Abstract
All viruses rely on host cell proteins and their associated mechanisms to complete the viral life cycle. Identifying the host molecules that participate in each step of virus replication could provide valuable new targets for antiviral therapy, but this goal may take several decades to achieve with conventional forward genetic screening methods and mammalian cell cultures. Here we describe a novel genome-wide RNA interference (RNAi) screen in Drosophila1 that can be used to identify host genes important for influenza virus replication. After modifying influenza virus to allow infection of Drosophila cells and detection of influenza virus gene expression, we tested an RNAi library against 13,071 genes (90% of the Drosophila genome), identifying over 100 whose suppression in Drosophila cells significantly inhibited or stimulated reporter gene (Renilla luciferase) expression from an influenza virus-derived vector. The relevance of these findings to influenza virus infection of mammalian cells is illustrated for a subset of the Drosophila genes identified above. That is, the human homologues of ATP6V0D1, COX6A1 and NXF1 are shown to have key functions in the replication of H5N1 and H1N1 influenza A viruses, but not vesicular stomatitis virus or vaccinia virus, in HEK 293 cells. Thus, we have demonstrated the feasibility of using genome-wide RNAi screens in Drosophila to identify previously unrecognized host proteins that are required for influenza virus replication. This could accelerate the development of new classes of antiviral drugs for chemoprophylaxis and treatment, which are urgently needed given the obstacles to rapid development of an effective vaccine against pandemic influenza and the likely emergence of strains resistant to available drugs.
Influenza, a highly contagious disease of birds and mammals, is caused by negative-strand RNA viruses of the family Orthomyxoviridae. Influenza outbreaks kill millions of people worldwide during pandemic years and hundreds of thousands during other years. Since their first lethal infection of humans in 1997, H5N1 influenza A viruses have spread throughout Asia and to Europe and Africa, posing a major risk for a new influenza pandemic2. To provide rational bases for improved treatment and control of influenza virus infection, we sought to advance understanding of viral infection mechanisms by elucidating previously unknown virus-host cell interactions. Many steps in the viral life cycle, including intracellular trafficking, gene expression, replication and virion assembly, depend on interactions with specific host cell gene products. Although the vast majority of such host molecules remain elusive, emerging results indicate that their identification and characterization can provide new insights into the mechanisms by which viruses complete their life cycle and hence illuminate potentially valuable targets for prophylactic and therapeutic intervention3-5.
Systematic, genome-wide RNAi analysis offers an exciting tool to identify host genes that function in viral replication. Such analysis is facilitated by well-developed model systems such as Drosophila, whose genome contains only ∼14,000 genes that can nearly all be specifically targeted for high efficiency mRNA depletion by double-stranded RNA (dsRNA) libraries1. Because of its powerful genetics and conservation with vertebrates, Drosophila has been used to make numerous critical contributions to mammalian cell biology 6-9. Thus, in principle, Drosophila RNAi studies could accelerate identification of host interactions essential for influenza virus replication.
Since Drosophila D-Mel2 cells do not express the human influenza virus receptor α2,6-linked sialic acid (Supplementary Fig. S1), we predicted that wild-type human influenza virus would not be able to infect them. Indeed, we failed to detect viral protein expression by immunofluorescence assays in Drosophila D-Mel2 cells inoculated with influenza virus A/WSN/33 (H1N1; WSN) (data not shown). To bypass this block to wild-type (wt) influenza virus entry, we generated a genetically modified virus, Flu-VSV-G-GFP (FVG-G), in Madin-Darby canine kidney (MDCK) cells, by replacing the receptor-binding hemagglutinin (HA) and neuraminidase (NA) genes with genes encoding vesicular stomatitis virus (VSV) glycoprotein G and enhanced green fluorescence protein (eGFP), respectively10, 11 (Fig. 1A). Since the envelopes of the resulting virions bear VSV G, which mediates entry into mammalian, Drosophila and other cells12, FVG-G virions should readily infect Drosophila cells. Twenty-four hours after infection, GFP fluorescence was detected in FVG-G-infected Drosophila D-Mel2 cells (Supplementary Fig. S2). We also confirmed by real-time PCR that influenza virus RNA replication occurred in Drosophila cells (Supplementary Fig. S3). However, Drosophila cells infected with FVG-G did not release detectable virions into the medium, as assayed by infectivity tests on MDCK cells and by electron microscopy (data not shown). This was due, at least in part, to failure of Drosophila cells to express some viral proteins required for virion assembly and infectivity (Supplementary Fig. S4). Thus, Drosophila cells can support influenza virus replication from post-entry to at least the protein expression phase of the viral life cycle. This span encompasses multiple other steps in the life cycle, including cytoplasmic release of genomic RNA-containing viral ribonucleoprotein complexes (vRNPs), vRNP import into the nucleus, mRNA synthesis from the negative-strand viral RNA genome, mRNA export to the cytoplasm, and translation.
