Abstract
Agents that activate expression of specific genes to probe cellular pathways or alleviate disease would go beyond existing approaches for controlling gene expression. Duplex RNAs complementary to promoter regions can repress or activate gene expression. The mechanism of these promoter-directed antigene RNAs (agRNAs) has been obscure. Other work has revealed noncoding transcripts that overlap mRNAs. The function of these noncoding transcripts is also not understood. Here we link these two sets of enigmatic results. We find that antisense transcripts are the target for agRNAs that activate or repress expression of progesterone receptor (PR). agRNAs recruit Argonaute proteins to PR antisense transcripts and shift localization of the heterogeneous nuclear ribonucleoprotein-k, RNA polymerase II and heterochromatin protein 1γ. Our data demonstrate that antisense transcripts have a central role in recognition of the PR promoter by both activating and inhibitory agRNAs.
agRNAs complementary to target sequences within gene promoters can either activate or inhibit gene expression in mammalian cells1–7 (we use the terminology agRNA to distinguish them from duplex RNAs that target mRNA). Gene activation and inactivation are sequence specific. agRNAs that differ by just a few bases can either activate or silence gene expression and compete for recognition at overlapping target sites7. Although these data demonstrate that agRNA activity requires complementarity to promoter sequences, the identity of their molecular target was not known.
Characterizing the target for agRNAs is essential for understanding their mechanism of action, for investigating the existence of endogenous agRNAs and for developing agRNAs as therapeutics. We and others have shown that agRNAs recruit Argonaute (AGO) proteins to promoter DNA4,5 and that reducing levels of AGO blocks agRNA activity4,6. AGO proteins are known to mediate recognition of mRNA by small RNAs during post-transcriptional RNA interference (RNAi)8,9. Involvement of AGO in the activity of agRNAs suggested that agRNAs might also bind to RNA. There were, however, no known RNA transcripts at the PR promoter and therefore no obvious RNA targets for agRNAs.
Here we characterize the transcriptional landscape of the PR promoter in human cancer cells and identify previously undiscovered transcripts that span 70,000 nucleotides across the coding region of the gene and the promoter. Our data demonstrate that antisense transcripts are essential for agRNA-mediated gene activation at the PR promoter and that binding of agRNAs to antisense transcripts mediates formation of complexes with proteins and chromosomal DNA.
RESULTS
Antisense transcripts overlap the PR promoter
agRNAs (Supplementary Table 1 online) activate or inhibit PR expression in different cellular contexts. agRNAs inhibit transcription of PR in T47D breast cancer cells3,4, which express high levels of PR. Similar agRNAs activate PR expression in MCF7 breast cancer cells, which express low levels of PR, and in T47D cells grown under conditions that repress PR expression7.
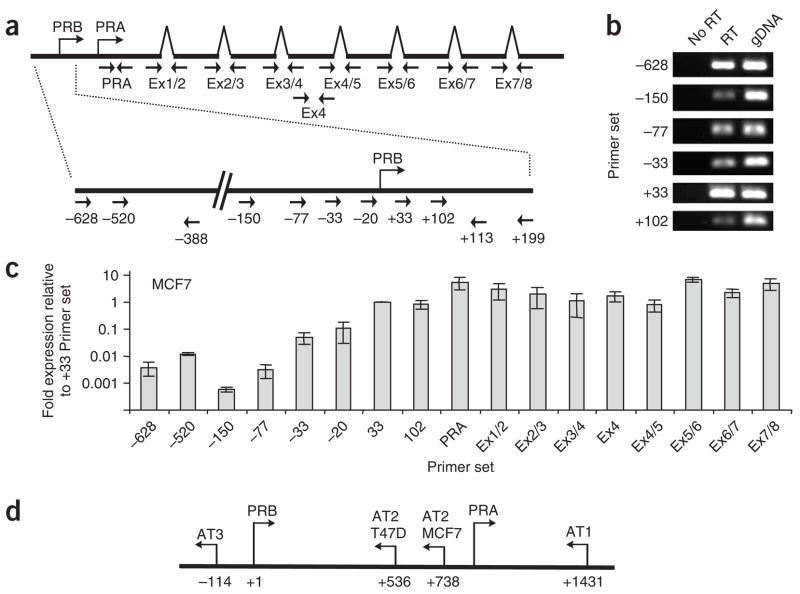
To investigate the existence of RNA transcripts that might act as targets for agRNAs, we used 5′ rapid amplification of cDNA ends (RACE) to search for undiscovered transcripts that initiate upstream from the transcription start site for PR. We did not identify any transcripts initiating upstream of the previously determined transcription start site10 (Supplementary Fig. 1a,b,f,g online). 5′ RACE of MCF7 cells treated with activating agRNA PR11 (targeted to the −11/+8 sequence at the PR promoter)7 revealed a shift to a similar distribution of transcription start sites to that defined in T47D cells (Supplementary Fig. 1c). Although we did not detect any transcripts initiating upstream of the established +1 site for PR, we were able to use reverse-transcription PCR (RT-PCR) to detect RNA overlapping the PR promoter (Fig. 1a,b). Quantitative PCR (qPCR; Supplementary Table 2 online) revealed that RNA levels at the PR promoter in both MCF7 and T47D cells were 10-fold to 1,000-fold below PR mRNA levels (Fig. 1c and Supplementary Fig. 1d).
Figure 1.
Antisense transcription at the PR promoter. (a) Locations of PCR primers used for quantifying the presence of transcripts throughout the PR gene. (b) PCR detection of RNA at the PR promoter without addition of reverse transcriptase (No RT), RNA treated with reverse transcriptase (RT) and genomic DNA (gDNA). (c) Levels of RNA evaluated using qPCR from polyadenylated RNA purified from MCF7 cells. (d) Location of antisense transcripts within the PR promoter in T47D or MCF7 cells. Error shown is s.d., calculated from four independent isolations of poly-A RNA and duplicate qPCR analysis of each sample. PRA, transcription start site for the A isoform of PR. PRB, transcription start site for the B isoform of PR.
