Abstract
CD22 is a B cell specific sialic-acid-binding immunoglobulin-like lectin (Siglec) whose function as a regulator of B cell signaling is modulated by its interaction with glycan ligands bearing the sequence NeuAcα2-6Gal. To date, only highly multivalent polymeric ligands (n=450) have achieved sufficient avidity to bind to CD22 on native B cells. Here we demonstrate that a synthetic bi-functional molecule comprising a ligand of CD22 linked to an antigen (nitrophenol; NP) can use a monoclonal anti-NP-IgM as a decavalent protein scaffold to efficiently drive assembly of IgM-CD22 complexes on the surface of native B cells. Surprisingly, anti-NP antibodies of lower valency, IgA (n =4) and IgG (n =2), were also found to drive complex formation, though with lower avidity. Ligands bearing alternate linkers of variable length and structure were constructed to establish the importance of a minimal length requirement, and versatility in the structural requirement. We show that the ligand drives assembly of IgM complexes exclusively on the surface of B cells and not other classes of white blood cells that do not express CD22, which lends itself to the possibility of targeting B cells in certain hematopoietic malignancies.
Introduction
Glycan-binding proteins (GBP) participate in the communication of cells with their extracellular environment, mediating diverse biology including host-pathogen interactions, cell-cell adhesion and modulation of cell signaling receptors 1-3. GBPs typically exhibit low affinity (e.g. Kd ∼ 10-1000 μM) for their ligands, and require multivalent interactions to mediate biological effects 4, 5. Because of the low intrinsic affinity, the design of ligand-based probes of GBPs has largely relied on highly multivalent display of ligands on synthetic polymers or ‘neoglycoproteins’ to provide the avidity needed to stabilize binding 4-7. Rational design of lower valency constructs or dendrimers has met with limited success, with several notable exceptions 8-12. To achieve sufficient avidity while maintaining lower valency, protein scaffolds have shown promise due to the opportunity for defined architecture that can, in some cases, benefit from symmetrical oligomerization. Exemplary is the demonstration that bivalent glycan ligands tethered to a central core with pentameric radial symmetry could bridge two pentameric Shiga toxins. Optimal spacing of low affinity glycan ligands (IC50=2-5 mM) gave up to 107-fold enhancement of affinity 8, 10. Using a similar strategy, bi-functional ligands were found to drive assembly of a face-to-face complex of a toxin with another pentameric protein, serum amyloid protein (SAP) 13, 14. Complex formation could be achieved with either covalently tethered or non-tethered bi-functional ligands. In the latter case, SAP effectively becomes a pentameric scaffold for presentation of low affinity monomeric glycan ligands to the pentameric toxin 13-15.
Use of a bi-functional ligand-antigen molecule to ‘decorate’ a target cell or microbe with an antigen has been proposed by several groups for targeting the immune response 12, 16-18, and has been successfully demonstrated for targeting mammalian cells that contain the folate receptor or αvβ3 integrin16, 17. These receptors bind with high affinity to their ligands, folate and RGD peptide, respectively (Kd= 10-9-10-10). Thus, the corresponding bi-functional ligand-antigen conjugates bind tightly to the surface of the cell, which display the conjugated antigen for subsequent recognition and binding by antibody. This powerful approach has not yet been exploited for mammalian GBPs, largely due to the constraints of the low affinity for their ligands.
Members of the siglec family of GBPs are expressed on cells of the immune system, exhibiting roles in cell signaling and cell adhesion that are modulated by interaction with their sialic acid containing glycan ligands 1. Exemplary is CD22, a known regulator of B cell signaling that has been established as a target for immunotherapy of B cell lymphomas and autoimmune disease 19, 20. CD22 exhibits low intrinsic affinity for its ligands (Kd = ∼0.2 mM), and their abundance on B cells provide cis (on the same cell) interactions that effectively mask the ligand-binding site 21, 22. To date, only highly multivalent synthetic ligands have been demonstrated to compete with cis ligands and bind to native B cells23.
Motivated by reports of bi-functional ligand-mediated targeting of multivalent protein scaffolds to proteins of interest, we investigated the possibility of designing such a ligand that would dock decavalent IgM to CD22. Although CD22 is a monovalent GBP and does not share decameric symmetry with IgM, it is concentrated in clathrin rich microdomains and forms multimers in situ 23-26, suggesting that the radially symmetric IgM, with binding sites spread over a diameter of 350Å, might bridge the distance between clustered CD22 monomers. Accordingly, we constructed a bi-functional ligand that would simultaneously bind to CD22 and a monoclonal antibody specific for the hapten nitrophenol (NP; Figure 1). The CD22 ligand comprises the preferred glycan sequence recognized by CD22 (NeuAcα2-6Galβ1-4GlcNAc) with a 9-biphenyl carbonylamido (BPC) substituent on the sialic acid, known to increase affinity for CD22 by 100 fold 23, 27. We show herein that this ligand is able to efficiently and specifically assemble complexes between anti-NP antibodies with CD22 on the surface of a B cell.
Figure 1. Design of a bi-functional ligand to drive self-assembly of IgM-CD22 complexes.
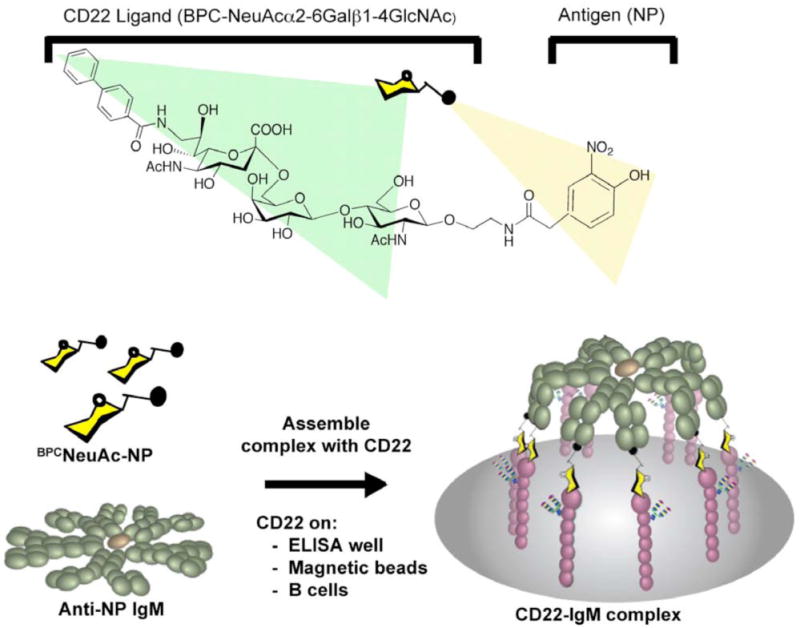
(a) Structure of the bi-functional ligand, BPCNeuAc-NP. (b) Schematic of the ligand-driven complex formation between the decavalent scaffold, anti-NP IgM, and the B cell surface lectin CD22 on either B cells or on solid supports decorated with the extracellular domain of CD22.
Experimental Section
General Methods for synthesis of bi-functional ligands
2-(4-hydroxy-3-nitrophenyl)acetic acid, N-hydroxysuccinimide ester, was purchased from Biosearch Technologies. 2-Acetamido-3,4,6-tri-O-acetyl-2-deoxy-β-D-glucopyranosyl azide was purchased from TCI North America. All other reagents and solvents were purchased from Aldrich Chemicals and used without further purification. Enzymes including the β1-4-galactosyltransferase-/UDP-4′-Gal Epimerase fusion protein28 (GalE-GalT), Nesseria meningitidis CMP-NeuAc synthetase, and human ST6Gal I sialyltransferase (hST6Gal 1) were prepared as previously reported28-30. Solvent concentration was performed under reduced pressure at < 40 °C bath temperature. All 1H and 13C NMR spectra were recorded at 30 °C using a Varian Unity Inova 400 or a Bruker DRX 600 Spectrometer. Chemical shifts are reported in parts per million from high to low field and referenced to the solvents. Standard abbreviations indicating multiplicity were used as follows: s = singlet, d = doublet, t = triplet, q = quadruplet, and m = multiplet. MALDI-FTMS spectrometry was recorded with an IonSpec Ultima FTMS (IonSpec Corp., Irvine, CA,) using dihydroxybenzoic acid as the matrix, and carried out by services at The Scripps Research Institute. Thin layer chromatography was performed on Silica Gel 60F from Fisher. After development with appropriate eluents, spots were visualized by dipping in 5% sulfuric acid in ethanol, followed by charring. Flash chromatography was performed on 230-400 mesh silica gel under positive air pressure.
Synthesis of 5-Acetamido-9-(biphenyl-4-carboxamido)-3,5,9-trideoxy-D-glycero-α-D-galacto-non-2-ulopyranosonic acid sodium salt (9-BPC-Neu5Ac)
5-Acetamido-9-amino-3,5,9-trideoxy-d-glycero-d-galacto-2-nonulo-pyranosyl-onate (9-amino-NeuAc)24 (200 mg) in methanol was kept basic with triethylamine, and to the solution biphenylacetyl chloride dissolved in methylene chloride (2M) was added dropwise until the reaction was complete as monitored by TLC. The solvents were evaporated and the residue was purified by flash chromatography on silica gel eluted with methylene chloride/methanol (10:1 to 1:2) to afford 9-BPC-Neu5Ac (180 mg) in 75% yield. Rf: 0.5 (4:3:2 i-PrOH:MeCN:MeOH). 1H NMR (CD3OD) δ 7.92 (d, 2H, J = 8.4 Hz), 7.71 (d, 2H, J = 8.4 Hz), 7.64 (d, 2H, J = 7.2 Hz), 7.45 (t, 2H, J = 7.5 Hz), 7.37 (m, 1H), 4.05 (m, 2H), 3.91 (m, 2H), 3.55 (t, 1H, J = 6.3Hz), 3.52 (m, 2H), 2.60 (m, 1H), 2.36 (t, J = 6.9Hz), 2.16 (dd, 1H, J = 9.0, 4.2 Hz), 1.95 (s, 3H), 1.87 (t, 1H, J = 12.0 Hz); MS: (C24H28N2O9): calculated (MH+): 489.1867, found: 489.1876.
