Abstract
Behavioral genetic mapping studies in model organisms predominantly use crosses originating from a single pair of inbred lines to determine the location of alleles that confer genetic variation in the trait of interest, and they often make sweeping generalizations about the genetic architecture of the trait based on these results. A previous study fine mapped mate preference variation between one pair of Drosophila pseudoobscura lines and identified 2 strong-effect behavioral quantitative trait loci (QTLs). Here, we replicated the previous study's mapping design to examine the extent of variation at these behavioral QTLs across 6 pairs of lines, but we were unable to detect effects of either QTL region in the pairs of lines studied. We suggest that the low-discrimination alleles at these 2 QTLs may occur at low frequency within D. pseudoobscura, although other explanations for the inconsistency are possible. These results underscore the need to examine multiple strains across a species when describing the genetic variation underlying behavioral traits.
Keywords: Drosophila, QTL mapping, sexual isolation, species discrimination
Most behavioral genetic mapping studies in model organisms use crosses stemming from a single pair of inbred lines in a single environmental condition to identify the location of alleles conferring genetic variation in the trait of interest. This approach is especially common when “recombinant inbred lines” (RILs) are used to precisely map such variation, and the resultant study may confidently make a sweeping general claim that “species A possesses a few large-effect genes that contribute to the genetic variance in trait B.” In principle, this approach may successfully capture most variation if 2 isolated populations or species are examined (e.g., if species A differs by numerous fixed differences causing behavioral divergence from species B), but it may capture only a (perhaps grossly unrepresentative) snapshot of variation when performed using strains derived from a large panmictic population. Correspondingly, although some studies have successfully replicated major-effect quantitative trait loci (QTLs) in crosses derived from multiple independent lines (e.g., Shaw et al. 2007), others have failed to identify effects of the same regions (e.g., Ortiz-Barrientos et al. 2004).
Ortiz-Barrientos et al. (2004) used 2 independent male-parent backcrosses to investigate which chromosomes contribute to variation among populations of Drosophila pseudoobscura in female mating discrimination against Drosophila persimilis males. In this system, females derived from populations where the 2 species coexist exhibit higher discrimination against D. persimilis males than do females derived from populations where D. pseudoobscura is found by itself (Singh 1983; Noor 1995; Ortiz-Barrientos et al. 2004; Lorch and Servedio 2005; Noor and Ortiz-Barrientos 2006). This pattern is consistent across strains and experimental design (choice vs. no choice). However, Ortiz-Barrientos et al. (2004) found that the whole chromosomal contributions to this difference in discrimination differed between the 2 crosses. As different populations were surveyed in the 2 crosses, this difference could reflect variation within or between populations. They fine mapped the mate preference variation to 2 strong-effect QTLs in one pair of lines, one on the right arm of the X chromosome (Coy-1) and one on the fourth chromosome (Coy-2), but it is unknown how much these 2 QTLs contribute to mate preference variation across strains within populations of this species.
In this study, we replicated the Ortiz-Barrientos et al. (2004) mapping design using 6 pairs of lines to determine whether and how often the same 2 QTLs are identified in other pairings. Although one of the original lines used by Ortiz-Barrientos et al. (2004) has gone extinct, we focused on the other line they used and other strains derived from the same 2 populations. We find little or no statistically significant effect associated with either of these 2 QTLs in any pair of lines we surveyed, and we infer that the low-discrimination alleles at these 2 QTLs are likely at low frequency within this species.
Materials and Methods
Drosophila Strains and Crosses
The D. pseudoobscura strains used in this study were Flagstaff1993b (originally derived from crosses between multiple isofemale lines collected from Flagstaff, AZ, in 1993, including some of the same lines used to create the original Flagstaff1993, but subsequently inbred), Flagstaff14, Flagstaff16 (both isofemale lines collected from Flagstaff, AZ, in 1997), Mather10, Mather17, Mather48, and Mather52 (all isofemale lines collected from Mather, CA, in 1997). The Mather17 strain was used for fine mapping by Ortiz-Barrientos et al. (2004). The D. persimilis strain used is Mount St Helena, 1993, collected from Robert Louis Stevenson State Park in California in 1993 and was also used in the assays by Ortiz-Barrientos et al. (2004). Stocks and crosses were maintained at 20 °C with 12:12 h light:dark cycle on standard sugar-yeast-agar medium. For pure strain assays, 50–200 females were assayed per strain, whereas for genetic mapping assays, 500–600 female backcross progeny were assayed per cross. Backcross progeny between D. pseudoobscura strains were produced by mating females from a Flagstaff strain to males from a Mather strain, and resultant F1 females were crossed to males from the Flagstaff strain. The cross was done in this direction because the higher discrimination by Mather females was found to be consistently genetically dominant (Noor 1999; Ortiz-Barrientos et al. 2004).