Detection of RNA at the PR promoter, combined with our inability to detect sense transcripts, suggested that transcription might be occurring in the antisense direction. Using 5′ RACE we identified antisense RNA transcripts AT1, AT2-T47D (found only in T47D cells) and AT2-MCF7 (found only in MCF7 cells) as overlapping the region targeted by agRNAs (Fig. 1d and Supplementary Fig. 2 online), making them potential targets for direct physical interactions with agRNAs. The AT2 transcripts are spliced and polyadenylated, contain repetitive elements (long and short interspersed nuclear elements (LINEs and SINEs) and long terminal repeats (LTRs)) and are transcribed over a 70-kb region of genomic DNA (Supplementary Fig. 1e).
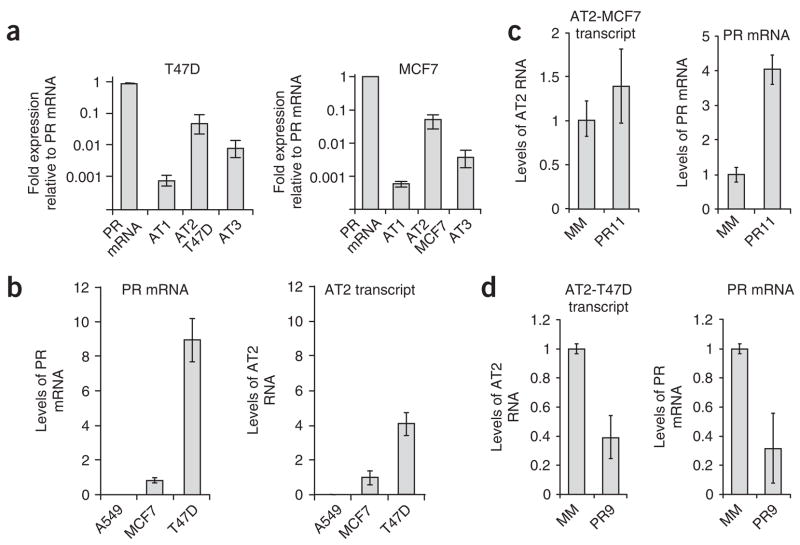
AT2-T47D and AT2-MCF7 antisense transcripts were the most highly expressed transcripts (Fig. 2a), and we chose them for further study (Fig. 2b–d). The physiological levels of AT2 transcript parallel levels of PR mRNA. Both PR mRNA and PR antisense transcript levels are highest in T47D cells, lower in MCF7 cells and undetectable in A549 lung cancer cells (Fig. 2b). Addition of activating agRNA PR11 to A549 cells does not increase PR expression, supporting the suggestion that expression of the antisense transcript is necessary for agRNA activity. Recognition of RNA targets by fully complementary duplex RNAs is often associated with target-strand cleavage by AGO and reduced levels of the target RNA. Addition of activating RNA PR11 to MCF7 cells, however, did not affect levels of the antisense transcript (Fig. 2c), suggesting that the agRNA does not induce transcript cleavage.
Figure 2.
Expression levels of antisense transcript AT2. (a) Expression of AT1, AT2-MCF7, AT2-T47D and AT3 relative to PR mRNA in T47D or MCF7 cells. (b) Relative levels of PR mRNA and AT2 in MCF7, T47D and A549 cells. (c,d) Effect on expression of antisense transcript AT2 and PR mRNA of adding activating agRNA PR11 to MCF7 cells (c) or silencing agRNA PR9 to T47D cells (d). These data use primers spanning exon boundary 5/6 of AT2, and identical results were seen using primers spanning other exon boundaries as well as bracketing the transcription start site (PR-33). Error bars indicate s.d. calculated from four independent isolations of total RNA and duplicate qPCR analysis of each sample.
Antisense transcripts are necessary for gene activation
To assess involvement of antisense transcripts in the regulation of gene expression by agRNAs, we obtained single-stranded oligonucleotides (Supplementary Table 3 online) complementary to sequences shared by antisense transcripts AT2-MCF7 and AT2-T47D. These single-stranded oligonucleotides are ‘gapmers’ containing a central DNA portion designed to recruit RNase H to cleave their RNA target and flanking 2′-methoxyethyl RNA regions to enhance affinity to target sequences11. Gapmers are effective gene-silencing agents and show promise in clinical trials11. The goal for these experiments was to use gapmers to test the effect of reducing antisense transcript levels on the activity of agRNAs.
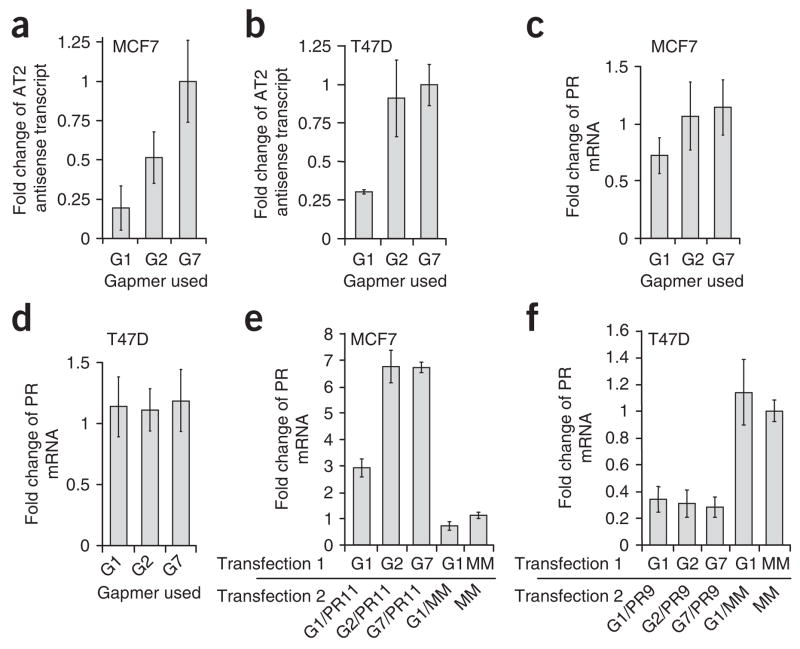
We tested ten gapmers (G1–G10; Supplementary Table 3) for their ability to reduce levels of antisense transcript AT2 in both MCF7 and T47D cells. Gapmers G1–G3 were complementary to spliced AT2; G4–G10 were not. We identified one gapmer, G1, capable of reducing levels of AT2 in both MCF7 and T47D cells (Fig. 3a,b and Supplementary Fig. 3 online). We also identified a less active gapmer, G2, capable of reducing transcript levels in MCF7 cells. Reduced levels of the antisense transcript after the addition of gapmers did not change the basal expression of PR in MCF7 (Fig. 3c) or in T47D cells (Fig. 3d), suggesting that inhibiting expression of the antisense transcript was not sufficient to activate or repress endogenous PR. Gapmer G7, which is not complementary to AT2, was used as a negative control.
