Summary
The Escherichia coli tolC encodes a major outer membrane protein with multiple functions in export (e. g., diverse xenobiotics, hemolysin) and as an attachment site for phage and colicins. tolC is regulated in part by MarA, SoxS and Rob, three paralogous transcriptional activators which bind a sequence called the marbox and which activate multiple antibiotic and superoxide resistance functions. Two previously identified tolC promoters, p1 and p2, are not regulated by MarA, SoxS or Rob but p2 is activated by EvgAS and PhoPQ which also regulate other functions. Using transcriptional fusions and primer extension assays, we show here that tolC has two additional strong overlapping promoters, p3 and p4, which are downstream of p1, p2 and the marbox and are activated by MarA, SoxS and Rob. p3 and p4 are configured so that a single marbox suffices to activate transcription from both promoters. At the p3 promoter, the marbox is separated by 20 bp from the −10 hexamer for RNA polymerase but at the p4 promoter, the same marbox is separated by 30 bp from the −10 hexamer. The multiple tolC promoters may allow the cell to respond to diverse environments by coordinating tolC transcription with other appropriate functions.
Keywords: gene regulation, outer membrane protein, transcriptional start sites, efflux pumps, antibiotic resistance
Introduction
TolC is an outer membrane protein (Koronakis et al., 2004) with diverse functions including efflux of multiple antibiotics, colicin uptake, phage adsorption and export of hemolysin and enterobactin. A major function of TolC is to interact with certain inner membrane transporters (e.g., AcrB) and periplasmic membrane fusion proteins (e.g., AcrA) to form a tripartite structure which makes it possible for substrates to be pumped directly out of the cell. At least eight such pairs of partners of TolC have been reported (Keseler et al., 2005). Basal levels of tolC expression are critical in providing wild-type E. coli with intrinsic resistance to many xenobiotics including clinically important antibiotics and bile salts; higher levels of expression are found in certain multidrug resistant mutants (Piddock, 2006). TolC-mediated efflux also seems to play a role in pathogenesis. Interestingly, recent evidence suggests that non-efflux functions of TolC may be involved in pathogenesis (Baucheron et al., 2005; Buckley et al., 2006; Piddock, 2006; Imuta et al., 2008; Virlogeux-Payant et al., 2008). Thus, TolC plays important roles in xenobiotic resistance and virulence.
tolC is a member of the marA/soxS/rob regulon (Fralick, 1996; Aono et al., 1998) which includes over 40 genes that promote resistance to multiple antibiotics, numerous other xenobiotics and to superoxides (Martin and Rosner, 2003; for comprehensive reviews, see articles in White et al., 2005). These genes are transcriptionally activated by three paralogous proteins, MarA, SoxS and Rob. marA and soxS expression can be increased by treating cells with salicylate (Cohen et al., 1993) and paraquat (Demple, 1996), respectively, and the activity of Rob can be increased post-translationally by treatment with 2,2′-dipyridyl, bile salts or decanoate (Rosenberg et al., 2003; Rosner et al., 2002).
MarA, SoxS and Rob bind as monomers to an asymmetrical and degenerate 19 bp DNA sequence known as the marbox (Martin et al., 1999; Wood et al., 1999). Because of marbox degeneracy, there are about 10,000 such binding sites per genome (Griffith et al., 2002; Martin et al., 2002). From an analysis of the promoters that are activated by these proteins, it is clear that the orientation of the marbox and its distance from the −10 hexamer of the promoter are critical in determining whether MarA, SoxS and Rob can activate the promoter.
Three functional promoter configurations have been found (Martin et al., 1999; Wood et al., 1999). At Class II promoters (e. g., fumC), the marbox is separated by ~20 bp from the −10 hexamer, overlaps the −35 hexamer and is in the “forward” orientation. At Class I promoters, the marbox is separated by ~39 (e. g., marRAB) or 50 bp (e. g., acrAB) from the −10 hexamer and is in the “backward” orientation. At a subset of the Class I promoters, Class I* (e. g., zwf), the marbox is separated by ~30 bp from the −10 hexamer but can be functional in either orientation.