Synthesis of 2-aminoethyl 4-O-{6-O-[5-Acetamido-9-(biphenyl-4-carboxamido)-3,5,9-trideoxy-d-glycero-α-d-galacto-non-2-ulopyranosyl-2-onic acid sodium salt]-β-d-galactopyranosyl}-2-acetamido-2-deoxy-β-d-glucopyranoside (6)
To a solution of sodium cacodylate buffer (50 ml, 100 mM, pH 8.0) containing MgCl2 (20 mM), MnCl2 (20 mM), CTP (150 mg), 9-BPC-Neu5Ac (50 mg), UDP-Glucose (150 mg), and 2-aminoethyl 2-acetamido-2-deoxy-β-D-glucopyranoside31 (1, 40 mg), was added GalE-GalT (20 U), CMP-NeuAc synthetase (10 U) and hST6Gal 1 (2U). The reaction mixture was slowly stirred at room temperature for 3 days. Enzymes were removed by passing the reaction mixture through a Centricon filter (MWCO 10,000 Dalton). The filtrate was lyophilized and the residue was dissolved in a minimum amount of water, loaded onto a P-2 gel chromatography column and eluted with water. Appropriate fractions were pooled and lyophilized to afford the desired product (55 mg) in 70% yield. 1H NMR (CD3OD) δ 7.82 (d, 2H, J = 8.4Hz), 7.61 (d, 2H, J = 8.4Hz), 7.55 (d, 2H, J = 7.2 Hz), 7.36 (t, 2H, J = 7.8Hz), 7.27 (m, 1H), 4.46 (d, 1H, J = 8.4 Hz), 4.23 (d, 1H, J = 7.2 Hz), 3.56-4.1 (m, 10H), 3.30 (d, 1H, J = 7.8 Hz), 3.20 (m, 4H), 3.01 (m, 1H), 2.64 (dd, 1H), 1.91 (s, 3H), 1.88 (s, 3H), 1.57 (t, 1H, J = 12Hz); 13C NMR (CD3OD) δ: 174.87, 174.63, 169.83, 145.41, 141.18, 134.42, 130.07, 129.11, 129.04, 128.11, 128.02, 105.04, 102.36, 101.54, 81.80, 76.47, 75.67, 74.66, 74.07, 73.42, 72.57, 72.33, 71.65, 70.32, 69.77, 66.51, 64.91, 62.01, 59.60, 52.28, 56.73, 53.93, 52.80, 51.23, 44.64, 42.35, 40.98, 23.49, 22.88, 17.66, 15.89; MS(C40H56N4O19) Calculated (MNa+) 919.3436, found 919.3473
Synthesis of N-(2-acetamido-2-deoxy- β-d-glucopyranosyl)-2-(4-hydroxy-3-nitro-phenyl)acetamide (7)
Glycosylamine 2, prepared according to a published procedure32, (200 mg, 0.91 mmol) was dissolved in DMF (7.2 mL). To this was added HBTU (0.69 g, 1.82 mmol), HOBt (245 mg, 1.82 mmol) and 2-(4-hydroxy-3-nitrophenyl)acetic acid (197 mg, 1 mmol). The mixture was stirred at room temperature for 5 min and Et3N (11 μL, 1.82 mmol) was added. The reaction mixture was stirred at room temperature for 1 h. The solvent was removed under reduced pressure. Flash chromatography on the crude material (Chloroform : Methanol 4:1) yielded the desired product as a yellow powder (360 mg, 99%). 1H MHR (CD3OD, 600 MHz) δ 7.91 (d, J = 1.8 Hz, 1H), 7.43 (dd, J = 3.6, 8.4 Hz), 7.07 (t, 1H, J = 3.0, 5.4 Hz), 4.93 (d, 1H, J = 10.2 Hz), 3.76 (dd, 1H, J = 1.8, 12.6 Hz), 3.65 (m, 2H), 3.52 (2d, J = 15 Hz), 3.46 (t, J = 8.4 Hz), 3.38 (m, 2H), 1.70 (s, 3H); 13C NMR (CD3OD, 175 MHz) δ 175.34, 175.21, 153.44, 139.02, 134.98, 127.86, 126.46, 120.94, 120.80, 79.33, 78.54, 74.96, 70.22, 61.35, 55.14, 41.96, 22.67. MS (C16H21N3O9) Calculated (MH+) 400.1356, found 400.1355.
4-O-{6-O-[5-Acetamido-9-(biphenyl-4-carboxamido)-3,5,9-trideoxy-d-glycero-α-d-galacto-non-2-ulopyranosyl-2-onic acid sodium salt]-β-D-galactopyranosyl}-2-acetamido-2-deoxy-β-D-glucopyranosyl azide (8)
2-Acetamido-3,4,6-tri-O-acetyl-2-deoxy-β-d-glucopyranosyl azide (1 g, 2.7 mmol) was dissolved in methanol (100 mL). To this solution, sodium (30 mg, 1.3 mmol) was added. The reaction was stirred at room temperature for 30 minutes. The pH was adjusted to 7 with Dowex H+ resins. Solvent was then removed under reduced pressure to give deprotected glycosyl azide as a white powder (630 mg, 95%). This material, compound 3, (25 mg, 0.1 mmol) was dissolved in sodium cacodylate buffer (20 mL, 100 mM) containing MgCl2 (20 mM), MnCl2 (20 mM), CTP (112.8 mg), 9-BPC-Neu5Ac (92 mg), UDP-Glucose (100 mg). To this solution, GalE-GalT (1 U), CMP-NeuAc synthetase (600 U) and hST6Gal 1(1.6 U) were added. The reaction was stirred at room temperature for 24 h while the pH was constantly kept at 7.5. The reaction mixture was then centrifuged at 3000 rpm for 15 min and the supernatant was separated and lyophilized. The product was then first purified from a G15 size exclusion column (1 × 75 cm, 5% n-Butanol in water) to give compound 8 as a white powder (50 mg, 56%). 1H NMR (CD3OD, 600 MHz) δ 7.79 (m, 3H), 7.39 (m, 5H), 7.21 (m, 4H), 4.58 (d, 1H, J = 8.4 Hz), 4.22 (d, 1H, J = 8.4 Hz), 3.92 (m, 3H), 3.77 (m, 5H), 3.62 (m, 9H), 3.49 – 3.38 (m, 7H), 2.53 (bd, 1H, J = 8.4 Hz), 1.93 (s, 3H), 1.85 (s, 3H), 1.71 (t, 1H, J = 12 Hz); 13C NMR (CD3OD, 175 MHz) δ 176.02, 172.32, 171.23, 144.72, 139.63, 133.02, 129.89, 129.09, 128.55, 127.76, 104.31, 100.14, 89.15, 81.05, 77.19, 74.33, 73.60, 73.24, 73.14, 71.44, 70.85, 70.39, 69.18, 68.30, 64.20, 60.96, 55.12, 52.62, 44.11, 39.89, 23.08, 22.91. MS: (C38H50N6O18): calculated (MH+): 879.3254, found: 879.3245.
Synthesis of 2-(6-aminohexanoylamide)-ethyl 4-O-{6-O-[5-Acetamido-9-(biphenyl-4-carboxamido)-3,5,9-trideoxy-d-glycero-α-d-galacto-non-2-ulopyranosyl-2-onic acid sodium salt]-β-D-galactopyranosyl}-2-acetamido-2-deoxy-β-D-glucopyranoside (10)
Compound 1 (200 mg, 0.76 mmol), 6-(Fmoc-amino)caproic acid (320 mg, 0.9 mmol, 1.2 equiv.), HBTU (576 mg, 1.52 mmol, 2 equiv.) and HOBt (205.2 mg, 1.52 mmol, 2 equiv.) were mixed in 4 mL DMF. To this triethylamine (220 uL, 1.52 mmol, 2 equiv.) was added. The reaction mixture was stirred at room temperature overnight. TLC showed a complete conversion to the product. The solvent was removed under reduced pressure. The product was purified by flash chromatography (Chloroform: Methanol 4:1) to give Fmoc protected hexanoic acid derivative as a white powder (326 mg, 72%). Part of this material (25 mg, 0.042 mmol) was treated with 20% piperidine solution in DMF (416 uL) for 4 h and the solvent was removed under reduced pressure. The residue, compound 5, was suspended in sodium cacodylate buffer (10 mL, 100 mM) containing MgCl2 (20 mM), MnCl2 (20 mM), CTP (56.4 mg), 9-BPC-Neu5Ac (46 mg), UDP-Glucose (49 mg). To this GalE-GalT (0.5 U), CMP-NeuAc synthetase (300 U) and hST6Gal 1(0.8 U) was added. The reaction was stirred at room temperature for 24 h while the pH was constantly kept at 7.5. The reaction mixture was then centrifuged at 3000 rpm for 15 min and the supernatant was separated and lyophilized. The product was then purified from a G15 size exclusion column (1 × 75 cm, 5% n-Butanol in water) to give compound 10 as a white powder (25 mg, 60%). 1H NMR (CD3OD, 600 MHz) δ 7.69 (m, 8H), 7.42 (m, 3H), 7.35 (m, 1H), 4.33 (d, 1H, J= 8.4 Hz), 4.30 (d, 1H, J = 8.4 Hz), 3.98 – 3.80 (m, 5H), 3.76 – 3.64 (m, 6H), 3.59 (dd, 1H, J = 7.2, 13.8 Hz), 3.53 (m, 3H), 3.47 – 3.38 (m, 6H), 3.20 (t, 1H, J = 4.8 Hz), 2.85 (t, 1H, J = 7.2 Hz), 2.55 (dd, 1H, J = 4.8, 12.6 Hz), 2.10 (m, 2H), 1.97 (s, 3H), 1.87 (s, 3H), 1.58 (t, 1H, J = 12.6 Hz), 1.55 (m, 2H), 1.47 (m, 2H), 1.22 (m, 1H); 13C NMR (CD3OD, 175 MHz) δ 178.12, 174.98, 173.40, 171.87, 144.41, 141.27, 133.04, 125.92, 125.85, 118.57, 112.11, 104.23, 101.73, 101.14, 81.28, 75.27, 74.51, 73.34, 73.31, 73.26, 71.54, 71.11, 70.98, 70.87, 70.64, 69.87, 69.18, 69.01, 68.95, 68.08, 64.27, 63.30, 61.15, 55.62, 53.15, 52.70, 44.20, 43.81, 40.97, 40.24, 40.10, 36.21, 27.23, 25.90, 25.55, 23.17, 22.86. MS: (C24H28N2O9): calculated (MH+): 1010.4458, found: 1010.4438.