Figure 3.
Gene activation by agRNAs was reversed by single-stranded oligonucleotides that target the antisense transcript. (a, b) Levels of antisense transcript in MCF7 (a) or T47D (b) cells after treatment with gapmer oligonucleotides G1, G2 or G7. Data were normalized relative to the results of treatment with gapmer G7. (c, d) Levels of PR mRNA after adding gapmers G1, G2 and G7 in MCF7 (c) or T47D (d) cells. Data were normalized relative to transfection with mismatch RNA. (e) Levels of PR mRNA in MCF7 cells after co-treatment with gapmers G1, G2 or G7 and activating agRNA PR11. (f) Levels of PR mRNA in T47D cells after co-treatment with gapmers G1, G2 or G7 and inhibitory agRNA PR9. Data were normalized relative to results of double-transfection with mismatch RNA. Gapmer G7 contains a sequence not found in the antisense transcript and represents a noncomplementary negative control. Error bars indicate s.d. calculated from triplicate independent transfection experiments with duplicate qPCR measurements for each.
When gapmer G1 was added to cells, the gene activation by agRNA PR11 was reversed (Fig. 3e). This result supports the hypothesis that the antisense transcript is involved in RNA-mediated gene activation. Addition of the less active gapmer G2 or gapmer G7, which was in the sense orientation (that is, gapmer G7 possessed the same sequence as the antisense transcript and was not complementary to AT2), did not reduce activation of PR expression. Addition of gapmer G1 to T47D cells has little effect on gene silencing by inhibitory agRNA PR9 (targeted to the –9/+10 sequence at the PR promoter)3,4 (Fig. 3f). One explanation for the inability of gapmer G1 to reverse gene silencing is that the antisense transcript is more prevalent in T47D cells than in MCF7 cells (Fig. 2b), making it difficult to reduce the level of the antisense transcript sufficiently in T47D cells to inhibit agRNA activity.
agRNAs bind to antisense transcripts
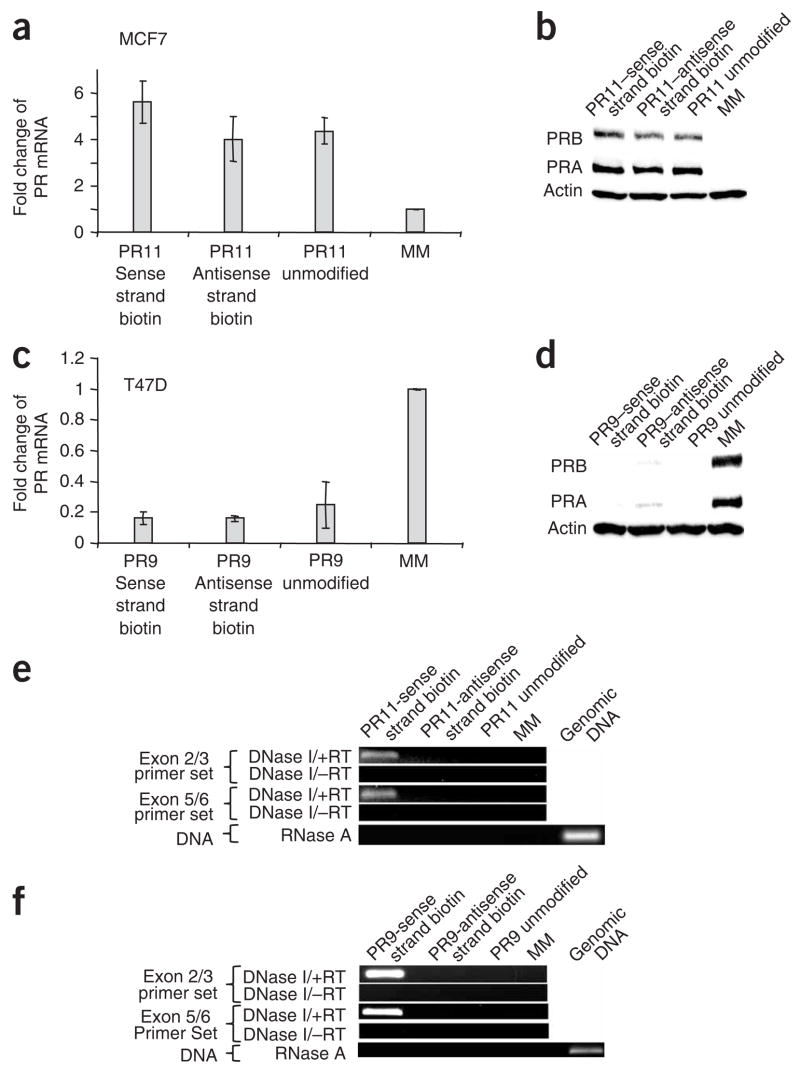
To investigate the potential for physical interactions between agRNAs and antisense transcripts, we modified the 3′ termini of either agRNA strand with biotin (Supplementary Table 1). Biotin labeling did not affect agRNA activity. Biotinylated agRNAs activated PR expression in MCF7 cells (Fig. 4a,b) and inhibited PR expression in T47D cells (Fig. 4c,d) with efficiencies similar to those shown by analogous unmodified agRNAs.
