Abstract
The vertebrate lens provides an excellent model to study the mechanisms that regulate terminal differentiation. Although fibroblast growth factors (FGFs) are thought to be important for lens cell differentiation, it is unclear which FGF receptors mediate these processes during different stages of lens development. Deletion of three FGF receptors (Fgfr1-3) early in lens development demonstrated that expression of only a single allele of Fgfr2 or Fgfr3 was sufficient for grossly normal lens development, while mice possessing only a single Fgfr1 allele developed cataracts and microphthalmia. Profound defects were observed in lenses lacking all three Fgfrs. These included lack of fiber cell elongation, abnormal proliferation in prospective lens fiber cells, reduced expression of the cell cycle inhibitors p27kip1 and p57kip2, increased apoptosis and aberrant or reduced expression of Prox1, Pax6, c-Maf, E-cadherin and α-, β- and γ-crystallins. Therefore, while signaling by FGF receptors is essential for lens fiber differentiation, different FGF receptors function redundantly.
Keywords: apoptosis, cell cycle, FGF receptor, lens development, lens fiber differentiation, redundancy, conditional knockout
INTRODUCTION
Normal development of an organism requires elaborate control over proliferation, cell cycle exit and differentiation. The ocular lens is an excellent tissue in which to study these basic processes of development. Murine lens morphogenesis begins with the formation of a lens placode in the surface ectoderm in response to inductive signals from several tissues including the underlying optic vesicle (reviewed in Fisher and Grainger, 2004). The lens placode subsequently invaginates and separates from surface ectoderm giving rise to the lens vesicle, composed of a single layer of proliferating epithelial cells. Cells located in the posterior half of the lens vesicle exit the cell cycle, rapidly elongate and differentiate into primary fiber cells. The epithelial cells maintain the capacity to proliferate, and ultimately fuel the lifelong growth of the lens. Epithelial cells near the lens equator stop dividing, elongate and differentiate into secondary fiber cells. This process is characterized by a dramatic increase in the expression of β- and γ-crystallins, which are found exclusively or preferentially in fiber cells, and an abrupt decrease in lens epithelial cell-specific gene expression.
Over the past decade, significant progress has been made in identifying the signals that control lens induction. Bone Morphogenetic Proteins (BMPs) are essential for lens induction and converge with FGFs to regulate the expression of Pax6 (Faber et al., 2001; Furuta and Hogan, 1998; Wawersik et al., 1999), a critical transcription factor for lens formation (Ashery-Padan et al., 2000). A genetic cascade including Pax6, Mab2111 and FoxE3 is then initiated. These factors are required for the proliferation and maintenance of the lens placode and lens epithelial cells (Ashery-Padan et al., 2000; Blixt et al., 2000; Dimanlig et al., 2001; Yamada et al., 2003). Heparan sulfate proteoglycans are essential for FGF signaling and deletion of Ndst1, encoding an enzyme involved in heparan sulfate synthesis, prevents the formation of the lens and retina by interfering with FGF receptor (Fgfr) signaling (Pan et al., 2006). Furthermore, mutation of two tyrosines that are essential for the docking of Shp2 to the FGF receptor adaptor protein Frs2α, impairs the formation of the lens and retina (Gotoh et al., 2004).
In contrast, little is known about the signals that control lens fiber cell differentiation. Accumulating evidence suggests that FGF signaling plays an important role in this process. Multiple FGF ligands are expressed in ocular tissues and promote fiber differentiation in vitro and in vivo (reviewed in Robinson, 2006). The developing lens expresses all four members of the Fgfr gene family (Fgfr1–4) in distinctive spatio-temporal patterns (de Iongh et al., 1997; de Iongh et al., 1996; Kurose et al., 2005). Transgenic mice expressing secreted dimers or truncated versions of Fgfrs showed defects in lens growth and differentiation, suggesting the importance of Fgfr signaling during lens development (Chow et al., 1995; Govindarajan and Overbeek, 2001; Robinson et al., 1995a; Stolen and Griep, 2000). We showed by chimera analysis and tissue-specific knockout that Fgfr1 is dispensable for lens development (Garcia et al., 2005; Zhao et al., 2006) and retroviral transduction of chicken embryonic lens epithelial cells with a dominant-negative Fgfr1 gene did not affect fiber cell differentiation (Huang et al., 2003). Defective placental development leads to embryonic lethality in Fgfr2 null embryos before the onset of eye development (Arman et al., 1998; Xu et al., 1998), but Fgfr2-deficient embryos where the placental defect is rescued, survive to birth and undergo lens fiber cell differentiation (Li et al., 2001). Lens fiber cells also form after targeted inactivation of either or both of the splice variants of Fgfr2 (Fgfr2IIIb or Fgfr2IIIc) (Eswarakumar et al., 2002; Garcia et al., 2005; Revest et al., 2001). Mice deficient in Fgfr3 and Fgfr4 do not show obvious defects in lens development (Deng et al., 1996). Therefore, no single Fgfr is required for lens formation or fiber cell differentiation.
Given the complexity conferred by the existence of genes encoding 22 Fgf ligands in the mouse genome, many of which are expressed in the eye (reviewed in Robinson, 2006), we are deleting all four of the Fgfrs using germ line and conditional gene targeting. In the present study, MLR10 transgenic mice, which express Cre recombinase in lens fiber and epithelial cells beginning at the lens pit stage (Zhao et al., 2004), were used to inactivate Fgfr1 and Fgfr2 in a lens-specific manner. Mice lacking Fgfr3 are viable and fertile. Lens development was not compromised in mice deficient in any two Fgfrs. However, mice lacking all three of these Fgfrs in the lens displayed profound defects, involving cell cycle exit, cell survival and fiber cell differentiation. This demonstrates that signaling by these three Fgfrs is essential for lens fiber cell differentiation, but that different Fgfrs play redundant roles in this process.
MATERIALS AND METHODS
Mice
MLR10 transgenic mice expressing Cre in the lens from the lens pit stage were described previously (Zhao et al., 2004). Mice with a conditional allele of Fgfr1 were a generous gift of Janet Rossant and Juha Partanen (Trokovic et al., 2003b). The conditional mutation in Fgfr2 has been described (Yu et al., 2003). Fgfr3 null mice were described previously (Colvin et al., 1996; Deng et al., 1996) and were the gift of Michael Weinstein and Chu-Xia Deng. All animal procedures were approved by the IACUCs of either Columbus Children’s Research Institute or Miami University.
In situ hybridization and Immunohistochemistry
Embryos, neonatal and adult eyes were fixed in 4% paraformaldehyde overnight at 4°C, processed and embedded in paraffin and sectioned at 5 μm. Radioactive in situ hybridization was carried out according to previously described methods (Robinson et al., 1995b). Non-radioactive in situ hybridization was carried out using a digoxigenin-probe labeling system according to manufacturer instructions (Roche Diagnostics, Indianapolis, IN). Riboprobe vectors for Pax6 (nucleotides 709–962 of GenBank accession no. NM_013267), Six3 (nucleotides 951-1616 of GenBank accession no. NM_011381), Prox1 (nucleotides 419-2999 of GenBank accession no. NM_008937), p57kip2 (nucleotides 142-654 of GenBank accession no. NM_009876), c-Maf (nucleotides 950-1221 of GenBank accession no. NM_001025577, and Sox1(nucleotides 1446-2376 of GenBank accession no. X94162) were kindly provided by Dr. Paul Overbeek (Baylor College of Medicine, Houston, TX). A riboprobe vector for FoxE3 was the generous gift of Dr. Milan Jamrich (Baylor College of Medicine, Houston, TX).
For immunohistochemistry, tissue sections were incubated 0.3% H2O2 for 15 minutes at room temperature followed by blocking with Power Block (BioGenex, San Ramon, CA) for 20 minutes at room temperature. The slides were then incubated with primary antibody at 4°C overnight. After brief washes, the slides were incubated with biotinylated secondary antibody (ScyTek Laboratories, Inc., Logan, UT) at room temperature for 30 minutes, followed by UltraTek HRP (ScyTek Laboratories, Inc) and visualized by diaminobenzidine (Vector Laboratory, Burlingame, CA) according to manufacturer’s instructions. For immunofluorescence, tissue sections were incubated with 0.5% bovine serum albumin and 1% Triton X-100 for 30 minutes at room temperature, followed by incubation with primary antibody at 4°C overnight. After brief washes, the slides were incubated with Cy-3 labeled secondary antibodies (Jackson Immunoresearch Laboratories, West Grove, PA) or Alexafluor 546-labeled secondary antibodies (Invitrogen, Carlsbad, CA) for 1 hour at room temperature. Then the sections were counterstained with DAPI (Vector Laboratories). Antibodies to α-, β-, and γcrystallins were the gift of Dr. Samuel Zigler (Johns Hopkins University, Baltimore, MD). Polyclonal antibodies for Pax6 and Prox1 were purchased from Covance Research Products, Inc., Berkeley, CA. Polyclonal antibodies for c-Maf, p57Kip2, and cyclin D2 were purchased from Santa Cruz Biotechnology, Inc., Santa Cruz, CA. Polyclonal antibodies for phospho (p-44/42) Erk (#9101) were purchased from Cell Signaling Technology, Danvers, MA. Other antibodies were: cyclin D1 (Biocare Medical, Walnut Creek, CA), p27Kip1 (Beckman Coulter, Inc., Miami, FL), PCNA (Zymed Laboratories, Inc., South San Francisco, CA) and E-cadherin (DAKO, Carpinteria, CA).
Prior to detection of phospho (p-44/42) Erk, antigen retrieval was performed by treatment with 0.01 M sodium citrate (pH 6.0) at 100 °C in a rice steamer for 30 minutes, followed by rinsing with distilled water after cooling to ambient temperature.
BrdU and TUNEL analysis
Bromodeoxyuridine (BrdU) (0.1mg/gram body weight) was administered IP into pregnant females 2 hours prior to embryo isolation. S-phase cells were visualized using an anti-BrdU monoclonal antibody (DAKO). The TUNEL assay was performed using FragEL™ DNA Fragmentation Detection Kit (Oncogene Research Products, San Diego, CA) according to manufacturer’s instructions. Quantification of cell proliferation and apoptosis was performed by determining the fraction of labeled nuclei over the total number of nuclei present on a given section. A minimum of 3 different embryos were analyzed per genotype/time point. For this analysis, MLR10-mutant embryos were compared to littermates lacking the MLR10 Cre transgene, but homozygous for conditional mutations in Fgfr1 and Fgfr2 and a null mutation in Fgfr3. MLR10-mutant and control lens BrdU incorporation rates were analyzed by comparing the total MLR10-mutant lens BrdU incorporation rate (S-phase index) with the control lens epithelial S-phase index. Mean values of S-phase index or TUNEL positive percentage data were arcsine/square root transformed before analysis by two tailed Student’s t test. Significance was accepted at P ≤0.050.
