Abstract
As our understanding of biological pathways and the genes that regulate these pathways increases, consideration of these biological pathways has become an increasingly important part of genetic and molecular epidemiology. Pathway-based genetic association studies often involve genotyping of variants in genes acting in certain biological pathways. Such pathway-based genetic association studies can potentially capture the highly heterogeneous nature of many complex traits, with multiple causative loci and multiple alleles at some of the causative loci. In this paper, we develop two nonparametric test statistics that consider simultaneously the effects of multiple markers. Our approach, which is based on data-adaptive U-statistics, can handle both qualitative data such as case-control data and quantitative continuous phenotype data. Simulations demonstrate that our proposed methods are more powerful than standard methods, especially when there are multiple risk loci each with small genetic effects. When the number of disease-predisposing genes is small, the data-adaptive weighting of the U-statistics over all the markers produces similar power to commonly used single marker tests. We further illustrate the potential merits of our proposed tests in the analysis of a data set from a pathway-based candidate gene association study of breast cancer and hormone metabolism pathways. Finally, potential applications of the proposed tests to genome-wide association studies are also discussed.
Keywords: Genetic heterogeneity, Global tests, Genetic pathways, Breast cancer
1 Introduction
Since most complex diseases are due to the disruption of normal biological processes, pathways or networks, the genetic basis of many common genetic traits is expected to be highly heterogeneous, with multiple causative loci and multiple alleles at some of the causative loci, each with small and weak marginal effects (Zondervan and Cardon, 2004; Schaid et al., 2005). For example, if the pathway activity levels determine the phenotype of interest, it is expected that different mutations in different genes within this pathway can lead to similar phenotypes. Such genetic heterogeneity, which refers to the presence of a variety of genetic defects in the same or different genes that cause the same disease, is common in many complex human diseases such as breast cancer (Ford et al., 1998; Easton, 1999) and Alzheimer’s disease (Lambert and Amouyel, 2007). Instead of evaluating single candidate genes, pathway-based genetic association studies consider entire pathways comparing a dozen or more genes or multiple pathways that link up or compete in complex genetic networks. However, such genetic heterogeneity can lead to loss of power to detect genetic associations (Slager et al., 2000; Schaid et al., 2005) when single marker-based analysis is used due to weak marginal effect and the issues of adjusting for multiple testing. An alternative approach is based on haplotype association tests; however, since genes within a given pathway are often from different chromosomes, haplotype analysis of functional variants does not make biological sense. In addition, tests based on haplotypes often have large degrees of freedom, resulting in loss of power. As an alternative to haplotype analysis, new multilocus association tests have also been developed for tagSNPs within a region of interest (Kwee et al., 2008).
Genetic heterogeneity among genes within pathways suggests that one may want to develop tests for joint testing between multiple genes or SNPs with complex phenotypes and to draw an overall conclusion as to whether the set of SNPs is related to the disease risk. One can use linear/logistic regression to simultaneously test the main effects (and possibly interactions) of multiple SNPs. Although this approach can be more powerful than testing each marker separately (Longmate, 2001), it still suffers from weak power because of the large number of degrees of freedom. Schaid et al. (2005) proposed a nonparametric test of association of multiple SNPs and disease status using U-statistics (Hoeffding, 1948) and presented several interesting choices of kernel functions. Their approach first measures a score over all markers for pairs of subjects and then compares the averages of these scores between cases and controls. The power of the proposed tests depends on the choice of the kernel used in the U-statistics. When there are both protective and disease-predisposing genes in the gene set, use of the wrong kernel can result in a loss in power, especially for the allele-match kernel. This is due to the fact that comparing average similarities between cases and controls is influenced by how much the allele frequencies depart from equality within a group and thereby potentially eliminating a signal when summing these allele-match kernels across markers (Schaid et al., 2005). The linear dosage kernel, which is defined as the sum of the number of the minor allele for a pair of genotypes, suffers the same potential loss of power when the minor alleles across multiple markers are both protective and disease predisposing, as indicated by their simulations (Schaid et al., 2005).
In this paper, we propose an alternative U-statistics-based nonparametric test of the association between multiple SNPs and qualitative traits using data-adaptive U-statistics. U-statistics (Hoeffding, 1948) is defined as an average of the kernel functions over all unordered subsets of the observed samples, when the kernel function is often chosen as a measurement of similarity among the samples. The use of U-statistics often requires fewer statistical assumptions and leads to more robust statistical tests, as compared to the statistics derived from parametric models. Following Sen (2006), we consider defining our test statistics based on both the within-group and between-group U-statistics, instead of simply considering the contrast between case and control genotype U-statistics scores. Also different from Schaid et al. (2005), our proposed test can be applied to qualitative traits of more than two categories and is more robust in power to misspecification of the genetic models. We also propose a nonparametric test of association between multiple SNPs and quantitative traits by extending the idea of Wei and Johnson (1985). We propose to weight the U-statistics across different markers using the negative of the logarithm of the single marker p-values, which makes the final test statistics data-adaptive. Such weighting increases the test power, especially when there are only one or two disease-associated markers in the marker set. Both tests are based on U-statistics that do not require a particular parametric model of dependence imposed on the SNPs or model to relate the genotypes to the phenotypes and therefore are robust to misspecfication of the underlying genetic models.
The rest of the paper is organized as follows: in the Statistical Methods section, we describe the U-statistics-based tests for both qualitative and quantitative traits. To illustrate the properties of our methods, we perform simulations. We also apply our methods to a study of candidate genes for breast cancer risk and age of onset of breast cancer, to illustrate their utility and interpretation. Finally, we give a brief discussion of the methods.
2 Statistical Methods
2.1 U-statistics-based test of association for qualitative traits
We first introduce notation. Suppose that we have K SNPs from genes in a given pathway or from genes with similar molecular functions, each with two alleles 0 and 1, where without loss of generality, we assume that allele 1 is the minor allele. At each SNP, there are three genotypes, coded as G = {00, 10, 11}. We consider a qualitative trait, taking C different possible categorical values. For example, for case-control studies, there are two trait groups with C = 2. Let nc be the number of individuals in the cth phenotype group. Let Xci = (Xci1, …, XciK) be the observation vector over the K SNPs for the ith individual in the cth group, for i = 1, …, nc, where Xcik is the genotype of the ith individual in the cth group at the kth SNP that takes one of the three possible genotype values in G. The probability law of Xci is denoted by πc = {πc(g):g ∈ G × G … × G}, where πc(g) is the probability of observing genotype g in phenotype group c. We are interested in testing the null hypothesis of homogeneity of the πc, c = 1, 2, …, C.
