Abstract
The heart is formed from cardiogenic progenitors expressing the transcription factors Nkx2-5 and Isl1 1,2. These multipotent progenitors give rise to cardiomyocyte, smooth muscle, and endothelial cells, the major lineages of the mature heart 3,4. Here we identify a novel cardiogenic precursor marked by expression of the transcription factor Wt1 and located within the epicardium, an epithelial sheet overlying the heart. During normal heart development, a subset of these Wt1+ precursors differentiated into fully functional cardiomyocytes. Wt1+ proepicardial cells arose from progenitors that express Nkx2-5 and Isl1, suggesting that they share a developmental origin with multipotent Nkx2-5+/Isl1+ progenitors (Suppl. Fig 1). These results identify Wt1+ epicardial cells as previously unrecognized cardiomyocyte progenitors, and lay the foundation for future efforts to harness the cardiogenic potential of these progenitors for cardiac regeneration and repair.
Epicardial cells migrate from the proepicardium (PE), an outgrowth of the septum transversum, and spread over the surface of the heart5,6. A subset of epicardial cells transition to a mesenchymal phenotype, migrate into the subjacent myocardium, and differentiate into smooth muscle cells (SMCs) and endothelial cells (ECs) 7-13. Wt1 was expressed in PE and epicardium, but not in myocardium (Fig. 1a-c). In order to trace the fate of Wt1-expressing PE and epicardial cells, we knocked a GFPCre cDNA 14 into the endogenous Wt1 start codon (Suppl. Fig. 2). GFP and Cre expression in Wt1GFPCre/+ embryos co-localized with Wt1, indicating that the knockin strategy placed GFPCre under control of endogenous Wt1 regulatory elements (Fig. 1d-f). In the heart, GFPCre expression was confined to PE and epicardium from E9.5 to E15.5, and not found in the myocardium (Fig. 1d-g).
Fig. 1. Cardiac Wt1 and Wt1-driven GFPCre expression.
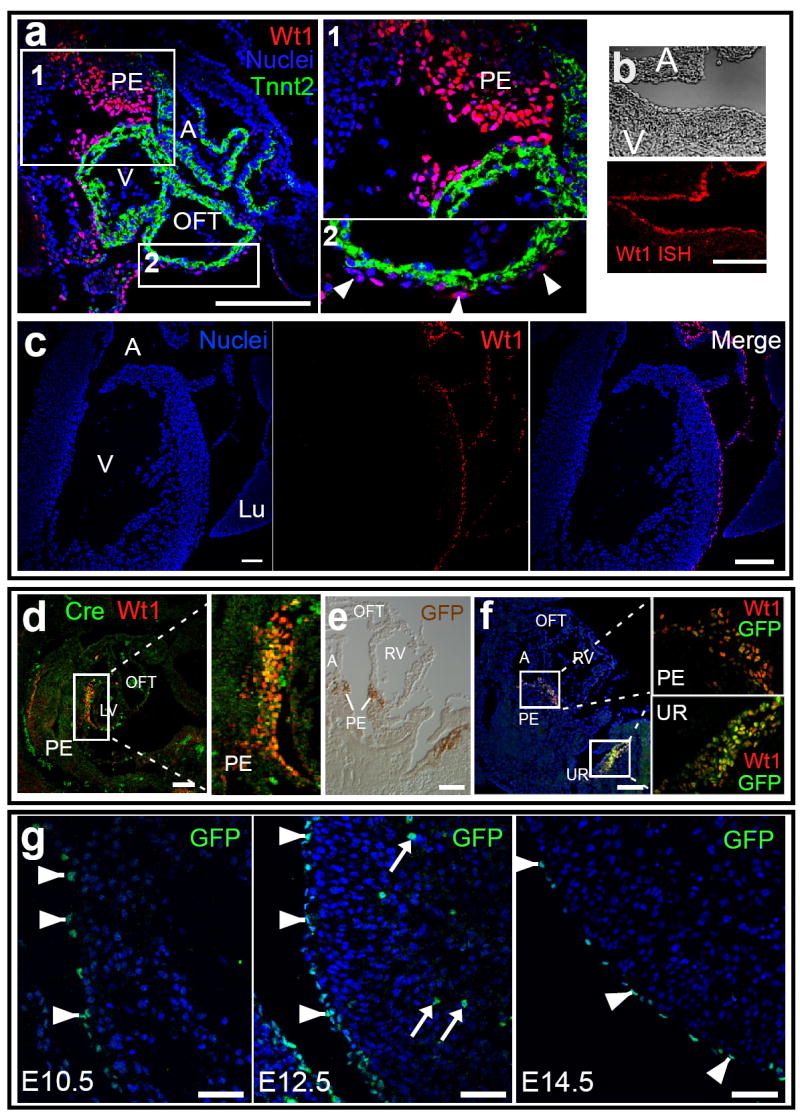
a. At E9.5, Wt1 was expressed in proepicardium (PE) and in scattered pericardial cells over the surface of the heart (arrowheads, a2). b-c. At E15.5, Wt1 expression was confined to the epicardium, as determined by in situ hybridization (ISH, b) and immunohistochemistry. d-f. Co-expression of Wt1 and GFPCre expression in Wt1GFPCre/+ E9.5 embryos. Wt1 and GFPCre were co-expressed in PE and urogenital ridge (UR). g. Wt1-driven GFPCre was confined to epicardium (arrowheads), and not detected within myocardium, at E9.5-E15.5 (representative images for E10.5, E12.5, and E14.5 are shown). Arrows indicate autofluorescent red blood cells. All images except b show immunohistochemical staining. A, atrium. V, ventricle. LV and RV, right and left ventricle. Lu, lung. OFT, outflow tract. Scale bars: 50 ⌠m.