RNAse protection
RNAse protection assays were performed using the RPAIII kit (Ambion, Austin, TX) according to manufacturer’s instructions. Probes were generated by in vitro transcription of a 415 bp fragment of Fgfr1 cDNA (nucleotides 1990-2404 of GenBank accession number NM010206), a 370 bp fragment of Fgfr2 cDNA (nucleotides 1701-2070 of GenBank accession number X55441) and a 264 bp fragment of Fgfr3 cDNA (nucleotides 596-859 of GenBank accession number NM008010). Total RNA loading was assessed using a 126 bp riboprobe derived from mouse Hprt cDNA (nucleotides 116-241 of GenBank accession number NM013556).
Microscopy and photography
Embryos, neonatal and adult mice were photographed using a Nikon CoolPix-5700 digital camera (Nikon Instruments, Melville, NY). Lenses were photographed using SMZ1000 zoom stereomicroscope (Nikon Instruments) equipped SPOT digital camera system (Diagnostic Instruments, Sterling Heights, MI). Tissue sections were photographed using a Nikon Eclipse E800 microscope (Nikon Instruments) with a SPOT digital camera system.
RESULTS
Lens fiber cell differentiation is not affected in mice with combined deletion of two FGF receptor genes
Though none of the Fgfrs expressed in the lens is individually essential for lens development, this does not mean that Fgfr signaling is dispensable for normal lens formation. To address possible functional redundancy among different Fgfrs, we made double deletions of Fgfrs1-3 in the lens. Due to early embryonic lethality associated with null mutations of Fgfr1 and Fgfr2, we deleted loxP-flanked (floxed) alleles of these genes using transgenic mice (MLR10) that express Cre recombinase from the lens pit stage (Zhao et al., 2004). Previous studies showed that both conditional alleles of these genes can be efficiently inactivated by Cre-mediated recombination (Hebert et al., 2003; Pirvola et al., 2002; Trokovic et al., 2003a; Trokovic et al., 2003b; Yu et al., 2003).
All Fgfr double mutant mice were viable and developmentally normal, except that mice homozygous for the Fgfr3-null allele exhibited skeletal phenotypes typical of Fgfr3 deficiency (Colvin et al., 1996; Deng et al., 1996). Lens morphology examined at birth (P0), 7 days (P7) and 1 month after birth (Adult) in different Fgfr double mutant strains was similar to age-matched control lenses (Supplemental Fig. 1A–L).
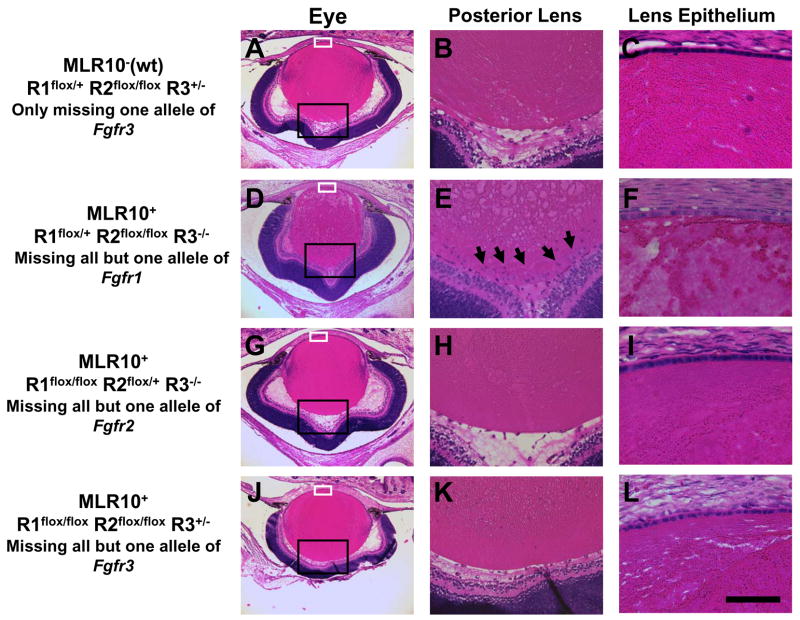
To further define the quantitative requirement for Fgfr signaling, we produced mice lacking five of the six Fgfr1-3 alleles in the lens. Lenses retaining only a single wild type allele of either Fgfr2 or Fgfr3 appeared normal and retained clear lenses through at least six months of age (Fig 1G–L). In contrast, lenses possessing only one wild type allele of Fgfr1 were microphthalmic with cataractous lenses. Although lens fibers clearly formed, gross lens abnormalities in these mice were evident at birth (Fig. 1D–F) and included a sparsely populated lens epithelium, evidence of lens fiber degeneration and the accumulation of nucleated cells at the lens posterior pole. Therefore, morphologically normal lens development required at least one wild type allele of either Fgfr2 or Fgfr3, suggesting that, of the three Fgfrs examined, Fgfr1 plays the least important role in lens development.
Figure 1.
Lens development when five of the six Fgfr1-3 alleles are missing. Sections from newborn eyes from animals missing just one allele of Fgfr3 (A–C) are compared with those missing all FGF receptors, except one allele of Fgfr1 (D–F), all except one allele of Fgfr2 (G–I) or all except one allele of Fgfr3 (J–L). Regions boxed in black and white in the first column (A, D, G, J) are shown at higher magnification in the second (B, E, H, K) and third (C, F, I, L) columns, respectively. Notice that there is an accumulation of nucleated cells at the posterior region of the lens containing only one allele of Fgfr1 (arrows, E). This genotype is also typified by fiber cell degeneration and a lower than normal density of lens epithelial cells (F). MLR10-designates mice in which the MLR10 transgene was not present. R1, R2 and R3 represent Fgfr1, Fgfr2 and Fgfr3 respectively. The conditional, null and wild type (wt) alleles of these genes are represented by flox, −, and + respectively. The scale bar in (L) represents 50 μm in (C, F, I, L), 125 μm in (B, E, H, K) and 500 μm in (A, D, G, J).
FGF receptor signaling is essential for lens fiber cell elongation
To determine if fiber cell differentiation required signaling from Fgfr1-3, we produced mice lacking all six alleles of these receptors in the lens. The expected Mendelian ratio of triple Fgfr mutant mice (MLR10/Fgfr1flox/flox/Fgfr2flox/flox/Fgfr3−/−), designated here MLR10-mutant were produced by interbreeding single and double mutant lines. The control littermates used for analysis were non-transgenic mice homozygous for Fgfr1flox and Fgfr2flox alleles and wild type for Fgfr3 (Fgfr1flox/flox/Fgfr2flox/flox/Fgfr3+/+), unless otherwise indicated.
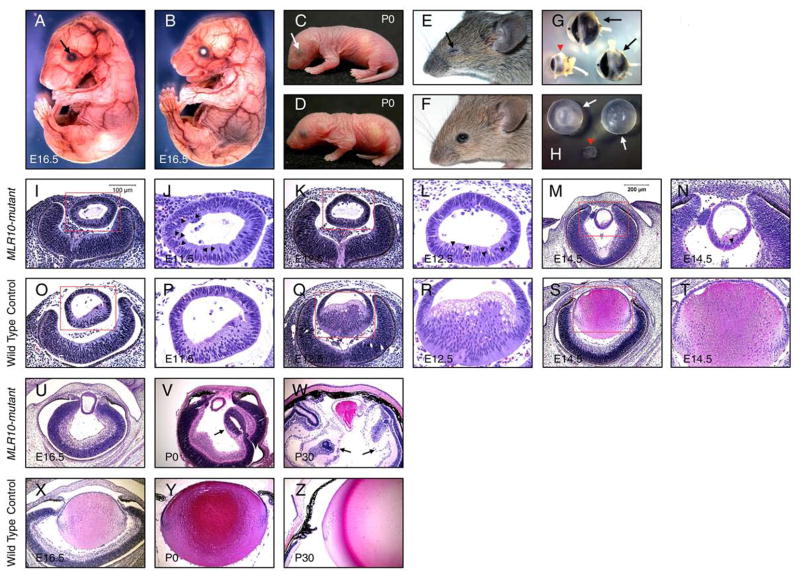
Visual inspection of MLR10-mutant embryonic day 16.5 (E16.5) embryos revealed severe microphthalmia, which became easily identifiable at birth and most pronounced after eye opening (Fig. 2A–F). In adults, MLR10-mutant eyelids were closed and the anterior chamber of the eye was missing (Fig. 2E, G). Lenses dissected from newborn MLR10-mutant eyes were significantly smaller than those of control littermates (Fig. 2H). MLR10-mutant mice exhibited profound defects in lens development. Although a lens vesicle formed by E11.5, the initial elongation of primary lens fiber cells that is typically seen at this stage did not occur (Fig. 2I, J, O, P). The arrest of fiber cell elongation became more obvious at E12.5, when control embryos displayed elongated lens fiber cells that began filling the lumen of the lens vesicle (Fig. 2K, L, Q, R). E14.5, E16.5, and P0 MLR10-mutant lenses remained as hollow structures without anterior-posterior polarity or secondary lens fiber cell differentiation (Fig. 2M, N, S, T, U, V, X, Y). Numerous pyknotic nuclei were detected in mutant lenses as early as E11.5, concurrent with the arrest of fiber elongation (Fig. 2J, L, N, P, R, T). As development progressed, the MLR10-mutant lenses exhibited severe growth retardation. Consequently, only a rudimentary lens was present in adult MLR10-mutants (Fig. 2W, Z). Other ocular defects included failure to form an anterior chamber, vascularization of the corneal stroma, rudimentary ciliary body and iris development, and retinal folding (Fig. 2V, W, Y, Z), features typical of eyes with severe defects in early lens development.
Figure 2.
