Abstract
The NMR structure of budding yeast chaperone Chz1 complexed with histones H2A.Z-H2B has been determined. Chz1 forms a long irregular chain capped by two short α-helices, and uses both positively and negatively charged residues to stabilize the histone dimer. A molecular model that docks Chz1 onto the nucleosome has implications for its potential functions.
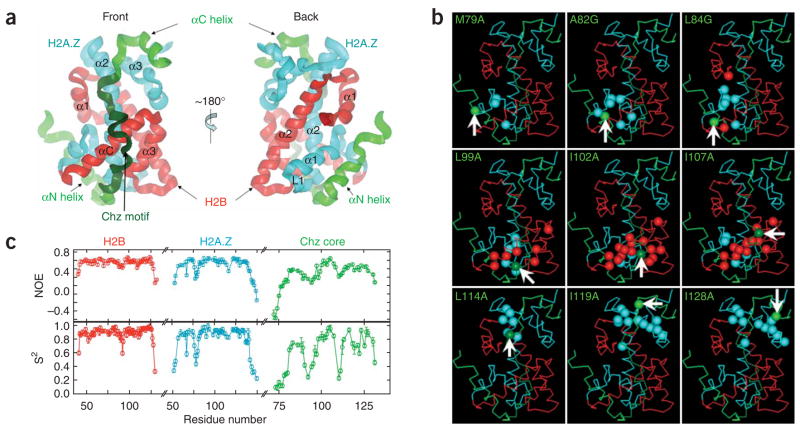
Histone chaperones have important roles in many biological events1. Chz1 complexed with H2A.Z-H2B provides H2A.Z-H2B for the SWR1-catalyzed replacement of nucleosomal H2A-H2B2–4. Chz1 alone is unfolded but forms a stable heterotrimer with H2A.Z-H2B, and a highly conserved region of Chz1 (termed the Chz motif) is crucial for its stabilization3. We set out to determine the structure of the Chz1–H2A.Z-H2B complex by NMR and identified a folded core including residues 71–132 of Chz1 (Chz core), residues 37–131 of H2B and residues 29–125 of H2A.Z (Supplementary Fig. 1 online). We linked the folded regions of the two histones together to make a single-chain histone (sH2B_H2A.Z) and used it to form a complex with Chz core (Chz core–sH2B_H2A.Z), which is referred to as the CZB complex (~28 kDa). The determination of the CZB structure (Fig. 1a, Supplementary Table 1 and Supplementary Methods online) was also aided by the analysis of chemical shift changes of backbone amide 1H and 15N of sH2B_H2A.Z from 11 single-site amino acid substitutions throughout the Chz core (Fig. 1b and Supplementary Fig. 2 online). For each mutation, those residues with large chemical shift changes in sH2B_H2A.Z were used to define the backbone topology of the Chz core and the binding surface of sH2B_H2A.Z.
Figure 1.
Structure and dynamics of the CZB complex. (a) Ribbon representation. (b) Determination of the binding surface on sH2B_H2A.Z and the chain topology of the Chz core. Arrows and green balls indicate the Cαatoms of mutated residues in the Chz core. Other balls (cyan in H2A.Z and red in H2B) are the Cαatoms of the residues that show large chemical shift changes. (c) Backbone dynamics10.
In the CZB complex, sH2B_H2A.Z retains the same histone fold as observed in the nucleosome5 (Supplementary Fig. 3 online). Notably, the Chz1 core forms a long irregular chain capped by two short α-helices and makes contact with broad regions of both H2A.Z and H2B (Fig. 1a). The N-terminal helix region (81–93) of the Chz core binds to the α1 helix and L1 loop of H2A.Z. The long irregular chain, representing mainly the conserved Chz motif (94–115), contacts the surface of the α2 helix of H2A.Z, and both the α3 and αC helices of H2B. The C-terminal helix region (116–132) binds to the α2 and α3 helices of H2A.Z. The backbone amide 15N-{1H} NOEs that probe picosecond-timescale motions and order parameters (S2) that report on amplitudes of amide bond motions (ranging from 0 (unrestricted motion) to 1 (fully restricted motions)) establish that Chz core is much more dynamic than sH2B_H2A.Z (Fig. 1c).
