Abstract
While a number of strategies have been developed to reduce data collection requirements for multidimensional NMR based on non-Fourier methods of spectrum analysis, there is an increasing awareness that the principal differences in the performance of these methods is attributable to the sampling strategies employed, and not the method of spectrum analysis per se. The ability of maximum entropy reconstruction to utilize essentially arbitrary sampling schemes makes it an useful platform for comparative analysis of sampling strategies. Here we use maximum entropy reconstruction to demonstrate that artifacts characteristic of sparse sampling result from regularity in the sampling pattern, and that they can be substantially reduced by introducing a degree of randomness to an otherwise regular sampling scheme, without requiring additional sampling.
Introduction
A key limiting factor in the application of multidimensional NMR spectroscopy is the time required to collect sufficient data to achieve high resolution in the indirect dimensions. Using conventional methods of spectrum analysis, the data must be sampled at regular intervals that are short enough to avoid aliasing. These dual constraints make it difficult to realize the potential resolution afforded by high-field magnets in a reasonable amount of time. To avoid this problem, a host of methods that can provide high-resolution spectral estimates from short data records have been developed. These include (but are not limited to) reduced dimensionality (RD) [1], GFT [2], and back-projection reconstruction (BPR) techniques [3], maximum entropy (MaxEnt) reconstruction [4; 5], Bayesian [6] and maximum likelihood [7] methods, filter diagonalization [8], multi-way decomposition [9], and covariance NMR [10].
Although they appear quite different, RD, GFT, and BPR approaches all utilize the concept of coupled evolution periods to selectively sample the data, resulting in sampling along radial vectors in the indirect time dimensions. We recently showed that radial sampling can be approximated by samples selected from a conventional Cartesian grid of samples determined by the Nyquist constraints, and that BPR and MaxEnt reconstruction yield similar results when applied to the same data [11]. The restriction to the Nyquist grid using MaxEnt is necessitated by practical, rather than fundamental reasons, as efficient MaxEnt algorithms utilize the fast Fourier transform (FFT) to compute gradients. The similarity of results obtained using different methods applied to similar data suggests that the differences among the methods derive mainly from the sampling strategy, and not the method of spectrum estimation.
Prominent ridge artifacts are characteristic of BPR spectra constructed from small numbers of radial samples [12]. These artifacts diminish with the collection of additional radial samples, however at the cost of additional experiment time. Using the ability of MaxEnt to process data collected at essentially arbitrary times (provided only that they fall on the Nyquist grid), we demonstrate here that the ridge artifacts can be suppressed by randomizing the sampling, in effect “blurring” the radial sampling vector, without collecting additional samples. This suggests that a general strategy for optimizing sparse sampling strategies is to maximize the “incoherence”. As a final example we illustrate the power of incoherent sampling applied to deliberately undersampled data.
Randomness has been and continues to be useful in other fields of NMR. For example, before the advent of pulsed field gradients, one of the most effective ways to eliminate zero-quantum artifacts from NOESY spectra was to add a small amount of random "dither" to the NOE mixing period [13]. Randomization of evolution times remains a useful technique for minimizing "t1 noise" in relaxation and other lengthy multidimensional experiments [14]. For very weak signals, added noise can improve sensitivity by increasing the frequency with which weak signals generate voltages that exceed the least significant bit of the digitizer. The use of random excitation, as opposed to random sampling, is the basis for stochastic NMR [15], which offers a number of advantages over conventional multiple-pulse experiments.
Random sampling in the time domain was introduced as a strategy for reducing experiment time for multidimensional NMR experiments by Barna et al. [4; 16] and subsequently employed by many investigators [17; 18; 19; 20]. In this manuscript we demonstrate the benefits of combining randomness with other sampling strategies.
