Abstract
Protein-protein interactions have key importance in various biological processes and modulation of particular protein-protein interactions has been shown to have therapeutic effects. However, disrupting or modulating protein-protein interactions with low-molecular-weight compounds is extremely difficult due to the lack of deep binding pockets on protein surfaces. Herein we describe the development of an unprecedented lead synthesis and discovery method that generates only biologically active compounds from a library of reactive fragments. Using the protein Bcl-XL, a central regulator of programmed cell death, we demonstrated that an amidation reaction between thio acids and sulfonyl azides is applicable for Bcl-XL-templated assembly of inhibitory compounds. We have demonstrated for the first time that kinetic target-guided synthesis can be applied not only on enzymatic targets but also for the discovery of small molecules modulating protein-protein interactions.
Protein-protein interactions are central to many biological processes and hence represent a large and important class of potential targets for human therapeutics.1 Recent discovery of a variety of low-molecular-weight compounds interfering with protein-protein complexes launches and validates viable routes for a large number of new therapies.2 However, disrupting or modulating protein-protein interactions with low-molecular-weight compounds remains challenging. Most protein-protein interfaces lack deep pockets that might provide binding sites for small molecules and they are composed of two relatively large protein surfaces that are complementary with respect to shape and electrostatics.3 Moreover, the adaptive and flexible nature of amino acid residues on protein surfaces creates additional challenges for lead compound design and discovery.4
The Bcl-2 family consists of both anti- and pro-apoptotic members, which are central regulators of programmed cell death.5 The anti-apoptotic proteins like Bcl-2, Bcl-XL, and Mcl-1 heterodimerize with the pro-apoptotic constituents, which include the multidomain molecules Bax and Bak, and BH3-only proteins such as Bim, Bad, Bid, Noxa or Puma, through the conserved BH3 domain.5 The relative ratios of pro- and anti-apoptotic Bcl-2 family proteins determine the ultimate sensitivity or resistance of cells to a wide variety of apoptotic signals.6 Meanwhile, small molecules have been reported to modulate the extent of heterodimerization between anti- and pro-apoptotic Bcl-2 family members inducing apoptosis in cancer cells.5 Using a combination of SAR by NMR screening, parallel synthesis and structure-guided lead design, the Abbott laboratories developed ABT-737 and a large series of analogues displaying inhibition constants in the nanomolar or subnanomolar range.7
Herein, we report our progress toward the development of lead synthesis and discovery methods that generate only biologically active compounds targeting protein-protein interactions. In the last decade, various fragment-based lead discovery approaches have been reported in which the biological target is actively engaged in the assembly of its own multidentate inhibitor from a pool of smaller reactive fragments. These approaches can roughly be divided into three different categories: (1) dynamic combinatorial chemistry (DCC)8, (2) catalyst/reagent-accelerated target-supported assembly9, and (3) kinetic target-guided synthesis (TGS)10. While DCC utilizes reversible reactions to create a library of all possible heterodimeric compounds, kinetic TGS approaches employ irreversible reactions generating only the biologically active bidentate members of the entire virtual library. In situ click chemistry10c, in particular, has shown to produce potent triazole inhibitors of the enzymes acetylcholine esterase11, carbonic anhydrase12, and HIV protease13. So far, kinetic TGS approaches have been applied only for the discovery of enzyme inhibitors but not for protein-protein interaction modulators (PPIMs). Based on studies by Abbott laboratories, we investigated whether a kinetic TGS approach is a viable route for the discovery of PPIMs targeting the anti-apoptotic protein Bcl-XL.