Fig. 1. Overview of genome-wide RNAi screen to identify host factors involved in influenza virus replication in Drosophila cells.
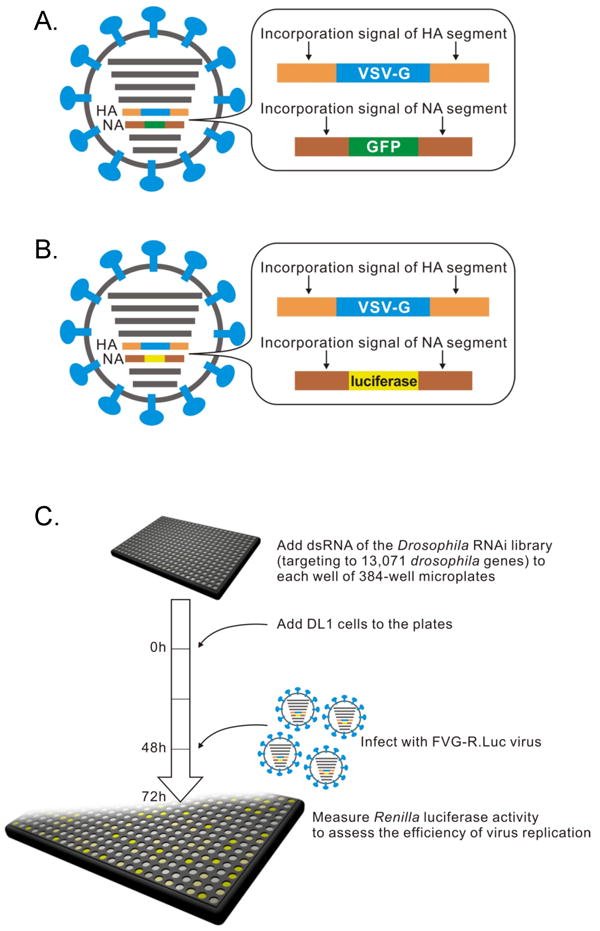
Schematic diagrams showing recombinant influenza viruses, (A) FVG-G, in which genes encoding the HA and NA proteins were replaced with the VSV-G and eGFP genes, respectively, and (B) FVG-R, in which the genes encoding the HA and NA were replaced with the VSV-G and Renilla luciferase genes, respectively. (C) Schematic diagram of systematic analysis of host genes affecting influenza virus replication and gene expression in Drosophila cells. Experimental details are given in Methods.
For high-throughput, functional genomics analysis of influenza virus replication in Drosophila cells, we engineered Flu-VSV-G-R.Luc (FVG-R), in which VSV-G and Renilla luciferase genes replaced the viral HA and NA ORFs (Fig. 1B). FVG-R virions were then used with an RNAi library (Ambion) against 13,071 Drosophila genes (∼90% of all genes) to identify host genes affecting influenza virus-directed Renilla luciferase expression (Fig. 1C). Two independent tests of the entire library were performed (Supplementary Table S1). For 176 genes whose dsRNAs inhibited FVG-R-directed luciferase expression in both replicates, repeated secondary tests using alternate dsRNAs to control for possible off-target effects confirmed the effects of 110 genes (Supplementary Tables S2 and S3). This confirmation rate is comparable to that in a Drosophila screen with a natural Drosophila-infecting virus5. Cell viability testing identified six genes with potentially significant cytotoxic effects, which were excluded from further consideration (Supplementary Information and Table S3). Secondary tests of candidate genes whose dsRNAs increased FVG-R-directed luciferase expression produced a much lower confirmation rate, suggesting a higher rate of off-target or other false positive effects in this class (Supplementary Information and Table S4).