Figure 4.
Binding of biotinylated agRNAs to PR antisense transcript AT2. (a,b) qPCR (a) and western analysis (b) showing activation of PR expression in MCF7 cells upon addition of unmodified duplex agRNA PR11 or duplex biotinylated on either the sense or the antisense strand. (c,d) qPCR (c) and western analysis (d) showing reduction of PR expression in T47D cells upon addition of unmodified duplex agRNA PR9 or duplex biotinylated on either the sense or the antisense strand. Data in a and c were normalized relative to the results from treatment with mismatched RNA (MM), and error bars represent s.d. calculated from three independent experiments. (e,f) PCR detection of antisense transcript AT2 RNA after streptavidin purification from MCF7 (e) or T47D (f) cells. Data in parts e and f are representative of data from three independent experiments. RT, reverse transcriptase. PRA, A isoform of PR. PRB, B isoform of PR.
We harvested cell nuclei, purified biotinylated material using streptavidin-coated beads and eluted bound material. We analyzed the eluate by qPCR and detected the antisense transcript AT2-MCF7 from MCF7 cells transfected with activating agRNA PR11 biotinylated on the strand complementary to AT2-MCF7 (Fig. 4e). Similarly, antisense transcript AT2-T47D could be purified from T47D cells transfected with inhibitory agRNA PR9 biotinylated on the strand complementary to AT2-T47D (Fig. 4f). The identities of the amplified products were verified by sequencing (Supplementary Fig. 4 online).
Control experiments support the conclusion that agRNAs are binding to antisense transcripts. The antisense transcript was not detected after treatment with agRNAs that lacked biotin, and no amplified product was observed when the biotinylated strand was not complementary to AT2 (Fig. 4e,f). No product was detected using conditions designed to amplify genomic DNA, suggesting that there is no direct interaction between biotinylated agRNAs and chromosomal DNA.
agRNAs recruit AGO protein to antisense transcripts
Models for how transcribed RNA can act as a scaffold for protein complexes that affect heterochromatin formation in Schizosaccharomyces pombe and Drosophila melanogaster have been described previously12,13. We proposed that the antisense transcripts in human cells may act as scaffolds for organizing proteins at promoters and reasoned that AGO proteins would be involved.
Chromatin immunoprecipitation (ChIP) using a well-characterized antibody that recognizes all four human AGO proteins14 revealed a five-fold increase in the association of AGO with the PR promoter in MCF7 cells treated with activating agRNA PR11 (Supplementary Fig. 5a online). We observed a similar increase in AGO association in T47D cells treated with silencing agRNA PR9 (Supplementary Fig. 5b). In parallel, we used ChIP to evaluate binding of RNA polymerase II (RNA Pol II) at the PR promoter. We observed that activated agRNA PR11 enhanced association of RNA Pol II, whereas inhibitory agRNA PR9 reduced association (Supplementary Fig. 5c,d).
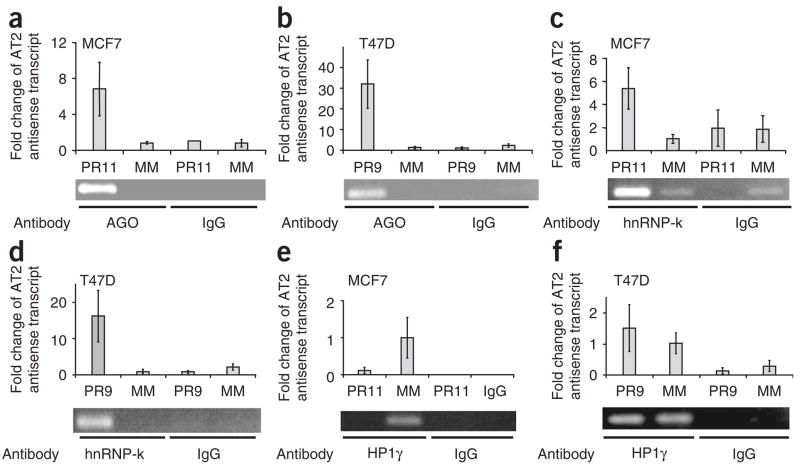
To investigate the potential for interactions between antisense transcript AT2 and AGO proteins we used RNA immunoprecipitation (RIP)15. This method is similar to ChIP but has been modified to detect RNA associated with proteins. Addition of activating agRNA PR11 promoted the association of AGO protein with antisense transcript AT2-MCF7 (Fig. 5a and Supplementary Fig. 4b). We performed RIP experiments using T47D cells and observed that silencing agRNA PR9 also promoted association of AGO to antisense transcript AT2-T47D (Fig. 5b).
Figure 5.
Association of AGO proteins, hnRNP-K and HP1γ with PR antisense transcript AT2 examined by RIP. Fold changes in qPCR values are relative to the results of treatment with mismatched RNA. (a) RIP of PR antisense transcript in MCF7 cells using an anti-AGO antibody after treatment with mismatch-containing RNA (MM) or activating agRNA PR11. (b) RIP of PR antisense transcript in T47D cells using an anti-AGO antibody after treatment with mismatch-containing RNA or inhibitory agRNA PR9. (c) RIP of PR antisense transcript in MCF7 cells using anti–hnRNP-k antibody after treatment with mismatch-containing RNA or activating agRNA PR11. (d) RIP of PR antisense transcript in T47D cells using an anti–hnRNP-k antibody after treatment with mismatch-containing RNA or inhibitory agRNA PR9. (e) RIP of PR antisense transcript in MCF7 cells using an anti-HP1γ antibody after treatment with mismatch-containing RNA or activating RNA pdPR11. (f) RIP of PR antisense transcript in T47D cells using an anti-HP1γ antibody after treatment with mismatch-containing RNA or inhibitory RNA agPR9. RIP data are from a representative experiment chosen from three similar data sets. Error bars for RIP data represent s.d. calculated from quadruplicate qPCR measurements for each.