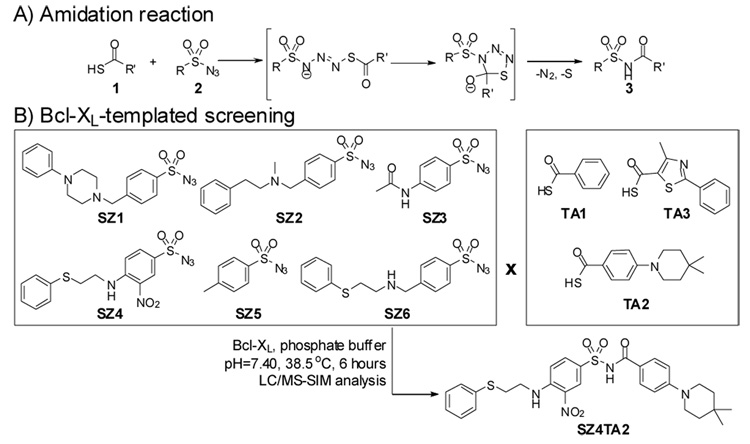
The reaction combining two fragments in the presence of a biological template into a larger molecule plays a crucial role for kinetic TGS. For example, the slow nature and the bio-orthogonality of the Huisgen cycloaddition, the compatibility of the reactants with water, and the stability of the triazole products are distinct characteristics of in situ click chemistry.10c Recently, Williams and co-workers developed an amidation reaction displaying a reactivity profile suitable for kinetic TGS applications.14 This particular amidation reaction between thio acids 1 and sulfonyl azides 2 to give corresponding acylsulfonamides 3 is effective at room temperature in both organic and aqueous solvents (Scheme 1-A).
Scheme 1.
Bcl-XL-templated assembly of acylsulfonamide SZ4TA2 from fragments decorated with thio acid or sulfonyl azide functionalities.
To probe if this amidation reaction is suited for kinetic TGS applications targeting protein-protein interactions, building blocks, of which some are structurally related to fragments of ABT-737, were decorated with sulfonyl azide or thio acid groups (Scheme 1-B). Thio acids TA1–TA3 and sulfonyl azides SZ1–SZ6 were incubated as binary mixtures in the presence of buffer solution containing Bcl-XL for 6 hours at 38°C. As a control, 18 identical binary building block combinations were kept under the same incubation conditions in buffer without Bcl-XL. All incubations were then analyzed by HPLC with product detection by electrospray ionization in the positive selected ion mode (LC/MS-SIM).11b From all the screened samples, only one incubation sample led to an increased amount of acylsulfonamide SZ4TA27e in the Bcl-XL-containing sample compared to the incubation without the protein. We synthesized this compound for hit validation.15 Comparison of the LC/MS-SIM traces of the Bcl-XL incubation mixture with the trace of the corresponding synthesized SZ4TA2 clearly confirmed that Bcl-XL templates the formation of the characterized hit compound (Figure 1. A – C).
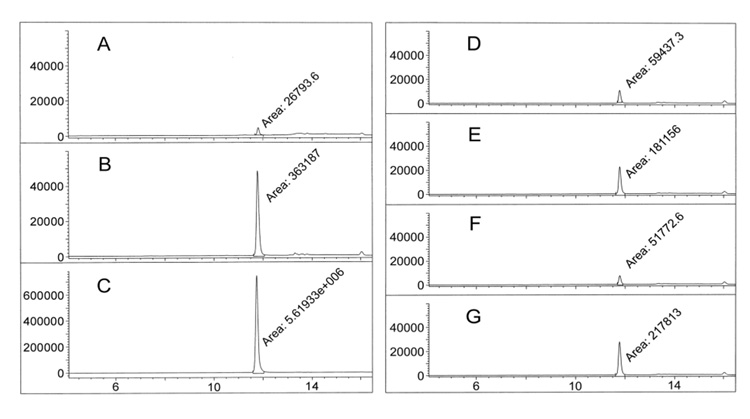
Figure 1.
Hit identification of acylsulfonamide SZ4TA2 by LC/MS-SIM. (A) Incubation of SZ4 and TA2 in buffer without Bcl-XL. (B) Bcl-XL-templated reaction after 6 hours of incubation. (C) Synthesized SZ4TA2 as reference. (D) Suppression of Bcl-XL-templated reaction by Bak BH3 peptide. (E) Bcl-XL-templated incubation in presence of mutant Bak BH3 peptide. (F) Suppression of Bcl-XL-templated reaction by Bim BH3 peptide. (G) Bcl-XL-templated incubation in presence of mutant Bim BH3 peptide.