Results and Discussion
MaxEnt reconstructions were computed using the Rowland NMR Toolkit and an automated protocol for determining the parameters governing the reconstruction [21]. Figure 1 illustrates a 1H-13C plane (15N chemical shift 121.96 ppm) extracted from the HNCO spectrum of ubiquitin using radial sampling corresponding to 3 (0°, 45°, 90°), 5 (0°, 22.5°, 45°, 67.5°, 90°), and 9 (0°, 11.25°, 22.5°, 33.75°, 45°, 56.25°, 67.5°, 78.75°, 90°) projections. Contours are plotted at multiples of 1.4 starting from 3 per cent of the height of the highest peak (white) and 0.5 per cent of the highest peak (blue). While the high contour levels show that the major peaks are reasonably well characterized even for 3 projections, the lower (blue) contours reveal that the dynamic range, and thus the ability to detect weak peaks, is severely compromised. Utilizing additional projections diminishes the sampling artifacts, but at the cost of additional instrument time. The numbers of samples in the indirect dimensions for the 3, 5 and 9 projection data sets are 382, 634, and 1132, respectively.
Figure 1.

1H/13C plane (15N chemical shift 121.96 ppm) from the HNCO spectrum of Ubiquitin, using data collected at 9.4 T (400 MHz for 1H) on a Varian Inova instrument. Spectra were computed using MaxEnt reconstruction and radial sampling corresponding to 3, 5, and 9 projections (left to right). Top: sampling schedule. Middle/Bottom: MaxEnt spectrum. White contour levels are plotted at multiples of 1.4 starting with 3% of the height of the highest peak. Blue contours start with 0.5% of the height of the highest peak. One-dimensional cross-sections at the frequency indicated by the horizontal line on the contour plots are shown at the bottom. Data used in this and subsequent figures fall on the same Nyquist grid.
Figure 2 illustrates the use of blurred radial sampling. Blurring is achieved by adding random numbers from a two-dimensional distribution characterized by the root-mean-square (RMS) variation measured in points. The top panels illustrate the two-dimensional sampling schedules (corresponding to the indirect dimensions of the HNCO experiment) with RMS blurring of 0, 0.625, and 1.25. The bottom panels depict the corresponding MaxEnt spectra, with contour levels determined as in Figure 1. Blurring clearly diminishes the sampling artifacts, without requiring additional experiment time or additional post-processing.
Figure 2.

As in Figure 1, but using 5 projections with different amounts of random “blurring” of the sampling schedule (RMS zero (none), 0.625 and 1.25, left to right). Top: sampling schedules. Middle/Bottom: MaxEnt spectra; the one-dimensional cross-sections are multiplied by 2 relative to Fig. 1.
Blurring can improve spectral estimates obtained from virtually any sampling scheme, not just radial sampling. To demonstrate this, Figure 3 illustrates the results of blurring applied to deliberately-undersampled data. The top panels depict sampling schedules starting with every fourth point along both time dimensions, with increasing levels of blurring. MaxEnt spectra obtained for synthetic data consisting of four exponentially decaying sinusoids are shown in the bottom panels. The aliased peaks in the spectrum rapidly diminish with additional blurring. However too much blurring can result in diminishing returns, as illustrated by the results obtained using completely random sampling.
Figure 3.

MaxEnt spectra for synthetic two-dimensional data consisting of five exponentially decaying sinusoids plus noise. The left-most panels depict deliberate undersampling selecting every fourth point along both dimensions. The center and right panels depict blurred undersampling, RMS 0.625 and 1.25, respectively. Contour levels were chosen as for Figs. 1 and 2.
One way to characterize the incoherence of a sampling schedule is to examine the discrete Fourier transform of a time series consisting of the values 1 for every sampled interval and zero for intervals not sampled. The resulting spectrum is called a point-spread function (PSF) in image processing, but we suggest the term “sampling spectrum” as more evocative in the present context. To a very good approximation the artifacts that appear in spectra obtained from NUS data can be described as resulting from convolution of the sampling spectrum with the true spectrum.