Eguchi and coworkers have described two tolC promoters, p1 and p2 (Eguchi et al., 2003). The p2 promoter is activated by the joint action of the two-component systems EvgAS and PhoPQ but the regulation of the p1 promoter is not known. It has been assumed that the tolC promoter is activated by MarA, SoxS and Rob via a marbox identified only by its activity in chimeric promoters (Martin et al., 1999). However, the tolC transcription start sites (TSS) for p1 and p2 (Eguchi et al., 2003), were inconsistent with that interpretation because they lie upstream of the marbox (Fig. 1, top). On reinvestigation, we found two new TSSs for tolC (corresponding to promoters p3 and p4) that lie downstream of the marbox and are activated by MarA, SoxS and Rob. The single tolC marbox is uniquely configured so that it can be used for activation of both promoters.
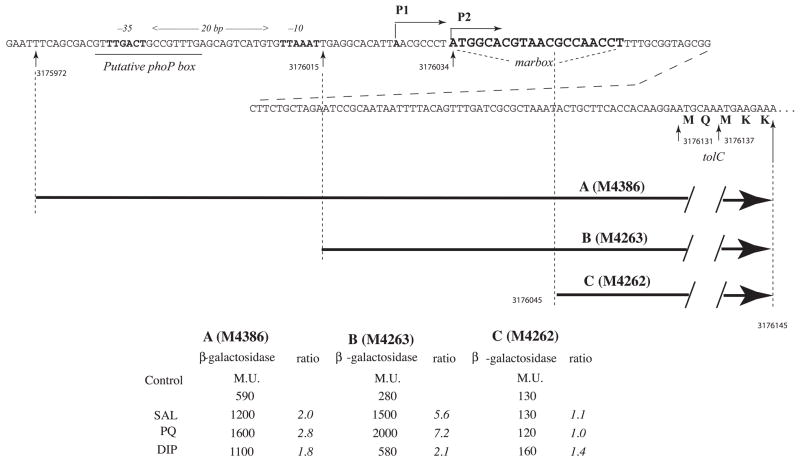
Fig. 1.
The sequence of the promoter region upstream of the structural gene for tolC is indicated with P1 and P2 transcription start sites (TSSs) and the putative −35, −10 and phoP box sites as suggested by Eguchi et al. (2003), and the presumptive marbox (Martin et al., 1999). Seven digit numbers refer to positions on the E. coli chromosome (Blattner et al., 1997). Below are indicated the fragments of this region incorporated into various promoter::lacZ fusions with strain numbers in parentheses and the β-galactosidase activities (Miller Units) obtained for these strains after treatment with 5 mM Na salicylate (SAL), 50 μM paraquat (PQ) or 5 mM 2,2′-dipyridyl (DIP) for 1 hr at 32°C. The ratio is the activity of the treated cells divided by that of the untreated cells.
Results and Discussion
tolC transcription start sites
Transcription of tolC is activated by MarA, SoxS and Rob (Aono et al., 1998) but the relevant tolC promoter has not yet been defined. Two transcription start sites (TSS) corresponding to two promoters had been reported for tolC by nuclease S1 digestion (Eguchi et al., 2003): a principal site, P1 (position 3176026, Fig. 1) and a secondary site, P2 (position 3176034), whose promoter, p2, is activated by EvgA and PhoPQ. Activation by the monomeric proteins MarA, SoxS and Rob requires that they bind to a marbox located in a particular orientation upstream of the TSS. However, a sequence (positions 3176034 to 3176052) that fits the 19 bp consensus marbox sequence and that has been found to have marbox activity in chimeras (Martin et al., 1999) lies downstream of both P1 and P2. Since no activator-responsive regulon marbox has yet been found downstream of its associated TSS (Martin and Rosner, 2002), we considered it unlikely that promoter p1 or p2 could be activated by this marbox.
To locate the tolC promoter(s) stimulated by MarA, SoxS and Rob and to identify the marbox required for tolC activation, three tolC::lacZ fusions (Fig. 1) were constructed in the bacteriophage λRS45 vector (Simons et al., 1987) and analyzed in single copy lysogens. Fusion B, commencing only 19 bp upstream of the putative marbox and thus lacking the −35 and −10 signals for p1 and p2 (Eguchi et al., 2003), had 50% of the basal activity of fusion A (200 vs 400 MU) which begins 43 bp farther upstream than fusion B and contains those promoters. Both fusions terminate at position 3176145, which is 91 bp downstream of the marbox, and both were significantly activated by agents known to activate marA and soxS transcription or Rob function (salicylate, paraquat and 2,2′-dipyridyl, respectively). Fusion C which lacks promoters p1 and p2 and 11 bp of the 19 bp marbox had 30% of the basal activity of fusion A (120 vs 400 MU) and was not activated by these agents. We conclude that the tolC promoter(s) activated by MarA, SoxS and Rob must lie downstream of p1 and p2.