Among the over 100 candidate genes found to be important for influenza virus replication in Drosophila cells, we selected several encoding components in host pathways/machineries that are known to be involved in the life cycle of influenza virus, for example, ATP6V0D1 (endocytosis pathway), COX6A1 (mitochondrial function), and NXF1/TAP (mRNA nuclear export machinery), for further analysis in mammalian cells to assess the relevance of our Drosophila results 13-17. ATP6V0D1 encodes subunit D of vacuolar (H+)-ATPase (V-ATPase), a proton pump that functions in the endocytosis pathway (i.e., the acidification and fusion of intracellular compartments18). COX6A1 encodes a subunit of cytochrome c oxidase (COX), an enzyme of the mitochondrial electron transport chain that catalyzes electron transfer from cytochrome c to oxygen19. NXF1/TAP encodes a nuclear export factor critical for exporting the majority of cellular mRNAs containing exon-exon junctions20, 21.
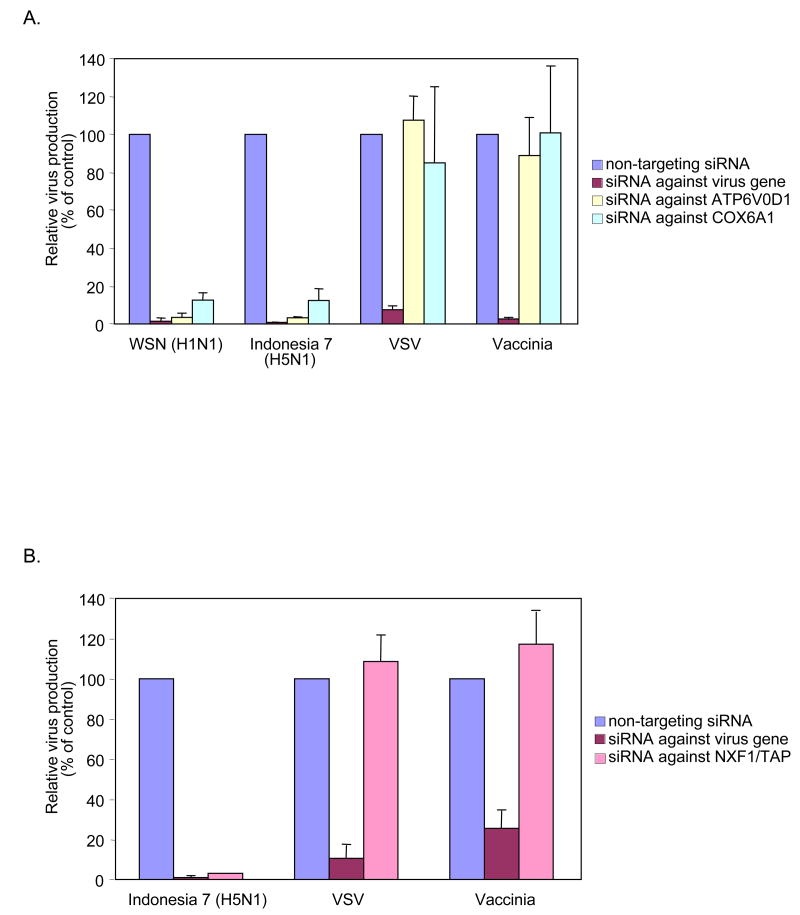
As a first test for the possible contribution of these gene products to influenza virus replication in mammalian cells, we treated human HEK 293 cells twice at 24-h intervals with siRNAs (siGENOME, Dharmacon) against the human homologue of each selected Drosophilia gene. 24 h after the second siRNA treatment, the cells were infected with FVG-R virus and, 2 days later, Renilla luciferase activity was measured to assess viral replication and gene expression. siRNA against ATPV0D1 or COX6A1 dramatically decreased Renilla luciferase activity (Fig. 2A) but not cell viability (Supplementary Fig. S5A), implying that these genes play important roles in influenza virus replication in mammalian cells, as in Drosophila cells. Inhibition was not due to off-target effects since, for each gene, each of four distinct siRNAs inhibited FVG-R-directed expression of Renilla luciferase (Supplementary Table S6). Since COX6A1 encodes a subunit of mitochondrial electron transport chain complex IV, COX, we used specific inhibitors to test whether in 293 cells influenza virus also required other complexes in this chain (Fig. 2C). Inhibitors of complexes III, IV and V selectively inhibited FVG-R-directed Renilla luciferase expression by 50- to 100-fold, while complex I and II inhibitors had little or no effect. Thus, in mammalian cells, influenza virus depends on multiple late stages but not early stages in the mitochondrial electron transport chain.
