Abstract
Diacylglycerol-lactone (DAG-lactone) libraries generated by a solid-phase approach using IRORI technology produced a variety of unique biological activities. Subtle differences in chemical diversity in two areas of the molecule, the combination of which generates what we have termed “chemical zip codes”, are able to transform a relatively small chemical space into a larger universe of biological activities, as membrane-containing organelles within the cell appear to be able to decode these “chemical zip codes”. It is postulated that after binding to protein kinase C (PKC) isozymes or other non-kinase target proteins that contain diacylglycerol responsive, membrane interacting domains (C1 domains), the resulting complexes are directed to diverse intracellular sites where different sets of substrates are accessed. Multiple cellular bioassays show that DAG-lactones, which bind in vitro to PKCα to varying degrees, expand their biological repertoire into a larger domain, eliciting distinct cellular responses.
Introduction
Protein kinase C isozymes (PKCs) are key signaling molecules, responding to the second messenger diacylglycerol (DAG) which is generated from phosphoinositides upon activation of many cellular receptors. The activation of conventional protein kinases (cPKCs: α, βI, βII, γ) and novel protein kinases (nPKCs: δ, ε, η, θ), as well as other proteins containing similar C1 membrane targeting domains, typically requires recruitment to membranes and allosteric activation by DAG, which results from the specific molecular interactions of DAG with the C1 domains and the modulation of the physical properties of membranes by the DAG alkyl chains. The combination of these two factors contributes to the localization of an isozyme to specific intracellular sites where different substrates are accessed. Molecular interactions revealed by the crystal structure of the PKCδ C1b domain bound to phorbol-13-acetate1 showed the existence of a network of hydrogen bonds between phorbol ester and conserved amino acids Thr242, Leu251 and Gly253, which were reproduced precisely by computer modeling when DAG was inserted into the same C1 domain.2 Other highly conserved hydrophobic amino acids of the δ-C1b domain, such as Pro231, Phe243, Leu250, and Trp252, which form a contiguous hydrophobic ring around the top of the C1 domain, are likely to interact with the membrane by sensing changes in the physical properties of the lipid bilayer induced by DAG, including fluidity, curvature, hydrocarbon volume, and head group separation, all of which may facilitate the insertion of the enzyme into membranes.3–6 It has become increasingly clear that these type of physical perturbations of lipid membranes contribute to the modulation of PKC activity.7
The cellular distribution of activated PKC isozymes is additionally controlled by interactions with other signaling or scaffolding proteins through the formation of multiprotein complexes that facilitate signal transduction and confer specificity for individual PKC isoforms by regulating their activity and cellular localization allowing the modulation of biological functions within the cell.8–10 Furthermore, these signaling complexes are not confined to the plasma membrane alone, but are found on various cellular membranes, including Golgi, mitochondrial, and nuclear membranes.11, 12 Recent studies have also shown that lipid-protein interactions can similarly provide the necessary specificity and affinity to achieve a particular subcellular localization of signaling proteins.13 Thus, membrane lipids may help compartmentalize signaling complexes and regulate the spatio-temporal dynamics of PKC activation through interacting membrane microdomains, some of which can be formed transiently during signaling after the release of DAG.14–16 An additional level of control where lipids play a critical role in enzyme activation is that of disrupting intramolecular, interdomain interactions between C1 and C2 domains, as demonstrated with PKCα,17, 18 and between the C1 domain and at least four different regions of the protein in β2-chimaerin (the N-terminus, the SH2 and RacGAP domains, and the linker between C1 and SH2) as elegantly described by Hurley et al.19
The most useful pharmacological probes for C1 domains, the family of phorbol esters (e.g. TPA), cannot distinguish between PKC isoforms, and PKC is only one of multiple families of proteins that have appropriate C1 domains to act as DAG/phorbol receptors.20, 21 There are other proteins with typical C1 domains that are subject to DAG modulation including the PKD family, a distinct class of serine/threonine kinases;22 the chimaerin family, inhibitors of p21Rac;23 the munc-13 family, proteins involved in synaptic vesicle priming;24 the RasGRP family, guanyl nucleotide exchange factors for Ras and Rap1;25, 26 and the DAG kinase family, which function to abrogate DAG signaling, thus providing negative feedback on DAG signaling pathways.27 Of these C1 domain-containing families, the PKC family is the best-studied mediator in the DAG signaling pathways and an important target for drug development if one could learn how to selectively impact PKC regulation in cells subject to proliferating or differentiating stimuli.
Because the actual DAG binding site is a complex of lipid bilayer and C1 domain, for which the C1 domain is only a half-site, the interactions with the membrane and other important protein components are difficult to study. We surmise that the specific cellular localization of the activated PKC isozyme, or C1 domain containing protein, ought to be determined in part by the different lipid composition of the membranes, the targeting information intrinsic to the individual C1 domain bound to DAG, and the binding to scaffolding or signaling proteins. This multifaceted control of diversity suggests that in order to improve our chance of developing DAG-like molecules capable of translocating these proteins to specific cellular targets, we need to gain access to the chemical space surrounding the DAG binding site. Because structure-activity analyses that incorporate the lipid bilayer and other proteins into the model are still rudimentary, we explored this aspect indirectly by combinatorial chemistry.
Combinatorial libraries built on our potent DAG-lactone template28 using carefully chosen R and R′ groups (Figure 1) that function as “chemical zip codes” were expected to create a unique microenvironment surrounding the binding site. These “chemical zip codes” result from the exclusive combination of two diversity elements on the DAG-lactone at the sn-1 (R′) and sn-2 (R) positions (Figure 1).
Figure 1.
“Chemical zip codes” for the DAG-lactones Libraries 1 and 2. For Library 1 (top) the rows correspond to a set of R groups adjacent to the sn-2 carbonyl, which are derived from a collection of aldehydes. The columns correspond to R′ groups which are derived from a set of acid chlorides and are connected to the sn-1 carbonyl. For library 2 (bottom), the orientation is reversed and the rows display the R′ groups at sn-1, whereas the columns display the R groups at sn-2. The red numbers in each library correspond to measured Ki values (nM); the blue numbers correspond to calculated log P values; and the green numbers in parentheses correspond to the observed m/z for MH+ (or M•+).
The chosen elements of our “chemical zip codes” were selected to exploit membrane-ligand interactions and protein-ligand interactions that operate outside the C1 domain in the ternary complex (C1-domain–DAG–membrane) through various mechanisms for which the side chains would be expected to be critical, such as Coulombic interactions between cationic residues and anionic lipids; interactions between aromatic residues and zwitterionic lipids; and hydrophobic interactions between conserved amino acids along the rim of the C1 domain and membrane lipids. By applying this approach, we have been able to identify combinations of R and R′ groups that produce DAG-lactones capable of eliciting distinct, specific cellular responses, thus providing strong proof-of-principle for the use of “chemical zip codes” on DAG-lactones to develop novel therapeutic strategies for PKC agonists with unique biology.
Chemistry
In a previous publication we reported the development of a solid-phase method using a PL-DHP (3,4-dihydro-2H-pyran) resin for the synthesis of DAG-lactones with an exploratory set of R and R′ groups.29 This method was tested with the synthesis of a small, nine-member array with the idea of forecasting the reliability of the biological data that could be obtained when using larger chemical libraries synthesized under the same conditions. From that study, we concluded that the biological data acquired using crude samples directly obtained from the resin provided valuable information as the difference in binding affinities (Ki) for PKCα between pure and crude materials varied only by a factor between 1.5 and 3.7.
The synthetic approach utilized here was essentially the same (Scheme 1), except that we chose two starting lactones (1b and 1d) with different protecting groups (Scheme 2), which were designed to overcome incompatibilities between some of the chemical groups selected for the library and the conditions for deprotection. Using this method, two exploratory libraries were investigated (Figure 1).
Scheme 1.
Scheme 2.

The basic concept behind the design of Library 1 was to explore the effect of swapping similar groups – containing simple polar substituents that are either electron-withdrawing or electron-donating and non-polar alkyl or aryl moieties – between the two available positions on the DAG-lactone scaffold. Hence, in Library 1 the R groups chosen for the rows of the matrix (A-H) were the same as the R′ groups of the first eight columns (1–8). Four additional acid chlorides (columns 9–12) were added to explore the effect of branching on the aromatic moiety. The reason for selecting branched chains was based on the excellent activity achieved earlier with DAG-lactones containing aliphatic, branched acyl chains.28 In addition, the chosen groups allowed us to explore a range of log P values, which is also an important parameter for activity and selectivity.
Based on a previous study where we found differences between DAG-lactones bearing identical functionalized phenyl groups at either the sn-2 (R) or the sn-1 (R′) positions, we concluded that DAG-lactones prefer to bind to the C1 domain with the R′ acyl chain oriented toward the interior of the membrane and the R alkylidene or arylidene group directed to the surface of the C1 domain, adjacent to the lipid interface.30 Keeping that concept in mind, we decided to expand the chemical R space by designing Library 2 with a larger aldehyde-derived set (columns 1–12) consisting exclusively of aromatic moieties. The smaller, acid chloride-derived rows were comprised mostly of alkyl groups (A, B, C, F, G, H) and aryl-alkyl groups (D and E), all of which are non-polar and able to interact effectively with the membrane.
The selection of these two libraries was intended to showcase the potential of this approach as every member of the library was expected to bind to the C1 domain, but where differences between the various bioassays and cellular activities would reflect the manner in which the ternary complex, C1-domain–DAG-lactone–membrane, could sense the different composition of the “chemical zip codes”. All of the crude compounds synthesized in Libraries 1 and 2 were racemates and consisted of variable mixtures of geometrical E- and Z-isomers. The identity of individual DAG-lactone library members and the overall library quality were initially assessed by FAB/MS analysis employing a modified combine-and-analyze strategy31 (see Experimental Section).
Biological Assays
Library 1, PKCα
PKCα binding affinities were ascertained with partially purified mouse enzyme. The Ki values were determined by competition with [3H]PDBU as described previously, except that values were calculated from a single ligand concentration.32 The groups chosen for Library 1 covered a useful range of log P values between 2.42 to 7.30 calculated according to the atom-based program MOE SLog P.33 There appears to be a parabolic dependence between Ki and log P, which peaks near a log P value of 4.8, as observed for compound L1-G2. Column12, which contains the most hydrophobic compounds with the highest log P values (between 6 and 7), did not produce very potent compounds, except for L1-G12.
Remarkably, the entire G row, which contains the trifluoromethylphenyl R group, is rather unique and for the most part contains very active ligands, including the most potent compound of the entire library (L1-G2). Because of this finding, the trifluoromethylphenyl group was more thoroughly studied in Library 2, which included positional isomers and multiple CF substitutions (vide infra).
An interesting observation that supports the binding mode described before is the effect of exchanging the positions of polar and non-polar groups. For example, a comparison between compounds L1-B5, L1-B6, L1-B7 and L1-B8, with polar dimethylamino, methoxy, trifluoromethyl, and nitro groups as part of R′ (sn-1), vis-à-vis compounds L1-E2, L1-F2, L1-G2 and L1-H2 where the same polar groups become part of R (sn-2), showed that the latter arrangement produced better ligands. Comparing the Ki values for L1-B8 and L1-H2 between crude and pure samples (E-isomers) corroborated the proposed binding mode, as the 10-fold difference in binding affinity observed between the crude samples was magnified to a 100-fold difference for the pure compounds (Table 1, rows 3 and 12).
Table 1.
Comparison of Ki values for PKCα between crude library samples and pure compounds in Library 1.
| Row # | Compound | Library Sample Ki (nM) | Pure Sample Ki (nM) | Ratio†Ki crude/Ki pure |
|---|---|---|---|---|
| 1 | L1-A12 | 126. ± 38 | 2.5 ± 0.1* | 50.4 |
| 2 | L1-B5 | 466 ± 55 | 133.8 ± 8.7* | 3.4 |
| 3 | L1-B8 | 460 ± 46 | 334 ± 29* | 1.4 |
| 4 | L1-E2 | 96 ± 12 | 12.9 ± 0.6 | 7.4 |
| 5 | L1-E5 | 1080 ± 190 | 102 ± 14 | 10.6 |
| 6 | L1-E6# | 890 ± 170 | 102.8 ± 5.5 | 8.7 |
| 7 | L1-E8# | 1230 ± 230 | 213 ± 20 | 5.8 |
| 8 | L1-F5 | 359 ± 25 | 48.0 ± 7.6 | 7.5 |
| 9 | L1-F6 | 332 ± 40 | 129 ± 19 | 2.6 |
| 10 | L1-F11 | 118.5 ± 9.5 | 3.7 ± 0.6 | 32.0 |
| 11 | L1-F12 | 197 ± 37 | 8.0 ± 1.6 | 24.6 |
| 12 | L1-H2 | 41 ± 2.8 | 3.10 ± 0.40 | 13.2 |
| 13 | L1-H2 | 41 ± 2.8 | 73.5 ± 7.3* | 0.56 |
| 14 | L1-H5# | 362 ± 37 | 127 ± 30 | 2.9 |
| 15 | L1-H5# | 362 ± 37 | 1076 ± 89* | 0.34 |
| 16 | L1-H6# | 281 ± 35 | 103 ± 17 | 2.7 |
| 17 | L1-H8 | 660 ± 220 | 127 ± 16 | 5.2 |
= samples dissolved in DMSO for testing;
= Z-isomer;
= ratio calculated relative to the unresolved library mixture of E- and Z-isomers.
For the compounds that were synthesized in pure form (Table 1), the differences in affinity between crude and pure samples were variable. Pure compounds L1-A12, L1-B5, and L1-B8 were obtained as Z-isomers, whereas the rest of the compounds with α-arylidene moieties were obtained almost exclusively as E-isomers (vide infra). The majority of the compounds showed Ki crude/Ki pure ratios of 10-fold, except L1-A12, L1-F11, L1-F12 and L1-H2. Obviously, factors such as purity of the crude samples (see Analysis of Chemical Libraries in the Experimental Section), solubility, and operational variation can explain these differences. Nevertheless, for the most part, the results with the crude samples mimicked well the real trends of the pure samples and aided in the selection of compounds for additional studies.
Library 1, RasGRP
The high conservation of the C1 domains of PKC has been a challenge for the development of selective ligands. Furthermore, the identification of other families of signaling molecules containing DAG-responsive C1 domains makes achieving specificity an even more daunting task. The PKCα binding assay of the purified members of Library 1 (Table 1) showed that several compounds produced unusually shaped dose-response curves. These curves were shallow and could be modeled as two binding components with different affinities and with each representing half of the total binding sites, suggesting that the compounds might show selectivity between the C1a and C1b domains. To test that model, we assayed these compounds for RasGRP1/3 binding since it only has a single C1 domain. This led to the discovery of two compounds, L1-H5 and L1-H6, which displayed much higher binding affinities for RasGRP1/3 than for PKCα and other PKC isozymes.34 This finding provided strong support for the hypothesis that the complexes formed between the ligand, the C1 domain, membrane phospholipids, and intervening proteins can decipher the “chemical zip codes” and show specificity. Compound L1-H5, also known as 130C037,34 served as the basis for the development of a small library comprised of all of its possible isomers (Table 2).
Table 2.
Ki (nM) values for binding to RasGRP3 (blue) and PKCα (red) competing with [3H]PDBu (Ki values correspond to pure samples).
| p-NMe2 | m-NMe2 | o-NMe2 | ||
|---|---|---|---|---|

|
p-NO2 | 3.8±0.1 | 2.3±0.6 | 1.8±0.3 |
| 1.27±30 | 31.8±1.9 | 38.5±6.5 | ||
| (L1-H5, 130C037) | ||||
| m-NO2 | 8.0±1.1 | 5.3±1.1 | 3.9±0.4 | |
| 72.4±4.8 | 63±16 | 44±10 | ||
| o-NO2 | 14.4±1.4 | 16.4±1.8 | 9.4±1.9 | |
| 54.5±5.3 | 11.5±9.7 | 134±18 |
A remarkable point about this mini-library was the relative small variance in Ki values as a function of the substitution pattern on either aromatic moiety. Furthermore, Ki(PKCα)/Ki(RasGRP3) ratios varied only from 4-fold to 33-fold with the original p,p-isomer (L1-H5) showing the greatest selectivity for RasGRP3. In addition, as previously reported, the inverted p,p-isomer, known as 130C045 (not shown),34 where the dimethylaminophenyl group and the nitrophenyl group were switched, showed a similar selectivity to that of L1-H5 with a Ki of 7.8 ± 0.94 nM for RasGRP3 and a Ki of 215 ± 14 nM for PKCα.34 Not surprisingly, in the present study, we also found that pure Library 1 compounds L1-E5 and L1-H8 with identical p-dimethylaminophenyl or p-nitrophenyl groups at both sn-1 and sn-2 positions displayed similar affinities for RasGRP3 with Ki values of 4.68 ± 0.39 nM and 2.22 ± 0.08 nM, respectively. Furthermore, the bis-p-nitro compound (L1-H8) now appears to be the ligand with the highest selectivity ratio [Ki(PKCα)/Ki(RasGRP3) = 57], suggesting that these types of compounds, irrespective of the orientation of the polar moieties, are able to form productive electrostatic interactions at the lipid interface when bound to the C1 domain of RasGRP. The different decoding of the “chemical zip codes” for these compounds was also demonstrated in living cells where L1-H5 effectively translocated RasGRP3 while failing to translocate PKCα.34
Because RasGRP1/3 are enzymes that contain a solitary C1 domain, L1-H5 was also assayed with individual C1 domains for PKC isozymes α and δ. The results showed that only the δC1b domain bound effectively to L1-H5 in the same nanomolar range as with intact RasGRP1/3, and that it was exclusively translocated in living cells.34 Figure 2 shows the close structural similarity between (A) the empty δC1b domain (obtained from the crystal structure in complex with phorbol 13-acetate,1 (B) the RasGRP1 and (C) RasGRP3 C1 domains (built from the coordinates of the δC1b by homology modeling) in contrast to the subtle structural differences in the C1 domains of (D) δC1a, (E) αC1a and (F) αC1b, which do not translocate in response to L1-H5.34 This illustrates that while the binding sites of all six C1 domains are highly hydrophobic, even small differences in sequence and surface shape can affect binding affinity to DAG-lactones and the targeting of the ternary complex (C1-domain–DAG–membrane) resulting in large differences in biological effects.
