Abstract
Marine actinomycetes have generated much recent interest as a potentially valuable source of novel antibiotics. Like terrestrial actinomycetes the marine actinomycetes are shown here to produce mycothiol as their protective thiol. However, a novel thiol, U25, was produced by MAR2 strain CNQ703 upon progression into stationary phase when secondary metabolite production occurs and became the dominant thiol. MSH and U25 were maintained in a reduced state during early stationary phase but become significantly oxidized after 10 days in culture. Isolation and structural analysis of the monobromobimane derivative identified U25 as a homolog of mycothiol in which the acetyl group attached to the nitrogen of cysteine is replaced by a propionyl residue. This N-propionyl-desacetyl-mycothiol was present in 13 of the 17 strains of marine actinomycetes examined, including 5 strains of Salinispora and representatives of the MAR2, MAR3, MAR4 and MAR6 groups. Mycothiol and its precursor, the pseudodisaccharide 1-O-(2-amino-2-deoxy-α-D-glucopyranosyl)-D-myo-inositol, were found in all strains. High levels of mycothiol S-conjugate amidase activity, a key enzyme in mycothiol-dependent detoxification, was found in most strains. The results demonstrate that major thiol/disulfide changes accompany secondary metabolite production and suggest that mycothiol-dependent detoxification is important at this developmental stage.
Keywords: mycothiol, N-propionyl-desacetyl-mycothiol, marine actinomycetes, Salinispora arencola, Mca
Introduction
There has been a growing interest in recent years in the study of marine actinomycetes (Bull et al. 2005; Fenical and Jensen 2006; Bull and Stach 2007). A major motivation underlying such studies is the promise that they represent a source of new antibiotics, and a number of potentially important compounds have been isolated from marine-derived actinomycetes (Feling et al. 2003; Fiedler et al. 2005; Jensen et al. 2005b). Phylogenetic analyses based on sequences of 16S rRNA have provided evidence that marine actinomycetes are distinct from their terrestrial counterparts (Mincer et al. 2002). Although characterization of secondary metabolites produced by marine actinomycetes has received considerable attention, little is known about the levels of common metabolic intermediates in these new species. In the present study we set out to determine the low-molecular-weight thiol distribution in marine actinomycetes.
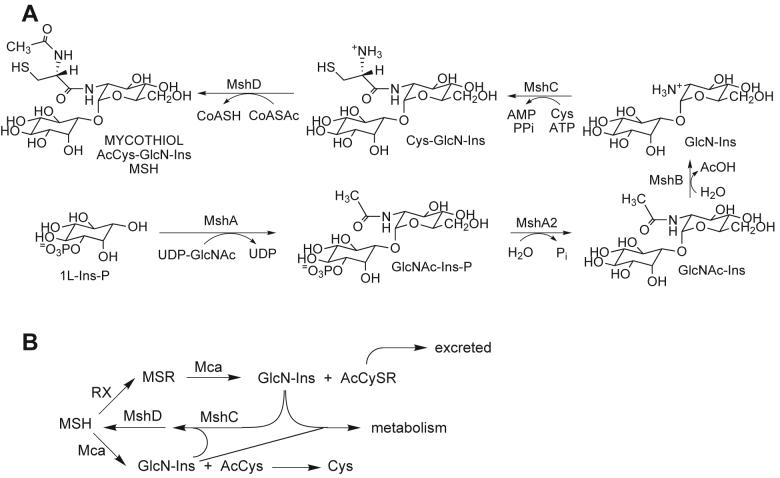
The diverse biochemistry associated with major thiols produced by microorganisms and by various marine organisms is the subject of a review in which mycothiol emerges as a thiol of major significance in the gram positive actinomycetes (Hand and Honek 2005). The main protective thiol found in gram negative bacteria and most eukaryotes is glutathione but glutathione is not found in most Gram positive bacteria (Fahey et al. 1978; Fahey and Sundquist 1991). Mycothiol (MSH, Fig. 1A) is the protective thiol produced by most terrestrial actinomycetes and it has thus far not been found outside of these bacteria (Newton et al. 1996; Newton and Fahey 2002). Mycothiol biosynthesis is initiated by the glycosyltransferase MshA (Newton et al. 2003) whose substrates have been identified as UDP-GlcNAc and 1L-inositol-1-P (Newton et al. 2006). An as yet unidentified phosphatase, MshA2, a deacetylase MshB (Newton et al. 2000a), an ATP-dependent cysteine:GlcN-Ins ligase MshC (Sareen et al. 2002), and the mycothiol synthase MshD (Koledin et al. 2002) complete the biosynthesis of mycothiol as summarized in Fig. 1A.
Fig. 1.
(A) Structure and biosynthesis of mycothiol (MSH). (B) Cleavage of mycothiol S-conjugate (MSR) by the amidase Mca, MSH resynthesis from GlcN-Ins, and degradation of MSH by Mca as a source of energy and metabolic intermediates.
MSH forms S-conjugates (MSR) with a wide range of toxins, Fig. 1B (Newton et al. 2000b), including the antibiotic rifamycin S (Steffek et al. 2003). The enzyme mycothiol S-conjugate amidase (Mca) cleaves such S-conjugates to produce a mercapturic acid, AcCySR, which can be excreted from the cell, and the pseudo-disaccharide GlcN-Ins (Fig. 1B), which can be recycled for MSH biosynthesis (Newton et al. 2000b). This detoxification process may be important for protection of antibiotic producing bacteria against their own antibiotics (Newton and Fahey 2002). Mycobacterial Mca has lower activity with MSH itself and under limiting nutrient conditions the AcCys produced is rapidly deacetylated to liberate Cys (Bzymek et al. 2007). Both Cys and GlcN-Ins can be further degraded to generate energy and important metabolic intermediates. Mycobacteria deficient in Mca or the enzymes of MSH biosynthesis exhibit enhanced sensitivity to certain antibiotics (Newton et al. 1999; Rawat et al. 2002; Buchmeier et al. 2003) and production of MSH is required for growth of Mycobacterium tuberculosis (Sareen et al. 2003; Buchmeier and Fahey 2006).
Because marine actinomycetes are an important source of novel antibiotics and since mycothiol in conjunction with Mca has been shown to play a role in antibiotic detoxification in terrestrial actinomycetes, it was of interest to examine the mycothiol and Mca content in marine actinomycetes. In the present study we show that mycothiol is generally found in marine actinomycetes and the level of Mca activity is quite high. However, in stationary phase when secondary metabolites are produced the thiol composition was found to change dramatically and a new thiol, shown to be a homolog of mycothiol, predominates over mycothiol.
Materials and methods
Bacterial strains and culture
The organisms studied are listed in Table 1. These strains were isolated and phylogenetically characterized as previously described (Mincer et al. 2002b; Jensen et al. 2005a). Each strain was cultured in a 1 liter of TCG medium (3 g tryptone (Fisher), 5 g casitone (Difco), 4 g glucose, 1 liter natural seawater) for 2-10 days while shaking at 215 rpm at 25-27°C.
Table 1.