Since the space of the alternative hypotheses is very large, the standard multi-way contingency table analysis to test for global association suffers loss of power. Instead, following Sen (2006), we consider defining a test statistic based on the U-statistics (Hoeffding, 1948). We first define a symmetric kernel between a pair (i, j) of observations Xi = {Xi1, …, XiK} and Xj = {Xj1, …, XjK} as
| (1) |
where wk is a SNP-specific weight. This kernel function can be regarded as a weighted Hamming distance between individuals i and j over the K SNPs. Note that the definition of this kernel does not depend on particular specifications of the high- or low-risk alleles. The weight can be defined based on prior knowledge of the importance of the K SNPs. Alternatively, we can take a data-adaptive weight as wk = −log(Pk) where Pk is the p-value based on a univariate test for the kth SNP. Using this weight, the SNPs with smaller p-values are given larger weights.
Instead of simply considering the difference of the kernel (1) between cases and controls as in Schaid et al. (2005), we propose to derive a test statistic following Sen (2006) by considering both the within-group and the between-group U-statistics. Specifically, for phenotype group c, we define the within-group U-statistic as
| (2) |
where nckg is the number of individuals in the cth group for which at the kth SNP the observed genotype label is g. Note that if the within-group genotypes are all the same, then Ucc = 0. Similarly, for phenotype group c and c′, we define the between-group U-statistic as
| (3) |
From this equation, we note that a larger difference in genotype distribution between the cth and the c′th group corresponds to a larger value of Ucc′.
Let n = n1 + n2 + …+ nC be the total number of individuals across all the C phenotype groups and let U0 be the pooled group U-statistic corresponding to the same kernel φ, which can be written as
| (4) |
which can then be decomposed into within-group component W and between-group component B, where Ucc and Ucc′ are defined as in equations (2) and (3). Under the null hypothesis, B has zero expectations and it is positive under the alternative. We define the following statistic for testing the association between K genotypes and a discrete phenotype,
which is the ratio of the between-group contribution versus the within-group contribution to the pooled U-statistic. For data-adaptive weights wk = −log(Pk), which depends on the data, the asymptotic distribution of Td is unclear. We therefore determine the critical region of the test statistic Td by permutations. Specifically, we permute the discrete trait labels M times, and for each permutation m, we calculate the test statistic and obtain the permutation-based p-value as .
2.2 Nonparametric tests for quantitative traits
In this section, we consider constructing a test for testing the association between a group of SNPs and a quantitative trait phenotype Y based on the U-statistics. Let Yi be the observed trait value for the ith individual for i = 1, …, n. Let Xi = (Xi1, …, XiK) be the observation genotype vector over the K SNPs for the ith individual for i = 1, …, n, where Xik is the genotype of the ith individual at the kth SNP that takes one of the three possible genotype values G = {00, 10, 11}, where we assume that allele 1 is the minor allele. The hypothesis that we wish to test is H0: F (Y|X) = H(Y), where F (Y|X) is the conditional distribution function of Y given X, and H(Y) is the marginal distribution function of Y.
To define the U-statistics, for marker k, we define the set Sgk = {i: Xik = g, i = 1, …, n} the individuals with genotype g at the kth marker for g ∈ G and k = 1, …, K and let mgk = |Sgk| be the number of such individuals. Consider a kernel function between two trait values Yi and Yj as
| (5) |
We define the following U-statistics for SNP k,
which compare the quantitative trait values between every two genotype groups at the SNP k, where θk0 = E(φ(Yi, Yj) for i ∈ S10k, j ∈ S11k and θk1 and θk2 are similarly defined. Under the null hypothesis, θkj = 0 for j = 0, 1, 2 and let Ukj = Ukj0, j = 1, 2, 3. In order to combine these three U-statistics, we assume that the quantitative trait value is a monotone function of the number of the minor allele at the trait-associated SNPs and further define
To define a statistic over K SNPs, we consider the multivariate U-statistic (U1, …, UK)′, which has limiting normal distribution with zero mean, and limiting covariance matrix Σ = ((σkl)). It is easy to show that Σ can be consistently estimated by Σ̂ (see Appendix). In order to draw an overall conclusion on association between the K SNPs and the quantitative trait, we consider a linear combination of the statistics Uk0 defined as the test statistic
where wk is a data-adaptive weight. We consider the data-adaptive weight wk = −log(Pk)sign(rk) where Pk is the p-value based on a univariate test for the kth SNP, and rk = corr(Y, gk) is the correlation between the observed trait values Y = {Y1, …, Yn} and the genotypes gk at the kth SNP coded by counting the numbers of minor alleles. The rationale of using the sign of the correlation in the weight is to account for the fact that the minor alleles across all of the K SNPs can either increase or decrease the trait phenotype. We then define a statistic for testing the association between K genotype and a continuous trait as
| (6) |
Using the data-adaptive weight vector, the asymptotic distribution of the test statistic Tc is no longer the standard normal distribution. Its significance level is again estimated using permutations by randomly permuting the continuous trait values across all the individuals.
Finally, if we can make an assumption on the mode of inheritance as dominant or recessive, we can similarly define a U-statistics-based test statistic based on comparing two genotype groups, {00} vs. {10,11} for the dominant model or {00, 10} vs. {11} for the recessive model.
3 Simulation Studies
We performed simulations to evaluate the power of the proposed U-statistics-based tests and to compare with some of the standard methods. Since significance levels of the proposed test statistics are determined by permutations of the phenotypes, the type 1 errors of these tests are automatically controlled and we therefore did not report the results of the type 1 error evaluations.
3.1 Simulation studies for qualitative traits
For the first simulation study, we generated the data set as described in Schaid et al. (2005). In this simulation, the genotypes for 10 independent markers were simulated. Of these 10, the number of markers associated with disease ranged from 1 to 10. The frequency of each high-risk allele, for all markers, was set to 0.15. Hardy-Weinberg proportions were used to generate the genotypes for the controls, and the genotypes for cases were generated by assuming that the high-risk allele had a multiplicative effect on the odds ratio. The effect per allele was set at an odds ratio of 1.5. The total sample size was set to 500 individuals, of which half were cases and half were controls. All simulations were based on 500 replicates. For each replicate, 50,000 permutations were used to estimate the p-values.