Synthesis of (4-hydroxy-3-nitrophenyl)methylcarbamidoethyl 4-O-{6-O-[5-acetamido-9-(biphenyl-4-carboxamido)-3,5,9-trideoxy-d-glycero-α-d-galacto-non-2-ulopyranosyl-2-onic acid sodium salt]-β-d-galactopyranosyl}-2-acetamido-2-deoxy-β-d-glucopyranoside (11)
To a flask with methanol (5 mL) were added 2-(4-hydroxy-3-nitrophenyl)acetic acid, N-hydroxysuccinimide ester (50 mg) and compound 6 (20 mg). The solution kept basic with addition of triethylamine and stirred at room temperature for two days. The solvent was evaporated and the residue was purified by flash chromatography on silica gel to afford the desired product (18 mg) in 75% yield. 1H MHR (CD3OD) δ7.85 (d, 1H, J = 2.4 Hz), 7.86 (d, 2H, J = 8.4 Hz), 7.60 (d, 2H, J = 8.4Hz), 7.54 (d, 2H, J = 7.2 Hz), 7.35 (dd, 1H, J = 6.3, 1.8 Hz), 7.32 (t, 2H, J = 7.2 Hz), 7.25 (m, 1H), 6.93 (d, 1H, J = 8.7 Hz), 4.40 (d, 1H, J = 8.1 Hz), 4.20 (d, 1H, J = 6.9 Hz), 3.95 (m, 2H), 3.34 (s, 2H), 3.80-3.40 (m, 8H), 3.15-3.3 (m, 4H), 2.64 (dd, 1H, J = 4.5, 12.0 Hz), 1.87 (s, 3H), 1.86 (s, 3H), 1.60 (t, 1H, J = 12.0 Hz), 13C NMR (CD3OD) δ: 174.83, 174.50, 173.51, 170.03, 145.52, 141.26, 138.71, 134.38, 130.10, 130.01, 129.98, 129.05, 129.00, 128.10, 128.03, 126.47, 105.18, 102.49, 101.54, 76.42, 73.98, 72.42, 69.10, 64.89, 62.13, 42.09, 40.90, 26.11, 23.44, 9.51; MS(C48H61N5O23) Calculated (MNa+) 1098.3649, found 1098.3649.
Synthesis of N-{4-O-{6-O-[5-Acetamido-9-(biphenyl-4-carboxamido)-3,5,9-trideoxy-d-glycero-α-d-galacto-non-2-ulopyranosyl-2-onic acid sodium salt]-β-D-galactopyranosyl}-2-acetamido-2-deoxy-β-D-glucopyranosyl} 2-(4-hydroxy-3-nitro-phenyl)acetamide (12)
Compound 7 (20 mg, 0.05 mmol) was dissolved in sodium cacodylate buffer (10 mL, 100 mM) containing MgCl2 (20 mM), MnCl2 (20 mM), CTP (56 mg), 9-BPC-Neu5Ac (46 mg), and UDP-Glucose (50 mg). To this GalE-GalT (0.5 U), CMP-NeuAc synthetase (300 U) and hST6Gal 1(0.8 U) were added. The reaction was stirred at room temperature for 24 h while the pH was kept constant at 7.5. The reaction mixture was then centrifuged at 3000 rpm for 15 min and the supernatant was separated and lyophilized. The product was then first purified from a G15 size exclusion column (1 × 75 cm, 5% n-Butanol in water) to give compound 12 as a yellow powder (34 mg, 65%). 1H NMR (CD3OD, 600 MHz) δ 7.75 (2d, 4H, J = 9 Hz), 7.66 (dd, 2H, J = 1.2, 7.2 Hz), 7.60 (d, 1H, J = 2.4 Hz), 7.44 (t, 2H, J = 7.2 Hz), 7.04 (dd, 1H, J = 2.4, 9.0 Hz), 6.63 (d, 1H J = 9.0 Hz), 4.88 (d, 1H, J = 9.6 Hz), 4.30 (d, 1H, J = 7.8 Hz), 3.95 (m, 1H), 3.86 – 3.40 (m, 12H), 2.55 (dd, 1H, J = 4.2, 12 Hz), 1.89 (s, 3H), 1.70 (s, 3H), 1.58 (t, 1H, J = 12 Hz). 13C NMR (CD3OD, 175 MHz) δ 183.41, 176.44, 175.81, 175.44, 174.18, 171.61, 144.85, 140.50, 137.63, 137.32, 133.42, 130.03, 129.23, 128.73, 128.07, 127.99, 127.07, 126.85, 104.23, 101.14, 80.73, 79.11, 77.04, 74.46, 73.54, 73.38, 73.23, 71.58, 71.11, 70.94, 69.17, 68.98, 54.33, 52.71, 44.17, 42.27, 41.20, 22.84, 22.65. MS: (C46H57N5O22): calculated (MH+): 1032.3568, found: 1032.3556.
Synthesis of N1-{4-O-{6-O-[5-Acetamido-9-(biphenyl-4-carboxamido)-3,5,9-trideoxy-d-glycero-α-d-galacto-non-2-ulopyranosyl-2-onic acid sodium salt]-β-D-galactopyranosyl}-2-acetamido-2-deoxy-β-D-glucopyranosyl}-4-[2-(4-hydroxy-3-nitro-phenyl)acetamidomethyl]-[1,2,3]-triazol (13)
Compound 8 (∼44 mg, 0.05 mmol) was dissolved in water (1 mL). 4-hydroxy-3-nitro-phenylethyloyl-propargylamine (15 mg, 0.06 mmol) in t-Butanol (1 mL) was added. To this sodium ascorbate (9.1 mg) and copper sulfate (3.8 mg) were added and the reaction mixture was stirred at room temperature overnight. The solvent was then lyophilized and the residue solid was purified by a G15 size exclusion column (1 × 75 cm, 5% n-Butanol in water) to give compound 13 as a yellow crystal (39 mg, 70%). 1H NMR (CD3OD, 600 MHz) δ 7.81 (d, 1H, J = 2.4 Hz), 7.74 (m, 2H), 7.61 (d, 2H, J = 8.4 Hz), 7.55 (d, 2H, J = 7.8 Hz), 7.38 (m, 3H), 6.98 (d, 2H, J = 9 Hz), 5.66 (d, 1H, J = 10.2 Hz), 4.37 (d, 1H, J = 7.8 Hz), 4.30 (m, 2H), 4.06 (t, 1H, J = 10.2 Hz), 3.96 – 3.91 (m, 2H), 3.86 (d, 1H, J = 3 Hz), 3.81 – 3.71 (m, 5H), 3.68 – 3.58 (m, 5H), 3.44 (m, 4H), 2.55 (dd, 1H, J = 4.8, 12.6 Hz), 1.89 (s, 3H), 1.69 (t, 1H, J = 12.0 Hz), 1.65 (s, 3H); 13C NMR (CD3OD, 175 MHz) δ 175.81, 174.83, 174.20, 172.46, 171.63, 153.52, 146.16, 144.98, 139.98, 139.24, 134.56, 133.15, 129.98, 129.22, 128.61, 128.09, 127.91, 127.82, 126.18, 122.84, 120.75, 118.57, 116.84, 104.06, 100.07, 86.85, 79.95, 78.26, 74.29, 73.62, 73.25, 72.89, 71.56, 70.96, 70.45, 69.01, 68.40, 64.09, 60.77, 55.49, 52.59, 43.99, 41.54, 39.86, 35.00. MS: (C49H60N8O22): calculated (MH+): 1113.3895, found: 1113.3895.
Synthesis of N1-{4-O-{6-O-[5-Acetamido-9-(biphenyl-4-carboxamido)-3,5,9-trideoxy-d-glycero-α-d-galacto-non-2-ulopyranosyl-2-onic acid sodium salt]-β-D-galactopyranosyl}-2-acetamido-2-deoxy-β-D-glucopyranosyloxyethyl}-4-[2-(4-hydroxy-3-nitro-phenyl)acetamidomethyl]-[1,2,3]-triazol (14)
Compound 9 (synthesized as described previously33) (20 mg, 0.02 mmol) and N-propargyl-2-(4-hydroxy-3-nitro-phenyl)acetamide (5.2 mg, 0.022 mmol) were mixed in water (1 mL) and t-Butanol (1 mL). To this sodium ascorbate (6.8 mg) and copper sulfate (2.8 mg) were added. The reaction mixture was stirred at room temperature overnight. The solvent was then lyophilized and the residue solid was purified by a G15 size exclusion column (1 × 75 cm, 5% n-Butanol in water) to give compound 14 as a yellow crystal (20 mg, 80%). 1H NMR (CD3OD, 600 MHz) δ 7.73 (s, 1H), 7.67 (t, 2H, J = 7.8 Hz), 7.54 (m, 3H), 7.32 (m, 3H), 6.92 (d, 1H, J = 8.4 Hz), 4.37 – 4.26 (m, 4H), 4.17 (d, 1H, J = 7.8 Hz), 4.03 (dd, 1H, J = 1.8, 12.0 Hz), 3.93 (m, 1H), 3.88 (t, 1H, J = 8.4 Hz), 3.82 (s, 1H), 3.74 (m, 3H), 3.66 (d, 1H, J = 13.2 Hz), 3.60 – 3.35 (m, 9H), 3.29 (m, 1H), 2.52 (dd, 1H, J = 1.2, 12.0 Hz), 1.88 (s, 3H), 1.73 (s, 3H), 1.63 (t, 1H, J = 12.6 Hz); 13C NMR (CD3OD, 175 MHz) δ 175.72, 175.04, 174.24, 173.13, 171.28, 153.23, 144.94, 144.67, 139.91, 139.24, 134.48, 133.01, 130.01, 129.94, 129.20, 128.56, 126.22, 125.01, 120.75, 120.68, 104.20, 103.70, 101.72, 101.65, 100.35, 81.13, 75.28, 74.29, 73.55, 73.32, 73.22, 73.13, 73.06, 71.51, 71.10, 70.32, 70.21, 69.06, 68.56, 68.51, 64.06, 61.07, 55.38, 55.28, 52.58, 51.01, 44.19, 44.10, 41.42, 40.36, 35.44, 22.87. MS: (C51H64N8O23): calculated (MH+): 1157.4157, found: 1157.4127.