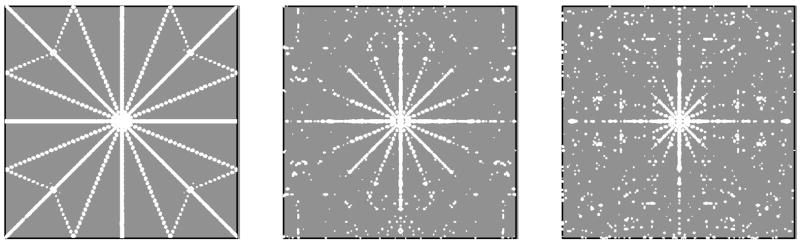
Figure 4 depicts the sampling spectra computed for the schedules used in Figure 2. A useful measure of the incoherence of the schedules is the largest absolute values outside the central (zero frequency) peak, relative to the absolute value of the central peak. Large peaks other than the central peak effectively limit the useful dynamic range of the sampling schedule, as real peaks smaller than this (relative to the strongest peak) become difficult to distinguish from sampling artifacts. The normalized absolute values diminish with increasing sample size and with randomization; for the sampling schedules shown in Fig. 2 they monotonically decrease in the order 21.7, 11.5, 10.3 with rms blurring of zero (non-blurred), 0.625, and 1.25, respectively.
Figure 4.
Two-dimensional sampling spectra (PSFs) for the sampling schedules used in Fig. 2. Contours are plotted at multiples of 1.4 starting with 3% of the height of the central (zero, zero) frequency peak.
Conclusions
NUS is a very general method for obtaining high-resolution spectral estimates from relatively short data records in multidimensional NMR, and should enable the potential resolution afforded by high field magnets to be routinely achieved in the indirect dimensions. All of the various methods proposed for spectral analysis of NUS data are nonlinear, and additional investigation to fully characterize the nature and extent of the nonlinearities, especially for noisy and high dynamic-range data, is needed. Characterization of the sampling schedules is applicable to each of the methods, however, and the degree of incoherence of the sampling schedules as derived from the PSF is one of many possible metrics for assessing the relative performance of NUS strategies. We will report the results of a comprehensive survey of metrics for NUS strategies in a future publication.
Acknowledgments
We thank Drs. Alan Stern and Mehdi Mobli for useful discussions. This work was supported by grants from the National Institutes for Health (RR020125, GM47467, GM072000).
References
- 1.Ding K, Gronenborn AM. Novel 2D triple-resonance NMR experiments for sequential resonance assignments of proteins. J Magn Reson. 2002;156:262–8. doi: 10.1006/jmre.2002.2537. [DOI] [PubMed] [Google Scholar]
- 2.Kim S, Szyperski T. GFT NMR, a new approach to rapidly obtain precise high-dimensional NMR spectral information. J Amer Chem Soc. 2003;125:1385–1393. doi: 10.1021/ja028197d. [DOI] [PubMed] [Google Scholar]
- 3.Kupce E, Freeman R. Projection-reconstruction of three-dimensional NMR spectra. J Am Chem Soc. 2003;125:13958–9. doi: 10.1021/ja038297z. [DOI] [PubMed] [Google Scholar]
- 4.Barna JCJ, Laue ED. Conventional and exponential sampling for 2D NMR experiments with application to a 2D NMR spectrum of a protein. J Magn Reson. 1987;75:384–389. [Google Scholar]
- 5.Stern AS, Li KB, Hoch JC. Modern spectrum analysis in multidimensional NMR spectroscopy: comparison of linear-prediction extrapolation and maximum-entropy reconstruction. J Am Chem Soc. 2002;124:1982–93. doi: 10.1021/ja011669o. [DOI] [PubMed] [Google Scholar]