Our data suggest that agRNAs promote the association of AGO, antisense transcripts and chromosomal DNA in close proximity to the PR promoter. Data obtained using biotinylated agRNAs suggest a direct interaction between agRNAs and the antisense transcripts. The association between AGO and chromosomal DNA is probably mediated through proteins, making it indirect. Our ChIP and RIP protocols include a step that cross-links protein and nucleic acid. It is likely that cross-linked proteins provide a bridge between AGO proteins, the antisense transcript and chromosomal DNA.
agRNAs shift localization of hnRNP-k from DNA to RNA
We proposed that formation of RNA–protein complexes at promoters would include interactions between RNA binding proteins, the anti-sense transcript and proteins that interact with the PR promoter. We chose to examine heterogeneous nuclear ribonucleoprotein-k (hnRNP-k)16 as a candidate for participating in agRNA-mediated complexes because it interacts with RNA and DNA and forms interactions with a wide variety of proteins. The PR promoter contains potential binding sites for hnRNP-k, providing another reason to test its involvement in the mechanism of agRNA action.
We used ChIP with an anti–hnRNP-k antibody to characterize the association of hnRNP-k at the PR promoter. Transfection of cells with activating agRNA PR11 or inhibitory agRNA PR9 reduced levels of hnRNP-k at the PR promoter (Supplementary Fig. 5e,f). We then used RIP to determine whether hnRNP-k associates with PR antisense transcripts upon addition of agRNAs. We performed RIP experiments in MCF7 cells treated with activating agRNA PR11 or in T47D cells treated with inhibitory agRNA PR9. Addition of either agRNA PR11 or agRNA PR9 recruited hnRNP-k to antisense transcript AT2 (Fig. 5c,d). The ChIP and RIP data showed that addition of agRNAs shifted localization of hnRNP-k from chromosomal DNA to the antisense transcript, suggesting that agRNAs can induce the remodeling of protein interactions at gene promoters.
Involvement of heterochromatin protein 1 (HP1)
In S. pombe and D. melanogaster, Swi6 localization and heterochromatic gene silencing are affected by expression of proteins involved in RNAi. In S. pombe, interactions between silencing RNAs and chromosomal DNA are mediated by the RNA induced transcriptional silencing (RITS) complex, and Swi6 seems to be required for this process13,17. HP1 proteins are the mammalian homolog of Swi6. They were initially characterized for their association with heterochromatic gene silencing in mammalian cells. A RITS-associated role for HP1, however, has not been demonstrated in human cells. HP1 exists in three isoforms (α, β and γ). We examined the involvement of HP1γ because it had been relatively well characterized in many roles in human cells, and, unlike HP1α and HP1β, HP1γ has been found in both heterochromatin and euchromatin18–20.
We performed ChIP and RIP experiments using an anti-HP1γ antibody to assess binding of HP1γ to the PR promoter or association with the AT2 transcript in T47D and MCF7 cells transfected with silencing agRNA PR9 or activating agRNA PR11, respectively. Transfection of MCF7 cells with activating agRNA PR11 reduced levels of HP1γ at the PR promoter (Supplementary Fig. 5g) and at the antisense transcript (Fig. 5e). Addition of inhibitory agRNA PR9 had little effect on levels of HP1γ at the PR promoter (Supplementary Fig. 5h) or at the antisense transcript (Fig. 5f). The observed reduction of HP1 levels during gene activation, but not gene silencing, suggests a potentially important mechanistic difference between the two processes.
DISCUSSION
The ability to use small RNAs to sequence-specifically activate a gene to alleviate a disease or probe a biological problem would supply a capability that is not provided by traditional RNA-mediated approaches focusing on gene silencing. Although the promise of gene activation is clear, the ability of agRNAs to activate or inhibit gene expression has been controversial2,21, in large part because agRNAs seemed to have no clear molecular target. We have found that agRNAs complementary to the PR promoter form a complex with AGO protein and bind an antisense RNA transcript that overlaps the promoter (Fig. 6). The antisense transcript–agRNA–AGO complex then acts as a scaffold for recruiting or redirecting other factors, such as hnRNP-k and HP1. The agRNA–antisense RNA transcript–protein complex forms in proximity to the promoter, affecting the balance of gene regulation.
Figure 6.
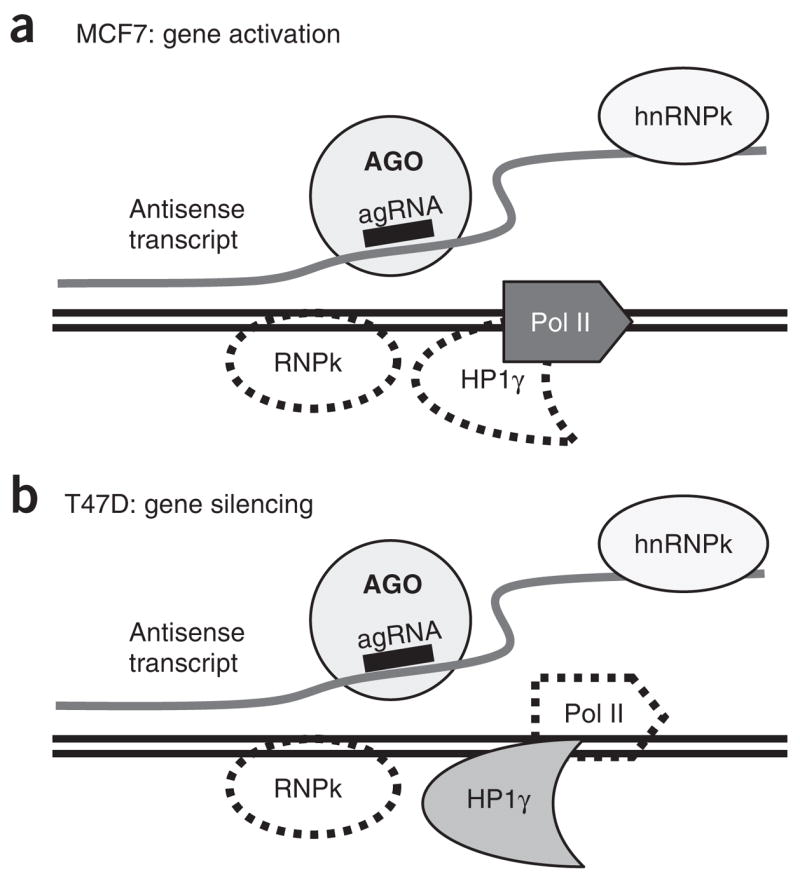
Factors that associate with gene promoters during agRNA-mediated gene activation or silencing. Solid lines denote factors that associate with promoter DNA upon addition of agRNA. Dashed lines denote factors that dissociate from promoter DNA upon addition of agRNA. For both activation and inhibition, the agRNA recruits AGO to an antisense transcript and causes the localization of hnRNP-k to shift from the chromosome to the antisense transcript. For gene activation, addition of agRNA recruits RNA Pol II, whereas during gene silencing RNA Pol II leaves the transcript. HP1γ levels are reduced upon gene activation and unchanged during silencing.
