SUMMARY
We conducted a genome-wide scan in 46 pedigrees, with 671 phenotyped adults, from the independent nation of Samoa to map quantitative trait loci (QTLs) for adiposity-related phenotypes, including body mass index (BMI), abdominal circumference (ABDCIR), percent body fat (%BFAT), and fasting serum leptin and adiponectin. A set of 378 autosomal and 14 X chromosomal microsatellite markers were genotyped in 572 of the adults. Significant genetic correlations (0.82–0.96) were detected between pairs of BMI, ABDCIR, %BFAT and leptin. Suggestive linkages were found on 13q31 (LOD = 2.30 for leptin, LOD = 2.48 for %BFAT, LOD = 2.04 for ABDCIR, and LOD = 2.09 for BMI) and on 9p22 (LOD = 3.08 for ABDCIR and LOD = 2.53 for %BFAT). Furthermore, bivariate linkage analyses indicated that the genetic regions on 9p22 (bivariate LOD 2.35–3.10, LODeq (1df) 1.88–2.59) and 13q31 (bivariate LOD 1.96–2.64, LODeq 1.52–2.21) might harbor common major genes with pleiotropic effects. Other regions showing suggestive linkage included 4q22 (LOD = 2.95) and 7p14 (LOD = 2.64) for %BFAT, 2q13 for adiponectin (LOD = 2.05) and 19q12 for BMI-adjusted leptin (LOD = 2.03). Further fine mapping of these regions may help identify the genetic variants contributing to the development of obesity in Samoan adults.
Keywords: adiposity, linkage analysis, variance components, Samoa
INTRODUCTION
The prevalence of obesity and overweight has increased rapidly in the last 10–20 years in developed market economies and in developing countries (Popkin et al. 2006). In the U.S., approximately 65% of adults are overweight (BMI ≥ 25 kg/m2) and 31% are obese (BMI ≥ 30 kg/m2) (Hedley et al. 2004). The public health burden of overweight and obesity is substantial due to their significant risk for developing type 2 diabetes and cardiovascular diseases (Must et al. 1999, Grundy 2004). While environmental, cultural, nutritional, and lifestyle factors are important, there is considerable evidence that familial and genetic factors play important roles in the etiology of obesity (Friedman 2004, Bell et al. 2005). Complex interaction among multiple genes and their variants together with environmental factors contribute to the difficulties in identifying the genes associated with obesity related phenotypes. Nonetheless, numerous linkage studies, candidate gene and more recently genome-wide association studies have identified many chromosomal regions and genetic variants associated with obesity and related traits (Bell et al. 2005, Rankinen et al. 2006, Herbert et al. 2006, Frayling et al. 2007).
Population isolates on account of their reduced genetic variation are likely to offer considerable advantages in mapping genes associated with complex traits (Peltonen et al. 2000). The Samoans of Polynesia, with an evolutionary history of approximately 3,000 years, relative isolation, large family sizes and their recent exposure to modernization and the nutrition transition, provide a unique opportunity to identify susceptibility loci associated with adiposity-related traits (McGarvey 1991, 1994, Galanis et al. 1999, Tsai et al. 2004, Dai et al. 2007). In addition to the independent nation of Samoa (formerly known as Western Samoa), the Samoan islands of Polynesia also include the U.S. territory of American Samoa. There is substantial economic disparity between the two polities, Samoa with a largely rural agricultural economy has a per capita GDP of $2,556, while American Samoa benefits from direct and indirect aid from the U.S. and has a per capita GDP of $5,800. However, Samoans from both polities form a single socio-cultural unit with extended family relationships across the borders (Baker et al. 1986).
Genetically the Samoans of Polynesia represent a single homogenous population (Deka et al. 1994, Tsai et al. 2004). Compared to most other populations, the levels of overweight and obesity are remarkably high among Samoans. Body composition studies for Polynesians prescribe BMI values of 26–32 kg/m2, and > 32 kg/m2 to define overweight and obesity, respectively (Swinburn et al. 1999). Using these standards, in American Samoa 30% of men and 21% of women are overweight and 59% of men and 71% of women are obese, while in Samoa 39% of men and 31% of women are overweight, and 29% of men and 53% of women are obese (Keighley et al. 2006). Both the contemporary high prevalence of overweight and obesity among all Samoan adults and the differences between the two polities likely reflect an interaction between genetic susceptibility and differential exposure to the process of modernization (Keighley et al. 2006, McGarvey 1991, 1994).
Our recent genome-wide scan for adiposity-related traits among adults from American Samoa showed significant evidence for a major susceptibility locus on 6q32 influencing serum leptin levels and a region on 16q21 with significant pleiotropic effects on multiple adiposity traits (Dai et al. 2007). In the present study, we report findings from a genome-wide linkage scan on adiposity-related traits, assessed by BMI, abdominal circumference (ABDCIR), percent body fat (%BFAT), fasting serum leptin and serum adiponetin levels, among adults from Samoa, who, as described above, practice an agricultural life style to a greater extent and have less financial means available in comparison to American Samoans. Our results show suggestive linkage (LOD ≥ 1.9) with several adiposity-related traits on 2q13, 4q22, 7q14, 9p22, 13q31 and 19q12-q13 and support for pleiotropic effects on chromosome 9p22 and 13q31.
MATERIALS AND METHODS
Subjects
Recruitment in Samoa in 2003 was first based on finding individuals in Samoa who were members of American Samoa pedigrees who had been recruited in 2002. We then selected samples from villages throughout Samoa to assess geographic and economic diversity, and chose families based on available number of adult siblings. Probands and families were unselected for obesity or related phenotypes. The average size of the 46 pedigrees included in this study is 35.09 individuals, (ranging from 3 to 222) and the average number of generations is 4.28 (ranging from 2 to 8). In order to investigate the sensitivity of our linkage results to different pedigree structures, we also decomposed the 46 intact pedigrees into 196 nuclear pedigrees and performed linkage analyses using these pedigrees. The average size of the nuclear pedigrees is 4.61 with a range from 3 to 14 individuals. However, unless stated otherwise, the results discussed below use the 46 intact pedigrees.
Standard anthropometric techniques were used to measure stature, weight, abdominal circumference, and to calculate BMI (Dai et al. 2007). Bioelectrical impedance was assessed with a BIA-101Q device (RJL Systems Inc., Clinton, MI) and fat-free mass and body fat percentage (%BFAT) was calculated using equations established from body composition studies in Samoans (Swinburn et al. 1999, Keighley et al. 2006). In Samoa both impedance devices failed and %BFAT is missing for about 30% of the participants.