To identify the tolC promoters activated by MarA and SoxS, primer extension analyses were carried out with total cellular RNA isolated from strains M3710 and M3944. These ΔmarRAB, rob::kan strains carry plasmid pRGM9818 (pMarA) and pJLR70 (pSoxS) (Martin et al., 2000) which express marA and soxS, respectively, from a plac promoter but also carry an F’ lacIq so that only in the presence of IPTG is MarA or SoxS made. To analyze activation of tolC by Rob, a different approach was used since in vivo Rob protein has to be post-translationally activated for it to be a potent transcriptional activator (Rosner et al., 2002; Rosenberg et al., 2003). To achieve this, we compared the RNA from two strains, M4110 and M4113, treated with a Rob activator, 2,2′-dipyridyl (DIP), for 1 hr during exponential growth. Both strains have Δmar sox::cat rob::kan null mutations but M4110 carries the vector pTA108 whereas M4113 carries pTA108:rob which expresses rob from a lac promoter. Assays of β-galactosidase due to the expression of micF::lacZ fusions in strains M3710 and M3944 and inaA::lacZ fusions in strains M4110 and M4113 showed 19-, 16- and 61-fold increases of fusion expression due to the upregulation of MarA, SoxS and Rob, respectively.
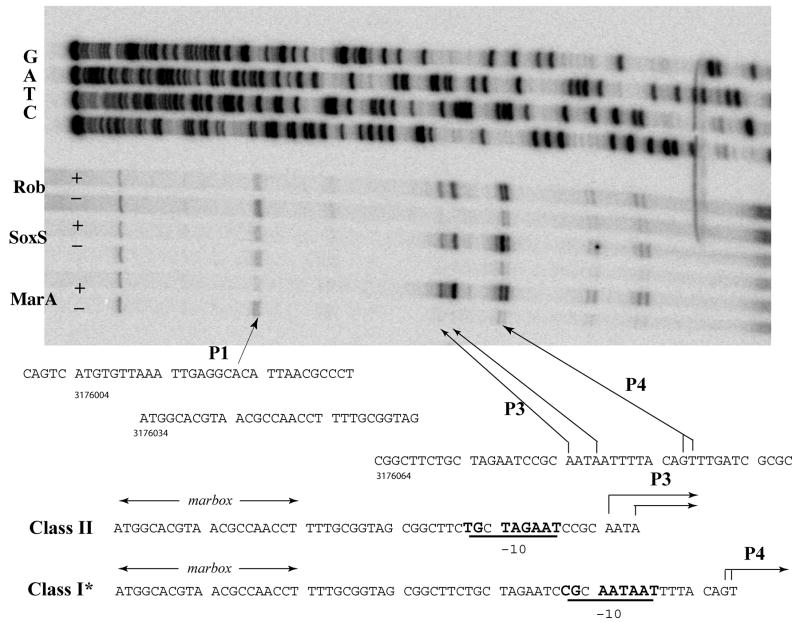
In the absence of IPTG (marA and soxS repressed) or Rob (M4110), six extension products were apparent (Fig. 2, top): at the adjacent G and T residues, positions 3176096 and 3176097, respectively; at two A residues at positions 3176084 and 3176087 (apparent on longer exposure); at a residue upstream near position 3176021; and at one near the top of the gel. The first four were greatly enhanced on overexpression of MarA and SoxS (growth in IPTG) or on activation of Rob in strain M4113 by DIP whereas the latter two products were diminished. (Overexpression of MarA and SoxS, and activation of Rob resulted in the appearance of two weak bands at positions 317616 and 317617 which we interpret as premature terminations of the reverse transcription assay or as mRNA degradation products.)
Fig. 2.