Origin and reference data of strains studied.
| Strain # | Phylotype (Groupa) | Origin | Depth (meters) | 16S RNA Accession No. |
|---|---|---|---|---|
| CNR107 | Salinispora arenicola (MAR1) | Guamb | 40 | AY4646534 |
| CNR003 | Salinispora pacifica (MAR1) | Guamb | 184 | DQ224161 |
| CNR643 | Salinispora arenicola (MAR1) | Bahamasc | 1 | AY040619 |
| CNH725 | Salinispora arenicola (MAR1) | Red Seac | 20 | AY040621 |
| CNQ976 | Salinispora arenicola (MAR1) | Guambb | 72 | AY464533 |
| CNR252 | Streptomycetaceae (MAR2) | Guamb | 3 | AY464536 |
| CMQ695 | Streptomycetaceae (MAR2) | Guamb | 500 | AY464537 |
| CNQ703 | Streptomycetaceae (MAR2) | Guamb | 75 | AY464538 |
| CNQ857 | Streptomycetaceae (MAR3) | Guamb | 10 | AY464545 |
| CNQ719 | Streptomycetaceae (MAR3) | Guamb | <1 | AY464544 |
| CNQ698 | Streptomycetaceae (MAR3) | Guamb | 3 | AY464543 |
| CNQ766 | Streptomycetaceae (MAR3) | Guamb | 500 | AY464546 |
| CNQ687 | Streptomycetaceae (MAR3) | Guamb | <1 | AY464542 |
| CNQ530 | Streptomycetaceae (MAR4) | Guamb | 45 | AY464541 |
| CNQ525 | Streptomycetaceae (MAR4) | San Diego | 160 | EF177816 |
| CNR363 | Thermomonosporaceae (MAR5) | Guamb | 3 | AY464547 |
| CNR431 | Thermomonosporaceae (MAR6) | Guamb | 500 | AY464548 |
Assays performed
For thiol (RSH) analysis, cells were extracted in warm acetonitrile containing monobromobimane (mBBr). The acetonitrile denatures and precipitates most proteins. The thiol-labeling agent, mBBr, rapidly alkylates thiols and produces fluorescent derivatives for analysis by HPLC. Separate cell extractions and labeling of amines with AccQ-Fluor allowed determination of the GlcN-Ins content. A third extract was prepared and used to assay for Mca activity with MSmB as substrate. For determination of disulfide forms (RSS) an extract prepared in the presence of N-ethylmaleimide to block thiols and then subsequently reduced with dithiothreitol and labeled with monobromobimane was analyzed by HPLC to determine RSS levels and allow calculation of the thiol redox status (RSH/RSS).
Assay of cellular thiols
Cells were collected by centrifugation at 4000 × g for 30 min at 4°C. The cells were split into 6 equal pellets (pellets A-F), the wet weight was determined using preweighed 15 ml polypropylene centrifuge tubes and the tubes stored at −70°C prior to extraction. Two pellets, typically between 0.5 and 2 g wet weight, were used for duplicate thiol analysis (pellets A and B). Pellets A and B were extracted in an equal volume of warm acetonitrile containing 4 mM monobromobimane (mBBr; Molecular Probes) and 20 μl 1 M HEPES, pH 8.0, per ml of extraction. The cells suspensions were incubated at 60°C, vortexed every 5 min for 15 min, and then cooled on ice. To quench the reaction and slow the formation of mBBr hydrolysis products, 5 μl of 5 M methanesulfonic acid was added per ml of total extract. The thiol control sample (pellet C) was treated similarly, except that the extraction buffer contained 10 mM N-ethylmaleimide (NEM) in place of mBBr. The sample was incubated at 60°C for 10 min after which mBBr was added to a final concentration of 2 mM and the sample incubated for an additional 15 min at 60°C. The control was cooled on ice and 5 μl of 5M methansulfonic acid was added per ml of extract. The extracts were clarified by centrifugation for 30 min at 10,000 × g. The pellets were dried to constant weight in a vacuum oven and weighed to provide the residual dry weight used to calculate results. The supernatant was removed to a separate tube and was usually two phases due to the saline content of the cells. The supernatant was reduced to half of the starting volume in a Savant Speed Vac, producing a single phase.
The samples were assayed using three analytical HPLC protocols developed for analysis of a wide variety of thiols. Coelution of a thiol-bimane derivative with a standard on two or more HPLC protocols was considered confirmation of the identity for that thiol. HPLC methods 1 and 2 were the primary methods used and the retention times for various biological thiols have been reported for these protocols (Fahey and Newton 1987). The retention times reported for unknown thiols (U-retention time) were those recorded using method 1M, a shortened version of method 1 as described by Koledin, et al. (Koledin et al. 2002). Quantitation was based on known standards when the identity of the thiol was confirmed and the fluorescent factor for the bimane derivative of cysteine (CySmB) was used for quantitation of unknown thiols. The control sample, in which thiols were blocked by NEM prior to reaction with mBBr, was utilized to verify that peaks resulting from nonthiol fluorescent components of the extract did not interfere with the analysis.
Assay of GlcN-Ins
Duplicate pellets labeled D and E were extracted in an equal volume of 60°C acetonitrile containing 20 μl per ml 1 M HEPES pH 7.5. The cells were vortexed every 5 min and incubated at 60°C for 15 min. The extract was cooled on ice and centrifuged. The supernatant was reduced to half the original volume, as above, to produce one phase. A sample of this extract was reacted with AccQ-Fluor (Waters) amine labeling reagent and the extract was assayed for GlcN-Ins according to Buchmeier et al (Buchmeier et al. 2003). The retention time for AccQ-GlcN-Ins was typically 22-24 min using the chromatographic protocol (method 4) described by Anderberg et al (Anderberg et al. 1998).
Mycothiol S-conjugate amidase (Mca) activity
The final cell pellet (pellet F) from each culture was extracted in neutral aqueous buffer for estimation of the Mca activity. Each pellet was suspended in 5 ml per g wet weight of 50 mM NaCl, 50 mM HEPES pH 7.5 containing 3 mM 2-mercaptoethanol and 35 μM each of N-α-p-tosyl-L-phenylalanylchloromethyl ketone and N-α-p-tosyl-L-lysinechloromethyl ketone to inhibit proteolytic degradation. The cells were disrupted by sonicating 5 times for 30 sec using a 50% pulsed sonication at maximum power (Branson Sonifier). The cells were iced between sonications and the extract was maintained between 5 and 15°C at all times. The cell free supernatant was collected by centrifugation at 12,000 × g for 30 min at 4°C and the protein content was assayed by the Bradford dye binding assay (Biorad). For the amidase reaction, extraction buffer containing MSmB derived from M. smegmatis (Newton et al. 2000b) was preheated at 37°C. The reaction was started by the addition of 12.5 μl of extract to produce a final volume of 250 μl containing 0.1 mM MSmB. At 0, 3, 6, and 12 min a 50 μl sample was removed and mixed with 200 μl of 40 mM methanesulfonic acid to rapidly terminate the reaction. The sample was centrifuged for 3 min at 13,000 × g and was analyzed by HPLC without dilution for the formation of AcCySmB as described previously (Newton et al. 2000b). The amount of extract was adjusted as necessary to produce accurate initial rate estimates and the activity is reported as specific activity, nmol/min/mg protein.