Figure 2.

Binding site surface of the crystal structure of the C1b domain of PKCδ1 (A) compared to homology models of RasGRP1 (B), RasGRP3 (C), δC1a (D), αC1a (E), and αC1b (F). Residues are colored according to hydrophobicity,35 from highly hydrophobic residues in red to hydrophilic residues in blue. C1 domains in the top row (A, B, C) are capable of binding and translocating L1-H5 whereas those in the bottom row (D, E, F) are not.
One advantage of performing the syntheses of pure samples of DAG-lactones with α-arylidene moieties was the isolation of small amounts of the minor Z-isomers in cases where they were formed. When that was possible, the distinct chemical shifts for the vinyl protons in the 1H NMR spectra confirmed the identity of both isomers. In the case of the Z-isomers of L1-H5 and L1-H2, the compounds displayed characteristic multiplets at δ 6.93–7.04, while the corresponding signal for the E-isomers appeared more downfield at δ 7.58–7.59. The relative affinities of the L1-H5 geometric isomers for PKCα and RasGRP3 revealed that the Z-isomer was a poor ligand for both enzymes. The Ki value for PKCα was 1076 ± 89 nM, which corresponds to a 8-fold drop in binding affinity relative to the E-isomer (see Table 1), while the affinity for RasGRP3 decreased even more; 21-fold from 3.8 ± 0.1 nM (Table 2) to 79 ± 17 nM. The situation for the Z-isomer of L1-H2 was similar, displaying a 24-fold drop in binding affinity for PKCα (Ki = 73.5 ± 7.2 nM, Table 1) relative to the E-isomer (Ki = 3.10 ± 0.40 nM, Table 1). This preference for the E-isomer is exactly opposite to what we observed with α-alkylidene moieties where the Z-isomers generally showed greater binding affinities.28
Library 1, Activation of α-secretase
It is known that activation of α-secretase upon PKC stimulation by various ligands causes increased degradation of the amyloid precursor protein (APP), resulting in enhanced secretion of sAPPα and reduced deposition of β-amyloid peptide (Aβ). Formation of this β-amyloid peptide is a key factor implicated in the pathogenesis of Alzheimer’s disease.36, 37 The release of sAPPα and a second fragment (C83) which is further processed to the N-terminally truncated Aβ variant (p3), are not involved in the pathogenesis of the disease and instead show neuroprotective effects.
One of our previously synthesized, branched DAG-lactones (8, Figure 3) was earlier found to efficiently increase α-secretase activity in a dose-dependent manner.38 Hence, the compounds in Library 1 offered the opportunity to optimize the scaffold of this DAG-lactone with more drug-like substituents with reduced lipophilicities. α-Secretase activity was measured in W4 cells (a human APP695 transfected rat neuroblastoma cell line).39, 40 The amount of secreted sAPPα, the hydrolysis product of APP upon cleavage by α-secretase, was measured by gel electrophoresis and immunoblot analysis with monoclonal antibody 6E10 that recognizes the N-terminus of the Aβ peptide.41 The augmented amount of sAPPα reflects an increase in α-secretase activity. The intensity of the sAPPα band in the experimental groups was analyzed by densitometry and compared to that of untreated cells, which were normalized to 100%. A preliminary screening of Library 1 revealed 14 active compounds (L1-A9, L1-A10, L1-B2, L1-B10, L1-B11, L1-C2, L1-D2, L1-D10, L1-D11, L1-D12, L1-E2, L1-E6, L1-F2 and L1-F6) that increased basal enzymatic activity to a level equal to or above that induced by control DAG-lactone (8). These compounds were then assayed in triplicate (Figure 3). As compounds L1-A10 and L1-C2 were chemically more similar to the parent DAG-lactone 8, compounds belonging to rows E and F, particularly L1-E6 and L1-F6, were of special interest despite their weak PKCα binding affinity (Table 1) because of their polar “chemical zip codes”. As a crude sample, L1-E6 equaled the activity of control DAG-lactone 8 (177%) relative to untreated cells (100%). Under more rigorous conditions, pure samples of L1-E6 and L1-F6 increased enzymatic activity to 154% and 162% above untreated cells (data not shown). Such potent activities were achieved despite a >2 orders of magnitude reduction in log P values and weaker PKCα binding affinities compared to DAG-lactone 8.28 These compounds, as well as the compounds previously identified as selective ligands for RasGRP3, demonstrate that the DAG-lactone scaffold accepts a variety of novel substitution patterns and that good PKCα binding affinity is not the exclusive predictor of α-secretase activity.
Figure 3.
Densitometric analysis of Western blots of secreted sAPPα over basal activity (100%) for the 14 active compounds (average of 3 experiments). All compounds were assayed at 1 μM and the control was the cell culture medium.
Library 2, PKCα and isozyme selectivity
The groups chosen for Library 2 also cover a range of log P values between 2.36 and 7.12, similar to Library 1. For this library, the acid chlorides in the R′ rows were limited to non-polar alkyl groups (A, B, C, F, G, H) and aryl-alkyl groups (D and E), all of which were expected to interact effectively with membranes. For the aldehyde columns (R), we expanded the important trifluoromethylphenyl group discovered in Library 1 to include positional isomers as well as multiple CF3 substitutions (columns 1–6). The rest of the groups included mostly common halogens such as Cl, Br and F in various substitution patterns. The affinity for the most potent ligand from Library 1, compound L1-G2, which in Library 2 corresponded to compound L2-C4, gave an almost identical Ki value (15.5 nM versus 13.1 nM), reflecting the reproducibility of our assay. Because we wanted to limit the number of isomers to just the racemic DAG-lactone, plus the inevitable mixture of E- and Z- isomers, we selected the similar compound L2-B4 (Ki =16.6 nM) with a symmetric acyl moiety for further synthesis. Although intuitively this selection made sense, we found later that even these two very similar branched acyl chains could be discriminated by the cell in some specific bioassays (see next section). The selected compound, L2-B4, and all the members of column 4, had good binding affinities and potencies that varied by less than 5-fold. The o-isomers (column 1) were less effective, while the m-isomers (column 2) were very similar to the p-isomers (column 4). The presence of an extra trifluoromethyl group on the aromatic ring does not seem to improve binding affinity despite an increase in log P (column 3). In terms of PKCα binding affinity, the most potent compounds from this library are clustered in the upper right-hand corner in rows B and C. Again, for the same reason stated above, the symmetric acyl chain of row B was preferred for further synthesis.
Because the compounds in Library 2 are more homogeneous in terms of their physicochemical properties, the quality of the library was better (see Analysis of Chemical Libraries in the Experimental Section), and this was reflected in the improved match of the Ki values between crude and pure samples (Table 3). For these compounds, the differences in Ki crude/Ki pure ratios averaged 3.6. As with Library 1, we were also able to obtain significant amounts of Z-isomers in two cases (L2-C11 and L2-F11) and, surprisingly, in the case of L2-A1 only the Z-isomer was obtained. As before, the 1H NMR spectra confirmed the chemical structures, and, in the case of the E-isomer of L2-F11, a crystal structure further corroborated the assignment (Figure 4). Again, as opposed to the DAG-lactones with an α-alkylidene side chain where the Z-isomer outperforms the E-isomer, α-arylidene DAG-lactones showed that binding affinity increases in favor of the more abundant E-isomer. Indeed, for compounds L2-C11 and L2-F11, the differences in potency favoring the E-isomer were 2.1 and 4.6-fold, respectively (Table 3).
Table 3.
Comparison of Ki values for PKCα between crude library samples and pure compounds in Library 2.
| Row # | Compound | Library Sample Ki (nM) | Pure Sample Ki (nM) | Ratio†Ki crude/Ki pure |
|---|---|---|---|---|
| 1 | L2-A1 | 157.6 ± 7.3 | 310± 38* | 0.51 |
| 2 | L2-B4 | 16.6 ± 0.4 | 2.96 ± 0.34 | 5.6 |
| 3 | L2-B7 | 17.1 ± 1.4 | 5.53 ± 0.59 | 3.0 |
| 4 | L2-B10 | 9.3 ± 1.2 | 2.45 ± 0.65 | 3.8 |
| 5 | L2-B11 | 21.5 ± 1.9 | 3.38 ± 0.15 | 6.3 |
| 6 | L2-B12 | 8.1 ± 1.2 | 2.66 ± 0.44 | 3.0 |
| 7 | L2-C8 | 12.7 ± 0.3 | 2.90 ± 0.15 | 4.3 |
| 8 | L2-C11 | 12.4 ± 0.2 | 4.89 ± 0.69 | 2.5 |
| 9 | L2-C11 | 12.4 ± 0.2 | 10.1 ± 1.3* | 1.2 |
| 10 | L2-C12 | 6.1 ± 0.5 | 2.07 ± 0.9 | 2.9 |
| 11 | L2-D6 | 108± 12 | 18.3 ± 3.8 | 5.8 |
| 12 | L2-F10 | 20.5 ± 1.2 | 10.9 ± 0.4 | 1.8 |
| 13 | L2-F11 | 28.8 ± 1.8 | 7.9 ± 3.8 | 3.6 |
| 14 | L2-F11 | 28.8 ± 1.8 | 36.73 ± 0.71* | 0.77 |
= Z-isomer;
= ratio calculated relative to the unresolved library mixture of E- and Z-isomers,
Figure 4.

Crystal structure of L2-F11 represented as one enantiomer (displacement ellipsoid plot drawn at the 50% probability level).
The discovery of the highly potent PKCα ligand, DAG-lactone L2-C12 (Ki = 2 nM) prompted the study of this compound in more detail, particularly in terms of cellular translocation of various PKC isozymes. Translocation of PKCs from cytosol to membrane compartments provides one measure of their activation. This process can be detected in real time by over-expressing different green fluorescent protein-tagged (GFP-tagged) PKC isoforms in Chinese hamster ovary (CHO) cells and following their localization before and after ligand treatment using confocal microscopy. All of the isozymes tested translocated very quickly at low doses of L2-C12 (300 nM) and there was no evidence of selectivity between the conventional isozymes α and β, and the novel calcium-independent isozymes δ and ε (Figure 5). The behavior of this ligand is akin to that of the potent phorbol esters, which are unable to distinguish between PKC isoforms.20, 21
Figure 5.
Translocation of PKCα, β, δ and ε in living CHO cells by L2-C12 (300 nM). Results for PKCα and δ are representative of 3 independent experiments; those for PKCβ and ε are representative of 2 independent experiments.
Library 2, PKCα activation and cellular motility
Some of the critical issues in developing PKC agonists, which are evident with the use of the prototypic activator 12-O-tetradecanoyl-phorbol-13-O-acetate (TPA), are the appearance of phenotypic changes associated with malignancy, including an increase in metastatic behavior. In light of observations that correlate over-expression and activation of PKCα with metastatic behavior, we screened Library 2 in a cell motility assay using MCF-10A cells.42 These cells are immortalized, non-transformed, and non-tumorigenic human breast epithelial cells that show enhanced cell motility after treatment with TPA. Such enhanced motility is believed to be the consequence of PKCα activation. To examine the effects of the DAG-lactones in this system, we screened the entire library using a semi-high throughput method for quantifying rates of cell movement. The assay measures the movement of cells that have been plated in a concentrated circular area on a slide, enabled by a manifold that accommodates 10 cell samples. After cell attachment, the manifold is removed and time-resolved radial movement of the cells can be captured via photomicroscopy.43 From the image data, the extent of movement can be calculated by the time-dependent increase in area or radius after a 20 h exposure. The concentration of each member of the library was adjusted to 10 μM and the same DAG-lactone (8) used in the α-secretase assay was employed as a positive control, while the negative control was the vehicle DMSO. When the results were analyzed, there was clear discordance between PKCα binding affinity and cell motility (Figure 6).
Figure 6.

Plot of radius versus log (1/Ki) for Library 2. Ki values were taken from Figure 1. Each motility assay was done in triplicate at 10 μM DAG-lactone and compared with a positive control (compound 8) and a negative control (1% DMSO v/v). Each measurement was done after 20 h of exposure to achieve the greatest radius traveled by the cells (in microns). The y-axis is the ratio of the radius induced by the sample to that radius produced by the positive control. For highly motile cells, including those receiving the positive control, the radius was ≥ 4 microns and the error was typically within 20%. Red circles indicate selected compounds for further studies.
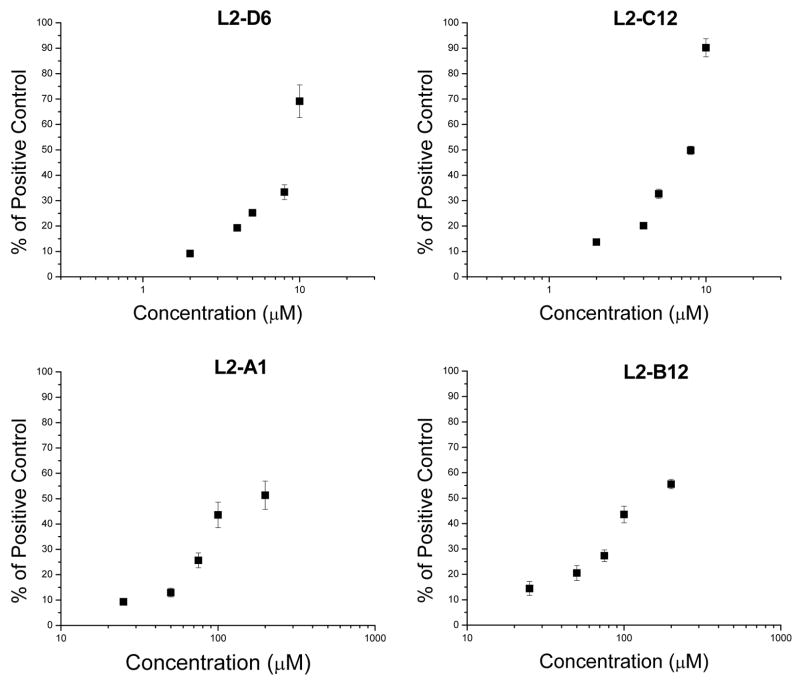
Because of the lack of correlation that showed either very potent PKCα activators (large log 1/Ki) with little or no effect on cell motility, as well as weak PKCα activators with significant effects on cell motility, we selected four compounds at each extreme of the figure for testing as pure, single agents (L2-A1, L2-B12, L2-C12 and L2-D6). A dose response experiment where the total area rather than the radius measurement was used gave much better statistics without qualitatively changing the results (Figure 7).
Figure 7.
Migration results as % of positive control and DAG-lactone concentration. Each DAG-lactone concentration was tested in triplicate and the increase in area occupied by the cells was measured in cm2. The error of these measurements was within 10%. The results are representative of two or more independent experiments.
The results for the pure compounds were consistent with the raw data obtained from the crude library and confirmed that while compounds L2-B12 and L2-C12 are potent PKCα ligands (Table 3), only L2-C12, which possesses an unsymmetrical acyl chain, induced cell motility. On the other hand, L2-D6, which behaves as a weaker PKCα ligand (Table 3) mimics L2-C12 in terms of inducing cell motility. Compound L2-A1 represents the case where both PKCα binding affinity and induction of cell motility are low; interestingly, this was the only compound for which the pure isolated product was obtained exclusively as the Z-isomer.
These striking results for DAG-lactones L2-B12 and L2-C12 also argue in favor of mechanisms through which very minor structural changes in the “chemical zip codes” can be decoded differently in the cellular milieu despite their similar potencies in binding to PKCα in vitro. This finding appears to be consistent with other contrasting cellular responses observed previously with DAG-lactones bearing slightly different branched patterns, which probably have an impact on lipid organization in membranes.44 Analysis of the dose-response curves on cell motility (Figure 7) also supports this concept as the corresponding pairs of active and less potent DAG-lactones engendered similar curves in terms of cell motility. The similar sharp rise in the curves for L2-C12 and L2-D6 at 10 μM is very clear and suggests perhaps the existence of off-target effects beyond this concentration.
Library 2, AP-1 activation
The transcription factor AP-1 is a heterodimer composed of a Jun and a Fos family member,45 and the association between the Jun and Fos proteins is required for binding to DNA. AP-1 binds to DNA sequences (TPA response elements) in the promoter region of many genes, which are involved in regulating cell proliferation and oncogenesis. Several signaling pathways and factors, including PKC, may regulate AP-1-mediated gene expression as the activation of PKC results in an increase in nuclear AP-1 DNA binding activity, as well as enhanced AP-1 dependent transcription. Expectedly, TPA induces AP-1 activation but at the same time it is a transforming agent that induces tumor formation.46 Thus, our library strategy provided an opportunity to search for DAG-lactones that could activate AP-1 without the tumor-promoting activity characteristic of TPA.
The Balb/C JB6 model is a well characterized model of genetic variants for a neoplastic transformation response to tumor promoters and is suited for studying tumor promotion and promotion-relevant molecular events.47 AP-1 activity measured in JB6 cells transfected with the AP-1-driven luciferase reporter gene is strongly induced by TPA, a response that can be used as a measure of AP-1 dependent transcription.48 Concomitantly, a transformation assay that measures anchorage independent colony induction can give an estimation of the tumor promoting activity of TPA.48 After a preliminary screening of the entire Library 2, several compounds were identified as capable of inducing AP-1 activation without transforming JB6 cells (Figure 8). Responses in each experiment were normalized to those of TPA. In these assays, JB6 cells transfected with an AP-1 luciferase reporter48 were treated with DMSO, TPA (10 ng/mL) or pure synthesized DAG-lactone (100 ng/mL) and AP-1 dependent luciferase activity was determined 18 h later. For the transformation assays in soft agar, 104 cells were suspended in top agar containing DMSO, TPA (10 ng/mL) or DAG-lactone (100 ng/mL) and layered over bottom agar also containing the compound of interest. Transformed colonies that grow anchorage independently were counted 14 days later using automated image analysis.48
Figure 8.

