Abstract
The laboratory mouse has long been an important tool in the study of the biology and genetics of human cancer. With the advent of genetic engineering techniques, DNA microarray analyses, tissue arrays, and other large-scale, high-throughput data generating methods, the amount of data available for mouse models of cancer is growing exponentially. Tools to integrate, locate and visualize these data are crucial to aid researchers in their investigations. The Mouse Tumor Biology database (http://tumor.informatics.jax.org) seeks to address that need.
It has long been acknowledged that genetics has a significant role in the initiation and progression of many forms of cancer1. In the early 1900s scientists recognized that different strains of mice varied in their susceptibilities to specific forms of cancer and that these phenotypes were tied to each strain's distinct genetic background2,3. Inbred strains of mice have served as powerful genetic tools to explore the underlying mechanisms of variable susceptibility4,5. The relatively recent emergence of genetic engineering as a means to manipulate the mouse genome has led to the development of mouse models specifically designed to provide insight into the roles of specific genes and pathways in the development and progression of cancer6,7. Early genetically engineered mouse models, such as transgenic mice that overexpressed oncogenes, proved to be an effective method for generating reproducible neoplasms, but such models did little to recapitulate the complexities of the genetics of cancer development and progression. More recent genetically engineered mouse models use techniques such as conditional, retrovirus-delivered and 'latent' mutant alleles that provide a genetic picture that is more similar to the processes observed in developing human neoplasms8–12. As data continue to be generated from models old and new at an ever-increasing rate (FIG. 1), the need for a repository for this information continues to grow as well. The Mouse Tumor Biology database (MTB) provides a publicly accessible electronic information resource for cancer researchers worldwide to both explore and contribute to this ever-expanding base of cancer biology data13–15.
Figure 1. Publications for mouse models of cancer.
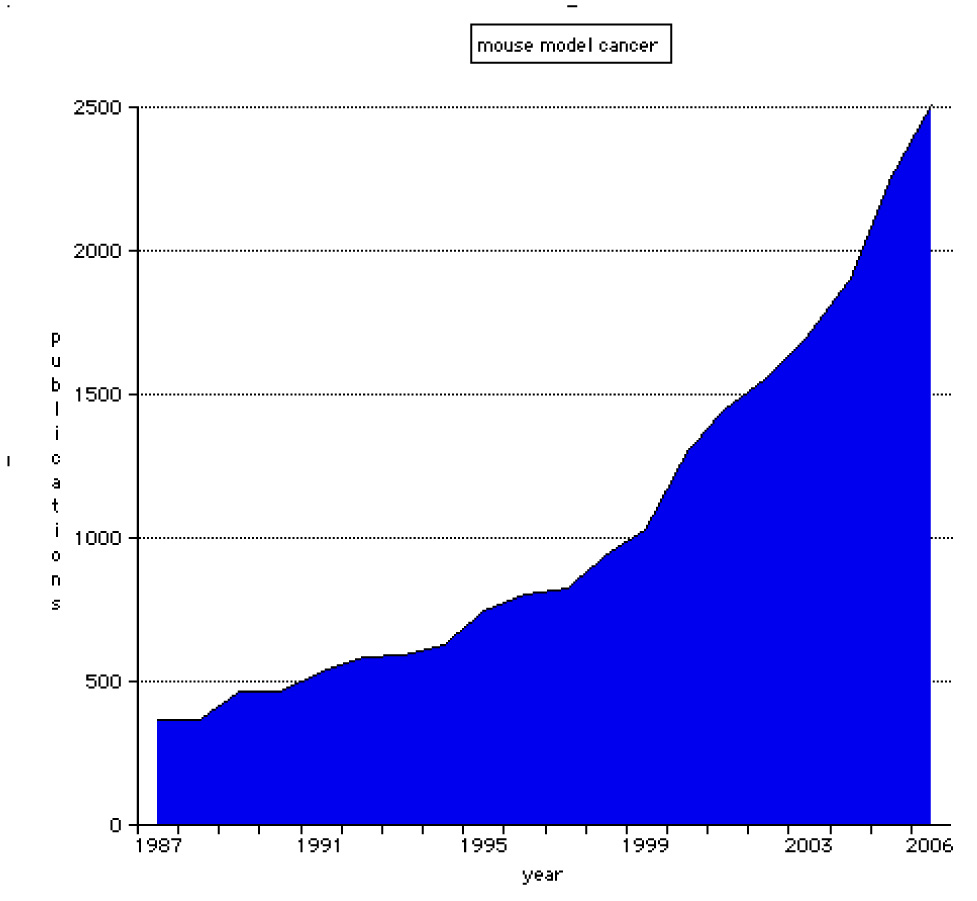
This graph was generated from data held in PubMed. A search was performed using the terms 'mouse', 'model' and 'cancer', and the numbers of matching references for each publication year were recorded.
What is MTB?
The MTB database provides web-based access to primary research data on the pathobiology of cancer in genetically defined strains of laboratory mice. Other databases on the pathobiology of cancer that provide information on mouse tumours are available and comparative examples are shown in BOX 1. The MTB project was initiated in 1997 with funding from the National Cancer Institute (NCI). The design and prototype phase of the project followed an initial review of the relevant biomedical literature and interviews with cancer geneticists. An early prototype was presented to the attendees of "The Mouse in Mammary Carcinogenesis Research" conference in October 1997 at The Jackson Laboratory (TJL) and a questionnaire was circulated requesting feedback. The database was first made public online in the autumn of 1998. Since that time the database and web interface have undergone various enhancements to respond to the demands of the cancer genetics research community. Examples of enhancements include visualization tools, a pathology data image submission interface and user help documentation13–15.
MTB in context
The Mouse Tumor Biology (MTB) database provides integrated access to data on the pathobiology and genetics of cancers in mouse models of human disease. All data are attributed to a source, either published or personal submission. Listed below are other resources available online that offer data related to that found in MTB.
Cancer Models (caMOD)35. This resource provides data on the pathobiology, often including images, and genetics of experimental models of human cancers developed in mouse, rat, hamster and zebrafish. References are provided for most of the animal models represented in caMOD, but not all data are directly attributed to a reference. Of the 4,675 mouse model records in caMOD, 4,019 were obtained directly from MTB.