Fig. 2. Effect of selected siRNAs and inhibitors on Renilla luciferase expression in FVG-R-infected human cells.

Renilla luciferase activity was measured in FVG-R-infected 293 cells treated with siRNAs against (A) ATP6V0D1, COX6A1, (B) NXF1, or (C) the indicated mitochondrial electron transport chain inhibitors. Inhibitors of complexes III, IV and V inhibited FVG-R-directed Renilla luciferase expression significantly, while complex I and II inhibitors had little or no effect. In contrast, the inhibitors had no significant effects on cell viability and Gaussia luciferase expression of a murine leukemia virus derivative (MLV-GL) that, like FVG-R, depended on VSV G-mediated entry. All experiments (A-C) were conducted three times in duplicate, with the results reported as means ± SD.
Treatment for 4 days with siRNA against NXF1 significantly decreased mammalian cell viability (data not shown), as predicted by the critical role of NXF1 in general host cell metabolism. Accordingly, the total incubation time with siRNA against NXF1 was shortened to 36 h by transfecting cells with the siRNA twice at a 12-h interval, infecting with FVG-R virus 12 h later, and assaying for Renilla luciferase at 12-h postinfection. Under these conditions, cell viability was not detectably affected (Supplementary Fig. S5B) while Renilla luciferase activity was reduced by nearly 5-fold (Fig. 2B). While recent results indicated that influenza virus protein NS1 binds to NXF1 to inhibit host mRNA export 17, these results imply that influenza virus RNAs and/or proteins are transported by an NXF1-dependent pathway (see also Supplementary Information).
To test the effects of these genes on authentic influenza viruses, we infected siRNA-treated 293 cells with WSN virus or H5N1 influenza A/Indonesia/7/05 (Indonesia 7; isolated from a patient) or with VSV or vaccinia virus as controls. Progeny viruses were collected from the medium at 24 h (Indonesia 7, VSV or vaccinia virus) or 48 h (WSN) postinfection and titrated. Depleting ATP6V0D1 and COX6A1 did not affect VSV or vaccinia virus replication, but decreased the WSN and Indonesia 7 virus yields by ∼10-fold or more (Fig. 3A). Thus, ATP6V0D1 and COX6A1 are required for replication of influenza viruses but not VSV and vaccinia virus. Depleting the NXF1 nuclear export factor, again using an accelerated 36-h timeline, reduced Indonesia 7 virus titers by 20-fold (Fig. 3B), indicating that NXF1 plays a critical role in H5N1 influenza virus replication. Unfortunately, because WSN grows in 293 cells more slowly than does Indonesia 7 virus, the WSN virus yield at 12-h postinfection was insufficient to test for an effect of NXF1 depletion (data not shown). In comparison to Indonesia 7 virus, NXF1 depletion had no effect on VSV or vaccinia virus yields (Fig. 3B). Moreover, siRNA against NXF1 inhibited FVG-G, but not adenovirus, which, like influenza virus, depends on nuclear steps for genome transcription and replication (Supplementary Fig. S6). Thus, multiple genes identified in Drosophila cells correspond to important, selective host factors for influenza virus replication in mammalian cells.