DAG-lactones that retain AP-1 activation but lack potential for inducing transformation. Values were normalized to TPA induced activity. For soft agar transformation an average of 4 individual assays is shown. For AP-1 induced luciferase, an average of 6 individual assays is shown. Methods employed for TPA induced transformation and AP-1 luciferase activities have been previously described.48
It is interesting that compounds L2-B4 and L2-B7, with similar Ki values for PKCα (Table 3), acted correspondingly in the AP-1 and anchorage independent transformation assays showing very strong AP-1 transcription factor activation and minimal transforming properties in dramatic contrast to TPA. The other three compounds, L2-C8 and L2-C11 (E- and Z-isomers), showed that the addition of fluorine ortho to the chlorine is a critical factor in differentiating AP-1 activation from oncogenic transformation. Compound L2-C8, although less potent than TPA, showed significant transforming activity, while both geometric isomers of L2-C11 were comparable to L2-B4 and L2-B7. The apparent equal potency of both geometric isomers of L2-C11 in this assay contrasts with their 2-fold difference in PKCα binding affinities (Table 3). The conclusion from these results is that some DAG-lactones are indeed capable of inducing AP-1 luciferase activity to similar levels as TPA, but without its transforming activity. Although a detailed interpretation could be complicated by the rather different conditions of the two assays, it is clear that rather subtle chemical changes in the nature of the “chemical zip codes” have marked effects on outcome.
Library 2, Apoptosis and radiosensitization
As discussed in the previous section, TPA-mediated activation of PKC has been shown to be associated with cellular transformation and proliferation. TPA-induced PKC activation protects many cell types from apoptosis induced by TNFa, 49, chemotherapy,50 growth factor withdrawal,51 and radiation.52 However, in other cells, it has also been shown that TPA can induce apoptosis via PKC activation. Because the subcellular distribution or translocation of PKC isozymes to various membranes of cellular organelles is a critical determinant, we evaluated as apoptosis inducers the two non-transforming DAG-lactones (L2-B4 and L2-B7) that were obtained as single geometrical isomers and which also behaved as potent AP-1 activators. Garcia-Bermejo et al. reported that both PKCα and PKCδ mediate TPA-induced apoptosis in LNCaP cells and showed that DAG-lactone 8 (Figure 4) was selective in inducing only PKCα-mediated apoptosis in these cells.53 More recently, Haimovitz-Friedman et al. have shown that treatment with TPA and the same DAG-lactone (8) leads to increased ceramide generation via ceramide synthetase (CS).54 The proposed pathway is that PKCα activation leads to down-regulation of ATM (ataxia telengiectasia mutated gene), a protein that functions to constrain CS activation.55 A key finding of that work was that PKCδ, an isoform known to be involved in TPA-induced apoptosis in LNCaP cells, had no effect on ATM levels.54 Because reduced ATM levels enhance radiation-induced CS activation, it was not surprising that PKCα activation alone could sensitize these cells to ionizing radiation. Indeed, while prostate-tumor (LNCaP) bearing nude mice treated with DAG-lactone 8 significantly decreased ATM levels and delayed tumor growth, the combination of radiation and DAG-lactone treatment greatly potentiated tumor growth delay.54
An initial comparison between DAG-lactones L2-B4 and L2-B7 in terms of their capacity to induce apoptosis in LNCaP cells at two different doses (10 and 20 μM) 24 h post radiation (20 Gy) showed induction of apoptosis by both compounds, with the more potent PKCα activator of the two (L2-B4, Table 3) being more effective (Figure 9).
Figure 9.
Effect of DAG-lactones L2-B4 and L2-B7 on PKCα-mediated apoptosis. LNCaP cells were pretreated for 16 h with 10 μM of DAG-lactones L2-B4 and L2-B7 followed by 20 Gy irradiation. Cells were fixed 24 h post radiation, stained with bis-benzimide and scored for apoptosis. The results are the averages of two separate experiments with standard deviation shown.
To explore the potential of L2-B4 for tumor radiosensitization, the orthotopic LNCaP model was employed using Swiss nude mice. LNCaP tumors reaching a size of ~100 mm3 were injected i.p. with L2-B4 (12 mg/kg). Two injections were delivered at 24 h intervals with prostate resection at 48 h after the first injection. ATM levels in tumor extracts were significantly decreased following L2-B4 treatment, dropping to a level between 7% and 3% of vehicle control. These results compare favorably with TPA treatment that reduced ATM levels to 12–27% of baseline (data not shown).
We next tested whether the reduction in ATM correlated with radiation response using a fractionated radiation scheme employing 1Gy delivered daily 5 times per week (Monday-Friday) to a total dose of 10 Gy. Mice received daily i.p. injections of L2-B4 (12 mg/kg) 16 h prior to each radiation treatment during the first week, whereas during the second week only two additional injections were delivered at a 48 h interval. Tumor response was assessed indirectly by measuring serum PSA levels, shown to correlate with tumor weight.56 PSA doubling time in our orthotopically-implanted tumors was approximately 10 days (data not shown). Figure 10 shows that fractionated radiotherapy alone resulted in an initial tumor growth response. However by week 4, PSA started to rise and by week 8 when mice were sacrificed the tumor had increased to 1 cm3. In contrast, the combination of L2-B4 and fractionated radiation completely inhibited tumor growth for the duration of the experiment, and no elevation of serum PSA was detected during this period. These data showed that L2-B4 significantly enhances the LNCaP tumor response to radiotherapy, indicating that it might serve as a potent radiosensitizer in vivo.
Figure 10.
Effect of DAG-lactone L2-B4 (12 mg/Kg) in the orthotopic prostate cancer model. Control non-irradiated mice (n = 14, red); L2-B4 treated mice (n = 13, blue); radiation only (XRT) treated mice (n = 14, black); combination L2-B4 plus radiation treated mice (n = 12, green). Values represent the mean.
Library 2, Immunostimulatory activity
PKC activators can not only induce apoptosis but also regulate the host immune response. One measure of the effects of these compounds on their ability to modulate the host immune response is their effect on the production of a key immunoregulatory cytokine, interferon-gamma (IFN-γ), produced by T cells and NK cells.57, 58 Thus, a human cell line, NK92, was utilized to analyze if the different DAG-lactone derivatives were able to alter IFN-γ gene expression. As it has been previously shown that IL-12 synergizes strongly with TPA to induce IFN-γ,59 we tested the induction of IFN-γ by the DAG-lactone library members both alone and in combination with IL-12. As shown in Figure 11, some selected compounds induced IFN-γ independently and also strongly synergized with IL-12 to induce IFN-γ. Thus, these compounds may also have an important immunostimulatory activity in addition to their pro-apoptotic activity.
Figure 11.
NK92 IFN-γ production in response to TPA and selected DAG-lactone derivatives plus IL12. The human NK cell line, NK92, (106 cells/mL) was incubated at 37 °C for 6 hours, in the presence or absence of the derivatives (1μg/mL) in 10% RPMI plus 10% FCS, glutamine and Pen/Strep. Duplicate wells were set up with derivatives + IL12 (100u/106 cells). Cell supernatants were analyzed for IFN-γ by Elisa (R & D Systems, Minneapolis, MN). Data are representative of multiple experiments. Standard deviation within the Elisa ranged from 0.001–0.49.
Discussion
The preliminary data obtained with only two 96-member DAG-lactone libraries, which are characterized by a modest array of “chemical zip codes”, demonstrate that the compounds obtained are indeed a source of exciting biological activities. A primary focus of our work has been to understand the basic mechanisms by which 1,2-diacylglycerol (DAG) and the newly developed DAG-lactones activate specific downstream subsets of cellular responses which occur through the activation of PKC isozymes, RasGrRP, and the other families of C1 domain target proteins. The multiplicity of cellular responses, often antagonistic, which result from PKC activation, or DAG binding to other C1 domain target proteins, can be explained by two still inadequately understood events: (1) the diversity of substrates for PKC and C1 domain proteins which have different roles in downstream events, and (2) the localization of these ligand-activated complexes into specific cellular compartments, which aside from the plasma membrane include the endoplasmic reticulum, the Golgi and the nucleus, thus allowing access to unique substrates. Therefore, gaining knowledge about how the regulation of PKC isozymes and the non-kinase protein targets can be controlled by the specific binding of DAG-lactones to C1 domains, and finding which structural elements influence the translocation of the ligand-protein complexes to specific cellular compartments represent important goals of our work. The ultimate objective is to design DAG-lactones that could selectively direct the flow of information from the DAG signaling pathways through their multiple signal transducers – the PKC isoforms and other signaling proteins that along with the PKCs possess DAG-responsive C1 domains – in order to achieve a specific cellular response with a potential therapeutic value.
The disadvantages of the weak affinity of endogenous DAGs and the complicated structures and restricted supply of exogenous, natural product ligands for the C1 domains – the diterpenes (phorbol, ingenol, and daphnane esters), the macrocyclic lactones (bryostatins), the indole alkaloids (teleocidin, lynbyatoxin), and the polyacetates (aplysiatoxin) – prompted us to devise a strategy to enhance the affinity of the endogenous ligand through appropriate constraint of the DAG by means of an optimized DAG-lactone. Because of the rudimentary nature of current structure-activity analyses that incorporate the lipid bilayer into the model, we decided to explore this binding site combinatorially by generating DAG-lactone libraries where arrangements of R and R′ groups produced a series of “chemical zip codes” capable of creating a different PKC-lipid microenvironment that would direct the activated complex to different sub-cellular sites.
Our preliminary results provide a strong proof-of-principle for this concept which could form the basis for therapeutic strategies targeting specific pathways. One key difference between a conventional combinatorial library approach and our targeted DAG-lactone libraries is that every member of the library was expected to bind, to a certain degree, to C1 domain containing proteins. This was confirmed by the Ki values obtained for the PKCα isozyme with Libraries 1 and 2. On the other hand, in the more specific bioassays, the cellular activities that were measured reflected the manner in which the activated complex “C1-domain–DAG-lactone–membrane” was able to induce a unique biological response according to the nature of the “chemical zip codes”.
The design of Library 2 evolved from the initial results obtained with Library 1. The more polar DAG-lactones in Library 1 showed good activity and selectivity for RasGRP and α-secretase, despite having low PKCα binding affinity. Then, from Library 1 we selected the most potent PKCα binding ligand (L1-G2) and explored its chemical space with less polar groups to generate Library 2. Therefore, the selection of bioassays for each of the libraries was different and depended on the intrinsic affinities of the compounds for PKCα. The Ki values that were obtained for PKCα in both libraries simply established a benchmark against a known C1 domain containing protein in our standard assay. Aside from the general observation that non-polar groups correlate better with activity when present as R′ acyl groups (sn-1) and polar groups are better when present as R groups at sn-2, as exemplified by compounds L1-B8 and L1-H2, we found it difficult to establish a clear structure-activity relationship for this enzyme. Some differences were also noted for the same chemical functionality depending on its location on the molecule. For example, when the trifluoromethylphenyl group was present as an sn-2, α-arylidene R group (Library 2, column 4), it correlated consistently with more potent ligands than when the same functional group appeared as an sn-1, R′ group (Library 1, column 7). An interesting observation that was gathered from all of the various biological and cellular assays was the lack of clear correlation between potency as a PKCα ligand and activity in other biological assays, bearing in mind that all the compounds were designed to have activity on PKCα. This is entirely consistent with the expectation that other isozymes, as well as other C1 domain containing targets, are likely involved in directing specific biological responses. This fact was exemplified for compound L2-C12, which was identified as the most potent ligand for PKCα in both libraries. L2-C12 failed to show any specificity in the cellular translocation of PKC isozymes α, β, δ and ε (Figure 5), and while the compound was able to stimulate cell migration effectively as expected for a PKCα activator, this activity was matched by a much weaker PKCα ligand, L2-D6 (Figures 6 and 7). Another dramatic result was the specific and potent activity associated with DAG-lactones bearing polar substituents simultaneously at sn-1 and sn-2 positions for non-PKC targets like RasGRP, and the effect of some of these compounds as inducers of α-secretase activity. This clearly implies that membrane associations with these ligand–C1 domain complexes can occur by different mechanisms, most likely involving Coulombic interactions with phospholipid headgroups.
The triad of compounds L2-B4, L2-B7, L2-C8 represents another interesting example of the lack of a correlation between PKCα binding affinity and cellular activity. These compounds have Ki values of 2.96, 5.53 and 2.90 nM, respectively, as re-synthesized pure samples (Table 3). All were able to induce AP-1 activation, but only L2-C8 with a Ki value identical to L2-B4 was able to induce cellular transformation. The L2-C11 isomers (both E and Z), which had up to a 3-fold difference in binding affinity for PKCα relative to L2-B4, matched the activity of L2-B4 in AP-1 activation, as well as in their lack of induction of cellular transformation (Figure 8). The small variation in chemical structure from L2-C8 to L2-C11 involved the simple addition of a fluorine atom. Surprisingly, this change was sufficient to abrogate the effect of L2-C8 on cellular transformation. Another important minor structural change associated with a dramatic difference in biological behavior is represented by compounds L2-B12 and L2-C12, which differ only in the level of branching of the R acyl chain. The DAG-lactone L2-B12 with a symmetrical branched chain had a very weak effect on cell motility, while L2-C12 with the asymmetric branched chain was very effective despite both compounds having nearly identical Ki values of ca. 2 nM as PKCα ligands (Table 3). This is possibly due to changes in membrane organization induced by both compounds which affect in a different fashion membrane curvature and other parameters associated with cell motility. This finding is in agreement with studies on DAGs of different acyl chain lengths and degrees of unsaturation that are capable of differentially perturbing membranes.5
Finally, the radiosensitization properties of PKC ligands associated with transforming activity, as in the case of the phorbol ester TPA, were dissociated in the non-transforming DAG-lactones L2-B4 and L2-B7. These two compounds were strong apoptosis inducers whose activities were augmented by ionizing radiation as demonstrated in vivo in an orthotopic prostate cancer model that measured PSA levels associated with tumor growth. The more potent compound, L2-B4, is an interesting drug candidate and represents a prototypic DAG-lactone that has specific targeting properties without the side effects that are typically associated with the phorbol esters. This compound and others described in this article provide a strong proof-of-principle for the concept of “chemical zip codes” and strongly support the continued search for additional sets of DAG-lactones with more specific targeting properties.
Experimental Section
General Techniques
All reagents and solvents purchased were of the highest commercial quality and used without further purification unless otherwise stated. MicroKan™ (IRORI) reactors were purchased from Discovery Partners International, San Diego, CA (Now Nexus Biosystems, Poway, CA). DHP (3,4-Dihydro-2H-pyran) resin (1.7 mmol/g, 150–300 μm) was purchased from Polymer Laboratories, Ltd., Amherst, MA. All solid-phase reactions were performed in MicroKan™ reactors filled with 14.7 mg (0.025 mmol) of DHP resin per Kan. Final crude products were obtained after cleavage from the resin and evaporation of the solvent. Selected, individual samples of pure materials were synthesized through conventional solution-phase methods as described previously30, 44 and were fully characterized by FT-IR, 1H NMR, FAB-MS and elemental analysis or HRMS.
Analysis of inhibition of [3H]PDBU binding by non-radioactive ligands
Enzyme-ligand interactions were analyzed by competition with [3H]PDBU binding for the single isozyme PKCα essentially as described previously.32 The ID50 values were determined by fitting a theoretical sigmoidal competition curve to the binding data. The Ki was calculated from the ID50 values according to the relationship:
where L is the concentration of free [3H]PDBU at the ID50 and Kd is the dissociation constant for [3H]PDBU under the assay conditions. Values represent the mean ± SEM. All values represent a minimum of three independent experiments. For the purified compounds, complete dose response curves with 6–7 concentrations of ligand were performed. For the unpurified library compounds, a single ligand concentration (1 μM) was used. In either case, triplicate determinations were made at each ligand concentration in each experiment.
Molecular modeling
Homology models of the C1 domains of RasGRP1 and RasGRP3, the C1a and C1b domains of PKCα, and the C1a domain of PKCδ, shown in Figure 2, were built on the backbone coordinates of the crystal structure of the C1b domain of PKCδ.1 Side chains for the models were constructed using the program SCWRL,60 which uses a backbone-dependent rotamer library to place residues in their most likely conformation given the backbone phi-psi angles at that position. Homologous residues were left unchanged from their crystallographic positions. The models were refined with a small energy minimization, with phorbol-13-O-acetate left in position from the crystal structure to prevent the binding site loops from closing during the minimization. Harmonic positional restraints on the backbone atoms were gradually relaxed over the course of the minimization to eliminate steric clashes in the sidechains without inducing deformations in the backbone.
Activation of α-secretase activity
The protocol for the α-secretase activation assay was followed as previously described.38
Translocation of GFP-tagged PKC isoforms in CHO cells
Translocation of GFP-tagged PKCα, β, δ and ε was followed in Chinese Hamster Ovary (CHO) cells as described previously.61 The GFP-tagged PKC isoforms were constructed by subcloning the PKC isoforms into a modified pEGFP-N1 plasmid (Clontech, Palo Alto, CA) as described by Wang et al.62
Cellular motility assay
Cell culture serum, growth factors, and media were purchased from Invitrogen, Inc. (Carlsbad, CA). Mid-passage MCF-10A human breast epithelial cells were obtained from the Barbara Ann Karmanos Cancer Institute (Detroit, MI). MCF-10A cells were cultured as previously described.42
Motility of MCF-10A cells was analyzed by monitoring the extent of cell movement using a computer-assisted digital camera (Moticam 2000) attached to an inverted Nikon Diaphot microscope. The cells were applied to a 10-well slide through a 10-hole manifold (CSM, Inc., Phoenix, AZ) that restricts sedimentation of cells to a small, circumscribed area. Upon removal of the manifold (t = 0), the DAG-lactone was added to the cells to a final concentration of 10 μM. Cells receiving no reagent were treated with an equivalent volume of DMSO to a final concentration of 1% (v/v). Following addition of the DAG-lactone, cells moved radially over a 20-h period while incubated at 37 °C, 5% CO2. The extent of movement was determined by measuring either a change in radius (in microns) or a change in total area (in cm2) occupied by the cells using Motic Images Plus 2.0 software. Each reported value is the average of triplicate measurements for which the error was typically within 10%.
AP-1 luciferase assay
On day 1, 104 JB6 P+ cells were seeded. Cells were transfected with a 3 to 1 ratio of Fugene (Roche Applied Science) to 4x AP-1 luciferase reporter DNA (0.2 μg). On day 3, DMSO, TPA (10 ng/mL) or DAG-lactone (100 ng/mL) were added to cells and 18 h later cells were harvested and luciferase activity determined.48 Six independent luciferase assays are reported.
