Summary
The replicon model devised to explain replication control in bacteria has served as the guiding paradigm in the search for origins of replication in the more complex genomes of eukaryotes. In S. cerevisiae, this model has proved to be extremely useful, leading to the identification of specific genetic elements (replicators) and the interacting initiator proteins that activate them. However, replication control in organisms ranging from S. pombe to mammals is far more fluid: only a small number of origins seem to represent classic replicators, while the majority correspond to zones of inefficient, closely-spaced start sites none of which are indispensable for origin activity. In addition, it is apparent that the epigenetic state of a given sequence largely determines its ability to be used as a replication initiation site. These conclusions were arrived at over a period of three decades, and required the development of several novel replicon mapping techniques, as well as new ways of examining the chromatin architecture of any sequence of interest. Recently, methods have been elaborated for isolating all of the active origins in the genomes of higher eukaryotes en masse. Microarray analyses and more recent high-throughput sequencing technology will allow all the origins to be mapped onto the chromosomes of any organism whose genome has been sequenced. With the advent of whole-genome studies on gene expression and chromatin composition, the field is now positioned to define both the genetic and epigenetic rules that govern origin activity.
DNA fiber autoradiographic studies on yeast and mammalian genomes showed that origins of replication are bidirectional (Newlon et al., 1974; Huberman and Riggs, 1968) and are spaced, on average, 30 kb and 100 kb apart, respectively. Not surprisingly, the replicon model proposed in 1963 to explain the regulation of DNA replication in bacteria and their episomes (Jacob and Brenner, 1963) has been the guiding paradigm for virtually all subsequent studies directed toward the isolation and characterization of origins from eukaryotic genomes. In this model, a replicon was defined as a functional unit that contains two important elements: a gene encoding a trans-acting initiator and a mutable genetic replicator that directs the initiator (and initiation) to itself. Obviously, the model has had to be adjusted to accommodate the fact that eukaryotic chromosomes contain hundreds to thousands of individual origins (Newlon et al., 1974; Huberman and Riggs, 1968) that apparently are activated in each case by a single set of initiation factors.
The replicon model was so successful in guiding explorations in yeast that it promised to hold throughout the eukaryotic world. However, there has been a serious paradigm shift in the last decade or so because many aspects of the model do not adequately describe regulation of initiation in eukaryotic organisms more evolved than S. cerevisiae. In this article, we will summarize the relevant data that have led to this paradigm shift. Our focus will be largely on the localization and characterization of DNA sequences that function as origins of replication in complex genomes - whether they be true replicators or not. There also has been an explosion of information in recent years concerning initiators in higher eukaryotic organisms, as well as epigenetic requirements for determining the activity of origins as well as the time of firing in the S-period. We refer the reader to excellent recent reviews that cover these important concepts (Bell and Dutta, 2002; Aladjem et al., 2006; Zhou et al., 2005), although we will briefly describe the state-of-the-art in these areas where relevant.
Identifying eukaryotic replicators phenotypically
A critical advance in the eukaryotic replication field was the isolation of autonomously-replicating sequence (ARS) elements from the S. cerevisiae and S. pombe genomes(Stinchcomb et al., 1979; Chan and Tye, 1980; Wright et al., 1986). In S. cerevisiae, ARS elements are ∼100 bp in length and are characterized by a required consensus sequence (5′-[A/T]TTTAT[A/G]TTT[A/T]-3′) as well as two redundant, non-conserved, auxiliary elements (recently reviewed in Aladjem et al., 2006). Identification of an ARS consensus allowed the isolation of the ORC complex (Bell and Stillman, 1992), which functions at origins as the initiator in this and apparently all other eukaryotes (although it has other non-origin targets). ARS elements from S. pombe are much larger (>500 bp) and consist of clusters of As and Ts that are asymmetrically distributed. Deletion of these elements in selected origins does, in fact, lower origin activity (reviewed in Dai et al., 2005; Kim and Huberman, 2001); however, there is no recognizable consensus element analogous to the S. cerevisiae ARS element, and unrelated AT-rich sequences can substitute in ARS assays without inactivating origin function. Thus, although ARS elements in S. cerevisiae and S. pombe both represent true, mutable, replicators, those in S. pombe clearly deviate from the original replicator paradigm.
Two-dimensional (2-D) gel replicon mapping methods that take advantage of the unique physical properties of restriction fragments containing either internal replication bubbles (i.e., origins), single forks, or termination structures (Brewer and Fangman, 1987; Nawotka and Huberman, 1988) have shown that most S. cerevisiae ARS elements correspond to legitimate chromosomal origins (reviewed in Aladjem et al, 2006). In both yeasts, most origins are not active 100% of the time, with some being activated in fewer than 10% of cell cycles. Thus, bona fide termini defining individual replicons are not likely to be common in these genomes. A corollary to this inefficiency is that closely-spaced potential initiation sites are unlikely to interfere with each other by firing on the same template, as has been reported for very active origins juxtaposed on plasmids (Brewer and Fangman, 1993).
The ARS assay, when applied to complex mammalian genomes, has not led to the identification of verifiable genetic replicators. For example, when a cloned human genomic library was transfected into a human cell line (Krysan et al., 1989), virtually every cloned fragment replicated autonomously to some degree (even those of bacterial origin), but the larger fragments were much more efficient. This result suggested either that origins of replication are distributed at very frequent intervals in mammalian genomes, or that virtually any sequence, when taken out of context, can serve as a template for initiation at a measurable frequency. These suggestions were compatible with observations that initiation occurs at closely-spaced, apparently random, sequences in the early cleavage stages of Xenopus laevis and Drosophila melanogaster development (e.g., Blumenthal et al., 1974). Furthermore, virtually any DNA sequence (even from viruses and bacterial plasmids) can replicate in vitro in frog egg extracts prior to the mid-blastula transition when transcription begins (Harland and Laskey, 1980).
Identifying origins and potential replicators by biochemical methods that localize start sites
Given the lack of a useful ARS assay for rescuing higher eukaryotic replicators, identification of origins has relied on the biochemical approach. The several origin mapping strategies all have required that the region under study be cloned and mapped. As a consequence, the majority of chromosomal regions were characterized initially because of previous interest in their respective genes. Methods for origin localization fall into several categories (reviewed in Hamlin, 1992): 1) labeling nascent strands in early S-phase with radioactive thymidine or BUdR, which are then used to detect the earliest fragments synthesized in a region of interest; 2) monitoring the direction of fork movement through a defined region; 3) determining the position at which either leading or lagging strands switch from one template to the other; or 4) monitoring the development and expansion of replication bubbles around a nascent strand start site.