Defective lens fiber elongation in MLR10-mutant (MLR10/Fgfr1flox/flox Fgfr2flox/flox Fgfr3 −/−) mice. A–F: MLR10-mutant mice (A, C, E) were compared with control mice (B, D, F) at E16.5 (A, B), P0 (C, D) and P30 (E, F). Triple Fgfr-deficient mice were characterized by severe microphthalmia (arrows A, C, E); G: Eyes of MLR10-mutant (arrowhead, G) were placed together with control eyes (arrows, G). The anterior chamber (*) is absent in the mutant eye; H: Lenses from MLR10-mutant mice (arrowhead, H) were much smaller than control lenses (arrows, H); I-Z: Histological analysis of MLR10-mutant (I–N, U–W) and control (O–T, X–Z) eyes. Developmental stages studied include E11.5 (I, J, O, P), E12.5 (K, L, Q, R), E14.5 (M, N, S, T) E16.5 (U, X), P0 (V, Y) and P30 (W, Z). The boxed regions in I, O, K, Q, M, S are shown at higher magnification in J, P, L, R, N, T respectively and pyknotic nuclei are indicated by arrowheads (J, L, N). Arrows in V and W indicate abnormal folds of neural retina in the mutant eyes.
Prospective fiber cells in MLR10-mutants fail to withdraw from the cell cycle
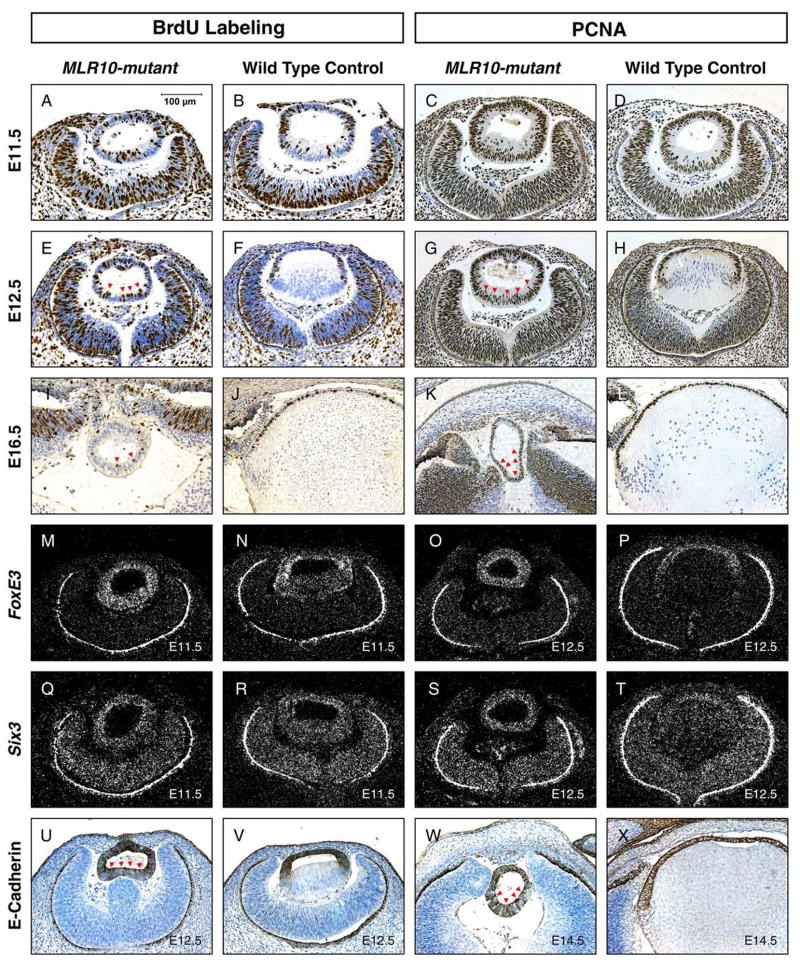
Lens fiber cell differentiation involves the conversion of actively-proliferating epithelial cells to post-mitotic, terminally-differentiated fiber cells. We examined cell proliferation in MLR10-mutant lenses by measuring the bromodeoxyuridine (BrdU) labeling index and by detecting immunostaining for proliferating cell nuclear antigen (PCNA). At E11.5, just before the onset of lens fiber differentiation, cells incorporating BrdU were detected in the posterior of the lens vesicle in MLR10-mutant and control embryos (Fig. 3A, B). At E12.5 and E16.5, BrdU-positive cells were restricted to the anterior epithelium in the control lenses, while in MLR10-mutant lenses, BrdU-positive cells were detected in the prospective fiber cells at the posterior of the lens (Fig. 3E, F, I, J). The BrdU labeling index did not differ in anterior and posterior halves of MLR10-mutant lenses at E12.5 (P = 0.064). However, by E14.5, there was more proliferation in the anterior than the posterior hemisphere of MLR10-mutant lenses (P = 0.044). The BrdU labeling index for the whole MLR10-mutant lens (27.9%) did not differ significantly from the rate of proliferation in the lens epithelial cells of control lenses (28.4%) at E12.5 (P = 0.950). By E14.5, the BrdU labeling index of control lens epithelial cells (34.1%) was significantly higher than that of total MLR10-mutant lens cells (14.5%) (P = 0.004). Similarly, at E11.5, PCNA was detected in all nuclei of control and MLR10-mutant lens vesicles (Fig. 3C, D). At E12.5 and E16.5, PCNA expression was observed in the control lens epithelium and newly formed lens fiber cells, but not in more mature fiber cells. At these stages, MLR10-mutant lenses exhibited PCNA staining in nuclei of cells in all areas of the lens (Fig. 3G, H).
Figure 3.
Defects in cell cycle exit and persistent expression of lens epithelial markers in MLR10-mutant lenses. A-L: BrdU-incorporation (A, B, E, F, I, J) and PCNA expression (C, D, G, H, K, L) analyses were performed on mutant (A, E, I, C, G, K) and control (B, F, J, D, H, L) embryos at E11.5 (A–D), E12.5 (E–H) and E16.5 (I–L). Brown nuclear staining indicated cells that were in the S-phase of cell cycle (1st and 2nd column, A–L), during BrdU labeling, or that express PCNA (3rd and 4th column, A-L), typical of proliferating cells. Red arrowheads mark BrdU-incorporating or PCNA-expressing mutant cells in the posterior portion of lenses. M–T: In-situ hybridization analyses of lens epithelial markers FoxE3 (M–P), Six3 (Q–T) at E11.5 (M, N, Q, R) and E12.5 (O, P, S, T) were performed on mutant (M, Q, O, S) and control (N, R, P, T) lenses. Bright-appearing silver grains in the dark field photos indicate expression of these genes. The retinal pigmented epithelium (RPE) appears as a bright line surrounding the optic cup in the darkfield illumination, due to light scattering by pigment granules. U–X: Both mutant (U, W) and control (V, X) lenses at E12.5 day (U, V) and E14.5 day (W, X) were analyzed for the expression of the lens epithelial marker E-cadherin by immunohistochemistry. Dark brown staining indicates E-cadherin expression in junctions between lens epithelial cells. Red arrowheads mark cells expressing E-cadherin in the posterior of the MLR10-mutant lens.
To test if the cells of MLR10-mutant lenses retained the characteristics of lens epithelial cells, we investigated the expression of FoxE3, Six3 and E-cadherin, genes normally expressed in the lens epithelium but not in lens fibers. In control E11.5 lenses, FoxE3 transcripts were abundant in the anterior lens cells; with lower expression in the prospective fiber cells at the posterior of the lens vesicle. FoxE3 appeared to be uniformly expressed in the MLR10-mutant lenses at this stage (Fig. 3M, N). At E12.5, FoxE3 mRNA was not detected in the primary fiber cells of wild type lenses, but was detected throughout the MLR10-mutant lenses at a level similar to that observed in control lens epithelial cells (Fig. 3O, P). Likewise, Six3 mRNA was detected in lens epithelial cells and decreased in elongating primary fiber cells of control lenses by E12.5. In contrast, in MLR10-mutant eyes, Six3 mRNA was detected at similar levels in all lens cells (Fig. 3Q–T). E-cadherin protein was confined to the lens epithelial cells in control lenses at E12.5 and E14.5, while no such distinction was seen between epithelial and fiber cells in MLR10-mutant lenses (Fig. 3U-X). These analyses suggested that the MLR10-mutant lens was composed entirely of cells with the characteristics of lens epithelial cells.
FGF receptor signaling is required for the proper expression of p27kip1, p57kip2 and Prox1 during lens fiber cell differentiation
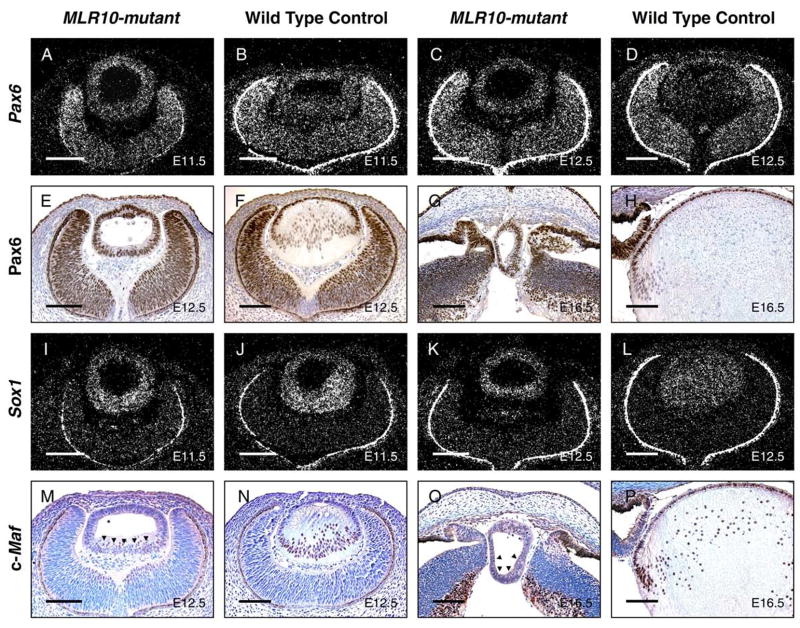
To gain more insight into the abnormal cell cycle regulation exhibited by MLR10-mutant lenses, we examined the expression of regulators of the G1/S transition, cyclins D1, D2, the cyclin dependent kinase inhibitors (CKIs) p27kip1 and p57kip2 and the transcription factor Prox1. In control lenses, cyclins D1 and D2 were expressed at high levels in the anterior epithelium and equatorial lens fiber cells, but at reduced levels in more mature lens fiber cells (Fig. 4B, D, F), and by E16.5, cyclin D2 protein is restricted to the control lens epithelium (Fig. 4H). However, in MLR10-mutant lenses, cyclin D1 expression appeared uniformly distributed at both E12.5 and E16.5 (Fig. 4A, C). At E12.5, cyclin D2 was expressed in the posterior of the mutant lens at a level close to that observed in the newly-differentiating equatorial fiber cells in control lenses (Fig. 4E, F), and cyclin D2 was only weakly expressed throughout the mutant lens at E16.5 (Fig. 4G, H).