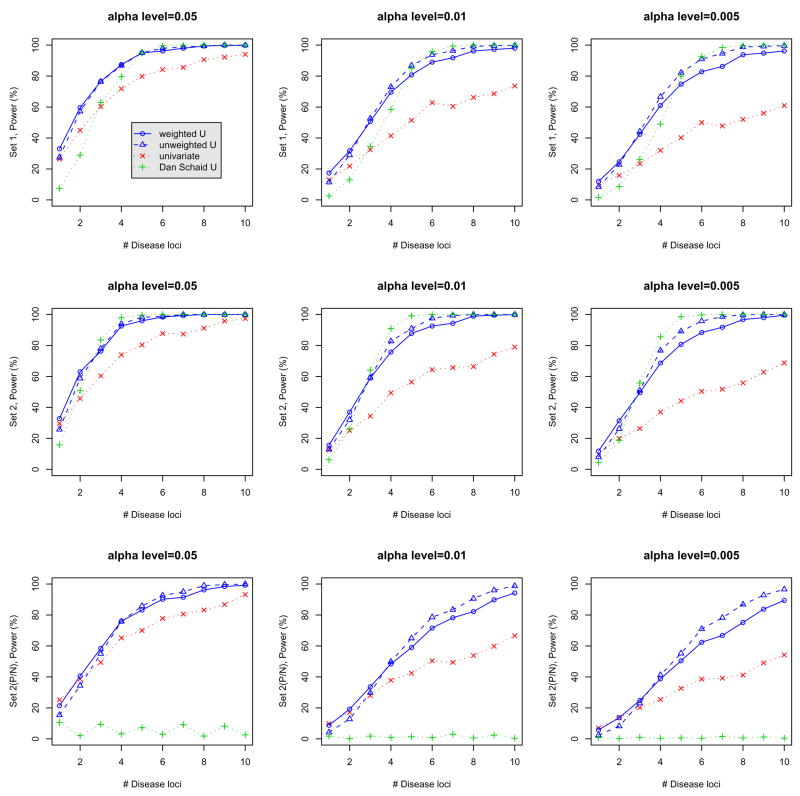
The top panel of Figure 1 shows the power of the four different tests, including the unweighted U-statistics-based test, weighted U-statistics-based test, the maximum of univariate χ2-test with Bonferroni correction for multiple testing, and the test proposed by Schaid (2005) using “linear-dosage” kernel. Each evaluation considered three different α-levels of 0.05, 0.01 and 0.005, and different number of disease genes ranging from 1 to 10. These figures illustrate that, as the number of high-risk SNPs increases, there is a gain in power of the proposed U-statistic-based tests, and the gain is greater when the number of true disease-related SNPs increases. When there are only one or two disease SNPs, the unweighted U-statistic-based test performs similarly in power when compared with the single marker analysis, but the weighted test provides slightly higher power than the single-SNP test. As expected, when the number of disease SNPs is high, the weighted test is less powerful than the unweighted test. We also observed that the proposed tests have almost the same power as Schaid’s test.
Figure 1.
Comparison of power for different alpha-levels (0.05, 0.01, and 0.005) when the 250 case-control pairs were simulated to have a marginal risk ratio of 1.5 (top panel), to have a fixed disease prevalence of 5% (middle panel) or to have both high-risk and protective markers (bottom panel). Weighted (unweighted) U: our poposed weighted (unweighted) U-statistics-based tests; univariate: maximum χ2 test with Bonferonni correction; Schaid U: U-statitics-based test of Schaid et al. (2005) using lienar kenels.
For the second simulation study, we fixed the disease prevalence at 5%. Briefly, we generated genotypes for 10 independent markers, with the number of markers associated with the disease loci ranging from 1 to 10. All markers had minor allele frequency 0.3 and the genotypes were generated following Hardy-Weinberg proportions in the general population. The minor alleles were designated as the high-risk alleles. We assigned penetrance as , where gi ∈ {0, 1, 2} is the number of risk alleles at disease locus i and D ∈ {1, …, 10} is the number of disease loci. This is equivalent to assuming multiplicative effects across disease loci on the odds scale. The parameters βi were chosen so that the locus-specific sibling recurrence risk ratio λs = 1.02, corresponding to genotype relative risks of 1.34 and 1.79 for having one and two copies of the risk alleles, respectively. The intercept β0 was chosen so that the population disease prevalence was 5%. The second panel of Figure 1 shows the power of the three different tests for three different α-levels and a different number of disease genes ranging from 1 to 10. Similar patterns were observed as in previous simulations.
For the last set of simulations, we considered the model where the minor alleles correspond to both disease-predisposing and protective loci among the SNPs considered. The simulation set-up was the same as the second simulation study except that for markers 2, 4, 6, 8 and 10, the corresponding βs were negative so that the minor alleles were protective. Similar patterns were observed as in previous simulations for the proposed U-statistics-based tests. However, the bottom panel of Figure 1 shows that Schaid’s test using a “linear-dosage” kernel can have very low power under these conditions when there are both disease-predisposing and protective minor alleles. This is expected, since in Schaid’s U-statistics, the scores derived from both disease-predisposing and protective minor alleles can potentially cancel each other out and hence can eliminate any potential signal for the association.
3.2 Simulation studies for quantitative traits
To evaluate the performance of the proposed U-statistics-based test for quantitative traits, we simulated the trait values based on the following model,
| (7) |
where Xk = 0, 1, 2 for the three genotypes at the kth disease gene, and ε is error term following N(0, 1). We considered the scenarios when there are 1–10 disease genes. For each disease gene, we chose the minor allele frequency and the regression coefficient to explain 1% of the total trait variance when considered individually. Specifically, for minor allele frequencies of 0.1, 0.3 and 0.5, the corresponding βs are 0.24, 0.16 and 0.14, respectively. For each model, 500 individuals were simulated for each replicate and a total of 500 replicates were performed. For each simulation, 50,000 permutations were used to estimate the p-values.