Mating Assays
Mating assays generally followed the procedure of Ortiz-Barrientos et al. (2004). Briefly, virgin female D. pseudoobscura were collected under light anesthesia within 8 h of eclosion and confined until assay (7 days posteclosion) in a food-containing vial. Drosophila persimilis males were also collected under light anesthesia and confined. One day before the mating assays, males were separated into individual food-containing vials to reduce crowding-mediated courtship inhibition (Noor 1997).
Mating assays were conducted between 8:30 AM and 10:30 AM, corresponding with the time of the incubator lights turned on each day. On the morning of the mating assay, individual D. pseudoobscura females were aspirated without anesthesia into a vial containing a D. persimilis male. The cotton plug was pushed down to allow only a few cubic centimeters of interaction space, and the vials were observed for 10 min. Although this is a “no-choice” mate preference design, this mimics the design followed previously (Ortiz-Barrientos et al. 2004), and comparable results were obtained from “female-choice” designs using these species (Noor and Ortiz-Barrientos 2006).
Assay starting time, the time of first courtship, and the time of copulation were recorded. Following Ortiz-Barrientos et al. (2004), vials in which no courtship occurred were discarded. All remaining backcross females were kept for genotyping.
Genotyping and Analysis
DNA was extracted from flies individually using the rapid Drosophila squish-prep protocol (Gloor and Engels 1992). For genotyping, one primer was designed with an M13 tail at the 5′-end, and polymerase chain reaction (PCR) was performed in a 10-ml reaction volume with 0.5 picomoles of each primer, 0.4 picomoles fluorescent dye–labeled M13, 200 mM dNTPs, 1 μl 10× PCR buffer (100 mM, Tris pH 8.3, 500 mM KCl, 15 mM MgCl2), 1 U Taq polymerase, and 1 μl from a 50-μl squish-prep. PCR was executed using a touchdown cycle (Palumbi 1996). After PCR, 3 μl of LiCor (Lincoln, NE) stopping buffer was added to the reactions, and 1 μl of the PCR reaction was loaded onto an acrylamide gel on a LiCor 4200 DNA sequencer for visualization.
Primers were selected that flanked microsatellites within or very near to (160 kb or less) the markers in the Coy-1 and Coy-2 regions mostly strongly associated with mate preference variation (Ortiz-Barrientos et al. 2004). Primer sequences and marker positions in the D. pseudoobscura genome assembly are presented in Supplementary 1. For 4 pairings, markers closely flanking the Coy-1 region were also surveyed, but no greater association was found with these markers to mate preference variation than for the marker closest to the focal region.
Analyses were all performed as Fisher's Exact tests (mating status × genotype) for simplicity, but other measures gave identical results.
Results
Pure Strain Mating Assays
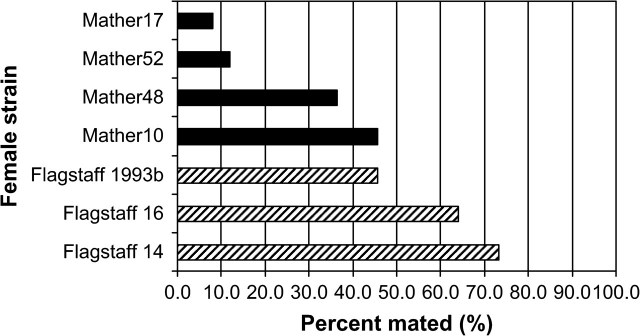
Mating rate in pairings involving pure strains are presented in Figure 1. Consistent with the extensive previous research in this system, we observed that females from strains derived from Mather, CA (where both species co-occur), were generally more reluctant to mate with D. persimilis males than were females from strains derived from Flagstaff, AZ (where D. pseudoobscura occurs alone, t = 2.9, P = 0.032). With respect to the specific pairings to be used in crosses below, we found a statistically significant difference in female interspecies mating propensity between the following pairs: Flagstaff1993b–Mather17 (Fisher's Exact test, P < 0.0001), Flagstaff1993b–Mather52 (P < 0.0001), Flagstaff14–Mather17 (P < 0.0001), and Flagstaff16–Mather17 (P < 0.0001). Flagstaff1993b did not differ significantly from Mather10 (P > 0.9) or Mather48 (P = 0.06) in female interspecies mating propensity.
