ constructs with domains: 8&9, 10, or 12&13, as labeled. Top panel shows polyacrylamide gel stained with Coomassie blue to visualize total protein. Strong bands represent GST-tagged PDZ domain constructs. Bottom panel shows immunoblots of the same samples probed with anti-myc to detect recombinant Tech protein, indicated by arrow.
Based on these findings, we proceeded to check whether PDZ domains 10 and 13 are necessary for the interaction of MUPP1 with Tech by examining the effect of mutating or deleting these PDZ domains in the full-length construct. We found that mutation of the entire GLGL sequence to RPVR in domain 10 of the full-length MUPP1 construct did not impair its co-precipitation with Tech (Figure 6A), consistent with our previous findings suggesting that domain 13 may also bind to Tech. To assess the role of PDZ domain 13 in the context of the full-length MUPP1 protein, we tested two MUPP1 constructs for co-precipitation with Tech following co-transfection in hEK293 cells. One construct consisted of wild type MUPP1 that was truncated after domain 12 (MUPP1Δ13); in the other, we truncated the m10 construct after domain 12 (MUPP1m10Δ13). We found that truncation of domain 13 alone did not impair binding to Tech, however, disruption of both domains 10 and 13 markedly reduced the level of Tech in the immunoprecipitate (Figure 6B). In contrast, deletion of domains 7 and 10 did not reduce Tech binding (data not shown).
Figure 6. Role of MUPP1 PDZ domains 10 and 13 in Tech interaction.
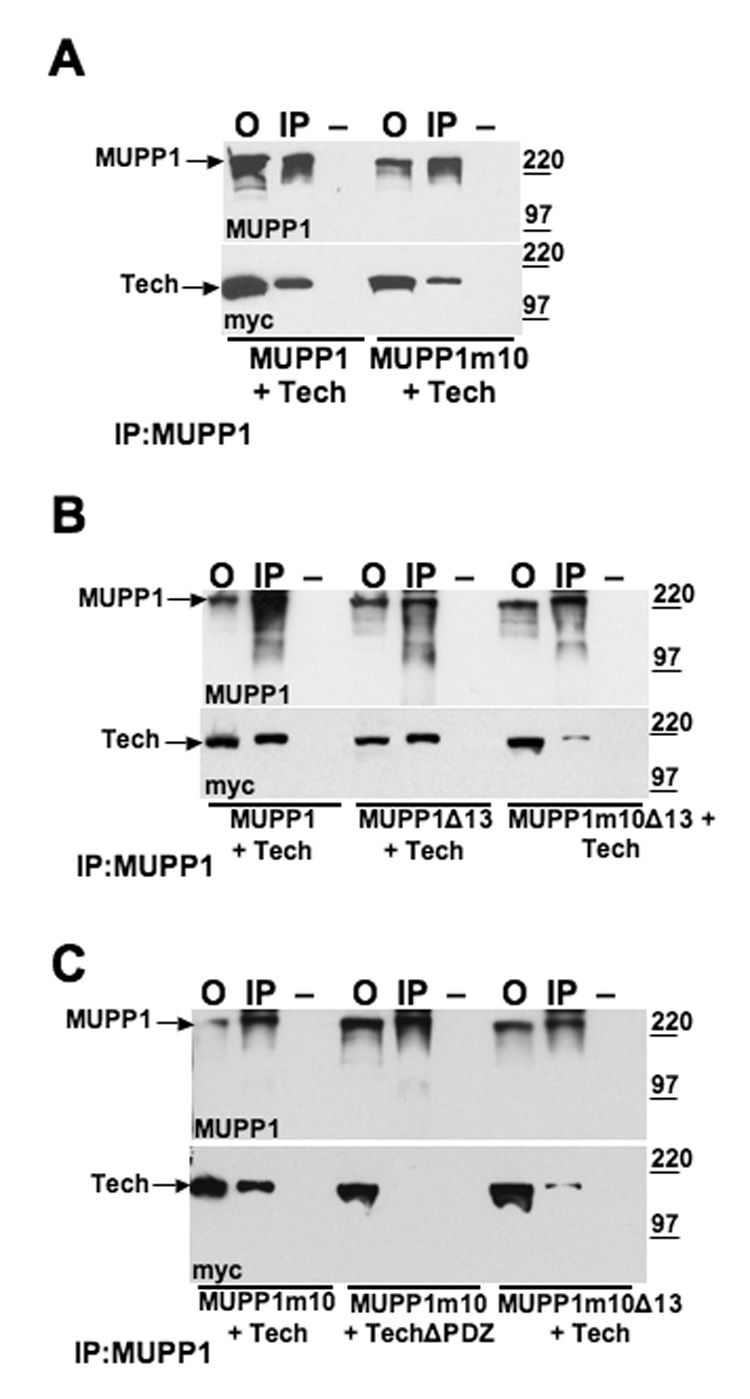
(A) HEK 293 cells were co-transfected with myc-tagged Tech and either wild-type MUPP1 or MUPP1m10, which has a four-residue mutation in the GLGL sequence of PDZ domain 10 (mutated to RPVR). From left to right, immunoblots show offered sample (O), immunoprecipitation with anti-MUPP1 (IP) and minus primary antibody control (−). Immunoblots were probed with anti-MUPP1 (top) and ant-imyc (bottom) to detect Tech. (B) HEK 293 cells were transfected, as labeled on the bottom of the blots, with myc-Tech and a MUPP1 construct, either wild-type MUPP1, MUPP1Δ13, which is truncated after PDZ domain 12, or MUPP1m10Δ13, which has both a mutation in domain 10 and is missing domain 13. From left to right, immunoblots show offered sample (O), immunoprecipitation with MUPP1 antibody (IP) and a minus antibody control (−). Blots were probed with anti-MUPP1 (top), or anti-myc (bottom) to detect Tech. (C) HEK 293 cells were co-transfected with the MUPP1 and Tech constructs indicated at the bottom of the blots.