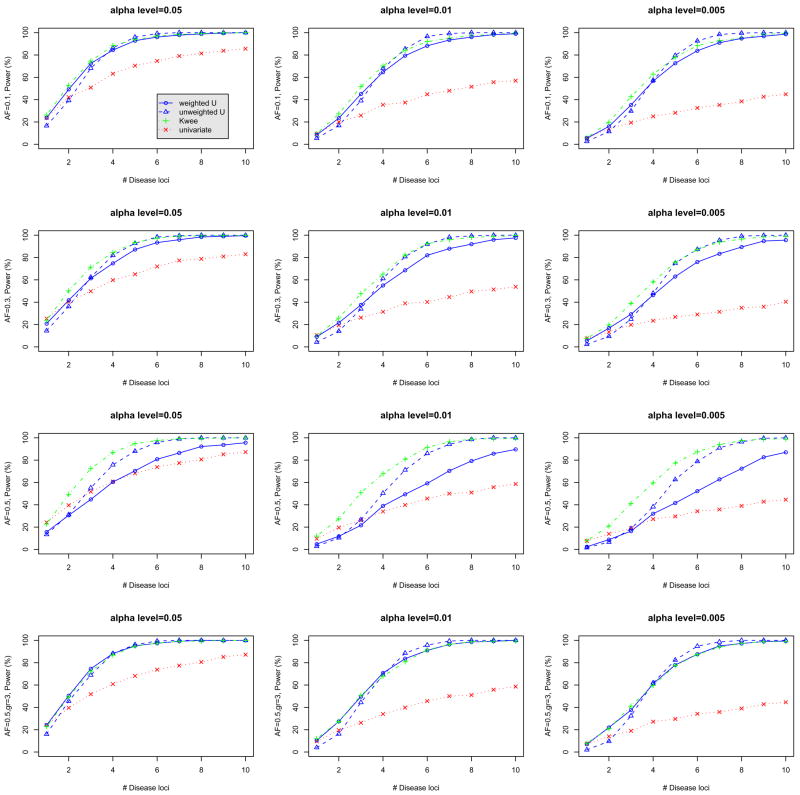
Figure 2 shows the power of the three different tests for α-levels of 0.05, 0.01 and 0.005. The U-statistics of the top three panels were derived by assuming a dominant model for each of the markers. Clearly, we observed substantial increases in power comparing the single marker tests with Bonferroni corrections, especially when the number of disease markers was large. In addition, we observed a very small loss of power when there were only one or two disease markers. We also observed that when the minor allele frequency is 0.1, the number of individuals in the 11 genotype group is small and the resulting U-statistic test based on three genotype groups is not as powerful as the test based on two genotype groups by assuming dominant models (results not shown). However, when the minor allele frequency is not too small, the U-statistics tests using three genotype groups can lead to a gain in power (see the fourth row of Figure 2).
Figure 2.
Comparison of power for different alpha-levels (0.05, 0.01, and 0.005) and for a minor allele frequency of 0.1, 0.3 and 0.5 (top, middle and bottom two panels) when each disease gene can marginally explain 1% of the trait variance. U-statistics assuming a dominant model were used for the top three panels and the general three-group U-statistic was used for the last panel. AF: allele frequency. Weighted (unweighted) U: our poposed weighted (unweighted) U-statistics-based tests; univariate: maximum χ2 test with Bonferonni correction; Kwee: multilocus test of Kwee et al. (2008).
We also compared the power of the proposed tests with the multilocus association test of Kwee et al. (2008) (see plots in Figure 2). The powers were very comparable between these two tests, except for the case when the minor allele frequency was large and a dominant model was assumed (see the third penal of Figure 2). This is not surprising since there were indeed three genotype groups with different trait values. When the test based on the three-group U-statistics was used, the power was essentially the same as the test of Kwee et al. (2008) (see the third penal of Figure 2).
In summary, these simulations indicate that our proposed U-statistics-based tests for multiple SNPs have similar or better power than some of the recently developed statistical tests. The new tests have much better power than the simple single marker tests with multiple-comparison adjustments. These simulations also indicate that the p-values weighted tests do not gain any power over the unweighted tests when there are indeed multiple disease-associated SNPs in the set. We would therefore recommend the use of unweighted tests in practice.
3.3 Simulation based on LD
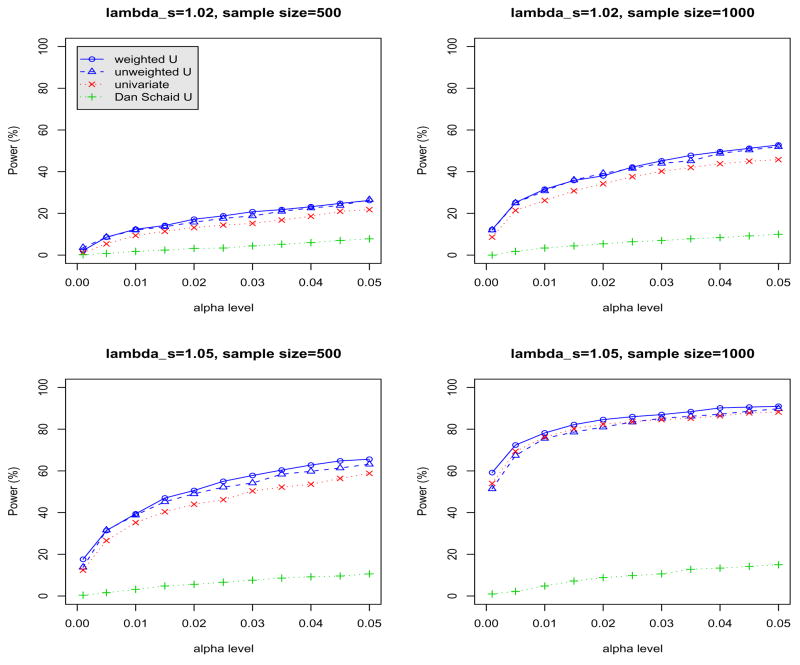
We also evaluated whether the proposed tests can gain power in the analysis of SNP data that are in LD with the disease variants. To simulate such data, we used the algorithm of Durrant et al. (2004). We downloaded the phased genotype data for 60 CEU (CEPH samples with ancestry from northern and western Europe) founder subjects from HapMap release #21 (www.hapmap.org). As the reference data, we picked the haplotypes of 11 SNPs on chromosome 6, SNP 6 (MAF = 0.25) was assigned as the disease locus, and the minor allele was designated as the risk allele with locus-specific sibling recurrence risk ratio λs = 1.02 and 1.05. The disease locus displayed moderate to strong LD with the other SNPs in the CEU samples, with r2 values ranging from 0.47 to 0.83. We simulated m cases and m controls (m = 500, 1000). For each individual, we first generated genotypes at the pre-determined disease locus and assigned one allele to each of the two haplotypes carried by that individual. The remaining genotypes of each haplotype were generated as followings: let d denote the disease locus. For each haplotype, given the allele at d, the algorithm starts by picking, at random, a five-SNP haplotype from the 120 CEU haplotypes at markers [d −2, d + 2] that has the same allele at d. The algorithm then gradually grows the haplotype as follows: for markers on the right side of the disease locus, it generates an allele at locus d+i given the haplotype at [d+i−4, d+i−1] for i −3; the conditional probabilities for the alleles at locus d+i given the haplotype at [d+i−4, d+i−1] are determined based on the CEU phased data. Similarly, for markers on the left side of the disease locus, the algorithm generates an allele at locus d−i given the haplotype at [d−i+1, d−i+4] for i ≥ 3. By generating haplotypes this way, the simulated haplotypes are not exact copies of those in the original HapMap samples. Instead, the 120 CEU founder haplotypes are used to generate plausible haplotypes that may be representative of a wider population. The disease locus genotypes were removed prior to data analysis. For each simulation, 50,000 permutations were used to estimate the p-values.