Fasting blood specimens were drawn after a 10-hour overnight fast. Serum leptin was assayed by RIA (ALPCO, Windham NH), serum insulin by RIA (Diagnostic Products, Inc), serum glucose using an automatic analyzer, Beckman CX4, and serum adiponectin using RIA kits (Linco, Inc., St. Charles, MI).
This study was approved by the Brown University Institutional Review Board, and the Government of Samoa, Ministry of Health, Health Research Committee.
Genotyping
Genotyping was performed using an ABI PRISM 3130XL genetic Analyzer (Applied Biosystems Inc., Foster City, CA) and internal size standard GeneScanTM 500 LIZ ® (Applied Biosystems Inc., Foster City, CA). Details regarding genotyping and quality control have been described previously (Dai et al. 2007). In total, we genotyped 378 autosomal microsatellite markers with an average intermarker spacing of 9.51 (8.06 –12.54) cM (Haldane), and 14 microsatellite markers on the X chromosome with an average intermarker spacing of 12.25 cM, in 572 Samoan adults (278 males and 294 females).
The relationship-testing programs RELPAIR (Duren et al. 2004) and PREST (McPeek and Sun 2000) were used to check for genetic relationship errors and to conservatively adjust the pedigree structure to minimize the number of pedigree structure errors. For the nuclear pedigree structure we used PEDCHECK (O’Connell and Weeks 1998) to search for Mendelianly inconsistent genotypes. All genotypes for a specific marker within a nuclear pedigree were removed if inconsistencies were found. LOKI (Heath 1997) was used to remove a small set of genotypes so as to generate an internally consistent set of genotypes for all family members. Mega2 (Mukhopadhyay et al. 2005) and the statistical software R (Version 2.5.1, The R Project for Statistical Computing) were used interactively to set up the input files for all analyses.
Statistical Analyses
We used LOKI to estimate allele frequencies for each marker and compute multipoint identity-by-descent (IBD) sharing matrices, which were then imported into the Sequential Oligogenic Linkage Analysis Routines (SOLAR) program (Almasy and Blangero 1998) to carry out autosomal variance component linkage analyses, in which maximum likelihood techniques were implemented to partition the phenotypic variance of a given trait into components attributable to an additive genetic component, a residual polygenic component, and other nongenetic (e.g., environmental) components (Amos 1994). Since LOKI employs Markov chain Monte Carlo methodology to compute multipoint IBD matrices that vary depending on the initial random seed, this results in variation of the LOD scores SOLAR estimates. To address this issue, we performed 10 independent SOLAR/LOKI autosomal scans for each trait and reported the averages of the maximum LOD scores as well as their ranges in magnitude and location over the 10 runs. For more details, please see our previous paper (Dai et al. 2007) – here we again used exactly the same statistical methodology.
Before linkage analyses, we estimated the heritability of traits of interest, in which we screened two sets of covariates for inclusion (p ≤ 0.10) while modeling familial relationship using SOLAR. These sets are [i] the demographic covariate set including age and sex, and [ii] the environmental covariate set which in addition to age and sex also includes farm work, education and cigarette smoking. Residuals after adjusted for significant covariates were generated and used for subsequent linkage analyses. Autosomal univariate linkage analyses were performed first on the intact pedigrees and then on the smaller nuclear pedigrees, in order to investigate the influence of pedigree structure on the linkage results. A LOD score ≥ 3.3 was taken as evidence of significant linkage, a LOD score ≥ 1.175 and a LOD score ≥ 1.9 were considered to show evidence of potential linkage and suggestive linkage, respectively (Lander and Kruglyak 1995).
Autosomal bivariate linkage analyses were performed on the intact pedigrees for chromosomes where we observed a maximum LOD score ≥ 1.9 and clustering of linkages to multiple phenotypes in the univariate linkage analyses. Genetic and environmental correlations between the traits were also assessed using bivariate genetic analysis techniques as implemented in SOLAR (Almasy and Blangero 1998, Almasy et al. 1997). Likelihood-ratio tests were conducted to test the null hypotheses of complete pleiotropy or coincident linkage, with a p-value cutoff of 0.05 for the rejection of null hypotheses (Almasy et al. 1997).
We performed the same X-linked multipoint linkage analyses on nuclear pedigrees as we described in our previous work (Dai et al. 2007). Mendel (Lange et al. 2001) was used here because it incorporates a proper variance-components model for mapping X-linked QTLs (Lange and Sobel 2006), yet limits our analyses to nuclear pedigrees. For the X chromosome we included all covariates from a covariate set regardless of the significance of each specific covariate.
RESULTS
Demographic statistics of the 671 phenotyped study participants are shown in Table 1. As observed previously in the American Samoan sample (Dai et al. 2007), the study sample from Samoa has remarkably high mean values for BMI, %BFAT and ABDCIR, as well as low serum adiponectin levels (Table 1). Women have higher mean serum leptin level than men, which was also observed previously in American Samoans. The mean level of education is slightly lower among males than among females, while smoking and farm work are more common among males. The gender-specific differences for some adiposity-related traits may reflect the fact that 83% of men and only 31% of the women in Samoa participate in farm work.
Our pedigree structures contain a large number of relative pairs (≥ 1,633, trait %BFAT), including ≥ 251 phenotyped sib pairs and ≥ 503 phenotyped cousin pairs that are informative for linkage analysis (Table A1, supplementary material). After adjustment of different sets of significant covariates (p-value ≤ 0.1), the residual heritability estimates (hr2) of the adiposity-related traits range from 0.26 to 0.43 (Table 2). All heritability estimates are significantly different from zero at p-value < 5 × 10−3.
Autosomal univariate multipoint linkage results with (average of 10 runs) maximum LOD scores ≥ 1.5 are listed in Table 3. Note that the ranges of magnitude in LOD scores and the ranges in location for the maximum LOD score from p-terminus are displayed as well. Differences in maximum LOD score up to 0.32 and in location up to 4.9 cM were observed over the 10 runs. In Figure 1, we plot the univariate multipoint LOD scores for chromosomes 4, 7, 9, and 13 where we observed LOD scores that reached the suggestive linkage level (LOD ≥ 1.9). Genome-wide multipoint linkage results for the primary traits, BMI, %BFAT, ABDCIR, leptin and adiponectin are displayed in Supplementary Figures A1 and A2 (supplementary material). As shown in Table 3, the highest (average) LOD score that we observed was 2.30 for leptin, close to marker D13S265 in 13q31.3. In this region we also detected suggestive linkage to BMI (LOD = 2.09) and potential linkage to %BFAT (LOD = 1.62) and ABDCIR (LOD = 1.66). Suggestive linkages were also observed for five other trait/region pairs including, %BFAT (LOD = 2.09) near marker D4S414 in 4q22.1, %BFAT (LOD = 2.19) near D7S484 in 7p14.3, ABDCIR (LOD = 2.14) near D9S285 in 9p22.3-p22.2, adiponectin (LOD = 1.96) near marker D2S160 on 2q13, and BMI-adjusted leptin in 19q12-q13.13 (LOD = 2.03) (Table 3).