Time course analysis of strain CNQ703
A culture of CNQ703 was inoculated in 2 liters TCG medium with shaking (250 rpm) at 28°C. Samples (200 ml) were removed at 2, 3, 6, 8, and 10 days. The cells were pelleted by centrifugation into 4 pellets of equal size. Three of the pellets were extracted in 1 ml of 4 mM mBBr in acetonitrile plus 20 μl of 1M HEPES pH 8.0 and processed as described above for thiol samples. The fourth pellet was extracted in 1 ml 10 mM NEM plus 20 μl of 1M HEPES pH 8.0 and incubated at 60°C for 15 min. The sample was cooled on ice and the cell debris removed by centifugation for 5 minutes at 13,000 × g. The supernatant was reduced in a speed vac to generate a 0.5 ml one phase supernatant. An aliquot (100 μl) of the NEM supernatant was reacted with 2 mM mBBr for 15 min at 60° C to serve as a control for the thiol samples. The balance of the NEM sample was reacted with 10 mM dithothreitol (DTT) for 20 min at 23° C to reduce disulfides and then with 20 mM mBBr for an additional 15 min to derivitze the DTT released thiols. A redox ratio was calculated as the mean of the tripicate thiol analyses divided by the DTT reduced soluble disulfides.
Isolation and structure elucidation of U25
U25 was isolated as the bimane derivative from a 20 g pellet derived from 5 liters of a 3-day CNQ703 culture. The cells were collected by centrifugation at 4000 × g for 30 min at 4°C and extracted in 200 ml of 50% acetonitrile (20 mM HEPES pH 8.0) containing 2 mM mBBr at 60°C. After centrifugation the supernatant was reduced to 25 ml using a rotary evaporator and diluted to 100 ml with water. The bimane derivatives were concentrated by solid phase extraction on two Sep Pak C-18 (Waters, 5 g each) columns. The columns were washed with aqueous 0.1% TFA, and eluted with 0.1% aqueous TFA containing 10%, 25%, and 50% methanol. U25mB eluted in the 10-25% methanol fraction and this was concentrated on a Savant Speed Vac. U25mB was purified on a Vydac preparative HPLC column (#218TP1022) using aqueous 0.1% TFA and methanol as solvents and a linear 0-40% methanol gradient. U25mB (10 mg) eluted in about 20% methanol and was further purified in a second pass on the same column. For structural studies residual water and TFA were removed in a Savant Speed Vac.
NMR studies of U25-mB
The structure of U25-mB was determined by a combination of 1D and 2D NMR experiments. All spectra were obtained on a Varian INOVA spectrometer with 1H NMR spectra being recorded at 500 MHz and 13C NMR spectra recorded at 125 MHz, using residual solvent as an internal reference (CD3OD, δH 3.30 ppm, δC 49.0 ppm). Carbon multiplicities were determined using a multiplicity edited HSQC experiment, while 1H-1H and 1H-13C correlations were established by gradient homonuclear correlated spectroscopy (gCOSY) and gradient homonuclear multiple bond coherence (gHMBC) NMR experiments.
Results
Initial findings
We set out to sample a broad spectrum of marine actinomycetes representative of the main MAR groupings (Table 1). Mycothiol was detected in all of the strains initially studied but we soon found that an additional thiol that could not be assigned as a known biological thiol (Fahey and Newton 1987; Newton and Fahey 1995) was also present. It was designated U25 based upon the retention time (25 min) under HPLC analysis using HPLC method 1M (Koledin et al. 2002), one of three protocols used to analyze thiols. In some cases the U25 content exceeded the MSH content. One MAR2 strain, CNQ703, appeared to produce U25 in large amounts and was selected for more detailed study.
Thiol composition and redox status of MAR2 Strain CNQ703
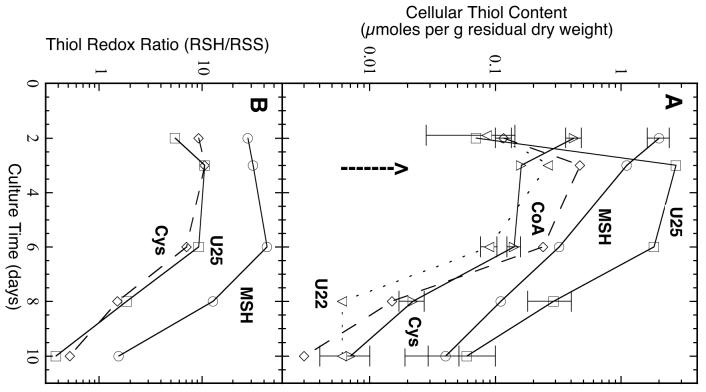
We determined the thiol content (Fig. 2A) and the thiol redox ratio (Fig. 2B) from day 2 to day 10 of the growth of CNQ703. The culture could not be monitored by optical density owing to the production of intensely absorbing secondary metabolites beginning on day 3, but the yield of residual dry weight per liter of culture remained constant at 4.1 ± 1.0 g per liter from day 2 to day 10 indicating that the culture was in stationary phase. During this period the MSH content fell 50-fold (Fig. 2A). However, a novel thiol (U25) was also produced whose content increased almost 40-fold from day 2 to day 3 and U25 was the dominant thiol from day 3 to day 10. A second unidentified thiol, U22, remained at a low level throughout. The penultimate intermediate in MSH biosynthesis, Cys-GlcN-Ins (Fig. 1), was also observed on day 3 (0.037 ± 0.007 μmol per g) but was not observed above background (≤ 0.001 μmol per g) at any other time.
Fig. 2.
Variation in thiol content (A) and thiol redox ratio (B) with time for MAR 2 strain CNQ703: ◯, MSH; ◻, U25 (N-propionyl-desacetyl-mycothiol; PdAMSH); ◇, CoA; Δ, Cys; ▽, U22. Arrow indicates the onset of secondary metabolite production. Error bars represent the range of values, where larger than the symbol, from duplicate determinations.
Thiol components present in disulfide forms (RSS) were measured and this permitted calculation of the thiol redox ratios (RSH/RSS) that are shown in Fig. 2B. The only terrestrial actinomycetes for which redox ratios have been determined are M. smegmatis and M. tuberculosis. In M. smegmatis the MSH/MSS ratio was found to fall from ∼1000 in exponential phase to ∼200 in stationary phase (Newton et al. 2005) whereas in M. tuberculosis it drops from ∼180 in exponential phase to ∼50 in stationary phase (Buchmeier et al. 2006). The values from Fig. 2B (MSH/MSS = 30-50) at days 2-6 are comparable to the stationary phase values for M. tuberculosis. The sharp fall on days 8 and 10 reflects both a fall in MSH and an increase in MSS. The redox state of U25 is comparable to that of cysteine (Fig. 2) and is several fold more oxidized than that of MSH.
Structure of U25
The bimane derivative of unknown thiol U25 was purified from mBBr labeled cell extracts of CNQ703 by solid phase extraction and HPLC. Electrospray positive ion mass spectroscopy produced a molecular ion peak at m/z 713.2 (MNa+) and a weaker one at m/z 691.1 (MH+), corresponding to molecules of 14 mass units greater than that of MSH. A strong fragmentation ion at m/z 511.1 reflects the loss of a 180 Da fragment from the MH+ peak at m/z 691.1. This can be assigned to loss of a C6H12O6 inositol unit as was seen in the mass spectrum of the bimane derivative of mycothiol, MSmB (Newton et al. 1995).