Although disruption of both domains 10 and 13 in MUPP1 greatly reduced its interaction with Tech, we reliably detected a residual Tech band in the immunoprecipitated pellet which was not present in the immunoprecipitate obtained in the minus antibody control sample. To assess whether this residual binding was PDZ dependent, we compared the results of performing parallel immunoprecipitation on lysates from hEK293 cells co-transfected with the MUPP1 m10 construct and either full-length Tech or Tech ΔPDZ constructs (Figure 6C). Since these studies indicate that deletion of the PDZ ligand motif abolishes binding of Tech to MUPP1, the presence of a residual Tech band following co-transfection with the MUPP1m10Δ13 construct, implies that one or more additional PDZ domains, besides 10 and 13, are able to mediate binding to Tech.
To narrow down which of the other 11 PDZ domains might be involved, we first tested a MUPP1 construct with deletions of domains 7, 10 and 13, MUPP1Δ7Δ10Δ13 (data not shown). Tech still shows residual binding to this construct, indicating that this weak interaction is not mediated by domain 7 or mutant domain 10. As these results still left 10 candidate PDZ domains in MUPP1 that might mediate this residual interaction with Tech, we tested a series of MUPP1 truncation constructs that enabled us to evaluate multiple PDZ domains efficiently. In particular, we found that truncation of MUPP1 after domain 7 (MUPP1Δ8-13) abolished its interaction with Tech, which allowed us to eliminate domains 1 through 7 from consideration (Figure 7A). Working from the other end of MUPP1, we also tested a construct which contains a mutant domain 10 and is truncated after domain 11 (MUPP1m10Δ12-13). As this construct still yields a residual Tech band in the immunoprecipitate, these results, taken together, imply that either domains 8, 9 or 11 are responsible for this residual interaction. Accordingly, we proceeded to test a MUPP1 construct that is truncated after domain 9 (Figure 7B). As this truncation abolishes the residual interaction, we conclude that domains 1 through 9 are not involved, but that domain 11 can mediate the weak interaction detected in the absence of domains 10 and 13.