Figure 3 shows the power of the three tests for various α-levels (x-axis). Again showing that both weighted and unweighted U-statistics-based tests resulted in better power in detecting the associations between the SNP markers of the diseases than a single SNP test with Bonferroni corrections, although the increase is not substantial. However, it is important to note that the Schaid’s test using a linear kernel gives very low power when directions of the LDs between the SNPs and the true disease variant are different. This agrees with our previous simulations when there are both predisposing and protective minor alleles.
Figure 3.
Comparison of power for different α-levels (x-axis) and sample size of 500 and 1000 when only one disease gene is simulated. The tests are based on the 10 SNP markers that are in LD with the disease variant, which is removed from the analysis. Weighted (unweighted) U: our poposed weighted (unweighted) U-statistics-based tests; univariate: maximum χ2 test with Bonferonni correction; Schaid U: U-statitics-based test of Schaid et al. (2005) using lienar kenels.
4 Application to Real Data Sets
In this section, we present applications of the proposed methods for analysis of an association between the genes in the hormone metabolism pathway and the risk of breast cancer and breast cancer age of diagnosis.
4.1 Application to breast cancer case-control data set
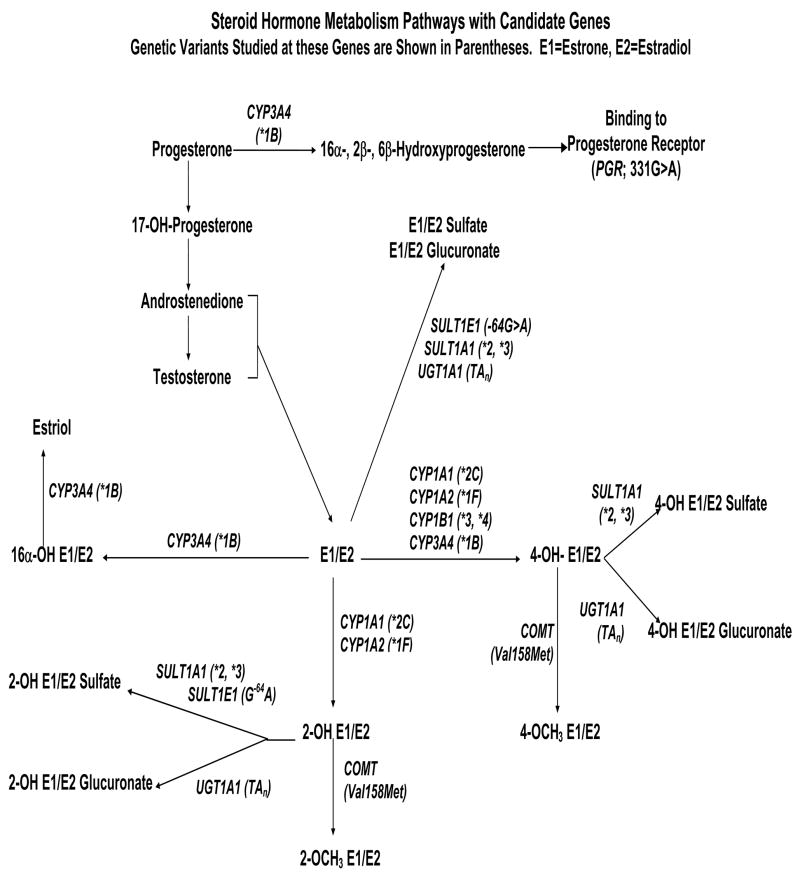
It has long been recognized that female hormones, whether endogenous or exogenous, can be risk factors for female cancers (Davis and Sieber, 1997). In order to explore the cause of susceptibility to these hormone-associated cancers, we undertook a population-based association study of genetic variants in candidate steroid hormone metabolism genes and cancer risk. The Women’s Insights and Shared Experiences (WISE) study used incident breast cancer cases and frequency-matched controls selected from the community using random digit dialing (RDD). Additional details of our study design can be found in Strom et al. (2006), Rebbeck et al. (2006) and Bunin et al. (2006). Genomic DNA was obtained from each participant. Eleven variants in nine genes were selected for study based on their role in the downstream metabolism of steroid hormones (Table 1 and Figure 4), where the binary codings of the SNP genotypes were determined by the functionality of the SNPs. For genes PGR, SULT1A1 and SULT1E1, two different codings (dominant on A allele and dominant on G allele) are considered. For gene UGT1A1, alleles *1 or *33 are low-risk alleles and allele *24 or *34 are high-risk alleles. Details of the genotype analyses can be found in Rebbeck et al. (2006). Table 1 presents the p-value for each SNP based on the univariate logistic regression, indicating that the two polymorphisms in CYP1B1, the SNP in CYP3A4 and one polymorphism in SULT1A1 are associated with the risk of breast cancer. After Bonferroni correction for multiple testing, CYP3A4 A729G and SULT1A1 A667G remain significant at the 0.01 level. Both of these associations are biologically plausible: these genotypes are associated with altered estrogen and catecholestrogen metabolism, and would be predicted to alter breast cancer risk (Raftogianis et al., 1999; Amirimani et al., 2003).
Table 1.
Steroid hormone metabolism pathways with 11 candidate genes for breast cancer in WISE study. Genetic variants studied at these 11 genes are shown in the second column, where the binary codings of the SNP genotypes were determined by the functionality of the SNPs. The numbers are the p-values based on the univariate logistic regression for case-control data (column BCA) and linear regression analysis for age of diagnosis data for each variant. For genes PGR, SULT1A1 and SULT1E1, two different codings (A-dominant: dominant on A allele, G-dominant: dominant on G allele) are considered. For gene UGT1A1, allele *1 or *33 is a low-risk allele and allele *24 or *34 is a high-risk allele, and the number of high-risk alleles is used in the regression analysis.