Figure 1.
Percent of Drosophila pseudoobscura females from various strains that mated with a Drosophila persimilis male during 10-min observation.
Female mate preferences are susceptible to uncontrollable environmental influences in some cases, so we also analyzed the dataset using equal numbers of pairings for each comparison per day. With this reanalysis, the percentages mated and statistical significance were unchanged from above.
Effects of Coy-1 and Coy-2 Regions on Variation in Female Interspecies Mating Propensity
We generated 500–600 backcross progeny between Mather and Flagstaff strains, assayed their female interspecies mating propensity, and genotyped them for markers in and near the Coy-1 and Coy-2 regions. Ortiz-Barrientos et al. (2004) detected statistically significant effects of these regions (reducing mating propensity by 10% or more), and resampling their larger backcross with the same number of females assayed here (or even fewer) still identifies the effect of the Coy-1 region consistently and often the Coy-2 region as statistically significant. Our goal was to determine the extent to which variation in these regions of the genome explains the more generally observed phenotypic difference across strains rather than just the pair of inbred lines studied previously. Although the original Flagstaff1993 strain used by Ortiz-Barrientos et al. (2004) has been lost, we were able to use the same high-discrimination Mather17 strain and the same strain of D. persimilis for courting males.
In the 6 sets of backcross progeny generated using different pairs of strains, essentially none exhibited substantial differences in female interspecies mating propensity associated with either of these regions (Table 1). No general trend was observed, even when considering the nonsignificant effects. A marginally significant (Fisher's Exact test, P = 0.036) effect of the Coy-2 region was detected in the cross between strains Flagstaff1993b and Mather52, but this was opposite in direction to expectation (flies homozygous for the Flagstaff allele had higher discrimination). The data were also analyzed using just the first 5 min of courtship, and results were unchanged.
Table 1.
Effects of genotype at the Coy-1 and Coy-2 regions on female interspecies mating propensity in backcrosses of various Drosophila pseudoobscura lines derived from Flagstaff and Mather
| Flag/Flag | Mather/Flag | P | |
| Ortiz-Barrientos et al. (2004): | |||
| Coy-1, Flagstaff1993/Mather17 | 39 | 27 | <0.0001 |
| Coy-2, Flagstaff1993/Mather17 | 39 | 29 | 0.0078 |
| This study: | |||
| Coy-1, Flagstaff14/Mather17 | 70 | 67 | 0.55 |
| Coy-2, Flagstaff14/Mather17 | 70 | 67 | 0.49 |
| Coy-1, Flagstaff16/Mather17 | 62 | 62 | 0.89 |
| Coy-2, Flagstaff16/Mather17 | 61 | 64 | 0.49 |
| Coy-1, Flagstaff1993b/Mather10 | 40 | 43 | 0.53 |
| Coy-2, Flagstaff1993b/Mather10 | 39 | 43 | 0.54 |
| Coy-1, Flagstaff1993b/Mather17 | 55 | 50 | 0.35 |
| Coy-2, Flagstaff1993b/Mather17 | 51 | 53 | 0.68 |
| Coy-1, Flagstaff1993b/Mather48 | 51 | 53 | 0.76 |
| Coy-2, Flagstaff1993b/Mather48 | 49 | 50 | 0.84 |
| Coy-1, Flagstaff1993b/Mather52 | 44 | 40 | 0.52 |
| Coy-2, Flagstaff1993b/Mather52 | 36 | 46 | 0.036 |
Numbers reported are percentage of females with a particular genotype from the specified cross and marker that mated with a Drosophila persimilis male.
Discussion
Ortiz-Barrientos et al. (2004) mapped variation between 2 D. pseudoobscura strains (Flagstaff1993 and Mather17) in female interspecies mating propensity to 2 regions, dubbed Coy-1 or Coy-2. Backcross females heterozygous for the high- and low-discrimination alleles at these loci displayed a low number of interspecies matings (high discrimination) when compared with the females homozygous for the low-discrimination allele. Using other strains derived from these same populations, we were unable to identify effects of the 2 behavioral QTLs in any of the many pairs of lines surveyed (except for one marginally significant association in one pairing). The strains we surveyed still displayed the same mating preference patterns as those of the original Ortiz-Barrientos et al. (2004) lines; however, the backcross lines did not exhibit statistically significant effects of these 2 QTLs. These results suggest that, across strains, different loci contribute allelic variation conferring a phenotypic difference distinguishing 2 D. pseudoobscura populations.