Figure 1.
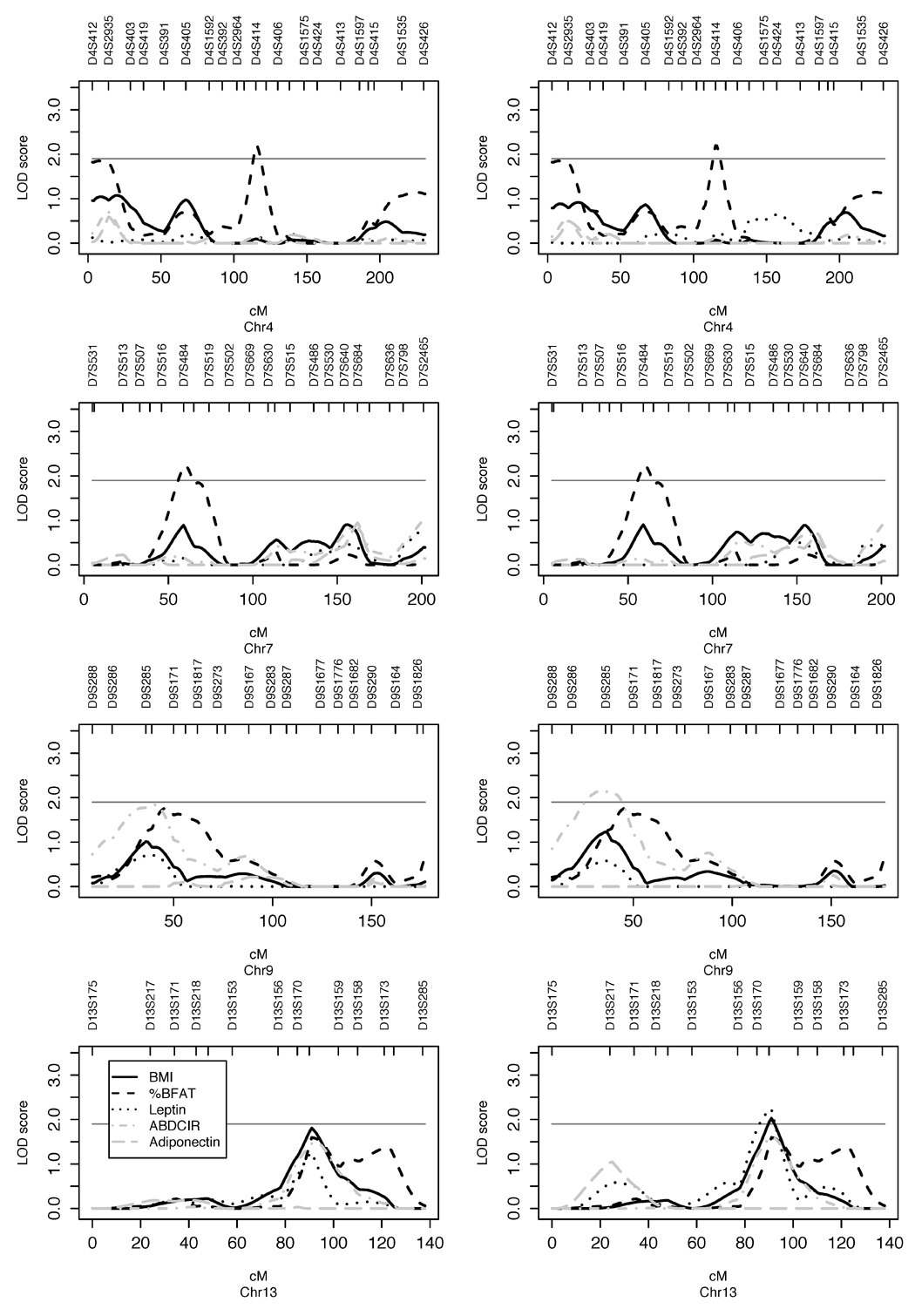
As shown in Table 4, we observed significant positive genetic correlations (0.82–0.96) and environmental correlations (0.71–0.92) between BMI, %BFAT, ABDCIR and leptin. There are negative genetic correlations between adiponectin and other adiposity-related traits, however only the correlation between adiponectin and ABDCIR (−0.48) are statistically significant.
Bivariate linkage analyses revealed a bivariate LOD score of 3.10 (p-value 2.9 × 10−4) for %BFAT-ABDCIR in 9p22.2-p21.3, where for BMI-%BFAT we obtained a bivariate LOD score of 2.79 and for BMI-ABDCIR a score of 2.35 (Table 5). On chromosome 13q31.3, right at marker D13S365, bivariate LOD scores ranged from 1.96 to 2.69 for pairs of BMI, %BFAT, ABDCIR and leptin. Table 5 also displays the “equivalent-univariate” LOD scores (LODeq), which are directly comparable to univariate LOD scores (Almasy et al. 1997). Figure 2 shows plots with the LODeq scores for chromosome 9 and 13. For both chromosomal regions studied by bivariate linkage, no tests for complete pleiotropy (Table 5, p-valuee), but all tests for coincident linkage (Table 5, p-valuef), were strongly rejected for the phenotype pairs studied. In addition, the phenotypic correlations due to QTL effects ( ρq ) range from 0.95 to 1.00, all of which are very significantly different from zero with p-values less than 0.01 (Table 5). We also performed linkage analyses of principal components from a weighted linear combination of these highly correlated phenotypes, no big difference of LOD scores were detected (data not shown)
Figure 2.