Pathbase32. This resource provides access to histopathology, including images, of normal and abnormal tissues from genetically engineered mice and rats. Each record is attributed to the submitting laboratory. Several standard ontologies are incorporated into the data for each record. Unlike MTB, Pathbase does not provide information on frequencies of cancers in different strains of mice.
Cancer Genome Anatomy Project. This resource provides data on gene expression profiles from a variety of normal, precancerous and cancerous tissues from both mice and humans.
Biology of the Mammary Gland. This resource offers information on the genetics and pathobiology of mammary gland cancers, including genetic mutations and approximately 115 experimental models. Other data available include a histology atlas, information on tools and technologies, and several instructive tutorials.
UC Davis Center for Comparative Medicine Image Archive. This resource offers access to whole-slide scanned images for normal and neoplastic tissues from both mouse and human from a subset of organs. The archive does not support online submission of pathology data from the community.
The data in the MTB database are accumulated from several sources including the peer-reviewed scientific literature, pathology reports from the TJL mouse colonies and submissions from the cancer research community. All data entered into the database are attributed to a source, either published or personal communication. Data incorporated into MTB are reviewed by scientific curators with expertise in the areas of cancer biology and mouse genetics. The literature curation process begins with the identification of appropriate publications. The entire Mouse Genome Informatics (MGI) curatorial staff routinely reviews over 150 scientific journals that are determined to regularly carry articles involving some aspect of mouse biology and genetics, screening them for relevant papers. In addition, MTB staff carry out monthly PubMed searches to identify relevant articles published in other journals not covered in the primary screening process. Each paper is reviewed by a curator and information relevant to the database is entered. The scope of the data in MTB includes all endogenous tumours in mice, both spontaneous and induced, from genetically defined strains of mice (inbred, hybrid, mutant and genetically engineered strains of mice). These data include standardized tumour names and classifications, pathology reports and images, mouse genetics, genomic changes occurring in the tumour, strain names, tumour frequency and latency, and literature citations. An important part of the curation process is ensuring that data such as genes, alleles and strains all adhere to the accepted nomenclature for those data types16 (BOX 2). This requires that curators must sometimes translate published, non-conforming terminology into synonymous approved nomenclature terms.
What's in a name?
Adherence to official nomenclature for data types such as genes and strains is vital for clarity of communication. Without adherence to these standards, confusion may easily arise. For example, a researcher may have made a knockout of the ‘p75’ gene. Is this a knockout of nerve growth factor receptor (often referred to as p75)? Or is this a knockout of tumor necrosis factor receptor superfamily, member 1b (also often referred to as p75)? Without contextual clarification as to which gene is meant, the reader may be unable to determine which one was actually targeted.
The Mouse Tumor Biology (MTB) database adheres to the official nomenclature for genes, alleles and strains as determined by the guidelines of the International Committee on Standardized Genetic Nomenclature for Mice16. These data are synchronized with the corresponding data held in the Mouse Genome Database. To facilitate searching on alternate names, MTB has used a system to capture synonyms. For example, ‘cadherin 1’ is the official gene name for a gene commonly known as ‘E-cadherin’. Searching MTB for ‘E-cadherin’ in the ‘Gene/Marker’ field of the ‘Genetics’ search form will return a record for the cadherin 1-knockout strain annotated in MTB.
The use of controlled vocabularies in MTB contributes to the assurance of accuracy and completeness of database searches. For example, a researcher interested in mouse models of lung cancer might turn to PubMed to identify relevant publications on this topic. However, the search terms used and the writing style of different authors will directly affect the information that is returned from a literature search. A recent test search of PubMed for ‘mouse model lung tumor’ and ‘mouse model pulmonary tumor’ returned 2,372 and 1,452 results respectively. Only about 600 citations appeared in both searches. The remainder of the citations returned by one search would be missed by someone searching with the alternate terminology. This situation is avoided in MTB by the use of standardized terminology for cells, tissues and organs. Because of this standardization, searches on the term ‘lung’ will return all results for lesions of the lung regardless of the varying terminology used in published articles to describe them (‘lung tumor’, ‘pulmonary tumor’, ‘lung carcinoma’, ‘pulmonary adenoma’ and so on). MTB curators ensure that synonymous terms are entered into the database in a systematic manner.
Annotated histopathology images of tumours from mouse colonies held at TJL, including data from The Jackson Laboratory Aging Center's large-scale study of spontaneous diseases in commonly used inbred strains as they age, are also present in the database. Additionally, submission of mouse tumour data and photomicrographs of lesions are encouraged from the broader cancer research community, including visiting investigators at TJL. A custom-designed data submission tool is available on the MTB web site for contribution of such data.
The MTB database does not currently include data on tumour cell lines or human tumour xenografts. The project's focus on the importance of genetic background in the aetiology of cancer makes this database a particularly useful resource for scientists studying mouse models of human cancers and its expanding image archive appeals to pathologists. It can also serve as a powerful training tool for the next generation of cancer researchers.
How to use MTB
The MTB web site is designed to provide users with a variety of search options that accommodate a broad range of perspectives from within the cancer research community. This design allows users to search the database using a query designed to address their own particular area of interest. The following are examples of some of the types of queries that may be asked of and answered by the database.
First-time MTB users
First-time users of MTB might not be sure where to start in their exploration of the database. In this case the ‘Quick Organ/Tissue’ search form, located on the MTB home page, provides a simple introduction to the data and the interface. This search form consists of a pick list that contains an extensive vocabulary of mouse anatomical terms. Choosing one of the terms from the list and clicking on the ‘Search’ button will launch a search for records in which the organ of origin of the tumour matches the chosen term, regardless of tumour type. For example to answer the question “Do mice get tumours of the adrenal gland?” one could first search for ‘adrenal gland’. This returns over 200 records of adrenal gland tumours in mice; some are spontaneous tumours and others are those induced following some sort of treatment (chemical, hormonal or radiation). These lesions arose in a variety of strain types (inbred, hybrid, fostered, targeted mutant, chimeric and transgenic). Data provided on the results page include tumour name, organ affected, treatment type and agents (if applicable), strain name and type, tumour frequency and metastasis information as well as links to any related images (if available). Clicking on the ‘Summary’ hyperlink for a particular tumour record will display the ‘Tumor Summary’ page, which presents additional information regarding the selected tumour. These data may include information such as reproductive status of the affected mice, tumour frequency details (colony size and number of mice affected), tumour latency, pathology notes, tumour genetics and additional notes as well as links to the appropriate references.