Figure 7. MUPP1 PDZ domains 1–9 do not mediate its interaction with Tech.
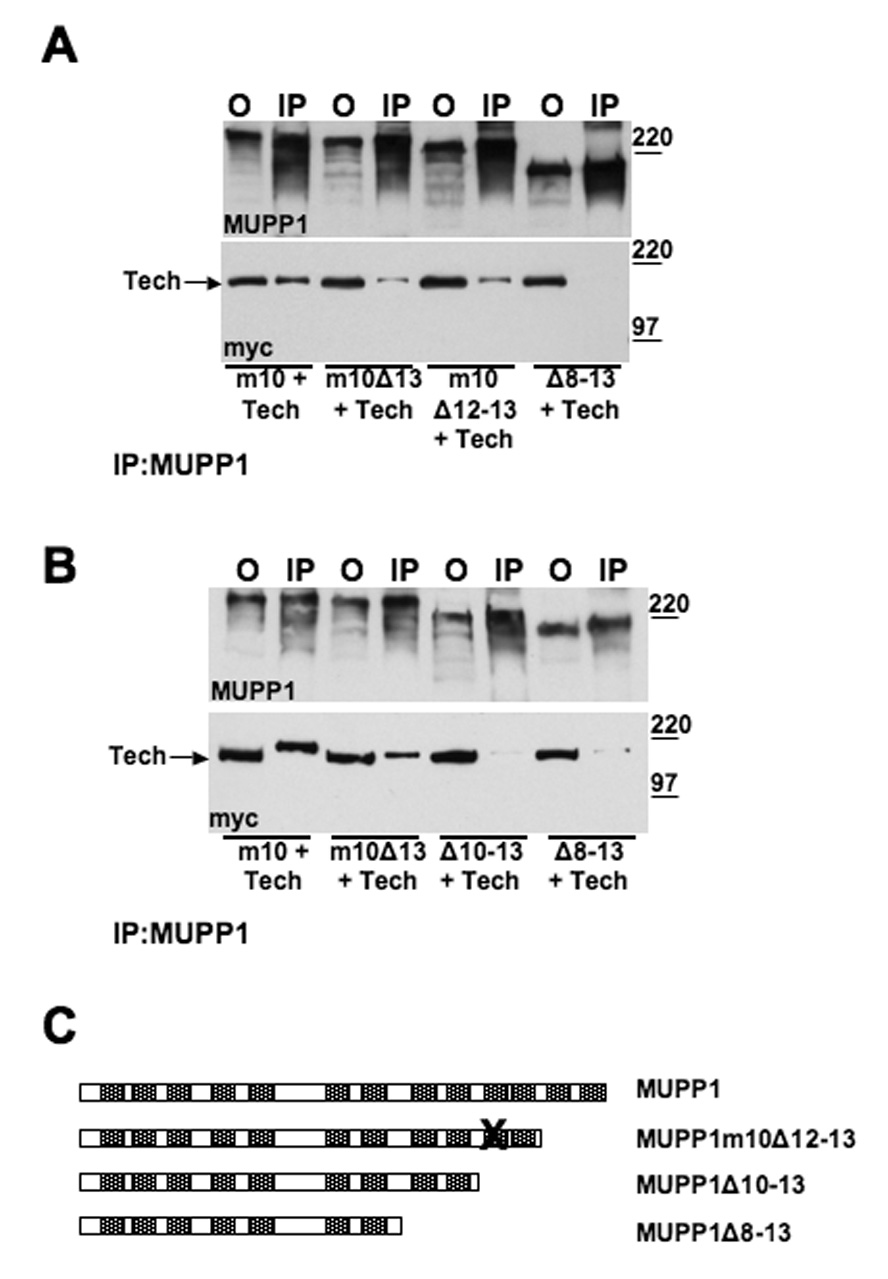
Panels A and B present immunoprecipitation analysis of the interaction of Tech with the MUPP1 constructs listed below each set of blots. In panel A, note that truncation of domains 12 and 13 in a construct that contains a mutant domain 10 does not eliminate binding. However, truncation of domains 8 through 13 does. As shown in panel B, truncation of domains 10 through 13 also abolishes binding to Tech. The trace Tech bands detected with the MUPP1Δ10-13 and MUPP1Δ8-13 constructs represent non-specific binding as trace binds of comparable intensity are detected in control samples in which the immunoprecipitating antibody was omitted. (C) Schematic diagram illustrating the MUPP1 constructs used in these blots.