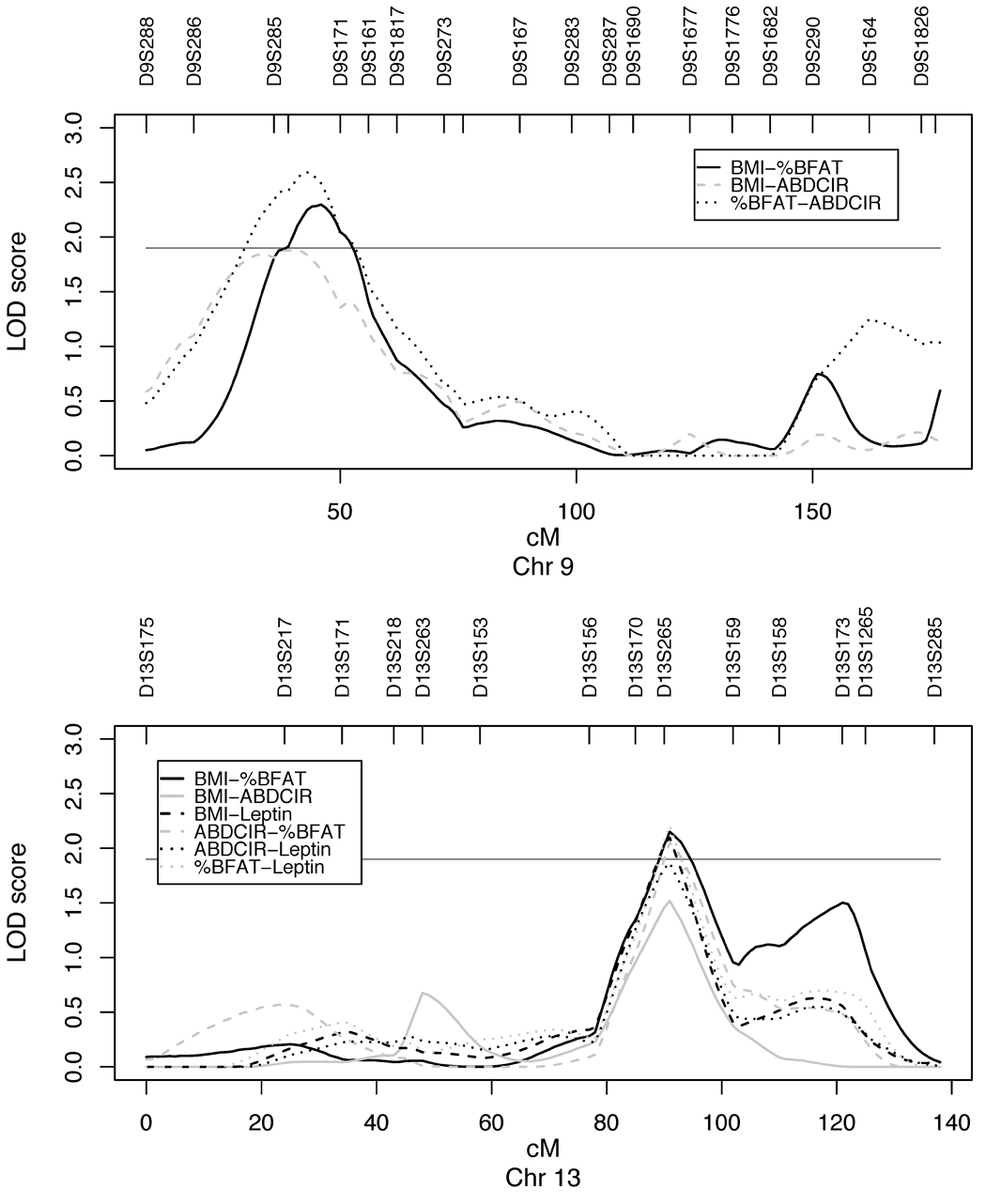
When performing a genome-wide linkage study using a nuclear pedigree structure instead of the intact pedigree structure we detected five chromosomal regions with maximum multipoint LOD scores (≥ 1.5) (Table A2, supplementary material). We detected two QTLs for %BFAT with suggestive linkage on chromosome 11q13.2 (LOD = 2.22) and on 12q23.1 (LOD = 2.18) and a QTL for ABDCIR (LOD = 1.99) on 9p22.2-p21.3. The QTL for ABDCIR was observed also when using the intact pedigree structure (Table 3).
No multipoint LOD scores > 1.0 were detected anywhere across the X chromosome. Modeling either X-linked polygenic and autosomal polygenic backgrounds together or only the latter in the X-linked analysis, as well as adjusting for environmental covariates or not (Dai et al. 2007), gave only marginal changes in the LOD scores (data not shown). X-linked linkage results from using model 4 in Table A3 (supplementary material) are plotted in Figure A1 and A2 (supplementary material).
DISCUSSION
Susceptibility chromosomal regions for adiposity-related traits
Our genome-wide linkage study using 46 Samoan pedigrees detected six chromosomal regions 2q13, 4q22.1, 7p14.3, 9p22.3-p22.2, 13q31.3 and 19q12-q13.13, with at least suggestive linkage evidences (LOD ≥ 1.9) for adult adiposity-related traits. In two of these regions, 9p22.3-p22.2 and 13q31.3, we found significant evidence for bivariate linkage and for pleiotropic effects. When these 46 pedigrees were divided into 196 nuclear families, the genome-wide linkage study showed suggestive linkage to the region on chromosome 9p22.2-p21.3 as well as to two additional chromosomal regions, 11q13.2 and 12q23.1,
Our strongest support for linkage was detected on chromosome 9p22.2-p21.3 where we found LODeq 2.59 (bivariate LOD = 3.10) for the bivariate trait %BFAT-ABDCIR and LODeq 2.30 for BMI-%BFAT (The LODeq, is comparable to univariate LOD scores). Our result indicated that a locus with significant pleiotropic effects that influence all studied trait pairs might be located on chromosome 9p22.2-p21.3. In this region we found suggestive univariate linkage to ABDCIR when using the environmental covariate set, and potential linkage to ABDCIR and to %BFAT when using the demographic covariate set, but no univariate linkage to BMI. In addition, we found suggestive linkage to 9p22.2-p21.3 for ABDCIR when using the nuclear pedigree structure. Strong evidence of linkage (LOD = 3.4) to high-density lipoprotein (HDL-C) levels has been found in Mexican Americans (Arya et al. 2002) within this 9p region and it is known that obesity is associated with lower HDL-C levels, but to our knowledge no previous studies have shown linkage to ABDCIR, %BFAT or to BMI. One possible candidate gene in this region is adipose differentiation-related protein (ADFP) gene (9p22.1, MIM 103195), which may have a role in cellular fatty acid uptake and storage (Tobin et al. 2006).