If a researcher is interested primarily in spontaneous tumours in inbred mice, another useful option is the Tumor Frequency Grid (FIG. 2, Tumor Frequency Grid movie in Further information). This is a display and dynamic search tool that can be accessed by clicking on the ‘Tumor Frequency Grid’ hyperlink. The current version of the grid derives from all the current data held in MTB for spontaneous tumours in inbred mice and clicking on one of the data cells will open a ‘Tumor Results’ page as described above. The grid may be particularly useful when taking genetic background into consideration in planning the construction of a new mutant strain, choosing strains for a study of complex traits or designing other experiments.
Figure 2. The Tumor Frequency Grid.
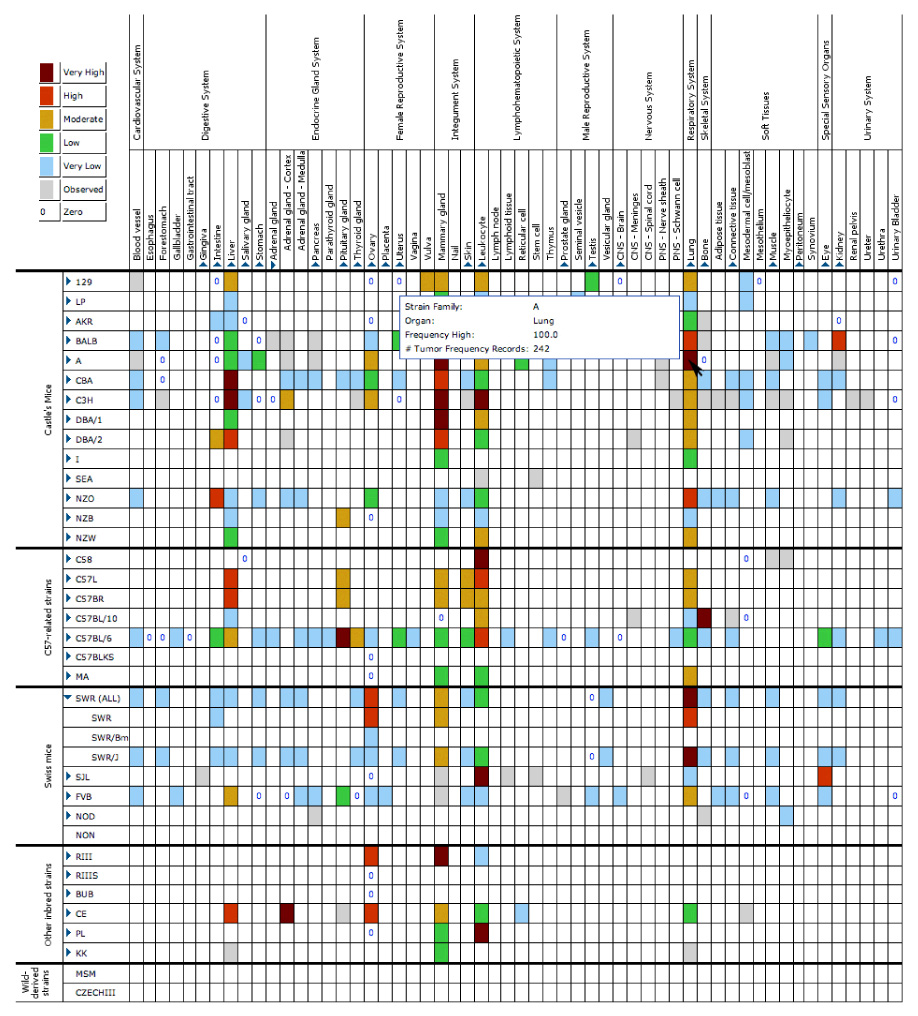
The grid provides a visual summary of the data held in the Mouse Tumor Biology (MTB) database regarding spontaneous tumours in standard inbred strains of mice. Each cell in the grid is colour-coded based on the highest reported frequency of a tumour in each particular inbred strain. Columns and rows for substrains or organ substructures can be viewed by clicking on the associated triangles beside the strain or organ to expand the selected region of the grid (note expansion shown for substrains of the SWR strain and for substructures of the adrenal gland). All of the coloured cells that contain data are also hyperlinked. Clicking on any one of them will launch a search of the database for the records of spontaneous tumours matching the combined strain and organ criteria designated by that cell and will present these results in a ‘Tumor Results’ page. The grid is updated dynamically as data are entered into MTB.
Repeat MTB visitors
After familiarizing themselves with the results of the broad search provided by the ‘Quick Organ/Tissue Search’ option, a user may decide they would like to refine their search parameters. The ‘Tumor’ search form allows users to ask more complex questions, such as “Which strains of mice spontaneously develop adrenal gland pheochromocytomas that metastasize?”. This search may be performed by choosing ‘adrenal gland’ from the ‘Organ/Tissue of Origin’ menu as before and then adding ‘pheochromocytoma’ from the ‘Tumor Classification’ menu and ‘None (spontaneous)’ from the ‘Treatment Type’ menu. Finally, the question can be restricted further by checking the box beside ‘Restrict search to metastatic tumors only’. This search will return five records for spontaneous, metastatic pheochromocytomas, all of which arose in targeted mutant mice. As with the other ‘Tumor Results’ pages, the user can view more detailed information for each tumour record by clicking on the ‘Summary’ hyperlink for the record of interest.