We used Wt1GFPCre and the Cre-activated reporters Rosa26fsLz15 and Z/Red 16 to analyze the fate of Wt1-expressing cells in the heart. Upon Cre-mediated recombination, these reporters heritably express ®-galactosidase (®-gal) or red fluorescent protein (RFP), respectively. Using two different reporters minimized potential artefacts related to unanticipated behavior of Cre-activated reporters, or to false-positive immunostaining. While Wt1 and GFPCre expression were confined to the epicardium, descendants of Wt1-expressing progenitors (hereafter called Wt1-derived cells), marked by β-gal, were found in a mosaic pattern throughout the myocardium (Fig. 2a). Consistent with prior reports showing that epicardially-derived mesenchyme predominately differentiates into SMCs in mammals, most Wt1-derived cells adopted a SMC fate, and a minority differentiated into ECs (Suppl. Fig. 3) 8,9.
Fig. 2. Wt1-derived cells differentiate into cardiomyocytes.
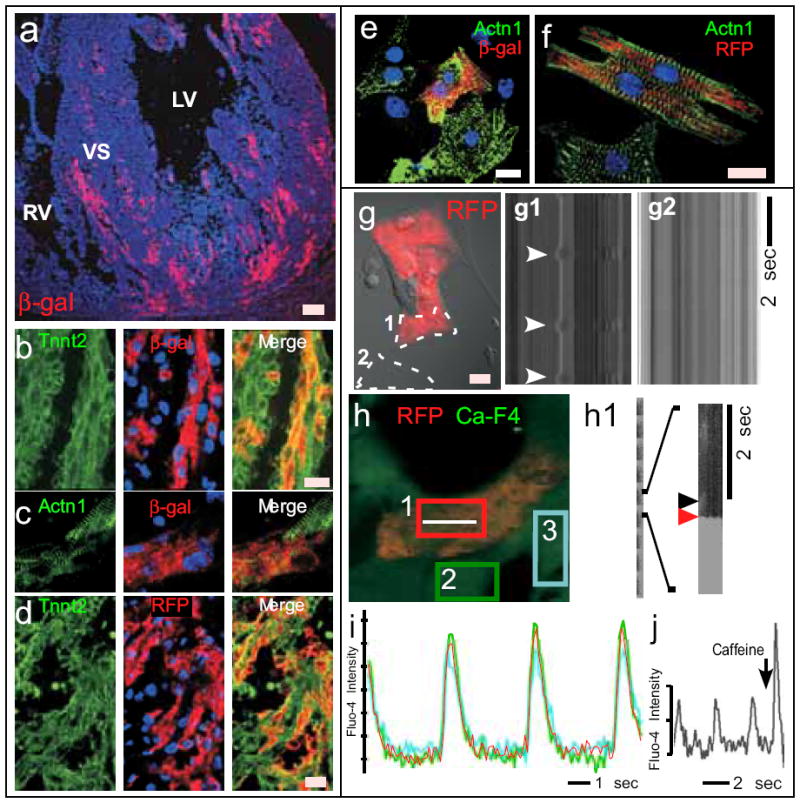
The fate of Wt1-derived cells, marked by ®-gal expression in Wt1GFPCre/+; Rosa26fsLz hearts (a-c,e), or RFP expression in Wt1GFPCre/+;Z/Red hearts (d,f,g-j), was analyzed by immunohistochemistry (a-f) and live cell imaging (g-j). a. Overview of contribution of Wt1-derived cells to myocardium. b-d. Cryosections of E15.5 heart stained for genetic lineage tracers (β-gal or RFP) and cardiomyocyte-specific markers Tnnt2 and Actn1. e-f. Co-expression of genetic lineage tracers with cardiomyocyte markers in dissociated E15.5 heart culture. g-j. Live cell imaging of Wt1-derived cells, identified by RFP fluorescence. g. Spontaneous contraction of an RFP+ cells. Transmission line scan along the paths indicated by dashed lines. Arrowheads indicate contraction of an RFP+ cell (g1). A non-contractile cell is shown in g2. h. Fluo-4 AM calcium imaging of a cluster of beating cells, one of which was RFP+. Calcium oscillations in the RFP+ cell, measured by line scan along the white line, showed calcium sparks (black arrowhead, h1) preceding calcium waves (red arrowhead). i. Calcium oscillations in the RFP+ cell was synchronized with adjacent cardiomyocytes, indicating electrical coupling. Plots correspond to scans within colored boxes shown in h. j. Caffeine augmentation of the amplitude of calcium oscillations in an RFP+ cell. Blue staining, a-f, DAPI. Scale bars: a, 50 ⌠m, others 10 μm.