The strongest univariate linkage signal (LOD = 2.30) was detected after adjustment of measured “environmental” effects as well as for age and sex effects (the environmental covariate set), for leptin on chromosomal region 13q31.3. When only age and sex effects were adjusted for, (the demographic covariate set), a lower signal (LOD = 1.47) was detected for this trait (Figure 1, bottom left, dotted line). Furthermore, in this region we detected potential linkage to BMI, %BFAT and ABDCIR using the demographic covariate set and a slightly higher potential linkage to BMI and ABDCIR using the environmental covariate set. Similar results were observed in this region for ABDCIR in the 196 nuclear pedigrees whether we used the demographic or the environmental covariate set.
Furthermore, we found support for linkage to adiposity-related traits within chromosomal region 13q31.3 from our bivariate linkage study of pairs of leptin, BMI, %BFAT and ABDCIR with LODeq ranging from 1.52 to 2.21. We also found significant evidence for pleiotropy in this region. Chromosome 13q31.3 and its 1-LOD-drop support interval have been reported in other studies as linked to obesity-related traits (Kraja et al. 2000, Dong et al. 2005). For example, a genome-wide parent-of-origin linkage analysis by Dong et al. (2005) found strong evidence (LOD of 3.72 for BMI) for an obesity susceptibility locus with paternal effect in 13q32 in a European American sample. Recently, Saunders et al. (2007) reported suggestive evidence of linkage to BMI in a meta-analysis of 37 genome-wide linkage studies in this region (13q13.2-q33.1). In addition, this meta-analysis detected a region located on 12q23-q24 as linked to BMI in the total sample set as well as a region located on 11q13-q22 as linked to BMI-defined obesity. Interestingly, both of these regions showed suggestive linkage to %BFAT, but not to BMI, when the nuclear pedigree structure was investigated in the present study.
The chromosomal region in 7p14.3, near marker D7S484, appears to be suggestively linked to %BFAT in our present study (LOD = 2.19). One possible positional candidate gene, the neuropeptide Y gene (NPY) (7p15.1, MIM 162640) located in this region has been reported to be linked to (Bray et al. 1999a) and associated (Bray et al. 1999b) with both obesity (BMI > 32 kg/m2) and other obesity-related traits in Mexican Americans, yet its role in the etiology of common forms of obesity in this population is unclear. Another chromosomal region exhibiting suggestive linkage (LOD = 2.09) to %BFAT is 4q22.1. To our knowledge this region has not been reported as linked to adiposity-related phenotypes previously.
The chromosomal region 2q13 may contain a QTL for variation of adiponectin, with a LOD score of 1.96 near marker D2S160. At the flanking 2q14 region, Deng et al. (2002) obtained a maximum LOD score of 4.44 for BMI in their genome-wide linkage scan of obesity phenotypes. However within the current study we were not able to detect any linkage signal of interest for BMI within this region.
There is evidence of suggestive linkage to BMI-adjusted leptin (LOD = 2.03) in 19q12-q13.3. In addition, potential linkage to adiponectin (LOD = 1.87) was found within this region. The nearby 19q13 region contains two prominent candidate genes for obesity: apolipoprotein E (APOE) (19q13.32, MIM 107741) and transforming growth factor beta 1 (TGFB1) (19q13.2, MIM 190180). APOE codes a glycoprotein that plays a central role in lipid metabolism and several studies have reported positive associations of APOE with obesity phenotypes (Oh et al. 2001, Nicklas et al. 2002). The TGFB1 peptide is a multifunctional cytokine with roles in cell differentiation, and immune modulation in many cell types including adipocyte precursor cells (Petruschke et al. 1994). Long et al. (2003) have reported positive associations between APOE and TGFB1 and obesity phenotype variation in a large sample of Europeans.
Effects of genetic and environmental influences
Both in this study and in our previous study of American Samoans (Dai et al. 2007), all heritability estimates of adiposity-related traits are significantly different from zero, which demonstrates that the traits are highly heritable and that genetics plays an important role in mediating phenotypic variation. We observed significant pair-wise genetic correlations between the traits BMI, %BFAT, leptin and ABDCIR (Table 4). This implies that there might be shared genes influencing the phenotypic variation of these traits. Additional support for such pleiotropic effects is given by the bivariate linkage tests that suggest promising susceptibility loci for multiple adiposity-related traits on chromosome 9p and 13q in Samoa and on 16q in American Samoa (Dai et al. 2007). However, since the involved genetic correlations also are significantly different (p-value < 0.01) from 1 and −1, there are also distinct genetic influences on each trait.
Furthermore, for traits with significant genetic correlations, the environmental correlations between them are also statistically significant (Table 4) with similar values to the genetic correlations, which indicate the equal importance of shared genetics and shared environments in influencing the phenotypic variation of these traits.
Differences in environmental exposure demands a need for additional covariate adjustments
Despite the overall genetic homogeneity in the Samoan islands, there is still considerable variation in environmental exposures (e.g., diet, exercise, etc.) across the Samoan islands from more economically developed American Samoa to rural Samoa, which partly may be reflected in phenotypic heterogeneity observed in the two polities (Keighley et al. 2006). In general, BMI, %BFAT, ABDCIR and serum leptin levels tend to be lower and serum adiponectin levels tend to be higher in the individuals from Samoa (Table 1). Furthermore, the Samoa adult study sample, which is less influenced by modernization, tends to perform more farm work and smoke less than adults from American Samoa. In our previous studies of the influence of socioeconomic factors on cardiovascular disease risk factors in the Samoan archipelago, we have also demonstrated significant differences (p-value < 0.0001) between adults from Samoa and American Samoa in sex-stratified socioeconomic factors like basic education level, wage employment, material lifestyle score (e.g., table 2 in Ezeamama et al. 2006).
Minimizing phenotypic or genetic heterogeneity by incorporating covariates into linkage analysis could potentially increase the power to detect genetic effects. In our linkage analyses we initially considered two different models of covariate adjustments involving age, sex (the demographic covariate set), and additional environmental covariates including farm work, cigarette smoking and education (the environmental covariate set). As we were finalizing this manuscript, comments from a reviewer motivated us to consider two more complicated covariate sets including interaction between the primary covariates. Adjustments were made for sex, age, age^2, age*sex and age^2*sex (the higher-term demographic covariate set), and additional environmental covariates including farm work, education, cigarette smoking and their possible interaction terms with age, sex, age^2 (the higher-term environmental covariate set). Again only those covariates with significant effects at p-value ≤ 0.10 were retained in the polygenic models. New heritability estimates of the primary adiposity-related traits and new genome-wide linkage results are provided in table A4 and A5, respectively (supplementary material). When using the two higher-term covariate sets, we observed increased heritability estimates with more of the total variance explained by adjusted covariates (Table A4 vs. Table 2), which reinforces the importance of appropriately adjusting for environmental effects in genetic mapping analyses.