Geneticists and developers of mouse models of human cancers
Many researchers will be primarily interested in a particular strain (either inbred or mutant) rather than in a particular type of tumour. In this case the ‘Strain’ search form is a good starting point. For example, searching for records in which the ‘Strain Name’ contains ‘Kras’ and the ‘Strain Type’ is ‘targeted mutation (knockout)’ returns seven records for strains carrying a knockout allele of Kras, either singly or in combination with some other mutation, on a variety of genetic backgrounds. Clicking on one of the hyperlinked strain names brings the user to the ‘Strain Tumor Overview’ page on which are listed all the tumour types analysed in those particular mice. The default view for this page is the ‘Collapsed View’, which shows data on the tumour type, organ(s) affected treatment type, and includes a link to additional information on the tumours. There are also links to other resources citing these mice (when applicable). These resources include places where mice may be obtained, such as the JAX® Mice web site and the Mouse Models of Human Cancers Consortium's (MMHCC's) Mouse Repository as well as data resources such as the Mouse Phenome Database (MPD), the Biology of the Mammary Gland Web Site, and Festing's Listing of Inbred Strains of Mice (see URLs in Further information). Clicking on the ‘Strain Tumor Overview Expanded View’ link at the top of the page will expand the provided table to include metastasis data and specific related reference identifiers with hyperlinks to additional reference information. On either of the views, clicking on the hyperlinked numbers in the right-hand column will present the user with a ‘Tumor Summary’ page for that tumour type in those particular mice.
Researchers interested in investigating the role of a specific gene or genes in tumorigenesis can search MTB for specific genes using the ‘Genetics’ search. For example, a researcher studying Cdkn2a may want to explore all of the strains and tumours in MTB that have been reported to harbour a mutation in this gene. By choosing the ‘Genetics’ search form, entering Cdkn2a in the ‘Gene/Marker’ field, and clicking on the ‘Search’ button, the subsequent results page provides the user with data regarding mutant strains in which the mutation involves Cdkn2a (in this case there are both targeted mutant and transgenic strains) as well as data regarding tumours that have been reported to harbour a mutation in the Cdkn2a gene. These somatic mutations are changes such as point mutations, deletions and loss of heterozygosity. The strain section of the results page provides the user with information regarding the mutation type, mouse chromosome, gene symbol and name, and genotype of the mice. The hyperlinked numbers beside each genotype will provide users with a list of all the strains in MTB carrying the associated genotype. The tumour section of the results page provides the user with information regarding mouse chromosome, gene symbol and name, and the type of genetic change observed in the tumour. Hyperlinked numbers beside the type of genetic change will link the user to a page with additional information on the specific mutations as well as on the number and classification of the tumours found to carry those mutations. From there users may click on a link to view details of one or more of those tumour records.
Pathologists and pathology students
One of the sections of the database many users find most valuable is the pathology image archive. Researchers characterizing neoplasms from a newly developed mutant strain may wish to compare the tumours they have observed with other examples of the same tumour type from other strains of mice. This type of data can be accessed using the ‘Pathology Image’ search form. For example, one might want to compare the histopathology of mammary fibroadenomas in mice. A query for ‘mammary gland’ in the ‘Organ/Tissue of Origin’ menu and ‘fibroadenoma’ in the ‘Tumor Classification’ menu returns two pathology reports. Each report lists the tumour name, organ(s) affected, treatment type and agents (if applicable), strain name, sex, reproductive status, tumour frequency, age at necropsy and a short description. Under the report summary are thumbnail images, each with its own caption, written by a pathologist. Clicking on any of the thumbnail images will present the user with an enlarged version of the image along with all the associated data relevant to that image (FIG. 3). Many of the images currently held in MTB are displayed using the dynamic Zoomify viewer (see Zoomify and Zoomify movie URLs in Further information). This viewer allows users to zoom in and out and pan around high-resolution image files so that the image can be viewed in greater detail than would be available from a handful of static JPEG images.
Figure 3. Pathology Image Detail page.
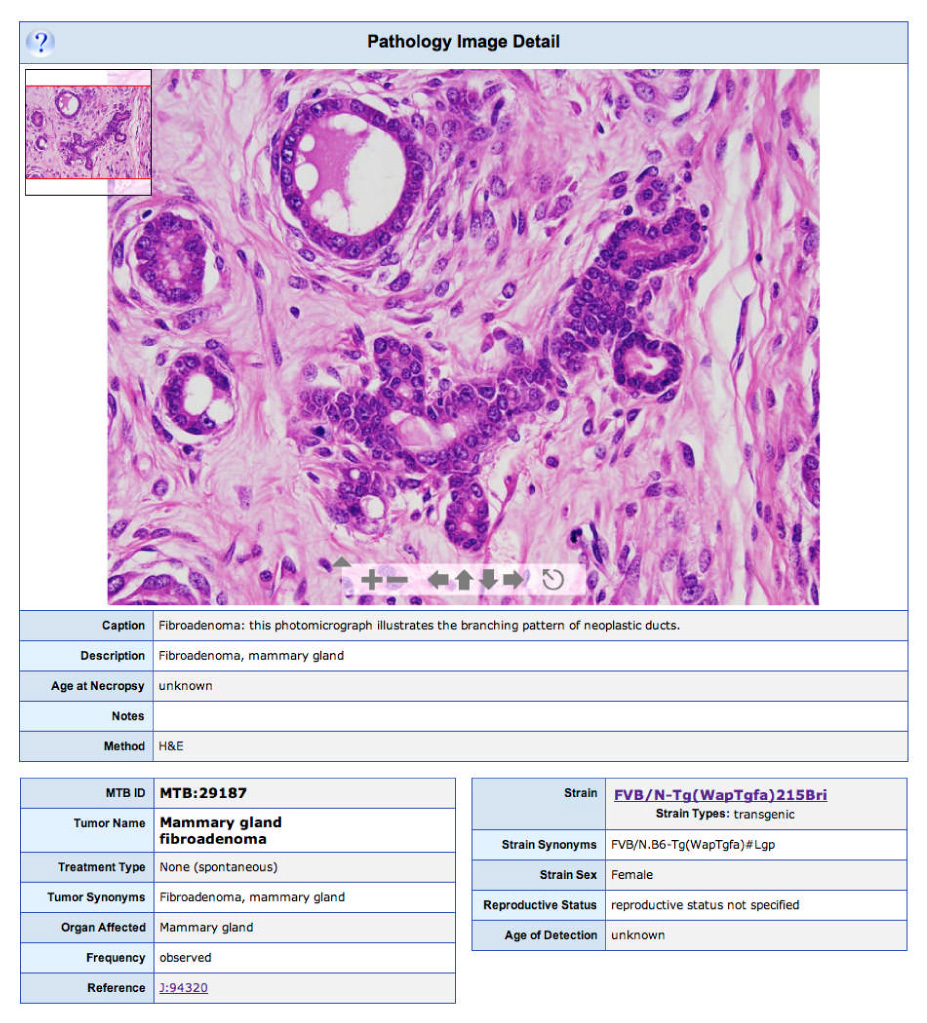
Each pathology image page shows a histopathological image; in this example a mammary gland fibroadenoma observed in a female FVB/N-Tg( WapTgfa)215Bri mouse. The embedded Zoomify interface allows users to change the magnification of the image and to scan the entire image at a range of image resolutions.