The first mammalian origin was identified in the 240 kb amplified dihydrofolate reductase (DHFR) domain of the methotrexate-resistant CHO cell line, CHOC 400. This locus has been analyzed by early-labeled fragment (ELF) assays, by leading and lagging strand template bias assays, by PCR-based small nascent strand abundance assays, and by both the neutral/neutral and neutral/alkaline 2-D gel assays (reviewed in Dijkwel and Hamlin, 1996). Virtually all of the studies on this complex origin have converged on a model in which initiation can occur at more than 30 sites within the 55 kb spacer between the convergently-transcribed DHFR and 2BE2121 genes, with the efficiencies of utilization varying from a minimum near the 3′ ends of the convergently-transcribed genes to maxima at two different sites (termed ori-_ and ori-_), which are spaced about 20 kb apart (e.g., Leu and Hamlin, 1989). A quantitative estimate suggests that, at most, 20% of initiations in the 55-kb spacer occur within the 2 kb encompassing the most active ori-_ region, and the spacer as a whole sustains an initiation event in only15-20% of cell cycles. Thus, even the most active region in this origin (ori-_) is very inefficient, firing in only 3-4% of cell cycles.
How do these results on the prototypic DHFR origin compare to other origins that have been identified by locus-centric approaches in higher eukaryotes? By one (usually) or more (rarely) mapping methods, about 30 origins have now been identified in organisms ranging from fruit flies to man (see (Aladjem et al., 2006 for original references). Origins have been localized in the amplified D. melanogaster chorion, Xenopus laevis and human rDNA, and Syrian hamster CAD and murine adenosine deaminase domains. Single-copy human origins have been identified in the c-myc, _-globin,, IGFII, HSP70, MCM4/PRKDC, RPE, TOP1, and YWHAH domains. Other single-copy origins have been localized in Drosophila histone and polymerase-_ domains, in African green monkey cells, in the murine _-globin and IGH loci, as well as in the Chinese hamster rhodopsin, APRT, GNA13, GADD45A (Delgado et al., 1998), RSP14, and TK1 domains.
Many of these origins have been shown on 2-D gels, or by extensive analysis of the surrounding region by the small nascent strand abundance assay, to represent zones of inefficient initiation sites (although certain subregions can be preferred. Importantly, most of the origins that have been identified in the last 8-10 years were discovered with the PCR-based small nascent strand abundance assay (e.g., Giacca et al., 1994), and most of these have not been characterized with any other state-of-the-art assay such as neutral/neutral or neutral/alkaline 2-D gel analysis, either of which can readily distinguish isolated single initiation sites from sites residing within initiation zones. Indeed, our laboratory has shown that the small nascent strand abundance assay is insensitive to low levels of initiation that can be detected on 2-D gels (Dijkwel et al., 2002). Thus, the jury is still out as to how many of the 30 higher eukaryotic origins actually reside in initiation zones.
Apparently, the vast majority of active origins in S. cerevisiae (Raghuraman et al., 2001) and in S. pombe (Gomez and Antequera, 1999) reside in the spacers between genes. All but five of the mammalian origins cited above (Syrian hamster CAD; Chinese hamster GADD45A, and RSP14, as well as human DMMT1 and YWHAH) reside in intergenic regions (see Aladjem et al., 2006 for original references). This is consistent with the observation that in early cleavage stages in Xenopus, initiation can occur in both the transcription units and spacers when the genes are silent prior to the mid-blastula transition, but only in the spacers after transcription commences (Hyrien et al., 1995). In concert with older electron microscopic studies on early cleavage Xenopus and Drosophila embryos (e.g., Buongiorno-Nardelli et al., 1976; Blumenthal, Kriegstein, and Hogness, 1974), these data suggest that, at least in early development in these organisms, the entire genome constitutes a potential substrate for initiation.
However, other higher eukaryotic origins are clearly more circumscribed and come closer to the bacterial paradigms that generated the replicon model in the first place. These include the human lamin B2 (Giacca et al., 1994) and _-like globin (Aladjem et al., 1998) origins, possibly the Chinese hamster GNA13 origin (Toledo et al., 1998), and the ardB&ardC (Benard et al., 1996) and rDNA origins in Physarum (Benard et al., 1995). Of these, the human globin and GNA13 origins, as well as the Physarum rDNA origin, constitute mini-zones of initiation, while the lamin B2 (Abdurashidova et al., 2000) and ardB&C (Benard et al., 1996) origins appear to correspond most closely to single sites. Interestingly, the latter two reside within the promoters of the nearby genes. With the exception of the Physarum origins and Chinese hamster GNA13, however, none of these has been subjected to 2-D gel analysis.
Thus, there appears to be a spectrum of origin types ranging from quite circumscribed to extremely broad initiation zones. Likewise, the efficiency of origin firing apparently can range from nearly 100% (e.g., human _-globin (Kitsberg et al., 1993) and Physarum ardB&C (Benard et al., 1996) to less than 5% (e.g., the CHO DHFR; Dijkwel et al., 2002).
Recently, several groups have devised methods for isolating origins from complex genomes en masse, in an attempt to discover how analyses of the 30 or so origins cited (most of which were localized by virtue of proximity to one's favorite gene) reflects the spectrum of origins in the genome as a whole. The first such study isolated nascent strands 800-3,000 nt in length and prepared a small library that was shown to be enriched ∼7-fold over the starting total genomic DNA (Todorovic et al., 2005). Although several new origins were identified in this study, there apparently was a high degree of contamination with non-origin material. A second approach takes advantage of the circular nature of restriction fragments containing internal initiation sites (i.e., replication bubbles or eye forms) by trapping them in gelling agarose (Mesner et al., 2006). Contaminating simple Y structures, linear fragments, and X-shaped termination structures then can be electrophoresed out of the agarose plus, leaving purified bubble-containing fragments behind. In a pilot project, this method yielded thousands of clones from CHO cells, virtually all of which appear to correspond to initiation sites in the hamster genome (Mesner et al., 2006), as demonstrated by 2-D gel analysis of their respective genomic positions. Importantly, >90% of the cloned genomic regions that were analyzed in early S-phase cells display the composite pattern characteristic of initiation sites residing in initiation zones (i.e., a complete bubble arc and a complete, more prominent single fork arc; L.D. Mesner, unpublished; (Mesner et al., 2006). However, the bubble-to-fork-arc ratios varied greatly among them, suggesting that they arise from both large and small initiation zones. The 2-D gel analysis also suggested that most if not all origins probably fire in less than 30% of cell cycles.