When adjusting for the higher-term covariate sets, we identified increased LOD scores on most of the regions shown in Table 3 (Table A5, supplementary material). Most prominent among these is the QTL for ABDCIR on chromosomal region 9p22.2-p21.3 where the LOD score increased from suggestive linkage of 2.14 to near significant linkage of 3.08. In addition, higher suggestive evidence of linkage was detected on 4q22.1 (LOD = 2.95), 7p14.3 (LOD = 2.64), and 13q31.3 (LOD = 2.48) to %BFAT, on 12p13.31 (LOD 2.10), 13q31.1 (LOD = 2.04) to ABDCIR, on 2q13 (LOD = 2.05), 18q22.3 (LOD = 1.94) to adiponectin. However, not all previously suggestive linkage signals increased. The LOD scores for the suggestive QTLs for BMI and for leptin in chromosomal region 13q31.1 both decreased to less than 1.5 when adjusting for the higher-term covariate set. One new suggestive linkage to %BFAT (LOD = 1.91) was obtained on chromosome 4p14.
While it might be interesting to explore multiple genetic models including various covariate adjustments, serious caution must be taken in choosing covariates prior to exploring the actual linkage results. As we have shown, adjusting for new environmental covariates and/or new interaction terms of current covariates might provide a better-fitting polygenic model, however such models should only be advocated when there is biological sense to do so. More importantly, we should not solely rank the success of a model according to the magnitude of the LOD scores because we really do not know whether there is true linkage or not in our data. In practice, adjusting for more covariates might result in a smaller sample with complete records if there are a lot of missing data in the covariate measurements. To avoid this problem, it might be possible to further develop and use a propensity score model, currently designed for bivariate traits and fully observed covariates (Doan et al. 2006), to combine multiple covariates into one covariate for adjustment.
Overlap between the Samoan and the American Samoan study
Due to its unique population history and rapid nutritional transition in the population from the Samoan islands, susceptibility loci for adiposity found in this study may or may not be identified in multiple independent studies elsewhere (Rankinen et al. 2006). Despite there being no evidence of population substructure (Tsai et al. 2004) in the population from the Samoan islands there are few overlaps of linkage signals between this scan and our previous scan of American Samoa (Table 6). Although we screened for inclusion of the same covariates from the demographic and the environmental covariate sets in the two studies, the significant covariates for the majority of phenotypes are different. It is therefore possible that adjustment for environmental variance of adiposity phenotypes was not equally successful in the two studies, which might be why we the residual heritability estimates of the traits differed in the two studies (Table 2 vs. Table 2 in Dai et al. 2007).
Another possibility of lack of overlap between the two studies of the Samoan polities could be due to variable dependence on environment factors (e.g., amount of farm work) of underlying genes. Suppose a gene only influences obesity if the calorie intake markedly exceeds the calorie burning rate. If so, it would only be detectable in a low-exercise group, such as American Samoans. Furthermore, a covariate might have a different effect size depending on how extreme the environmental variable is (i.e., differential gene expression in different environments), which again could hamper detection of genetic effects in individuals from certain genetic risk groups. In addition, unmeasured environmental variables (such as food intake or differences in material life standard) might be different between the two polities, which might have strong effect on the investigated traits. Finally, the relatively larger family size in the American Samoan study (Dai et al. 2007) vs. the Samoan study might play a role in explaining the lack of reproducibility due to variation in statistical power.
Pooling data across studies is one way to increase power in linkage analysis of complex disease (Lander and Kruglyak 1995, Wu et al. 2002). We are currently carrying out a genome-wide linkage scan for adiposity phenotypes based on a combined sample set of the Samoan families analyzed in this study and the previously analyzed American Samoan families (Dai et al. 2007). Because two different platforms were used for genotyping the American Samoan and Samoan family samples, and also because several families have members from both polities, efficient alignment of allelic fragments across the two study samples remains challenging.
In summary, we report several chromosomal regions with evidence of suggestive linkage that may harbor susceptibility genes for adiposity in adults from the independent nation of Samoa. Among these, the chromosomal region on 9p22.2-p21.3, which showed suggestive linkage to adiposity-related traits regardless of which pedigree structure was used and regardless of which covariate set was applied may be the most promising region. The current study as well as our previous study (Dai et al. 2007) identified chromosomal regions that appear to harbor genes that have pleiotropic effects on multiple adiposity traits. Furthermore, the study samples from Samoa and American Samoa with their homogenous population history but with their heterogeneous environmental settings offers a unique possibility to study gene by environmental interactions that should be taken advantage of. Further exploration of our implicated susceptibility regions by additional linkage and association studies is hence warranted. We are currently applying for a grant to perform genome-wide association analysis of the same adiposity phenotypes on this valuable dataset, which, if funded, may allow for identification of candidate genes and better understanding of biological pathways that are involved in the variation of adiposity phenotypes.
Acknowledgments
Our work is supported by NIH grant R01-DK59642 (STM PI). KÅ was supported by the Sweden-America Foundation and the Swedish Research Council. We thank the leaders of the Ministry of Health for their support and the local political officials for their permission to work in the villages. We are grateful to the families and study participants for their contributions to this research.