To assess whether the Bcl-XL-templated reactions occur at the BH3 binding pocket or randomly elsewhere on the protein surface, control experiments have been performed, in which the reactive fragments SZ4 and TA2 were incubated with Bcl-XL and various pro-apoptotic BH3-containing peptides.15 Bak BH3 and Bim BH3 peptides bind to Bcl-XL through their BH3 domain and theoretically compete with the reactive building blocks for binding during these incubations. In comparison, their mutants exhibit lower affinity towards Bcl-XL and hence do not suppress the Bcl-XL-templated acylsulfonamide formation to the same extent. Comparison of the LC/MS-SIM traces (Figure 1. D–G) between the Bcl-XL incubations with and without these peptides suggests that the generation of SZ4TA2 occurs at the BH3 binding site on Bcl-XL.
An advantage of kinetic TGS approaches is the increased throughput screening capability with incubations containing more than two reactive building blocks. Experiments were undertaken to test whether our amidation kinetic TGS screening can be performed as incubations containing more than two complimentary reacting building blocks. Best results were obtained with reactions containing one thio acid and six sulfonyl azides.15 Samples of Bcl-XL with all 9 reactive building blocks at the same time failed at giving clear results. Although the multi-component screening of the entire library failed, these experiments prove that the amidation kinetic TGS approach can be tested in an enhanced screening throughput.
To investigate the ability of the hit compound to disrupt the interaction between Bcl-XL and Bak, we performed the well-established fluorescence polarization competition assay using Bcl-XL and fluorescein-labeled Bak BH3 peptide.16 Abbott laboratories reported that SZ4TA2 is a good Bcl-XL PPIM with a Ki constant of 19 nM, as determined by a competitive fluorescence polarization assay using a fluorescein-labeled Bad-BH3 peptide.7e Consistently, compound SZ4TA2 is validated again as a Bcl-XL inhibitor with an IC50 constant of 78.8 nM by our assay.15 To compare the activity of acylsulfonamides, not assembled by Bcl-XL, with that of the hit compound SZ4TA2, compounds SZ2TA1, SZ2TA2, SZ2TA3, SZ4TA1, SZ5TA1 and SZ5TA2 have been synthesized and tested for their binding to Bcl-XL. The IC50 values of these compounds have been determined to be 5 µM or higher.15 Finally, we also determined the IC50 constants for the corresponding reactive building blocks SZ4 and TA2 to be higher than 100 µM. Taken together, these results indicate that the hit compound SZ4TA2 identified through the kinetic TGS screening is indeed a respectable ligand of the biological target, which underscores the utility of kinetic TGS as a valuable approach to PPIM discovery and optimization.
In a proof-of-concept study, we have shown for the first time that kinetic TGS can be applied not only for enzymatic targets but also for protein-protein interaction disruption by using the recently-reported amidation reaction between sulfonyl azides and thio acids. In the future, we will study in great detail the kinetics and mechanism of the kinetic TGS approach. Additionally, we will investigate the scope and limitations of the herein reported lead discovery and optimization method targeting other protein-protein interactions related to various diseases.
Supplementary Material
Synthetic procedures, LC/MS-SIM traces and determination of IC50 values. This material is available free of charge via the Internet at http://pubs.acs.org.
Acknowledgment
We are grateful to the James and Esther King Biomedical Research Program (NIR Grant 07KN-08 to R.M.) and the National Cancer Institute, National Institutes of Health (Grant P01CA118210 to H-G.W.) for financial support.