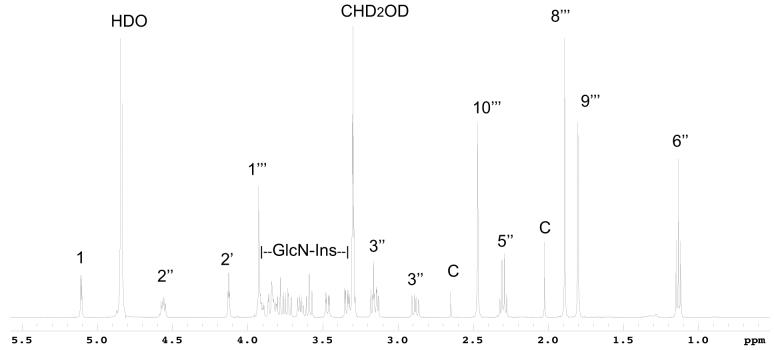
Further structural analysis of the bimane derivative of U25 (U25mB) was achieved by NMR studies. The 500 MHz proton spectrum (Fig. 3) was very similar to that of MSmB (Newton et al. 1995) but contained additional signals ascribable to a CH3CH2CO residue (δ 1.14 ppm (t, J = 7.5 Hz); δ 2.3 ppm (q, J = 7.5 Hz)) and it lacked the singlet resonance at 2.09 ppm for the acetyl moiety of AcCys in MSmB. The myo-inositol and glucosamine residues were established by a combination of gCOSY NMR correlations and coupling constants analysis (Table 2). The structure and the numbering of the carbons for N-propionyl-desacetyl-mycothiol is shown in Fig. 4. The amide linkage between the anomeric carbon of glucosamine (C1) and carbonyl carbon C1” of cysteine was determined based on a HMBC NMR correlation between H1 (δ 5.11 ppm (d, J = 4.0 Hz)) and C1” (δ 172.5 ppm), while the N-propionyl residue was established to be at C2” based on a HMBC NMR correlation between H2” (δ 4.56 ppm (dd, J = 8.5, 5.0 Hz)) and C4” (δ 177.4 ppm). These studies establish that U25 is a close relative of MSH with the N-acetyl group replaced by an N-propionyl residue, i.e., 1-O-[2-[[(2R)-2-(propionylamino)-3-mercapto-1-oxopropyl]amino]-2-deoxy-α-D-glucopyranosyl]-D-myo-inositol (propionyl-Cys-GlcN-Ins). We propose the common name N-propionyl-desacetyl-mycothiol (PdAMSH) for this homolog of mycothiol.
Fig. 3.
500 MHz proton NMR spectrum of U25-mB; HDO and CHD2OD, solvent impurities; C, contaminant.
Table 2.
NMR Data for U25-mB recorded in CD3OD
| Carbon # | 1H δ (mult., #, J (Hz)) | 13C δ | HMBC correlations |
|---|---|---|---|
| D-glucosamine | |||
| 1 | 5.11 (d, 1H, J = 4.0) | 100.2 | H2, H1′ |
| 2 | 3.90 (dd, 1H, J = 10.5, 4.0) | 55.7 | H1, H3 |
| 3 | 3.73 (dd, 1H, J = 10.5, 9.5) | 73.0 | H2, H4 |
| 4 | 3.32 (t, 1H, J = 9.5) | 72.4 | H3, H5, H6 |
| 5 | 3.82 (m, 1H) | 74.4 | H4, H6 |
| 6 | 3.92 (m, 1H) | 62.8 | H4, H5 |
| 3.65 (dd, 1H, J = 11, 2.5Hz) | |||
| D-myo-inositol | |||
| 1′ | 3.47 (dd, 1H, J = 9.5, 3.0) | 80.5 | H1, H2′, H6′ |
| 2′ | 4.12 (t, 1H, J = 3.0) | 73.4 | H1′, H3′ |
| 3′ | 3.34 (dd, 1H, J = 9.5, 3.0) | 73.2 | H2′, H6′ |
| 4′ | 3.59 (t, 1H, J = 9.5) | 74.0 | H3′, H5′ |
| 5′ | 3.16 (t, 3H, J = 9.5) | 76.5 | H4′, H6′ |
| 6′ | 3.78 (t, 1H, J = 9.5) | 74.1 | H1′, H5′ |
| N-propionyl cysteine | |||
| 1″ | - | 172.5 | H2, H2″. H3″ |
| 2″ | 4.56 (dd, 1H, J = 8.5,5.0) | 54.2 | H3″ |
| 3″ | 2.89 (dd, 1H, J = 13.5, 8.5) | 35.1 | H2″, H1‴ |
| 3.14 (dd, 1H, J = 13.5, 5.0) | |||
| 4″ | - | 177.4 | H5″, H6″ |
| 5″ | 2.30 (q, 2H, J = 7.5) | 29.9 | H6″ |
| 6″ | 1.14 (t, 3H, J = 7.5) | 10.2 | H5″ |
| Bimane | |||
| 1‴ | 3.92 (s, 2H) | 26.2 | |
| 2‴ | 114.9 | H1‴, H8‴ | |
| 3‴ | 148.9 | H1‴, H8‴ | |
| 4‴ | 162.3 | H8‴ | |
| 5‴ | 162.8 | H9‴ | |
| 6‴ | 149.7 | H9‴, H10‴ | |
| 7‴ | 112.9 | H9‴, H10‴ | |
| 8‴ | 1.89 (s, 3H) | 7.0 | |
| 9‴ | 1.80 (s, 3H) | 6.5 | |
| 10‴ | 2.47 (s, 3H) | 11.8 |
Fig. 4.
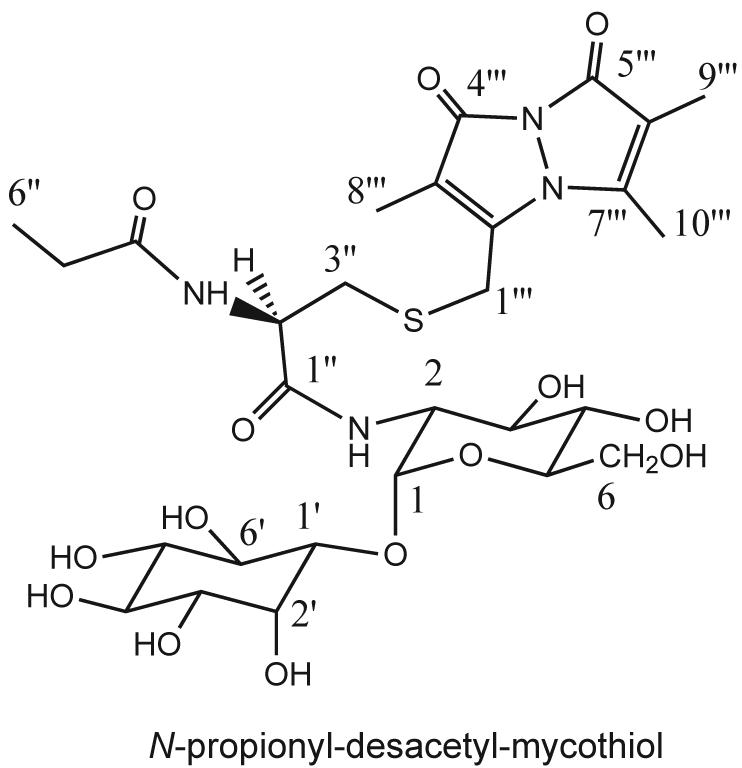
Structure of N-propionyl-desacetyl-mycothiol (PdAMSH) with numbering of carbons as listed in Table 2.