Remarkably, we found that some Wt1-derived cells differentiated into cardiomyocytes (CMs) during normal heart development, as demonstrated by co-expression of lineage tracers and CM markers cardiac troponin T (Tnnt2) and sarcomeric 〈-actinin (Actn1) (Fig. 2b-d). The Wt1-derived CMs also expressed cardiac transcription factors Gata4 and Nkx2-5 (Suppl. Fig. 4). Wt1-derived CMs were located in the myocardium of all four cardiac chambers and in the interventricular septum, constituting 7-10% of CMs in ventricles and 18% in atria (Suppl. Fig. 5). We further confirmed co-expression of CM and lineage tracers in isolated cells by immunostaining of dissociated fetal heart cultures (Fig. 2e-f). 4% of CMs in dissociated fetal heart cultures were Wt1-derived (Suppl. Fig. 5), comparable to the frequency observed in tissue sections.
To determine if Wt1-derived CMs had functional properties of CMs, we analyzed dissociated cells from Wt1GFPCre/+; Z/Red fetal hearts. A subset of red fluorescent cells exhibited spontaneous contractile activity (Fig. 2g; Suppl. Movie 1). In addition, these contracting RFP+ cells exhibited calcium oscillations with kinetics, amplitude, and frequency characteristic of CMs (Fig. 2h-j; Suppl. Movie 2). Also characteristic of CMs were calcium sparks preceding calcium waves (Fig. 2h1) and caffeine augmentation of calcium transient amplitude (Fig. 2j), consistent with calcium release from CM ryanodine receptors. Calcium transients of RFP+ cells were indistinguishable from and synchronous with adjacent RFP- cells (Fig. 2i), suggesting electrical coupling between Wt1-derived and non-Wt1-derived CMs. Consistent with this finding, the gap junction protein connexin 43 (Cx43) localized to the membrane between Wt1-derived and neighboring CMs (Suppl. Fig. 6a). A similar pattern of Cx43 expression was observed in Wt1-derived CMs in tissue sections of E15.5 hearts (Suppl. Fig. 6b), suggesting that Wt1-derived CMs are also electrically coupled to other CMs in vivo. Collectively, these data indicate that during heart development a subset of Wt1-expressing cells differentiate into CMs.
To further characterize the Wt1-expressing CM precursors, we utilized several independent methods to control the temporal and spatial window during which these precursors were labeled. To temporally regulate Cre-labeling, we knocked a cDNA encoding a Cre-modified estrogen ligand binding domain (CreERT2) into the Wt1 locus (Suppl. Fig. 7). CreERT2 fusion protein recombinase activity requires tamoxifen 17. Maternal injection of tamoxifen at E10.5 and E11.5 induced Cre activity and resulted in ®-gal expression within Wt1CreERT2; Rosa26fsLz myocardium (Fig. 3a-e), while Wt1CreERT2 did not activate Rosa26fsLz in the absence of tamoxifen (Fig. 3b and Suppl. Fig. 7). The frequency of Wt1CreERT2-labeled cells in epicardium and myocardium (Suppl. Fig. 7-8) was reduced compared to constitutive labeling by Wt1GFPCre, likely due to inefficient CreERT2 activation by tamoxifen concentrations compatible with maintenance of pregnancy. Co-staining for differentiation markers showed that β-gal+ cells differentiated into CM, EC, and SMC lineages (Fig. 3c-e). The distribution of labeled cells between these lineages was comparable between pulse and constitutive labeling approaches. We verified co-expression of CM markers and pulse-labeled lineage tracers at the single cell level by staining cardiomyocytes dissociated from E16.5 Wt1CreERT2/+; Z/Red hearts, pulsed with tamoxifen at E10.5 and E11.5 (Fig. 3f-h). We consistently observed these pulse-labeled CMs, although the frequency (0.02 ± 0.01%) was notably less than with constitutive labeling with Wt1GFPCre. Within the window of the tamoxifen pulse, cardiac Wt1 expression was confined to the epicardium (Fig. 1). Based on these data, we conclude that a subset of epicardial cells expressing Wt1 differentiate into CMs.