Discussion
Using immunoprecipitation to assess the interaction of Tech and MUPP1, we have demonstrated that recombinant versions of these proteins co-precipitate from hEK293 cells and that the endogenous proteins co-precipitate from brain extracts. Furthermore, we have found in immunostaining studies that these proteins co-localize in dendrites in the vicinity of synaptic zones. Taken together, these findings suggest that Tech is involved in regulating RhoA signaling in this compartment. In previous studies, we found that expression of a constitutively active form of Tech, which would be expected to induce global activation of RhoA throughout the transfected neurons, triggers a marked reduction in their dendritic complexity (Marx et al., 2005). As RhoA activation has been linked to retraction of dendrites or spines (Luo, 2000; Neumann et al, 2002; Ahnert-Hilger et al, 2004), we hypothesize that localized activation of Tech might play an important role in modifying neuronal morphology selectively in the affected dendritic segments. In addition to its prominent effects on neuronal morphology, RhoA has also been implicated in a variety of other cellular functions ranging from receptor trafficking to transcriptional regulation (Etienne-Manneville and Hall 2002; Kaneko et al. 2005; Tabuchi et al. 2005). Accordingly, it is also conceivable that Tech might be involved in regulating RhoA signaling pathways involved in these processes.
Our findings add Tech to the roster of proteins that bind to MUPP1 which include cell surface receptors, i.e. 5-HT2C (Becamel et al., 2001) and GABA B receptors (Balasubramanian et al., 2007), and intracellular signaling molecules, such as synGAP (Krapivinsky et al., 2004) and kalirin-7 (Penzes et al., 2001), which like Tech, also regulate the activity of small G proteins. SynGAP is a synaptic protein that has GAP activity for both Ras and Rap (Kim et al., 1998; Chen et al., 1998; Krapivinsky et al., 2004). Kalirin-7 is a Rac GEF that has been shown to regulate spine morphology (Penzes et al, 2001). Accordingly, MUPP1 may serve as a scaffold protein that coordinates the activity of multiple small G protein signaling pathways in response to synaptic stimuli. As found for Tech, approximately 40 percent of RhoGEF family members contain C-terminal PDZ ligand motifs (Garcia-Mata and Burridge, 2007) suggesting that PDZ domain scaffold proteins, such as MUPP1, play a general role in organizing small G protein signaling.
Evidence that the interaction of Tech with PDZ domain containing proteins is of physiological importance has been provided from studies of Tech’s function in endothelial cells. In these cells, Tech has been shown to interact with GIPC, also called synectin, a small adapter protein that contains a single PDZ domain which interacts with numerous proteins via their C-terminal PDZ ligand motif (Liu and Horowitz, 2006). As a result, Tech has also been called Syx, synectin interacting exchange protein. Of note, overexpression of Tech in endothelial cells enhances their migratory rate under basal or stimulated conditions. This response is not shared by an alternatively spliced form of Tech that differs only in lacking the PDZ ligand motif. Furthermore, this response is also blocked in endothelial cells treated with GIPC siRNA. Thus, these findings indicate that Tech’s PDZ interaction with GIPC plays a critical role in mediating its effects on endothelial cell migration. Consistent with these findings in endothelial cells, we also identified GIPC as a putative Tech interactor in our yeast two hybrid screen. Furthermore, we have confirmed that GIPC co-precipitates with Tech from hippocampal extracts. In addition, double immunostaining of cultured hippocampal neurons for GIPC and Tech revealed that about one-third of Tech puncta co-localize with GIPC puncta (J.A.H., unpublished observations). Recent studies suggest that GIPC regulates endocytic trafficking of multiple receptors including extra-synaptic NMDA receptors (Yi et al., 2007; Varsano et al., 2006; Naccache et al., 2006). Thus, Tech, via its interaction with GIPC may modulate trafficking of cell surface receptors. Although Tech immunostaining indicates that Tech is locaiized in the post-synaptic compartment in the vicinity of synapses, based on our current studies, we cannot conclude that it binds to MUPP1 or GIPC in the post-synaptic density. Preliminary experiments assessing whether Tech interacts with these proteins in hippocampal extracts prepared under conditions that solubilize PSD proteins did not provide evidence for co-precipitation of Tech with either of these proteins. Accordingly, it is certainly possible that Tech may bind to GIPC or MUPP1 present on endocytic vesicles located in the vicinity of synapses.