References
- Almasy L, Blangero J. Multipoint quantitative trait linkage analysis in general pedigrees. Am J Hum Genet. 1998;62:1198–1211. doi: 10.1086/301844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almasy L, Dyer TD, Blangero J. Bivariate quantitative trait linkage analysis, pleiotropy versus co-incident linkages. Genet Epidemiol. 1997;14:953–958. doi: 10.1002/(SICI)1098-2272(1997)14:6<953::AID-GEPI65>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Amos CI. Robust variance-components approach for assessing genetic linkage in pedigrees. Am J Hum Genet. 1994;54:535–543. [PMC free article] [PubMed] [Google Scholar]
- Arya R, Duggirala R, Almasy L, Rainwater DL, Mahaney MC, Cole S, Dyer TD, Williams K, Leach RJ, Hixson JE, MacCluer JW, O'Connell P, Stern MP, Blangero J. Linkage of high-density lipoprotein-cholesterol concentrations to a locus on chromosome 9p in Mexican Americans. Nat Genet. 2002;30:102–105. doi: 10.1038/ng810. [DOI] [PubMed] [Google Scholar]
- Baker PT, Baker TS, Hanna JM, editors. The Changing Samoans, Behavior and Health in Transition. New York: Oxford Press; 1986. [Google Scholar]
- Bell CG, Walley AJ, Froguel P. The genetics of human obesity. Nat Rev Genet. 2005;6:221–234. doi: 10.1038/nrg1556. [DOI] [PubMed] [Google Scholar]
- Bray MS, Boerwinkle E, Hanis CL. Linkage analysis of candidate obesity genes among the Mexican-American population of Starr County, Texas. Genet Epidemiol. 1999a;16:397–411. doi: 10.1002/(SICI)1098-2272(1999)16:4<397::AID-GEPI6>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- Bray MS, Boerwinkle E, Hanis CL. Sequence variation within the neuropeptide Y gene and obesity in Mexican Americans. Obes Res. 1999b;8:219–226. doi: 10.1038/oby.2000.25. [DOI] [PubMed] [Google Scholar]
- Dai F, Keighley ED, Sun G, Indugula SR, Roberts ST, Åberg K, Smelser D, Tuitele J, Jin L, Deka R, Weeks DE, McGarvey ST. Genome-wide scan for adiposity-related phenotypes in adults from American Samoa. Int J Obes. 2007;31:1832–1842. doi: 10.1038/sj.ijo.0803675. [DOI] [PubMed] [Google Scholar]
- Deng HW, Deng H, Liu YJ, Liu YZ, Xu FH, Shen H, Conway T, Li JL, Huang QY, Davies KM, Recker RR. A genomewide linkage scan for quantitative-trait loci for obesity phenotypes. Am J Hum Genet. 2002;70:1138–1151. doi: 10.1086/339934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deka R, McGarvey ST, Ferrell RE, Kamboh MI, Yu LM, Aston CE, Jin L, Chakraborty R. Genetic characterization of American and Western Samoans. Hum Biol. 1994;66:805–822. [PubMed] [Google Scholar]
- Doan BQ, Sorant AJ, Frangakis CE, Bailey-Wilson JE, Shugart YY. Covariate-based linkage analysis, application of a propensity score as the single covariate consistently improves power to detect linkage. Eur J Hum Genet. 2006;14:1018–1026. doi: 10.1038/sj.ejhg.5201650. [DOI] [PubMed] [Google Scholar]
- Dong C, Li WD, Geller F, Lei L, Li D, Gorlova OY, Hebebrand J, Amos CI, Nicholls RD, Price RA. Possible genomic imprinting of three human obesity-related genetic loci. Am J Hum Genet. 2005;76:427–437. doi: 10.1086/428438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duren WL, Epstein M, Li M, Boehnke M. RELPAIR, A Program that Infers the Relationships of Pairs of Individuals Based on Marker Data. Version 2.0.1. 2004. [Google Scholar]
- Ezeamama AE, Viali S, Tuitele J, McGarvey ST. The influence of socioeconomic factors on cardiovascular disease risk factors in the context of economic development in the Samoan archipelago. Soc Sci Med. 2006;63:2533–2545. doi: 10.1016/j.socscimed.2006.06.023. [DOI] [PubMed] [Google Scholar]
- Frayling TM, Timpson NJ, Weedon MN, Zeggini E, Freathy RM, Lindgren CM, Perry JR, Elliott KS, Lango H, Rayner NW, Shields B, Harries LW, Barrett JC, Ellard S, Groves CJ, Knight B, Patch AM, Ness AR, Ebrahim S, Lawlor DA, Ring SM, Ben-Shlomo Y, Jarvelin MR, Sovio U, Bennett AJ, Melzer D, Ferrucci L, Loos RJ, Barroso I, Wareham NJ, Karpe F, Owen KR, Cardon LR, Walker M, Hitman GA, Palmer CN, Doney AS, Morris AD, Smith GD, Hattersley AT, McCarthy MI. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316:889–894. doi: 10.1126/science.1141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman JM. Modern science versus the stigma of obesity. Nature Med. 2004;6:563–569. doi: 10.1038/nm0604-563. [DOI] [PubMed] [Google Scholar]
- Galanis D, McGarvey ST, Quested C, Sio B, Afele-Fa’amuli S. Dietary intake among modernizing Samoans, Implications for risk of cardiovascular disease. J Am Diet Assoc. 1999;99:184–190. doi: 10.1016/s0002-8223(99)00044-9. [DOI] [PubMed] [Google Scholar]
- Grundy SM. Obesity, metabolic syndrome, and cardiovascular disease. J Clin Endocrinol Metabol. 2004;89:2595–2600. doi: 10.1210/jc.2004-0372. [DOI] [PubMed] [Google Scholar]
- Keighley ED, McGarvey ST, Turituri P, Viali S. Farming and adiposity in Samoan adults. Am J Hum Biol. 2006;18:112–122. doi: 10.1002/ajhb.20469. [DOI] [PubMed] [Google Scholar]
- Kraja AT, Rao DC, Weder AB, Cooper R, Curb JD, Hanis CL, Turner ST, de Andrade M, Hsiung CA, Quertermous T, Zhu X, Province MA. Two major QTLs and several others relate to factors of metabolic syndrome in the family blood pressure program. Hypertension. 2005;46:751–757. doi: 10.1161/01.HYP.0000184249.20016.bb. [DOI] [PubMed] [Google Scholar]
- Heath SC. Markov chain Monte Carlo segregation and linkage analysis for oligogenic models. Am J Hum Genet. 1997;61:748–760. doi: 10.1086/515506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedley AA, Ogden CL, Johnson CL, Carroll MD, Curtin LR, Flegal KM. Prevalence of overweight and obesity among US children, adolescents, and adults, 1999–2002. JAMA. 2004;291:2847–2850. doi: 10.1001/jama.291.23.2847. [DOI] [PubMed] [Google Scholar]