In a more recent study, libraries of millions of clones from human cells have been prepared and used to probe microarrays of the 30 MB of genomic DNA being analyzed in the NHGRI ENCODE project (N. Karnani, L.D. Mesner, C. Taylor, A. Malhotra, J. L. Hamlin, and A. Dutta, unpublished). This method, too, suggests that more than 50% of the cloned initiation sites arise from initiation zones. This number is likely to be even larger, because many of the smaller zones will be confined to a single restriction fragment, as would be the case with the human c-myc and _-globin origins, both of which are small zones of initiation. Thus, our current thinking is that most mammalian origins correspond to initiation zones rather than single sites analogous to the lamin B2 origin. By hybridizing these libraries to microarrays and comparing their distributions to the other information being compiled for the 30 MB ENCODE region, it soon should be possible to paint a relatively detailed picture of the activity of origins vis-a-vis their times of synthesis, proximity to active genes, chromosomal milieu (i.e., histone and DNA covalent modifications), etc.
In search of classic genetic replicators in higher eukaryotic genomes
Huberman and Riggs coined the operational term origin to denote replication initiation sites in complex genomes (Huberman and Riggs, 1968). The term replicator arose directly from the replicon model of Jacob and Brenner, and is defined as a mutable genetic controlling element (Jacob and Brenner, 1963). Origins necessarily coincide with replicators in simple genomes with fixed initiation sites (bacteria, viruses and plasmids, budding yeast), and it has been assumed that the multiple origins in complex genomes would do the same. However, as pointed out above, the ARS assay has not been successful in identifying replicators from complex genomes more evolved than S. pombe. Therefore, small fragments containing origins identified by biochemical means have been analyzed for the presence of required genetic elements in two other ways: 1) by positioning them (or mutated versions) at random or fixed ectopic chromosomal sites to ask whether initiation competence travels with the fragment (the negative control is usually a nearby site in the accompanying vector), and 2) by performing in loco mutagenesis by homologous recombination to determine whether initiation sites are required genetic elements.
By the first approach, mutable “replicator” activity has been detected in the ACE origin from the Drosophila chorion puff (Heck and Spradling, 1990; Delidakis and Kafatos, 1987), the human _-globin origin (Aladjem et al., 1998), the human c-myc origin (Liu et al., 2003), the human lamin B2 origin (Paixao et al., 2004), and the CHO ori-_ locus (e.g., Handeli et al., 1989; Gray et al., 2007). Of these, both the human _-globin origin (Aladjem et al., 1998) and the human c-myc origin (Liu et al., 2003) appear to have redundant sequences that must both be removed to inhibit origin activity; additionally, the _-globin, c-myc, ori-_, and ACE sequences all derive from zones of initiation sites in their native chromosomal contexts. Note, however, that initiation does not occur within ACE itself. Thus, only the very AT-rich lamin B2 origin, which is in a very narrow intergenic spacer, appears to correspond to a single, mutable origin/replicator by this criterion. Furthermore, a subregion of this fragment has been shown in ChIP assays to bind to hORC2 (reviewed in Aladjem et al., 2006). Interestingly, the same techniques used to identify a relatively fixed origin containing mutable genetic elements in the human _-like globin locus detected no such circumscribed start sites or potential replicators in the homologous region in the murine genome (see Aladjem et al., 2006). Thus, the mechanisms of initiation at these loci seem not to have been conserved between the two species.
The DHFR initiation zone constitutes one of the few origins that has been mutagenized systematically in loco to search for required genetic elements (replicators). Although the activity of the ori-_ locus can be compromised by mutagenesis when placed by itself at ectopic (Gray et al., 2007), deletion from its in loco position has no effect on initiation in the remainder of the 55 kb initiation zone nor on the time of replication of the locus as a whole (Kalejta et al., 1998). In fact, the central 45 kb of the intergenic spacer, which encompasses >95% of the usual nascent strand start sites in this zone, can be deleted without a significant change in the early-replicating properties of the DHFR locus (Mesner et al., 2003b); this was achieved by greatly increasing the frequency of initiation in the truncated spacer that remained. However, it remains to perform the same systematic genetic analysis that was carried out at ectopic positions on the ori-_ region in loco. If the same results were obtained, one could conclude that, ori-_ is, indeed, a genetic replicator, but it must be redundant with many other replicators spread throughout the intergenic spacer. Alternatively, a responsible replicator could conceivably reside outside of the actual initiation zone itself.
Interestingly, when tested directly, a fragment containing ori-_ proved to replicate autonomously in human or CHO cells no better than similarly-sized fragments that presumably do not contain replicators (P. Foreman, J.D. Milbrandt, and J.L. Hamlin, unpublished; Caddle and Calos, 1992). Furthermore, a fragment from the DHFR gene that never initiates replication in the native locus is an efficient template for initiation when positioned at ectopic chromosomal sites, as are the adjacent bacterial vector sequences (Lin et al., 2005a). [Parenthetically, the notion that matrix attachment regions (MARs) might coincide with genetic replicators (Razin et al., 1986) was tested directly by deleting the prominent MAR that lies in the approximate center of the DHFR initiation zone (Mesner et al., 2003a). However, its removal had no effect on initiation per se; rather, the daughter chromatids, which are usually separated from one another within minutes of replication, failed to separate until just prior to mitosis. Thus, at least in this locus, the local MAR is within the origin but is not the element responsible for origin activity.]
Epigenetic factors that regulate origin activity and the time of firing
Is it possible that virtually any sequence in complex genomes can serve as a template for initiation provided that it is in a non-permissive environment? There are several examples of the plasticity of origins of replication that support this view. For example, when nucleotide pools are lowered in CHO cells by inhibitors (Anglana et al., 2003), or when the transcription patterns of neighboring genes are altered during development, the number and distribution of initiation sites can change dramatically (e.g., Norio et al., 2005). These findings are compatible with the idea that there are many more potential initiation sites in the genome than are normally used in a given cell cycle, but their usage can be negatively or positively regulated by environmental effects such as local gene activity and/or chromatin architecture.