References
- 1.(a) Arkin MR, Wells JA. Nat. Rev. Drug Discov. 2004;3:301–317. doi: 10.1038/nrd1343. [DOI] [PubMed] [Google Scholar]; (b) Berg T. Angew. Chem. Int. Ed. Engl. 2003;42:2462–2481. doi: 10.1002/anie.200200558. [DOI] [PubMed] [Google Scholar]
- 2.(a) Clackson T, Wells JA. Science. 1995;267:383–386. doi: 10.1126/science.7529940. [DOI] [PubMed] [Google Scholar]; (b) Arkin M. Curr. Opin. Chem. Biol. 2005;9:317–324. doi: 10.1016/j.cbpa.2005.03.001. [DOI] [PubMed] [Google Scholar]; (c) Bogan AA, Thorn KS. J. Mol. Biol. 1998;280:1–9. doi: 10.1006/jmbi.1998.1843. [DOI] [PubMed] [Google Scholar]; (d) DeLano WL. Curr. Opin. Struct. Biol. 2002;12:14–20. doi: 10.1016/s0959-440x(02)00283-x. [DOI] [PubMed] [Google Scholar]
- 3.(a) Preissner R, Goede A, Frommel C. J. Mol. Biol. 1998;280:535–550. doi: 10.1006/jmbi.1998.1878. [DOI] [PubMed] [Google Scholar]; (b) Mccoy AJ, Epa VC, Colman PM. J. Mol. Biol. 1997;268:570–584. doi: 10.1006/jmbi.1997.0987. [DOI] [PubMed] [Google Scholar]; (c) Lo Conte L, Chothia C, Janin J. J. Mol. Biol. 1999;285:2177–2198. doi: 10.1006/jmbi.1998.2439. [DOI] [PubMed] [Google Scholar]; (d) Nooren IMA, Thornton JM. J. Mol. Biol. 2003;325:991–1018. doi: 10.1016/s0022-2836(02)01281-0. [DOI] [PubMed] [Google Scholar]
- 4.(a) DeLano WL, Ultsch MH, de Vos AM, Wells JA. Science. 2000;287:1279–1283. doi: 10.1126/science.287.5456.1279. [DOI] [PubMed] [Google Scholar]; (b) Wodak SJ, Janin J. Protein Modules and Protein-Protein Interactions. 2003;61:9–73. doi: 10.1016/s0065-3233(02)61001-0. [DOI] [PubMed] [Google Scholar]
- 5.Walensky LD. Cell Death Differ. 2006;13:1339–1350. doi: 10.1038/sj.cdd.4401992. [DOI] [PubMed] [Google Scholar]
- 6.(a) Danial NN, Korsmeyer SJ. Cell. 2004;116:205–219. doi: 10.1016/s0092-8674(04)00046-7. [DOI] [PubMed] [Google Scholar]; (b) Huang DCS, Strasser A. Cell. 2000;103:839–842. doi: 10.1016/s0092-8674(00)00187-2. [DOI] [PubMed] [Google Scholar]; (c) Kelekar A, Thompson CB. Trends Cell Biol. 1998;8:324–330. doi: 10.1016/s0962-8924(98)01321-x. [DOI] [PubMed] [Google Scholar]
- 7.(a) Oltersdorf T, et al. Nature. 2005;435:677–681. doi: 10.1038/nature03579. [DOI] [PubMed] [Google Scholar]; (b) Wendt MD, et al. J. Med. Chem. 2006;49:1165–1181. doi: 10.1021/jm050754u. [DOI] [PubMed] [Google Scholar]; (c) Park CM, Oie T, Petros AM, Zhang H, Nimmer PM, Henry RF, Elmore SW. J. Am. Chem. Soc. 2006;128:16206–16212. doi: 10.1021/ja0650347. [DOI] [PubMed] [Google Scholar]; (d) Petros AM, et al. J. Med. Chem. 2006;49:656–663. doi: 10.1021/jm0507532. [DOI] [PubMed] [Google Scholar]; (e) Wendt MD, et al. J. Med. Chem. 2006;49:1165–1181. doi: 10.1021/jm050754u. [DOI] [PubMed] [Google Scholar]; (f) Bruncko M, et al. J. Med. Chem. 2007;50:641–662. doi: 10.1021/jm061152t. [DOI] [PubMed] [Google Scholar]; (g) Tse C, et al. Cancer Res. 2008;68:3421–3428. doi: 10.1158/0008-5472.CAN-07-5836. [DOI] [PubMed] [Google Scholar]