Consistent with our finding that Tech and MUPP1 co-precipitate from brain extracts, these proteins also co-precipitate from endothelial cell lysates (Liu and Horowitz, personal communication). Accordingly, it may be interesting to assess whether the effects of Tech on migration of endothelial cells are also dependent on its interaction with MUPP1.
Our detailed analysis of which PDZ domains within MUPP1 mediate its interaction with Tech indicate that several are involved. Consistent with the yeast two hybrid results, fragments containing domains 10 and higher appear to be sufficient to mediate this interaction. Deletion of both domains 10 and 13 were required to produce a substantial reduction in the intensity of the Tech band contained in the immunoprecipitate. While, ultimately, truncation of domains 10 through 13 was needed to reduce the interaction to the level of "non-specific" binding detected with the TechΔPDZ construct. Interestingly, deletion or truncation of domains 10, 12 and 13 did not eliminate this interaction, indicating that domain 11 is able to mediate a weak interaction with Tech. In retrospect, this observation fits with the low level of Tech binding that remains in the dyad MUPP1 10 & 11 construct after mutating domain 10. For completeness, it should be pointed out that we have not eliminated the possibility that domain 12 may also interact with Tech as we have not examined the impact of mutating domain 11 in the context of an intact domain 12.
Previous studies indicate that, like Tech, TAPP1 and 2 are also able to bind to PDZ domains 10 and 13 of MUPP1 (Kimber et al., 2002). Of note, we have also found that, like Tech, TAPP2 also retains binding to MUPP1 constructs in which both these domains have been inactivated (Estévez M., unpublished observations). Accordingly, Tech’s ability to interact with at least 3 PDZ domains in MUPP1 is also shared by TAPP2. The ability of multiple proteins to bind to PDZ domains 10 and 13 indicate that these interactions may occur in different cell types or distinct subcellular compartments within the same cell. In addition, competiton among MUPP1 interacting proteins to occupy specific PDZ domains may be regulated in response to activity or developmental cues. For example, stimulation of the serotonin 2C receptor triggers phosphorylation of two serine residues located just prior to the C-terminal valine residue. Substitution of aspartate residues for these serines reduces the interaction of the serotonin 5-HT2C receptor with PDZ domain 10, as does stimulation of the receptor with serotonin (Parker et al., 2003). Conceivably, phosphorylation of the serine residue at the minus 2 position of Tech may also regulate its interaction with MUPP1 as well.
In addition to Tech, several other RhoA GEFs and GAPs have been localized to neurons, such as Lfc, a RhoA specific GEF (Ryan et al., 2005), and p250GAP, which acts on multiple Rho family subtypes (Nakazawa et al., 2003). At present, little is known about which types of physiological synaptic stimuli trigger Rho A signaling pathways, however, several reports indicate that exogenous glutamate receptor agonists are able to activate this pathway (Iida et al., 2007; Semenova et al., 2007; Schubert et al., 2006). Accordingly, it will be interesting in future studies to determine how Tech and the other RhoA GEFs and GAPs expressed in neurons act in concert to regulate activation of RhoA signaling pathways in response to synaptic stimulation.