Images of tumours can also be added to MTB. These can be images from a published paper (if allowed by copyright) or they can be additional images collected during a study but not published. MTB has designed an online image submission system to facilitate community pathology image contribution through the inclusion of the ‘Submit Pathology Images’ link. Each mouse record in the submission system may be associated with one or more diagnoses, and each diagnosis may be associated with one or more images (FIG. 4). Submission of photomicrographs of cancer case material to MTB by the scientific community are much encouraged.
Figure 4. Pathology Submission Tool.
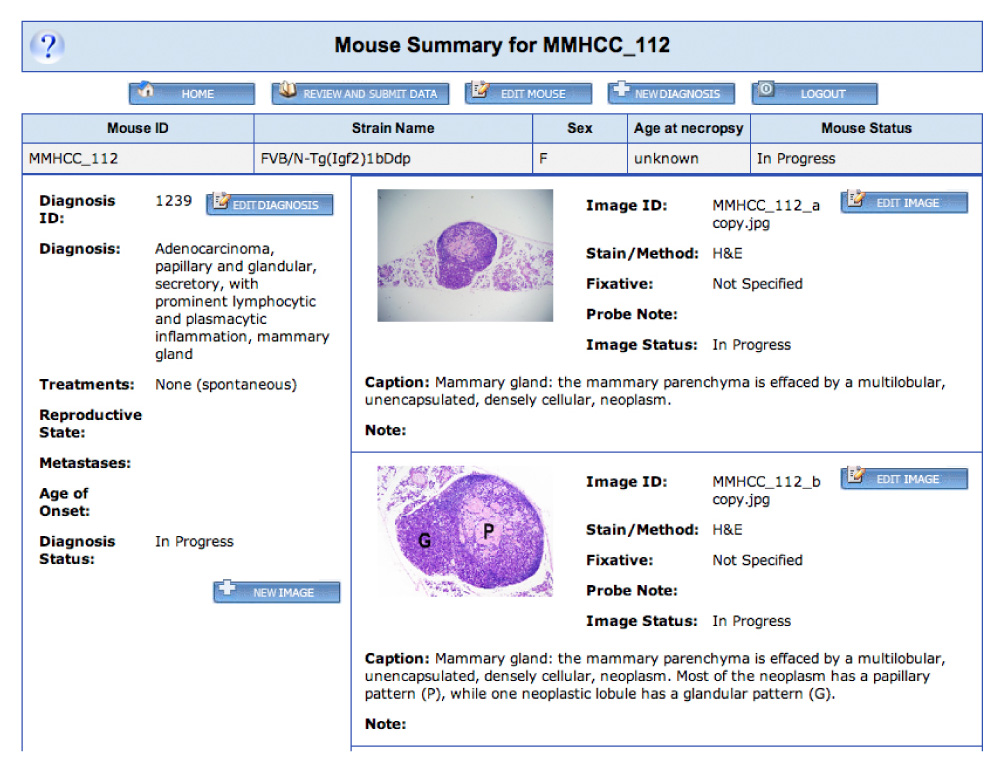
Clicking on the ‘Submit Pathology Images’ link in the left-hand menu on a Mouse Tumor Biology (MTB) web page brings the user to this submission system. New users may request a log-in and password by clicking on the ‘Register for Pathology Image Submission’ link. This opens up a new e-mail that may be filled in and sent to the MTB staff. A log-in and password will then be returned by e-mail to the user. Submissions are only accessible to the originating user, and that user has control over when his/her data are ready for release to the public MTB web site. Users may enter records for one or more mice. When a set of data is ready for submission to MTB the user clicks the ‘Review and Submit Data’ button. This will notify the MTB staff that the data are ready for release. Users may also return to submitted data for future editing or addition of subsequent images. The staff may contact the submitting researcher if clarification is needed for specifics such as strain nomenclature or the genotype of the mice. Using this password-protected interface, multiple diagnoses may be entered for each mouse, and each diagnosis may be associated with multiple images.
Experienced MTB users
Although the search forms already discussed provide the user with multiple avenues to delve into the data in the database, many users may find it most useful to perform a combination search, one that allows the user to specify both strain-specific and tumour-specific search terms. This type of search may be done using the ‘Advanced’ search form. For example, it may be useful to a researcher to search for transgenic mice that were developed on an FVB strain background and identify those mice that develop metastatic mammary adenocarcinomas (FIG. 5). Such a search returns the first 25 records of a total of 92 that are found. It is worth noting that the user may return to the search form and choose the ‘No Limit’ option to retrieve all 92 records. The results returned from this search are formatted in the same way as results from a ‘Tumor’ or ‘Quick Organ/Tissue’ search, and links to images (where available) as well as to the additional data given on the subsequent ‘Tumor Summary’ pages are also displayed.
Figure 5. The Advanced Search Form.