Synthesis of N1-{4-O-{6-O-[5-Acetamido-9-(biphenyl-4-carboxamido)-3,5,9-trideoxy-d-glycero-α-d-galacto-non-2-ulopyranosyl-2-onic acid sodium salt]-β-D-galactopyranosyl}-2-acetamido-2-deoxy-β-D-glucopyranosyloxyethyl}-4-[2-(4-hydroxy-3-nitro-phenyl)acetamidohexyl]-[1,2,3]-triazol (15)
Compound 10 (11 mg, 0.0109 mmol) was dissolved in DMF (0.28 mL). To this 2-(4-hydroxy-3-nitrophenyl)acetic acid N-hydroxysuccinimide ester (3.85 mg, 0.013 mmol) and Et3N (3.85 uL, 0.028 mmol) were added. The reaction was stirred at room temperature for 2 h. The solvent was then removed under reduced pressure. The residue solid was purified with a G15 size exclusion column (1 × 30 cm, 5% n-butanol in water) to give compound 15 as a yellow powder (8 mg, 62%). 1H NMR (CD3OD, 600 MHz) δ 7.80 – 7.59 (m, 6H), 7.38 (m, 4H), 6.97 (m, 2H), 4.35 (d, 1H, J = 7.8 Hz), 4.29 (d, 1H, J = 7.2 Hz), 3.95 (m, 1H), 3.90 – 3.35 (m, 16H), 3.25 – 3.03 (m, 5H), 2.54 (dd, 1H, J = 2.4, 12 Hz), 2.04 (m, 1H), 1.90 (s, 3H), 1.88 (s, 3H), 1.62 (t, 1H, J = 12 Hz), 1.38 (m, 3H), 1.13 (m, 3H); 13C NMR (CD3OD, 175 MHz) δ 177.61, 175.65, 175.28, 174.28, 173.59, 171.29, 153.24, 144.78, 139.14, 134.58, 133.14, 130.04, 129.34, 128.61, 127.95, 125.95, 120.82, 104.22, 101.60, 81.20, 75.37, 74.44, 73.52, 73.36, 71.75, 71.04, 70.66, 69.17, 68.59, 64.11, 61.14, 55.74, 52.56, 47.49, 44.02, 43.66, 41.80, 41.74, 40.68, 40.13, 40.02, 38.09, 36.38, 29.27, 28.72, 28.62, 26.32, 25.73, 24.03, 23.47, 23.16, 22.85. MS: (C54H72N6O24): calculated (MH+): 1189.4671, found: 1189.4698.
Antibodies and Cells
Cells expressing the B1-8 anti-NP IgM were kindly provided by R.G. Corley and maintained in RPMI (Gibco) with 5% low IgG sera (HyClone). Anti-NP IgG and IgA were purchased from AbD Serotec. Secondary antibodies to anti-NP antibodies were FITC -conjugated F(ab′)2 goat anti-mouse IgM + IgG, goat anti-human IgA (α-chain specific), and goat anti-human IgG (Fcγ-specific) (Jackson Immunoresearch). Antibodies for lymphocyte labeling were Phycoerythrin (PE)-conjugated mouse anti-human CD3, CD4, CD8, CD19, and CD56 (BD Pharmingen). CD22-Fc was produced by transient transfection of COS cells with a plasmid encoding hCD22-Fc, a gift of Ajit Varki (UCSD)34. Cells maintained in DMEM were transfected by lipofectamine (Invitrogen) and then cultured for 3 additional days in OptiMEM. Culture supernatant was used without purification. Native BJAB cells were maintained in RPMI 1640 with 10% FBS and 50 μM 2-mercaptoethanol. Asialo BJAB cells were prepared by culturing BJAB cells in serum-free HYQ-SFM4MAb media (HyClone) for several days. Peripheral blood from healthy human donors was obtained from Scripps Normal Blood Donor Services after collection into heparanized tubes.
Purification of anti-NP IgM and labeling for immunomicroscopy
The secreted anti-NP IgM produced by B1-8 hybridoma cells was isolated by dialysis of the culture supernatant against water overnight with two changes of water. Precipitated IgM was pelleted by centrifugation at 13000 × g for 1 h at 4 °C and resuspended in phosphate-buffered saline. Anti-NP IgM was further purified using a Hi Trap IgM purification HP column (GE Healthcare). IgM was then labeled using the Alexa Fluor® 488 Protein Labeling Kit (Invitrogen-Molecular Probes) according to manufacturer's instructions. In short, the 1 mg/mL anti-NP IgM was combined with 10 mg of labeling reagent and 200 μM 2-(4-hydroxy-3-nitrophenyl)acetic acid (NP) to protect the binding site. After allowing the labeling to proceed for 1 h, protein was purified by gel filtration.
Elisa assays
ELISA type assays used High Binding microtiter plates (Nunc Brand, Fisher) coated with Protein A (1 μg/well in 50 μl sodium bicarbonate pH 9.5) overnight at 4° C. Wells were washed with 200 μl of Hank's buffered saline solution containing 5 mg/mL bovine serum albumin (HBSS/BSA), and culture supernatant containing hCD22Fc chimera was incubated for 2 h at room temperature. Wells were then washed with 2 × 200 μl of HBSS/BSA. A mixture of anti-NP IgM (5 μg) and the indicated concentration of bi-functional ligand was added in 50 μl and allowed to bind for 30 min. Wells were washed with 2 × 200 μl of HBSS/BSA and then incubated with a 1:3000 dilution of the alkaline phosphatase conjugated anti-IgM for 30 min at RT. Wells were washed 3 × 200 μl and developed with p-nitrophenyl phosphate. Data are the average +/- S.D. of triplicates, and are representative of 3 independent experiments.
CD22-Fc Bead Binding
hCD22-Fc, which comprises a chimera of the constant fragment of a human IgG fused to the C-terminus of the extracellular portion of CD22, was immobilized onto Protein A conjugated magnetic beads (Dynal Biotech) by adding 5 mL of culture supernatant containing hCD22-Fc chimera to 20 μL of bead slurry and incubating overnight at 4° C with end-over-end rotation. Beads were isolated by exposure to a magnet, washed three times with HBSS/BSA, and then resuspended in 200 μL of HBSS/BSA. Aliquots of 2 μL of beads were added to the indicated concentrations of anti-NP IgM diluted into HBSS/BSA and containing 1 μM of either NP or BPCNeuAc-NP, for a final volume of 100 μL. Binding proceeded for 1 hour at 4 °C. Beads were then isolated and washed with 300 μL HBSS/BSA containing 50 nM of NP or BPCNeuAc-NP. Beads were then incubated for 30 min at 4 °C in 100 μl HBSS/BSA containing 50 nM NP or BPCNeuAc-NP and FITC-anti-IgM to detect bound IgM. Beads were isolated and washed with 300 μl HBSS/BSA containing 50 nM NP or BPCNeuAc-NP and then resuspended in 200 μl of the same buffer and read by flow cytometry using a BD FACS Calibur Flow Cytometer, using CellQuest software for analysis. For ligand titration experiments, two-fold dilutions of ligand were prepared in 96-well plates (50 μL/well). To each well, 50-μL aliquots of a solution containing 10 μg/mL anti-NP Ig and approximately 1-2 × 105 beads/mL were added. Plates were incubated for at least 14 hours at 4 °C, 21 °C, or 37 °C to allow complex formation to reach equilibrium. Using a magnetic block to immobilize the beads at the bottoms of the wells (Dynal MPC®-96 S, Invitrogen), beads were washed with HBSS/BSA containing 100 nM BPCNeuAc-NP, then stained with FITC-labeled anti-IgG, IgA, or IgM. Beads were analyzed from the plates on a BD FACS Calibur HTS and data was analyzed using FlowJo software. With the exception of data in Figure S1, data points for all ligand titrations represent the average +/- S.D. of duplicates. Curves were generated by non-linear regression analysis of the data using the equation for log (agonist) vs. response – variable slope in GraphPad Prism software, version 5.0a.
IgM binding to BJAB cells
BJAB cells, cultured with or without exogenous sialic acid (4 mM), were resuspended in HBSS/BSA or RPMI medium containing 10% FBS and 50 μM 2-mercaptoethanol at 2 × 106 cells/mL and the indicated amount of anti-NP IgM, IgA, or IgG and NP or BPCNeuAc-NP were added to the cells and allowed to bind for 1 hour at 4 °C. Cells were then washed with 0.2 mL of HBSS/BSA containing 100 nM ligand and then incubated with FITC labeled anti-IgM, IgA, or IgG for 30 min on ice. After washing again, the cells were read by flow cytometry. For ligand titrations, 2-fold dilutions of BPCNeuAc-NP were prepared in the RPMI medium above at 50 μL/sample. BJAB cells were suspended at 4 × 106 cells/mL, in the same medium, also containing anti-NP Ig at 10 μg/mL. Aliquots of 50 μL of the cells were added to the ligand dilutions, followed by incubation for 7 hours at 4 °C. Data were analyzed as described above for titrations on CD22-Fc beads. For immunomicroscopy, BJAB cells were incubated on ice for 1 hr with 4 μg Alexa488-anti-NP IgM in the presence of either 4 μM NP or BPCNeuAc-NP. After washing, cells were fixed, stained for nuclei with Hoechst dye, and visualized by fluorescence microscopy.
IgM binding to lymphocytes from human peripheral blood
Lymphocytes were isolated by overlaying four 14-mL batches of freshly drawn heparinized human blood diluted 1:1 with Dulbecco's phosphate-buffered saline (DPBS) onto 10.5-mL aliquots of Ficoll-Paque Plus (GE Healthcare) and processing according to the manufacturer's protocol. Briefly, lymphocytes were collected at the interface by centrifugation (400 × g, 30 minutes, 18-20 °C), then washed and resuspended in DPBS at 2 × 106 cells/mL for staining. Cells were aliquoted at 2 × 105 cells/tube, and labeled with 8 μL PE-conjugated anti-CD3, CD4, CD8, CD19, or CD56 to identify cell type, or with 10 μg/mL Alexa488-anti-NP IgM and 2 μM BPCNeuAc-NP for complex formation. Alexa488-anti-NP IgM and BPCNeuAc-NP were also combined with cell type specific antibodies for double labeling. Labeling proceeded for 1 hour at 37 °C. Cells were washed with HBSS/BSA containing 100 nM BPCNeuAc-NP and fluorescence was measured using a FACS Calibur flow cytometer with CellQuest Pro software for analysis.
Results and Discussion
Synthesis of the bi-functional ligands
The exemplary CD22 ligand, BPCNeu5Acα2-6-Galβ1-4-GlcNAc-ethylamine was synthesized in a one-pot enzymatic reaction employing glycosyltransferases to sequentially transfer galactose and 9-BPC-Neu5Ac to N-acetyl-glucosamine conjugated to an ethylamine linker (Scheme 1). The product was then coupled directly to a commercially available N-hydroxysuccinamide activated 2-(4-hydroxy-3-nitrophenyl)acetic acid to yield the desired bi-functional ligand (BPCNeuAc-NP 11). Ligands with alternate linkers were synthesized using related strategies modified as appropriate to merge the chemical and enzymatic steps. The shortest linker was constructed by first coupling the glycosylamine 2 to 2-(4-hydroxy-3-nitrophenyl)acetic acid under standard conditions to give compound 7, which underwent one-pot enzymatic transformations to afford bi-functional ligand 12. Compound 15, with the longest linear linker, was prepared by first coupling compound 1 to Fmoc-protected 6-aminohexanoic acid. Removal of the Fmoc protecting group and enzymatic transformation yielded the trisaccharide 10, which was then coupled with 2-(4-hydroxy-3-nitrophenyl)acetic acid N-hydroxysuccinimide ester to give desired ligand 15. Alternate ligands with a 1,2,3-triazol linker modification were prepared by using a copper(I)-catalyzed azide-alkyne cycloaddition reaction35 to install the triazole linkers (Scheme 1). CD22 ligands with an azide group at the reducing end were either readily prepared from commercially available 2-Acetamido-3,4,6-tri-O-acetyl-2-deoxy- -D-glucopyranosyl azide (ligand 8) or synthesized as previously described (ligand 9)33. The nitrophenyl moiety was first coupled with propargylamine to install an alkyne group that was then coupled with the azide group of the corresponding CD22 ligands.