Gene activation and gene silencing
Both activating and silencing agRNAs bind to antisense transcripts and recruit AGO and hnRNP-k proteins. In view of these similarities, what mechanism can explain the ability of closely related RNAs to activate expression of a gene in one context and inhibit it in another? Our studies are at an early stage, and we cannot present any definitive answers for this central question. Our data, however, do provide several important clues. The first is that basal levels of expression affect whether activation or silencing is observed. Under growth conditions that lead to high PR expression in T47D cells, agRNAs repress gene expression. However, under growth conditions that lead to low PR expression in T47D cells, agRNAs can activate gene expression7. Thus, different outcomes can be achieved in the same cell line depending on basal expression levels and on the status of the promoter when the agRNA–AGO complex is recruited to the gene.
Another important clue is provided by our earlier observation that inactive RNAs can block gene activation by PR11 and other activating agRNAs, even though their target sequences differ by just a few bases7. These data suggest that agRNAs and related inactive RNAs compete for binding to target sequences. Relatively small differences in the location of target sequences dictate whether activation will occur.
The ability of agRNAs to activate gene expression in one context and repress expression in another is reminiscent of hormone-mediated regulation at gene promoters. In both MCF7 and T47D cell lines, expression levels of PR are poised to change upon addition of small-molecule ligands or by altering cell-culture conditions. For example, addition of estrogen will increase PR expression in MCF7 cells22, whereas removal of hormone-like compounds will reduce PR expression in T47D cells23. Small molecules alter expression by changing the recruitment of proteins at the promoter. The fact that small molecules can remodel the protein machinery at the PR promoter and affect transcription provides a precedent for the ability of RNA to also trigger or repress gene expression.
Our data demonstrate that agRNAs target noncoding transcripts that overlap gene promoters. For PR, the transcript is in the antisense orientation, but it is possible that sense transcripts may also serve as platforms for agRNA recognition of other genes. Endogenous miRNAs have been found with near-perfect complementarity to gene promoters in either the sense or antisense orientations (S.T.Y., unpublished data). Genomic studies have revealed that both sense and antisense transcripts commonly overlap gene promoters24–26, providing a wide selection of possible targets for agRNAs. Antisense transcripts have been studied considerably in lower eukaryotes, notably ‘aberrant’ transcripts in yeast, which can be suppressed by histone deacetylase complexes27,28.
Noncoding RNAs that act as scaffolds for protein recruitment or nucleic acid interactions may be a general strategy for controlling gene expression29 that encompasses various regulatory mechanisms. Steitz has suggested that miRNAs can recruit AGO protein to AU-rich elements within the 3′ UTR to activate translation30. In contrast to our results, mRNA was targeted, and activation required that the cells undergo cell-cycle arrest. Inhibition of dihydrofolate reductase (DHFR) expression by a sense transcript synthesized from an upstream promoter has been reported, and the authors postulate that the transcript directly binds to the DHFR promoter by triple-helix formation31. A single-stranded RNA labeled at the 5′ terminus with biotin and complementary to the promoter for elongation factor 1α (EF1a) has been reported to associate with an extended 5′ UTR variant of EF1a mRNA32, and a related duplex RNA can control EF1a expression. In contrast to these studies, we observe that (i) double-stranded agRNAs recognize antisense transcripts that originate within the target gene, (ii) interactions with the antisense transcript can lead to gene activation as well as gene silencing, regardless of cell-cycle stage, and (iii) agRNAs can recruit proteins to antisense transcripts and shift the localization of proteins from promoter DNA.
Endogenous agRNAs?
Our findings provide a mechanism for gene activation and gene silencing by agRNAs. The central role of antisense transcripts is especially intriguing because of the recent demonstration that networks of noncoding RNAs are ubiquitous features of genes24–26.
Gene activation or gene silencing of PR by agRNAs is a robust phenomenon, consistent with the possibility that agRNAs tap into pre-existing natural mechanisms. Antisense or sense transcripts that overlap promoters might act as targets for natural miRNAs. Computational approaches have revealed that many known miRNAs have substantial complementarity to sequences within gene promoters (S.T.Y., unpublished data). Endogenous RNA-mediated recognition of RNA transcripts at promoters by proteins would provide an additional mechanism for sequence-specifically delivering proteins to key regulatory regions. Because RNA can evolve new specificities for nucleic acid recognition more readily than proteins, a mechanism for RNA-mediated regulation of gene promoters would have evolutionary advantages.
METHODS
Cellular delivery of agRNAs
We used Oligofectamine or RNAi-max (Invitrogen) to deliver pdRNAs (Supplementary Table 1) or single-stranded gapmer oligonucleotides (Supplementary Table 3) into MCF7 or T47D cells33.
Quantitative PCR
We performed qPCR on an ABI7900 real-time PCR (Applied Biosystems) using Sybr Mastermix (Qiagen). Primers were designed using primer3 software (http://fokker.wi.mit.edu/primer3/input.htm) with the exception of primers for GAPDH, which were supplied as a control (Applied Biosystems) (Supplementary Table 2). Only those primer sets that show linear amplification over several orders of magnitude were used. RNA was treated with DNase before reverse transcription.