- 8.(a) Huc I, Lehn J-M. Proc. Natl. Acad. Sci. U.S.A. 1997;94:2106–2110. doi: 10.1073/pnas.94.6.2106. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Corbett PT, Leclaire J, Vial L, West KR, Wietor J-L, Sanders JKM, Otto S. Chem. Rev. 2006;106:3652–3711. doi: 10.1021/cr020452p. [DOI] [PubMed] [Google Scholar]
- 9.(a) Nicolaou KC, Hughes R, Cho SY, Winssinger N, Smethurst C, Labischinski H, Endermann R. Angew. Chem. Int. Ed. Engl. 2000;39:3823–3828. doi: 10.1002/1521-3773(20001103)39:21<3823::AID-ANIE3823>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]; (b) Nicolaou KC, Hughes R, Cho SY, Winssinger N, Labischinski H, Endermann R. Chem.-Eur. J. 2001;7:3824–3843. doi: 10.1002/1521-3765(20010903)7:17<3824::aid-chem3824>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 10.(a) Inglese J, Benkovic SJ. Tetrahedron. 19991;47:2351–2364. [Google Scholar]; (b) Nguyen R. Huc. Angew. Chem. Int. Ed. Engl. 2001;40:1774–1776. [PubMed] [Google Scholar]; (c) Sharpless KB, Manetsch R. Expert Opinion on Drug Discovery. 2006;1:525–538. doi: 10.1517/17460441.1.6.525. [DOI] [PubMed] [Google Scholar]
- 11.(a) Lewis WG, Green LG, Grynszpan F, Radic Z, Carlier PR, Taylor P, Finn MG, Sharpless KB. Angew. Chem. Int. Ed. Engl. 2002;41:1053–1057. doi: 10.1002/1521-3773(20020315)41:6<1053::aid-anie1053>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]; (b) Manetsch R, Krasinski A, Radic Z, Raushel J, Taylor P, Sharpless KB, Kolb HC. J. Am. Chem. Soc. 2004;126:12809–12818. doi: 10.1021/ja046382g. [DOI] [PubMed] [Google Scholar]; (c) Krasinski A, Radic Z, Manetsch R, Raushel J, Taylor P, Sharpless KB, Kolb HC. J. Am. Chem. Soc. 2005;127:6686–6692. doi: 10.1021/ja043031t. [DOI] [PubMed] [Google Scholar]
- 12.(a) Mocharla VP, Colasson B, Lee LV, Roeper S, Sharpless KB, Wong C-H, Kolb HC. Angew. Chem. Int. Ed. Engl. 2005;44:116–120. doi: 10.1002/anie.200461580. [DOI] [PubMed] [Google Scholar]; (b) Wang J, Sui G, Mocharla VP, Lin RJ, Phelps ME, Kolb HC, Tseng H. Angew. Chem. Int. Ed. Engl. 2006;45:5276–5281. doi: 10.1002/anie.200601677. [DOI] [PubMed] [Google Scholar]
- 13.Whiting M, Muldoon J, Lin Y-C, Silverman SM, Lindstrom W, Olson AJ, Kolb HC, Finn MG, Sharpless KB, Elder JH, Fokin VV. Angew. Chem. Int. Ed. Engl. 2006;45:1435–1439. doi: 10.1002/anie.200502161. [DOI] [PubMed] [Google Scholar]
- 14.(a) Shuangguan N, Katukojvala S, Greenberg R, William LJ. J. Am. Chem. Soc. 2003;125:7754–7755. doi: 10.1021/ja0294919. [DOI] [PubMed] [Google Scholar]; (b) Kolakowski RV, Shuangguan N, Sauers RR, Greenberg R, William LJ. J. Am. Chem. Soc. 2006;128:5695–5702. doi: 10.1021/ja057533y. [DOI] [PubMed] [Google Scholar]
- 15.Please see supporting information for details.
- 16.Yin H, Lee G, Sedey KA, Rodriguez JM, Wang H-G, Sebti SM, Hamilton AD. J. Am. Chem. Soc. 2005;127:5463–5468. doi: 10.1021/ja0446404. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Synthetic procedures, LC/MS-SIM traces and determination of IC50 values. This material is available free of charge via the Internet at http://pubs.acs.org.