In support of these ideas are the following examples. Transcription through S. cerevisiae ARS1 (Snyder et al., 1988), as well as through cloned human fragments (Haase et al., 1994), inhibits their ability to replicate autonomously. In the CHO DHFR locus, deletion of the 3′ processing signals from the DHFR gene allows RNA polymerase to traverse almost the entire former intergenic spacer, completely eliminating replication initiation in its path (Mesner and Hamlin, 2005). Conversely, when a fragment from the body of the DHFR gene, which never initiates in loco, is inserted at ectopic (presumably non-transcribed) chromosomal sites, it now serves as a perfectly good template for replication initiation (Lin et al., 2005). Interestingly, deletion of a functional promoter from the DHFR gene, which prevents transcription by RNA polymerase, allows the former 55 kb initiation zone to spread into the body of the now inactive gene (Saha et al., 2004). These data again suggest that potential initiation sites are spread throughout the genome - even in the bodies of potential genes, but their use is inhibited by transcription through the template. As noted above, however, at least five mammalian origins reside in the coding regions of active genes. It remains to be determined whether both alleles at these loci are actively transcribing. Indeed, whole-genome microarray approaches that identified initiation sites in S. cerevisiae, S. pombe, and D. melanogaster, suggest that most sites reside in non-transcribed spacers (e.g., Raghuraman et al., 2001)
In contrast to the negative effects of transcription through a DNA sequence on replication initiation, several recent studies suggest that proximity to an active promoter can establish a chromatin ambience that facilitates origin activity without requiring transcription itself. In S. cerevisiae, both RNA Polymerase II and III transcription factors can stimulate initiation at the nearby ARS1, apparently by facilitating chromatin remodeling (Bodmer-Glavas et al., 2001). In Xenopus laevis eggs, an artificial origin can be created in a plasmid simply by targeting Gal4-VP16 to binding sites positioned next to a TATA box (Danis et al., 2004). Transcription is not required for this effect, and origin activation is accompanied by local histone acetylation. Recent studies on the c-myc replicator suggest that local promoter elements are required for origin activity, but that very active transcription from that promoter inhibits origin firing (Ghosh et al., 2004).
Additional studies on D. melanogaster ACE (the amplification control element) show that origins of replication in this system bind well-known transcription factors, suggesting a direct role for these factors in initiation of replication (Beall et al., 2002; Bosco et al., 2001). The lamin B2 origin resides in a very narrow intergenic zone, and elements that enhance transcription (e.g., a CpG island) also enhance origin activity (Paixao et al., 2004). In fact, sequences encompassing CpG islands are greatly enriched in small nascent strands in mammalian cells (Delgado et al., 1998), again suggesting that transcription factors can modulate both gene and local origin activity. Various attempts have been made to categorize the elements residing in or near origins of replication in mammalian cells, and although there is no consistent pattern among the various well-studied examples, transcription factor binding sites appear regularly in or near several origins (Aladjem et al., 2006). In most cases, however, the precision with which replication initiation sites have been mapped does not allow the conclusion that these sites have anything to do with origin activity per se.
Another clear-cut example of promoter/origin interactions is manifested in the CHO DHFR locus. The DHFR promoter lies 26 kb upstream from the nearest initiation site in the intergenic spacer, yet deletion of critical elements that eliminate transcription greatly diminishes the efficiency of initiation of replication in the spacer (Saha et al., 2004). It has not yet been determined whether inhibition of origin activity by the promoter deletion results from the absence of transcription per se, as opposed to some transcription-independent change in chromatin architecture that depends on an intact promoter and extends all the way into the intergenic zone. This effect may be manifested on a more global scale by studies in which nuclei from CHO cells at various positions in the cell cycle are allowed to replicate in Xenopus egg extracts. These nuclei have been shown to pass through a regulatory Origin Decision Point (ODP) in early G1 that converts a totally random pattern of initiation throughout the DHFR locus (and presumably the whole genome) to the more focused intergenic pattern displayed by cultured cells (e.g., Keezer and Gilbert, 2002). There is some evidence to suggest that the critical difference between early- and late-G1 nuclei is the dramatic increase in transcription of neighboring genes that occurs during this interval. However, transcription commences in this system early in G1, well before the ODP itself. Clearly, something else has to happen in addition to transcription per se to activate the nearby origin.
Thus, there appear to be both local and more far-reaching effects of transcription competence on origin activity, neither of which may require transcription itself under most circumstances. Data from a genome-wide microarray study on replication timing, transcription patterns, and the distribution of ORC in the Drosophila genome may suggest how this is achieved (MacAlpine et al., 2004). Based on the interaction of these different, large, data sets, it appears that transcription (or a competent promoter) may have a local effect by influencing origin selection, while at the same time regulating origin activation over domains at least 100 kb in length (in the latter case by determining the activation of or the time of activation of origins within the domain). In fact, a single molecule analysis of the multicopy rDNA locus in S. cerevisiae reveals that the intergenic rDNA origins fire in clusters (Pasero et al., 2002), which could result from packaging several tandem copies of the repeating unit into distinct architectural chromatin domains. Presumably, in all of these cases, transcription factors are mediating chromatin remodeling toward a permissive configuration. The question then arises as to what constitutes a permissive chromatin ambience. There is little doubt that most active origins will find themselves in euchromatin, which has long been known to replicate early in S-phase (Goldman et al., 1984). Recent genome-wide studies on tlinecovalent histone modifications in Drosophila define a binary pattern in which active genes are hyperacetylated on histones H3 and H4 and hypermethylated at H3/K4 and H3/K79; inactive genes, on the other hand, are hypomethylated and deacetylated at the same lysines (Schubeler et al., 2004). These modifications are tightly linked to RNA polymerase activity itself. However, this suggestion is not consistent with studies cited above in which the act of transcription (rather than transcription factor loading at the promoter) is not required to activate the local origins. Furthermore, most activating marks are prevalent only in the promoter and immediately downstream (Pattenden et al., 2005).This suggests that acetylation of histones in promoters could be responsible for activating those origins that lie within a short distance (e.g., c-myc, _-globin, lamin B2), but cannot readily explain activation of origins that lie considerable distances from a local promoter (e.g., the DHFR origin). Therefore, no unifying rules have been uncovered to explain the complex and subtle effects of transcription (or active promoters) on establishing a permissive environment for initiation.
Altogether, these data are consistent with earlier studies showing that actively transcribed genes in somatic metazoan cells are very often replicated in early S-phase when most of the origins are likely to be activated (e.g., Holmquist, 1987). In budding yeast, the default time of activation of most origins generally appears to be early S-phase, although the activity and/or time of firing of ARS elements can be regulated by chromosomal context effects such as proximity to telomeres (Ferguson and Fangman, 1992).
Indeed, there is a well-established literature suggesting that, at a population level, origins of replication in budding yeast (e.g., Raghuraman et al., 2001), fission yeast (e.g. Segurado et al., 2003), fruit flies (e.g., MacAlpine and Bell, 2005), and mammals (e.g., Jeon et al., 2005) fire in predetermined orders within the S-phase. However, in a recent analysis of replication timing of single DNA fibers in S. cerevisiae, it is quite clear that the timing of replication of given chromosomal segments is essentially probabilistic within an individual cell and is not strictly determined (Czajkowsky et al., 2008); however, the averaged replication times for all molecules examined recapitulates the ensemble-averaged patterns observed in the population. This same phenomenon may be reflected in the fact that the single-copy, so-called early-firing, mammalian origins that have been examined on 2-D gels clearly fire at any time during a 2-3 hr window - presumably also in a probabilistic manner (Dijkwel and Hamlin, 1992). In the few documented cases of mammalian mid- or late-firing origins that have been examined on 2-D gels, the window of activation is equally broad (e.g., Larner et al., 1999). Even in synchronized populations of budding yeast, the activation times of early- and mid-firing origins, as judged from 2-D gels, clearly overlap (Ferguson and Fangman, 1992), suggesting the absence of any strictly-defined temporal organization.