Fig. 3. Wt1-expressing epicardial cells differentiate into cardiomyocytes.
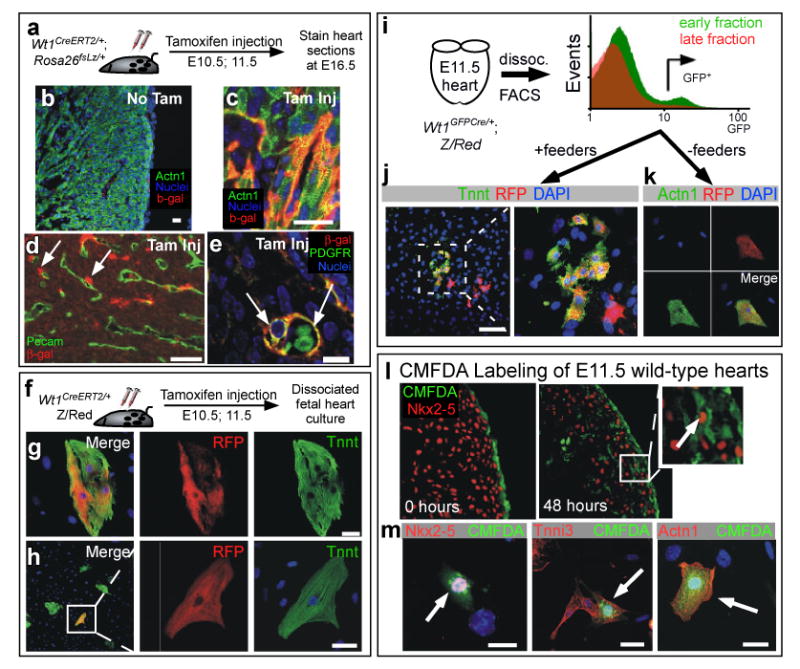
a-h. Temporal restriction of Cre labeling with Wt1CreERT2. Wt1CreERT2 did not recombine the fate mapping reporter Rosa26fsLz in the absence of tamoxifen (b). Tamoxifen treatment at E10.5 and E11.5 induced Wt1CreERT2 labeling of cardiomyocytes, as determined in both tissue sections (c) and in dissociated heart culture (g-h). Also labeled were SMCs adjacent to endothelial tubes (d-e). i-k. E11.5 Wt1GFPCre/+ heart cells actively expressing Wt1, as determined by GFP fluorescence, differentiated into cardiomyocytes. Wt1+ epicardial cells were enriched in early digestion fractions compared to late digestion fractions. GFP+ cells from early fractions were plated either with mitotically inactivated feeders (j) or without feeders (k). l-m. CMFDA dye, selectively incorporated into E11.5 epicardium, was found in CMs at 48 hours. Brief incubation of E11.5 heart explants with the dye CMFDA resulted in selective labeling of epicardium (0 hours culture). After 48 hours in explant culture, dye labeled cells were present in the myocardial wall. A subset of dye-labeled cells co-expressed the CM markers Nkx2-5, cardiac troponin I (Tnni3), and Actn1. Arrows indicate co-expression. Scale bars: 10 ⌠m.
To further delimit the location of Wt1-expressing cells that differentiate into CMs, we microdissected E11.5 Wt1GFPCre/+; Z/Red fetal hearts. Serial enzymatic digestion of intact hearts yielded epicardial cells preferentially in the early fractions, due to their location on the exterior of the heart. Early (epicardial) and late (negative control) digestion fractions were sorted for GFP fluorescence, yielding a population enriched for active GFP expression (Fig. 3i). These GFP+ cells were plated on either mitotically inactivated cardiac feeders or untreated tissue culture dishes (Fig. 3j-k). In both conditions a subset of the sorted Wt1GFPCre/+; Z/Red cells differentiated into CMs, identified by co-expression of the RFP lineage tracer and CM markers (Fig. 3j-k). These data provide further evidence that a subset of heart cells actively expressing Wt1, confined within epicardium at E10-11.5 (Fig. 1), differentiated into cardiomyocytes.