Figure 4.
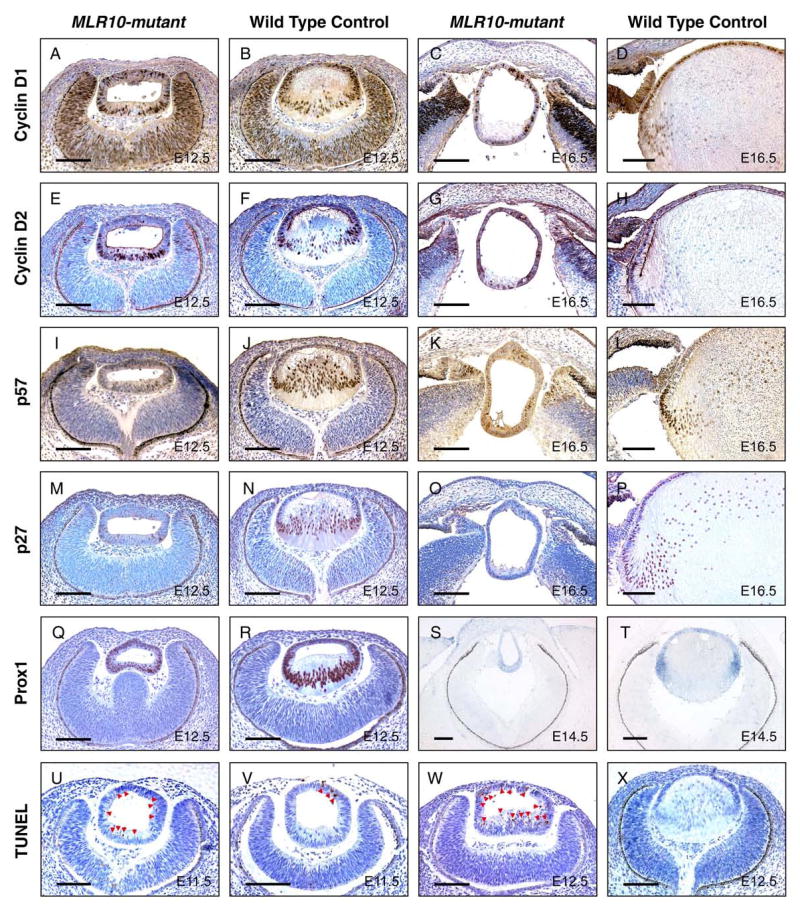
Analysis of the expression of cell cycle regulators and cell death in MLR10-mutant lenses. Cyclin D1 (A–D), cyclin D2 (E–H), p57kip2 (I-L), p27kip1 (M–P) and Prox1 (Q–R) levels were analyzed by immunohistochemistry in the lenses of MLR10-mutant (A, E, I, M, C, G, K, O, Q) and control (B, F, J, N, D, H, L, P, R) mice. Prox1 mRNA expression was also examined by in situ hybridization in MLR10-mutant and control lenses (S and T, respectively). TUNEL assays were conducted on mutant (U, W) and control (V, X) embryonic lenses. Developmental stages studied included E11.5 (U,V) E12.5 (A, B, E, F, I, J, M, N, Q, R, W, X), E14.5 (S, T) and E16.5 (C, D, G, H, K, I, O, P). Brown nuclear staining in A–D, I–L, U–W, purplish nuclear staining in E–H and M–R and dark blue staining in S and T indicated positive staining for the relevant protein or mRNA. Arrowheads in U, V, W marked apoptotic cells in MLR10-mutant and control lenses. Note that TUNEL-positive cells were detected throughout the mutant lens. All scale bars = 100 μm.
Consistent with previous reports, p57kip2 expression increased in control lenses as cells begin the process of differentiation into primary (Fig. 4J) and secondary (Fig. 4L) fiber cells. A second CKI, p27kip1, is strongly expressed in fiber cells and minimally in epithelial cells (Fig. 4N, P). In contrast, the expression of both p27kip1 and p57kip2 was greatly reduced or absent in the MLR10-mutant lenses (Fig. 4I, K, M, O).
The defective cell cycle regulation and morphological features in MLR10-mutant lenses appeared similar to those of Prox1-null mice, suggesting that Prox1 might be a downstream target for Fgfr signaling. In E12.5 control embryos, Prox1 expression was observed in the anterior lens epithelial cells and significantly increased in the nuclei of posterior primary lens fiber cells. In contrast, Prox1 protein was uniformly expressed in MLR10-mutant lenses at a level close to that of the control lens epithelium (Fig. 4Q, R). At E14.5, newly differentiated secondary lens fiber cells at the control lens equator had a high level of Prox1 transcript expression. MLR10-mutant lenses lacked this increased expression of Prox1 mRNA with hybridization signals in the posterior half of the mutant lens being similar to that of the control lens epithelium (Fig. 4S, T). The expression of Prox1 gene products during fiber cell differentiation closely resembled the distribution of Prox1 mRNA, suggesting that regulation of Prox1 expression occurs at the transcriptional level.
The smaller size and increased number of pyknotic nuclei in MLR10-mutant lenses suggested higher levels of apoptosis (Fig. 2J, L, N). This was confirmed by TUNEL analysis. At E11.5, when primary fiber elongation is about to commence, more TUNEL-positive nuclei were present in MLR10-mutant than in control lenses (Fig. 4U, V). At E12.5, the percentage of TUNEL-positive nuclei in MLR10-mutant lenses was greater (24.2%) than in control (1.5%) lenses (P = 0.005), with more apoptosis in both the anterior and posterior regions of the mutant lenses (Fig. 4W, X). Similar results were obtained at E14.5 (10.9% in MLR10-mutant vs. 0.3% in control lenses, P = 0.011). Thus, Fgfr signaling plays an important role in lens cell survival.
Impaired fiber-specific gene expression in MLR10-mutant lenses
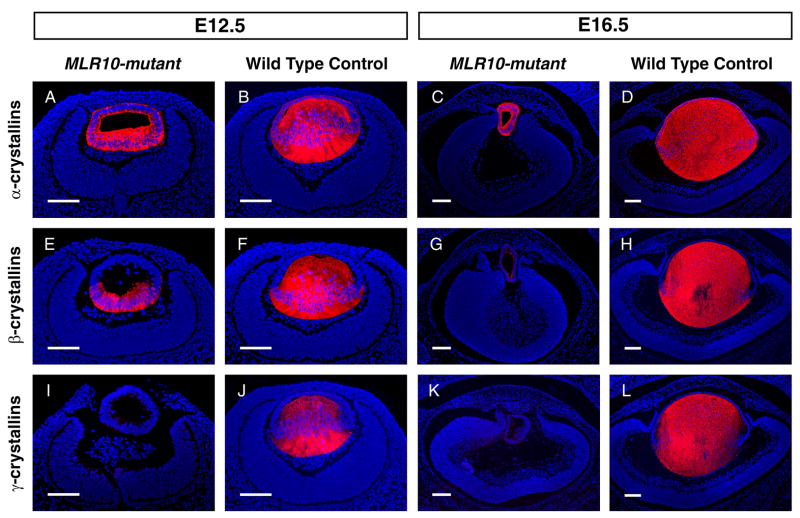
Lens fiber cell differentiation is characterized by the temporal and spatial expression of crystallin genes (reviewed in Duncan et al., 2004). However, the characteristics of MLR10-mutant lenses suggested that fiber cell differentiation was compromised. To test this hypothesis, we analyzed the accumulation of α-, β- and γ-crystallins. At E12.5, α-crystallins were readily detected throughout control and MLR10-mutant lenses, with increased accumulation in the posterior (lens fiber cell) compartment. At E16.5, α-crystallin was distributed in a similar manner in the epithelium and fiber cell compartment of MLR10-mutant and control lenses, although, due to the increased number and volume of wild type fiber cells, much more total α-crystallin was present in these cells (Fig. 5A–D). At E12.5, β-crystallins were present only in the posterior of the MLR10-mutant lenses. Control lenses at this stage appeared to accumulate significantly more β-crystallins in their fiber cells than mutant lenses (Fig. 5E, F). At E16.5, a low level of β-crystallin staining was seen in all cells of MLR10-mutant lenses, whereas control embryos showed a similar pattern of β-crystallin expression as that seen at E12.5 (Fig. 5G, H). Expression of γ-crystallins was abundant in the fiber cells of control lenses at E12.5 and E16.5, but not detected in MLR10-mutant lenses at either stage (Fig. 5I–L). These results suggested that signaling through Fgfr1-3 is not required for expression of α- and β-crystallins, but is required for γ-crystallin expression and may be needed for the maximal expression of all crystallins.
Figure 5.
Impaired crystallin expression in MLR10–mutant lenses. Immunofluorescence analyses of α-crystallins (A–D), β-crystallins (E–H) and γ-crystallins (I–L) were carried out in MLR10–mutant (A, C, E, G, I, K) and control (B, D, F, H, J, L) animals. Developmental stages studied included E12.5 (A, B, E, F, I, J) and E16.5 (C, D, G, H, K, L). Red-fluorescence indicates positive antibody staining. DAPI stained nuclei are blue. All scale bars = 100 μm.
FGF receptor signaling regulates c-Maf expression in the lens
Previous studies showed that crystallin expression is primarily regulated at the transcriptional level. Therefore, we examined the expression of Pax6, Sox1 and c-Maf, transcription factors known to be important for lens formation and crystallin expression. Pax6 mRNA and protein was present in all control lens cells from E11.5 to E16.5, with reduced expression in more mature fiber cells in the older lenses. In contrast, Pax6 was uniformly expressed throughout MLR10-mutant lenses at these stages (Fig. 6A–H). At E11.5 and E12.5, mRNA encoding Sox1, a transcription factor required for γ-crystallin expression (Nishiguchi et al., 1998), was present in all lens cells from both MLR10-mutant and control lenses, with increased accumulation in the posterior (fiber) cells (Fig. 6I–L). At E12.5, c-Maf accumulation was significantly increased in the nuclei of the primary fiber cells of control embryos. Little c-Maf was detected in the cells at the posterior of MLR10-mutant lenses, with most nuclei being negative. (Fig. 6M, N). At E16.5, when secondary fiber cell differentiation is underway, c-Maf staining was again abundant in the nuclei of elongating lens fiber cells but was only rarely seen in nuclei of MLR10-mutant lenses (Fig. 6O, P). c-Maf staining was uniformly weaker in the MLR10-mutant lenses and likely represents a total reduction in c-Maf protein rather than simply a failure of c-Maf to accumulate in the nucleus. Cytoplasmic staining of c-Maf in the MLR10-mutant lenses was not more intense than in the control lenses. Thus, signaling through Fgfr1-3 is required for c-Maf accumulation during lens development.