Mapping of tolC transcription start sites. Above: Primer extension analysis was carried out using total RNA isolated from the cells of strain M3710 (pMarA) or M3944 (pSoxS) grown for 1 hr during exponential phase without (−) or with (+) IPTG treatment to overexpress MarA and SoxS, respectively, as described in Materials and Methods. For activation by Rob, the RNA from M4110 (−) and M4113 (+) treated with 2,2′-dipyridyl for 1 hr during exponential phase was used. Sequencing ladders are also shown. The TSSs P3 and P4 from MarA-, SoxS- and Rob-dependent promoters (p3 and p4) are indicated by arrows as is a new position suggested for P1, 5 nt upstream of that previously suggested (Eguchi et al., 2003). Below: two alignments of the marbox with respect to promoters p3 and p4; their TSSs (P3 and P4); and potential extended −10 signals are underlined.
Corresponding to these four TSSs are two potential promoters. A potential extended −10 signal (TGnaATAAT) is spaced 6 nt upstream of the TSSs labeled P4 and another potential extended −10 signal (cGnTAgAAT) is spaced 7 nt upstream of the stronger P3 TSSs (Fig. 2, bottom). Extended −10 signals are frequently observed for promoters that have very poor −35 signals (Keilty and Rosenberg, 1987; Schultzaberger et al., 2007), as is the case for these promoters. We conclude that tolC has two overlapping promoters, p3 and p4 (Fig. 2): p3 has its marbox in the Class II configuration (Martin et al., 1999; Wood et al., 1999) separated by 20 bp from the −10 signal with TSSs 5 and 8 nt downstream of the −10 signal (positions 3176084 and 3176087); p4 has the Class I* configuration with the marbox separated by 30 bp from the −10 signal with TSSs 7 and 8 nt downstream of the −10 signal (positions 3176096 and 3176097). That one marbox can activate transcription from two different promoters is unique among the known marA/soxS/rob regulon promoters.
Similar results were obtained in strains in which SoxS was overproduced or Rob was activated by treatment with 2,2′-dipyridyl. Quantitation of the products indicated that MarA and Rob utilized p3 and p4 to about equal extents whereas SoxS utilized p4 to a greater extent than p3 (approximately 2-fold).Under the experimental conditions employed, we did not expect to observe transcripts starting at P2 because they require high levels of EvgA. We should, however, have seen P1 at position 3176026 (Eguchi et al., 2003) but instead observed an apparent TSS 5 nt upstream at position 3176021. Since the only apparent 10 signal in this region, and the one suggested by Eguchi et al. (2003), TTAAAT, is 6 nt distant from the TSS we observed but unusually distant (11 nt) from the TSS they reported, we suspect that our P1 TSS assignment is correct.
The multiplicity of tolC promoters may reflect the need to adapt the expression of tolC to different environments. The role of TolC in the efflux of xenobiotics has been well studied. Recently, a role in adherence to vertebrate cells has been reported for the TolC of Salmonella enterica serovar Typhimurium (Buckley et al., 2006; Virlogeux-Payant et al., 2008). Since acrB mutants were not similarly defective in adherence, this may suggest that the outer membrane TolC surface itself has important functions in colonization. It might be beneficial to regulate these different functions differently. Promoter p2 appears to be activated by phosphorylated EvgA (in a strain with a mutant evgS) and PhoPQ together (Eguchi et al., 2003). Concomitant with upregulation of tolC, EvgA upregulated several multidrug resistance genes (acrAB, emrKY, mdfA and yhiUV) and showed increased antibiotic resistance that was fully dependent on TolC but only partially dependent on AcrA. The environmental signal for EvgAS is not known but PhoPQ is activated by low Mg++ concentrations such as exists in the phagosome (Groisman, 2001).
The four tolC promoters respond to at least five different environmental signals. The p1 promoter is not responsive to EvgAS, PhoPQ, MarA, SoxS or Rob but seems to have significant constitutive expression under laboratory conditions (Eguchi et al., 2003; and Figs. 1 and 2). The p2 promoter is responsive to EvgAS and PhoPQ but not the others. As shown here, p3 and p4 are activated by MarA, SoxS and Rob, with each activator being regulated by different environmental signals. The nesting of the p3 and p4 promoters with a single activator binding site might facilitate even further levels of control as intimated by the different relative extents to which p3 and p4 are utilized by these activators. These three activators also upregulate acrAB and other genes which are critical for increased multidrug and superoxide resistances whereas EvgAS and PhoPQ regulate different sets of genes. Thus, tolC has multiple transcriptional regulatory elements which respond to different environments and possibly tailor the response to the particular TolC function (efflux, protein transport, adherence) that is adaptive.