We obtained additional independent evidence that epicardial cells differentiate into cardiomyocytes by selective dye labeling of epicardium in E11.5 explanted hearts (Fig. 3l). Explanted hearts were briefly incubated in culture media containing the dye CMFDA and then place in culture media without dye. This resulted in selective labeling of epicardium, as confirmed in sections of hearts fixed immediately after CMFDA incubation (Fig. 3l, 0 hours). After two days of explant culture, labeled epicardial cells were found within myocardium, and a subset expressed the CM markers Nkx2-5 (Fig. 3l, 48 hours). Presence of dye and the CM markers Nkx2-5, Tnnt2, and Actn1 within the same cell was further demonstrated in single cells isolated by dissociating heart explants two days after labeling (Fig. 3m). Collectively, these data indicate that precursors actively expressing Wt1 within E10.5-E11.5 epicardium differentiate into CMs.
Reported cardiac precursors derive from multipotent Isl1+/Nkx2-5+ progenitors 1-4. We used Cre-based lineage tracing to ask if Wt1+ PE cells are related to these progenitors, or represent a different cardiogenic lineage. Using an Nkx2-5IRES-Cre knockin allele 18, we found that Nkx2-5-driven Cre activated Rosa26fsLz in a subset of PE cells (Fig. 4a-b), suggesting descent of Wt1+ PE cells from Nkx2-5-expressing cells.
Fig. 4. Proepicardium arises from Nkx2-5+ precursors.
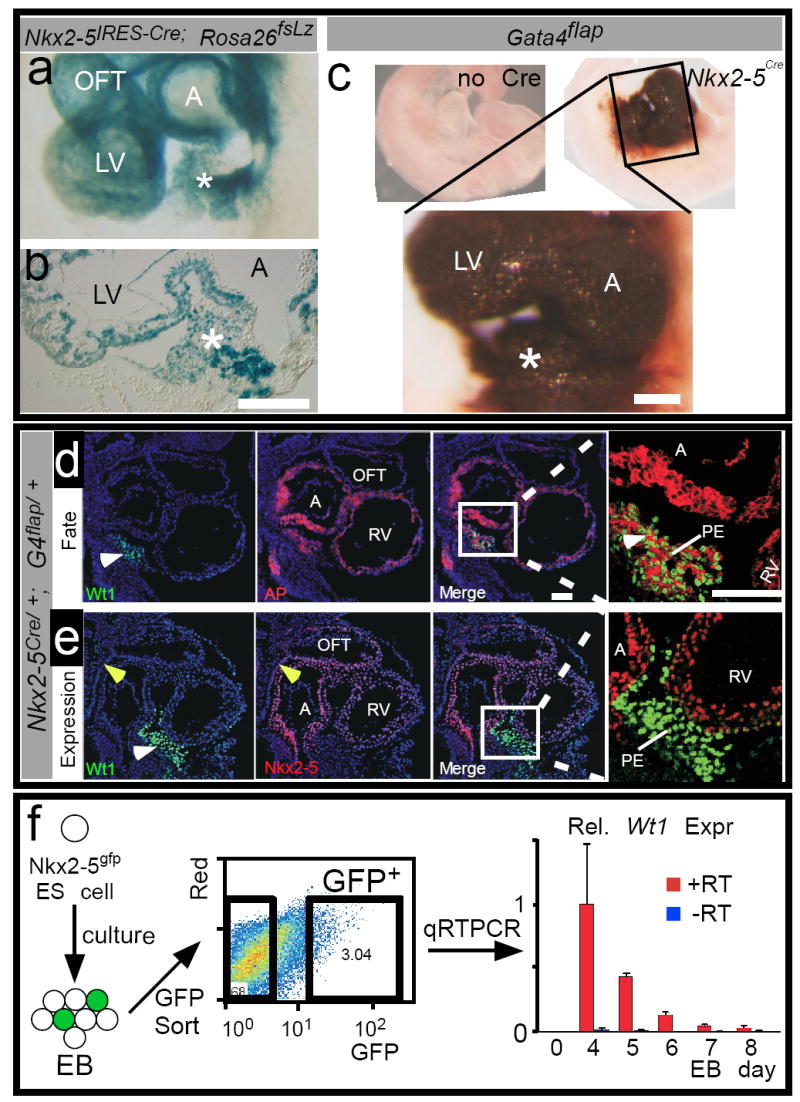
a-c. The Nkx2-5 fate map was determined in Nkx2-5IRES-Cre; Rosa26fsLz (a-b) or Nkx2-5Cre; Gata4flap (c) embryos. Nkx2-5-driven Cre activated lineage tracer expression in PE (asterisk). d. Nkx2-5 fate, marked by expression of alkaline phosphatase (AP, red membrane staining) compared to Wt1 immunohistochemistry (green nuclear staining). Wt1 was expressed in Nkx2-5-derived cells in PE (white arrowhead). e. Immunohistochemistry comparing Nkx2-5 (red) and Wt1 expression (green). Yellow arrowhead indicates anterior heart field, which did not express Wt1. f.Nkx2-5gfp ES cells were differentiated in embryoid body (EB) culture for the indicated number of days. The cells were then dissociated, and GFP+ cells were isolated by flow cytometry. Wt1 expression in GFP+ cells was measured by quantitative reverse transcription PCR. Bars indicate mean ± sd. n=3. Scale bars: 50 ⌠m. A, atrium . RV, right ventricle. LV, left ventricle. OFT, outflow tract.