- Herbert A, Gerry NP, McQueen MB, Heid IM, Pfeufer A, Illig T, Wichmann HE, Meitinger T, Hunter D, Hu FB, Colditz G, Hinney A, Hebebrand J, Koberwitz K, Zhu X, Cooper R, Ardlie K, Lyon H, Hirschhorn JN, Laird NM, Lenburg ME, Lange C, Christman MF. A common genetic variant is associated with adult and childhood obesity. Science. 2006;312:279–283. doi: 10.1126/science.1124779. [DOI] [PubMed] [Google Scholar]
- Lander E, Kruglyak L. Genetic dissection of complex traits, guidelines for interpreting and reporting linkage results. Nat Genet. 1995;11:241–247. doi: 10.1038/ng1195-241. [DOI] [PubMed] [Google Scholar]
- Lange K, Cantor R, Horvath S, Perola M, Sabatti C, Sinsheimer J, Sobel E. Mendel version 4.0, a complete package for the exact genetic analysis of discrete traits in pedigree and population data sets. Am J Hum Genet. 2001;69 Suppl:A1886. [Google Scholar]
- Lange K, Sobel E. Variance component models for X-linked QTLs. Genet Epidemiol. 2006;30:380–383. doi: 10.1002/gepi.20158. [DOI] [PubMed] [Google Scholar]
- Long JR, Liu PY, Liu YJ, Lu Y, Xiong DH, Elze L, Recker RR, Deng HW. APOE and TGF-beta1 genes are associated with obesity phenotypes. J Med Genet. 2003;40:918–924. doi: 10.1136/jmg.40.12.918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGarvey ST. Obesity in Samoans and a perspective on the etiology in Polynesians. Am J Clin Nutr. 1991;53 Suppl:1586–1594. doi: 10.1093/ajcn/53.6.1586S. [DOI] [PubMed] [Google Scholar]
- McGarvey ST. The thrifty gene concept and adiposity studies in biological anthropology. J Polyn Soc. 1994;103:29–42. [Google Scholar]
- McPeek MS, Sun L. Statistical tests for detection of misspecified relationships by use of genome-screen data. Am J Hum Genet. 2000;66:1076–1094. doi: 10.1086/302800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay N, Almasy L, Schroeder M, Mulvihill WP, Weeks DE. Mega2, data-handling for facilitating genetic linkage and association analyses. Bioinformatics. 2005;21:2556–2557. doi: 10.1093/bioinformatics/bti364. [DOI] [PubMed] [Google Scholar]
- Must A, Spadano J, Coakley EH, Field AE, Colditz G, Dietz WH. The disease burden associated with overweight and obesity. JAMA. 1999;282:1523–1529. doi: 10.1001/jama.282.16.1523. [DOI] [PubMed] [Google Scholar]
- Nicklas BJ, Ferrell RE, Bunyard LB, Berman DM, Dennis KE, Goldberg AP. Effects of apolipoprotein E genotype on dietary-induced changes in high-density lipoprotein cholesterol in obese postmenopausal women. Metabolism. 2002;51:853–858. doi: 10.1053/meta.2002.33337. [DOI] [PubMed] [Google Scholar]
- O'Connell JR, Weeks DE. PedCheck, a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet. 1998;63:259–266. doi: 10.1086/301904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh JY, Barrett-Connor E. Apolipoprotein E polymorphism and lipid levels differ by gender and family history of diabetes, the Rancho Bernardo Study. Clin Genet. 2001;60:132–137. doi: 10.1034/j.1399-0004.2001.600207.x. [DOI] [PubMed] [Google Scholar]
- Peltonen L, Palotie A, Lange K. Use of population isolates for mapping complex traits. Nat Rev Genet. 2000;1:182–189. doi: 10.1038/35042049. [DOI] [PubMed] [Google Scholar]
- Petruschke T, Rohrig K, Hauner H. Transforming growth factor beta (TGF-beta) inhibits the differentiation of human adipocyte precursor cells in primary culture. Int J Obes Relat Metab Disord. 1994;18:532–536. [PubMed] [Google Scholar]
- Popkin BM. Global nutrition dynamics, the world is shifting rapidly toward a diet linked with noncommunicable diseases. Am J Clin Nutr. 2006;84:289–298. doi: 10.1093/ajcn/84.1.289. [DOI] [PubMed] [Google Scholar]
- Rankinen T, Zuberi A, Chagnon YC, Weisnagel SJ, Argyropoulos G, Walts B, Pérusse L, Bouchard C. The Human Obesity Gene Map, The 2005 Update. Obesity. 2006;14:529–644. doi: 10.1038/oby.2006.71. [DOI] [PubMed] [Google Scholar]
- Saunders CL, Chiodini BD, Sham P, Lewis CM, Abkevich V, Adeyemo AA, de Andrade M, Arya R, Berenson GS, Blangero J, Boehnke M, Borecki IB, Chagnon YC, Chen W, Comuzzie AG, Deng HW, Duggirala R, Feitosa MF, Froguel P, Hanson RL, Hebebrand J, Huezo-Dias P, Kissebah AH, Li W, Luke A, Martin LJ, Nash M, Ohman M, Palmer LJ, Peltonen L, Perola M, Price RA, Redline S, Srinivasan SR, Stern MP, Stone S, Stringham H, Turner S, Wijmenga C, A.Collier D. Meta-analysis of genome-wide linkage studies in BMI and obesity. Obesity. 2007;15:2263–2275. doi: 10.1038/oby.2007.269. [DOI] [PubMed] [Google Scholar]
- Swinburn BA, Ley SJ, Carmichael HE, Plank LD. Body size and composition in Polynesians. Int J Obes. 1999;23:1178–1183. doi: 10.1038/sj.ijo.0801053. [DOI] [PubMed] [Google Scholar]
- Tobin KA, Harsem NK, Dalen KT, Staff AC, Nebb HI, Duttaroy AK. Regulation of ADRP expression by long-chain polyunsaturated fatty acids in BeWo cells, a human placental choriocarcinoma cell line. J Lipid Res. 2006;47:815–823. doi: 10.1194/jlr.M500527-JLR200. [DOI] [PubMed] [Google Scholar]
- Tsai HJ, Sun GY, Smelser D, Viali S, Tufa J, Jin L, Weeks DE, McGarvey ST, Deka R. Distribution of genome-wide linkage disequilibrium based on microsatellite loci in the Samoan population. Hum Genomics. 2004;1:327–334. doi: 10.1186/1479-7364-1-5-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Cooper RS, Borecki I, Hanis C, Bray M, Lewis CE, et al. A combined analysis of genomewide linkage scans for body mass index from the National Heart, Lung, and Blood Institute Family Blood Pressure Program. Am J Hum Genet. 2002;70:1247–1256. doi: 10.1086/340362. [DOI] [PMC free article] [PubMed] [Google Scholar]