Anchorage independent transformation
104 cells were suspended in 0.33% top agar with DMSO, TPA 10 ng/mL or DAG-lactone at 100 ng/mL and were layered over 0.5% bottom agar also containing the compound of interest. After 14 days anchorage independent colonies were counted. Colonies with more than 8 cells were counted as transformed.48 Four independent plates were seeded and 6 independent assays were scored. The average number of colonies is reported with TPA used as the 100% value.
Apoptosis and radiosensitization
Cell culture products were obtained through Mediatech (800-Cellgro, VA), except for fetal calf serum, which was purchased from Gemini Bio-products (CA). The DNA-binding fluorochrome bis-benzimide trihydrochloride was obtained from Sigma-Aldrich (St. Louis, MO).
The human prostate cancer cell line LNCaP was obtained from ATCC. Cells were cultured at 37°C in a humidified 5% CO2 atmosphere in RPMI-1640 supplemented with 10% fetal calf serum, 2 mM L-glutamine, 100 U/mL penicillin and 100 mg/mL streptomycin, and 10 mM HEPES (pH 7.2). For experiments, cells were cultured in the same medium except 0.2% human albumin (HA) was substituted for FCS 24 h before the beginning of the experiment. LNCaP cells were treated with L2-B4 and L2-B7 (10 or 20 μM) for 16 h prior to irradiation.
Morphologic changes in nuclear chromatin of cells undergoing apoptosis were detected by staining with the DNA-binding fluorochrome bis-benzimide trihydrochloride, as described.63 Cells were visually inspected using an Olympus BH2 fluorescence microscope equipped with a Dich mirror cube filter (BH2-DMU2UV). Cells displaying at least three apoptotic bodies were scored as apoptotic.
Orthotopic prostate cancer model
Eight to ten week-old male Swiss nude (nu/nu) mice were implanted orthotopically with 3.0×106 LNCaP cells. Immediately before tumor implantation, cultured LNCaP cells were trypsinized and resuspended in RPMI 1640 with 10% FBS, and viability determined by trypan blue exclusion. Only single cell suspensions with >90% viability were used for in vivo injection. Mice were anesthetized with 12.5 mg Ketamine+1.25 mg Xylazine (i.p., intraperitoneal), and orthotopic tumor implantation was performed as described.64 Briefly, a low midline abdominal incision was made with a #15 blade (Bard Parker). The peritoneal cavity was entered by sharply incising the linea alba. The bladder and seminal vesicles were identified and gently raised, thus exposing the dorsal lobes of the mouse prostate. LNCaP cells were injected in 0.1 mL of medium using a 26 gauge needle. Proper implantation of cell suspension was indicated by blebbing under the prostatic capsule. Visceral contents were then replaced into the abdominal cavity, and the wound closed with surgical autoclips (Becton Dickinson). Mice were monitored during the postoperative period according to animal care facility guidelines. Injected mice were housed (5 mice/cage) in a pathogen-free environment, using filtered, laminar airflow hoods in standard vinyl cages with air filter tops. Cages, bedding, and water were autoclaved before use.
Orthotopically-transplanted LNCaP tumors in nude mice secrete PSA that can be detected histochemically in tumor cells and by radioimmunassay in mouse serum.56 To assess LNCaP tumor take and tumor volume after intra-prostatic transplantation, mice were anesthetized with 12.5 mg Ketamine + 1.25 mg Xylazine (i.p.). Phlebotomy was performed by accessing the retro-orbital venous plexus with a microcapillary pipette (Fisher). Serum PSA determinations were performed by radioimmunoassay (Hybritech) according to the recommendations of the manufacturer. Using serum PSA determinations as an indicator of tumor size, mice were only utilized in experiments when PSA levels ranged from 5.0–15.0 ng/mL representing tumors with a mass in the range of 70–150 mg. Mice were divided into four treatment groups: a) Control, DMSO i.p.; b) DAG-Lactone L2-B4 at 12 mg/kg/day in DMSO (MTD dose) i.p.; c) DMSO 16 h before10×1Gy; d) DAG-Lactone L2-B4 at 12 mg/kg/day 16 h before10×1Gy. Mice were locally irradiated by exposure to a small pelvic RT field using a specially designed Lucite jig. All procedures and post-operative care with animals were in accordance with NIH guidelines and received prior approval by the Institutional Animal Care and Use Committee at Memorial Sloan-Kettering Cancer Center.
Analysis of immunostimulatory activity
The human NK cell line NK92 (106 cells/mL) was incubated in 10% RPMI plus 10% FCS, glutamine and Pen/Strep at 37 °C for 6 h in the presence or absence of derivatives (1μg/mL) alone, or in the presence of derivatives plus IL12 (100 u/million cells). Cell supernatants were analyzed for IFN-γ expression by Elisa (R & D Systems, Minneapolis, MN).
Analysis of Chemical Libraries
For a typical DAG-lactone derivative, the FAB mass spectrum consisted of either MH+ or M•+ to designate the molecular weight plus several structure-indicating fragment ions derived from both the acyl and the alkylidene portions of the molecule.65 Use of a 3-nitrobenzyl alcohol (NBA) matrix was found to offer significant advantages in terms of speed, simplicity and information content for the analysis of these lipophilic derivatives. Furthermore, since these DAG-lactones are all structurally alike, their surface activity in the FAB matrix, and hence FAB ionization efficiency, were expected to be similar.66
The issue of library quality and individual library component purity was investigated further with pure, individually synthesized DAG-lactone standards. Thus we constructed standard curves over the range of linear mass spectral response using the absolute intensity of diagnostic ions (e.g., R′-C≡O+, MH+) from the analysis of measured amounts of individually synthesized standards and used these to measure the amount of DAG-lactone in similar amounts of crude combinatorial library product.67 As shown for Library 2 (Table 4), the average purity of the DAG-lactones was 38%.
Table 4.
Mass Spectrometrically Determined Combinatorial Purity (Library 2). Comparison of the semi-quantitative compound quality rating derived from an individual mass spectrum of the crude solid-phase combinatorial product and the quantitatively determined synthetic purity. Library Quality Rating: +++ - MH+ ≥1 V (spectrum similar to that of a pure or standard compound); ++ - MH+ ≥250 mV (spectrum contains diagnostic fragment ions and may indicate presence of minor amounts of synthetic byproducts); + - MH+ ≥25 mV (overall spectrum is weak and/or contains evidence of substantial by-product formation); 0 – (no mass spectral evidence for the desired compound). Library components are mixtures of E/Z isomers.
| Library Component | Library Quality Rating | Library Purity (%) | Measurements (n) |
|---|---|---|---|
| L2-A1 | +++ | 25 ± 1.2 | 3 |
| L2-B4 | +++ | 47 ± 1.2 | 3 |
| L2-B7 | +++ | 40 ± 1.8 | 3 |
| L2-B10 | +++ | 43 ± 1.3 | 4 |
| L2-B11 | +++ | 50 ± 2.0 | 4 |
| L2-B12 | ++ | 19 ± 3.4 | 4 |
| L2-C8 | +++ | 62 ± 2.1 | 3 |
| L2-C11 | ++ | 37 ± 2.3 | 3 |
| L2-D6 | +++ | 56 ± 1.3 | 3 |
| L2-F10 | + | 6 | 1 |
| L2-F11 | ++ | 35 | 1 |
Library 1
For Library 1, column 6 and row H were individually analyzed (Figure 1S in Supporting Information) to see whether the desired DAG-lactone products were present. The compounds in column 6 all have the same p-methoxyphenyl (PMP) acyl moiety but different alkylidene groups, so their mass spectra exhibited the same characteristic acylium ion (MeOC6H4-C≡O+, m/z 135) but different masses for MH+. The compounds of row H all have the same p-nitrophenyl alkylidene moiety but each has a different acyl group, so the masses observed for both the acylium ion and MH+ varied in concert. The initial analytical evaluation of Library 1 gave clear evidence for the presence of 94 out of the 96 desired DAG-lactone analogues. For the mixture analysis, mixtures were made using compounds of the same row so that the acylium ions representing the different acyl groups (R′) would not overlap. For example, the three, 4-component mixtures representing row A of Library 1 were composed of L1-A1, L1-A4, L1-A7 and L1-A10; L1-A2, L1-A5, L1-A8 and L1-A11; and L1-A3, L1-A6, L1-A9 and L1-A12. Those compounds that were not observed during mixture analysis were subsequently analyzed individually (e.g. L1-B11 and L1-B12). One compound, L1-H10, was questionable since its MH+ peak was present, but below the absolute intensity threshold required for a minimal rating. There was no mass spectral evidence for the presence of DAG-lactone L1-H8.
Library 2
A similar analysis strategy was followed for Library 2. The components of column 4 and row H were chosen for individual mass spectral analysis to assess the suitability of this library for the combine-and-analyze strategy (Figure 2S in Supporting Information). Thus, the anticipated acylium ion (R′-C≡O+) varied in concert with MH+ for each member of column 4, all of which have the same p-trifluoromethyl-phenyl alkylidene (R) moiety; while the expected acylium ion (C8H17-C≡O+, m/z 141) remained constant for all the compounds of row H, which possess the same acyl group. For the mixture analysis, mixtures were made using compounds of the same column so that the acylium ions representing the different acyl groups (R′) would not overlap. For example, for the analysis of column 4, two mixtures were generated consisting of components L2-A4, L2-C4, L2-E4 and L2-G4 (Mix 4A) and compounds L2-B4, L2-D4, L2-F4 and L2-H4 (Mix 4B), respectively.
X-ray Crystal Structure of L2-F11
Single-crystal X-ray diffraction data on compound L2-F11 were collected at 103 °K using MoKα radiation and a Bruker APEX 2 CCD area detector. A 0.35 × 0.31 × 0.04 mm3 crystal was prepared for data collection coating with high viscosity microscope oil (Paratone-N, Hampton Research). The oil-coated crystal was mounted on a MicroMesh mount (MiTeGen, Ithaca, NY) and transferred immediately to the cold stream (−170 °C) on the diffractometer. The crystal was monoclinic in space group P21/c with unit cell dimensions a = 13.986(7) b = 15.392(8) Å, c = 8.137(4) Å, and β = 103.17(1)°. Corrections were applied for Lorentz, polarization, and absorption effects using the program SADABS (Bruker, SADABS v2.10, 2000a, Bruker AXS Inc., Madison, WI). Data were 98.6% complete to 28.29° θ (approximately 0.75 Å) with an average redundancy of 3.03. The structure was solved by direct methods and refined by full-matrix least squares on F2 values using the programs found in the SHELXTL suite (Bruker, SHELXTL v6.14, 2000b, Bruker AXS Inc., Madison, WI). Parameters refined included atomic coordinates and anisotropic thermal parameters for all non-hydrogen atoms. Hydrogen atoms on carbons were included using a riding model [coordinate shifts of C applied to H atoms] with C-H distance set at 0.96 Å. The asymmetric unit contains a single molecule. Atomic coordinates for compound L2-F11 are included in the Supporting Information Available from this journal and have also been deposited with the Cambridge Crystallographic Data Centre (CCDC 680078). Copies of the data can be obtained, free of charge, on application to CCDC, 12 Union Road, Cambridge, CB2 1EZ, UK [fax: +44(0)-1223-336033 or e-mail: deposit@ccdc.cam.ac.uk.
Preparation of starting materials
5-(Hydroxymethyl)-5-[(4-methoxyphenoxy)methyl]-3,4,5-trihydrofuran-2-one (1b)
This compound was prepared by the treatment of 5-[(4-methoxyphenoxy)methyl]-5-[(phenylmethoxy)methyl]-3,4,5-trihydrofuran-2-one (1a)68 with boron trichloride as described.30
5-(Hydroxymethyl)-5-[(tert-butyldiphenylsilyloxy)methyl]-3,4,5-trihydrofuran-2-one (1d)
A stirred solution of 1b (4.0 g, 15.9 mmol, 1.0 equiv) in dichloromethane (160 mL) was treated with tert-butyldipenylsilyl (TBDPS) chloride (8.2 mL, 31.7 mmol, 2.0 equiv) in the presence of imidazole (2.2 g, 31.7 mmol, 2.0 equiv) at room temperature for 1.5 h. The reaction was quenched with water and the separated organic phase was washed once with water, dried (Na2SO4) and concentrated to dryness. The crude product (15.9 mmol, 1.0 equiv) was dissolved in acetonitrile:water (4:1, 125 mL) and treated with ammonium cerium (IV) nitrate (CAN, 26.2 g, 47.7 mmol, 3.0 equiv) at 0 °C. After stirring for 30 min at this temperature, the reaction was quenched with a 10% aqueous sodium thiosulfate solution. After most of the acetonitrile was removed under vacuum, the final solution was extracted with ethyl acetate (3 ×), dried (Na2SO4) and concentrated to dryness. The residue was purified by silica gel flash column chromatography with ethyl acetate:hexane (1:2) as eluant to afford a light yellow oil (5.24g, 86% after two steps) which solidified upon standing; mp 82–83 °C (hexane); IR (neat) 3451 (OH), 1774 (C=O) cm−1; 1H NMR (400 MHz, CDCl3) δ 7.61–7.64 (m, 4 H, ArH) 7.35–7.46 (m, 6 H, ArH), 3.72 (d, J = 12.3 Hz, 2H, HOCH2), 3.63 (ABq, J = 11.6 Hz, 2 H, SiOCH2), 2.52–2.72 (m, 2 H, H3a,b), 2.05–2.22 (m, 3 H, H4a,b and OH), 1.04 (s, 9 H, C(CH3)3); FAB-MS (m/z, relative intensity) 385 (MH+, 14), 135 (100); Anal (C13H16O5) C, H.
Library 1: “PMP approach”
Procedure for loading 5-(hydroxymethyl)-5-[(4-methoxyphenoxy)methyl]-3,4,5-trihydrofuran-2-one (1b) onto the DHP resin in 96 MicroKan™ reactors
Ninety six MicroKan™ reactors, each containing DHP resin (0.0147 g, 0.025 mmol) and a radio frequency tag capsule, were placed in a 500 mL round bottom flask equipped with a stir bar. DAG-lactone 1b (6.1 g, 24 mmol, 10 equiv), pyridinium p-toluenesulfonate (6.0 g, 24 mmol, 10 equiv) and 1,2-dichloroethane (240 mL) were introduced into the flask. The reactors were degassed at −30 °C in vacuo for 2 min. The temperature was then raised to 60 °C and the solution was vigorously stirred for 24 h. After cooling to room temperature, the solution was decanted into another flask to recover the excess of unreacted DAG-lactone (1b). The reactors containing polymer-bound lactone 2b were washed sequentially with dichloromethane (1), 1:1 DMF/water (3 ×), DMF (3 ×), methanol (2 ×), dichloromethane (3 ×), and finally dried in vacuo.
Procedure for the aldol reaction of polymer-bound lactone 2b with aldehydes
The 96 MicroKan™ reactors were sorted into 8 vessels to react with 8 different aldehydes. Each vessel contained 12 reactors. The 8 parallel reactions were performed following an identical, general procedure: MicroKan™ reactors containing polymer-bound lactone 2b (12 reactors, 0.3 mmol, 1 equiv) were suspended in THF (30 mL) in a Schlenk vessel equipped with a stir bar. After degassing at −78 °C in vacuo for 2 min, LDA (1.5 mL, 2.0 M in heptane/THF/ethylbenzene, 3.0 mmol, 10 equiv) was added while the temperature was maintained at −78 °C. After stirring 6 h at −78 °C, the solution was treated with ZnCl2 (7.2 mL, 0.5 M in THF, 3.6 mmol, 12 equiv) and the reaction was allowed to warm to −45 °C for 30 min. A solution of the corresponding aldehyde (3.6 mmol, 12.0 equiv) in THF (2 mL) was then added to each of the vessels and stirring continued at −45 °C for 3 h. The reactions were quenched with a saturated solution of ammonium chloride and allowed to reach room temperature. All MicroKan™ reactors containing polymer-bound aldol product 3b were pooled together and washed sequentially with 1:1 DMF/water (3 ×), DMF (3 ×), methanol (2 ×), dichloromethane (3 ×) and dried in vacuo.
Procedure for the triflation of polymer-bound aldol products 3b followed by DBU-assisted elimination
MicroKan™ reactors containing polymer-bound aldol product 3b (96 MicroKan™ reactors, 2.4 mmol, 1 equiv) were placed in a round bottom flask equipped with a stir bar and suspended in dichloromethane (240 mL). After degassing at −78 °C in vacuo for 2 min, pyridine (3.9 mL, 48 mmol, 20 equiv), 4-(dimethylamino)pyridine (4-DMAP, 0.29 g, 2.4 mmol, 1 equiv) and trifluoromethylsulfonic anhydride (4.0 mL, 24 mmol, 10 equiv) were subsequently introduced into the vessel at the same temperature. The resulting mixture was stirred at −20 °C for 20 h, after which time the reaction was quenched with a saturated solution of ammonium chloride and allowed to reach room temperature. The MicroKan™ reactors were washed sequentially with 1:1 DMF/water (3 ×), DMF (3 ×), methanol (2 ×), dichloromethane (3 ×), and dried in vacuo. All reactors were then suspended in toluene (240 mL), and after degassing at −78 °C in vacuo for 2 min, DBU (7.2 mL, 48 mmol, 20 equiv) was added at 0 °C. The MicroKan™ reactors were stirred at the same temperature for 6 h, after which time the reaction was quenched with a saturated solution of ammonium chloride. The MicroKan™ reactors containing polymer-bound α-alkylidene-lactone 4b were washed with 1:1 DMF/water (3 ×), DMF (3 ×), methanol (2 ×), dichloromethane (3 ×), and dried in vacuo.
Procedure for removal of the p-methoxyphenyl (PMP) group from polymer-bound α-alkyl/arylidene-lactones 4b
MicroKan™ reactors containing polymer-bound α-alkyl/arylidene-lactone 4b (96 MicroKan™ reactors, 2.4 mmol, 1.0 equiv) were suspended in THF (192 mL) in a round bottom flask equipped with a stir bar. After degassing at −78 °C in vacuo for 2 min, the solution was warmed to 0 °C. Water (48 mL) and ammonium cerium (IV) nitrate (CAN, 26.3 g, 48 mmol, 20 equiv) were subsequently introduced into the vessel and stirring continued at the same temperature for 6 h after which time the reaction was quenched with a 5% aqueous solution of sodium thiosulfate. Finally, the MicroKan™ reactors containing polymer-bound diol-α-alkyl/arylidene-lactone 5 were washed with a 5% aqueous solution of sodium thiosulfate (2 ×), 1:1 DMF/water (2 ×), DMF (3 ×), methanol (2 ×), dichloromethane (3 ×) and dried in vacuo.