We independently corroborated this result using a different Nkx2-5 knockin allele, Nkx2-5Cre 19, and a novel Cre-activated reporter, Gata4flap. Gata4 is expressed in CM, SMC, and EC compartments of the myocardium, as well as in PE (Suppl. Fig. 9) 20-22. Therefore, within this domain endogenous Gata4 regulatory elements can be used to drive expression of a Cre-dependent reporter gene, alkaline phosphatase (AP). We generated such a reporter, Gata4flap (Suppl. Fig. 10). In the absence of Cre, Gata4flap did not express AP (Suppl. Fig. 10). In the presence of well-characterized Cre transgenes, Gata4flap expressed AP in patterns consistent with the expected sites of Cre activity (cardiac troponin Cre (cTNTCre) and myosin heavy chain α-Cre (MHCαCre), myocardium; or Tie2Cre, endothelium; Suppl. Fig. 10). Quantitative analysis revealed that MHCαCre activated Gata4flap in a greater percentage of cardiomyocytes than Rosa26fsLz (Gata4flap 93 ± 3%, versus Rosa26fsLz 72 ± 6%, p < 0.005, n=4), suggesting that Gata4flap has greater sensitivity to Cre recombination than Rosa26fsLz. Therefore, we asked if Gata4flap would show a greater contribution of Nkx2-5+ cells to PE than suggested by Nkx2-5IRES-Cre; Rosa26fsLz.
Gata4flap demonstrated a robust contribution of Nkx2-5-expressing progenitors to PE (asterisk, Fig. 4c). The Nkx2-5Cre-labeled PE cells expressed Wt1 (Fig. 4d), indicating that Wt1+ cells in PE are derived from Nkx2-5-expressing precursors. Gata4flap also showed a robust contribution of Isl1-expressing precursors to the Wt1+ cells in PE (Suppl. Fig. 11b). Supporting this finding, at E8.0 Wt1 and Isl1 were expressed in adjacent regions, and a subset of cells were positive for both markers (Suppl. Fig. 12a-b).
While Wt1+ cells in PE were labeled by Nkx2-5-driven Cre, they did not actively co-express Nkx2-5 at E9.5 (Fig. 4e). In E8.0 embryos, Nkx2-5 and Wt1 were expressed in adjacent cells, but were not co-expressed (Suppl. Fig. 12c), suggesting that Nkx2-5 and Wt1 are expressed sequentially, or transiently co-expressed. To further investigate the relationship of Nkx2-5 and Wt1 expression, we studied the expression of Wt1 in Nkx2-5+ cells during embryoid body differentiation of embryonic stem (ES) cells. Using transgenic Nkx2-5gfp ES cells 4 and FACS, we isolated Nkx2-5-expressing cells at several time points during embryoid body differentiation. Wt1 was transiently upregulated in Nkx2-5+ cells during ES cell differentiation (Fig. 4f). This result was specific, as we did not detect significant Wt1 expression in parallel experiments with Mef2c-AHF-GFP ES cells 23 (data not shown). In embryos, the activity domain of the Mef2c-AHF enhancer (on in anterior heart field, off in PE) did not overlap with the Wt1 expression domain 24 (yellow arrowhead, Fig. 4e). Collectively, these data suggest that Nkx2-5 and Wt1 are sequentially expressed, or transiently co-expressed, in a subset of PE precursors.
We have shown that Wt1+ PE/epicardial cells contribute to the CM lineage during normal heart development (Suppl. Fig. 1). Wt1+ cells located on the heart at E10.5-E11.5 differentiate into functional CMs. Although differentiation of PE cells into CMs was previously noted in vitro 25, prior fate-mapping studies of PE cells, using retroviral labels in chick or transgene labels in mice, did not describe PE contribution to the CM lineage in vivo 8-10,12,13. This may be attributable to differences in methodology, species, or domains of transgene activity. Consistent with the capacity of Wt1-expressing cells to differentiate into CMs, Wt1+ PE/epicardial cells are derived from progenitors that express Nkx2-5 and Isl1, suggesting that they share a common developmental origin with previously described multipotent cardiogenic progenitors 1,2. These experiments identify a previously unrecognized CM progenitor population in the developing heart that may be of use for cardiac regeneration or repair.