Acknowledgments
This work was supported in part by grants from NIH (DA-0266) and the Hopkins-Weizmann Collaborative Program. We thank A. Lanahan and P. Worley for providing a brain cDNA library. We also thank M. Deak and R. Javier for providing cDNA constructs, and W. Tang for expert technical assistance.
References
- Ahnert-Hilger G, Höltje M, Grosse G, Pickert G, Mucke C, Nixdorf-Bergweiler B, Boquet P, Hofmann F, Just I. Differential effects of Rho GTPases on axonal and dendritic development in hippocampal neurones. J. Neurochem. 2004;90:9–18. doi: 10.1111/j.1471-4159.2004.02475.x. [DOI] [PubMed] [Google Scholar]
- Becamel C, Figge A, Poliak S, Dumuis A, Peles E, Bockaert J, Lübbert H, Ullmer C. Interaction of serotonin 5-hydroxytryptamine type 2C receptors with PDZ10 of the multi-PDZ domain protein MUPP1. J. Biol. Chem. 2001;276:12974–12982. doi: 10.1074/jbc.M008089200. [DOI] [PubMed] [Google Scholar]
- Benarroch EE. Rho GTPases: Role in dendrite and axonal growth, mental retardation, and axonal regeneration. Neurology. 2007;68:1315–1318. doi: 10.1212/01.wnl.0000259588.97409.8f. [DOI] [PubMed] [Google Scholar]
- Doyle DA, Lee A, Lewis J, Kim E, Sheng M, MacKinnon R. Crystal structures of a complexed and peptide-free membrane protein-binding domain: molecular basis of peptide recognition by PDZ. Cell. 1996;85:1067–1076. doi: 10.1016/s0092-8674(00)81307-0. [DOI] [PubMed] [Google Scholar]
- Etienne-Manneville S, Hall A. RhoGTPases in cell biology. Nature. 2002;420:629–635. doi: 10.1038/nature01148. [DOI] [PubMed] [Google Scholar]
- Garcia-Mata R, Burridge K. Catching a GEF by its tail. Trends Cell Biol. 2007;17:36–43. doi: 10.1016/j.tcb.2006.11.004. [DOI] [PubMed] [Google Scholar]
- Garner CC, Kindler S, Gundelfinger ED. Molecular determinants of presynaptic active zones. Curr. Opin. Neurobiol. 2000;10:321–327. doi: 10.1016/s0959-4388(00)00093-3. [DOI] [PubMed] [Google Scholar]
- Govek EE, Newey SE, Akerman CJ, Cross JR, Van der Veken L, Van Aelst L. The X-linked mental retardation protein oligophrenin-1 is required for dendritic spine morphogenesis. Nat. Neurosci. 2004;7:364–372. doi: 10.1038/nn1210. [DOI] [PubMed] [Google Scholar]
- Iida J, Ishizaki H, Okamoto-Tanaka M, Kawata A, Sumita K, Ohgake S, Sato Y, Yorifuji H, Nukina N, Ohashi K, Mizuno K, Tsutsumi T, Mizoguchi A, Miyoshi J, Takai Y, Hata Y. S-SCAMalpha is a scaffold to mediate NMDA receptor-dependent RhoA activation in dendrites. Mol. Cell. Biol. 2007 doi: 10.1128/MCB.01901-06. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko T, Maeda A, Takefuji M, Aoyama H, Nakayama M, Kawabata S, Kawano Y, Iwamatsu A, Amano M, Kaibuchi K. Rho mediates endocytosis of epidermal growth factor receptor through phosphorylation of endophilin A1 by Rho-kinase. Genes Cells. 2005;10:973–987. doi: 10.1111/j.1365-2443.2005.00895.x. [DOI] [PubMed] [Google Scholar]
- Kim E, Sheng M. PDZ domain proteins of synapses. Nat. Rev. Neurosci. 2004;5:771–781. doi: 10.1038/nrn1517. [DOI] [PubMed] [Google Scholar]