| Gene | Polymorphsm | Genotype Coding | BCA | Age of diagnosis |
|---|---|---|---|---|
| COMT | G1947A | 1=T/T 0=C/T 0=C/C | 0.27 | 0.15 |
| CYP1A1 | A6750G | 0=A/A 1=A/G 1=G/G | 0.20 | 0.65 |
| CYP1A2 | C734A | 0=C/C 1=C/A 1=A/A | 0.62 | 0.018 |
| CYP1B1 | G1294C (C4326G) | 0=G/G 1=G/C 1=C/C | 0.013 | 0.73 |
| CYP1B1 | A1358G (A3290G) | 0=A/A 1=A/G 1=G/G | 0.0040 | 0.12 |
| CYP3A4 | A729G | 0=A/A 1=A/G 1=G/G | 4.90 × 10−4 | 0.086 |
| PGR | G331A | 0=GG 1=AG 1=AA | 0.59 | 0.17 |
| 1=GG 1=AG 0=AA | 0.19 | 0.50 | ||
| SULT1A1 | G638A | 1=AA 1=AG 0=GG | 0.12 | 0.60 |
| 0=AA 1=AG 1=GG | 0.28 | 0.51 | ||
| SULT1A1 | A667G | 0=AA 1=AG 1=GG | 8.34 × 10−6 | 0.041 |
| 1=AA 1=AG 0=GG | 0.0072 | 0.33 | ||
| SULT1E1 | G-64A | 0=G/G 1=A/A 1=A/G | 0.71 | 0.51 |
| UGT1A1 | TAn | *1 or *33 (low) | 0.71 | 0.088 |
| *24 or *34 (high) |
Figure 4.
Steroid hormone metabolism pathways with candidate genes for breast cancer in the WISE study. Genetic variants studied at these genes are shown in parentheses.
In order to demonstrate our proposed tests, we applied various statistical methods for testing the overall association between the 11 variants in the metabolism pathway and breast cancer risk. Table 2 shows the p-values based on various procedures. The maximum χ2 analysis with permutations or the minimum p-value with Bonferroni correlations for multiple testing all indicate that there are SNPs in the metabolism pathway that are significantly associated with the risk of developing breast cancer. The proposed U-statistics tests with and without weighting based on 100,000 permutations also indicate that overall the genes in the hormone metabolism pathway are significantly associated with breast cancer risk. Compared to single-marker analysis with Bonferroni corrections for multiple testing, our proposed tests provide a more significant assessment for such an association, as reflected by smaller overall p-values. It is also interesting to observe that G-dominant codings for PGR, SULT1A1 and SULT1E1 genes provided stronger evidence of association between the hormone metabolism pathway and breast cancer risk than A-dominant codings.
Table 2.
p-values from three different procedures for testing the association between the 11 SNPs on the hormone metabolism pathway and breast cancer risk or age of onset of breast cancer for the WISE data set. For genes PGR, SULT1A1 and SULT1E1, two different codings (A-dominant: dominant on A allele, G-dominant: dominant on G allele) are considered. U-stat: proposed U-statistics test with wk = 1; weighted U-stat: proposed U-statistics-based test with wk = − log(Pk) where Pk is the p-value from single-marker test for the kth marker; min P -value: minimum p-value over all 11 single-marker p-values with Bonferonni adjustment for multiple comparisons.
| Breast cancer risk
|
Age of onset
|
|||
|---|---|---|---|---|
| Test | A-dominant | G-dominant | A-dominant | G-dominant |
| U-stat | 0.00016 | 0.00 | 0.016 | 0.66 |
| weighted U-stat | 0.00063 | 0.00001 | 0.022 | 0.44 |
| min P-value* | 0.0054 | 0.000091 | 0.20 | 0.20 |
with Bonferonni adjusment.
The associations that have been identified here are biologically plausible. The statistically significant genotypes we identified are associated with altered estrogen and catecholestrogen metabolism, and would be predicted to alter breast cancer risk in a manner consistent with that observed in our analyses (Raftogianis et al., 1999; Amirimani et al., 2003). CYP3A4 is associated with the formation of 4-hydroxy estrone and estradiol (4-OH E1/E2; Fig 4). Increased 4-OH has been associated with genotoxicity and free radical generation and has been hypothesized to be associated with increased breast cancer risk (Yager and Liehr, 1996). Therefore, genotypes that may influence the formation of 4-OH E1/E2 are likely candidates for breast cancer susceptibility. The function of the CYP3A4 variant studied here is not resolved, but this variant has also been associated with increased prostate cancer risk as well as differences in hormone-related breast cancer risk factors such as age at menarche (Kadlubar et al., 2003). Similarly, SULT1A1 and SULT1E1 are involved in the sulfation of 2-OH E1/E2, 4-OH E1/E2 (Fig 4). The SULTs play a critical role in removing potentially genotoxic catecholestrogens by this sulfation mechanisms (Raftogianis et al., 1999). The SULT1A1 genotype studied here has been associated with increased estrogenicity and mutagenicity in the context of pathways that are involved in estrogen metabolism, and therefore the genotypes that are associated with variation in these pathways. The function of the SULT1E1 genotype remains unclear, but it is hypothesized to have a regulatory effect on the expression or activity of SULT1E1. Finally, PGR has two distinct isoforms (hPR-A and hPR-B) encoded from a single gene (PGR). Carriage of 331A allele is associated with the presence of the more biologically active progesterone receptor isoform is hypothesized to promote breast cell proliferation (De Vivo et al., 2002). Thus, our associations all involve genes and genetic variants that are biologically plausible causes of breast cancer.
4.2 Application to breast cancer age of diagnosis data set
We next examined whether the genetic variants in hormone metabolism pathway are associated with age of breast cancer diagnosis among the cases in the WISE data set. The last column of Table 1 shows the p-value from simple linear regression analysis for each SNP, indicating that CYP1A2 is associated with early onset among the breast cancer patients (p=0.018). However, the result is not statistically significant after the Bonferroni adjustment for multiple testing.
Table 2 presents the results based on the proposed U-statistics. The overall p-value is 0.016 using the unweighted test and 0.022 using the weighted test when the A-dominant codings are used for the SNPs in PGR, SULT1A1 and SULT1E1. This indicates that overall genetic variations in the hormone metabolism pathway are also related to age of breast cancer diagnosis, further demonstrating the benefit of the proposed global test for association. Finally, if G-dominant codings are used for the three polymorphisms, the results are not significant.
5 Discussion
Since many complex phenotypes are expected to be controlled by many genes each with small effects, single-marker tests of association can suffer a great loss of power due to genetic heterogeneity and multiple testing. A large body of biological knowledge suggests that genes often work as networks of pathways instead of acting alone to affect phenotype and disease risk. Since these pathways often have complex interactions and feedback loops, it would not be surprising to find that multiple genes within a biological pathway are associated with these complex phenotypes. This makes pathway-based genetic association analysis an attractive approach for identifying genes related to complex phenotypes. In this paper, we have proposed data-adaptive U-statistics-based tests for testing the association between multiple markers in a pathway and a phenotype. Our approach is quite general and does not require any parametric assumptions on the trait values or genetic models. This approach is particularly useful for pathway-based candidate gene association studies, where SNPs in a candidate gene can be tested simultaneously for association with the phenotype using knowledge of biological functions. Our simulation results demonstrate that our approach performs similarly to the U-statistic test defined by Schaid et al., (2005) or the multilocus test proposed by Kwee et al. (2008) and can be more powerful than standard single- marker-based methods under some conditions. However, our test statistic has better power than Schaid’s test when there are both high-risk and protective minor alleles of the SNPs among the SNP set. Application to the WISE breast cancer data sets illustrates the potential merits of our statistics over the standard single-SNP analysis.