Chromatin immunoprecipitation
We performed ChIP as described34. Anti–hnRNP-k antibodies were supplied by Sigma. Anti-HP1γ was acquired from Upstate. The anti-AGO antibody was developed in the Mourelatos laboratory (Z. Mourelatos, University of Pennsylvania)14. We harvested transfected cells immediately before ChIP analysis and used western analysis of PR protein levels to confirm either gene silencing or gene activation in the experiment. Primers used for ChIP are described in Supplementary Table 2.
RNA immunoprecipitation
We performed RIP essentially as described15. We grew MCF7 or T47D cells in 150 cm2 dishes and transfected duplex RNAs using RNAiMax (Invitrogen)33. Cells were cross-linked using 1% (v/v) formaldehyde solution and harvested. Cells were lysed and nuclei obtained. Antibody capable of binding the protein of interest (AGO14, hnRNP-k or HP1γ) was incubated with nuclei overnight. The antibody-treated material was then mixed with Protein A/G agarose Plus (Santa Cruz) and washed five times as described15. Complex was eluted and cross-linking reversed by adding 200 nM NaCl and heating to 65 °C for 2 h. Samples were amplified using primers complementary to antisense RNA transcripts (Supplementary Table 1).
5′ rapid amplification of cDNA ends
5′ and 3′ RACE was performed according to the manufacturer’s protocol using the GeneRacer kit (Invitrogen; http://tools.invitrogen.com/content/sfs/manuals/generacer_man.pdf). We chose this kit because it includes purification steps designed to yield full-length RNA with intact 5′ caps rather than truncated products. The 3′ RACE protocol in this kit selects for polyadenylated transcripts. Multiple primer sets (Supplementary Table 2) were used to maximize detection of transcripts and reduce the likelihood of bias from any one primer set. PCR products were cloned into a PCR-4 Topo vector and sequenced (McDermott sequencing core, University of Texas Southwestern). We used the Platinum Taq High Fidelity kit (Invitrogen) to produce product for cloning. We sequenced several clones from at least two independent experiments to confirm results. All 5′ and 3′ RACE products were further verified by PCR-based cloning of the full-length transcript using primers targeting the 5′ and 3′ ends of the transcripts.
Biotin pull-down of small RNA–transcript complexes
MCF7 or T47D were grown in six-well dishes and transfected with biotinylated RNA heteroduplexes (a 3′ biotin on either the sense or antisense strand, supplied by Sigma-Proligo) at a concentration of 100 nM for 24 h. Avidin-coated beads were prepared by preblocking with yeast tRNA and salmon sperm DNA. At 3 d after transfection, cells were harvested to obtain nuclei. The nuclei were mixed with avidin-coated beads at 4 °C for 2 h. We washed the beads exhaustively before elution of RNA using buffer (1.5% biotin, 4 M guanidine thiocyanate, 25 mM sodium citrate, 0.5% sodium N-lauroyl sarcosinate) for 2 h at 45 °C with periodic gentle agitation. Samples were treated with DNase to remove any contaminating DNA and then amplified by PCR using two different primer sets capable of amplifying the target antisense transcript. The biotin pull-down assay was modified from a previously reported protocol35, and the elution buffer was based on that described in ref. 36. Primer sequences for amplification of antisense transcript are described in Supplementary Table 2.
Supplementary Material
Note: Supplementary information is available on the Nature Structural & Molecular Biology website.
Acknowledgments
This work was supported by grants from the US National Institutes of Health (NIGMS 60642, 77253 and 73042 to D.R.C. and EB 05556 to J.C.S.) and Robert A. Welch Foundation (I-1244). We thank Z. Mourelatos (University of Pennsylvania) for providing anti-AGO antibody and D. Shames for helpful discussions.
References
- 1.Morris KV, Chan SW, Jacobsen SE, Looney DJ. Small interfering RNA-induced transcriptional silencing in human cells. Science. 2004;305:1289–1292. doi: 10.1126/science.1101372. [DOI] [PubMed] [Google Scholar]
- 2.Ting AH, Schuebel KE, Herman JG, Baylin SB. Short double-stranded RNA induces transcriptional gene silencing in human cells in the absence of DNA methylation. Nat Genet. 2005;37:906–910. doi: 10.1038/ng1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Janowski BA, et al. Inhibition of gene expression at transcription start sites using antigene RNAs (agRNAs) Nat Chem Biol. 2005;1:216–222. doi: 10.1038/nchembio725. [DOI] [PubMed] [Google Scholar]
- 4.Janowski BA, et al. Involvement of Ago1 and Ago2 in mammalian transcriptional silencing. Nat Struct Mol Biol. 2006;13:787–792. doi: 10.1038/nsmb1140. [DOI] [PubMed] [Google Scholar]
- 5.Kim DH, Villeneuve LM, Morris KV, Rossi JJ. Argonaute-1 directs siRNA-mediated transcriptional gene silencing in human cells. Nat Struct Mol Biol. 2006;13:793–797. doi: 10.1038/nsmb1142. [DOI] [PubMed] [Google Scholar]
- 6.Li LC, et al. Small dsRNAs induce transcriptional activation in human cells. Proc Natl Acad Sci USA. 2006;103:17337–17342. doi: 10.1073/pnas.0607015103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Janowski BA, et al. Activating gene expression in mammalian cells with promoter-targeted duplex RNAs. Nat Chem Biol. 2007;3:166–173. doi: 10.1038/nchembio860. [DOI] [PubMed] [Google Scholar]