Scheme 1. Synthesis of bi-functional ligands of CD22.
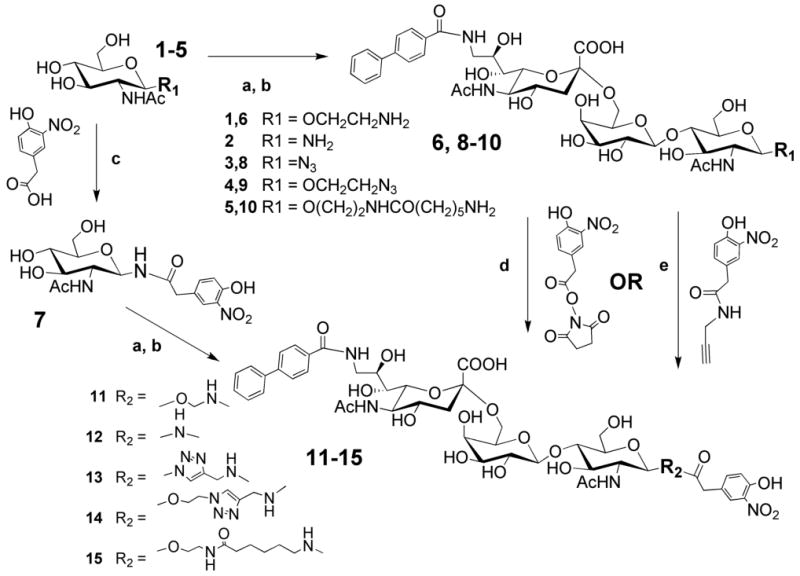
a) GalT-GalE, UDP-Gal; b) hST6Gal I, CMP-9-N-biphenylcarboxyl-9-deoxy-Neu5Ac; c) HBTU, HOBt, Et3N (for compound 7); d) MeOH, Et3N (for compounds 6 and 10); e) Cu2SO4, sodium ascorbate, water : t-BuOH = 1 : 1 (for compounds 8 and 9).
In vitro detection of bi-functional ligand-driven IgM-CD22 complex formation
The ability of the BPCNeuAc-NP (11) to assemble a complex between anti-NP-IgM and CD22 was initially tested in ELISA format with CD22-Fc chimera adsorbed to protein A coated ELISA wells (Figure 2a). Binding of IgM was observed with BPCNeuAc-NP in a dose-dependent manner, with no binding over background observed in the presence of 2-(4-hydroxy-3-nitrophenyl)acetic acid (NP) alone. The bi-functional ligand (1 μM) also assembled a complex with anti-NP IgM on CD22-Fc coated magnetic beads that could be observed by flow cytometry (Figure 2b), whereby varying the concentration of anti-NP IgM revealed detectable binding at 0.07 μg/ml and maximal binding at 5-10 μg/ml (∼ 7-13 nM IgM). CD22-Fc beads were also used to titrate the ligand concentration with a fixed concentration of anti-NP IgM (Figure S1, see also Figures S3, S4, 4 and 6). We found that the bi-functional ligand 11 becomes inhibitory at concentrations above ∼10 μM, presumably a result of the bi-functional ligand becoming a competitive inhibitor as it saturates the binding sites of both CD22 and the anti-IgM-NP, preventing a bridging of the two proteins.
Figure 2. Bi-functional ligand 11 drives complex formation of IgM with immobilized recombinant CD22-Fc and on native B cells.
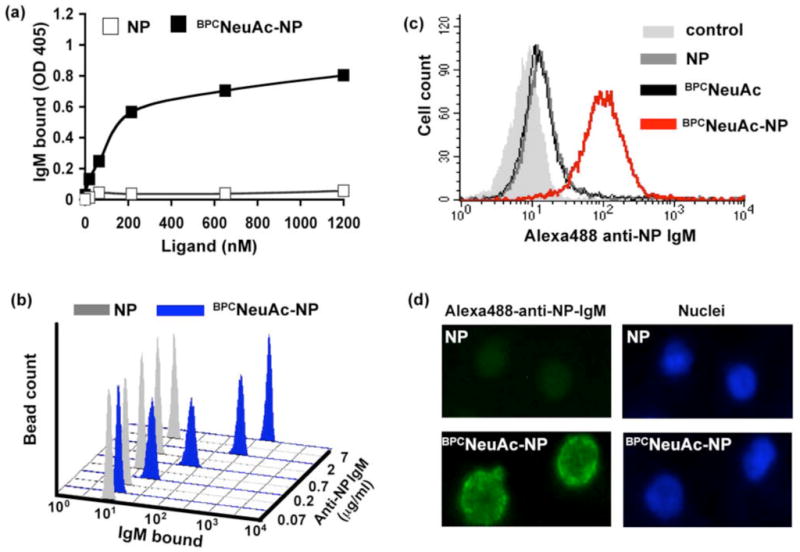
(a) Using ELISA, anti-NP IgM binding to CD22-Fc chimeras immobilized onto protein A coated wells was shown to be dose dependent with BPCNeuAc-NP, but not NP. (b) Flow cytometry was used to measure the anti-NP IgM dose dependence using CD22-Fc chimeras immobilized onto Protein A conjugated magnetic beads in the presence of 1 μM BPCNeuAc-NP and. The x-axis represents the degree of IgM binding in relative fluorescence units. (c) BPCNeuAc-NP dependent anti-NP IgM binding occurs on native BJAB cells as analyzed by flow cytometry. BJAB cells were incubated with 3 μM ligand and 10 μg/mL Alexa488-anti-NP IgM. (d) Immunofluorescence microscopy was used to visualize binding of Alexa488-anti-NP IgM to native BJAB cells.
Figure 4. Analysis of antibody valency in complex formation.
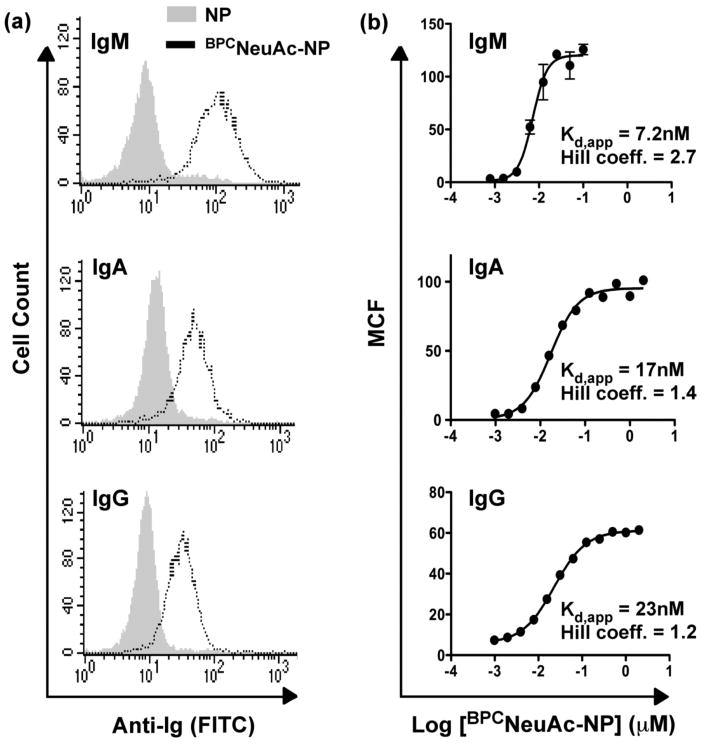
(a) Anti-NP antibodies of different valency were assessed for their binding to native BJAB cells in the presence of 2 μM BPCNeuAc-NP (11), using 10 μg/mL IgM (n=10; top), IgA (n=4; middle), or IgG (n=2; bottom). Ig binding was assessed by flow cytometry after staining with secondary antibodies (FITC). (b) Avidities of complex formation with anti-NP antibodies of different valency were assessed by titrating BPCNeuAc-NP (11) in the presence of CD22-immobilized magnetic beads and 5 μg/mL anti-NP IgM, IgA, or IgG, and measuring bead fluorescence by flow cytometry. Mean channel fluorescence (MCF) is plotted against ligand concentration.
Figure 6. Temperature dependence of complex formation.
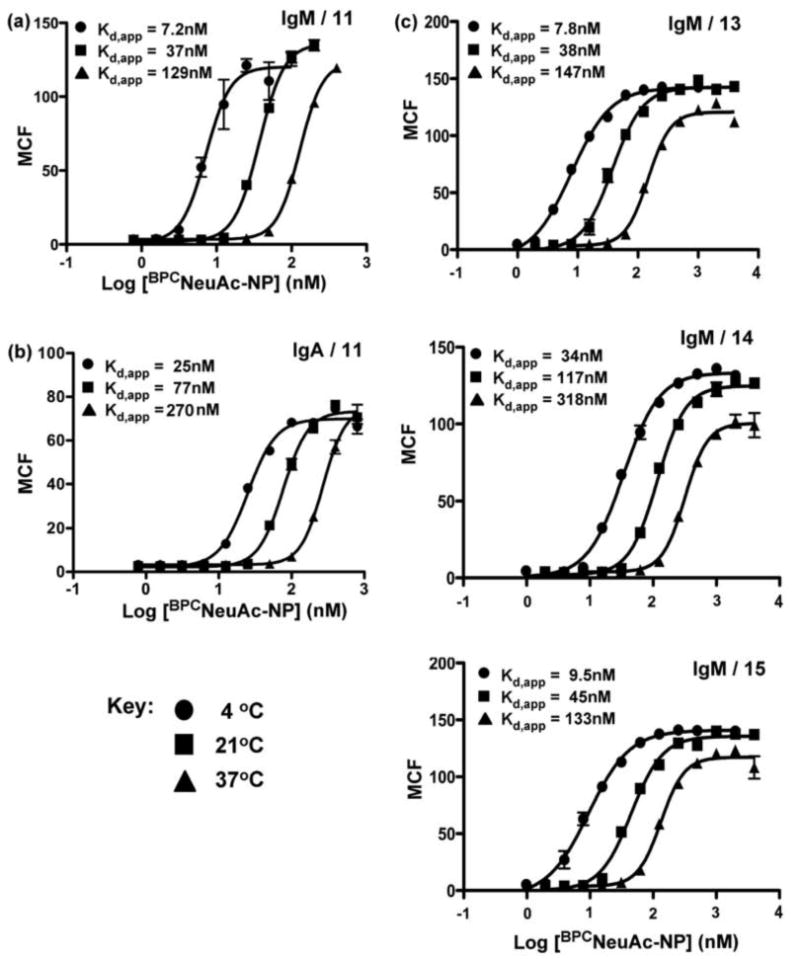
Bi-functional ligand 11 was compared for its ability to induce complex formation of of 5 μg/mL anti-NP IgM (a) or IgA (b) on CD22-immobilized magnetic beads at the indicated temperatures. (c) Bi-functional ligands 13-15 (top to bottom, respectively) were similarly compared for their ability to induce complex formation of anti-NP IgM on CD22 immobilized magnetic beads. Ig binding was assessed by flow cytometry following staining with secondary antibodies (FITC).