With these caveats in mind, a well-studied example of regulation of the time of firing is the _-globin domain in humans, in which there is a clearly-defined upstream Locus Control Region (LCR), whose loss in the Hispanic deletion renders the downstream origin late-firing (Aladjem et al., 1995). Another important contribution shows that tethering a histone acetyl transferase to a late-firing origin in S. cerevisiae can advance its time of firing in the S-period (Vogelauer et al., 2002). Thus, the transcription apparatus may serve only as a conveyor of the enzymes and factors required to adequately remodel or modify chromatin in the vicinity of a potential origin. Interestingly, there is another way of establishing replication timing, which was uncovered in a study of the EBV origin of replication. This origin utilizes the viral EBNA protein in addition to cellular proteins to effect initiation in a once-per-cell-cycle mode of replication (Zhou et al., 2006). The nucleosomes that flank the dyad symmetry (DS) element in this origin are remodeled by SNF2H in late G1 prior to S-phase entry. At the same time, and unlike other examples of origin activation, histones in the EBV origin are deacetylated by HDAC2. The authors point out that this origin is probably late-firing, so deacetylation may be the mark that delays origin firing in this system.
Regardless of how replication timing is established, a fascinating recent study suggests that transcriptional competence is established at the time of replication (i.e., exogenous genes are better able to support transcription when injected into early S-phase nuclei than when delivered in late S-phase (Zhang et al., 2002). These studies suggest how epigenetic states can be maintained from one cell generation to another. For a comprehensive summary of this fascinating aspect of origin regulation, the reader is directed to a recent review of replication timing related to imprinting and X-chromosome inactivation (Lande-Diner and Cedar, 2005). Lastly, another potential epigenetic influence that deserves further study is methylation of CpG islands. The jury is still out on the real involvement of this covalent DNA modification in origin activation.
How do these data fit the original replicon model, and does that model help or hinder our future ability to understand regulation of DNA synthesis in higher eukaryotes?
We would like to propose the following model for control of replication in higher eukaryotic cells, which is fluid enough to accommodate the overwhelming majority of the complex data sets cited above. We suggest that the genomes of metazoans are peppered at very frequent but random intervals (perhaps every few hundred base pairs) with a hierarchy of potential, degenerate, replicators. When activated, these replicators are suggested to control initiation only in their immediate environments. In the theoretical absence of any chromosomal context effects (i.e., in naked DNA), these sites could be more or less active as recognition elements for metazoan initiators in much the same way that micrococcal nuclease exhibits a hierarchic preference for certain sequences in naked DNA when enzyme is limiting. Clearly, when viewed in isolation, deletion or mutagenesis of such an initiation (or cleavage) site would lower or eliminate its ability to attract the initiation complex (or nuclease) and inhibit initiation (or cleavage) at that site, but not at neighboring sites. In general, most of these initiation sites are suggested to be highly inefficient, explaining why even broad initiation zones appear to sustain only one or fewer initiation events within the zone in a given cell cycle.
Thus, the probability that any given site will efficiently attract an initiation complex will depend upon whether it finds itself in a permissive environment. In a euchromatic chromatin domain, the overall architecture would be permissive, allowing a non-transcribed (intergenic) region to initiate with more or less efficiency depending on its complement of other proteins (e.g., transcription factors, modified histones, etc.), while sites within an active transcription unit in the same euchromatic domain would not. The non-permissive environment of heterochromatin would preclude initiation of replication, and such regions would have to wait to be replicated passively from active upstream or downstream euchromatic regions, probably by a preceding wave of heterochromatization.
In this general model, lengthy intergenic spacers such as DHFR and rhodopsin correspond to broad initiation zones because they contain many exposed degenerate replicators, some of them more efficient than others. Conversely, very narrow intergenic spacers might isolate only one or a few active initiation sites (as in the lamin B2 and human _-globin loci), even though other potential sites might reside in the bodies of the neighboring actively-transcribed genes that define the spacer. Any of these sites (intra- or intergenic) might behave as classic, mutable, replicators when positioned at a permissive ectopic site. This general model could be extended to suggest that the one or two solo sites in narrow spacers may have evolved particularly high affinities for the initiation complex in order to ensure that they are activated in every cell cycle.
Viewed in this way, one could question the value of mutagenizing a site such as lamin B2 or ori-_ and testing the effects on initiation only within the immediate environment of a cassette at an ectopic chromosomal site (Aladjem et al., 1998; Altman and Fanning, 2004). It is already clear that there are no recognizable consensus sequences among the known origins except for the usual presence of AT-rich elements and easily unwound elements in the neighborhood (reviewed in Gilbert, 2004; Aladjem et al., 2006). However, these mutagenic studies will likely define the nature of sequence characteristics that have increased affinity for ORC and/or other proteins involved in the initial melting of the helix. To be truly meaningful, however, these approaches will have to control for possible effects of mutations on other epigenetic phenomena that play important roles in origin activity (e.g., local transcription or chromatin modification).
Perhaps by modulating our thinking around this more inclusive general model, our experiments can be designed to give the maximum amount of information about this elemental but complex process.
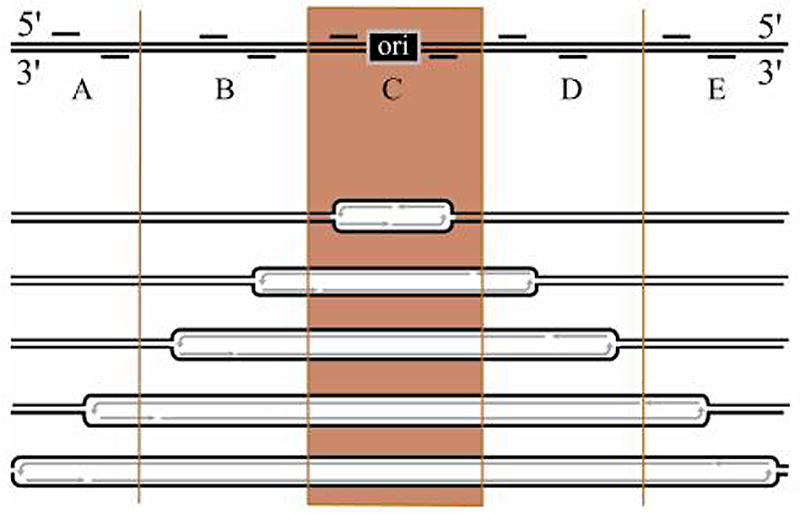
Figure 1.