Experimental procedures
Bacterial strains and plasmids
All strains are derivatives of Escherichia coli K-12. tolC::lacZ transcriptional fusions (see Fig. 1) were made using phage λRS45 as described (Simons et al., 1987). Strain GC4468 (Δlac) was infected with the phage, and lysogens with a single-copy prophage were used for β-galactosidase assays. Strains M3710 and M3944 are Δmar rob::kan micF::lacZ F’ lacIq carrying plasmid pRGM9818 or pJLR70 in which marA or soxS, respectively, are under the control of the tac promoter (Martin et al., 2000). Strains M4110 and M4113 are Δmar rob::kan sox::cat inaA::lacZ. Strain M4110 carries plasmid pTA108 (vector) and M4113 carries plasmid pTA108:rob in which rob is constitutively expressed from the lac promoter (Rosner et al., 2002).
Culture media and chemicals
LB (Lennox) media contained per liter: 10 gm Bacto-tryptone (DIFCO, Detroit, MI), 5 gm Bacto-yeast extract and 5 gm NaCl, pH adjusted to 7.5 with NaOH. Chemicals were purchased from Sigma Chemicals, St. Louis, MO
β-galactosidase assays
Bacteria were grown overnight in LB medium at 32°C, diluted 1000-fold in fresh medium and grown to an A600 of about 0.15 unless otherwise indicated. To test the transcriptional activation of tolC::lacZ fusions by MarA, SoxS or Rob, cells were grown to an A600 of about 0.15, diluted into an equal volume of LB medium containing sodium salicylate, paraquat or 2,2′-dipyridyl (final concentrations of 5mM, 50 μM and 5 mM, respectively) and aerated for 1 hr at 32°C. β-galactosidase was then measured using the CHCl3-SDS method of Miller (1972). All assays presented were performed at least twice in duplicate and the standard errors of the mean were <± 5%.
Primer extension analysis
Strains with wild-type tolC were grown in LB medium at 32°C to exponential phase (A600 = 0.4). Strains M3710 (pMarA) and M3944 (pSoxS) were then diluted two-fold into LB without (control) or with IPTG (0.5 mM final concentration) for 1 hr to induce plasmid-borne marA or soxS expression, respectively. Strains M4110 (pTA108 vector control) and M4113 (pTA108:Rob) were diluted into LB with 2,2′-dipyridyl (5 mM final concentration) for 1 hr to activate Rob. Total cellular RNA was isolated by using the TRIzol Reagent (Invitrogen, Carlsbad, CA). Primer extension assays (Zhang et al., 1998) were performed by incubating 20 μg of total RNA and 1 pmole of 5′-end 32P-labeled primer AZ#1006 (5′-CTC AGG CCG ATA AGA ATG G) at 65°C for 5 min, followed by incubation with 10 U of AMV Reverse Transcriptase (Life Sciences, Inc,, St. Petersburg, FL), 5 U of RNase Inhibitor (Invitrogen) in 1X RT buffer (Life Sciences, Inc.) in a 15-μl reaction volume at 42°C for 1 hour. The reactions were terminated by adding 7.5 μl Stop/Loading buffer and cDNA products were separated on an 8% polyacylamide gel containing 8 M urea in 1X TBE buffer at 70 watts for 70 min. To generate a sequence ladder, a tolC-specific PCR fragment was generated by using primers AZ#1007 (5′-CGC CGC ACC TCA TGA CTC AT) and AZ#1008 (5′-TGT GGC AGT AAT GGA CTG CG), and DNA sequencing reactions were carried out with AZ#1006 (above) and SequiTherm EXCEL™ II DNA sequencing Kit (Epicentre, Madison, WI). Bands on gels were quantitated using a Typhoon Trio Variable Mode Imager (GE Healthcare Life Sciences, Piscataway, NJ).
Acknowledgments
We thank G. Storz for discussions. This research was supported by the Intramural Research Program of the NIH.