Procedure for the acylation of polymer-bound diol lactones 5 with acid chlorides
The 96 MicroKan™ reactors were sorted into 12 vessels to react with 12 different acid chlorides. Each vessel contained 8 reactors. The 12 parallel reactions were performed following an identical, general procedure: MicroKan™ reactors containing polymer-bound diol-α-alkyl/arylidene-lactone 5 (8 reactors, 0.2 mmol, 1 equiv) were suspended in CH2Cl2 (20 mL) in a Schlenk vessel equipped with a stir bar. After degassing at −78 °C in vacuo for 2 min, triethylamine (0.56 mL, 4.0 mmol, 20 equiv), 4-DMAP (0.024 g, 0.2 mmol, 1 equiv) and the corresponding acid chloride (2.0 mmol, 10 equiv) were subsequently introduced into the vessel. The mixture was stirred at 0 °C for 6 h after which time the reactions were quenched with a saturated solution of ammonium chloride. All reactors were pooled together and washed with 1:1 DMF/water (3 ×), DMF (3 ×), methanol (2 ×), dichloromethane (3 ×) and dried in vacuo.
Procedure for TFA-assisted parallel cleavage
The 96 MicroKan™ reactors containing polymer-bound DAG-lactone 6 were sorted into 96 individual wells using the IRORI 96-cleavage station. To each well was added 1.5 mL of 1:10 TFA/dichloromethane. After shaking (15 min), the solution was filtered into 96 individual pre-weighed vials. An additional 1.5 mL of dichloromethane was introduced into each well for washing the MicroKan™ reactors. The combined organic solution in each vial was directly concentrated in vacuo and weighed
Library 1: “TBDPS approach”
With the exception of the loading step and the removal of the TBDPS group, all the other steps were performed in the same fashion as in Library 1 (PMP approach).
Procedure for loading 5-(hydroxymethyl)-5-[(tert-butyldiphenylsilyloxy)methyl]-3,4,5-trihydrofuran-2-one (1b) onto the DHP resin
One hundred MicroKan™ reactors, each containing DHP-resin (0.0147 g, 0.025 mmol) and a radiofrequency tag capsule, were placed in a 500 mL round bottom flask equipped with a stir bar. DAG-lactone 1d (4.8 g, 12.5 mmol, 5 equiv), pyridinium p-toluenesulfonate (2.5 g, 10 mmol, 4 equiv) and 1,2-dichloroethane (150 mL) were introduced into the flask. These reactors were degassed at −30 °C in vacuo for 2 min. The temperature was then raised to 60 °C and the solution was stirred for 24 h. After reaching the room temperature, the solution was decanted into another vessel to recover the unreacted lactone 1d. The reactors containing polymer-bound lactone 2d were washed sequentially with dichloromethane (1 ×), 1:1 DMF/water (3 ×), DMF (3 ×), methanol (2 ×), dichloromethane (3 ×), and finally dried in vacuo.
Procedure for removal of the t-butyldiphenylsilyl (TBDPS) group from polymer-bound α-alkyl/arylidene-lactones 4d
MicroKan™ reactors containing polymer-bound α-alkyl/arylidene-lactone 4d (96 MicroKan™ reactors, 2.4 mmol, 1 equiv) were suspended in THF (200 mL) in a round bottom flask equipped with a stir bar. After degassing at −78 °C in vacuo for 2 min, the solution was warmed to 0 °C and slowly treated with a solution of tetra-n-butylammonium fluoride (TBAF) in THF (1.0 M in THF, 48 mL, 20 equiv). Stirring continued at 0 °C for 6 h after which time the reaction was quenched with a saturated aqueous solution of ammonium chloride. Finally, the MicroKan™ reactors containing polymer-bound diol-α-alkyl/arylidene-lactone 5 were washed with 1:1 DMF/water (3 ×), DMF (3 ×), methanol (2 ×), dichloromethane (3 ×) and dried in vacuo.
Library 1: Hybrid “PMP” and “TBDPS” approach
Library components in Rows E and F were prepared through the “TBDPS” approach, whereas the others were synthesized through the “PMP” approach. The procedure was identical as described previously, except for the number of MicroKan™ reactors loaded with starting DAG-lactones 1b and 1d and the sorting of the polymer-bound reactors containing the different starting materials.
Procedure for sorting polymer-bound lactone 2b and 2d
MicroKan™ reactors containing polymer-bound lactone 2b were sorted into vessels 1, 2, 3, 4, 7, and 8, to respectively react with aldehydes A, B, C, D, G and H. MicroKan™ reactors containing polymer-bound lactone 2d were sorted into vessels 5 and 6 to react with aldehydes E and F, respectively. Each vessel contained 12 MicroKan™ reactors.
Procedure for scanning polymer-bound α-alkyl/arylidene-lactones 4b and 4d
The 96 MicroKan™ reactors were grouped into two vessels using the “decode tag” button in the UTILITIES menu. The 72 reactors containing polymer-bound α-alkyl/arylidene-lactone 4b, which were identified by aldehydes A, B, C, D, G and H, were placed into one vessel, whereas the 24 reactors containing polymer-bound α-alkyl/arylidene-lactone 4d, which were identified by aldehydes E and F, were placed into another vessel. The deprotection of the 72 MicroKan™ reactors containing α-alkyl/arylidene-lactones 4b (protected with the PMP group) and the 24 MicroKan™ reactors containing α-alkyl/arylidene-lactones 4d (protected with the TBDMS group) was separately performed as described previously.
Library 2: “PMP” approach
The loading of 5-(hydroxymethyl)-5-[(4-methoxyphenoxy)methyl]-3,4,5-trihydrofuran-2-one (1b) onto the DHP resin was performed as described for Library 1. The execution of this library was identical to the procedure previously described for Library 1, except that the matrix of this library consisted of 12 aldehydes and 8 acid chlorides.
(Z)-[4-Heptylidene-2-(hydroxymethyl)-5-oxo-2-2,3-dihydrofuryl]methyl 2,4,6-tris(methylethyl)benzoate (L1-A12, Library 1)
Oil; IR (neat) 3440 (OH), 2960 (CH), 2929 (CH), 2871 (CH), 1761 (C=O), 1734 (C=O), 1680 (C=C) cm−1; 1H NMR (400 MHz, CDCl3) δ 6.98 (s, 2 H, ArH), 6.72 (tt, J = 7.5, 2.8 Hz, 1 H, C=CH), 4.42 (AB q, J = 11.8 Hz, 2 H, CCH2OC), 3.71 (AB q, J = 12.1 Hz, 2 H, CCH2OH), 2.79 (m, 5 H, H3ab and C(O)C6H2(CH(CH3)2)3), 2.32 (v br s, 1 H, CCH2OH), 2.11 (m, 2 H, C=CHCH2CH2CH2CH2CH2CH3), 1.41 (m, 2 H, C=CHCH2CH2CH2CH2CH2CH3), 1.25 (m, 24 H, C=CHCH2CH2CH2CH2CH2CH3 and C(O)C6H2(CH(CH3)2)3), 0.85 (t, J = 6.9 Hz, 3 H, C=CHCH2CH2CH2CH2CH2CH3); FAB-MS (m/z, relative intensity) 473 (MH+, 20), 231 (100); Anal (C29H44O5) C,H.
(Z)- [4-(2-Ethylhexylidene)-2-(hydroxymethyl)-5-oxo-2-2,3-dihydrofuryl]methyl 4-(dimethylamino)benzoate (L1-B5, Library 1)
White solid, mp = 108–112 °C; IR (neat) 3417 (OH), 2962 (CH), 2924 (CH), 1728 (C=O), 1698 (C=O), 1609 (C=C) cm−1; 1H NMR (400 MHz, CDCl3) δ 7.85 and 7.84 [2 m (diastereomers), 2 H, ArH], 6.61 and 6.59 (2 m, 2 H, ArH), 6.56 and 6.53 [2 t, J = 2.8 Hz (diastereomers), 1 H, C=CH], 4.41 [2 AB q, J = 12.0 Hz (diastereomers), 2 H, CCH2OC], 3.71 (m, 2 H, CCH2OH), 3.02 (2 s, 6 H, N(CH3)2), 2.86 (app dt, J = 17.0 Hz, 1 H, H3a), 2.72 (app dt, J = 17.0 Hz, 1 H, H3b), 2.38 (m, 1 H, CCH2OH), 2.09 (m, 1 H, C=CHCH(CH2CH3)CH2CH2CH2CH3), 1.48 (m, 2 H, C=CHCH(CH2CH3)CH2CH2CH2CH3), 1.21 (m, 6 H, C=CHCH(CH2CH3)CH2CH2CH2CH3), 0.85 (m, 3 H, C=CHCH(CH2CH3)CH2CH2CH2CH3), 0.76 (m, 3 H, C=CHCH(CH2CH3)CH2CH2CH2CH3); FAB-MS (m/z, relative intensity) 404 (MH+, 49), 403 (M•+, 49), 148 (100); Anal (C23H33NO5) C, H, N.
(Z)-[4-(2-Ethylhexylidene)-2-(hydroxymethyl)-5-oxo-2-2,3-dihydrofuryl]methyl 4-nitrobenzoate (L1-B8, Library 1)
White solid, mp = 112–115 °C; IR (neat) 3392 (OH), 1716 (C=O) cm−1; 1H NMR (400 MHz, CDCl3) δ 8.26 (m, 2 H, ArH), 8.13 (m, 2 H, ArH), 6.59 and 6.56 [m (diastereomers), 1 H, C=CH], 4.52 [2 AB q, J = 11.9 Hz (diastereomers), 2 H, CCH2OC], 3.78 [2 AB q, J = 12.1 Hz (diastereomers), 2 H, CCH2OH], 2.90 [2 dd, J = 17.1, 3.0 Hz (diastereomers], 1 H, H3a), 2.74 [2 dd, J = 17.0, 2.7 Hz (diastereomers), 1 H, H3b], 2.20 (m, 1 H, CCH2OH), 2.08 (m, 1 H, C=CHCH(CH2CH3)CH2CH2CH2CH3), 1.47 (m, 2 H, C=CHCH(CH2CH3)CH2CH2CH2CH3), 1.27 (m, 4 H, C=CHCH(CH2CH3)CH2CH2CH2CH3), 1.10 (m, 2 H, C=CHCH(CH2CH3)CH2CH2CH2CH3), 0.84 (m, 3 H, C=CHCH(CH2CH3)CH2CH2CH2CH3), 0.72 (m, 3 H, C=CHCH(CH2CH3)CH2CH2CH2CH3); FAB-MS (m/z, relative intensity) 406 (MH+, 100); Anal (C21H27NO7) C, H, N.
(E)-(4-{[4-(Dimethylamino)phenyl]methylene}-2-(hydroxymethyl)-5-oxo-2-2,3-dihydrofuryl)methyl 2-ethylhexanoate (L1-E2, Library 1)
Light yellow solid, mp = 89–91 °C; IR (neat) 3371(OH), 2963 (CH), 2932 (CH), 2859 (CH), 1734 (C=O), 1712 (C=O), 1636 (C=C) cm−1; 1H NMR (400 MHz, CDCl3) δ 7.49 (t, J = 2.7, 1 H, C=CH), 7.41 (d, J = 8.9 Hz, 2 H, ArH), 6.85 (d, J = 8.2 Hz, 2 H, ArH), 4.28 [2 AB q, J = 11.9 Hz (diastereomers), 1 H, CCH2OC], 4.28 [2 AB q, J = 11.9 Hz (diastereomers), 1 H, CCH2OC], 3.71 (m, 2 H, CCH2OH), 3.14 (dd, J = 17.3, 2.8 Hz, 1 H, H3a), 3.04 (s, 6 H, C=CHC6H4N(CH3)2), 2.96 (dd, J = 17.3, 2.8 Hz, 1 H, H3b), 2.27 (m, 1 H, C(O)CH(CH2CH3)CH2CH2CH2CH3), 2.15 (v br s, 1 H, CCH2OH), 1.50 (m, 4 H, C(O)CH(CH2CH3)CH2CH2CH2CH3), 1.19 (m, 4 H, C(O)CH(CH2CH3)CH2CH2CH2CH3), 0.82 (m, 6 H, C(O)CH(CH2CH3)CH2CH2CH2CH3); FAB-MS (m/z, relative intensity) 404 (MH+, 74), 403 (M•+, 100); Anal (C23H33NO) C, H, N.
(E)-(4-{[4-(Dimethylamino)phenyl]methylene}-2-(hydroxymethyl)-5-oxo-2-2,3-dihydrofuryl)methyl 4-(dimethylamino)benzoate (L1-E5, Library 1)
Yellow solid, mp = 230–232 °C; IR (neat) 3386 (OH), 2898 (CH), 1713 (C=O), 1697 (C=O, 1606 (C=C) cm−1; 1H NMR (400 MHz, CD2Cl2) δ 7.82 (d, J = 9.1 Hz, 2 H, ArH), 7.45 (m, 3 H, ArH and C=CH), 6.76 (d, J = 8.9 Hz, 2 H, ArH), 6.63 (d, J = 9.1 Hz, 2 H, ArH), 4.42 (AB q, J = 11.8 Hz, 2 H, CCH2OC), 3.77 (irregular AB q, 2 H, CCH2OH), 3.19 (dd, J = 17.3, 2.8 Hz, 1 H, H3a), 3.09 (dd, J = 17.3, 2.6 Hz, 1 H, H3b), 3.03 (s, 12 H, N(CH3)2); FAB-MS (m/z, relative intensity) 425 (MH+, 77), 424 (M•+, 84), 148 (100); Anal (C24H28N2O5•0.3H2O) C, H, N.
(E)-(4-{[4-(Dimethylamino)phenyl]methylene}-2-(hydroxymethyl)-5-oxo-2-2,3-dihydrofuryl)methyl 4-methoxybenzoate (L1-E6, Library 1)
Light yellow solid, mp = 190–192 °C; IR (neat) 3370 (OH), 1705 (C=O), 1593 (C=C) cm−1; 1H NMR (400 MHz, CDCl3) δ 7.92 (d, J = 8.8 Hz, 2 H, ArH), 7.51 (t, J = 2.7 Hz, 1 H, C=CH), 7.40 (d, J = 8.9 Hz, 2 H, ArH), 6.87 (d, J = 8.9 Hz, 2 H, ArH), 6.76 (d, J = 8.7 Hz, 2 H, ArH), 4.48 (AB q, J = 11.9 Hz, 2 H, CCH2OC), 3.83 (s, 3 H, C(O)C6H4OCH3), 3.78 (app t, 2 H, CCH2OH), 3.21 (dd, J = 17.3, 2.8 Hz, 1 H, H3a), 3.06 (m, 7 H, H3b and C=CHC6H4N(CH3)2), 2.33 (t, J = 6.7 Hz, 1 H, CCH2OH); FAB-MS (m/z, relative intensity) 412 (MH+, 75), 411 (M•+, 75), 135 (100); Anal (C23H25NO6) C, H, N.
(E)-(4-{[4-(Dimethylamino)phenyl]methylene}-2-(hydroxymethyl)-5-oxo-2-2,3-dihydrofuryl)methyl 4-nitrobenzoate (L1-E8, Library 1)
Brown solid, mp = 191–193 °C; IR (neat) 3324(OH), 1721 (C=O), 1707 (C=O), 1594 (C=C) cm−1; 1H NMR (400 MHz, CDCl3) δ 8.22 (d, J = 9.0 Hz, 2 H, ArH), 8.10 (d, J = 9.0 Hz, 2 H, ArH), 7.52 (t, J = 2.7 Hz, 1 H, C=CH), 7.41 (d, J = 8.9 Hz, 2 H, ArH), 6.83 (br d, J ≈ 7.4 Hz, 2 H, ArH), 4.56 (AB q, J = 11.9 Hz, 2 H, CCH2OC), 3.84 (br AB q, J = 11.7 Hz, 2 H, CCH2OH), 3.23 (dd, J = 17.2, 2.8 Hz, 1 H, H3a), 3.08 (dd, J = 17.3, 2.5 Hz, 1 H, H3b), 3.04 (v br s, 6 H, C=CHC6H4N(CH3)2), 2.15 (s, 1 H, CCH2OH); FAB-MS (m/z, relative intensity) 427 (MH+, 100), 426 (M•+, 93); Anal (C22H22N2O7) C, H, N.
(E)-{2-(Hydroxymethyl)-4-[(4-methoxyphenyl)methylene]-5-oxo-1-2,3-dihydrofuryl}methyl 4-(dimethylamino)benzoate (L1-F5, Library 1)
White solid, mp = 180 °C; 1H NMR (300 MHz, CDCl3) δ 7.85 (d, J = 9.0 Hz, 2 H, ArH), 7.57 (m, 1 H, C=CH), 7.46 (d, J = 9.0 Hz, 2 H, ArH), 6.95 (d, J = 9.0 Hz, 2 H, ArH), 6.61 (d, J = 9.0 Hz, 2 H, ArH), 4.57 (d, J = 12.0 Hz, 1 H, CCH2OC), 4.38 (d, J = 12.0 Hz, 1 H, CCH2OC), 3.85 (s, 3 H, C=CHC6H4OCH3), 3.73–3.85 (m, 2 H, CCH2OH), 3.25 (dd, J = 17.4, 2.7 Hz, 1 H, H3a), 3.07 (dd, J = 17.4, 2.7 Hz, 1 H, H3b), 3.04 (s, 6 H, C(O)C6H4N(CH3)2); FAB-MS (m/z, relative intensity) 412 (MH+, 100); Anal (C23H25NO6) C, H, N.