Fig. 3. Effect of siRNAs against selected genes on the replication of influenza viruses, VSV or vaccinia virus in human 293 cells.
The titers of influenza viruses (WSN and Indonesia 7), VSV and vaccinia virus in 293 cells treated with siRNA against (A) ATP6V0D1, COX6A1 or (B) NXF1 are shown. Experimental details are given in Supplementary Methods. All experiments were conducted three times, with the results reported as means ± SD.
The method we have established, employing systematic analysis of a Drosophila RNAi library with confirmation in mammalian cells, can be used to identify host gene products that affect influenza virus replication. This utility was demonstrated in experiments with authentic influenza viruses, including an H5N1 virus isolated from a human patient. One of the candidate genes tested in mammalian cells, COX6A1, which encodes a subunit of COX, acts as a critical enzyme in cytochrome c-dependent electron transport in mitochondria. Influenza virus PB2 polymerase has a mitochondrial targeting signal14, and PB1-F2, the second protein encoded by the influenza PB1 gene, was reported to localize to inner and outer mitochondrial membranes, and to delay influenza virus clearance by host antiviral responses15. Thus, COX6A1 may be involved in PB2- and/or PB1-F2-mediated functions in mitochondria. Additionally, influenza vRNP export from the nucleus requires caspase 3 activation22, which can be induced by cytochrome c release from mitochondria. Moreover, Bcl2 inhibits both cytochrome c release and influenza vRNP export23, and COX function and changes in mitochondrial membrane potential have been linked to caspase activation24. These pathways may underlie our further findings (Fig. 2C) that influenza virus replication and expression in mammalian cells are strongly and selectively modulated by compounds that inhibit COX, cytochrome c-linked electron transport chain complex III, and normal ion transport across mitochondrial membranes (oligomycin and valinomycin). Potential roles of the other two host genes confirmed here in mammalian cells, ATP6V0D1 and NXF1, in influenza virus replication, are discussed further in the Supplementary Information.
The above results with multiple, diverse genes, including ATP6V0D1, NXF1, and COX6A1 as well as mitochondrial electron transport complexes III and V, demonstrate the feasibility and value of using Drosophila RNAi screening to identify novel host factors with important and potentially unsuspected roles in influenza virus replication. Simultaneously, the genome-wide results from our Drosophila RNAi screen provide over 100 additional candidate genes (many with unknown functions) to be tested in mammalian cells. We suggest that the same strategy could be applied to identify novel host factors involved in the replication of other viruses, whenever at least a portion of their replication cycle is supported by Drosophila cells.
Methods Summary
Cells and viruses
MDCK cells, 293 cells, baby hamster kidney (BHK) cells, D-Mel2 cells and DL1 cells were maintained as described in Methods. WSN, FVG-G, FVG-R and Indonesia 7 viruses were generated as previously described 10 and propagated in MDCK cells. VSV and vaccinia virus were grown in BHK and CV-1 cells, respectively. Gaussia luciferase-expressing murine leukemia virus (MLV-GL) and GFP-expressing adenovirus were provided by Dr. James Bruce and Dr. Robert Kalejta 25, respectively.
Drosophila RNAi library analysis
DsRNA of the Drosophila RNAi library (targeting 13,071 Drosophila genes) and DL1 cells were added to each well of 384-well microplates. After 2 days incubation, cells were infected with FVG-R virus. At 1 day postinfection, Renilla luciferase activity was measured as described below. Two independent analyses of the entire library were performed.
siRNA treatment of mammalian cells
293 cells were transfected with siRNA by TransIT-TKO (Mirus). Cells were incubated for 12 h in experiments with NXF1 or 24 h with other genes, transfected again under the same conditions, and inoculated with virus after 12 h for NXF1 or 24 h for other genes. After a further 12 h (for NXF1) or 48 h (other genes), the cells were harvested for the indicated analyses.