However, multiple reasons could explain the difference in results between these 2 studies. The simplest explanation is that a genetic survey of a single pair of lines from populations exhibiting a phenotypic difference gives only a snapshot of the underlying hereditary basis, and this snapshot may be inadequate to describing the full range of genetic variation contributing to this phenotypic difference in nature. This explanation is also consistent with the observed variance among strains derived from Flagstaff or Mather for degree of female interspecies mating propensity, despite the consistent difference between the 2 populations (see Figure 1). Alternatively, the same QTLs could contribute in all these line pairs, but our sample size was insufficient to detect their effects (see, e.g., Noor and Smith 2000). This explanation is inconsistent with our resampling of the original Ortiz-Barrientos et al. (2004) data with comparable (or smaller) sample sizes and observing statistical significance of these QTL effects, especially for Coy-1. Finally, uncontrolled genetic (epistatic) or environmental (G × E) effects (Boake et al. 2002) may have masked these QTLs. We have replicated the same conditions (including temperature, food media, and assay type) to the best of our ability, but this is a possibility impossible to exclude completely. Nonetheless, more than half of our assays had QTL genotypic effects opposite in direction (albeit nonsignificantly) to the phenotypic difference between these populations (Table 1), suggesting that strong epistasis or G × E would be required to create the observed difference between these studies.
We hypothesize that the low-discrimination alleles at these 2 QTLs most likely occur at low frequency within D. pseudoobscura because our study suggests that these lines have the same alleles at Coy-1 and Coy-2 as the originally surveyed high-discrimination Mather strain (17: Ortiz-Barrientos et al. 2004). A related study also suggested that strains of the sister-species D. persimilis bear the same high-discrimination allele at Coy-2 as D. pseudoobscura Mather17 (Ortiz-Barrientos and Noor 2005) and suggested as a possibility that it may have introgressed between these 2 species, consistent with a strong DNA sequence–based signature of introgression in this region of the 2 genomes (Machado et al. 2002). An alternative hypothesis to recent introgression of this allele is that the high-discrimination alleles are ancestral, and the low-discrimination alleles represent mutants that have arisen in D. pseudoobscura. Eventually identifying the genes within these Coy windows will allow us to test whether the low-discrimination alleles are ancestral or whether they represent mutant versions that may have arisen and persisted in populations where high discrimination is not needed, such as Flagstaff, AZ, because no closely related heterospecifics are found.
Our results strongly underscore the need for replication across multiple strains in behavior genetic mapping studies, particularly when studying intraspecies variation. These implications could extend to mapping studies outside of behavior, as well as interspecies phenotypes (e.g., Sweigart et al. 2007). Because of the difficulty associated with their initial construction, RIL mapping studies may be especially prone to nonreplication by investigators. Replicated linkage disequilibrium mapping and genome-wide association studies have also failed to identify effects of the same regions (e.g., Macdonald and Long 2004; Gruber et al. 2007; Ioannidis 2007), further underscoring the importance of replication before making conclusions (McCarthy et al. 2008). These problems are exacerbated further by detection issues when effect sizes of individual QTLs are small (Hsueh et al. 2001).
Supplementary Material
Supplementary Table 1 can be found at http://www.jhered.oxfordjournals.org/.
Funding
National Science Foundation (0509780, 0715484 to M.A.F.N.); National Institutes of Health (GM076051 to M.A.F.N.).
Supplementary Material
Acknowledgments
We thank W. Etges, A. Long, and S. McDermott for helpful comments on the manuscript and S. Bennett, R. Kommaraju, and S. Roth for technical assistance.