(E)-{2-(Hydroxymethyl)-4-[(4-methoxyphenyl)methylene]-5-oxo-1-2,3-dihydrofuryl}methyl 4-methoxybenzoate (L1-F6, Library 1)
White solid, mp = 169 °C; 1H NMR (300 MHz, CDCl3) δ 7.92 (d, J = 9.0 Hz, 2 H, ArH), 7.55–7.59 (m, 1 H, C=CH), 7.46 (d, J = 9.0 Hz, 2 H, ArH), 6.95 (d, J = 9.0 Hz, 2 H, ArH), 6.88 (d, J = 9.0 Hz, 2 H, ArH), 4.58 (d, J = 12.0 Hz, 1 H, CCH2OC), 4.42 (d, J = 12.0 Hz, 1 H, CCH2OC), 3.85 (s, 6 H, C=CHC6H4OCH3 and C(O)C6H4OCH3), 3.80–3.90 (m, 2 H, CCH2OH), 3.26 (dd, J = 17.4, 2.7, Hz, 1 H, H3a), 3.08 (dd, J = 17.4, 2.7 Hz, 1 H, H3b), 2.35–2.45 (m, 1 H, CCH2OH); FAB-MS (m/z, relative intensity) 399 (MH+, 100); Anal (C22H22O7) C, H.
(E)-{2-(Hydroxymethyl)-4-[(4-methoxyphenyl)methylene]-5-oxo-2-2,3-dihydrofuryl}methyl 2,4,6-trimethylbenzoate (L1-F11, Library 1)
White solid, mp = 56–58 °C; IR (neat) 3300 (OH), 1728 (C=O), 1649 (C=C) cm−1; 1H NMR (400 MHz, CDCl3) δ 7.47 (t, J = 2.7 Hz, 1 H, C=CH), 7.41 (d, J = 8.8 Hz, 2 H, ArH), 6.92 (d, J = 8.8 Hz, 2 H, ArH), 6.80 (s, 2 H, ArH), 4.49 (AB q, J = 11.9 Hz, 2 H, CCH2OC), 3.82 (s, 3 H, C=CHC6H4OCH3), 3.77 (AB q, J ≈ 12.2 Hz, 2 H, CCH2OH), 3.18 (dd, J = 17.5, 2.8 Hz, 1 H, H3a), 3.05 (dd, J = 17.5, 2.8 Hz, 1 H, H3b), 2.24 (s, 3 H, C(O)C6H2(CH3)3), 2.23 (s, 6 H, C(O)C6H2(CH3)3); FAB-MS (m/z, relative intensity) 411 (MH+, 51), 147 (100); Anal (C24H26O6•0.5H2O) C, H.
(E)-{2-(Hydroxymethyl)-4-[(4-methoxyphenyl)methylene]-5-oxo-2-2,3-dihydrofuryl}methyl 2,4,6-tris(methylethyl)benzoate (L1-F12, Library 1)
White solid, mp = 62–64 °C; IR (neat) 3687 (OH), 2966 (CH), 1731 (C=O), 1715 (C=O), 1647 (C=C) cm−1; 1H NMR (400 MHz, CDCl3) δ 7.49 (t, J = 2.7 Hz, 1 H, C=CH), 7.41 (d, J = 8.8 Hz, 2 H, ArH), 6.97 (s, 2 H, ArH), 6.92 (d, J = 8.8 Hz, 2 H, ArH), 4.49 (AB q, J = 11.8 Hz, 2 H, CCH2OC), 3.83 (s, 3 H, C=CHC6H4OCH3), 3.78 (AB q, J ≈11.9 Hz, 2 H, CCH2OH), 3.14 (dd, J = 17.5, 2.8 Hz, 1 H, H3a), 3.08 (dd, J = 17.6, 2.8 Hz, 1 H, H3b), 2.86 (sept., J = 6.9 Hz, 1 H, C(O)C6H2(CH(CH3)2)3), 2.76 (sept., J = 6.7 Hz, 2 H, C(O)C6H2(CH(CH3)2)3), 1.22 (d, J = 6.9 Hz, 6 H, C(O)C6H2(CH(CH3)2)3), 1.19 (d, J = 6.8 Hz, 6 H, C(O)C6H2(CH(CH3)2)3), 1.16 (d, J = 6.8 Hz, 6 H, C(O)C6H2(CH(CH3)2)3); FAB-MS (m/z, relative intensity) 495 (MH+, 25), 231 (100); HRMS (FAB) calcd for C30H38O6 (M+K+): 533.231, found: 533.230; HRMS (FAB) calcd for C30H38O6 (MH+): 495.2747, found: 495.2766.
(E)-{2-(Hydroxymethyl)-4-[(4-nitrophenyl)methylene]-5-oxo-2-2,3-dihydrofuryl}methyl 2-ethylhexanoate (L1-H2, Library 1)
White solid, mp = 98–100 °C; IR (neat) 3392 (OH), 1716 (C=O) cm−1; 1H NMR (400 MHz, CDCl3) δ 8.28 (d, J = 8.9 Hz, 2 H, ArH), 7.63 (d, J = 8.8 Hz, 2 H, ArH), 7.59 (t, J = 3.0 Hz, 1 H, C=CH), 4.28 (d AB q, J = 12.0 Hz, 2 H, CCH2OC), 3.77 (d AB q, J = 12.1, Hz, 2 H, CCH2OH), 3.28 (dd, J = 18.0, 3.1 Hz, 1 H, H3a), 3.03 (dd, J = 18.0, 2.6 Hz, 1 H, H3b), 2.24 (m, 2 H, CCH2OH and C(O)CH(CH2CH3)CH2CH2CH2CH3), 1.47 (m, 4 H, C(O)CH(CH2CH3)CH2CH2CH2CH3), 1.18 (m, 4 H, C(O)CH(CH2CH3)CH2CH2CH2CH3), 0.80 (m, 6 H, C(O)CH(CH2CH3)CH2CH2CH2CH3); FAB-MS (m/z, relative intensity) 406 (MH+, 86), 57 (100); Anal (C21H27NO7) C, H, N.
(Z)-{2-(Hydroxymethyl)-4-[(4-nitrophenyl)methylene]-5-oxo-2-2,3-dihydrofuryl}methyl 2-ethylhexanoate (L1-H2, Library 1)
Colorless oil; IR (neat) 3467 (OH), 2961 (CH), 2935 (CH), 2873 (CH), 1764 (C=O), 1738 (C=O), 1605 (C=C) cm−1; 1H NMR (400 MHz, CDCl3) δ 8.16 (d, J = 8.8 Hz, 2 H, ArH), 7.40 (d, J = 8.7 Hz, 2 H, ArH), 6.93 (app q, J = 1.4 Hz, 1 H, C=CH), 4.31 (2 AB q, J = 11.9 Hz, 2 H, CCH2OC), 3.75 (br s, 2 H, CCH2OH), 3.71 (br s, 2 H, H3ab), 2.22 (m, 1 H, C(O)CH(CH2CH3)CH2CH2CH2CH3), 1.46 (m, 4 H, C(O)CH(CH2CH3)CH2CH2CH2CH3), 1.21 (m, 4 H, C(O)CH(CH2CH3)CH2CH2CH2CH3), 0.83 (m, 6 H, C(O)CH(CH2CH3)CH2CH2CH2CH3); FAB-MS (m/z, relative intensity) 406 (MH+, 83), 57 (100); Anal (C21H27NO7) C, H, N.
(E)-{2-(Hydroxymethyl)-4-[(4-nitrophenyl)methylene]-5-oxo-2-2,3-dihydrofuryl}methyl 4-(dimethylamino)benzoate (L1-H5, Library 1)
Yellow solid, mp = 212–215 °C; IR (neat) 3383 (OH), 1703 (C=O) cm−1; 1H NMR (400 MHz, CDCl3) δ 8.25 (d, J = 8.8 Hz, 2 H, ArH), 7.78 (d, J = 8.8 Hz, 2 H, ArH), 7.58 (m, 3 H, C=CH and ArH), 6.58 (d, J = 9.0 Hz, 2 H, ArH), 4.47 (AB q, J = 12.1 Hz, 2 H, CCH2OC), 3.83 (dd, J = 12.2, 6.8 Hz, 1 H, CCH2OH), 3.77 (dd, J = 12.3, 6.7 Hz, 1 H, CCH2OH), 3.31 (dd, J = 18.0, 3.1 Hz, 1 H, H3a), 3.11 (dd, J = 18.0, 2.8 Hz, 1 H, H3b), 3.03 (s, 6 H, CC6H4N(CH3)2), 2.34 (t, J = 6.9 Hz, 1 H, CCH2OH); FAB-MS (m/z, relative intensity) 427 (MH+, 69), 426 (M•+, 51), 148 (100); Anal (C22H22N2O7) C, H, N.
(Z)-{2-(Hydroxymethyl)-4-[(4-nitrophenyl)methylene]-5-oxo-2-2,3-dihydrofuryl}methyl 4-(dimethylamino)benzoate (L1-H5, Library 1)
Yellow solid, mp = 188–191 °C; IR (neat) 3466 (OH), 2923 (CH), 1746 (C=O), 1736 (C=O), 1606 (C=C) cm−1; 1H NMR (400 MHz, CDCl3) δ 7.89 (d, J = 8.8 Hz, 2 H, ArH), 7.68 (d, J = 9.1 Hz, 2 H, ArH), 7.23 (d, J = 10.0 Hz, 2 H, ArH), 7.04 (t, J = 1.3 Hz, 1 H, C=CH), 6.56 (d, J = 9.1 Hz, 2 H, ArH), 4.76 (d, J = 12.0 Hz, 1 H, CCH2OC), 4.40 (d, J = 12.0 Hz, 1 H, CCH2OC), 3.83 (br s, 2 H, CCH2OH), 3.64 (app AB q, 2 H, H3ab), 3.04 (s, 6 H, C(O)C6H4N(CH3)2); FAB-MS (m/z, relative intensity) 427 (MH+, 100), 426 (M•+, 73); Anal (C22H22N2O7•0.3H2O) C, H, N.
(E)-{2-(Hydroxymethyl)-4-[(4-nitrophenyl)methylene]-5-oxo-2-2,3-dihydrofuryl}methyl 4-methoxybenzoate (L1-H6, Library 1)
White solid, mp = 202–204 °C; IR (neat) 3388 (OH), 1711 (C=O) cm−1; 1H NMR (400 MHz, CDCl3) δ 8.26 (d, J = 8.9 Hz, 2 H, ArH), 7.88 (d, J = 8.9 Hz, 2 H, ArH), 7.59 (m, 3 H, C=CH and ArH), 6.87 (d, J = 8.9 Hz, 2 H, ArH), 4.50 (AB q, J = 12.0 Hz, 2 H, CCH2OC), 3.87 (dd, J = 12.2, 6.7 Hz, 1 H, CCH2OH), 3.84 (s, 3 H, CC6H4OCH3), 3.79 (dd, J = 12.2, 6.8 Hz, 1 H, CCH2OH), 3.33 (dd, J = 18.0, 3.2 Hz, 1 H, H3a), 3.11 (dd, J = 18.0, 2.8 Hz, 1 H, H3b), 2.20 (t, J = 6.8 Hz, 1 H, CCH2OH); FAB-MS (m/z, relative intensity) 414 (MH+, 56), 135 (100); Anal (C21H19NO8) C, H, N.
(E)-{2-(Hydroxymethyl)-4-[(4-nitrophenyl)methylene]-5-oxo-2-2,3-dihydrofuryl}methyl 4-nitrobenzoate (L1-H8, Library 1)
White solid, mp = 205–208 °C; IR (neat) 3427 (OH), 1720 (C=O), 1606 (C=C) cm−1; 1H NMR (400 MHz, CD2Cl2) δ 8.26 (m, 3 H, ArH), 8.11 (m, 1 H, ArH), 7.67 (d, J = 8.8 Hz, 2 H, ArH), 7.61 (t, J = 3.0 Hz, 1 H, C=CH), 4.59 (AB q, J = 12.0 Hz, 2 H, CCH2OC), 3.90 (AB q, J = 12.0 Hz, 2 H, CCH2OH), 3.37 (dd, J = 18.1, 3.2 Hz, 1 H, H3a), 3.18 (dd, J = 18.1, 2.8 Hz, 1 H, H3b); FAB-MS (m/z, relative intensity) 429 (MH+, 63), 89 (100); Anal calcd for (C20H16N2O9) C, H, N.
(Z)-(2-(Hydroxymethyl)-5-oxo-4-{[2-(trifluoromethyl)phenyl]-methylene}-2-2,3-dihydrofuryl)methyl 2-methylpropanoate (L2-A1, Library 2)
Oil; IR (neat) 3522 (OH), 2980 (CH), 2880 (CH), 1769 (C=O), 1745 (C=O), 1609 (C=C) cm−1; 1H NMR (400 MHz, CDCl3) δ 7.64 (d, J = 7.7 Hz, 1 H, ArH), 7.49 (t, J = 7.5 Hz, 1 H, ArH), 7.36 (m, 2 H, ArH), 6.68 (t, J = 1.6 Hz, 1 H, C=CH), 4.28 (AB q, J = 11.9 Hz, 2 H, CCH2OC), 3.77 (br s, 2 H, CCH2OH), 3.69 (br s, 2 H, H3ab), 2.52 (br s, 1 H, CCH2OH), 2.46 (sept, J = 7.0 Hz, 1 H, C(O)CH(CH3)2), 1.06 (t, J = 6.8 Hz, 6 H, C(O)CH(CH3)2); FAB-MS (m/z, relative intensity) 373 (MH+, 100); Anal (C18H19F3O5) C, H.
(E)-(2-(Hydroxymethyl)-5-oxo-4-{[4-(trifluoromethyl)phenyl]methylene}-2-2,3-dihydrofuryl)methyl 2-propylpentanoate (L2-B4, Library 2)
White solid, mp = 70–72 °C; IR (neat) 3381 (OH), 2935 (CH), 1727 (C=O), 1655 (C=C) cm−1; 1H NMR (400 MHz, d6-DMSO) δ 7.67 (d, J = 8.2 Hz, 2 H, ArH), 7.57 (d, J = 8.4 Hz, 2 H, ArH), 7.55 (t, J = 3.0 Hz, 1 H, C=CH), 4.26 (AB q, J = 12.0 Hz, 2 H, CCH2OC), 3.75 (AB q, J = 12.2 Hz, 2 H, CCH2OH), 3.25 (dd, J = 17.9, 3.1 Hz, 1 H, H3a), 3.01 (dd, J = 17.9, 2.8 Hz, 1 H, H3b), 2.33 (m, 2 H, CCH2OH and C(O)CH(CH2CH2CH3)2), 1.48 (m, 2 H, C(O)CH(CH2CH2CH3)2), 1.34 (m, 2 H, C(O)CH(CH2CH2CH3)2), 1.17 (m, 4 H, C(O)CH(CH2CH2CH3)2), 0.79 (t, J = 7.3 Hz, 3 H, C(O)CH(CH2CH2CH3)2), 0.75 (t, J = 7.3 Hz, 3 H, C(O)CH(CH2CH2CH3)2); FAB-MS (m/z, relative intensity) 429 (MH+, 100); Anal (C22H27F3O5) C, H.
(E)-{4-[(4-Fluorophenyl)methylene]-2-(hydroxymethyl)-5-oxo-2-2,3-dihydrofuryl}methyl 2-propylpentanoate (L2-B7, Library 2)
Oil; IR (neat) 2965 (CH), 1742 (C=O) cm−1; 1H NMR (400 MHz, d6-DMSO) δ 7.50 (t, J = 2.9 Hz, 1 H, C=CH), 7.46 (m, 2 H, ArH), 7.11 (m, 2 H, ArH), 4.26 (AB q, J = 12.0 Hz, 2 H, CCH2OC), 3.73 (AB q, J = 12.1 Hz, 2 H, CCH2OH), 3.19 (dd, J = 17.6, 2.9 Hz, 1 H, H3a), 2.96 (dd, J = 17.6, 2.7 Hz, 1 H, H3b), 2.33 (m, 2 H, CCH2OH and C(O)CH(CH2CH2CH3)2), 1.49 (m, 2 H, C(O)CH(CH2CH2CH3)2), 1.34 (m, 2 H, C(O)CH(CH2CH2CH3)2), 1.17 (m, 4 H, C(O)CH(CH2CH2CH3)2), 0.77 (overlap t, J = 7.3 Hz, 6 H, C(O)CH(CH2CH2CH3)2); FAB-MS (m/z, relative intensity) 379 (MH+, 100); Anal (C21H27FO5) C, H.
(E)-{4-[(3-Chloro-4-fluorophenyl)methylene]-2-(hydroxymethyl)-5-oxo-2-2,3-dihydrofuryl}methyl 2-propylpentanoate (L2-B10, Library 2)
Oil; IR (neat) 3508(OH), 2959 (CH), 2872 (CH), 1741 (C=O), 1663 (C=C) cm−1; 1H NMR (400 MHz, CDCl3) δ 7.24 (m, 1 H, C=CH), 7.02 (m, 3 H, ArH), 4.06 (AB q, J = 12.0 Hz, 2 H, CCH2OC), 3.54 (AB q, J = 12.1 Hz, 2 H, CCH2OH), 2.99 (dd, J = 17.8, 3.0 Hz, 1 H, H3a), 2.76 (dd, J = 17.8, 2.8 Hz, 1 H, H3b), 2.23 (v br s, 1 H, CCH2OH), 2.14 (m, 1 H, C(O)CH(CH2CH2CH3)2), 1.29 (m, 2 H, C(O)CH(CH2CH2CH3)2), 1.15 (m, 2 H, C(O)CH(CH2CH2CH3)2), 0.97 (m, 4 H, C(O)CH(CH2CH2CH3)2), 0.60 (t, J = 6.5 Hz, 3 H, C(O)CH(CH2CH2CH3)2), 0.57 (t, J = 6.5 Hz, 3 H, C(O)CH(CH2CH2CH3)2); FAB-MS (m/z, relative intensity) 413 (MH+, 100); Anal (C21H26ClFO5) C, H.