There are several issues that deserve further investigation. First, we studied only the kernel function φ (., .) defined using the Hamming distance (see equation (1)) for the qualitative phenotype, and the kernel defined by trait value difference for the quantitative phenotype. These kernels are chosen without making strong assumptions on genetic models and trait distribution and tend to be more robust in power as compared to for example the linear kernel used by Schaid et al. (2005). However, other kernel functions can be considered in the definition of the U-statistics. For example, for the quantitative phenotype, rank-based kernel defined by φ (x, y) = 1 if y > x and 0 otherwise, can be used. For the qualitative phenotype, Schaid et al. (2005) presented several interesting kernels that can be applied in combination with our definitions of the U-statistics. However, some of these kernels are sensitive to model assumption, which can lead to lower power if the assumption is not met. Second, while a significant global test of a set of SNPs suggests that some variants in the set are associated with the phenotype, it is not immediately clear which SNPs have led to the statistical significance. To identify signal contributing SNPs, one possible approach, as suggested by Schaid et al. (2005), is to use a stepwise removal procedure that involves the following steps: 1) remove the marker with the smallest p-value, 2) recompute the adjusted global U-statistics-based test after that SNP is removed, 3) repeat steps 1) and 2) until the adjusted global U-statistics-based test is no longer significant. Alternatively, one may perform the standard step-wise regression analysis to identify the most relevant SNPs. Finally, since the proposed U-statistics-based tests are not model-based and are nonparametric, covariates cannot be naturally handled in these tests. For quantitative traits, one can first perform regression analysis to adjust for possible covariate effects and then apply our proposed test on the residuals.
The proposed methods also have potential applications in genome-wide association studies (GWAS). GWAS often involve genotyping of hundreds of thousands of SNPs. For example, the Illumina 550K array can be used to type approximately 550,000 SNP markers on each individual. To account for allelic heterogeneity, one may want to perform joint tests of all the SNPs in both intragenic and regulatory regions of a given gene using the proposed test statistics. This gene-based association analysis makes more biological sense since genes, not the SNPs, are the true functional unit of biology (Neale and Sham, 2003). Additionally, one can also consider using pathway databases to perform pathway-based analysis for GWAS. Our simulations and analysis of breast cancer dataset indicate that by jointly considering multiple contributing factors in the same pathway, one can potentially identify sets of associated SNPs that would be missed by single SNP analysis. Recently studies (Wang et al., 2007; Dinu et al., 2007) have also clearly demonstrated the potential insights that one can gain by integrating pathways information into analysis of GWAS data. An interesting direction for future research is to develop methods for analysis of data from GWAS, where the SNP data have natural hierarchical structures, i.e., genes belong to pathways, and SNPs belong to genes. When there are many pathways under consideration, our proposed tests can be applied to each of the pathways and the false discovery rate (Benjamini and Hochberg, 1995; Efron, 2004) procedure can be used for correcting for multiple pathways. Alternatively, a recently developed non-parametric pathway-based regression (Wei and Li, 2006) can be used for selecting the relevant pathways. Detailed comparisons of these different approaches deserve further investigation.
In summary, we have proposed two U-statistics-based tests that provide a simultaneous test of association of multiple genetic markers with complex phenotypes (R codes are available upon request). The tests can be applied to pathway-based association analysis and have potential applications in gene-based genetic association analysis in genome-wide genetic association studies.
Acknowledgments
This research was supported by NIH grants R01-ES009911, U19-AG023122 and P01-CA77596. The authors wish to thank Edmund Weisberg, MS for editorial assistance and to acknowledge the contributions of the WISE study collaborators: Andrea B. Troxel, Yiting Wang, Amy H. Walker, Saarene Panossian, Stephen Gallagher, Ekaterina G. Shatalova, Rebecca Blanchard, Sandra Norman, Greta Bunin, Angela DeMichele, Stephen C. Rubin, Mona Baumgarten, Michelle Berlin, Rita Schinnar, Jesse A. Berlin, Anita Weber, Elene Turzo, Shawn Fernandez, Desiree Burgh, J. A. Grisso, and Brian L. Strom.
Appendix
We provide some details on estimating the covariance matrix under the null hypothesis that the markers are not associated with the phenotype for the proposed test statistic Tc defined in equation (6). For SNP k and l, we have
We provide some details on estimating E(Uk1Ul1). Other terms can be estimated similarly. When p = 1, q = 1, we have
where i ∈ S10k, j ∈ S11k, i′ ∈ S10l, j′ ∈ S11l and
For the quadruplet (i, j, i′, j′), we have
where N is the total sample size. Therefore,
where
These expectations can be estimated by their empirical means to obtain the estimate of the covariance matrix Σ̂, which is used in our definition of the test statistic Tc defined in equation (6).