Figure 6.
Analysis of the expression of transcription factors required for crystallin expression in MLR10–mutant lenses. In-situ hybridization (A–D, I–L) and immunohistochemistry (E–H, M–P) revealed the expression of Pax6 (A–H), Sox1 (I–L) and c-Maf (M-P) in mutant (A, C, E, G, I, K, M, O) and control (B, D, F, H, J, L, N, P) lenses. Developmental stages studied included E11.5 (A, B, I, J), E12.5 (C, D, E, F K, L, M, N) and E16.5 (G, H, O, P). For pictures of immunohistochemistry, brown nuclear staining in E–H and purplish nuclear staining in M–P indicate positive staining. Bright silver grains reveal hybridization signals from relevant transcripts. The retinal pigmented epithelium (RPE) appears as a bright line surrounding the optic cup, due to light scattering by pigment granules. Note that the Pax6 staining in the nuclei of both the anterior and posterior cells of the MLR10-mutant lenses (E and G) while Pax6 staining in the wild type lens is largely restricted to anterior lens epithelial cells (F and H). Black arrowheads in M and O represent sparse nuclei expressing c-Maf in the MLR10-mutant lens. All scale bars = 100 μm.
Erk activation is reduced in MLR10-mutant lenses
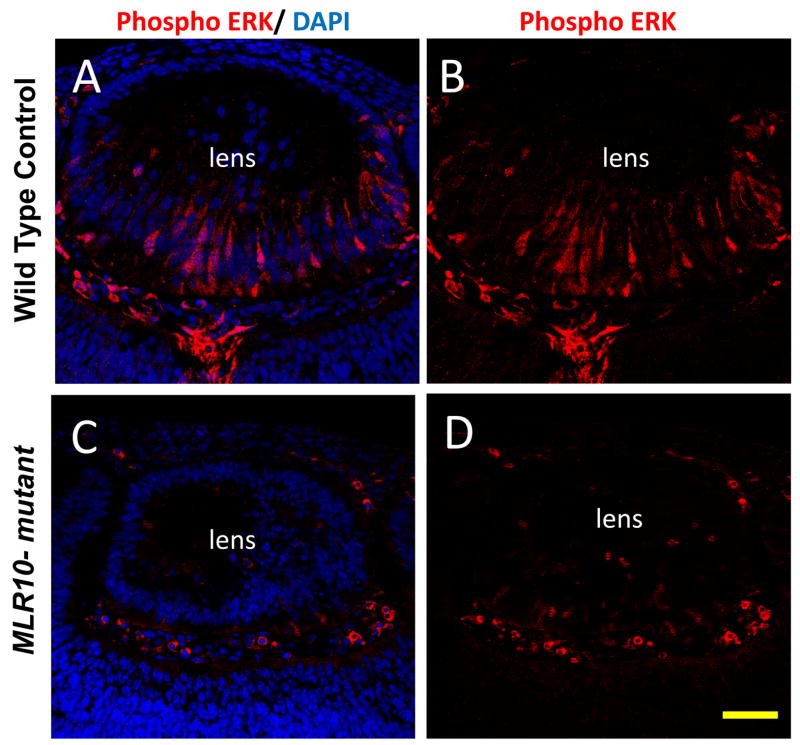
Fgfr activation is known to activate the ERK-MAP-kinase pathway in the lens, resulting in increased levels of phosphorylated Erk1 and Erk2 (Iyengar et al., 2007; Lovicu and McAvoy, 2001). We tested the level and distribution of phosphorylated Erk1/2 in MLR10-mutant and Cre-negative (control) littermates by immuofluoresence using a phospho-specific antibody. At E12.5, phosphorylated Erk1/2 staining was evident in elongating primary fiber cells, but not the epithelial cells, of control lenses (Fig 7A, B). However, phosphorylated Erk1/2 staining was weak or undetectable in the prospective fiber cells in the posterior region of MLR10-mutant lenses. Similar levels of phospho Erk1/2 were seen outside the lens in MLR10-mutant and control eyes (not shown).
Figure 7.
Deletion of Fgfrs leads to reduction in phosphorylated Erk1/2 in the lens. Phosphorylated (active) forms of Erk1 and Erk2 were not evident in lens epithelial cells, but were readily detected in elongating primary fiber cells in control lenses (A, B). Phospho-Erk1/2 staining was dramatically reduced in the MLR10-mutant lenses (C and D) at E12.5. Nuclei were counterstained with DAPI (blue), with phosphorylated Erk1/2 staining appearing red. Scale bar = 50 μm.
Discussion
FGF signaling is essential for lens fiber cell differentiation
Several Fgfs can promote fiber differentiation from lens epithelial cells when over expressed in transgenic mice (Lovicu and Overbeek, 1998; Robinson et al., 1998; Robinson et al., 1995b), but conclusive evidence to show that Fgf signaling is required for fiber cell differentiation in vivo was lacking. Previous studies showed that no single Fgfr is needed for the differentiation of normal-appearing fiber cells (reviewed in Robinson, 2006). The present study evaluated the quantitative requirement of Fgfr-signaling in mouse lens development. These experiments revealed that both alleles of any one the three receptors tested, and even a single allele of Fgfr2 or Fgfr3, was sufficient for normal lens development.
Given our previous results, in which deletion of Fgfr2 using the LeCre transgene (Garcia et al., 2005), compromised later lens development, it was surprising that a single allele of Fgfr3 was able to rescue normal lens development when both Fgfr2 alleles were deleted by MLR10. In LeCre mice, Cre recombinase is expressed in all surface ectoderm-derived ocular tissues at E9.0–9.5. This is at least 24 hours before lens-specific Cre expression is initiated in MLR10 mice. The difference in the timing of Fgfr2 deletion may account for the different phenotypes observed using the different Cre strains. If this interpretation is correct, Fgfr2 signaling early in lens formation contributes to later lens cell differentiation and survival. Alternatively, one of the Cre constructs may selectively enhance or suppress the Fgfr2 knockout phenotype.
Studies using in situ hybridization failed to detect Fgfr3 at the lens placode or the lens vesicle stage (de Iongh et al., 1997). However, the present genetic analysis shows that sufficient Fgfr3 is expressed by the lens vesicle stage to compensate for the loss of Fgfr2. It is possible that Fgfr3 transcripts are induced by the loss of Fgfr1 and/or Fgfr2. If so, this is a transient effect, since RNAase protection assays in postnatal lenses lacking Fgfr2 revealed no increase in Fgfr3 transcripts (Supplemental Fig. 2). Also, since both the previous and current studies were conducted on mice of mixed genetic backgrounds, it is possible that there is some genetic variation in the quantitative Fgfr requirement for normal lens development.
In lenses lacking Fgfr1-3 from the lens pit stage onward, prospective primary and secondary fiber cells continued to proliferate, remained as a cuboidal or columnar epithelium, did not activate the expression of γ-crystallins and expressed transcripts typically found in lens epithelial cells, but not in fiber cells. Two of the transcription factors essential for fiber cell differentiation, c-Maf (Kawauchi et al., 1999; Kim et al., 1999; Ring et al., 2000) and Prox1 (Wigle et al., 1999), were absent or expressed at reduced levels in prospective fiber cells. The accumulation of the CKIs, p27Kip1 and p57Kip2, factors required for cell cycle exit during fiber cell differentiation (Zhang et al., 1998), was also not initiated in the Fgfr1-3 deficient lenses. Therefore, Fgfr signaling is required for multiple aspects of lens fiber cell differentiation.
However, some of the events that characterize early lens fiber cell differentiation occurred despite the deletion of Fgfr1-3. Expression of the β-crystallins began in the proper cells and at the proper time in the knockout lenses. The concentration of β- and α-crystallins also appeared to increase in prospective fiber cells during the early stages of lens development. Despite the presence of abnormal proliferating cells in prospective lens fiber cells in MLR10-mutant eyes, the percentage of S-phase cells in the posterior compartment of mutant lens is reduced when compared to that of the antieror lens epithelial cells by E14.5. There are several possible explanations for activation of β-crystallin expression in the knockout lenses. As the Cre transgene is expressed only one day prior to the onset of fiber cell differentiation, it is possible that a sufficient number of Fgfrs remained in the E11.5 lens to respond to early Fgf signals. This minimal stimulation may not have been sufficient to elicit the full spectrum of fiber cell characteristics, but was enough to activate β-crystallin expression. It is also possible that exposure of the lens vesicle to other differentiation factors, like Bmps or Fgfs that bind to Fgfr4, is sufficient to activate β-crystallin expression but unable to activate the full fiber differentiation program. Previous studies showed that dominant negative constructs of the Bmp receptor, Alk6 (Faber et al., 2002), or exposure of the lens to increased levels of the Bmp antagonist, noggin (Belecky-Adams et al., 2002), delayed primary fiber cell elongation. Targeted null mutations in Fgfr4 exist (Weinstein et al., 1998), and tests of the expression and functions of Fgfr4 in lens induction and fiber cell differentiation are in progress using mice lacking all four Fgfrs. Fgfr4 likely plays a more prominent role in lens development in some species, such as zebrafish, than it appears to play in mice (Nakayama et al., 2008).
FGF signaling promotes the survival of lens cells
Defective lens fiber elongation, similar to that seen in MLR10-mutant lenses, was observed previously in Prox1 (Wigle et al., 1999), Sox1 (Nishiguchi et al., 1998) and c-Maf mutant mice (Kawauchi et al., 1999; Kim et al., 1999; Ring et al., 2000). However, comparison of MLR10-mutants to these mouse strains revealed a more severe reduction in the size of lenses lacking the Fgfrs. The small size of the MLR10-mutant lens is also at odds with the increased cellular proliferation seen in these lenses. Our studies suggest that their smaller size could be attributed to increased apoptosis observed throughout the MLR10-mutant lenses.