Survey of marine actinomycetes
Diverse marine-derived actinomycetes belonging to the families Micromonosporaceae, Streptomycetaceae, and Thermomonosporaceae were selected to test the generality of production of MSH, PdAMSH and related compounds production (Table 1). These strains include four representatives of the newly described marine genus Salinispora (Maldonado et al. 2005), as well as examples of the clearly defined clade within the Streptomycetaceae that has tentatively been called MAR2. An earlier study of the terrestrial Streptomyces clavuligerus had established that a thiol, later identified as mycothiol (Newton et al. 1995), is produced at substantial and roughly stable levels during exponential growth and into stationary phase when secondary metabolites are produced (Newton et al. 1993). For the present survey the various marine actinomycetes strains were grown and harvested 3-10 days after innoculation, following entry into stationary phase when secondary metabolite production became evident. This provided ample material from a one-liter culture to prepare samples for determination of thiols and GlcN-Ins in duplicate and to assay Mca activity.
In the Salinispora strains (MAR1), MSH was the major thiol with most values for MSH content between 2 and 4 μmol per g residual dry weight (Table 3). One strain, CNH725, gave a relatively low value (0.8 μmol per g residual dry weight) value for MSH content. The level of PdAMSH was 2-20% that of MSH in these samples. Significant levels (0.3-1.5 μmol per g) of an additional unknown, U18, were found in all strains except CNR107 and lower levels of U27 (0.14-0.38 μmol per g) were present in all strains except CNH725. The level of Mca activity was quite high in all Salinispora strains. Hydrogen sulfide was found at significant levels in all Salinispora strains (Table 3). This likely derives from iron-sulfur proteins, the labile sulfide being released and reacted with mBBr under our extraction conditions. The values for H2S are typical of those found in a wide range of bacteria (Newton et al. 1996).
Table 3.
GlcN-Ins, thiol, and Mca assays of marine actionmycetes.
| Cellular Content, μmoles per g residual dry weighta | ||||||
|---|---|---|---|---|---|---|
| Organism | Mca Activityb | |||||
| GlcN-Ins | Cys | MSH | CoA | PdAMSH (U25) | ||
| Salinispora MAR1 c | 0.3-1.5 | 0.4-1.8 | 0.8-4 | 0.1-0.4 | 0.03-0.8 | 30-133 |
| “Marinispora” MAR2d | 0.1-0.5 | 0.6-2.9 | 2.3-3.8 | <0.02-0.8 | 0.6-9 | 27-71 |
| Marine streptomycetaceae | ||||||
| MAR3e | 0.1-0.9 | 0.1-2.1 | 0.5-12 | <0.03-2.4 | <0.1-0.4 | 3-38 |
| MAR4f | 0.35, 0.40 | 0.6, 3.6 | 0.2, 33 | 0.5, 10 | <0.1, 0.44 | 29, 43 |
| Marine thermomonosporaceae | ||||||
| MAR5 CNR363g | 0.6 | 0.3 | 0.7 | 0.14 | <0.02 | 20 |
| MAR6 CNR431h | 0.11 | 1.1 | 27 | 1.0 | 0.43 | 1.2 |
Range of values from duplicate analyses of each strain; GSH and Cys-GlcN-Ins <0.005 in all extracts.
Assayed at 37°C in 0.1 mM MSmB, 50 mM NaCl, 1 mM 2-mercaptoethanol, 50 mM HEPES pH 7.5, nmol/min/mg protein.
Strains CNR107, CNR003, CNH643, CNH725 and CNQ976; also contained SSO3−2 (0.3-1.3) and H2S (1.5-2.5).
Strains CNR252, CNQ695 and CNQ703; also contained SSO3−2 (1.1-1.8) and H2S (2.2-6.6).
Strains CNQ687, CNQ698, CNQ719, CNQ766, CNQ857; also contained SSO3−2 (0.5-2.5) and H2S (0.7-6).
Strains CNR530 and CNQ525.
Also contained SSO3−2 (1.0) and H2S (1.1).
Also contained SSO3−2 (1.8) and H2S (7).
The three “Marinispora“ (Kwon et al. 2006) (MAR2) strains examined all had significant levels of GlcN-Ins, MSH and PdAMSH (Table 3). PdAMSH was the dominant thiol in strains CNQ695 and CNQ703 but MSH was 6-fold higher in strain CNR252. All three strains had a high level of Mca activity.
Five strains formed a coherent clade, designated MAR3, within the Streptomycetaceae. All were found to produce GlcN-Ins and MSH at measurable levels that were well reproduced in replicate samples of a given culture (Table 3). Significant HPLC peaks assignable to PdAMSH were found in three of the five strains but the level in strains CNQ687 and CNQ857 was undetectable (<0.1 μmol per g). Two strains of the MAR4 group gave widely different results, one strain (CNR530) producing the highest level of MSH of any strain tested and the other (CNR525) producing the lowest MSH content but both had similar amounts of GlcN-Ins (Table 3). GlcN-Ins is a dedicated intermediate in mycothiol biosynthesis and has no known function except MSH production. Strain CNR530 having the highest MSH content also had significant, but much lower PdAMSH whereas PdAMSH was undetectable in strain CNQ525.
One strain (CNR363) from the marine Thermomonosporaceae MAR5 group and a second (CNR431) from the MAR6 group differed 5-fold in the GlcN-Ins content and nearly 40-fold in MSH content but in opposite directions (Table 3). No PdAMSH (<0.002 μmol per g dry weight) was detected in strain CNR363 wherease strain CNR431 contained 0.43 μmol per g dry weight. Substantial Mca activity was found in both strains.
Discussion
The most intriguing feature of the present results is the replacement of MSH by PdAMSH during secondary metabolite production. It appears that this phenomenon is not restricted to MAR2 strain CNQ703. A survey of representative marine actinomycetes revealed that PdAMSH was detected in all five MAR1 strains, in all three MAR2 strains, in three of five MAR3 strains, in one of two MAR4 strains, in the one MAR6 strain but not in the one MAR5 strain. Whether all of the PdAMSH producing strains exhibit profiles similar to CNQ703 (Fig. 2) was not determined but it is noted that PdAMSH did exceed MSH in the data for another MAR2 strain, CNQ695. Moreover, we have recently found that PdAMSH is also produced in mycobacteria under some conditions (G. L. Newton, N. Buchmeier and R. C. Fahey, unpublished data). Thus, synthesis of PdAMSH is not limited to the marine actinomycetes and may prove to be a more general phenomenon among the mycothiol producing Actinobacteria.