(E)-{4-[(3-Chloro-4-fluorophenyl)methylene]-2-(hydroxymethyl)-5-oxo-2-2,3-dihydrofuryl}methyl 2-propylpentanoate (L2-B11, Library 2)
Oil; IR (neat) 3459(OH), 2958 (CH), 2872 (CH), 1735 (C=O) cm−1; 1H NMR (400 MHz, CDCl3) δ 7.50 (dd, J = 6.9, 2.0 Hz, 1 H, ArH), 7.42 (t, J = 2.8 Hz, 1 H, C=CH), 7.35 (m, 1 H, ArH), 7.19 (t, J = 8.6 Hz, 1 H, ArH), 4.26 (AB q, J = 12.0 Hz, 2 H, CCH2OC), 3.75 (AB q, J = 12.2 Hz, 2 H, CCH2OH), 3.19 (dd, J = 17.7, 2.7 Hz, 1 H, H3a), 2.95 (dd, J = 17.7, 2.5 Hz, 1 H, H3b), 2.57 (v br s, 1 H, CCH2OH), 2.34 (m, 1 H, C(O)CH(CH2CH2CH3)2), 1.49 (m, 2 H, C(O)CH(CH2CH2CH3)2), 1.34 (m, 2 H, C(O)CH(CH2CH2CH3)2), 1.18 (m, 4 H, C(O)CH(CH2CH2CH3)2), 0.80 (t, J = 6.3 Hz, 3 H, C(O)CH(CH2CH2CH3)2), 0.77 (t, J = 6.3 Hz, 3 H, C(O)CH(CH2CH2CH3)2); FAB-MS (m/z, relative intensity) 413 (MH+, 100); Anal (C21H26ClFO5) C, H.
(E)-{4-[(3-Bromo-4-fluorophenyl)methylene]-2-(hydroxymethyl)-5-oxo-2-2,3-dihydrofuryl}methyl 2-propylpentanoate (L2-B12, Library 2)
Oil; IR (neat) 3244 (OH), 1757 (C=O), 1658 (C=C) cm−1; 1H NMR (400 MHz, CDCl3) δ7.65 (dd, J = 6.5, 2.1 Hz, 1 H, ArH), 7.40 (t, J = 2.7 Hz, 1 H, C=CH), 7.38 (dd, J = 4.6, 2.2 Hz, 1 H, ArH), 7.16 (t, J = 8.3 Hz, 1 H, ArH), 4.25 (AB q, J = 12.0 Hz, 2 H, CCH2OC), 3.75 (AB q, J = 12.2 Hz, 2 H, CCH2OH), 3.20 (dd, J = 17.7, 2.9 Hz, 1 H, H3a), 2.95 (dd, J = 17.7, 2.7 Hz, 1 H, H3b), 2.73 (v br s, 1 H, CCH2OH), 2.32 (m, 1 H, C(O)CH(CH2CH2CH3)2), 1.48 (m, 2 H, C(O)CH(CH2CH2CH3)2), 1.33 (m, 2 H, C(O)CH(CH2CH2CH3)2), 1.16 (m, 4 H, C(O)CH(CH2CH2CH3)2), 0.79 (t, J = 6.4 Hz, 3 H, C(O)CH(CH2CH2CH3)2), 0.75 (t, J = 6.4 Hz, 3 H, C(O)CH(CH2CH2CH3)2); FAB-MS (m/z, relative intensity) 457 (MH+, 47), 57 (100); Anal (C21H26BrFO5) C, H, Br.
(E)-{4-[(4-Chlorophenyl)methylene]-2-(hydroxymethyl)-5-oxo-2-2,3-dihydrofuryl}methyl 2-ethylhexanoate (L2-C8, Library 2)
Light yellow solid, mp = 41–45 °C; IR (neat) 3384 (OH), 2959 (CH), 2932 (CH), 2873 (CH), 1733 (C=O), 1715 (C=O), 1650 (C=C) cm−1; 1H NMR (400 MHz, CDCl3) δ 7.48 (t, J = 2.9 Hz, 1 H, C=CH), 7.39 (m, 4 H, ArH), 4.26 [2 AB q, J = 12.0 Hz (diastereomers), 2 H, CCH2OC], 3.74 [2 AB q, J = 12.1 Hz (diastereomers), 2 H, CCH2OH], 3.19 (dd, J = 17.7, 3.0 Hz, 1 H, H3a), 2.96 (dd, J = 17.6, 2.6 Hz, 1 H, H3b), 2.40 (v br s, 1 H, CCH2OH), 2.24 (m, 1 H, C(O)CH(CH2CH3)CH2CH2CH2CH3), 1.46 (m, 4 H, C(O)CH(CH2CH3)CH2CH2CH2CH3), 1.15 (m, 4 H, C(O)CH(CH2CH3)CH2CH2CH2CH3), 0.79 (m, 6 H, C(O)CH(CH2CH3)CH2CH2CH2CH3); FAB-MS (m/z, relative intensity) 395 (MH+, 57), 57 (100); Anal (C21H27ClO5) C, H, Cl.
(E)-{4-[(4-Chloro-3-fluorophenyl)methylene]-2-(hydroxymethyl)-5-oxo-2-2,3-dihydrofuryl}methyl 2-ethylhexanoate (L2-C11, Library 2)
Oil; IR (neat) 3464 (OH), 2960 (CH), 2934 (CH), 2874 (CH), 1735 (C=O), 1657 (C=C) cm−1; 1H NMR (400 MHz, CDCl3) δ 7.42 (m, 2 H, C=CH and ArH), 7.22 (m, 2 H, ArH), 4.25 (2 AB q, J ≈11.9, 2 H, CCH2OC), 3.75 (AB q, J ≈12.2 Hz, 2 H, CCH2OH), 3.20 (dd, J = 17.8, 3.0 Hz, 1 H, H3a), 2.95 (dd, J = 17.8, 2.6 Hz, 1 H, H3b), 2.66 (v br s, 1 H, CCH2OH), 2.23 (m, 1 H, C(O)CH(CH2CH3)CH2CH2CH2CH3), 1.44 (m, 4 H, C(O)CH(CH2CH3)CH2CH2CH2CH3), 1.15 (m, 4 H, C(O)CH(CH2CH3)CH2CH2CH2CH3), 0.78 (m, 6 H, C(O)CH(CH2CH3)CH2CH2CH2CH3); FAB-MS (m/z, relative intensity) 413 (MH+, 100); Anal (C21H26ClFO5) C, H, Cl.
(Z)-{4-[(4-Chloro-3-fluorophenyl)methylene]-2-(hydroxymethyl)-5-oxo-2-2,3-dihydrofuryl}methyl 2-ethylhexanoate (L2-C11, Library 2)
Oil; IR (neat) 3461 (OH), 2960 (CH), 2934 (CH), 2874 (CH), 1766 (C=O), 1735 (C=O), 1582 (C=C) cm−1; 1H NMR (400 MHz, CDCl3) δ 7.31 (t, J = 7.9 Hz, 1 H, ArH), 7.01 (dd, J = 9.7, 1.9 Hz, 1 H, ArH), 6.95 (dd, J = 8.2, 1.4 Hz, 1 H, ArH), 6.90 (d, J = 1.3 Hz, 1 H, C=CH), 4.36 (dd, J = 11.9, 5.4 Hz, 1 H, CCH2OC), 4.25 (dd, J = 11.9, 7.9 Hz, 1 H, CCH2OC), 3.74 (app s, 2 H, CCH2OH), 3.56 (s, 2 H, H3ab), 2.21 (m, 2 H, CCH2OH and C(O)CH(CH2CH3)CH2CH2CH2CH3), 1.46 (m, 4 H, C(O)CH(CH2CH3)CH2CH2CH2CH3), 1.21 (m, 4 H, C(O)CH(CH2CH3)CH2CH2CH2CH3), 0.83 (m, 6 H, C(O)CH(CH2CH3)CH2CH2CH2CH3); FAB-MS (m/z, relative intensity) 413 (MH+, 21), 57 (100); Anal (C21H26ClFO5) C, H, Cl.
(E)-{4-[(3-Bromo-4-fluorophenyl)methylene]-2-(hydroxymethyl)-5-oxo-2-2,3-dihydrofuryl}methyl 2-ethylhexanoate (L2-C12, Library 2)
Oil; IR (neat) 3533 (OH), 2963 (CH), 2874 (CH), 1758 (C=O), 1739 (C=O), 1657 (C=C) cm−1; 1H NMR (400 MHz, CDCl3) δ 7.66 (dd, J = 6.5, 2.2 Hz, 1 H, ArH), 7.44 (t, J = 2.9 Hz, 1 H, C=CH), 7.40 (m, 1 H, ArH), 7.17 (t, J = 8.3 Hz, 1 H, ArH), 4.27 (2 AB q, J = 12.0 Hz, 2 H, CCH2OC), 3.75 (AB q, J = 11.9 Hz, 2 H CCH2OH), 3.20 (dd, J = 17.7, 3.0 Hz, 1 H, H3a), 2.95 (dd, J = 17.7, 2.7 Hz, 1 H, H3b), 2.33 (br s, 1 H, CCH2OH), 2.25 (m, 1 H, C(O)CH(CH2CH3)CH2CH2CH2CH3), 1.48 (m, 4 H, C(O)CH(CH2CH3)CH2CH2CH2CH3), 1.17 (m, 4 H, C(O)CH(CH2CH3)CH2CH2CH2CH3), 0.80 (m, 6 H, C(O)CH(CH2CH3)CH2CH2CH2CH3); FAB-MS (m/z, relative intensity) 457 (MH+, 50), 57 (100); Anal (C21H26BrFO5) C, H.
(E)-[2-(Hydroxymethyl)-5-oxo-4-({4-[4-(trifluoromethyl)phenyl]phenyl}methylene)-2-2,3-dihydrofuryl]methyl 2-phenylbutanoate (L2-D6, Library 2)
White solid, mp = 113–115 °C; IR (neat) 3729 (OH), 1745 (C=O), 1714 (C=O), 1648 (C=C) cm−1; 1H NMR (400 MHz, CDCl3) δ 7.71 (m, 2 H, ArH), 7.65 (m, 2 H, ArH), 7.51 (m, 5 H, C=CH and ArH), 7.12 (m, 4 H, ArH), 4.27 [2 AB q, J = 11.9 Hz (diastereomers), 2 H, CH2OC], 3.62 [2 AB q, J = 12.2 Hz (diastereomers), 2 H, CH2OH], 3.43 (m, 1 H, C(O)CHPhCH2CH3), 3.08 [m (diastereomers), 1 H, H3a], 2.84 [m (diastereomers), 1 H, H3b], 2.05 and 1.76 [m (diastereomers), 2 H, C(O)CHPhCH2CH3], 0.82 (m, 3 H, C(O)CHPhCH2CH3); FAB-MS (m/z, relative intensity) 525 (MH+, 100); Anal (C30H27F3O5) C, H.
(E)-{4-[(3-Chloro-4-fluorophenyl)methylene]-2-(hydroxymethyl)-5-oxo-2-2,3-dihydrofuryl}methyl 2,2-dimethylpropanoate (L2-F10, Library 2)
White solid, mp = 138–140 °C; IR (neat) 3495(OH), 2977 (CH), 1728 (C=O), 1717 (C=O), 1656 (C=C) cm−1; 1H NMR (400 MHz, CDCl3) δ 7.46 (m, 2 H, ArH), 7.22 (m, 2 H, C=CH and ArH), 4.25 (AB q, J = 12.0 Hz, 2 H, CCH2OC), 3.75 (AB q, J = 12.0 Hz, 2 H, CCH2OH), 3.21 (dd, J = 17.6, 2.4 Hz, 1 H, H3a), 2.96 (dd, J = 17.3, 1.8 Hz, 1 H, H3b), 1.13 (s, 9 H, C(O)C(CH3)3); FAB-MS (m/z, relative intensity) 371 (MH+, 74), 57 (100); Anal (C18H20ClFO5) C, H.
(E)-{4-[(4-Chloro-3-fluorophenyl)methylene]-2-(hydroxymethyl)-5-oxo-2-2,3-dihydrofuryl}methyl 2,2-dimethylpropanoate (L2-F11, Library 2)
White solid, mp = 188–190 °C; IR (neat) 3377(OH), 2977 (CH), 2875 (CH), 1714 (C=O), 1654 (C=C) cm−1; 1H NMR (400 MHz, CDCl3) δ 7.51 (dd, J = 6.9, 2.2 Hz, 1 H, ArH), 7.43 (t, J = 2.9 Hz, 1 H, C=CH), 7.35 (m, 1 H, ArH), 7.19 (t, J = 8.6 Hz, 1 H, ArH), 4.25 (AB q, J = 12.0 Hz, 2 H, CCH2OC), 3.75 (AB q, J = 12.2 Hz, 2 H, CCH2OH), 3.21 (dd, J = 17.6, 2.9 Hz, 1 H, H3a), 2.95 (dd, J = 17.7, 2.6 Hz, 1 H, H3b), 1.12 (s, 9 H, C(O)C(CH3)3); FAB-MS (m/z, relative intensity) 371 (MH+, 69), 57 (100); Anal (C18H20ClFO5) C, H, Cl.
(Z)-{4-[(4-Chloro-3-fluorophenyl)methylene]-2-(hydroxymethyl)-5-oxo-2-2,3-dihydrofuryl}methyl 2,2-dimethylpropanoate (L2-F11, Library 2)
White solid, mp = 97–99 °C; IR (neat) 3410 (OH), 1726 (C=O) cm−1; 1H NMR (400 MHz, CDCl3) δ 7.31 (t, J = 7.9 Hz, 1 H, ArH), 7.01 (dd, J = 9.7, 1.9 Hz, 1 H, ArH), 6.95 (dd, J = 8.2, 1.3 Hz, 1 H, ArH), 6.87 (t, J = 1.5 Hz, 1 H, C=CH), 4.30 (AB q, J = 11.9 Hz, 2 H, CCH2OC), 3.73 (AB q, J = 12.1 Hz, 2 H, CCH2OH), 3.56 (d, J = 1.4 Hz, 2 H, H3ab), 1.10 (s, 9 H, C(O)C(CH3)3); FAB-MS (m/z, relative intensity) 371 (MH+, 24), 57 (100); Anal (C18H20ClFO5) C, H.
(E)-{2-(Hydroxymethyl)-4-[(4-nitrophenyl)methylene]-5-oxo-2-2,3-dihydrofuryl}methyl 3-(dimethylamino)benzoate (Table 2)
Yellow solid, mp = 173–175 °C; IR (neat) 3421 (OH), 1735 (C=O) cm−1; 1H NMR (400 MHz, CDCl3) δ 8.24 (d, J = 8.9 Hz, 2 H, ArH), 7.56 (m, 3 H, C=CH and ArH), 7.25 (m, 3 H, ArH), 6.93 (br s, 1 H, ArH), 4.51 (br s, 2 H, CCH2OC), 3.85 (AB q, J = 12.2 Hz, 2 H, CCH2OH), 3.33 (dd, J = 18.0, 3.2 Hz, 1 H, H3a), 3.13 (dd, J = 18.0, 2.8 Hz, 1 H, H3b), 2.95 (s, 6 H, C(O)C6H4N(CH3)2), 2.27 (v br s, 1 H, CCH2OH); FA B-MS (m/z, relative intensity) 427 (MH+, 100), 426 (M•+, 84); Anal (C22H22N2O7•0.3H2O) C, H, N.
(E)-{2-(Hydroxymethyl)-4-[(4-nitrophenyl)methylene]-5-oxo-2-2,3-dihydrofuryl}methyl 2-(dimethylamino)benzoate (Table 2)
Yellow solid, mp = 57–60 °C; IR (neat) 3486(OH), 2880 (CH), 1755 (C=O), 1714 (C=O), 1597 (C=C) cm−1; 1H NMR (400 MHz, CDCl3) δ 8.24 (d, J = 8.8 Hz, 2 H, ArH), 7.59 (d, J = 8.8 Hz, 3 H, ArH), 7.56 (t, J = 2.8 Hz, 1 H, C=CH), 7.35 (m, 1 H, ArH), 6.95 (d, J = 8.2 Hz, 1 H, ArH), 6.80 (t, J = 7.4 Hz, 1 H, ArH), 4.48 (AB q, J = 12.0 Hz, 2 H, CCH2OC), 3.83 (m, 2 H, CCH2OH), 3.32 (dd, J = 18.0, 3.0 Hz, 1 H, H3a), 3.13 (dd, J = 18.0, 2.8 Hz, 1 H, H3b), 2.78 (s, 7 H, C(O)C6H4N(CH3)2 and CCH2OH); FAB-MS (m/z, relative intensity) 427 (MH+, 100), 426 (M•+, 28); HRMS (FAB) calcd for C22H22N2O7 (MH+): 427.1505, found: 427.1483.
(E)-{2-(Hydroxymethyl)-4-[(3-nitrophenyl)methylene]-5-oxo-2-2,3-dihydrofuryl}methyl 4-(dimethylamino)benzoate (Table 2)
Yellow solid, mp = 231–233 °C; IR (neat) 3379(OH), 1707 (C=O), 1608 (C=C) cm−1; 1H NMR (400 MHz, d6-DMSO) δ 8.41 (s, 1 H, ArH), 8.25 (d, J = 8.5 Hz, 1 H, ArH), 8.07 (d, J = 7.6 Hz, 1 H, ArH), 7.76 (t, J = 8.0 Hz, 1 H, ArH), 7.60 (m, 3 H, CH=CH, ArH), 6.65 (d, J = 8.8 Hz, 2 H, ArH), 5.74 (s, 2 H, CCH2OC), 5.40 (v br s, 1 H, CCH2OH), 4.40 (s, 2 H, CCH2OC), 3.70 (AB q, J = 11.9 Hz, 2 H, H3ab), 2.98 (s, 6 H, C(O)C6H4N(CH3)2); FAB-MS (m/z, relative intensity) 427 (MH+, 51), 426 (M•+, 38), 148 (100); Anal (C22H22N2O7•0.25H2O) C, H, N.
(E)-{2-(Hydroxymethyl)-4-[(3-nitrophenyl)methylene]-5-oxo-2-2,3-dihydrofuryl}methyl 3-(dimethylamino)benzoate (Table 2)
Yellow solid, mp = 160–162 °C; IR (neat) 3462(OH), 1717 (C=O), 2883 (CH), 2804 (CH), 1755 (C=O), 1718 (C=O), 1602 (C=C) cm−1; 1H NMR (400 MHz, CDCl3) δ 8.22 (s, 2 H, ArH), 7.71 (d, J = 7.8 Hz, 1 H, ArH), 7.58 (m, 3 H, ArH and C=CH), 7.22 (m, 3 H, ArH), 6.89 (s, 1 H, ArH), 4.51 (AB m, 2 H, CCH2OC), 3.86 (AB q, J = 11.2 Hz, 2 H, CCH2OH), 3.35 (dd, J = 17.9, 3.1 Hz, 1 H, H3a), 3.15 (dd, J = 18.0, 2.4 Hz, 1 H, H3b), 2.93 (s, 6 H, C(O)C6H4N(CH3)2), 2.35 (s, 1 H, CCH2OH); FAB-MS (m/z, relative intensity) 427 (MH+, 100), 426 (M•+, 79); HRMS (FAB) calcd for C22H22N2O7 (M+): 426.1427, found: 426.1439.