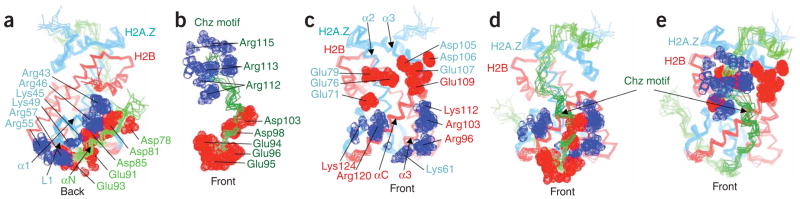
The stabilizing forces between Chz1 and sH2B_H2A.Z seem to be mainly electrostatic (Supplementary Fig. 4 online). Owing to the dynamic nature of the Chz core structure, we examined the electrostatic interactions by using an ensemble of structures. Five acidic residues in the αN helix region of the Chz core are close neighbors of six basic residues in the α1 helix and the L1 loop of H2A.Z (Fig. 2a). The Chz motif has a bipolar charge distribution with three basic residues at one end and five acidic residues at the other (Fig. 2b). Unexpectedly, the surface of sH2B_H2A.Z interacting with the Chz motif has a complementarily bipolar charge distribution with an acidic patch on one end and a basic region at the other (Fig. 2c). The acidic region of the Chz motif is surrounded by the basic region with five residues from H2B and one from H2A.Z (Fig. 2d). By contrast, the three basic residues of the Chz motif form a positively charged cloud on top of the acidic patch in the histone dimer, including six residues in H2A.Z and one in H2B (Fig. 2e). The αC helix region of the Chz core interacts with the α2 and α3 helices of H2A.Z through more dispersed electrostatic and hydrophobic interactions (Supplementary Fig. 4). Overall, these results indicate that both positively and negatively charged residues from the histones and the bipolar histone chaperone contribute to the stabilization of the complex, emphasizing the fact that histone chaperones are not merely negatively charged molecules for preventing nonproductive aggregation of histones with DNA phosphates.
Figure 2.
Electrostatic interactions between the Chz core and sH2B_H2A.Z. lysine and arginine are shown with blue dotted surfaces for the nitrogen and carbon atoms. (-COO)− of aspartic and glutamic acid are shown with red dotted surfaces for the carbon and oxygen atoms. (a) Electrostatic interactions between the αN helix of the Chz core and the α1 helix and the L1 loop of H2A.Z. (b) The bipolar distribution of charged residues in the Chz motif. (c) The bipolar distribution of the charged residues in the front surface of sH2B_H2A.Z. (d,e) Electrostatic interactions between the Chz motif and sH2B_H2A.Z. Green, Chz1, Cyan, H2A.Z, Red, H2B.
The docking of the Chz core on H2A.Z-H2B in the nucleosome based on the interactions in the CZB complex showed that the αN helix region of the Chz core occupies the DNA binding sites of H2A.Z-H2B in the nucleosome (Supplementary Fig. 5 online), indicating that the αN helix may prevent H2A.Z-H2B in the CZB complex from interacting with DNA directly, as anticipated for a histone chaperone. Notably, histone chaperone Asf1 binds to the H3-H4 dimer at the interface of the (H3-H4)2 tetramer6,7, preventing premature formation of the tetramer. Similarly, the long, irregular Chz motif and the αC helix of the Chz core can be modeled to sit on top of the exposed region of H2A-H2B in the nucleosome, including the acidic patch, which has been shown to bind to the basic peptide derived from Kaposi’s Sarcoma herpesvirus8 and the basic N-terminal tail of H4 (ref. 9). We have observed physical interactions between mononucleosomes and a truncated Chz core with the αN helix removed (Chz 96–132; Supplementary Fig. 6a–c online). These results raise the possibility that Chz1 might also have a role in the H2A-H2B eviction during the SWR1-catalyzed replacement reaction (Supplementary Fig. 6d).
Supplementary Material
Note: Supplementary information is available on the Nature Structural & Molecular Biology website.
Acknowledgments
We thank A. Bax, C. Klee and M. Lichten for comments. This work was supported by the intramural program of the US National Cancer Institute (C.W. and Y.B.) and a grant from the Canadian Institutes of Health Research (L.E.K.). D.F.H. is the recipient of a postdoctoral fellowship from the Danish Agency for Science, Technology and Innovation.
Footnotes
Accession codes. Protein Data Bank: Coordinates for the CZB complex have been deposited with accession code 2JSS.
Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions/
References
- 1.De Koning L, Corpet A, Haber JE, Almouzni G. Nat Struct Mol Biol. 2007;14:997–1007. doi: 10.1038/nsmb1318. [DOI] [PubMed] [Google Scholar]
- 2.Mizuguchi G, et al. Science. 2003;303:343–348. [Google Scholar]
- 3.Luk E, et al. Mol Cell. 2007;25:357–368. doi: 10.1016/j.molcel.2006.12.015. [DOI] [PubMed] [Google Scholar]
- 4.Wu WH, et al. Nat Struct Mol Biol. 2005;12:1064–1071. doi: 10.1038/nsmb1023. [DOI] [PubMed] [Google Scholar]
- 5.Suto RK, Clarkson MJ, Tremethick DJ, Luger K. Nat Struct Biol. 2000;7:1121–1124. doi: 10.1038/81971. [DOI] [PubMed] [Google Scholar]
- 6.English CM, Adkins MW, Carson JJ, Churchill ME, Tyler JK. Cell. 2006;127:495–508. doi: 10.1016/j.cell.2006.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Natsume R, et al. Nature. 2007;446:338–341. doi: 10.1038/nature05613. [DOI] [PubMed] [Google Scholar]
- 8.Barbera AJ, et al. Science. 2006;311:856–861. doi: 10.1126/science.1120541. [DOI] [PubMed] [Google Scholar]
- 9.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 10.Hansen DF, et al. J Am Chem Soc. 2007;129:11468–11479. doi: 10.1021/ja072717t. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Note: Supplementary information is available on the Nature Structural & Molecular Biology website.