- Kimber WA, Trinkle-Mulcahy L, Cheung PC, Deak M, Marsden LJ, Kieloch A, Watt S, Javier RT, Gray A, Downes CP, Lucocq JM, Alessi DR. Evidence that the tandem-pleckstrin-homology-domain-containing protein TAPP1 interacts with Ptd(3,4)P2 and the multi-PDZ-domain-containing protein MUPP1 in vivo. Biochem. J. 2002;361:525–536. doi: 10.1042/0264-6021:3610525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krapivinsky G, Medina I, Krapivinsky L, Gapon S, Clapham DE. SynGAP-MUPP1-CaMKII synaptic complexes regulate p38 MAP kinase activity and NMDA receptor-dependent synaptic AMPA receptor potentiation. Neuron. 2004;43:563–574. doi: 10.1016/j.neuron.2004.08.003. [DOI] [PubMed] [Google Scholar]
- Kuner R, Swiercz JM, Zywietz A, Tappe A, Offermanns S. Characterization of the expression of PDZ-RhoGEF, LARG and G(alpha)12/G(alpha)13 proteins in the murine nervous system. Eur. J. Neurosci. 2002;16:2333–2341. doi: 10.1046/j.1460-9568.2002.02402.x. [DOI] [PubMed] [Google Scholar]
- Lee SS, Glaunsinger B, Mantovani F, Banks L, Javier RT. Multi-PDZ domain protein MUPP1 is a cellular target for both adenovirus E4-ORF1 and high-risk papillomavirus type 18 E6 oncoproteins. J. Virol. 2000;74:9680–9693. doi: 10.1128/jvi.74.20.9680-9693.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Horowitz A. A PDZ-binding motif as a critical determinant of Rho guanine exchange factor function and cell phenotype. Mol. Biol. Cell. 2006;17:1880–1887. doi: 10.1091/mbc.E06-01-0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo L. Rho GTPases in neuronal morphogenesis. Nat. Rev. Neurosci. 2000;1:173–180. doi: 10.1038/35044547. [DOI] [PubMed] [Google Scholar]
- Marx R, Henderson J, Wang J, Baraban JM. Tech: a RhoA GEF selectively expressed in hippocampal and cortical neurons. J. Neurochem. 2005;92:850–858. doi: 10.1111/j.1471-4159.2004.02930.x. [DOI] [PubMed] [Google Scholar]
- Maystadt I, Rezsohazy R, Barkats M, Duque S, Vannuffel P, Remacle S, Lambert B, Najimi M, Sokal E, Munnich A, Viollet L, Verellen-Dumoulin C. The nuclear factor kappaB-activator gene PLEKHG5 is mutated in a form of autosomal recessive lower motor neuron disease with childhood onset. Am J Hum Genet. 2007;81:67–76. doi: 10.1086/518900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morais Cabral JH, Petosa C, Sutcliffe MJ, Raza S, Byron O, Poy F, Marfatia SM, Chishti AH, Liddington RC. Crystal structure of a PDZ domain. Nature. 1996;382:649–652. doi: 10.1038/382649a0. [DOI] [PubMed] [Google Scholar]
- Naccache SN, Hasson T, Horowitz A. Binding of internalized receptors to the PDZ domain of GIPC/synectin recruits myosin VI to endocytic vesicles. Proc Natl Acad Sci U S A. 2006;103:12735–12740. doi: 10.1073/pnas.0605317103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann H, Schweigreiter R, Yamashita T, Rosenkranz K, Wekerle H, Barde YA. Tumor necrosis factor inhibits neurite outgrowth and branching of hippocampal neurons by a Rho-dependent mechanism. J. Neurosci. 2002;22:854–862. doi: 10.1523/JNEUROSCI.22-03-00854.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penzes P, Johnson RC, Sattler R, Zhang X, Huganir RL, Kambampati V, Mains RE, Eipper BA. The neuronal Rho-GEF Kalirin-7 interacts with PDZ domain-containing proteins and regulates dendritic morphogenesis. Neuron. 2001;29:229–242. doi: 10.1016/s0896-6273(01)00193-3. [DOI] [PubMed] [Google Scholar]