References
- Amirimani B, Ning B, Deitz AC, Weber BL, Kadlubar F, Rebbeck TR. Transcriptional activity effects of a CYP3A4 promoter variant. Environmental and Molecular Mutagenesis. 2003;42(4):299–305. 57. doi: 10.1002/em.10199. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society, Ser B. 1995;57:289–300. [Google Scholar]
- Bunin GR, Baumgarten M, Norman SA, Strom BL, Berlin JA. Practical aspects of sharing controls between case-control studies. Pharmacoepidemiology and Drug Safty. 2005;14(8):523–30. doi: 10.1002/pds.1130. [DOI] [PubMed] [Google Scholar]
- Conti DV, Cortessis V, Molitor J, Thomas DC. Bayesian modeling of complex metabolic pathways. Human Heredity. 2003;56:8393. doi: 10.1159/000073736. [DOI] [PubMed] [Google Scholar]
- Davis DL, Sieber SM. Hormones, hormone metabolism, environment, and breast cancer: a workshop of the National Action Plan on Breast Cancer’s Etiology Working Group. Environmenal Health Perspectives. 1997;105(Suppl 3):557. doi: 10.1289/ehp.97105s3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vivo I, Guggins GS, Hankinson SE, Lescault PJ, Boezen M, Colditz GA, Hunter DJ. A functional polymorphism in the promoter of the progesterone receptor gene associated with endometrial cancer risk. Proc Natl Acad Sci U S A. 2002;99(19):12263–8. doi: 10.1073/pnas.192172299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinu V, Zhao H, Miller PL. Integrating domain knowledge with statistical and data mining methods for high-density genomic SNP disease association analysis. Journal of Biomedical Informatics. 2007;40(6):750–760. doi: 10.1016/j.jbi.2007.06.002. [DOI] [PubMed] [Google Scholar]
- Durrant C, Zondervan KT, Cardon LR, Hunt S, Deloukas P, Morris AP. Linkage disequilibrium mapping via cladistic analysis of single-nucleotide polymorphism haplotypes. American Journal of Human Genetics. 2004;75(1):35–43. doi: 10.1086/422174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Easton DF. How many more breast cancer predisposition genes are there? Breast Cancer Research. 1999;1(1):14–17. doi: 10.1186/bcr6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efron B. Large-scale simultaneous hypothesis testing: the choice of a null hypothesis. Journal of American Statistical Association. 2004;99:96–104. [Google Scholar]
- Ford D, Easton DF, Stratton M, Narod S, Goldgar D, Devilee P, Bishop DT, Weber B, Lenoir G, Chang-Claude J, Sobol H, Teare MD, Struewing J, Arason A, Scherneck S, Peto J, Rebbeck TR, Tonin P, Neuhausen S, Barkardottir R, Eyfjord J, Lynch H, Ponder BAJ, Gayther SA, Birch JM, Lindblom A, Stoppa-Lyonnet D, Bignon Y, Borg Y, Hamann U, Haites A, Scott RJ, Maugard CM, Vasen H, Seitz S, Cannon-Albright LA, Schoffeld A, Zelada-Hedman A Breast Cancer Linkage Consortium. Genetic Heterogeneity and Penetrance Analysis of the BRCA1 and BRCA2 Genes in Breast Cancer Families. The American Journal of Human Genetics. 1998;62(3):676–689. doi: 10.1086/301749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeffding W. A class of statistics with asymptotically normal distribution. Annals of Mathematical Statistics. 1948;22:165–179. [Google Scholar]
- Kadlubar FF, Berkowitz GS, Delongchamp RR, Wang C, Green BL, Tang G, Lamba J, Schuetz E, Wolff MS. The CYP3A4*1B variant is related to the onset of puberty, a known risk factor for the development of breast cancer. Cancer Epidemiology, Biomarkers & Prevention. 2003;12(4):327–31. [PubMed] [Google Scholar]
- Kwee LC, Liu D, Lin X, Ghosh D, Epstein MP. A powerful and fiexible multilocus association test for quantitative traits. American Journal of Human Genetics. 2008;82(2):386–97. doi: 10.1016/j.ajhg.2007.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert JC, Amouyel P. Genetic heterogeneity of Alzheimer’s disease: complexity and advances. Psychoneuroendocrinology. 2007;1(Suppl):S62–70. doi: 10.1016/j.psyneuen.2007.05.015. [DOI] [PubMed] [Google Scholar]
- Longmate JA. Complexity and power in case-control association studies. American Journal of Human Genetics. 2001;68:1229–1237. doi: 10.1086/320106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neale B, Sham P. The future of association studies: gene-based analysis and replication. American Journal of Human Genetics. 2004;75:353362. doi: 10.1086/423901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raftogianis RB, Wood TC, Otterness DM, Van Loon JA, Weinshilboum RM. Phenol sulfotransferase pharmacogenetics in humans: association of common SULT1A1 alleles with TS PST phenotype. Biochemistry and Biophysics Research Communication. 1997;239(1):298–304. doi: 10.1006/bbrc.1997.7466. [DOI] [PubMed] [Google Scholar]
- Rebbeck TR, Troxel AB, Wang Y, Walker AH, Panossian S, Gallagher S, Shatalova EG, Blan-chard R, Norman S, Bunin G, DeMichele A, Rubin SC, Baumgarten M, Berlin M, Schinnar R, Berlin JA, Strom BL. Estrogen sulfation genes, hormone replacement therapy, and endometrial cancer risk. Journal of the National Cancer Institute. 2006;98(18):1311–1320. doi: 10.1093/jnci/djj360. [DOI] [PubMed] [Google Scholar]
- Schaid DJ, McDonnell SK, Hebbring SJ, Cunningham JM, Thibodeau SN. Nonparametric tests of association of multiple genes with human disease. American Journal of Human Genetics. 2005;76(5):780–93. doi: 10.1086/429838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen PK. Robust statistical inference for high-dimensional data models with application to genomics. Austrian Journal of Statistics and Probability. 2006;35:197–214. [Google Scholar]
- Slager SL, Huang J, Vieland VJ. Effect of allelic heterogeneity on the power of the transmission disequilibrium test. Genetic Epidemiology. 2000;18:143–156. doi: 10.1002/(SICI)1098-2272(200002)18:2<143::AID-GEPI4>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- Strom BL, Schinnar R, Weber AL, Bunin GR, Berlin JA, Baumgarten M, DeMichele AM, Rubin SC, Berlin M, Troxel AB, Rebbeck TR. Protective effect of postmenopausal use of combined estrogen plus progestin hormone therapy on endometrial cancer risk. American Journal of Epidemiology. 2006;164(8):775. doi: 10.1093/aje/kwj316. [DOI] [PubMed] [Google Scholar]
- Thomas DC. The need for a systematic approach to complex pathways in molecular epidemiology. Cancer Epidemiology Biomarkers & Prevention. 2005;14:557–559. doi: 10.1158/1055-9965.EPI-14-3-EDB. [DOI] [PubMed] [Google Scholar]
- Wang K, Li M, Bucan M. Pathway-based approaches for analysis of genomewide association studies. American Journal of Human Genetics. 2007;81(6):1278–1283. doi: 10.1086/522374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei LJ, Johnson WE. Combining dependent tests with incomplete repeated measurements. Biometrika. 1985;72:359–364. [Google Scholar]
- Wei Z, Li H. Nonparametric pathway-based regression models for analysis of genomic data. Biostatistics. 2007;8(2):265–284. doi: 10.1093/biostatistics/kxl007. [DOI] [PubMed] [Google Scholar]
- Yager JD, Liehr JG. Molecular mechanisms of estrogen carcinogenesis. Annual Review of Pharmacology and Toxicology. 1996;36:203–32. doi: 10.1146/annurev.pa.36.040196.001223. [DOI] [PubMed] [Google Scholar]
- Zondervan KT, Cardon LR. The complex interplay among factors that influence allelic association. Nature Review Genetics. 2004;5:89–100. doi: 10.1038/nrg1270. [DOI] [PubMed] [Google Scholar]