References
- Aono R, Tsukagoshi N, Yamamoto M. Involvement of outer membrane protein TolC, a possible member of the mar-sox regulon, in maintenance and improvement of organic solvent tolerance of Escherichia coli K-12. J Bacteriol. 1998;180:938–944. doi: 10.1128/jb.180.4.938-944.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baucheron S, Mouline C, Praud K, Chaslus-Dancla E, Cloeckaert A. TolC but not AcrB is essential for multidrug-resistant Salmonella enterica serotype Typhimurium colonization of chicks. J Antimicrob Chemother. 2005;55:707–712. doi: 10.1093/jac/dki091. [DOI] [PubMed] [Google Scholar]
- Blattner FR, Plunkett G, III, Bloch CA, Perna NT, Burland V, Riley M, Collando-Vides J, Glasner JD, Rode CK, Mayhew GF, Gregor J, Davis NW, Kirkpatrick HA, Goeden MA, Rose DJ, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- Buckley AM, Webber MA, Cooles S, Randall LP, La Ragione RM, Woodward MJ, Piddock LJ. The AcrAB-TolC efflux system of Salmonella enterica serovar Typhimurium plays a role in pathogenesis. Cell Microbiol. 2006;8:847–856. doi: 10.1111/j.1462-5822.2005.00671.x. [DOI] [PubMed] [Google Scholar]
- Cohen SP, Levy SB, Foulds J, Rosner JL. Salicylate induction of antibiotic resistance in Escherichia coli: activation of the mar operon and a mar-independent pathway. J Bacteriol. 1993;175:7856–7862. doi: 10.1128/jb.175.24.7856-7862.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demple B. Redox signaling and gene control in the Escherichia coli soxRS oxidative stress regulon--a review. Gene. 1996;179:53–57. doi: 10.1016/s0378-1119(96)00329-0. [DOI] [PubMed] [Google Scholar]
- Eguchi Y, Oshima T, Mori H, Aono R, Yamamoto K, Ishihama A, Utsumi R. Transcriptional regulation of drug efflux genes by EvgAS, a two-component system in Escherichia coli. Microbiol. 2003;149:2819–2828. doi: 10.1099/mic.0.26460-0. [DOI] [PubMed] [Google Scholar]
- Fralick JA. Evidence that TolC is required for functioning of the Mar/AcrAB efflux pump of Escherichia coli. J Bacteriol. 1996;178:5803–5805. doi: 10.1128/jb.178.19.5803-5805.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith KL, Shah IM, Myers TE, O’Neill MC, Wolf RE., Jr Evidence for "pre-recruitment" as a new mechanism of transcription activation in Escherichia coli: the large excess of SoxS binding sites per cell relative to the number of SoxS molecules per cell. Biochem Biophys Res Commun. 2002;291:979–986. doi: 10.1006/bbrc.2002.6559. [DOI] [PubMed] [Google Scholar]
- Groisman EA. The pleiotropic two-component regulatory system PhoP-PhoQ. J Bacteriol. 2001;183:1835–1842. doi: 10.1128/JB.183.6.1835-1842.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imuta N, Nishi J, Tokuda K, Fujiyama R, Manago K, Iwashita M, Sarantuya J, Kawano Y. The Escherichia coli efflux pump TolC promotes aggregation of enteroaggregative E. coli 042. Infect Immun. 2008;76:1247–1256. doi: 10.1128/IAI.00758-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keilty S, Rosenberg M. Constitutive function of a positively regulated promoter reveals new sequences essential for activity. J Biol Chem. 1987;262:6389–6389. [PubMed] [Google Scholar]
- Keseler IM, Collado-Vides J, Gama-Castro S, Ingraham J, Paley S, Paulsen IT, Peralta-Gil M, Karp PD. EcoCyc: a comprehensive database resource for Escherichia coli. Nucl Acids Res. 2005;33(Database issue):D334–337. doi: 10.1093/nar/gki108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koronakis V, Eswaran J, Hughes C. Structure and function of TolC: the bacterial exit duct for proteins and drugs. Ann Rev Biochem. 2004;73:467–489. doi: 10.1146/annurev.biochem.73.011303.074104. [DOI] [PubMed] [Google Scholar]