One possible explanation for increased apoptosis in the fiber cell compartment could be from continued proliferation in the face of signals promoting terminal differentiation. Such conflicts are frequently accompanied by apoptosis. For example, mice deficient in p27kip1 and p57kip2 (Zhang et al., 1997; Zhang et al., 1998), Prox1 (Wigle et al., 1999) or Rb (Morgenbesser et al., 1994) exhibited increased cell proliferation and apoptosis in the posterior of the lens. However, it is important to recognize that no such conflict would exist unless the posterior lens cells had begun to undergo terminal differentiation. Therefore, if the increased cell death in the fiber compartment is accounted for by a conflict between simultaneous signals to proliferate and to differentiate, the cells of the lens vesicle in the MLR10-mutants must have received signals for terminal differentiation. Such signals could arise from residual Fgfrs remaining after Cre-mediated deletion, signaling by Bmps or through Fgfr4, or by other, as yet unknown, factors that promote fiber cell terminal differentiation. Alternatively, FGF signaling may be required for the survival of lens fiber cells, as well as for their terminal differentiation.
Cells in the epithelium of MLR10-mutant lenses also underwent apoptosis, although no increase in apoptosis was reported in the epithelial cells of mice lacking p27kip1 (Kiyokawa et al., 1996; Nakayama et al., 1996), p27kip1 and p57kip2 (Zhang et al., 1997), Prox1 (Wigle et al., 1999) or Rb (Morgenbesser et al., 1994). Thus, lens epithelial and, perhaps, fiber cells depend on continuous FGF signaling for their survival. In support of this view, previous studies showed that deletion of Fgfr2 at E9.5 resulted in increased apoptosis in the lens epithelium and fiber cells at later stages (Garcia et al., 2005). Increased cell death was also seen in transgenic studies in which a dominant negative Fgfr was expressed in the lens (Chow et al., 1995; Robinson et al., 1995a; Stolen and Griep, 2000), and FGFs have demonstrated protection against lens cell apoptosis both in transgenic mice (Stolen et al., 1997) and in culture (Renaud et al., 1994; Wang et al., 1999).
Signaling through Fgfr1-3 is not essential for the proliferation of newly-formed lens epithelial cells
Previous studies showed that FGFs could stimulate proliferation of postnatal rat lens epithelial cells in vitro (McAvoy and Chamberlain, 1989). Therefore, the relatively normal proliferation exhibited by lens epithelial cells in MLR10-mutant lenses was unexpected. No obvious decrease in BrdU-labeled cells was observed in MLR10-mutant lenses, relative to wild type lenses at E11.5 or E12.5, when defects in fiber differentiation were evident. The apparently normal expression levels of Pax6, FoxE3 and Six3, all of which are thought to be required for lens epithelial proliferation, was consistent with this observation (Blixt et al., 2000; Goudreau et al., 2002), Fgfr signaling promotes the phosphorylation of Erk1/2 in postnatal lens epithelial cells (Iyengar et al., 2007; Lovicu and McAvoy, 2001). However, we did not detect significant levels of phospho-Erk1/2 in the lens epithelial cells of E12.5 lenses. These observations agree with findings of Garcia, et al., who determined that the smaller Le-Cre; Fgfr2 mutant lenses at E12.5 had a BrdU labeling index in their epithelial cells that was indistinguishable from wild type (Garcia et al., 2005). In contrast, Faber, et al. found that over expression of a dominant-negative form of Fgfr1, which is expected to block signaling by all Fgfrs, reduced epithelial cell proliferation at E12.5 (Faber et al., 2001). Since lens epithelial proliferation at E12.5 was not dependent on the three Fgfrs tested, signaling through Fgfr4 or by other growth factors may mediate lens epithelial cell proliferation in the embryo. We are currently testing whether Fgfr4 contributes to the proliferation of lens epithelial cells. A significant decrease in BrdU incorporation was noted in the MLR10-mutant lenses by E14.5. Although uncertain at this point, the decreased proliferation rate at E14.5 could reflect a requirement for Fgf signaling to maintain proliferation at this later stage or it may be a secondary result of decreased overall cell survival and/or health in the knockout lenses.
Unlike epithelial cells, elongating lens fiber cells had substantial levels of phospho-Erk1/2 which decreased in the prospective fiber cells of MLR10-mutant lenses (Fig. 7). Although several other tyrosine kinase receptors known to result in phosphorylation of Erk1/2 are expressed in the lens, our results suggest that, at this stage, the majority of lens phospho-Erk1/2 is dependent on Fgfr signaling. This interpretation is also in agreement with the deletion of Ndst1, which also abrogates Fgfr signaling and results in the loss of lens phospho-Erk1/2 (Pan et al., 2006).
In summary, prospective lens fiber cells lacking Fgfr1-3 did not stop dividing or increase expression of p27kip1, p57kip2 or Prox1, events associated with withdrawal from the cell cycle during fiber cell differentiation (Wigle et al., 1999; Zhang et al., 1998). In addition, our results suggested that Fgf signaling is essential for the survival of lens epithelial and, perhaps, fiber cells. Lens fiber differentiation was decreased in the absence of Fgfr1-3, as evidenced by impaired crystallin expression, which might be ascribed to defective expression of c-Maf and Prox1. Finally, Fgf signaling appears to diminish E-cadherin, Six3 and FoxE3 expression. Overall, FGF signaling integrates cell cycle regulation with fiber cell differentiation pathways (Fig. 8). The molecular pathways elicited by Fgfr stimulation, and the manner in which these individual pathways impact various features of lens development, survival and differentiation will be important avenues for future research.
Figure 8.
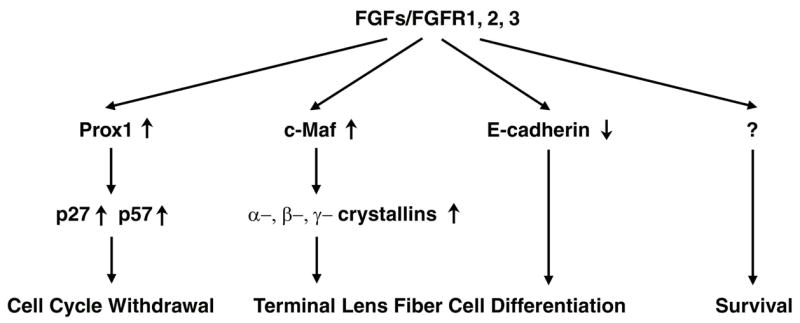
A model for the coordination of cell cycle withdrawal and lens fiber differentiation by FGF signaling. Prox1 is required for the increased expression of p27kip1 and p57kip2 that mediates the entry of lens fiber cells into G0. Deficient FGF receptor signaling led to reduced expression of Prox1 and decreased expression of p27kip1 and p57kip2, resulting in the abnormal proliferation of cells that would normally form lens fibers. The transcription factor c-Maf promotes the expression of α-, β- and γ-crystallins and fiber cell elongation. FGF signaling deficiency led to decreased crystallin gene expression and failure of fiber cell elongation. FGF signaling is also required for the decrease in E-cadherin expression that normally accompanies fiber cell differentiation. Finally, increased apoptosis in the MLR10-mutant lenses suggests that FGF signaling is required for lens epithelial cell survival.
The mouse lens expresses all four Fgfrs, but there appears to be considerable functional redundancy (at least among Fgfrs 1-3) subsequent to lens vesicle formation. Fgfr1 and Fgfr2 are known to function redundantly in cardiomyoblasts to promote myocardial growth (Lavine et al., 2006; Lavine et al., 2005). We previously demonstrated that Fgfr2 plays a non-redundant role in the lens at an earlier stage of development (Garcia et al., 2005) and suggest that, of the three Fgfrs examined, Fgfr1 plays the least important role. What is unclear is whether differential functionality of these receptors relates to the ligands present, variations in intracellular receptor signaling or simply the quantitative expression level or developmental expression pattern of different Fgfr genes. In this respect, mice carrying mutation (point mutations, isoform mutants) of different Fgfrs may help distinguish these possibilities. Given the redundancy exhibited by the different Fgfrs and the promiscuous binding of Fgfs to different Fgfrs (Ornitz et al., 1996; Zhang et al., 2006), it is likely that several Fgf ligands expressed in the ocular tissues surrounding the lens act redundantly to stimulate fiber differentiation. In agreement with this view, multiple Fgf ligands promote fiber differentiation in vitro and in vivo, while targeted disruption of many of these Fgf genes fail to reveal any abnormality in lens development. A future challenge will be to identify the ligands relevant to the process of lens fiber differentiation through Fgfr signaling.
Supplementary Material
Supplemental Figure 1. Lens development in mice with combined inactivation of two different Fgfrs. Postnatal (A–L) and E14.5-day (M–T) eyes were collected from Fgfr1,3 double mutants (MLR10/Fgfr1flox/flox Fgfr3 −/−)(A, E, I, M, Q), Fgfr2,3 double mutants (MLR10/Fgfr2flox/flox Fgfr3 −/−) (B, F, J, N, R), Fgfr1,2 double mutants (MLR10/Fgfr1flox/flox Fgfr2flox/flox)(C, G, K, O, S,) as well as wild type mice (D, H, L, P, T). Postnatal eyes collected at birth (P0) (A–D), 7 days after birth (P7) (E–H) or 1 month after birth (Adult) (I–L) were stained for histological evaluation. In-situ hybridization was conducted on embryonic eye sections to analyze the expression of lens epithelial markers FoxE3 (M–P) and Pax6 (Q–T). Some of the embryos analyzed were pigmented (N, R, T), and in sections from these mice the retina pigment epithelium (RPE) looks bright under dark-field illumination independent of probe hybridization. All scale bars = 100 μm.
Supplemental Figure 2. RNAse protection analysis of adult Fgfr lens transcripts after deletion of Fgfr2 mediated by MLR10 (lane 3) or in the presence of a null allele of Fgfr3 (lane 6). Note that Fgfr1 and Fgfr3 transcripts do not increase in response to Fgfr2 loss and Fgfr1 and Fgfr2 transcripts do not increase in response to Fgfr3 loss.
Acknowledgments
The authors thank Ying Yang, Lindsay Wallace, Brad D. Wagner, J. Tommy Barrs, and Florinda Jaynes for technical assistance. We are grateful to Drs. Milan Jamrich, Paul Overbeek and J. Samuel Zigler for probes, plasmids and antibodies. We are indebted to Drs. Michael Weinstein and Chu-Xia Deng for Fgfr3-null mice, Dr. David Cunningham for insightful discussions and suggestions, Michael Elnitsky and Timothy Muir for statistical consultation and Dr. Katia Del Rio-Tsonis for critical review of the manuscript. This work was supported by a grant from the National Eye Institute R01EY012995, a Miami University Undergraduate Research Award to Cornelius A. Thiels and by core facilities funded, in part, by The National Cancer Institute P30CA16058, Columbus Children’s Research Institute and The Department of Zoology at Miami University.