Assembly of CD22-IgM complexes on B cells
To evaluate the ability of IgM to bind to CD22 on B cells, Alexa488-anti-NP-IgM and BPCNeuAc-NP (11), BPCNeuAc, or NP were incubated with native human B lymphoma cells (BJAB K20) and assessed for binding by flow cytometry (Figure 2c). Only in the presence of the bi-functional ligand, anti-NP IgM is highly associated with BJAB cells. Complex formation was also visualized by immunofluorescence microscopy. Bright staining of the cells in the presence of the ligand (11), but not NP, demonstrated that the desired complex does indeed form on BJAB cells (Figure 2d). These results were somewhat surprising since cis ligands of CD22 are well documented to block the binding of all but highly multivalent polymeric ligands of CD22 22, 23. To demonstrate the specificity of complex formation for CD22, IgM was shown to bind to CHO cells transfected with a plasmid for CD22 expression only in the presence of ligand 11, while no binding was observed to non-transfected cells (Figure S2).
Effect of linker structure on complex assembly
To investigate the influence of linker structure on complex formation, bi-functional ligands with linkers of different length and/or with a heterocyclic triazole ring were also tested (ligands 12-15). With the exception of the shortest linker, 12, which may impose steric constraints between IgM and CD22, complex formation on native B cells was supported by each of the ligands (Figure 3), and thus is relatively tolerant to changes in the linker structure. These findings were confirmed in experiments examining complex formation of anti-NP IgM on CD22 beads upon titration of each of the bi-functional ligands (Figure S3). This analysis revealed very similar capacity of the ligands bearing medium and long linear and short triazole linkers to form complexes, and accommodation of the relatively rigid triazole ring validates the use of the copper catalyzed azide-alkyne cycloaddition reaction (“Click chemistry”)36 for the facile assembly of bi-functional ligands.
Figure 3. Influence of linker structure in bi-functional ligand driven assembly of IgM-CD22 complexes on B cells.
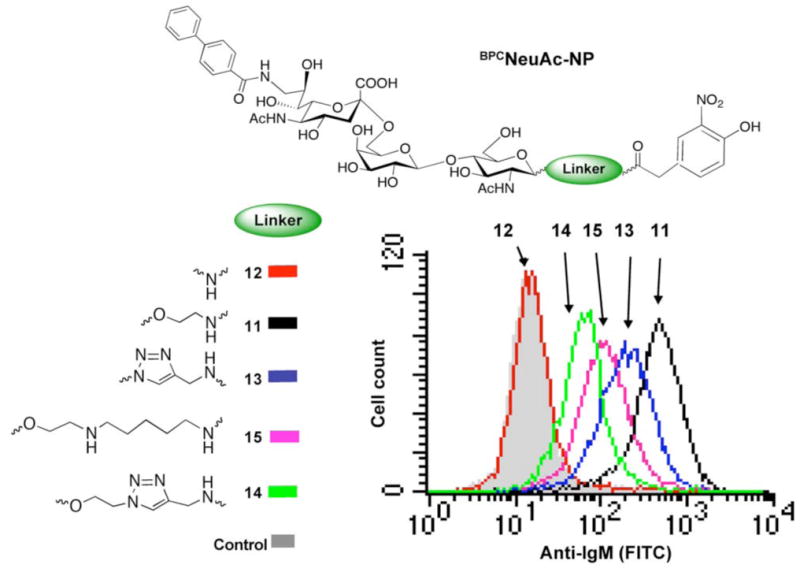
Ligands 11-15 or NP (2 μM) were tested in the self-assembly of IgM-CD22 complexes on native BJAB cells. Binding of anti-NP IgM (10 μg/mL) was analyzed by flow cytometry following staining with anti-IgM (FITC).
Assembly of immune complexes of lower valency
To better understand the importance of valency in complex formation, we evaluated the ability of the bi-functional ligand 11 (BPCNeuAc-NP) to induce complex formation between anti-NP antibodies of the IgM (decavalent), IgA (tetravalent), and IgG (bivalent) subclasses and CD22 on B cells and recombinant CD22 expressed on CHO cells37. All of these antibodies were able to form complexes on native B cells (Figure 4), and on CHO cells in both a ligand and CD22-dependent manner (data not shown, results similar to Figure S2). The binding of the lower valency antibodies were particularly surprising since cis ligands of CD22 are well documented to block the binding of all but highly multivalent polymeric ligands of CD22 22, 23.
For a more quantitative analysis, BPCNeuAc-NP titrations were carried out to examine Ig complex formation on CD22-immobilized magnetic beads. Clear differences in the concentration dependence for ligand-mediated complex formation is seen, with apparent Kd values of 7, 17 and 23 nM for IgM, IgA and IgG, respectively. The Hill plots of the data in Figure 4 reflect cooperativity in complex assembly, with Hill coefficients that are greater than 1. Bi-functional ligands 13-15 were also able to drive CD22-IgA complex formation with correspondingly lower avidity compared to IgM (Figure S4).
Of particular note, the apparent Kd values are ≥100 fold lower than the respective Kd values of the respective monovalent ligands for CD22 (Kd ≅ 2 μM)23 and the B1-8 anti-NP IgM (Kd ≅ 1 μM)38. Thus, at a concentration of 10-20 nM BPCNeuAc-NP, there is negligible occupancy of the binding sites of the free CD22 or anti-NP-IgM, providing strong evidence that the complexes self-assemble at the surface of the bead.
Effects of cis ligands in complex assembly on B cells
To establish the degree to which cis ligands modulate the assembly of Ig complexes with CD22 on B cells we took advantage of the fact that BJAB K20 B cell line is well suited to evaluate the effect of cis ligands, since it is deficient in the epimerase/kinase step of sialic acid biosynthesis 39. Thus, when the cells are cultured in the absence of sialic acids (asialo-BJAB) there are no cis ligands, but when grown in the presence of serum or sialic acid their glycoproteins are sialylated equivalently to wild type BJAB cells 24. The effect of cis ligands was clearly evident for CD22 complex formation with both IgM and IgA upon titration of BPCNeuAc-NP (11), revealing that complex formation on asialo-BJAB cells (Figure 5a) was achieved at much lower concentration than needed for fully sialylated BJAB cells (Figure 5b), as evidenced by the differences in apparent Kd values. All binding at 4° C is a result of surface complex formation, and not internalization, since it is readily displaced by brief treatment with buffer of low pH (data not shown). The avidities of complex formation on asialo-BJAB cells agree very well with those on CD22-Fc beads, which also do not bear cis ligands. The similarity between ligand induced binding to asialo B cells and CD22-Fc beads also suggests that lateral mobility is not necessary to achieve complex formation. The effect of cis ligands is even more pronounced for the lower-valency IgA, suggesting that increased valency is particularly important for competition with the high concentration of sialic acid bearing cis ligands at the cell surface.
Figure 5. Self-assembly of antibody-CD22 complexes on native B cells is in competition with cis ligands.
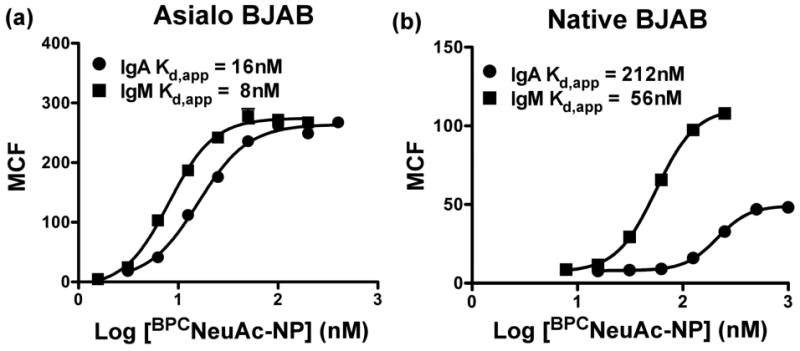
Titrations of BPCNeuAc-NP (11) in the presence of 5 μg/mL anti-NP IgM or IgA and BJAB cells cultured in the absence (a) or presence (b) of sialic acid reveal differences in concentrations of ligand required for complex formation.
These observations provide strong evidence that the bi-functional ligand drives dynamic assembly of the IgM-CD22 complex on the surface of the B cell, and does not require a preformed complex of the ligand with either CD22 or IgM. The CD22-binding portion of the bi-functional ligand, with a Kd of ∼ 2 μM 21, 23, is not by itself able to compete with cis ligands on BJAB cells to occupy a significant fraction of CD22 binding sites, since the same ligand in a multivalent polyacrylamide construct (n=15) fails to stably bind to B cells unless cis ligands are destroyed 23. Thus, the IgM scaffold is required for the BPCNeuAc-NP ligand to displace cis ligands and form a stable complex. As observed with complex formation on magnetic beads, the apparent dissociation constant of complex formation on cells is orders of magnitude lower than that of either of the individual binding interactions21, 38, suggesting that both IgM and CD22 mutually benefit from the multivalency afforded by one another.
The efficiency of the bi-functional ligand-mediated complex formation between anti-NP-IgM and CD22 is surprising not only because of the weak affinity of the CD22 ligand and the presence of cis ligands, but also because of the low copy number of CD22 on the B cell (∼10-20,000)40. Thus, the documented clustering of CD22 in clathrin rich microdomains may be a critical factor for bi-functional ligand mediated docking of IgM by providing a local high density of CD22 monomers 24, 26, 41, 42.