Methods Summary
Gene targeting and mouse lines are described in the Full Methods. Fetal hearts were dissociated by serial digestion with collagenase and trypsin. For calcium imaging, dissociated fetal heart cultures were loaded with Fluo-4 AM and imaged with an Olympus FV1000 confocal microscope. Cardiac feeders were prepared and mitotically inactivated as described 26. Nkx2-5gfp and Mef2c-AHF-GFP ES cells were differentiated in embryoid body culture and sorted for GFP fluorescence as described previously 4,23. Immunohistochemistry was performed according to standard methods, using primary and secondary antibodies listed in Supplementary Table 1.
Supplementary Material
Full Methods and any associated references are available in the online version of the paper at www.nature.com/nature.
Supplementary information is linked to the online version of the paper at www.nature.com/nature.
Acknowledgments
This work was funded by the National Heart, Lung, and Blood Institute of the National Institutes of Health, United States, and by a charitable donation from Edward P. Marram and Karen K. Carpenter. The authors thank the Schwartz, Harvey, Schneider, Yanagisawa, Evans, Soriano, Orkin, and Nagy labs for contributing mouse strains used in this study.
Footnotes
Author information. Reprints and permissions information is available at www.nature.com/reprints. The authors declare no competing financial interests.
References
- 1.Martin-Puig S, Wang Z, Chien KR. Lives of a heart cell: tracing the origins of cardiac progenitors. Cell Stem Cell. 2008;2:320–331. doi: 10.1016/j.stem.2008.03.010. [DOI] [PubMed] [Google Scholar]
- 2.Laugwitz KL, Moretti A, Caron L, Nakano A, Chien KR. Islet1 cardiovascular progenitors: a single source for heart lineages? Development. 2008;135:193–205. doi: 10.1242/dev.001883. [DOI] [PubMed] [Google Scholar]
- 3.Moretti A, et al. Multipotent embryonic isl1+ progenitor cells lead to cardiac, smooth muscle, and endothelial cell diversification. Cell. 2006;127:1151–1165. doi: 10.1016/j.cell.2006.10.029. [DOI] [PubMed] [Google Scholar]
- 4.Wu SM, et al. Developmental origin of a bipotential myocardial and smooth muscle cell precursor in the mammalian heart. Cell. 2006;127:1137–1150. doi: 10.1016/j.cell.2006.10.028. [DOI] [PubMed] [Google Scholar]
- 5.Manner J, Perez-Pomares JM, Macias D, Munoz-Chapuli R. The origin, formation and developmental significance of the epicardium: a review. Cells Tissues Organs. 2001;169:89–103. doi: 10.1159/000047867. [DOI] [PubMed] [Google Scholar]
- 6.Wessels A, Perez-Pomares JM. The epicardium and epicardially derived cells (EPDCs) as cardiac stem cells. Anat Rec A Discov Mol Cell Evol Biol. 2004;276:43–57. doi: 10.1002/ar.a.10129. [DOI] [PubMed] [Google Scholar]
- 7.Smart N, et al. Thymosin beta4 induces adult epicardial progenitor mobilization and neovascularization. Nature. 2007;445:177–182. doi: 10.1038/nature05383. [DOI] [PubMed] [Google Scholar]
- 8.Wilm B, Ipenberg A, Hastie ND, Burch JB, Bader DM. The serosal mesothelium is a major source of smooth muscle cells of the gut vasculature. Development. 2005;132:5317–5328. doi: 10.1242/dev.02141. [DOI] [PubMed] [Google Scholar]
- 9.Merki E, et al. Epicardial retinoid X receptor alpha is required for myocardial growth and coronary artery formation. Proc Natl Acad Sci U S A. 2005;102:18455–18460. doi: 10.1073/pnas.0504343102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gittenberger-de Groot AC, Vrancken Peeters MP, Mentink MM, Gourdie RG, Poelmann RE. Epicardium-derived cells contribute a novel population to the myocardial wall and the atrioventricular cushions. Circ Res. 1998;82:1043–1052. doi: 10.1161/01.res.82.10.1043. [DOI] [PubMed] [Google Scholar]
- 11.Manner J. Does the subepicardial mesenchyme contribute myocardioblasts to the myocardium of the chick embryo heart? A quail-chick chimera study tracing the fate of the epicardial primordium. Anat Rec. 1999;255:212–226. doi: 10.1002/(sici)1097-0185(19990601)255:2<212::aid-ar11>3.3.co;2-o. [DOI] [PubMed] [Google Scholar]