References
- Arman E, Haffner-Krausz R, Chen Y, Heath JK, Lonai P. Targeted disruption of fibroblast growth factor (FGF) receptor 2 suggests a role for FGF signaling in pregastrulation mammalian development. Proc Natl Acad Sci U S A. 1998;95:5082–7. doi: 10.1073/pnas.95.9.5082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashery-Padan R, Marquardt T, Zhou X, Gruss P. Pax6 activity in the lens primordium is required for lens formation and for correct placement of a single retina in the eye. Genes Dev. 2000;14:2701–11. doi: 10.1101/gad.184000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belecky-Adams TL, Adler R, Beebe DC. Bone morphogenetic protein signaling and the initiation of lens fiber cell differentiation. Development. 2002;129:3795–802. doi: 10.1242/dev.129.16.3795. [DOI] [PubMed] [Google Scholar]
- Blixt A, Mahlapuu M, Aitola M, Pelto-Huikko M, Enerback S, Carlsson P. A forkhead gene, FoxE3, is essential for lens epithelial proliferation and closure of the lens vesicle. Genes Dev. 2000;14:245–54. [PMC free article] [PubMed] [Google Scholar]
- Chow RL, Roux GD, Roghani M, Palmer MA, Rifkin DB, Moscatelli DA, Lang RA. FGF suppresses apoptosis and induces differentiation of fibre cells in the mouse lens. Development. 1995;121:4383–4393. doi: 10.1242/dev.121.12.4383. [DOI] [PubMed] [Google Scholar]
- Colvin JS, Bohne BA, Harding GW, McEwen DG, Ornitz DM. Skeletal overgrowth and deafness in mice lacking fibroblast growth factor receptor 3. Nature Genetics. 1996;12:390–397. doi: 10.1038/ng0496-390. [DOI] [PubMed] [Google Scholar]
- de Iongh RU, Lovicu FJ, Chamberlain CG, McAvoy JW. Differential expression of fibroblast growth factor receptors during rat lens morphogenesis and growth. Invest Ophthalmol Vis Sci. 1997;38:1688–1699. [PubMed] [Google Scholar]
- de Iongh RU, Lovicu FJ, Hanneken A, Baird A, McAvoy JW. FGF receptor-1 (flg) expression is correlated with fibre differentiation during rat lens morphogenesis and growth. Dev Dyn. 1996;206:412–26. doi: 10.1002/(SICI)1097-0177(199608)206:4<412::AID-AJA7>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Deng C, Wynshaw-Boris A, Zhou F, Kuo A, Leder P. Fibroblast Growth Factor Receptor 3 Is a Negative Regulator of Bone Growth. Cell. 1996;84:911–921. doi: 10.1016/s0092-8674(00)81069-7. [DOI] [PubMed] [Google Scholar]
- Dimanlig PV, Faber SC, Auerbach W, Makarenkova HP, Lang RA. The upstream ectoderm enhancer in Pax6 has an important role in lens induction. Development. 2001;128:4415–24. doi: 10.1242/dev.128.22.4415. [DOI] [PubMed] [Google Scholar]
- Duncan MK, Cvekl A, Kantorow M, Piatigorsky J. Lens Crystallins. In: Lovicu FJ, Robinson ML, editors. Development of the Ocular Lens. Cambridge University Press; New York, NY: 2004. pp. 119–150. [Google Scholar]
- Eswarakumar VP, Monsonego-Ornan E, Pines M, Antonopoulou I, Morriss-Kay GM, Lonai P. The IIIc alternative of Fgfr2 is a positive regulator of bone formation. Development. 2002;129:3783–93. doi: 10.1242/dev.129.16.3783. [DOI] [PubMed] [Google Scholar]
- Faber SC, Dimanlig P, Makarenkova HP, Shirke S, Ko K, Lang RA. Fgf receptor signaling plays a role in lens induction. Development. 2001;128:4425–38. doi: 10.1242/dev.128.22.4425. [DOI] [PubMed] [Google Scholar]
- Faber SC, Robinson ML, Makarenkova HP, Lang RA. Bmp signaling is required for development of primary lens fiber cells. Development. 2002;129:3727–37. doi: 10.1242/dev.129.15.3727. [DOI] [PubMed] [Google Scholar]
- Fisher M, Grainger RM. Lens induction and determination. In: Lovicu FJ, Robinson ML, editors. Development of the Ocular Lens. Cambridge University Press; New York, NY: 2004. pp. 27–47. [Google Scholar]
- Furuta Y, Hogan BL. BMP4 is essential for lens induction in the mouse embryo. Genes Dev. 1998;12:3764–75. doi: 10.1101/gad.12.23.3764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia CM, Yu K, Zhao H, Ashery-Padan R, Ornitz DM, Robinson ML, Beebe DC. Signaling through FGF receptor-2 is required for lens cell survival and for withdrawal from the cell cycle during lens fiber cell differentiation. Dev Dyn. 2005;233:516–27. doi: 10.1002/dvdy.20356. [DOI] [PubMed] [Google Scholar]
- Gotoh N, Ito M, Yamamoto S, Yoshino I, Song N, Wang Y, Lax I, Schlessinger J, Shibuya M, Lang RA. Tyrosine phosphorylation sites on FRS2alpha responsible for Shp2 recruitment are critical for induction of lens and retina. Proc Natl Acad Sci U S A. 2004;101:17144–9. doi: 10.1073/pnas.0407577101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goudreau G, Petrou P, Reneker LW, Graw J, Loster J, Gruss P. Mutually regulated expression of Pax6 and Six3 and its implications for the Pax6 haploinsufficient lens phenotype. Proc Natl Acad Sci U S A. 2002;99:8719–24. doi: 10.1073/pnas.132195699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govindarajan V, Overbeek PA. Secreted FGFR3, but not FGFR1, inhibits lens fiber differentiation. Development. 2001;128:1617–27. doi: 10.1242/dev.128.9.1617. [DOI] [PubMed] [Google Scholar]
- Hebert JM, Lin M, Partanen J, Rossant J, McConnell SK. FGF signaling through FGFR1 is required for olfactory bulb morphogenesis. Development. 2003;130:1101–11. doi: 10.1242/dev.00334. [DOI] [PubMed] [Google Scholar]
- Huang JX, Feldmeier M, Shui YB, Beebe DC. Evaluation of fibroblast growth factor signaling during lens fiber cell differentiation. Invest Ophthalmol Vis Sci. 2003;44:680–90. doi: 10.1167/iovs.01-1177. [DOI] [PubMed] [Google Scholar]
- Iyengar L, Wang Q, Rasko JE, McAvoy JW, Lovicu FJ. Duration of ERK1/2 phosphorylation induced by FGF or ocular media determines lens cell fate. Differentiation. 2007;75:662–8. doi: 10.1111/j.1432-0436.2007.00167.x. [DOI] [PubMed] [Google Scholar]
- Kawauchi S, Takahashi S, Nakajima O, Ogino H, Morita M, Nishizawa M, Yasuda K, Yamamoto M. Regulation of lens fiber cell differentiation by transcription factor c- Maf. J Biol Chem. 1999;274:19254–60. doi: 10.1074/jbc.274.27.19254. [DOI] [PubMed] [Google Scholar]
- Kim JI, Li T, Ho IC, Grusby MJ, Glimcher LH. Requirement for the c-Maf transcription factor in crystallin gene regulation and lens development. Proc Natl Acad Sci U S A. 1999;96:3781–5. doi: 10.1073/pnas.96.7.3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyokawa H, Kineman RD, Manova-Todorova KO, Soares VC, Hoffman ES, Ono M, Khanam D, Hayday AC, Frohman LA, Koff A. Enhanced growth of mice lacking the cyclin-dependent kinase inhibitor function of p27(Kip1) Cell. 1996;85:721–32. doi: 10.1016/s0092-8674(00)81238-6. [DOI] [PubMed] [Google Scholar]
- Kurose H, Okamoto M, Shimizu M, Bito T, Marcelle C, Noji S, Ohuchi H. FGF19-FGFR4 signaling elaborates lens induction with the FGF8-L-Maf cascade in the chick embryo. Dev Growth Differ. 2005;47:213–23. doi: 10.1111/j.1440-169X.2005.00795.x. [DOI] [PubMed] [Google Scholar]
- Lavine KJ, White AC, Park C, Smith CS, Choi K, Long F, Hui CC, Ornitz DM. Fibroblast growth factor signals regulate a wave of Hedgehog activation that is essential for coronary vascular development. Genes Dev. 2006;20:1651–66. doi: 10.1101/gad.1411406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavine KJ, Yu K, White AC, Zhang X, Smith C, Partanen J, Ornitz DM. Endocardial and epicardial derived FGF signals regulate myocardial proliferation and differentiation in vivo. Dev Cell. 2005;8:85–95. doi: 10.1016/j.devcel.2004.12.002. [DOI] [PubMed] [Google Scholar]
- Li C, Guo H, Xu X, Weinberg W, Deng CX. Fibroblast growth factor receptor 2 (Fgfr2) plays an important role in eyelid and skin formation and patterning. Dev Dyn. 2001;222:471–83. doi: 10.1002/dvdy.1205. [DOI] [PubMed] [Google Scholar]
- Lovicu FJ, McAvoy JW. FGF-induced lens cell proliferation and differentiation is dependent on MAPK (ERK1/2) signalling. Development. 2001;128:5075–84. doi: 10.1242/dev.128.24.5075. [DOI] [PubMed] [Google Scholar]
- Lovicu FJ, Overbeek PA. Overlapping effects of different members of the FGF family on lens fiber differentiation in transgenic mice. Development. 1998;125:3365–77. doi: 10.1242/dev.125.17.3365. [DOI] [PubMed] [Google Scholar]
- McAvoy JW, Chamberlain CG. Fibroblast growth factor (FGF) induces different responses in lens epithelial cells depending on its concentration. Development. 1989;107:221–8. doi: 10.1242/dev.107.2.221. [DOI] [PubMed] [Google Scholar]
- Morgenbesser SD, Williams BO, Jacks T, DePinho RA. p53-dependent apoptosis produced by Rb-deficiency in the developing mouse lens. Nature. 1994;371:72–4. doi: 10.1038/371072a0. [DOI] [PubMed] [Google Scholar]