Renilla luciferase and cell viability assays
Renilla luciferase activity and cell viability were measured with established Renilla luciferase and CellTiter-Glo assay systems (Promega) according to the manufacturer's instructions; signals were read with a GLOMAX 96 microplate luminometer.
Methods
Cells and viruses
MDCK cells, 293 cells, and BHK cells were maintained in minimum essential medium (MEM) containing 5% fetal calf serum (FCS) and antibiotics at 37°C in 5% CO2. D-Mel2 cells were maintained in Drosophila-SFM (GIBCO/Invitrogen) at 28°C. DL1 cells were maintained in Schneider's Drosophila (SD) medium containing 10% FBS at 28°C.
WSN, FVG-G, FVG-R and Indonesia 7 viruses were generated by a plasmid-based reverse genetics system10 and were grown and titrated in MDCK cells. MLV-GL was produced by replacing the CD4 ORF in pCMMP-CD4-eGFP 26 with that of Gaussia luciferase (J. Bruce, unpublished results). All experiments with Indonesia 7 virus were conducted in a biosafety level-3 containment laboratory approved for such use by the Centers for Disease Control and Prevention and the U.S. Department of Agriculture.
Drosophila RNAi library analysis
The Drosophila RNAi library (Ambion) contained 13,071 individual dsRNAs, each designed to specifically target a single Drosophila gene. Five microliters of dsRNA (40 ng/μl) were added to each well of 384-well plates, after which 2 × 104 DL1 cells in 10 microliters of SD medium were added to each well and incubated with the dsRNA at 28°C for 60 min. Twenty microliters of SD medium containing 20% FBS were then added to each well after incubation. Cells were treated with dsRNA for 2 days at 28°C and then inoculated with an amount of FVG-R virus corresponding to a multiplicity of infection (MOI) of 10 for MDCK cells, and transferred to 33°C. At 1 day postinfection, Renilla luciferase activity was measured as described below. Two independent analyses of the entire library were performed.
siRNA treatment of mammalian cells
Nontargeted siRNA was siCONTROL #1 (Dharmacon). Duplex siRNAs against candidate genes were obtained from siGENOME (Dharmacon, see Supplementary Table S7 for sequences); the effects of siRNA were evaluated by RT-PCR (Supplementary Fig. S7). The sequences of siRNA against the NP gene of influenza virus (GGA UCU UAU UUC CUUC GGA GUU) 27 and the E3L gene of vaccinia virus (AAU AUC GUC GGA GCU GUA CAC) 28 were reported previously. The sequence of siRNA against the gene encoding the L protein of VSV (CGA GUU AUC CAG CAA UCA UUU) was designed using BLOCK-iT™ RNAi designer (Invitrogen). A 293 cell suspension was seeded into the wells of a 24-well plate (2.0 × 104 cells / well), incubated for 1 h and transfected with siRNA (final concentration 10 nM; Dharmacon) using TransIT-TKO (Mirus), according to the manufacturer's instructions.
Inhibitor treatment of mammalian cells
Mitochondrial electron transport chain inhibitors were used at the following final concentrations: rotenone (0.5 μM), 3-nitroproprionic acid (3-NPA, 1 mM), antimycin A (25 μM), NaN3 (50 mM), oligomycin (25 μM), and valinomycin (0.9 μM). 293 cells were treated with the inhibitors for 5 h before being infected with FVG-R or MLV-GL, incubated 18 h, and assayed for Renilla or Guassia luciferase, respectively. Cell viability was assayed in parallel by a firefly luciferase-mediated ATP assay (CellTiter-Glo, Promega).