- Ryan XP, Alldritt J, Svenningsson P, Allen PB, Wu GY, Nairn AC, Greengard P. The Rho-specific GEF Lfc interacts with neurabin and spinophilin to regulate dendritic spine morphology. Neuron. 2005;47:85–100. doi: 10.1016/j.neuron.2005.05.013. [DOI] [PubMed] [Google Scholar]
- Schubert V, Da Silva JS, Dotti CG. Localized recruitment and activation of RhoA underlies dendritic spine morphology in a glutamate receptor-dependent manner. J. Cell. Biol. 2006;172:453–467. doi: 10.1083/jcb.200506136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semenova MM, Maki-Hokkonen AMJ, Cao J, Komarovski V, Forsberg KM, Koistinaho M, Coffey ET, Courtney MJ. Rho mediates calcium-dependent activation of p38α and subsequent excitotoxic cell death. Nature Neuroscience. 2007;10:436–443. doi: 10.1038/nn1869. [DOI] [PubMed] [Google Scholar]
- Parker LL, Backstrom JR, Sanders-Bush E, Shieh BH. Agonist-induced phosphorylation of the serotonin 5-HT2C receptor regulates its interaction with multiple PDZ protein 1. J Biol Chem. 2003;278:21576–21583. doi: 10.1074/jbc.M210973200. [DOI] [PubMed] [Google Scholar]
- Sitek B, Poschmann G, Schmidtke K, Ullmer C, Maskri L, Andriske M, Stichel CC, Zhu XR, Luebbert H. Expression of MUPP1 protein in mouse brain. Brain Res. 2003;970:178–187. doi: 10.1016/s0006-8993(03)02338-2. [DOI] [PubMed] [Google Scholar]
- Songyang Z, Fanning AS, Fu C, Xu J, Marfatia SM, Chishti AH, Crompton A, Chan AC, Anderson JM, Cantley LC. Recognition of unique carboxyl-terminal motifs by distinct PDZ domains. Science. 1997;275:73–77. doi: 10.1126/science.275.5296.73. [DOI] [PubMed] [Google Scholar]
- Tabuchi A, Estévez M, Henderson JA, Marx R, Shiota J, Nakano H, Baraban JM. Nuclear translocation of the SRF co-activator MAL in cortical neurons: role of RhoA signalling. J. Neurochem. 2005;94:169–180. doi: 10.1111/j.1471-4159.2005.03179.x. [DOI] [PubMed] [Google Scholar]
- Ullmer C, Schmuck K, Figge A, Lübbert H. Cloning and characterization of MUPP1, a novel PDZ domain protein. FEBS Lett. 1998;424:63–68. doi: 10.1016/s0014-5793(98)00141-0. [DOI] [PubMed] [Google Scholar]
- Varsano T, Dong MQ, Niesman I, Gacula H, Lou X, Ma T, Testa JR, Yates JR, 3rd, Farquhar MG. GIPC is recruited by APPL to peripheral TrkA endosomes and regulates TrkA trafficking and signaling. Mol Cell Biol. 2006;26:8942–8952. doi: 10.1128/MCB.00305-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi Z, Petralia RS, Fu Z, Swanwick CC, Wang YX, Prybylowski K, Sans N, Vicini S, Wenthold RJ. The role of the PDZ protein GIPC in regulating NMDA receptor trafficking. J Neurosci. 2007;27:11663–11675. doi: 10.1523/JNEUROSCI.3252-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Chittenden T, Simons M. Characterization of synectin expression and promoter activity. Gene. 2004;342:29–34. doi: 10.1016/j.gene.2004.07.028. [DOI] [PubMed] [Google Scholar]