Acknowledgments
We would like to apologize to many of our colleagues whose original references we could not cite owing to page limitations. However, we believe that their works are referenced in the several review articles that we referred to throughout the manuscript. Work in the authors' laboratory was supported by grants from the NIH to J.L.H. (RO1GM26108 and RO1HG_____). We thank the many members of the Hamlin who have contributed to these studies and brought many ideas to experimental fruition.
References
- Abdurashidova G, Deganuto M, Klima R, Riva S, Biamonti G, Giacca M, Falaschi A. Start sites of bidirectional DNA synthesis at the human lamin B2 origin. Science. 2000;287:2023–2026. doi: 10.1126/science.287.5460.2023. [DOI] [PubMed] [Google Scholar]
- Aladjem MI, Falaschi A, Kowalski D. In: Eukaryotic DNA Replication Origins. DePamphilis M, editor. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2006. pp. 31–62. [Google Scholar]
- Aladjem MI, Groudine M, Brody LL, Dieken ES, Fournier RE, Wahl GM, Epner EM. Participation of the human beta-globin locus control region in initiation of DNA replication. Science. 1995;270:815–819. doi: 10.1126/science.270.5237.815. [DOI] [PubMed] [Google Scholar]
- Aladjem MI, Rodewald LW, Kolman JL, Wahl GM. Genetic dissection of a mammalian replicator in the human beta-globin locus. Science. 1998;281:1005–1009. doi: 10.1126/science.281.5379.1005. [DOI] [PubMed] [Google Scholar]
- Anglana M, Apiou F, Bensimon A, Debatisse M. Dynamics of DNA replication in mammalian somatic cells: nucleotide pool modulates origin choice and interorigin spacing. Cell. 2003;114:385–394. doi: 10.1016/s0092-8674(03)00569-5. [DOI] [PubMed] [Google Scholar]
- Beall EL, Manak JR, Zhou S, Bell M, Lipsick JS, Botchan MR. Role for a Drosophila Myb-containing protein complex in site-specific DNA replication. Nature. 2002;420:833–837. doi: 10.1038/nature01228. [DOI] [PubMed] [Google Scholar]
- Bell SP, Dutta A. DNA replication in eukaryotic cells. Annu Rev Biochem. 2002;71:333–374. doi: 10.1146/annurev.biochem.71.110601.135425. [DOI] [PubMed] [Google Scholar]
- Bell SP, Stillman B. ATP-dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex. Nature. 1992;357:128–134. doi: 10.1038/357128a0. [DOI] [PubMed] [Google Scholar]
- Benard M, Lagnel C, Pallotta D, Pierron G. Mapping of a replication origin within the promoter region of two unlinked, abundantly transcribed actin genes of Physarum polycephalum. Mol Cell Biol. 1996;16:968–976. doi: 10.1128/mcb.16.3.968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benard M, Lagnel C, Pierron G. Site-specific initiation of DNA replication within the non-transcribed spacer of Physarum rDNA. Nucl Acids Res. 1995;23:1447–1453. doi: 10.1093/nar/23.9.1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumenthal AB, Kriegstein HJ, Hogness DS. The units of DNA replication in Drosophila melanogaster chromosomes. Cold Spring Harb Symp Quant Biol. 1974;38:205–223. doi: 10.1101/sqb.1974.038.01.024. [DOI] [PubMed] [Google Scholar]
- Bodmer-Glavas M, Edler K, Barberis A. RNA polymerase II and III transcription factors can stimulate DNA replication by modifying origin chromatin structures. Nucl Acids Res. 2001;29:4570–80. doi: 10.1093/nar/29.22.4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosco G, Du W, Orr-Weaver TL. DNA replication control through interaction of E2F-RB and the origin recognition complex. Nat Cell Biol. 2001;3:289–295. doi: 10.1038/35060086. [DOI] [PubMed] [Google Scholar]
- Brewer BJ, Fangman WL. The localization of replication origins on ARS plasmids in S cerevisiae. Cell. 1987;51:463–471. doi: 10.1016/0092-8674(87)90642-8. [DOI] [PubMed] [Google Scholar]
- Brewer BJ, Fangman WL. Initiation at closely spaced replication origins in a yeast chromosome. Science. 1993;262:1728–1731. doi: 10.1126/science.8259517. [DOI] [PubMed] [Google Scholar]
- Buongiorno-Nardelli M, Amaldi F, Lava-Sanchez PA. Electron microscope analysis of amplifying ribosomal DNA from Xenopus laevis. Exp Cell Res. 1976;98:95–103. doi: 10.1016/0014-4827(76)90467-5. [DOI] [PubMed] [Google Scholar]
- Caddle MS, Calos MP. Analysis of the autonomous replication behavior in human cells of the dihydrofolate reductase putative chromosomal origin of replication. Nucl Acids Res. 1992;20:5971–5978. doi: 10.1093/nar/20.22.5971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan CS, Tye BK. Autonomously replicating sequences in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1980;77:6329–6333. doi: 10.1073/pnas.77.11.6329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czajkowsky DM, Liu J, Hamlin JL, Shao Z. DNA combing reveals intrinsic temporal disorder in the replication of yeast chromosome VI. J Mol Biol. 2008;375:12–19. doi: 10.1016/j.jmb.2007.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai J, Chuang RY, Kelly TJ. DNA replication origins in the Schizosaccharomyces pombe genome. Proc Natl Acad Sci U S A. 2005;102:337–342. doi: 10.1073/pnas.0408811102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danis E, Brodolin K, Menut S, Maiorano D, Girard-Reydet C, Mechali M. Specification of a DNA replication origin by a transcription complex. Nat Cell Biol. 2004;6:721–730. doi: 10.1038/ncb1149. [DOI] [PubMed] [Google Scholar]
- Delgado S, Gomez M, Bird A, Antequera F. Initiation of DNA replication at CpG islands in mammalian chromosomes. EMBO J. 1998;17:2426–2435. doi: 10.1093/emboj/17.8.2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delidakis C, Kafatos FC. Amplification of a chorion gene cluster in Drosophila is subject to multiple cis-regulatory elements and to long-range position effects. J Mol Biol. 1987;197:11–26. doi: 10.1016/0022-2836(87)90605-x. [DOI] [PubMed] [Google Scholar]