Supplementary Material
Acknowledgments
We thank M. McGregor and K. Wells for technical assistance, M. Hatta, S. Yamada, and M. Ito for an H5N1 influenza virus, S. Watanabe and Y. Hatta for FVG-R and FVG-G viruses, R. Kalejta and J. Bruce for adenovirus and murine leukemia virus vectors, T. Noda for electron microscopy, J. Gilbert for editing the manuscript, and Y. Kawaoka for illustrations. This work was supported, in part, by U.S. National Institute of Allergy and Infectious Diseases Public Health Service research grants, by U.S. National Institute of General Medical Sciences grant GM35072, by a grant-in-aid for Specially Promoted Research and by a contract research fund for the Program of Funding Research Centers for Emerging and Reemerging Infectious Diseases from the Ministry of Education, Culture, Sports, Science and Technology, and by grants-in-aid from the Ministry of Health, Labor, Welfare of Japan. PA is Howard Hughes Medical Institute investigator.
Footnotes
Author Contribution: L.H., A.S., P.A. and Y.K. designed research; L.H., A.S., T.W. and C.A.N. performed research; L.H., A.S., T.W., E.S., M.A.N., P.A. and Y.K. analyzed data; L.H., A.S., T.W., P.A. and Y.K. wrote the manuscript.
References
- 1.Kuttenkeuler D, Boutros M. Genome-wide RNAi as a route to gene function in Drosophila. Brief Funct Genomic Proteomic. 2004;3:168–76. doi: 10.1093/bfgp/3.2.168. [DOI] [PubMed] [Google Scholar]
- 2.Webster RG, Govorkova EA. H5N1 influenza--continuing evolution and spread. N Engl J Med. 2006;355:2174–7. doi: 10.1056/NEJMp068205. [DOI] [PubMed] [Google Scholar]
- 3.Kushner DB, et al. Systematic, genome-wide identification of host genes affecting replication of a positive-strand RNA virus. Proc Natl Acad Sci U S A. 2003;100:15764–9. doi: 10.1073/pnas.2536857100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ludwig S, Planz O, Pleschka S, Wolff T. Influenza-virus-induced signaling cascades: targets for antiviral therapy? Trends Mol Med. 2003;9:46–52. doi: 10.1016/s1471-4914(02)00010-2. [DOI] [PubMed] [Google Scholar]
- 5.Cherry S, et al. Genome-wide RNAi screen reveals a specific sensitivity of IRES-containing RNA viruses to host translation inhibition. Genes Dev. 2005;19:445–52. doi: 10.1101/gad.1267905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hahn H, Wojnowski L, Miller G, Zimmer A. The patched signaling pathway in tumorigenesis and development: lessons from animal models. J Mol Med. 1999;77:459–68. doi: 10.1007/s001099900018. [DOI] [PubMed] [Google Scholar]
- 7.Dearolf CR. JAKs and STATs in invertebrate model organisms. Cell Mol Life Sci. 1999;55:1578–84. doi: 10.1007/s000180050397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lemaitre B, Nicolas E, Michaut L, Reichhart JM, Hoffmann JA. The dorsoventral regulatory gene cassette spatzle/Toll/cactus controls the potent antifungal response in Drosophila adults. Cell. 1996;86:973–83. doi: 10.1016/s0092-8674(00)80172-5. [DOI] [PubMed] [Google Scholar]
- 9.Takeda K, Kaisho T, Akira S. Toll-like receptors. Annu Rev Immunol. 2003;21:335–76. doi: 10.1146/annurev.immunol.21.120601.141126. [DOI] [PubMed] [Google Scholar]
- 10.Neumann G, et al. Generation of influenza A viruses entirely from cloned cDNAs. Proc Natl Acad Sci U S A. 1999;96:9345–50. doi: 10.1073/pnas.96.16.9345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Watanabe T, Watanabe S, Noda T, Fujii Y, Kawaoka Y. Exploitation of nucleic acid packaging signals to generate a novel influenza virus-based vector stably expressing two foreign genes. J Virol. 2003;77:10575–83. doi: 10.1128/JVI.77.19.10575-10583.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wyers F, Richard-Molard C, Blondel D, Dezelee S. Vesicular stomatitis virus growth in Drosophila melanogaster cells: G protein deficiency. J Virol. 1980;33:411–22. doi: 10.1128/jvi.33.1.411-422.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Perez L, Carrasco L. Involvement of the vacuolar H(+)-ATPase in animal virus entry. J Gen Virol. 1994;75(Pt 10):2595–606. doi: 10.1099/0022-1317-75-10-2595. [DOI] [PubMed] [Google Scholar]