- 8.Meister G, et al. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol Cell. 2004;15:185–197. doi: 10.1016/j.molcel.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 9.Liu J, et al. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- 10.Kastner P, et al. Two distinct estrogen-regulated promoters generate transcripts encoding the two functionally different human progesterone receptor forms A and B. EMBO J. 1990;9:1603–1614. doi: 10.1002/j.1460-2075.1990.tb08280.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Corey DR. RNAi learns from antisense. Nat Chem Biol. 2007;3:8–11. doi: 10.1038/nchembio0107-8. [DOI] [PubMed] [Google Scholar]
- 12.Grewal SIS, Elgin SCR. Transcription and RNA interference in the formation of heterochromatin. Nature. 2007;447:399–406. doi: 10.1038/nature05914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Buhler M, Verdel A, Moazed D. Tethering RITS to a nascent transcript initiates RNAi- and heterochromatin-dependent gene silencing. Cell. 2006;125:873–886. doi: 10.1016/j.cell.2006.04.025. [DOI] [PubMed] [Google Scholar]
- 14.Nelson PT, et al. A novel monoclonal antibody against human Argonaute proteins reveals unexpected characteristics of miRNAs in human blood cells. RNA. 2007;13:1787–1792. doi: 10.1261/rna.646007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gilbert C, Kristjuhan A, Winkler GS, Svejstrup JQ. Elongator interactions with nascent mRNA revealed by RNA immunoprecipitation. Mol Cell. 2004;14:457–464. doi: 10.1016/s1097-2765(04)00239-4. [DOI] [PubMed] [Google Scholar]
- 16.Bomsztyk K, Denisenko O, Ostrowski J. hnRNP K: one protein, multiple processes. Bioessays. 2004;26:629–638. doi: 10.1002/bies.20048. [DOI] [PubMed] [Google Scholar]
- 17.Zofall M, Grewal SL. Swi6/HP1 recruits a JmjC domain protein to facilitate transcription of heterochromatic repeats. Mol Cell. 2006;22:681–692. doi: 10.1016/j.molcel.2006.05.010. [DOI] [PubMed] [Google Scholar]
- 18.Vakoc CR, Mandat SA, Olenchock BA, Blobel GA. Histone H3 lysine 9 methylation and HP1γ are associated with transcription elongation through mammalian chromatin. Mol Cell. 2005;19:381–391. doi: 10.1016/j.molcel.2005.06.011. [DOI] [PubMed] [Google Scholar]
- 19.Eissenberg JC, Shilatifard A. Leaving a mark: the many footprints of the elongating RNA polymerase II. Curr Opin Genet Dev. 2006;16:184–190. doi: 10.1016/j.gde.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 20.Smallwood A, Black JC, Tanese N, Pradan S, Carey M. HP1-mediated silencing targets PolII coactivator complexes. Nat Struct Mol Biol. 2008;15:318–320. doi: 10.1038/nsmb.1385. [DOI] [PubMed] [Google Scholar]
- 21.Check E. RNA interference: hitting the switch. Nature. 2007;448:855–858. doi: 10.1038/448855a. [DOI] [PubMed] [Google Scholar]
- 22.Lee YJ, Gorski J. Estrogen-induced transcription of the progesterone receptor gene does not parallel estrogen receptor occupancy. Proc Natl Acad Sci USA. 1996;93:15180–15184. doi: 10.1073/pnas.93.26.15180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hurd C, et al. Hormonal regulation of the p53 tumor suppressor protein in T47D human breast carcinoma cell line. J Biol Chem. 1995;270:28507–28510. doi: 10.1074/jbc.270.48.28507. [DOI] [PubMed] [Google Scholar]
- 24.RIKEN Genome Exploration Research Group and Genome Science Group (Genome Network Project Core Group) and the FANTOM Consortium. Antisense transcription in the mammalian transcriptome. Science. 2005;309:1564–1566. doi: 10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]
- 25.ENCODE Project Consortium. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gingeras TR. Origin of phenotypes: genes and transcripts. Genome Res. 2007;17:682–690. doi: 10.1101/gr.6525007. [DOI] [PubMed] [Google Scholar]
- 27.Nicolas E, et al. Distinct roles of HDAC complexes in promoter silencing, antisense suppression and DNA damage protection. Nat Struct Mol Biol. 2007;14:372–380. doi: 10.1038/nsmb1239. [DOI] [PubMed] [Google Scholar]
- 28.Carrozza MJ, et al. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell. 2005;123:581–592. doi: 10.1016/j.cell.2005.10.023. [DOI] [PubMed] [Google Scholar]
- 29.Mattick JS. A new paradigm for developmental biology. J Exp Biol. 2007;210:1526–1547. doi: 10.1242/jeb.005017. [DOI] [PubMed] [Google Scholar]
- 30.Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318:1931–1934. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- 31.Martianov I, et al. Repression of the human dihydrofolate reducase gene by a noncoding interfering transcript. Nature. 2007;445:666–700. doi: 10.1038/nature05519. [DOI] [PubMed] [Google Scholar]
- 32.Han J, Kim D, Morris KV. Promoter-associated RNA is required for RNA-directed transcriptional gene silencing in human cells. Proc Natl Acad Sci USA. 2007;104:12422–12427. doi: 10.1073/pnas.0701635104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Janowski BA, Hu J, Corey DR. Antigene inhibition by peptide nucleic acids and duplex RNAs. Nat Protocols. 2006;1:436–443. doi: 10.1038/nprot.2006.64. [DOI] [PubMed] [Google Scholar]
- 34.Hardy DB, Janowski BA, Corey DR, Mendelson CR. Progesterone impairs the interleukin-1β stimulation of cyclooxygenase 2 (COX-2) gene expression in human myometrial cells. Mol Endocrinol. 2006;20:2724–2733. doi: 10.1210/me.2006-0112. [DOI] [PubMed] [Google Scholar]
- 35.Mizuno Y, et al. Increased specificity of reverse transcription priming of trehalose and oligo-blockers allows high efficiency window separation of mRNA display. Nucleic Acids Res. 1999;27:1345–1349. doi: 10.1093/nar/27.5.1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kraynack BA, Baker BF. Small interfering RNAs containing full 2′-O-methylribonucleotide-modified sense strands display Argonaute2/eIF2C2-dependent activity. RNA. 2006;12:163–176. doi: 10.1261/rna.2150806. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Note: Supplementary information is available on the Nature Structural & Molecular Biology website.