- Dijkwel PA, Hamlin JL. Initiation of DNA replication in the dihydrofolate reductase locus is confined to the early S period in CHO cells synchronized with the plant amino acid mimosine. Mol Cell Biol. 1992;12:3715–3722. doi: 10.1128/mcb.12.9.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dijkwel PA, Hamlin JL. Sequence and context effects on origin function in mammalian cells. J Cell Biochem. 1996;62:210–222. doi: 10.1002/(SICI)1097-4644(199608)62:2%3C210::AID-JCB9%3E3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- Dijkwel PA, Wang S, Hamlin JL. Initiation sites are distributed at frequent intervals in the Chinese hamster dihydrofolate reductase origin of replication but are used with very different efficiencies. Mol Cell Biol. 2002;22:3053–3065. doi: 10.1128/MCB.22.9.3053-3065.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson BM, Fangman WL. A position effect on the time of replication origin activation in yeast. Cell. 1992;68:333–339. doi: 10.1016/0092-8674(92)90474-q. [DOI] [PubMed] [Google Scholar]
- Ghosh M, Liu G, Randall G, Bevington J, Leffak M. Transcription factor binding and induced transcription alter chromosomal c-myc replicator activity. Mol Cell Biol. 2004;24:10193–10207. doi: 10.1128/MCB.24.23.10193-10207.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giacca M, Zentilin L, Norio P, Diviacco S, Dimitrova D, Contreas G, Biamonti G, Perini G, Weighardt F, Riva S. Fine mapping of a replication origin of human DNA. Proc Natl Acad Sci U S A. 1994;91:7119–7123. doi: 10.1073/pnas.91.15.7119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman MA, Holmquist GP, Gray MC, Caston LA, Nag A. Replication timing of genes and middle repetitive sequences. Science. 1984;224:686–692. doi: 10.1126/science.6719109. [DOI] [PubMed] [Google Scholar]
- Gomez M, Antequera F. Organization of DNA replication origins in the fission yeast genome. EMBO J. 1999;18:5683–5690. doi: 10.1093/emboj/18.20.5683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray SJ, Liu G, Altman AL, Small LE, Fanning E. Discrete functional elements required for initiation activity of the Chinese hamster dihydrofolate reductase origin beta at ectopic chromosomal sites. Exp Cell Res. 2007;313:109–120. doi: 10.1016/j.yexcr.2006.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haase SB, Heinzel SS, Calos MP. Transcription inhibits the replication of autonomously replicating plasmids in human cells. Mol Cell Biol. 1994;14:2516–2524. doi: 10.1128/mcb.14.4.2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamlin JL. Mammalian origins of replication. Bioessays. 1992;14:651–659. doi: 10.1002/bies.950141002. [DOI] [PubMed] [Google Scholar]
- Handeli S, Klar A, Meuth M, Cedar H. Mapping replication units in animal cells. Cell. 1989;57:909–920. doi: 10.1016/0092-8674(89)90329-2. [DOI] [PubMed] [Google Scholar]
- Harland RM, Laskey RA. Regulated replication of DNA microinjected into eggs of Xenopus laevis. Cell. 1980;20:761–771. doi: 10.1016/0092-8674(80)90439-0. [DOI] [PubMed] [Google Scholar]
- Heck MM, Spradling AC. Multiple replication origins are used during Drosophila chorion gene amplification. J Cell Biol. 1990;110:903–914. doi: 10.1083/jcb.110.4.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmquist GP. Role of replication time in the control of tissue-specific gene expression. Am J Hum Genet. 1987;40:151–173. [PMC free article] [PubMed] [Google Scholar]
- Huberman JA, Riggs AD. On the mechanism of DNA replication in mammalian chromosomes. J Mol Biol. 1968;32:327–341. doi: 10.1016/0022-2836(68)90013-2. [DOI] [PubMed] [Google Scholar]
- Hyrien O, Maric C, Mechali M. Transition in specification of embryonic metazoan DNA replication origins. Science. 1995;270:994–997. doi: 10.1126/science.270.5238.994. [DOI] [PubMed] [Google Scholar]
- Jacob F, Brenner S. Sur la Regulation de la synthese du DNA chez les bacteriers: l'hypothese du replicon. Comp Rendu Acad Sci. 1963;246:298–300. [PubMed] [Google Scholar]
- Jeon Y, Bekiranov S, Karnani N, Kapranov P, Ghosh S, MacAlpine D, Lee C, Hwang DS, Gingeras TR, Dutta A. Temporal profile of replication of human chromosomes. Proc Natl Acad Sci U S A. 2005;102:6419–6424. doi: 10.1073/pnas.0405088102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalejta RF, Li X, Mesner LD, Dijkwel PA, Lin HB, Hamlin JL. Distal sequences, but not ori-beta/OBR-1, are essential for initiation of DNA replication in the Chinese hamster DHFR origin. Molec Cell. 1998;2:797–806. doi: 10.1016/s1097-2765(00)80294-4. [DOI] [PubMed] [Google Scholar]
- Keezer SM, Gilbert DM. Sensitivity of the origin decision point to specific inhibitors of cellular signaling and metabolism. Exp Cell Res. 2002;273:54–64. doi: 10.1006/excr.2001.5421. [DOI] [PubMed] [Google Scholar]
- Kim SM, Huberman JA. Regulation of replication timing in fission yeast. EMBO J. 2001;20:6115–6126. doi: 10.1093/emboj/20.21.6115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitsberg D, Selig S, Keshet I, Cedar H. Replication structure of the human beta-globin gene domain. Nature. 1993;366:588–590. doi: 10.1038/366588a0. [DOI] [PubMed] [Google Scholar]
- Krysan PJ, Haase SB, Calos MP. Isolation of human sequences that replicate autonomously in human cells. Mol Cell Biol. 1989;9:1026–1033. doi: 10.1128/mcb.9.3.1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lande-Diner L, Cedar H. Silence of the genes--mechanisms of long-term repression. Nat Rev Genet. 2005;6:648–654. doi: 10.1038/nrg1639. [DOI] [PubMed] [Google Scholar]
- Larner JM, Lee H, Little RD, Dijkwel PA, Schildkraut CL, Hamlin JL. Radiation down-regulates replication origin activity throughout the S phase in mammalian cells. Nucl Acids Res. 1999;27:803–809. doi: 10.1093/nar/27.3.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leu TH, Hamlin JL. High-resolution mapping of replication fork movement through the amplified dihydrofolate reductase domain in CHO cells by in-gel renaturation analysis. Mol Cell Biol. 1989;9:523–531. doi: 10.1128/mcb.9.2.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin HB, Dijkwel PA, Hamlin JL. Promiscuous initiation on mammalian chromosomal DNA templates and its possible suppression by transcription. Exp Cell Res. 2005 doi: 10.1016/j.yexcr.2005.04.012. [DOI] [PubMed] [Google Scholar]