Thermodynamics of complex assembly
Once bound, IgM rapidly dissociates from the B cell upon repeated washing at 4° C, but is retained on the cell surface if at least 50 nM BPCNeuAc-NP ligand 11 is included in the wash buffer (Figure S5). These observations demonstrate that upon formation, the complex is in rapid equilibrium with the ligands in solution. The dissociation of IgM upon washing is even more pronounced at elevated temperatures, which is consistent with the temperature-dependence demonstrated for complex formation on magnetic beads with IgM (Figure 6a) or IgA (Figure 6b). Complex formation obeys a temperature dependence characterized by decreased affinity with increasing temperature, which is expected since the change in entropy (ΔS) upon complex formation is anticipated to be negative, and in fact this trend has been observed previously for CD22 binding to a glycoprotein ligand21. Complexes formed using bi-functional ligands with alternate linkers (13-15) also obey this temperature dependence (Figure 6c).
Specificity of IgM binding to primary B lymphocytes
B cell depletion therapy is now recognized to have therapeutic benefit for B cell leukemia as well as a number of chronic inflammatory diseases such as rheumatoid arthritis and systemic lupus erythamatosis43. CD22 is currently being evaluated in human clinical trials as a target for B cell depletion therapy due to its restricted expression on B cells, and efficient uptake of toxin-conjugated anti-CD22 antibody 19, 20. Driving the formation of immune complexes using bi-functional ligands comprising a ligand for CD22 coupled to an antigen, as reported here, may provide an alternative approach to targeting B cells. Haas et al. have shown that treatment of mice with an antibody targeted to the ligand-binding site of CD22, but not antibodies targeting other regions of the protein, resulted in depletion of both normal and malignant B cells44. The mechanism for depletion by targeting only the ligand binding site remains unclear, but may be addressed and exploited by IgM-mediated targeting of the CD22 ligand binding site with the native ligand. IgM antibodies are also well known activators of complement fixation, resulting in cell killing. To assess the specificity of targeting B cells using the bi-functional CD22 ligand, we measured the ability of anti-NP IgM to form complexes on mixed white blood cells from peripheral human blood. When cells were treated with Alexa488-anti-NP IgM in the presence of either NP or BPCNeuAc-NP (11), a population of IgM-binding cells was detected only in the presence of the bi-functional ligand (Figure 7a). To identify the cell type represented by this IgM-binding population, antibody markers for B cell, T cell, and natural killer cell subsets were used in a dual labeling experiment. Figure 7b shows that the cells that exhibit BPCNeuAc-NP (11) dependent binding of anti-NP IgM are B220+ B cells, and that all B cells bind the anti-NP IgM. Conversely, NK cells, CD4+ T cells and CD8+ T cells do not exhibit binding.
Figure 7. Specificity of BPCNeuAc-NP formation of immune complexes on B cells.
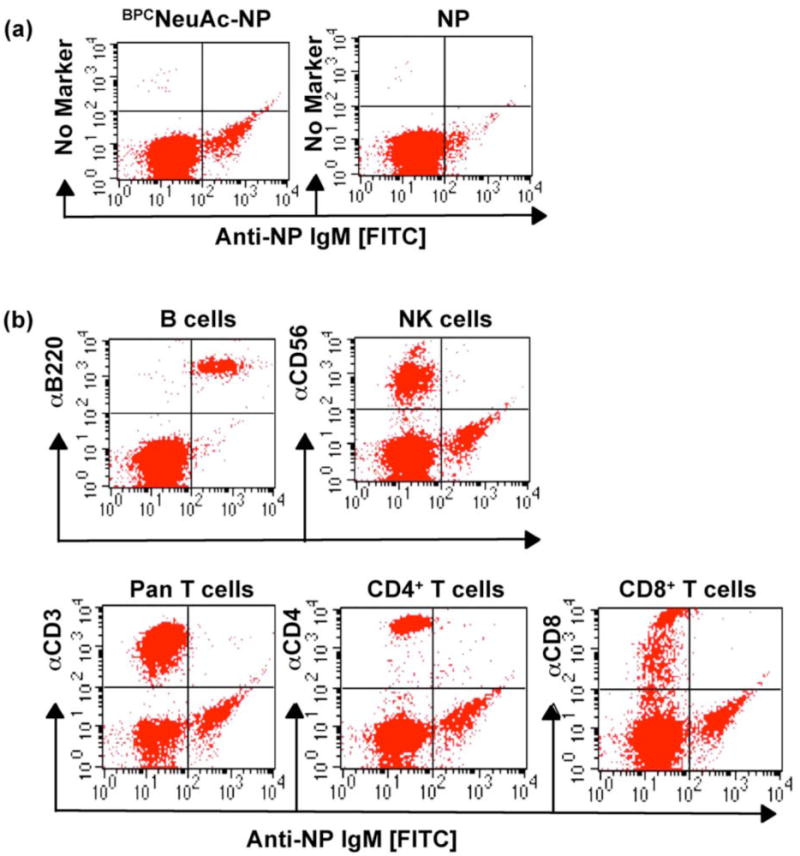
White blood cells isolated from peripheral human blood were (a) stained with Alexa488-anti-NP IgM in the presence of 2 μM NP or BPCNeuAc-NP (11) at 37 °C, or (b) subjected to double staining with Alexa488-anti-NP IgM (2 μM 11 included) and PE-labeled anti-CD3 (pan T cell), anti-CD4 (CD4+ T cells), anti-CD8 (CD8+ T cells), anti-CD19 (B cells), or anti-CD56 (natural killer cells). Binding was analyzed by flow cytometry.
Conclusions
Here we report the use of small molecule bi-functional ligands of CD22 that use antibodies as protein scaffolds to assemble antibody-CD22 complexes of low valency (n = 2-10) on B cells. This is in contrast to the failure of higher valency polymer-based ligands of CD22 to bind to native B cells due to masking by cis ligands22, 23, 45. We attribute the success of this approach to the unique geometry afforded by the antibody scaffold, allowing assembly of the complex with optimal spacing to allow simultaneous contact with multiple CD22 molecules. The results suggest a general approach in the rational design of ligand-based probes for glycan-binding proteins and other low-affinity receptors using soluble multivalent proteins as a scaffold, as an alternative to polymers, dendrimers or neo-glycoproteins. The relevant variables include ligand affinity, valency of the scaffold, density of the receptor, and linking chemistry of the bi-functional ligand. It is anticipated that design parameters for each variable will be better defined with each successful combination of protein scaffold and cell surface glycan-binding protein.
Supplementary Material
Supporting Information Available. Titration curve for compound 11 driving CD22-IgM complex formation, showing inhibition at higher concentrations. Flow cytometry data for IgM binding to CHO cells expressing CD22, and not to wild type CHO. Materials and methods for CHO cell binding assay. Titration curves for compounds 11 and 13-15 with anti-NP IgM and IgA on CD22 beads. Flow cytometry data demonstrating the maintenance of the IgM complex upon washing in the presence, but not in the absence, of compound 11.
Acknowledgments
We thank M. Pawlita, A. Varki, and R.B. Corley for generous gifts of the BJAB cell line, plasmid encoding hCD22, and the B1-8 IgM anti-NP hybridoma, respectively. We thank Dr. Ramya T. N. Chakravarthy for providing asialo BJAB cells. The authors wish to thank Ms. Anna Tran-Crie for her help with manuscript preparation. This work was funded in part by NIH grants GM060938 & AI050143 (J.C.P.), and Natural Science and Engineering Research Council and Alberta Ingenuity grants (D.R.B.). M.K.O. is supported by a postdoctoral fellowship from the American Cancer Society.
References
- 1.Crocker PR, Paulson JC, Varki A. Siglecs and their roles in the immune system. Nat Rev Immunol. 2007;7(4):255–66. doi: 10.1038/nri2056. [DOI] [PubMed] [Google Scholar]
- 2.Liu FT, Rabinovich GA. Galectins as modulators of tumour progression. Nat Rev Cancer. 2005;5(1):29–41. doi: 10.1038/nrc1527. [DOI] [PubMed] [Google Scholar]
- 3.Paulson JC, Blixt O, Collins BE. Sweet spots in functional glycomics. Nat Chem Biol. 2006;2(5):238–48. doi: 10.1038/nchembio785. [DOI] [PubMed] [Google Scholar]
- 4.Collins BE, Paulson JC. Cell surface biology mediated by low affinity multivalent protein-glycan interactions. Curr Opin Chem Biol. 2004;8(6):617–25. doi: 10.1016/j.cbpa.2004.10.004. [DOI] [PubMed] [Google Scholar]
- 5.Gestwicki JE, Cairo CW, Strong LE, Oetjen KA, Kiessling LL. Influencing receptor-ligand binding mechanisms with multivalent ligand architecture. J Am Chem Soc. 2002;124(50):14922–33. doi: 10.1021/ja027184x. [DOI] [PubMed] [Google Scholar]
- 6.Lees WJ, Spaltenstein A, Kingery-Wood JE, Whitesides GM. Polyacrylamides bearing pendant alpha-sialoside groups strongly inhibit agglutination of erythrocytes by influenza A virus: multivalency and steric stabilization of particulate biological systems. J Med Chem. 1994;37(20):3419–33. doi: 10.1021/jm00046a027. [DOI] [PubMed] [Google Scholar]
- 7.Bovin NV. Polyacrylamide-based glycoconjugates as tools in glycobiology. Glycoconj J. 1998;15(5):431–46. doi: 10.1023/a:1006963717646. [DOI] [PubMed] [Google Scholar]
- 8.Kitov PI, Sadowska JM, Mulvey G, Armstrong GD, Ling H, Pannu NS, Read RJ, Bundle DR. Shiga-like toxins are neutralized by tailored multivalent carbohydrate ligands. Nature. 2000;403(6770):669–72. doi: 10.1038/35001095. [DOI] [PubMed] [Google Scholar]
- 9.Lasala F, Arce E, Otero JR, Rojo J, Delgado R. Mannosyl glycodendritic structure inhibits DC-SIGN-mediated Ebola virus infection in cis and in trans. Antimicrob Agents Chemother. 2003;47(12):3970–2. doi: 10.1128/AAC.47.12.3970-3972.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang Z, Merritt EA, Ahn M, Roach C, Hou Z, Verlinde CL, Hol WG, Fan E. Solution and crystallographic studies of branched multivalent ligands that inhibit the receptor-binding of cholera toxin. J Am Chem Soc. 2002;124(44):12991–8. doi: 10.1021/ja027584k. [DOI] [PubMed] [Google Scholar]
- 11.Andre S, Pieters RJ, Vrasidas I, Kaltner H, Kuwabara I, Liu FT, Liskamp RM, Gabius HJ. Wedgelike glycodendrimers as inhibitors of binding of mammalian galectins to glycoproteins, lactose maxiclusters, and cell surface glycoconjugates. Chembiochem. 2001;2(11):822–30. doi: 10.1002/1439-7633(20011105)2:11<822::AID-CBIC822>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 12.Bertozzi C, Bednarski M. C-glycosyl compounds bind to receptors on the surface of Escherichia coli and can target proteins to the organism. Carbohydr Res. 1992;223:243–53. doi: 10.1016/0008-6215(92)80021-r. [DOI] [PubMed] [Google Scholar]