- 12.Dettman RW, Denetclaw WJ, Ordahl CP, Bristow J. Common epicardial origin of coronary vascular smooth muscle, perivascular fibroblasts, and intermyocardial fibroblasts in the avian heart. Dev Biol. 1998;193:169–181. doi: 10.1006/dbio.1997.8801. [DOI] [PubMed] [Google Scholar]
- 13.Mikawa T, Gourdie RG. Pericardial mesoderm generates a population of coronary smooth muscle cells migrating into the heart along with ingrowth of the epicardial organ. Dev Biol. 1996;174:221–232. doi: 10.1006/dbio.1996.0068. [DOI] [PubMed] [Google Scholar]
- 14.Le Y, Miller JL, Sauer B. GFPcre fusion vectors with enhanced expression. Anal Biochem. 1999;270:334–336. doi: 10.1006/abio.1999.4110. [DOI] [PubMed] [Google Scholar]
- 15.Mao X, Fujiwara Y, Orkin SH. Improved reporter strain for monitoring Cre recombinase-mediated DNA excisions in mice. Proc Natl Acad Sci U S A. 1999;96:5037–5042. doi: 10.1073/pnas.96.9.5037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vintersten K, et al. Mouse in red: red fluorescent protein expression in mouse ES cells, embryos, and adult animals. Genesis. 2004;40:241–246. doi: 10.1002/gene.20095. [DOI] [PubMed] [Google Scholar]
- 17.Feil R, Wagner J, Metzger D, Chambon P. Regulation of Cre recombinase activity by mutated estrogen receptor ligand-binding domains. Biochem Biophys Res Commun. 1997;237:752–757. doi: 10.1006/bbrc.1997.7124. [DOI] [PubMed] [Google Scholar]
- 18.Stanley EG, et al. Efficient Cre-mediated deletion in cardiac progenitor cells conferred by a 3’UTR-ires-Cre allele of the homeobox gene Nkx2-5. Int J Dev Biol. 2002;46:431–439. [PubMed] [Google Scholar]
- 19.Moses KA, DeMayo F, Braun RM, Reecy JL, Schwartz RJ. Embryonic expression of an Nkx2-5/Cre gene using ROSA26 reporter mice. Genesis. 2001;31:176–180. doi: 10.1002/gene.10022. [DOI] [PubMed] [Google Scholar]
- 20.Rivera-Feliciano J, et al. Development of heart valves requires Gata4 expression in endothelial-derived cells. Development. 2006;133:3607–3618. doi: 10.1242/dev.02519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heikinheimo M, Scandrett JM, Wilson DB. Localization of transcription factor GATA-4 to regions of the mouse embryo involved in cardiac development. Dev Biol. 1994;164:361–373. doi: 10.1006/dbio.1994.1206. [DOI] [PubMed] [Google Scholar]
- 22.Watt AJ, Battle MA, Li J, Duncan SA. GATA4 is essential for formation of the proepicardium and regulates cardiogenesis. Proc Natl Acad Sci U S A. 2004;101:12573–12578. doi: 10.1073/pnas.0400752101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qyang Y, et al. The Renewal and Differentiation of Isl1+ Cardiovascular Progenitors Are Controlled by a Wnt/B-Catenin Pathway. Cell Stem Cell. 2007;1:165–179. doi: 10.1016/j.stem.2007.05.018. [DOI] [PubMed] [Google Scholar]
- 24.Verzi MP, McCulley DJ, De Val S, Dodou E, Black BL. The right ventricle, outflow tract, and ventricular septum comprise a restricted expression domain within the secondary/anterior heart field. Dev Biol. 2005;287:134–145. doi: 10.1016/j.ydbio.2005.08.041. [DOI] [PubMed] [Google Scholar]
- 25.Kruithof BP, et al. BMP and FGF regulate the differentiation of multipotential pericardial mesoderm into the myocardial or epicardial lineage. Dev Biol. 2006;295:507–522. doi: 10.1016/j.ydbio.2006.03.033. [DOI] [PubMed] [Google Scholar]
- 26.Laugwitz KL, et al. Postnatal isl1+ cardioblasts enter fully differentiated cardiomyocyte lineages. Nature. 2005;433:647–653. doi: 10.1038/nature03215. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Full Methods and any associated references are available in the online version of the paper at www.nature.com/nature.
Supplementary information is linked to the online version of the paper at www.nature.com/nature.