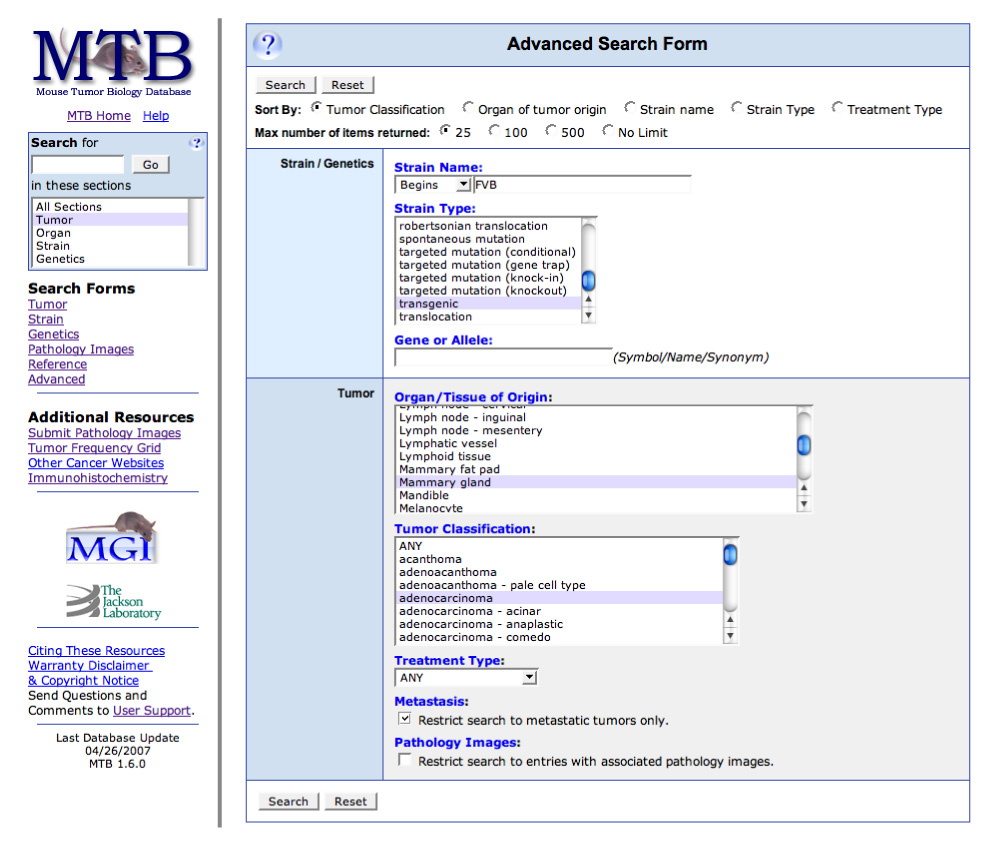
Users can use this form to perform specific or general queries, depending on their interests. Shown in this example are the selection criteria for a search for all metastatic mammary gland adenocarcinomas that arose in transgenic mice where the strain name begins with ‘FVB’. This can be done by entering ‘FVB’ in the ‘Strain Name’ field and changing the modifier from ‘Contains’ to ‘Begins’, choosing transgenic from the ‘Strain Type’ menu, ‘Mammary gland’ from the ‘Organ/Tissue of Origin’ menu, ‘adenocarcinoma’ from the ‘Tumor Classification’ menu, and clicking the check box in the ‘Metastasis’ section.
All MTB users
As many of the data in MTB are derived from the published literature, searching by reference can be a useful tool. This may be done using the ‘Reference’ search form. For example, searching for the word ‘myoepithelioma’ in the title field returns one result with a hyperlinked ID number that will access a ‘Reference Detail’ page. This page provides the user not only with details regarding the specified reference but also with a summary of the numbers of tumours, pathology images and strains that have been annotated to that reference in MTB as well as links to those data. This page also provides a link to the MGI reference page, from which users may access PubMed and, where available, download the original reference.
What's on the horizon for MTB?
After achieving the initial goal of building a prototype mouse tumour bioinformatics resource, MTB subsequently expanded in several areas such as pathology-related data and interfaces, graphic displays and visualization tools. It has also been transitioned to a multi-tier software architecture using Sybase as the relational database management system. Since the public launch of MTB in October 1998, both the amount of data held in the resource and the number of researchers using it has grown at a steady rate. After its first year online MTB held 76 pathology images. This number has recently grown to over 2,600. Over a recent 6 month period the MTB web site averaged nearly 50,000 web hits per month from approximately 1,200 unique IP addresses per month. This level of hits is 11-fold greater than that received in MTB's first 6 months online.
In order to continue to address the needs and interests of the broader cancer research community, MTB will expand to include a number of new data types in the near future. The integration of Quantitative Trait Loci (QTL) data is an area of acknowledged importance. Identification of the cancer susceptibility or resistance genes underlying these QTL in mice is an area of considerable research interest as it will probably lead to identification of human homologues that are influential in human cancers.4,17–19 Plans are also underway to incorporate and display various other types of tumour-associated genomic and genetic data, including fluorescence in situ hybridization20,21, spectral karyotyping20,22, comparative genome hybridization23–25 and gene expression array data26,27. Enhanced visualization tools for a variety of data are also under development, including the addition of improved flexibility to the Tumor Frequency Grid. This updated version will allow the user to specify the strains and organs mapped to the Grid and therefore will be configurable by the user to include data on mutant as well as inbred strains. There are also plans underway to link information on the strains in MTB with the International Mouse Strain Resource (see URL in Further information)28, thereby providing users with more comprehensive information on where to obtain a particular mutant strain of mice.
In order to better address the needs and interests of the bioinformatics and computational biology community, MTB will enhance its use of ontologies. MTB currently uses several controlled vocabularies to ensure accuracy and completeness for database searches. The integration and implementation of several standard ontologies would enhance this aspect of MTB and would also facilitate programming access to the resource. Several ontologies are under consideration for integration, including the Gene Ontology29, Cell Type Ontology (see URL in Further information), Mouse Anatomical Dictionary30, Mammalian Phenotype Ontology31, MPath Ontology (Pathbase; see URL in Further information)32 and the ontologies being developed by the MMHCC.
MTB has recently contributed to the wider cancer informatics community through participation in the NCI-initiated cancer Biomedical Informatics Grid (caBIG™)33,34, an initiative to develop an integrated data network of cancer research institutes, resources and data. Members of the MTB curatorial staff collaborate with the staff of one of the caBIG™ resources, the cancer Models (caMOD; see URL in Further information) database35, to assist in their incorporation of official standard gene and mouse strain nomenclature. Additionally, much of the mouse-related data in caMOD was acquired from MTB using exported data reports. Future plans for the MTB project include the development of programmatic access to MTB through an application programming interface to facilitate seamless integration of the data from MTB with caMOD as well as other software systems and databases. The inclusion of mouse model data in caBIG™ is a key component in the area of translational research, aiding in the application of lessons learned in experimental mouse models to clinical research.
Conclusion
For nearly a decade MTB has served as a valuable resource for the scientific community. Curation of the ever-expanding volume of mouse cancer pathobiological data remains a priority, as does the expansion of included data types and improved visualization tools. By focusing both on expanded data coverage and improved query and visualization tools, this resource will continue to assist cancer research efforts worldwide by facilitating the selection of experimental models for cancer research, the evaluation of mouse genetic models of human cancer, the review of patterns of mutations in specific cancers and the identification of genes that are commonly mutated across a range of cancers. Comments and suggestions from the cancer research community are encouraged.