PdAMSH is the first derivative of MSH to be found in wild type Actinobacteria. However analogs of MSH in which acyl groups other than acetyl or propionyl are linked to Cys-GlcN-Ins have been reported in mshD null mutants of M. smegmatis and M. tuberculosis (Newton et al. 2005; Buchmeier et al. 2006). In these mutants Cys-GlcN-Ins accumulates to such high levels (0.5 - 1.5 μmol per g dry weight) that transacylation chemical reaction with succinyl-CoA leads to the generation of succinyl-Cys-GlcN-Ins, a process that occurs some 20-fold faster than chemical production of MSH from acetyl-CoA or PdAMSH from propionyl-CoA (Newton et al. 2005). Formyl-Cys-GlcN-Ins is also produced from Cys-GlcN-Ins in these mutants, presumably by an enzymatic process. Chemical transacylation of Cys-GlcN-Ins by propionyl-CoA cannot be the source of PdAMSH in the marine actinomycetes since the level of Cys-GlcN-Ins in these organisms is very low or undetectable (<0.005 μmol per g dry weight). Production of PdAMSH must involve an enzymatic process. M. tuberculosis MshD is known to utilize propionyl-CoA at 20-25% the rate of acetyl-CoA (Vetting et al. 2006). If the marine actinomycete MshD has similar substrate flexibility then it seems likely that it is responsible for the enzymatic production of PdAMSH. However, replacement of MSH by PdAMSH would require that the level of propionyl-CoA substantially exceed the level of acetyl-CoA. Marine actinomycetes are known to produce a number of polyketide secondary metabolites (Kwon et al. 2006; Jensen et al. 2007; Udwary et al. 2007) and propionyl-CoA is used as an acyl starter unit and to generate methylmalonyl-CoA extender units in polyketide synthesis (Walsh 2003). Thus, the production of propionyl-CoA during generation of secondary metabolites is expected and PdAMSH synthesis did coincide with the appearance of the black-green secondary metabolite on day 3 (Fig. 2).
What function does production of PdAMSH serve? One possibility is that it functions as a detoxification pathway. Degradation of odd chain and branched fatty acids (Boshoff and Barry 2005) (Koburger et al. 2005) and of cholesterol (Pandey and Sassetti 2008) leads to the production of propionyl-CoA, as does growth in the presence of propionate (Brock and Buckel 2004). Accumulation of propionyl-CoA during degradative metabolism is potentially toxic owing to accumulation of intermediates produced by its metabolism via the methylcitrate cycle (Brock 2005). Diversion of propionyl-CoA into PdAMSH production could be a possible way of avoiding such toxicity.
It is also possible that production of PdAMSH serves a regulatory or protective function involving protein thiol groups. The redox status of PdAMSH is significantly more oxidized than is that of MSH and becomes more oxidized in the latter stages of secondary metabolite production (Fig. 2). The disulfide of PdAMSH may be an inefficient substrate relative to that of MSH for mycothiol disulfide reductase (Patel and Blanchard 2001). In glutathione producing organisms such changes in thiol redox status lead to the glutathionylation of proteins (Prot-SSG formation) which is thought be involved in regulation of protein function or protection against overoxidation of sensitive thiol groups (Ghezzi 2005; Hurd et al. 2005; Masip et al. 2006; Dalle-Donne et al. 2007). Analogous thiolation of proteins may be important when PdAMSH becomes the dominant thiol and could serve similar functions. Elaboration of the function of PdAMSH is being undertaken in studies of mycobacteria where availability of mutants defective in mycothiol biosynthesis and enzymes of mycothiol metabolism are available.
The values for MSH content determined here can be compared with those for terrestrial actinomycetes: 0.8 - 2.3 μmol per g residual dry weight for Streptomyces clavuligerus, increasing during the transition from exponential to stationary phase (Newton et al. 1993), and ∼12 μmol per g residual dry weight for Mycobacterium smegmatis mc2155, remaining constant during transition from exponential to stationary phase (Newton et al. 2005). While most of the values in Table 3 are comparable, the values for strains CNR530 and CNR431 are the highest we have measured. Since these bacteria are readily harvesed from liquid growth medium by filtration, such high-producing MSH strains may represent a suitable resource for practical production of MSH. Mycothiol is a difficult molecule to generate by chemical synthesis (Jardine et al. 2002; Nicholas et al. 2002; Lee and Rosazza 2004) and is more efficiently obtained by isolation from cells (Unson et al. 1998; Steenkamp and Vogt 2004).
The levels of Mca activity found are very high in most marine actinomycetes, especially in strain CNR003 which produced the highest activity, 133 nmol/min/mg protein. The only terrestrial actinomycete for which Mca activity has been measured is M. smegmatis where the activity of a crude extract was found to be ∼1 nmol/min/mg (Newton et al. 2000b) or 30-130-fold lower than that observed in Salinospora strains. A role for MSH and Mca in the detoxification of antibiotics has been identified in M. smegmatis (Steffek et al. 2003; Rawat et al. 2004) and in S. coelicolor (Park and Roe 2008). Thus, it appears likely that MSH and the high Mca activity are associated with secondary metabolite detoxification in the marine actinomycetes.
In summary, several unusual aspects of mycothiol metabolism have been identified in studies of marine actinomycetes. A novel variant of mycothiol, N-propionyl-desacetyl-mycothiol, was shown to replace mycothiol when nutrient depletion occurs and secondary metabolites are formed. Exceptionally high levels of mycothiol S-conjugate amidase activity were found in nearly all marine actinomycetes examined. Two strains, CNR530 (MAR4 group) and CNR431 (MAR6 group) produced record high levels of mycothiol and may have utility as a source of this thiol.
Acknowledgements
This work was supported by NIH grant AI49174 from the National Institute of Allergy and Infectious Diseases and grant MCB-0235705 from the National Science Foundation to RCF. Additional support was provided from the NIH, National Cancer Institute, under grant CA-44848 (to WF), and in part by the National Sea Grant College Program of the U.S. Department of Commerce's National Oceanic and Atmospheric Administration under NOAA Grant # NA040AR4170038, project # R/MP-96, through the California Sea Grant College Program; and in part by the California State Resources Agency to WF. The views expressed herein do not necessarily reflect the views of any of those organizations. We thank Nancy Buchmeier for critical review of the manuscript.