- Nakayama K, Ishida N, Shirane M, Inomata A, Inoue T, Shishido N, Horii I, Loh DY, Nakayama K. Mice lacking p27(Kip1) display increased body size, multiple organ hyperplasia, retinal dysplasia, and pituitary tumors. Cell. 1996;85:707–20. doi: 10.1016/s0092-8674(00)81237-4. [DOI] [PubMed] [Google Scholar]
- Nakayama Y, Miyake A, Nakagawa Y, Mido T, Yoshikawa M, Konishi M, Itoh N. Fgf19 is required for zebrafish lens and retina development. Dev Biol. 2008;313:752–66. doi: 10.1016/j.ydbio.2007.11.013. [DOI] [PubMed] [Google Scholar]
- Nishiguchi S, Wood H, Kondoh H, Lovell-Badge R, Episkopou V. Sox1 directly regulates the gamma-crystallin genes and is essential for lens development in mice. Genes Dev. 1998;12:776–81. doi: 10.1101/gad.12.6.776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ornitz DM, Xu J, Colvin JS, McEwen DG, MacArthur CA, Coulier F, Gao G, Goldfarb M. Receptor specificity of the fibroblast growth factor family. J Biol Chem. 1996;271:15292–7. doi: 10.1074/jbc.271.25.15292. [DOI] [PubMed] [Google Scholar]
- Pan Y, Woodbury A, Esko JD, Grobe K, Zhang X. Heparan sulfate biosynthetic gene Ndst1 is required for FGF signaling in early lens development. Development. 2006;133:4933–44. doi: 10.1242/dev.02679. [DOI] [PubMed] [Google Scholar]
- Pirvola U, Ylikoski J, Trokovic R, Hebert JM, McConnell SK, Partanen J. FGFR1 is required for the development of the auditory sensory epithelium. Neuron. 2002;35:671–80. doi: 10.1016/s0896-6273(02)00824-3. [DOI] [PubMed] [Google Scholar]
- Renaud F, Oliver L, Desset S, Tassin J, Romquin N, Courtois Y, Laurent M. Up-regulation of aFGF expression in quiescent cells is related to cell survival. J Cell Physiol. 1994;158:435–43. doi: 10.1002/jcp.1041580307. [DOI] [PubMed] [Google Scholar]
- Revest JM, Spencer-Dene B, Kerr K, De Moerlooze L, Rosewell I, Dickson C. Fibroblast growth factor receptor 2-IIIb acts upstream of Shh and Fgf4 and is required for limb bud maintenance but not for the induction of Fgf8, Fgf10, Msx1, or Bmp4. Dev Biol. 2001;231:47–62. doi: 10.1006/dbio.2000.0144. [DOI] [PubMed] [Google Scholar]
- Ring BZ, Cordes SP, Overbeek PA, Barsh GS. Regulation of mouse lens fiber cell development and differentiation by the Maf gene. Development. 2000;127:307–17. doi: 10.1242/dev.127.2.307. [DOI] [PubMed] [Google Scholar]
- Robinson ML. An essential role for FGF receptor signaling in lens development. Semin Cell Dev Biol. 2006;17:726–40. doi: 10.1016/j.semcdb.2006.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson ML, MacMillan-Crow LA, Thompson JA, Overbeek PA. Expression of a truncated FGF receptor results in defective lens development in transgenic mice. Development. 1995a;121:3959–3967. doi: 10.1242/dev.121.12.3959. [DOI] [PubMed] [Google Scholar]
- Robinson ML, Ohtaka-Maruyama C, Chan CC, Jamieson S, Dickson C, Overbeek PA, Chepelinsky AB. Disregulation of ocular morphogenesis by lens-specific expression of FGF-3/int-2 in transgenic mice. Dev Biol. 1998;198:13–31. doi: 10.1006/dbio.1998.8879. [DOI] [PubMed] [Google Scholar]
- Robinson ML, Overbeek PA, Verran DJ, Grizzle WE, Stockard CR, Friesel R, Maciag T, Thompson JA. Extracellular FGF-1 acts as a lens differentiation factor in transgenic mice. Development. 1995b;121:505–14. doi: 10.1242/dev.121.2.505. [DOI] [PubMed] [Google Scholar]
- Stolen CM, Griep AE. Disruption of lens fiber cell differentiation and survival at multiple stages by region-specific expression of truncated FGF receptors. Dev Biol. 2000;217:205–20. doi: 10.1006/dbio.1999.9557. [DOI] [PubMed] [Google Scholar]
- Stolen CM, Jackson MW, Griep AE. Overexpression of FGF-2 modulates fiber cell differentiation and survival in the mouse lens. Development. 1997;124:4009–17. doi: 10.1242/dev.124.20.4009. [DOI] [PubMed] [Google Scholar]
- Trokovic N, Trokovic R, Mai P, Partanen J. Fgfr1 regulates patterning of the pharyngeal region. Genes Dev. 2003a;17:141–53. doi: 10.1101/gad.250703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trokovic R, Trokovic N, Hernesniemi S, Pirvola U, Vogt Weisenhorn DM, Rossant J, McMahon AP, Wurst W, Partanen J. FGFR1 is independently required in both developing mid- and hindbrain for sustained response to isthmic signals. Embo J. 2003b;22:1811–23. doi: 10.1093/emboj/cdg169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, He H, Zigler JS, Jr, Iwata T, Ibaraki N, Reddy VN, Carper D. bFGF suppresses serum-deprivation-induced apoptosis in a human lens epithelial cell line. Exp Cell Res. 1999;249:123–30. doi: 10.1006/excr.1999.4450. [DOI] [PubMed] [Google Scholar]
- Wawersik S, Purcell P, Rauchman M, Dudley AT, Robertson EJ, Maas R. BMP7 acts in murine lens placode development. Dev Biol. 1999;207:176–88. doi: 10.1006/dbio.1998.9153. [DOI] [PubMed] [Google Scholar]
- Weinstein M, Xu X, Ohyama K, Deng CX. FGFR-3 and FGFR-4 function cooperatively to direct alveogenesis in the murine lung. Development. 1998;125:3615–23. doi: 10.1242/dev.125.18.3615. [DOI] [PubMed] [Google Scholar]
- Wigle JT, Chowdhury K, Gruss P, Oliver G. Prox1 function is crucial for mouse lens-fibre elongation. Nat Genet. 1999;21:318–22. doi: 10.1038/6844. [DOI] [PubMed] [Google Scholar]
- Xu X, Weinstein M, Li C, Naski M, Cohen RI, Ornitz DM, Leder P, Deng C. Fibroblast growth factor receptor 2 (FGFR2)-mediated reciprocal regulation loop between FGF8 and FGF10 is essential for limb induction. Development. 1998;125:753–65. doi: 10.1242/dev.125.4.753. [DOI] [PubMed] [Google Scholar]
- Yamada R, Mizutani-Koseki Y, Hasegawa T, Osumi N, Koseki H, Takahashi N. Cell-autonomous involvement of Mab21l1 is essential for lens placode development. Development. 2003;130:1759–70. doi: 10.1242/dev.00399. [DOI] [PubMed] [Google Scholar]
- Yu K, Xu J, Liu Z, Sosic D, Shao J, Olson EN, Towler DA, Ornitz DM. Conditional inactivation of FGF receptor 2 reveals an essential role for FGF signaling in the regulation of osteoblast function and bone growth. Development. 2003;130:3063–74. doi: 10.1242/dev.00491. [DOI] [PubMed] [Google Scholar]
- Zhang P, Liegeois NJ, Wong C, Finegold M, Hou H, Thompson JC, Silverman A, Harper JW, DePinho RA, Elledge SJ. Altered cell differentiation and proliferation in mice lacking p57KIP2 indicates a role in Beckwith-Wiedemann syndrome. Nature. 1997;387:151–8. doi: 10.1038/387151a0. [DOI] [PubMed] [Google Scholar]
- Zhang P, Wong C, DePinho RA, Harper JW, Elledge SJ. Cooperation between the Cdk inhibitors p27(KIP1) and p57(KIP2) in the control of tissue growth and development. Genes Dev. 1998;12:3162–7. doi: 10.1101/gad.12.20.3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Ibrahimi OA, Olsen SK, Umemori H, Mohammadi M, Ornitz DM. Receptor specificity of the fibroblast growth factor family. The complete mammalian FGF family. J Biol Chem. 2006;281:15694–700. doi: 10.1074/jbc.M601252200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Yang Y, Partanen J, Ciruna BG, Rossant J, Robinson ML. Fibroblast growth factor receptor 1 (Fgfr1) is not essential for lens fiber differentiation in mice. Mol Vis. 2006;12:15–25. [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Yang Y, Rizo CM, Overbeek PA, Robinson ML. Insertion of a Pax6 consensus binding site into the alphaA-crystallin promoter acts as a lens epithelial cell enhancer in transgenic mice. Invest Ophthalmol Vis Sci. 2004;45:1930–9. doi: 10.1167/iovs.03-0856. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. Lens development in mice with combined inactivation of two different Fgfrs. Postnatal (A–L) and E14.5-day (M–T) eyes were collected from Fgfr1,3 double mutants (MLR10/Fgfr1flox/flox Fgfr3 −/−)(A, E, I, M, Q), Fgfr2,3 double mutants (MLR10/Fgfr2flox/flox Fgfr3 −/−) (B, F, J, N, R), Fgfr1,2 double mutants (MLR10/Fgfr1flox/flox Fgfr2flox/flox)(C, G, K, O, S,) as well as wild type mice (D, H, L, P, T). Postnatal eyes collected at birth (P0) (A–D), 7 days after birth (P7) (E–H) or 1 month after birth (Adult) (I–L) were stained for histological evaluation. In-situ hybridization was conducted on embryonic eye sections to analyze the expression of lens epithelial markers FoxE3 (M–P) and Pax6 (Q–T). Some of the embryos analyzed were pigmented (N, R, T), and in sections from these mice the retina pigment epithelium (RPE) looks bright under dark-field illumination independent of probe hybridization. All scale bars = 100 μm.
Supplemental Figure 2. RNAse protection analysis of adult Fgfr lens transcripts after deletion of Fgfr2 mediated by MLR10 (lane 3) or in the presence of a null allele of Fgfr3 (lane 6). Note that Fgfr1 and Fgfr3 transcripts do not increase in response to Fgfr2 loss and Fgfr1 and Fgfr2 transcripts do not increase in response to Fgfr3 loss.