- 13.Liu J, Zhang Z, Tan X, Hol WG, Verlinde CL, Fan E. Protein heterodimerization through ligand-bridged multivalent pre-organization: enhancing ligand binding toward both protein targets. J Am Chem Soc. 2005;127(7):2044–5. doi: 10.1021/ja043817r. [DOI] [PubMed] [Google Scholar]
- 14.Solomon D, Kitov PI, Paszkiewicz E, Grant GA, Sadowska JM, Bundle DR. Heterobifunctional multivalent inhibitor-adaptor mediates specific aggregation between Shiga toxin and a pentraxin. Org Lett. 2005;7(20):4369–72. doi: 10.1021/ol051529+. [DOI] [PubMed] [Google Scholar]
- 15.Kitov PI, Lipinski T, Paszkiewicz E, Solomon D, Sadowska JM, Grant GA, Mulvey GL, Kitova EN, Klassen JS, Armstrong GD, Bundle DR. An entropically efficient supramolecular inhibition strategy for Shiga toxins. Angew Chem Int Ed Engl. 2008;47(4):672–6. doi: 10.1002/anie.200704064. [DOI] [PubMed] [Google Scholar]
- 16.Carlson CB, Mowery P, Owen RM, Dykhuizen EC, Kiessling LL. Selective tumor cell targeting using low-affinity, multivalent interactions. ACS Chem Biol. 2007;2(2):119–27. doi: 10.1021/cb6003788. [DOI] [PubMed] [Google Scholar]
- 17.Lu Y, Low PS. Folate targeting of haptens to cancer cell surfaces mediates immunotherapy of syngeneic murine tumors. Cancer Immunol Immunother. 2002;51(3):153–62. doi: 10.1007/s00262-002-0266-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shokat KM, Schultz PG. Redirecting the immune-response: Ligand-mediated immunogenicity. J Am Chem Soc. 1991;113:1861–1862. [Google Scholar]
- 19.Qu Z, Griffiths GL, Wegener WA, Chang CH, Govindan SV, Horak ID, Hansen HJ, Goldenberg DM. Development of humanized antibodies as cancer therapeutics. Methods. 2005;36(1):84–95. doi: 10.1016/j.ymeth.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 20.Steinfeld SD, Tant L, Burmester GR, Teoh NK, Wegener WA, Goldenberg DM, Pradier O. Epratuzumab (humanised anti-CD22 antibody) in primary Sjogren's syndrome: an open-label phase I/II study. Arthritis Res Ther. 2006;8(4):R129. doi: 10.1186/ar2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bakker TR, Piperi C, Davies EA, Merwe PA. Comparison of CD22 binding to native CD45 and synthetic oligosaccharide. Eur J Immunol. 2002;32(7):1924–32. doi: 10.1002/1521-4141(200207)32:7<1924::AID-IMMU1924>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 22.Razi N, Varki A. Masking and unmasking of the sialic acid-binding lectin activity of CD22 (Siglec-2) on B lymphocytes. Proc Natl Acad Sci U S A. 1998;95(13):7469–74. doi: 10.1073/pnas.95.13.7469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Collins BE, Blixt O, Han S, Duong B, Li H, Nathan JK, Bovin N, Paulson JC. High-affinity ligand probes of CD22 overcome the threshold set by cis ligands to allow for binding, endocytosis, and killing of B cells. J Immunol. 2006;177(5):2994–3003. doi: 10.4049/jimmunol.177.5.2994. [DOI] [PubMed] [Google Scholar]
- 24.Han S, Collins BE, Bengtson P, Paulson JC. Homomultimeric complexes of CD22 in B cells revealed by protein-glycan cross-linking. Nat Chem Biol. 2005;1(2):93–7. doi: 10.1038/nchembio713. [DOI] [PubMed] [Google Scholar]
- 25.Varki A, Angata T. Siglecs--the major subfamily of I-type lectins. Glycobiology. 2006;16(1):1R–27R. doi: 10.1093/glycob/cwj008. [DOI] [PubMed] [Google Scholar]
- 26.Grewal PK, Boton M, Ramirez K, Collins BE, Saito A, Green RS, Ohtsubo K, Chui D, Marth JD. ST6Gal-I restrains CD22-dependent antigen receptor endocytosis and Shp-1 recruitment in normal and pathogenic immune signaling. Mol Cell Biol. 2006;26(13):4970–81. doi: 10.1128/MCB.00308-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kelm S, Gerlach J, Brossmer R, Danzer CP, Nitschke L. The ligand-binding domain of CD22 is needed for inhibition of the B cell receptor signal, as demonstrated by a novel human CD22-specific inhibitor compound. J Exp Med. 2002;195(9):1207–13. doi: 10.1084/jem.20011783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Blixt O, Brown J, Schur MJ, Wakarchuk W, Paulson JC. Efficient preparation of natural and synthetic galactosides with a recombinant beta-1,4-galactosyltransferase-/UDP-4′-gal epimerase fusion protein. J Org Chem. 2001;66(7):2442–8. doi: 10.1021/jo0057809. [DOI] [PubMed] [Google Scholar]
- 29.Blixt O, Allin K, Pereira L, Datta A, Paulson JC. Efficient chemoenzymatic synthesis of O-linked sialyl oligosaccharides. J Am Chem Soc. 2002;124(20):5739–46. doi: 10.1021/ja017881+. [DOI] [PubMed] [Google Scholar]
- 30.Blixt O, Paulson JC. Biocatalytic preparation of N-glycolylneuraminic acid deaminoneuraminic acid (KDN) and 9-azido-9-deoxysialic acid oligosaccharides. Adv Synthesis & Catal. 2003;345:687–690. [Google Scholar]
- 31.Eklind K, Gustafsson R, Tiden AK, Norberg T, Arberg PM. A new simple synthesis of amino sugar b -D-glycosylamines. J Carbohydr Chem. 1996;15:1161–1178. [Google Scholar]
- 32.Likhosherstov LM, Novikova OS, Derevitskaya VA, Kochetkov NK. A new simple synthesis of amino sugar beta-D-glycosylamines. Carbohydrate Res. 1986;146(146):C1–C5. [Google Scholar]
- 33.Kaltgrad E, O'Reilly MK, Liao L, Han S, Paulson JC, Finn MG. On-virus construction of polyvalent glycan ligands for cell-surface receptors. J Am Chem Soc. 2008 Mar 15; doi: 10.1021/ja077801n. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brinkman-Van der Linden EC, Sjoberg ER, Juneja LR, Crocker PR, Varki N, Varki A. Loss of N-glycolylneuraminic acid in human evolution. Implications for sialic acid recognition by siglecs. J Biol Chem. 2000;275(12):8633–40. doi: 10.1074/jbc.275.12.8633. [DOI] [PubMed] [Google Scholar]
- 35.Kaltgrad E, Sen Gupta S, Punna S, Huang CY, Chang A, Wong CH, Finn MG, Blixt O. Anti-carbohydrate antibodies elicited by polyvalent display on a viral scaffold. Chembiochem. 2007;8(12):1455–62. doi: 10.1002/cbic.200700225. [DOI] [PubMed] [Google Scholar]
- 36.Kolb HC, Finn MG, Sharpless KB. Click Chemistry: Diverse Chemical Function from a Few Good Reactions. Angew Chem Int Ed Engl. 2001;40(11):2004–2021. doi: 10.1002/1521-3773(20010601)40:11<2004::AID-ANIE2004>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 37.Tateno H, Li H, Schur MJ, Bovin N, Crocker PR, Wakarchuk WW, Paulson JC. Distinct endocytic mechanisms of CD22 (Siglec-2) and Siglec-F reflect roles in cell signaling and innate immunity. Mol Cell Biol. 2007;27(16):5699–710. doi: 10.1128/MCB.00383-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dal Porto JM, Haberman AM, Shlomchik MJ, Kelsoe G. Antigen drives very low affinity B cells to become plasmacytes and enter germinal centers. J Immunol. 1998;161(10):5373–81. [PubMed] [Google Scholar]
- 39.Keppler OT, Hinderlich S, Langner J, Schwartz-Albiez R, Reutter W, Pawlita M. UDP-GlcNAc 2-epimerase: a regulator of cell surface sialylation. Science. 1999;284(5418):1372–6. doi: 10.1126/science.284.5418.1372. [DOI] [PubMed] [Google Scholar]
- 40.D'Arena G, Musto P, Cascavilla N, Dell'Olio M, Di Renzo N, Carotenuto M. Quantitative flow cytometry for the differential diagnosis of leukemic B-cell chronic lymphoproliferative disorders. Am J Hematol. 2000;64(4):275–81. doi: 10.1002/1096-8652(200008)64:4<275::aid-ajh7>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 41.Collins BE, Smith BA, Bengtson P, Paulson JC. Ablation of CD22 in ligand-deficient mice restores B cell receptor signaling. Nat Immunol. 2006;7(2):199–206. doi: 10.1038/ni1283. [DOI] [PubMed] [Google Scholar]
- 42.Zhang M, Varki A. Cell surface sialic acids do not affect primary CD22 interactions with CD45 and surface IgM nor the rate of constitutive CD22 endocytosis. Glycobiology. 2004;14(11):939–49. doi: 10.1093/glycob/cwh126. [DOI] [PubMed] [Google Scholar]
- 43.Browning JL. B cells move to centre stage: novel opportunities for autoimmune disease treatment. Nat Rev Drug Discov. 2006;5(7):564–76. doi: 10.1038/nrd2085. [DOI] [PubMed] [Google Scholar]
- 44.Haas KM, Sen S, Sanford IG, Miller AS, Poe JC, Tedder TF. CD22 ligand binding regulates normal and malignant B lymphocyte survival in vivo. J Immunol. 2006;177(5):3063–73. doi: 10.4049/jimmunol.177.5.3063. [DOI] [PubMed] [Google Scholar]
- 45.Yang ZQ, Puffer EB, Pontrello JK, Kiessling LL. Synthesis of a multivalent display of a CD22-binding trisaccharide. Carbohydr Res. 2002;337(18):1605–13. doi: 10.1016/s0008-6215(02)00270-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information Available. Titration curve for compound 11 driving CD22-IgM complex formation, showing inhibition at higher concentrations. Flow cytometry data for IgM binding to CHO cells expressing CD22, and not to wild type CHO. Materials and methods for CHO cell binding assay. Titration curves for compounds 11 and 13-15 with anti-NP IgM and IgA on CD22 beads. Flow cytometry data demonstrating the maintenance of the IgM complex upon washing in the presence, but not in the absence, of compound 11.