- Liu G, Malott M, Leffak M. Multiple functional elements comprise a Mammalian chromosomal replicator. Mol Cell Biol. 2003;23:1832–1842. doi: 10.1128/MCB.23.5.1832-1842.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacAlpine DM, Bell SP. A genomic view of eukaryotic DNA replication. Chromosome Res. 2005;13:309–326. doi: 10.1007/s10577-005-1508-1. [DOI] [PubMed] [Google Scholar]
- MacAlpine DM, Rodriguez HK, Bell SP. Coordination of replication and transcription along a Drosophila chromosome. Genes Dev. 2004;18:3094–3105. doi: 10.1101/gad.1246404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesner LD, Crawford EL, Hamlin JL. Isolating apparently pure libraries of replication origins from complex genomes. Mol Cell. 2006;21:719–726. doi: 10.1016/j.molcel.2006.01.015. [DOI] [PubMed] [Google Scholar]
- Mesner LD, Hamlin JL. Specific signals at the 3′ end of the DHFR gene define one boundary of the downstream origin of replication. Genes Dev. 2005;19:1053–1066. doi: 10.1101/gad.1307105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesner LD, Hamlin JL, Dijkwel PA. The matrix attachment region in the Chinese hamster dihydrofolate reductase origin of replication may be required for local chromatid separation. Proc Natl Acad Sci U S A. 2003a;100:3281–3286. doi: 10.1073/pnas.0437791100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesner LD, Li X, Dijkwel PA, Hamlin JL. The dihydrofolate reductase origin of replication does not contain any nonredundant genetic elements required for origin activity. Mol Cell Biol. 2003b;23:804–814. doi: 10.1128/MCB.23.3.804-814.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawotka KA, Huberman JA. Two-dimensional gel electrophoretic method for mapping DNA replicons. Mol Cell Biol. 1988;8:1408–1413. doi: 10.1128/mcb.8.4.1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newlon CS, Petes TD, Hereford LM, Fangman WL. Replication of yeast chromosomal DNA. Nature. 1974;247:32–35. doi: 10.1038/247032a0. [DOI] [PubMed] [Google Scholar]
- Norio P, Kosiyatrakul S, Yang Q, Guan Z, Brown NM, Thomas S, Riblet R, Schildkraut CL. Progressive activation of DNA replication initiation in large domains of the immunoglobulin heavy chain locus during B cell development. Mol Cell. 2005;20:575–587. doi: 10.1016/j.molcel.2005.10.029. [DOI] [PubMed] [Google Scholar]
- Paixao S, Colaluca IN, Cubells M, Peverali FA, Destro A, Giadrossi S, Giacca M, Falaschi A, Riva S, Biamonti G. Modular structure of the human lamin B2 replicator. Mol Cell Biol. 2004;24:2958–2967. doi: 10.1128/MCB.24.7.2958-2967.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasero P, Bensimon A, Schwob E. Single-molecule analysis reveals clustering and epigenetic regulation of replication origins at the yeast rDNA locus. Genes Dev. 2002;16:2479–2484. doi: 10.1101/gad.232902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattenden SG, Chandy MJ, Gutierrez JL, Workman JL. Chromatin dynamics rule the genome. Genome Biol. 2005;6:355. doi: 10.1186/gb-2005-6-11-355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghuraman MK, Winzeler EA, Collingwood D, Hunt S, Wodicka L, Conway A, Lockhart DJ, Davis RW, Brewer BJ, Fangman WL. Replication dynamics of the yeast genome. Science. 2001;294:115–121. doi: 10.1126/science.294.5540.115. [DOI] [PubMed] [Google Scholar]
- Razin SV, Kekelidze MG, Lukanidin EM, Scherrer K, Georgiev GP. Replication origins are attached to the nuclear skeleton. Nucl Acids Res. 1986;14:8189–8207. doi: 10.1093/nar/14.20.8189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha S, Shan Y, Mesner LD, Hamlin JL. The promoter of the CHO dihydrofolate reductase gene regulates the activity of the local origin and helps define its boundaries. Genes Dev. 2004;18:397–410. doi: 10.1101/gad.1171404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segurado M, de Luis A, Antequera F. Genome-wide distribution of DNA replication origins at A+T-rich islands in Schizosaccharomyces pombe. EMBO Rep. 2003;4:1048–1053. doi: 10.1038/sj.embor.7400008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder M, Sapolsky RJ, Davis RW. Transcription interferes with elements important for chromosome maintenance in Saccharomyces cerevisiae. Mol Cell Biol. 1988;8:2184–2194. doi: 10.1128/mcb.8.5.2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stinchcomb DT, Struhl K, Davis RW. Isolation and characterisation of a yeast chromosomal replicator. Nature. 1979;282:39–43. doi: 10.1038/282039a0. [DOI] [PubMed] [Google Scholar]
- Todorovic V, Giadrossi S, Pelizon C, Mendoza-Maldonado R, Masai H, Giacca M. Human origins of DNA replication selected from a library of nascent DNA. Mol Cell. 2005;19:567–575. doi: 10.1016/j.molcel.2005.07.005. [DOI] [PubMed] [Google Scholar]
- Toledo F, Baron B, Fernandez MA, Lachages AM, Mayau V, Buttin G, Debatisse M. oriGNAI3: a narrow zone of preferential replication initiation in mammalian cells identified by 2D gel and competitive PCR replicon mapping techniques. Nucl Acids Res. 1998;26:2313–2321. doi: 10.1093/nar/26.10.2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelauer M, Rubbi L, Lucas I, Brewer BJ, Grunstein M. Histone acetylation regulates the time of replication origin firing. Mol Cell. 2002;10:1223–1233. doi: 10.1016/s1097-2765(02)00702-5. [DOI] [PubMed] [Google Scholar]
- Wright AP, Maundrell K, Shall S. Transformation of Schizosaccharomyces pombe by non-homologous, unstable integration of plasmids in the genome. Curr Genet. 1986;10:503–508. doi: 10.1007/BF00447383. [DOI] [PubMed] [Google Scholar]
- Zhang J, Xu F, Hashimshony T, Keshet I, Cedar H. Establishment of transcriptional competence in early and late S phase. Nature. 2002;420:198–202. doi: 10.1038/nature01150. [DOI] [PubMed] [Google Scholar]
- Zhou J, Chau C, Deng Z, Stedman W, Lieberman PM. Epigenetic control of replication origins. Cell Cycle. 2005;4:889–892. doi: 10.4161/cc.4.7.1823. [DOI] [PubMed] [Google Scholar]
- Zhou J, Chau CM, Deng Z, Shiekhattar R, Spindler MP, Schepers A, Lieberman PM. Cell cycle regulation of chromatin at an origin of DNA replication. EMBO Journal. 2006;24:1406–1417. doi: 10.1038/sj.emboj.7600609. [DOI] [PMC free article] [PubMed] [Google Scholar]