Acknowledgements
For critical reading of the manuscript we thank K. Mills and B. Tennent of The Jackson Laboratory, Bar Harbor, USA. For sample images in FIG. 4 we thank J. Ward of the National Cancer Institute. The Mouse Tumor Biology Database is supported by grant CA089713 from the National Cancer Institute. Images generated from The Jackson Aging Center projects are supported by a grant from the Ellison Medical Foundation.
Biographies
DEBRA M. KRUPKE Debra Krupke received her B.A. in Biology from The Johns Hopkins University, Baltimore, Maryland, USA and did her M.S. graduate work in the laboratory of cancer biologist R. Evans at The Jackson Laboratory, Bar Harbor, Maine, USA. Since 1997 she has been working as a member of the Mouse Tumor Biology (MTB) database project, part of the Mouse Genome Informatics group at The Jackson Laboratory. She is currently a Senior Scientific Curator for MTB.
DALE A. BEGLEY Dale Begley received his B.Sc. in Biology and Ph.D. from The University of Michigan, USA studying gene regulation in the laboratory of S. Tsubota. He did his postdoctoral work at the UpJohn Company, Kalamazoo, Michigan, USA, in the laboratory of I. Abraham investigating multidrug resistance in cancer cells. He joined the Mouse Genome Informatics group at The Jackson Laboratory, Bar Harbor, Maine, USA in 1995 and is currently a Senior Scientific Curator for the Mouse Tumor Biology database project.
JOHN P. SUNDBERG John P. Sundberg is a Professor of pathology at The Jackson Laboratory, Bar Harbor, Maine, USA. He is director of the Pathology Program of The Jackson Aging Center as well as a Principal Investigator involved in developing mouse models for skin and adnexa diseases including cancer models. He has been involved with the development of the Mouse Tumor Biology database since its inception.
CAROL J. BULT Carol Bult, Ph.D., is an Associate Professor at The Jackson Laboratory, Bar Harbor, Maine, USA. Her research areas include data integration and developmental cancer genomics. She is a member of the Mouse Genome Informatics database consortium and serves as a Principal Investigator for the Mouse Genome Database, Mouse Tumor Biology Database and MouseCyc Biochemical Pathways database.
JANAN T. EPPIG Janan Eppig, Ph.D., is a Professor at The Jackson Laboratory, Bar Harbor, Maine, USA whose research interests include genome organization, model systems for human disease, bioinformatics resources and semantic standards for data integration. She is the Principal Investigator of the Mouse Genome Database and Mouse Tumor Biology Database and a member of the Mouse Genome Informatics database collaboration. She also heads the International Mouse Strain Resource.
Footnotes
FURTHER INFORMATION Biology of the Mammary Gland Web Site: http://mammary.nih.gov/
Cancer Biomedical Informatics Grid: https://cabig.nci.nih.gov
Cancer Genome Anatomy Project: http://cgap.nci.nih.gov/
Cancer Models: https://cancermodels.nci.nih.gov/
Cell Type Ontology: http://obofoundry.org/cgi-bin/detail.cgi?id=cell
Festing's Listing of Inbred Strains of Mice: http://www.informatics.jax.org/external/festing/search_form.cgi
International Mouse Strain Resource: http://www.findmice.org/
The Jackson Laboratory: http://www.jax.org/
JAX® Mice Web Site: http://jaxmice.jax.org/
MMHCC: http://emice.nci.nih.gov
MMHCC Mouse Repository: http://mouse.ncifcrf.gov/
Mouse Genome Informatics: http://www.informatics.jax.org
Mouse Nomenclature Home Page: http://www.informatics.jax.org/nomen/
Mouse Phenome Database: http://www.jax.org/phenome
Mouse Tumor Biology database: http://tumor.informatics.jax.org/
PathBase: http://www.pathbase.net
Tumor Frequency Grid movie: http://tumor.informatics.jax.org/media/MTB-TumorFrequencyGrid.mov
UC-Davis Center for Comparative Medicine Image Archive: http://imagearchive.compmed.ucdavis.edu/
Zoomify: http://www.zoomify.com/
Zoomify movie: http://tumor.informatics.jax.org/media/MTB-Zoomify.mov
References
- 1.Paigen K. One hundred years of mouse genetics: an intellectual history. I. The classical period (1902–1980) Genetics. 2003;163:1–7. doi: 10.1093/genetics/163.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tyzzer EE. A series of spontaneous tumors in mice with observations on the influence of heredity on the frequency of their occurrence. J. Med. Res. 1909;21:479–518. [PMC free article] [PubMed] [Google Scholar]
- 3.Lathrop AEC, Loeb L. Further investigations on the origin of tumors in mice. I. Tumor incidence and tumor age in various strains of mice. J. Exp. Med. 1915;22:646–673. doi: 10.1084/jem.22.5.646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lifsted T, et al. Identification of inbred mouse strains harboring genetic modifiers of mammary tumor age of onset and metastatic progression. Int. J. Cancer. 1998;77:640–644. doi: 10.1002/(sici)1097-0215(19980812)77:4<640::aid-ijc26>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 5.Demant P. Cancer susceptibility in the mouse: genetics, biology and implications for human cancer. Nature Rev. Genet. 2003;4:721–734. doi: 10.1038/nrg1157. [DOI] [PubMed] [Google Scholar]
- 6.Resor L, Bowen TJ, Wynshaw-Boris A. Unraveling human cancer in the mouse: recent refinements to modeling and analysis. Hum. Mol. Genet. 2001;10:669–675. doi: 10.1093/hmg/10.7.669. [DOI] [PubMed] [Google Scholar]
- 7.Frese KK, Tuveson DA. Maximizing mouse cancer models. Nature Rev. Cancer. 2007;7:645–658. doi: 10.1038/nrc2192. [DOI] [PubMed] [Google Scholar]