- 14.Carr SM, Carnero E, Garcia-Sastre A, Brownlee GG, Fodor E. Characterization of a mitochondrial-targeting signal in the PB2 protein of influenza viruses. Virology. 2006;344:492–508. doi: 10.1016/j.virol.2005.08.041. [DOI] [PubMed] [Google Scholar]
- 15.Zamarin D, Ortigoza MB, Palese P. Influenza A virus PB1-F2 protein contributes to viral pathogenesis in mice. J Virol. 2006;80:7976–83. doi: 10.1128/JVI.00415-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Momose F, et al. Cellular splicing factor RAF-2p48/NPI-5/BAT1/UAP56 interacts with the influenza virus nucleoprotein and enhances viral RNA synthesis. J Virol. 2001;75:1899–908. doi: 10.1128/JVI.75.4.1899-1908.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Satterly N, et al. Influenza virus targets the mRNA export machinery and the nuclear pore complex. Proc Natl Acad Sci U S A. 2007;104:1853–8. doi: 10.1073/pnas.0610977104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stevens TH, Forgac M. Structure, function and regulation of the vacuolar (H+)-ATPase. Annu Rev Cell Dev Biol. 1997;13:779–808. doi: 10.1146/annurev.cellbio.13.1.779. [DOI] [PubMed] [Google Scholar]
- 19.Gnaiger E, Lassnig B, Kuznetsov A, Rieger G, Margreiter R. Mitochondrial oxygen affinity, respiratory flux control and excess capacity of cytochrome c oxidase. J Exp Biol. 1998;201:1129–39. doi: 10.1242/jeb.201.8.1129. [DOI] [PubMed] [Google Scholar]
- 20.Cullen BR. Nuclear mRNA export: insights from virology. Trends Biochem Sci. 2003;28:419–24. doi: 10.1016/S0968-0004(03)00142-7. [DOI] [PubMed] [Google Scholar]
- 21.Reed R, Cheng H. TREX, SR proteins and export of mRNA. Curr Opin Cell Biol. 2005;17:269–73. doi: 10.1016/j.ceb.2005.04.011. [DOI] [PubMed] [Google Scholar]
- 22.Wurzer WJ, et al. Caspase 3 activation is essential for efficient influenza virus propagation. Embo J. 2003;22:2717–28. doi: 10.1093/emboj/cdg279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hinshaw VS, Olsen CW, Dybdahl-Sissoko N, Evans D. Apoptosis: a mechanism of cell killing by influenza A and B viruses. J Virol. 1994;68:3667–73. doi: 10.1128/jvi.68.6.3667-3673.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kadenbach B, Arnold S, Lee I, Huttemann M. The possible role of cytochrome c oxidase in stress-induced apoptosis and degenerative diseases. Biochim Biophys Acta. 2004;1655:400–8. doi: 10.1016/j.bbabio.2003.06.005. [DOI] [PubMed] [Google Scholar]
- 25.He TC, et al. A simplified system for generating recombinant adenoviruses. Proc Natl Acad Sci U S A. 1998;95:2509–14. doi: 10.1073/pnas.95.5.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bruce JW, Bradley KA, Ahlquist P, Young JA. Isolation of cell lines that show novel, murine leukemia virus-specific blocks to early steps of retroviral replication. J Virol. 2005;79:12969–78. doi: 10.1128/JVI.79.20.12969-12978.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ge Q, et al. RNA interference of influenza virus production by directly targeting mRNA for degradation and indirectly inhibiting all viral RNA transcription. Proc Natl Acad Sci U S A. 2003;100:2718–23. doi: 10.1073/pnas.0437841100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dave RS, et al. siRNA targeting vaccinia virus double-stranded RNA binding protein [E3L] exerts potent antiviral effects. Virology. 2006;348:489–97. doi: 10.1016/j.virol.2006.01.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.