- 6.Bretthorst GL. Bayesian Analysis I. Parameter estimation using quadrature NMR models. J Magn Reson. 1990;88:533–551. [Google Scholar]
- 7.Chylla RA, Markley JL. Theory and application of the maximum likelihood principle to NMR parameter estimation of multidimensional NMR data. J Biomol NMR. 1995;5:245–58. doi: 10.1007/BF00211752. [DOI] [PubMed] [Google Scholar]
- 8.Hu H, De Angelis AA, Mandelshtam VA, Shaka AJ. The multidimensional filter diagonalization method. J Magn Reson. 2000;144:357–66. doi: 10.1006/jmre.2000.2066. [DOI] [PubMed] [Google Scholar]
- 9.Orekhov VY, Ibraghimov IV, Billeter M. MUNIN: a new approach to multidimensional NMR spectra interpretation. J Biomol NMR. 2001;20:49–60. doi: 10.1023/a:1011234126930. [DOI] [PubMed] [Google Scholar]
- 10.Trbovic N, Smirnov S, Zhang F, Bruschweiler R. Covariance NMR spectroscopy by singular value decomposition. J Magn Reson. 2004;171:277–83. doi: 10.1016/j.jmr.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 11.Mobli M, Stern AS, Hoch JC. Spectral reconstruction methods in fast NMR: reduced dimensionality, random sampling and maximum entropy. J Magn Reson. 2006;182:96–105. doi: 10.1016/j.jmr.2006.06.007. [DOI] [PubMed] [Google Scholar]
- 12.Kupce E, Freeman R. Fast multidimensional NMR: radial sampling of evolution space. J Magn Reson. 2005;173:317–21. doi: 10.1016/j.jmr.2004.12.004. [DOI] [PubMed] [Google Scholar]
- 13.Macura S, Huang Y, Sutter Y, Ernst RR. Two-dimensional chemical exchange and cross-relaxation spectroscopy of coupled nuclear spins. J Magn Reson. 1981;43:259–281. [Google Scholar]
- 14.Bowyer PJ, Swanson AG, Morris GA. Randomized Acquisition for the Suppression of Systematic F1 Artifacts in Two-Dimensional NMR Spectroscopy. J Magn Reson. 1999;140:513–515. doi: 10.1006/jmre.1999.1882. [DOI] [PubMed] [Google Scholar]
- 15.Blümich B. Stochastic NMR Spectroscopy. Bull Magn Reson. 1985;7:5–26. [Google Scholar]
- 16.Barna JCJ, Laue ED, Mayger MR, Skilling J, Worrall SJP. Exponential sampling: an alternative method for sampling in two dimensional NMR experiments. J Magn Reson. 1987;73:69. [Google Scholar]
- 17.Kazimierczuk K, Zawadzka A, Kozminski W, Zhukov I. Random sampling of evolution time space and Fourier transform processing. J Biomol NMR. 2006;36:157–68. doi: 10.1007/s10858-006-9077-y. [DOI] [PubMed] [Google Scholar]
- 18.Pannetier N, Houben K, Blanchard L, Marion D. Optimized 3D-NMR sampling for resonance assignment of partially unfolded proteins. J Magn Reson. 2007;186:142–9. doi: 10.1016/j.jmr.2007.01.013. [DOI] [PubMed] [Google Scholar]
- 19.Schmieder P, Stern AS, Wagner G, Hoch JC. Application of nonlinear sampling schemes to COSY-type spectra. J Biomol NMR. 1993;3:569–76. doi: 10.1007/BF00174610. [DOI] [PubMed] [Google Scholar]
- 20.Schmieder P, Stern AS, Wagner G, Hoch JC. Improved resolution in triple-resonance spectra by nonlinear sampling in the constant-time domain. J Biomol NMR. 1994;4:483–90. doi: 10.1007/BF00156615. [DOI] [PubMed] [Google Scholar]
- 21.Mobli M, Maciejewski MW, Gryk MR, Hoch JC. Automatic maximum entropy spectral reconstruction in NMR. J Biomol NMR. 2007;39:133–9. doi: 10.1007/s10858-007-9180-8. [DOI] [PubMed] [Google Scholar]