- Martin RG, Rosner JL. Genomics of the marA/soxS/rob regulon of Escherichia coli: identification of directly activated promoters by application of molecular genetics and informatics to microarray data. Mol Microbiol. 2002;44:1611–1624. doi: 10.1046/j.1365-2958.2002.02985.x. [DOI] [PubMed] [Google Scholar]
- Martin RG, Rosner JL. Analysis of microarray data for the marA, soxS, and rob regulons of Escherichia coli. Meth Enzymol. 2003;370:278–280. doi: 10.1016/S0076-6879(03)70024-X. [DOI] [PubMed] [Google Scholar]
- Martin RG, Gillette WK, Rosner JL. Promoter discrimination by the related transcriptional activators, MarA & SoxS: differential regulation by differential binding. Mol Microbiol. 2000;35:623–634. doi: 10.1046/j.1365-2958.2000.01732.x. [DOI] [PubMed] [Google Scholar]
- Martin RG, Gillette WK, Martin NI, Rosner JL. Complex formation between activator and RNA polymerase as the basis for transcriptional activation by MarA and SoxS in Escherichia coli. Mol Microbiol. 2002;43:355–370. doi: 10.1046/j.1365-2958.2002.02748.x. [DOI] [PubMed] [Google Scholar]
- Martin RG, Gillette WK, Rhee S, Rosner JL. Structural requirements for marbox function in transcriptional activation of mar/sox/rob regulon promoters in Escherichia coli: sequence, orientation and spatial relationship to the core promoter. Mol Microbiol. 1999;34:431–41. doi: 10.1046/j.1365-2958.1999.01599.x. [DOI] [PubMed] [Google Scholar]
- Miller JH. Experiments in Molecular Genetics. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, N.Y.: 1972. [Google Scholar]
- Piddock LJ. Multidrug-resistance efflux pumps - not just for resistance. Nat Rev Microbiol. 2006;4:629–636. doi: 10.1038/nrmicro1464. [DOI] [PubMed] [Google Scholar]
- Rosenberg EY, Bertenthal D, Nilles ML, Bertrand KP, Nikaido H. Bile salts and fatty acids induce the expression of Escherichia coli AcrAB multidrug efflux pump through their interaction with Rob regulatory protein. Mol Microbiol. 2003;48:1609–1619. doi: 10.1046/j.1365-2958.2003.03531.x. [DOI] [PubMed] [Google Scholar]
- Rosner JL, Dangi B, Gronenborn AM, Martin RG. Posttranslational activation of the transcriptional activator Rob by dipyridyl in Escherichia coli. J Bacteriol. 2002;184:1407–1416. doi: 10.1128/JB.184.5.1407-1416.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shultzaberger RK, Chen Z, Lewis KA, Schneider TD. Anatomy of Escherichia coli σ70 promoters. Nucl Acids Res. 2007;35:771–788. doi: 10.1093/nar/gkl956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons RW, Houman F, Kleckner N. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene. 1987;53:85–96. doi: 10.1016/0378-1119(87)90095-3. [DOI] [PubMed] [Google Scholar]
- Virlogeux-Payant I, Baucheron S, Pelet J, Trotereau J, Bottreau E, Velge P, Cloeckaert A. TolC, but not AcrB, is involved in the invasiveness of multidrug-resistant Salmonella enterica serovar Typhimurium by increasing type III secretion system-1 expression. Int J Med Microbiol. 2008 doi: 10.1016/j.ijmm.2007.12.006. [Epub ahead of print, Feb 11, 2008, doi:10.1016/j.ijmm.2007.12.006] [DOI] [PubMed] [Google Scholar]
- White DG, Alekshun MN, McDermott PF. Frontiers in Antimicrobial Resistance. ASM Press; Washington, D.C.: 2005. [Google Scholar]
- Wood TI, Griffith KL, Fawcett WP, Jair KW, Schneider TD, Wolf RE., Jr Interdependence of the position and orientation of SoxS binding sites in the transcriptional activation of the class I subset of Escherichia coli superoxide-inducible promoters. Mol Microbiol. 1999;34:414–430. doi: 10.1046/j.1365-2958.1999.01598.x. [DOI] [PubMed] [Google Scholar]
- Zhang A, Altuvia S, Tiwari A, Argaman L, Hengge-Aronis R, Storz G. The oxyS regulatory RNA represses rpoS translation and binds Hfq (HF-I) EMBO J. 1998;17:6061–6068. doi: 10.1093/emboj/17.20.6061. [DOI] [PMC free article] [PubMed] [Google Scholar]