References
- Boake CRB, Arnold SJ, Breden F, Meffert LM, Ritchie MG, Taylor BJ, Wolf JB, Moore AJ. Genetic tools for studying adaptation and the evolution of behavior. Am Nat. 2002;160:S143–S159. doi: 10.1086/342902. [DOI] [PubMed] [Google Scholar]
- Gloor GB, Engels WR. Single-fly DNA preps for PCR. Drosoph Inf Serv. 1992;71:148–149. [Google Scholar]
- Gruber JD, Genissel A, Macdonald SJ, Long AD. How repeatable are associations between polymorphisms in achaete-scute and bristle number variation in Drosophila? Genetics. 2007;175:1987–1997. doi: 10.1534/genetics.106.067108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsueh WC, Goring HH, Blangero J, Mitchell BD. Replication of linkage to quantitative trait loci: variation in location and magnitude of the lod score. Genet Epidemiol. 2001;21:S473–S478. doi: 10.1002/gepi.2001.21.s1.s473. [DOI] [PubMed] [Google Scholar]
- Ioannidis JP. Non-replication and inconsistency in the genome-wide association setting. Hum Hered. 2007;64:203–213. doi: 10.1159/000103512. [DOI] [PubMed] [Google Scholar]
- Lorch PD, Servedio MR. Postmating-prezygotic isolation is not an important source of selection within and between species in Drosophila pseudoobscura and D. persimilis. Evolution. 2005;59:1039–1045. [PubMed] [Google Scholar]
- Macdonald SJ, Long AD. A potential regulatory polymorphism upstream of hairy is not associated with bristle number variation in wild-caught Drosophila. Genetics. 2004;167:2127–2131. doi: 10.1534/genetics.104.026732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machado CA, Kliman RM, Markert JA, Hey J. Inferring the history of speciation from multilocus sequence data: the case of Drosophila pseudoobscura and its close relatives. Mol Biol Evol. 2002;19:472–488. doi: 10.1093/oxfordjournals.molbev.a004103. [DOI] [PubMed] [Google Scholar]
- McCarthy MI, Abecasis GR, Cardon LR, Goldstein DB, Little J, Ioannidis JP, Hirschhorn JN. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat Rev Genet. 2008;9:356–369. doi: 10.1038/nrg2344. [DOI] [PubMed] [Google Scholar]
- Noor MA. Speciation driven by natural selection in Drosophila. Nature. 1995;375:674–675. doi: 10.1038/375674a0. [DOI] [PubMed] [Google Scholar]
- Noor MAF. Environmental effects on male courtship intensity in Drosophila pseudoobscura (Diptera: Drosophilidae) J Insect Behav. 1997;10:305–312. [Google Scholar]
- Noor MAF. Reinforcement and other consequences of sympatry. Heredity. 1999;83:503–508. doi: 10.1038/sj.hdy.6886320. [DOI] [PubMed] [Google Scholar]
- Noor MAF, Ortiz-Barrientos D. Simulating natural conditions in the laboratory: a re-examination of sexual isolation between sympatric and allopatric populations of Drosophila pseudoobscura and D. persimilis. Behav Genet. 2006;36:322–327. doi: 10.1007/s10519-005-9033-8. [DOI] [PubMed] [Google Scholar]
- Noor MAF, Smith KR. Recombination, statistical power, and genetic studies of sexual isolation in Drosophila. J Hered. 2000;91:99–103. doi: 10.1093/jhered/91.2.99. [DOI] [PubMed] [Google Scholar]
- Ortiz-Barrientos D, Counterman BA, Noor MAF. The genetics of speciation by reinforcement. PLoS Biol. 2004;2:e416. doi: 10.1371/journal.pbio.0020416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz-Barrientos D, Noor MAF. Evidence for a one-allele assortative mating locus. Science. 2005;310:1467. doi: 10.1126/science.1121260. [DOI] [PubMed] [Google Scholar]
- Palumbi SR. Nucleic acids II: the polymerase chain reaction. In: Hillis DM, Moritz C, Mable BK, editors. Molecular systematics. Sunderland (MA): Sinauer Associates; 1996. pp. 205–247. [Google Scholar]
- Shaw KL, Parsons YM, Lesnick SC. QTL analysis of a rapidly evolving speciation phenotype in the Hawaiian cricket Laupala. Mol Ecol. 2007;16:2879–2892. doi: 10.1111/j.1365-294X.2007.03321.x. [DOI] [PubMed] [Google Scholar]
- Singh RS. Genetic differentiation for allozymes and fitness characters between mainland and Bogota populations of Drosophila pseudoobscura. Can J Genet Cytol. 1983;25:590–604. [Google Scholar]
- Sweigart AL, Mason AR, Willis JH. Natural variation for a hybrid incompatibility between two species of Mimulus. Evolution. 2007;61:141–151. doi: 10.1111/j.1558-5646.2007.00011.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.