- 8.Holland EC, Hively WP, DePinho RA, Varmus HE. A Constitutively active epidermal growth factor receptor cooperates with disruption of G1 cell-cycle arrest pathways to induce glioma-like lesions in mice. Genes Dev. 1998;12:3675–3685. doi: 10.1101/gad.12.23.3675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johnson L, et al. Somatic activation of the K-ras oncogene causes early onset lung cancer in mice. Nature. 2001;410:1111–1116. doi: 10.1038/35074129. [DOI] [PubMed] [Google Scholar]
- 10.Jonkers J, Berns A. Conditional mouse models of sporadic cancer. Nature Rev. Cancer. 2002;2:251–265. doi: 10.1038/nrc777. [DOI] [PubMed] [Google Scholar]
- 11.Hahn WC, Weinberg RA. Modelling the molecular circuitry of cancer. Nature Rev. Cancer. 2002;2:331–341. doi: 10.1038/nrc795. [DOI] [PubMed] [Google Scholar]
- 12.Calbo J, Meuwissen R, van Montfort E, van Tellingen O, Berns A. Genotype-phenotype relationships in a mouse model for human small-cell lung cancer. Cold Spring Harb. Symp. Quant. Biol. 2005;70:225–232. doi: 10.1101/sqb.2005.70.026. [DOI] [PubMed] [Google Scholar]
- 13.Bult CJ, Krupke DM, Eppig JT. Electronic access to mouse tumor data: the Mouse Tumor Biology Database (MTB) project. Nucleic Acids Res. 1999;27:99–105. doi: 10.1093/nar/27.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Näf D, Krupke DM, Sundberg JP, Eppig JT, Bult CJ. The Mouse Tumor Biology Database: a public resource for cancer genetics and pathology of the mouse. Cancer Res. 2002;62:1235–1240. [PubMed] [Google Scholar]
- 15.Begley DA, et al. Mouse Tumor Biology Database (MTB): status update and future directions. Nucleic Acids Res. 2007;35:D638–D642. doi: 10.1093/nar/gkl983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eppig JT. In: The Mouse in Biomedical Research: History, Wild Mice, and Genetics. Fox JG, et al., editors. Amsterdam: Elsevier, Inc.; 2007. pp. 79–98. [Google Scholar]
- 17.Balmain A. Cancer as a complex genetic trait. Tumor susceptibility in humans and mouse models. Cell. 2002;108:145–152. doi: 10.1016/s0092-8674(02)00622-0. [DOI] [PubMed] [Google Scholar]
- 18.Dragani TA. 10 years of mouse cancer modifier loci: human relevance. Cancer Res. 2003;63:3011–3018. [PubMed] [Google Scholar]
- 19.Ewart-Toland A, Balmain A. The genetics of cancer susceptibility: from mouse to man. Toxicol. Pathol. 2004;32 Suppl. 1:26–30. doi: 10.1080/01926230490424716. [DOI] [PubMed] [Google Scholar]
- 20.Schröck E, Padilla-Nash H. Spectral karyotyping and multicolor fluorescence in situ hybridization revail new tumor-specific chromosomal aberrations. Semin. Hematol. 2000;37:334–347. doi: 10.1016/s0037-1963(00)90014-3. [DOI] [PubMed] [Google Scholar]
- 21.Kearney L. Multiplex-FISH (M-FISH): technique, developments and applications. Cytogenet. Genome Res. 2006;114:189–198. doi: 10.1159/000094202. [DOI] [PubMed] [Google Scholar]
- 22.Bayani JM, Squire JA. Applications of SKY in cancer cytogenetics. Cancer Invest. 2002;20:373–386. doi: 10.1081/cnv-120001183. [DOI] [PubMed] [Google Scholar]
- 23.Hodgson G, et al. Genome scanning with array CGH delineates regional alterations in mouse islet carcinomas. Nature Genet. 2001;29:459–464. doi: 10.1038/ng771. [DOI] [PubMed] [Google Scholar]
- 24.Hackett CS, et al. Genome-wide array CGH analysis of murine neuroblastoma reveals distinct genomic aberrations which parallel those in human tumors. Cancer Res. 2003;63:5266–5273. [PubMed] [Google Scholar]
- 25.O'Hagan RC, et al. Array comparative genome hybridization for tumor classification and gene discovery in mouse models of malignant melanoma. Cancer Res. 2003;63:5352–5256. [PubMed] [Google Scholar]
- 26.Desai KV, Kavanaugh CJ, Calvo A, Green JE. Chipping away at breast cancer: insights from Microarray studies of human and mouse mammary cancer. Endocr. Relat. Cancer. 2002;9:207–220. doi: 10.1677/erc.0.0090207. [DOI] [PubMed] [Google Scholar]
- 27.Malarkey DE, et al. Microarray data analysis of mouse neoplasia. Toxicol. Pathol. 2005;33:127–135. doi: 10.1080/01926230590888315. [DOI] [PubMed] [Google Scholar]
- 28.Strivens M, Eppig JT. Visualizing the laboratory mouse: capturing phenotypic information. Genetica. 2004;122:89–97. doi: 10.1007/s10709-004-1435-7. [DOI] [PubMed] [Google Scholar]
- 29.Ashburner M, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nature Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hayamizu TF, Mangan M, Corradi JP, Kadin JA, Ringwald M. The Adult Mouse Anatomical Dictionary: a tool for annotating and integrating data. Genome Biol. 2005;6:R29. doi: 10.1186/gb-2005-6-3-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smith CL, Goldsmith CA, Eppig JT. The Mammalian Phenotype Ontology as a tool for annotating, analyzing and comparing phenotypic information. Genome Biol. 2005;6:R7. doi: 10.1186/gb-2004-6-1-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schofield PN, et al. Pathbase: a database of mutant mouse pathology. Nucleic Acids Res. 2004;1:D512–D515. doi: 10.1093/nar/gkh124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nature editorial. Making data dreams come true. Nature. 2004;428:239. doi: 10.1038/428239b. [no author listed] [DOI] [PubMed] [Google Scholar]
- 34.caBIG Strategic Planning Workspace. The Cancer Biomedical Informatics Grid (caBIG): infrastructure and applications for a worldwide research community. Medinfo. 2007;12(Pt 1):330–334. [PubMed] [Google Scholar]
- 35.Xu F, et al. Mouse models of human cancer web-based resources. AMIA Annu. Symp. Proc. 2003:1056. [PMC free article] [PubMed] [Google Scholar]