References
- Anderberg SJ, Newton GL, Fahey RC. Mycothiol biosynthesis and metabolism: Cellular levels of potential intermediates in the biosynthesis and degradation of mycothiol. J Biol Chem. 1998;273:30391–30397. doi: 10.1074/jbc.273.46.30391. [DOI] [PubMed] [Google Scholar]
- Boshoff HI, Barry CE., 3rd Tuberculosis - metabolism and respiration in the absence of growth. Nat Rev Microbiol. 2005;3:70–80. doi: 10.1038/nrmicro1065. [DOI] [PubMed] [Google Scholar]
- Brock M. Generation and phenotypic characterization of Aspergillus nidulans methylisocitrate lyase deletion mutants: methylisocitrate inhibits growth and conidiation. Appl Environ Microbiol. 2005;71:5465–5475. doi: 10.1128/AEM.71.9.5465-5475.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brock M, Buckel W. On the mechanism of action of the antifungal agent propionate. Eur J Biochem. 2004;271:3227–3241. doi: 10.1111/j.1432-1033.2004.04255.x. [DOI] [PubMed] [Google Scholar]
- Buchmeier N, Fahey RC. The mshA gene encoding the glycosyltransferase of mycothiol biosynthesis is essential in Mycobacterium tuberculosis Erdman. FEMS Microbiol Lett. 2006;264:74–79. doi: 10.1111/j.1574-6968.2006.00441.x. [DOI] [PubMed] [Google Scholar]
- Buchmeier NA, Newton GL, Fahey RC. A mycothiol synthase mutant of Mycobacterium tuberculosis has an altered thiol-disulfide content and limited tolerance to stress. J Bacteriol. 2006;188:6245–6252. doi: 10.1128/JB.00393-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchmeier NA, Newton GL, Koledin T, Fahey RC. Association of mycothiol with protection of Mycobacterium tuberculosis from toxic oxidants and antibiotics. Mol Microbiol. 2003;47:1723–1732. doi: 10.1046/j.1365-2958.2003.03416.x. [DOI] [PubMed] [Google Scholar]
- Bull AT, Stach JE. Marine actinobacteria: new opportunities for natural product search and discovery. Trends Microbiol. 2007;15:491–499. doi: 10.1016/j.tim.2007.10.004. [DOI] [PubMed] [Google Scholar]
- Bull AT, Stach JE, Ward AC, Goodfellow M. Marine actinobacteria: perspectives, challenges, future directions. Antonie Van Leeuwenhoek. 2005;87:65–79. [PubMed] [Google Scholar]
- Bzymek KP, Newton GL, Ta P, Fahey RC. Mycothiol import by Mycobacterium smegmatis and function as a resource for metabolic precursors and energy production. J Bacteriol. 2007;189:6796–6805. doi: 10.1128/JB.00644-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalle-Donne I, Rossi R, Giustarini D, Colombo R, Milzani A. S-glutathionylation in protein redox regulation. Free Radic Biol Med. 2007;43:883–898. doi: 10.1016/j.freeradbiomed.2007.06.014. [DOI] [PubMed] [Google Scholar]
- Fahey RC, Brown WC, Adams WB, Worsham MB. Occurrence of glutathione in bacteria. J Bacteriol. 1978;133:1126–1129. doi: 10.1128/jb.133.3.1126-1129.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahey RC, Newton GL. Determination of low-molecular-weight thiols using monobromobimane fluorescent labeling and high-performance liquid chromatography. Methods Enzymol. 1987;143:85–96. doi: 10.1016/0076-6879(87)43016-4. [DOI] [PubMed] [Google Scholar]
- Fahey RC, Sundquist AR. Evolution of glutathione metabolism. Adv Enzymol Relat Areas Mol Biol. 1991;64:1–53. doi: 10.1002/9780470123102.ch1. [DOI] [PubMed] [Google Scholar]
- Feling RH, Buchanan GO, Mincer TJ, Kauffman CA, Jensen PR, Fenical W. Salinosporamide A: a highly cytotoxic proteasome inhibitor from a novel microbial source, a marine bacterium of the new genus Salinospora. Angew Chem Int Ed Engl. 2003;42:355–357. doi: 10.1002/anie.200390115. [DOI] [PubMed] [Google Scholar]
- Fenical W, Jensen PR. Developing a new resource for drug discovery: marine actinomycete bacteria. Nat Chem Biol. 2006;2:666–673. doi: 10.1038/nchembio841. [DOI] [PubMed] [Google Scholar]
- Fiedler HP, Bruntner C, Bull AT, Ward AC, Goodfellow M, Potterat O, Puder C, Mihm G. Marine actinomycetes as a source of novel secondary metabolites. Antonie Van Leeuwenhoek. 2005;87:37–42. doi: 10.1007/s10482-004-6538-8. [DOI] [PubMed] [Google Scholar]
- Ghezzi P. Regulation of protein function by glutathionylation. Free Radic Res. 2005;39:573–580. doi: 10.1080/10715760500072172. [DOI] [PubMed] [Google Scholar]
- Hand CE, Honek JF. Biological chemistry of naturally occurring thiols of microbial and marine origin. J Nat Prod. 2005;68:293–308. doi: 10.1021/np049685x. [DOI] [PubMed] [Google Scholar]
- Hurd TR, Costa NJ, Dahm CC, Beer SM, Brown SE, Filipovska A, Murphy MP. Glutathionylation of mitochondrial proteins. Antioxid Redox Signal. 2005;7:999–1010. doi: 10.1089/ars.2005.7.999. [DOI] [PubMed] [Google Scholar]
- Jardine MA, Spies HS, Nkambule CM, Gammon DW, Steenkamp DJ. Synthesis of mycothiol, 1D-1-O-(2-[N-acetyl-L-cysteinyl]amino-2-deoxy-alpha-D-glucopyranosyl)- myo- inositol, principal low molecular mass thiol in the actinomycetes. Bioorg Med Chem. 2002;10:875–881. doi: 10.1016/s0968-0896(01)00383-2. [DOI] [PubMed] [Google Scholar]
- Jensen PR, Gontang E, Mafnas C, Mincer TJ, Fenical W. Culturable marine actinomycete diversity from tropical Pacific Ocean sediments. Environ Microbiol. 2005a;7:1039–1048. doi: 10.1111/j.1462-2920.2005.00785.x. [DOI] [PubMed] [Google Scholar]
- Jensen PR, Mincer TJ, Williams PG, Fenical W. Marine actinomycete diversity and natural product discovery. Antonie Van Leeuwenhoek. 2005b;87:43–48. doi: 10.1007/s10482-004-6540-1. [DOI] [PubMed] [Google Scholar]
- Jensen PR, Williams PG, Oh DC, Zeigler L, Fenical W. Species-specific secondary metabolite production in marine actinomycetes of the genus Salinispora. Appl Environ Microbiol. 2007;73:1146–1152. doi: 10.1128/AEM.01891-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koburger T, Weibezahn J, Bernhardt J, Homuth G, Hecker M. Genome-wide mRNA profiling in glucose starved Bacillus subtilis cells. Mol Genet Genomics. 2005;274:1–12. doi: 10.1007/s00438-005-1119-8. [DOI] [PubMed] [Google Scholar]
- Koledin T, Newton GL, Fahey RC. Identification of the mycothiol synthase gene (mshD) encoding the acetyltransferase producing mycothiol in actinomycetes. Arch Microbiol. 2002;178:331–337. doi: 10.1007/s00203-002-0462-y. [DOI] [PubMed] [Google Scholar]
- Kwon HC, Kauffman CA, Jensen PR, Fenical W. Marinomycins A-D, antitumor-antibiotics of a new structure class from a marine actinomycete of the recently discovered genus “marinispora”. J Am Chem Soc. 2006;128:1622–1632. doi: 10.1021/ja0558948. [DOI] [PubMed] [Google Scholar]
- Lee S, Rosazza JP. First total synthesis of mycothiol and mycothiol disulfide. Org Lett. 2004;6:365–368. doi: 10.1021/ol0362008. [DOI] [PubMed] [Google Scholar]
- Maldonado LA, Fenical W, Jensen PR, Kauffman CA, Mincer TJ, Ward AC, Bull AT, Goodfellow M. Salinispora arenicola gen. nov., sp. nov. and Salinispora tropica sp. nov., obligate marine actinomycetes belonging to the family Micromonosporaceae. Int J Syst Evol Microbiol. 2005;55:1759–1766. doi: 10.1099/ijs.0.63625-0. [DOI] [PubMed] [Google Scholar]
- Masip L, Veeravalli K, Georgiou G. The many faces of glutathione in bacteria. Antioxid Redox Signal. 2006;8:753–762. doi: 10.1089/ars.2006.8.753. [DOI] [PubMed] [Google Scholar]
- Mincer TJ, Jensen PR, Kauffman CA, Fenical W. Widespread and persistent populations of a major new marine actinomycete taxon in ocean sediments. Appl Environ Microbiol. 2002;68:5005–5011. doi: 10.1128/AEM.68.10.5005-5011.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton GL, Arnold K, Price MS, Sherrill C, delCardayré SB, Aharonowitz Y, Cohen G, Davies J, Fahey RC, Davis C. Distribution of thiols in microorganisms: Mycothiol is a major thiol in most actinomycetes. J Bacteriol. 1996;178:1990–1995. doi: 10.1128/jb.178.7.1990-1995.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton GL, Av-Gay Y, Fahey RC. N-Acetyl-1-D-myo-inosityl-2-amino-2-deoxy-α-D-glucopyranoside deacetylase (MshB) is a key enzyme in mycothiol biosynthesis. J Bacteriol. 2000a;182:6958–6963. doi: 10.1128/jb.182.24.6958-6963.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton GL, Av-Gay Y, Fahey RC. A novel mycothiol-dependent detoxification pathway in mycobacteria involving mycothiol S-conjugate amidase. Biochemistry. 2000b;39:10739–10746. doi: 10.1021/bi000356n. [DOI] [PubMed] [Google Scholar]
- Newton GL, Bewley CA, Dwyer TJ, Horn R, Aharonowitz Y, Cohen G, Davies J, Faulkner DJ, Fahey RC. The structure of U17 isolated from Streptomyces clavuligerus and its properties as an antioxidant thiol. Eur J Biochem. 1995;230:821–825. doi: 10.1111/j.1432-1033.1995.0821h.x. [DOI] [PubMed] [Google Scholar]
- Newton GL, Fahey RC. Determination of biothiols by bromobimane labeling and high-performance liquid chromatography. Methods Enzymol. 1995;251:148–166. doi: 10.1016/0076-6879(95)51118-0. [DOI] [PubMed] [Google Scholar]
- Newton GL, Fahey RC. Mycothiol biochemistry. Arch Microbiol. 2002;178:388–394. doi: 10.1007/s00203-002-0469-4. [DOI] [PubMed] [Google Scholar]
- Newton GL, Fahey RC, Cohen G, Aharonowitz Y. Low molecular weight thiols in streptomycetes and their potential role as antioxidants. J Bacteriol. 1993;175:2734–2742. doi: 10.1128/jb.175.9.2734-2742.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton GL, Koledin T, Gorovitz B, Rawat M, Fahey RC, Av-Gay Y. The glycosyltransferase gene encoding the enzyme catalyzing the first step of mycothiol biosynthesis (mshA) J Bacteriol. 2003;185:3476–3479. doi: 10.1128/JB.185.11.3476-3479.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton GL, Ta P, Bzymek K, Fahey RC. Biochemistry of the initial steps of mycothiol biosynthesis. J Biol Chem. 2006;281 doi: 10.1074/jbc.M604724200. in press. [DOI] [PubMed] [Google Scholar]
- Newton GL, Ta P, Fahey RC. A mycothiol synthase mutant of Mycobacterium smegmatis produces novel thiols and has an altered thiol redox status. J Bacteriol. 2005;187:7309–7316. doi: 10.1128/JB.187.21.7309-7316.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton GL, Unson MD, Anderberg SJ, Aguilera JA, Oh NN, delCardayré SB, Davies J, Av-Gay Y, Fahey RC. Characterization of a Mycobacterium smegmatis mutant defective in 1-D-myo-inosityl-2-amino-2-deoxy-〈-D-glucopyranoside and mycothiol biosynthesis. Biochem Biophys Res Commun. 1999;255:239–244. doi: 10.1006/bbrc.1999.0156. [DOI] [PubMed] [Google Scholar]
- Nicholas GM, Kovac P, Bewley CA. Total synthesis and proof of structure of mycothiol bimane. J Amer Chem Soc. 2002;124:3492–3493. doi: 10.1021/ja017891a. [DOI] [PubMed] [Google Scholar]
- Pandey AK, Sassetti CM. Mycobacterial persistence requires the utilization of host cholesterol. Proc Natl Acad Sci U S A. 2008;105:4376–4380. doi: 10.1073/pnas.0711159105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J-H, Roe J-H. Mycothiol regulates and is regulated by a thiol-specific anti-sigma factor RsrA and sigmaR in Streptomyces coelicolor. Mol Microbiol. 2008 doi: 10.1111/j.1365-2958.2008.06191.x. in press. [DOI] [PubMed] [Google Scholar]
- Patel MP, Blanchard JS. Mycobacterium tuberculosis mycothione reductase: pH dependence of the kinetic parameters and kinetic isotope effects. Biochemistry. 2001;40:3119–3126. doi: 10.1021/bi0029144. [DOI] [PubMed] [Google Scholar]
- Rawat M, Newton GL, Ko M, Martinez GJ, Fahey RC, Av-Gay Y. Mycothiol-deficient Mycobacterium smegmatis mutants are hypersensitive to alkylating agents, free radicals and antibiotics. Antimicrob Agents Chemother. 2002;46:3348–3355. doi: 10.1128/AAC.46.11.3348-3355.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawat M, Uppal M, Newton G, Steffek M, Fahey RC, Av-Gay Y. Targeted mutagenesis of the Mycobacterium smegmatis mca gene, encoding a mycothiol-dependent detoxification protein. J Bacteriol. 2004;186:6050–6058. doi: 10.1128/JB.186.18.6050-6058.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sareen D, Newton GL, Fahey RC, Buchmeier NA. Mycothiol is essential for growth of Mycobacterium tuberculosis Erdman. J Bacteriol. 2003;185:6736–6740. doi: 10.1128/JB.185.22.6736-6740.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sareen D, Steffek M, Newton GL, Fahey RC. ATP-dependent L-cysteine:1D-myo-inosityl 2-amino-2-deoxy-α-D-glucopyranoside ligase, mycothiol biosynthesis enzyme MshC, is related to class I cysteinyl-tRNA synthetases. Biochemistry. 2002;41:6885–6890. doi: 10.1021/bi012212u. [DOI] [PubMed] [Google Scholar]
- Steenkamp DJ, Vogt RN. Preparation and utilization of a reagent for the isolation and purification of low-molecular-mass thiols. Anal Biochem. 2004;325:21–27. doi: 10.1016/j.ab.2003.10.011. [DOI] [PubMed] [Google Scholar]
- Steffek M, Newton GL, Av-Gay Y, Fahey RC. Characterization of Mycobacterium tuberculosis mycothiol S-conjugate amidase. Biochemistry. 2003;42:12067–12076. doi: 10.1021/bi030080u. [DOI] [PubMed] [Google Scholar]
- Udwary DW, Zeigler L, Asolkar RN, Singan V, Lapidus A, Fenical W, Jensen PR, Moore BS. Genome sequencing reveals complex secondary metabolome in the marine actinomycete Salinispora tropica. Proc Natl Acad Sci U S A. 2007;104:10376–10381. doi: 10.1073/pnas.0700962104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unson MD, Newton GL, Davis C, Fahey RC. An immunoassay for the detection and quantitative determination of mycothiol. J Immunol Meth. 1998;214:29–39. doi: 10.1016/s0022-1759(98)00034-9. [DOI] [PubMed] [Google Scholar]
- Vetting MW, Yu M, Rendle PM, Blanchard JS. The Substrate-induced Conformational Change of Mycobacterium tuberculosis Mycothiol Synthase. J Biol Chem. 2006;281:2795–2802. doi: 10.1074/jbc.M510798200. [DOI] [PubMed] [Google Scholar]
- Walsh C. Antibiotics: Actions, Origins, Resistance. ASM Press; Washington, D.C.: 2003. [Google Scholar]