(E)-{2-(Hydroxymethyl)-4-[(3-nitrophenyl)methylene]-5-oxo-2-2,3-dihydrofuryl}methyl 2-(dimethylamino)benzoate (Table 2)
Yellow solid, mp = 54–56 °C; IR (neat) 3480 (OH), 2885 (CH), 2833 (CH), 2801 (CH), 1757 (C=O), 1715 (C=O), 1597 (C=C) cm−1; 1H NMR (400 MHz, CDCl3) δ 8.29 (s, 1 H, ArH), 8.22 (d, J = 8.0 Hz, 1 H, ArH), 7.75 (d, J = 7.8 Hz, 1 H, ArH), 7.58 (m, 3 H, C=CH and ArH), 7.35 (m, 1 H, ArH), 6.94 (d, J = 8.2 Hz, 1 H, ArH), 6.80 (t, J = 7.4 Hz, 1 H, ArH), 4.49 (AB q, J = 12.0 Hz, 2 H, CCH2OC), 3.84 (br AB q, J = 12.2 Hz, 2 H, CCH2OH), 3.35 (dd, J = 17.9, 3.0 Hz, 1 H, H3a), 3.15 (dd, J = 17.9, 2.8 Hz, 1 H, H3b), 2.79 (s, 7 H, C(O)C6H4N(CH3)2 and CCH2OH); FAB-MS (m/z, relative intensity) 427 (MH+, 100), 426 (M•+, 18); Anal (C22H22N2O7•H2O) C, H, N.
(E)-{2-(Hydroxymethyl)-4-[(2-nitrophenyl)methylene]-5-oxo-2-2,3-dihydrofuryl}methyl 4-(dimethylamino)benzoate (Table 2)
Yellow solid, mp = 173–175 °C; IR (neat) 3409(OH), 2926 (CH), 1756 (C=O), 1699 (C=O), 1603 (C=C) cm−1; 1H NMR (400 MHz, CDCl3) δ 8.09 (dd, J = 8.2, 1.3 Hz, 1 H, ArH), 7.95 (t, J = 2.9 Hz, 1 H, C=CH), 7.82 (d, J = 9.1 Hz, 2 H, ArH), 7.64 (td, J = 7.6, 1.2 Hz, 1 H, ArH), 7.51 (m, 2 H, ArH), 6.69 (d, J = 9.0 Hz, 2 H, ArH), 4.42 (AB q, J = 12.1 Hz, 2 H, CCH2OC), 3.76 (AB q, J = 12.2 Hz, 2 H, CCH2OH), 3.16 (dd, J = 17.7, 3.2 Hz, 1 H, H3a), 3.04 (s, 6 H, C(O)C6H4N(CH3)2), 2.92 (dd, J = 17.7, 2.8 Hz, 1 H, H3b); FAB-MS (m/z, relative intensity) 427 (MH+, 53), 426 (M•+, 40), 148 (100); Anal (C22H22N2O7) C, H, N.
(E)-{2-(Hydroxymethyl)-4-[(2-nitrophenyl)methylene]-5-oxo-2-2,3-dihydrofuryl}methyl 3-(dimethylamino)benzoate (Table 2)
Yellow solid, mp = 62–64 °C; IR (neat) 3461(OH), 2880 (CH), 2813 (CH), 1756 (C=O), 1718 (C=O), 1604 (C=C) cm−1; 1H NMR (400 MHz, CDCl3) δ 8.07 (dd, J = 8.2, 1.2 Hz, 1 H, ArH), 7.91 (t, J = 2.9 Hz, 1 H, C=CH), 7.63 (td, J = 7.7, 1.3 Hz, 1 H, ArH), 7.52 (m, 2 H, ArH), 7.25 (m, 3 H, ArH), 6.91 (v br s, 1 H, ArH), 4.46 (AB q, J = 12.0 Hz, 2 H, CCH2OC), 3.80 (AB q, J = 12.2 Hz, 2 H, CCH2OH), 3.15 (dd, J = 17.8, 3.2 Hz, 1 H, H3a), 2.94 (m, 7 H, H3b and C(O)C6H4N(CH3)2), 2.45 (v br s, 1 H, CCH2OH); FAB-MS (m/z, relative intensity) 427 (MH+, 100), 426 (M•+, 69); Anal (C22H22N2O7•0.9H2O) C, H, N.
(E)-{2-(Hydroxymethyl)-4-[(2-nitrophenyl)methylene]-5-oxo-2-2,3-dihydrofuryl}methyl 2-(dimethylamino)benzoate (Table 2)
Yellow solid, mp = 47–49 °C; IR (neat) 3509 (OH), 2795 (CH), 1757 (C=O), 1597 (C=C) cm−1; 1H NMR (400 MHz, CDCl3) δ 8.07 (dd, J = 8.2, 1.3 Hz, 1 H, ArH), 7.88 (t, J = 2.9 Hz, 1 H, C=CH), 7.63 (m, 2 H, ArH), 7.52 (m, 1 H, ArH), 7.46 (d, J = 7.7 Hz, 1 H, ArH), 7.36 (m, 1 H, ArH), 6.96 (d, J = 8.3 Hz, 1 H, ArH), 6.85 (t, J = 7.4 Hz, 1 H, ArH), 4.44 (AB q, J = 12.0 Hz, 2 H, CCH2OC), 3.78 (AB m, 2 H, CCH2OH), 3.13 (dd, J = 17.8, 3.1 Hz, 1 H, H3a), 2.92 (dd, J = 17.7, 2.9 Hz, 1 H, H3b), 2.80 (s, 7 H, C(O)C6H4N(CH3)2 and CCH2OH); FAB-MS (m/z, relative intensity) 427 (MH+, 100), 426 (M•+, 17); Anal (C22H22N2O7•H2O) C, H, N.
Supplementary Material
Supporting Information Available. Crystal data and structural refinement, atomic coordinates, bond lengths, anisotropic displacement parameters, hydrogen coordinates and isotropic displacement parameters, torsion angles and hydrogen bonds for L2-F11; 1H NMR spectra, 13C NMR spectra and combustion analysis results. This material is available free of charge via the Internet at http://pubs.acs.org.
Acknowledgments
This research was supported in part by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research, and in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract N01-CO-12400. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government. This work was also supported in part by the Korea Health 21 R&D Grant (A020601) from the Ministry of Health and Welfare, by NIH Grant RO1 # CA105125-02 to A. H-F. and by NIH CA125632 to S.A.R. X-ray crystallographic studies were supported in part under contract Y1-DA6002 from the National Institute on Drug Abuse (NIDA).
References
- 1.Zhang GG, Kazanietz MG, Blumberg PM, Hurley JH. Crystal-Structure of the Cys2 Activator-Binding Domain of Protein-Kinase C-Delta in Complex with Phorbol Ester. Cell. 1995;81:917–924. doi: 10.1016/0092-8674(95)90011-x. [DOI] [PubMed] [Google Scholar]
- 2.Sigano DM, Peach ML, Nacro K, Choi Y, Lewin NE, Nicklaus MC, Blumberg PM, Marquez VE. Differential binding modes of diacylglycerol (DAG) and DAG lactones to protein kinase C (PK-C) J Med Chem. 2003;46:1571–1579. doi: 10.1021/jm020476o. [DOI] [PubMed] [Google Scholar]
- 3.Gómez-Fernández JC, Torrecillas A, Corbalán-García S. Diacylglycerols as activators of protein kinase C (Review) Mol Membr Biol. 2004;21:339–349. doi: 10.1080/09687860400010508. [DOI] [PubMed] [Google Scholar]
- 4.Goñi FM, Alonso A. Structure and functional properties of diacylglycerols in membranes. Prog Lipid Res. 1999;38:1–48. doi: 10.1016/s0163-7827(98)00021-6. [DOI] [PubMed] [Google Scholar]
- 5.Carrasco S, Mérida I. Diacylglycerol, when simplicity becomes complex. Trends Biochem Sci. 2007;32:27–36. doi: 10.1016/j.tibs.2006.11.004. [DOI] [PubMed] [Google Scholar]
- 6.Corbalán-García S, Gómez-Fernández JC. Protein kinase C regulatory domains: The art of decoding many different signals in membranes. Biochim Biophys Acta Mol Cell Biol Lipids. 2006;1761:633–654. doi: 10.1016/j.bbalip.2006.04.015. [DOI] [PubMed] [Google Scholar]
- 7.Goldberg EM, Zidovetzki R. Effects of dipalmitoylglycerol and fatty acids on membrane structure and protein kinase C activity. Biophys J. 1997;73:2603–2614. doi: 10.1016/S0006-3495(97)78290-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schechtman D, Mochly-Rosen D. Adaptor proteins in protein kinase C-mediated signal transduction. Oncogene. 2001;20:6339–6347. doi: 10.1038/sj.onc.1204778. [DOI] [PubMed] [Google Scholar]
- 9.Teicher BA. Protein kinase C as a therapeutic target. Clin Cancer Res. 2006;12:5336–5345. doi: 10.1158/1078-0432.CCR-06-0945. [DOI] [PubMed] [Google Scholar]
- 10.Poole AW, Pula G, Hers I, Crosby D, Jones ML. PKC-interacting proteins: from function to pharmacology. Trends Pharmacol Sci. 2004;25:528–535. doi: 10.1016/j.tips.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 11.Wong W, Scott JD. AKAP signalling complexes: Focal points in space and time. Nat Rev Mol Cell Biol. 2004;5:959–970. doi: 10.1038/nrm1527. [DOI] [PubMed] [Google Scholar]
- 12.Neri LM, Borgatti P, Capitani S, Martelli AM. Protein kinase C isoforms and lipid second messengers: a critical nuclear partnership? Histol Histopath. 2002;17:1311–1316. doi: 10.14670/HH-17.1311. [DOI] [PubMed] [Google Scholar]
- 13.Cho W, Stahelin RV. Membrane-protein interactions in cell signaling and membrane trafficking. Annu Rev Biophys Biomol Struct. 2005;34:119–151. doi: 10.1146/annurev.biophys.33.110502.133337. [DOI] [PubMed] [Google Scholar]
- 14.Simons K, Vaz WLC. Model systems, lipid rafts, and cell membranes. Annu Rev Biophys Biomol Struct. 2004;33:269–295. doi: 10.1146/annurev.biophys.32.110601.141803. [DOI] [PubMed] [Google Scholar]
- 15.Mukherjee S, Maxfield FR. Membrane domains. Annu Rev Cell Dev Biol. 2004;20:839–866. doi: 10.1146/annurev.cellbio.20.010403.095451. [DOI] [PubMed] [Google Scholar]
- 16.Carrasco S, Mérida I. Diacylglycerol-dependent binding recruits PKC theta and RasGRP1 C1 domains to specific subcellular localizations in living T lymphocytes. Mol Biol Cell. 2004;15:2932–2942. doi: 10.1091/mbc.E03-11-0844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stahelin RV, Wang JY, Blatner NR, Raftner JD, Murray D, Cho WH. The origin of C1A-C2 interdomain interactions in protein kinase C alpha. J Biol Chem. 2005;280:36452–36463. doi: 10.1074/jbc.M506224200. [DOI] [PubMed] [Google Scholar]
- 18.Slater SJ, Seiz JL, Cook AC, Buzas CJ, Malinowski SA, Kershner JL, Stagliano BA, Stubbs CD. Regulation of PKC alpha activity by C1-C2 domain interactions. J Biol Chem. 2002;277:15277–15285. doi: 10.1074/jbc.M112207200. [DOI] [PubMed] [Google Scholar]
- 19.Canagarajah B, Leskow FC, Ho JYS, Mischak H, Saidi LF, Kazanietz MG, Hurley JH. Structural mechanism for lipid activation of the Rac-specific GAP, beta 2-chimaerin. Cell. 2004;119:407–418. doi: 10.1016/j.cell.2004.10.012. [DOI] [PubMed] [Google Scholar]
- 20.Ron D, Kazanietz MG. New insights into the regulation of protein kinase C and novel phorbol ester receptors. FASEB J. 1999;13:1658–1676. [PubMed] [Google Scholar]
- 21.Brose N, Rosenmund C. Move over protein kinase C, you’ve got company: Alternative cellular effectors of diacylglycerol and phorbol esters. J Cell Sci. 2002;115:4399–4411. doi: 10.1242/jcs.00122. [DOI] [PubMed] [Google Scholar]
- 22.Rozengurt E, Sinnett-Smith J, Zugaza J. Protein kinase D: a novel target for diacylglycerol and phorbol esters. Biochem Soc Trans. 1997;25:565–571. doi: 10.1042/bst0250565. [DOI] [PubMed] [Google Scholar]
- 23.Ahmed S, Lee J, Kozma R, Best A, Monfries C, Lim L. A Novel Functional Target for Tumor-Promoting Phorbol Esters and Lysophosphatidic Acid - the P21rac-Gtpase Activating Protein N-Chimaerin. J Biol Chem. 1993;268:10709–10712. [PubMed] [Google Scholar]
- 24.Brose N, Rosenmund C, Rettig J. Regulation of transmitter release by Unc-13 and its homologues. Curr Opin Neurobiol. 2000;10:303–311. doi: 10.1016/s0959-4388(00)00105-7. [DOI] [PubMed] [Google Scholar]
- 25.Ebinu JO, Bottorff DA, Chan EYW, Stang SL, Dunn RJ, Stone JC. RasGRP, a Ras guanyl nucleotide-releasing protein with calcium- and diacylglycerol-binding motifs. Science. 1998;280:1082–1086. doi: 10.1126/science.280.5366.1082. [DOI] [PubMed] [Google Scholar]
- 26.Kawasaki H, Sprihtgett GM, Toki S, Canales JJ, Harlan P, Blumenstiel JP, Chen EJ, Bany IA, Mochizuki N, Ashbacher A, Matsuda M, Housman DE, Graybiel AM. A Rap guanine nucleotide exchange factor enriched highly in the basal ganglia. Proc Natl Acad Sci US A. 1998;95:13278–13283. doi: 10.1073/pnas.95.22.13278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luo B, Regier DS, Prescott SM, Topham MK. Diacylglycerol kinases. Cell Signal. 2004;16:983–989. doi: 10.1016/j.cellsig.2004.03.016. [DOI] [PubMed] [Google Scholar]
- 28.Marquez VE, Blumberg PM. Synthetic diacylglycerols (DAG) and DAG-lactones as activators of protein kinase C (PK-C) Acc Chem Res. 2003;36:434–443. doi: 10.1021/ar020124b. [DOI] [PubMed] [Google Scholar]
- 29.Duan DH, Lewin NE, Sigano DM, Blumberg PM, Marquez VE. Conformationally constrained analogues of diacylglycerol. 21. A solid-phase method of synthesis of diacylglycerol lactones as a prelude to a combinatorial approach for the synthesis of protein kinase C isozyme-specific ligands. J Med Chem. 2004;47:3248–3254. doi: 10.1021/jm030610k. [DOI] [PubMed] [Google Scholar]
- 30.Kang JH, Benzaria S, Sigano DM, Lewin NE, Pu YM, Peach ML, Blumberg PM, Marquez VE. Conformationally constrained analogues of diacylglycerol. 26. Exploring the chemical space surrounding the C1 domain of protein kinase C with DAG-lactones containing aryl groups at the sn-1 and sn-2 positions. J Med Chem. 2006;49:3185–3203. doi: 10.1021/jm060011o. [DOI] [PubMed] [Google Scholar]
- 31.Triolo A, Altamura M, Cardinali F, Sisto A, Maggi CA. Mass spectrometry and combinatorial chemistry: a short outline. J Mass Spectrom. 2001;36:1249–1259. doi: 10.1002/jms.238. [DOI] [PubMed] [Google Scholar]
- 32.Lewin NE, Blumberg PM. [3H]Phorbol 12,13-Dibutyrate Binding Assay for Protein Kinase C and Related Proteins. Methods Mol Biol. 2003;233:129–156. doi: 10.1385/1-59259-397-6:129. [DOI] [PubMed] [Google Scholar]
- 33.Wildman SA, Crippen GM. Prediction of physicochemical parameters by atomic contributions. J Chem Inf Comput Sci. 1999;39:868–873. [Google Scholar]
- 34.Pu Y, Perry NA, Yang D, Lewin NE, Kedei N, Braun DC, Choi SH, Blumberg PM, Garfield SH, Stone JC, Duan D, Marquez VE. A Novel Diacylglycerol-lactone Shows Marked Selectivity in Vitro among C1 Domains of Protein Kinase C (PKC) Isoforms {alpha} and {delta} as Well as Selectivity for RasGRP Compared with PKC{alpha} J Biol Chem. 2005;280:27329–27338. doi: 10.1074/jbc.M414132200. [DOI] [PubMed] [Google Scholar]
- 35.Eisenberg D, Schwarz E, Komaromy M, Wall R. Analysis of membrane and surface protein sequences with the hydrophobic moment plot. J Mol Biol. 1984;179:125–142. doi: 10.1016/0022-2836(84)90309-7. [DOI] [PubMed] [Google Scholar]
- 36.Wolfe MS. Secretase targets for Alzheimer’s disease: Identification and therapeutic potential. J Med Chem. 2001;44:2039–2060. doi: 10.1021/jm0004897. [DOI] [PubMed] [Google Scholar]
- 37.Kozikowski AP, Nowak I, Petukhov PA, Etcheberrigaray R, Mohamed A, Tan M, Lewin N, Hennings H, Pearce LL, Blumberg PM. New amide-bearing benzolactam-based protein kinase C modulators induce enhanced secretion of the amyloid precursor protein metabolite sAPP alpha. J Med Chem. 2003;46:364–373. doi: 10.1021/jm020350r. [DOI] [PubMed] [Google Scholar]
- 38.Lee J, Kang JH, Han KC, Kim Y, Kim SY, Youn HS, Mook-Jung I, Kim H, Han JHL, Ha HJ, Kim YH, Marquez VE, Lewin NE, Pearce LV, Lundberg DJ, Blumberg PM. Branched diacylglycerol-lactones as potent protein kinase C ligands and alpha-secretase activators. J Med Chem. 2006;49:2028–2036. doi: 10.1021/jm0509391. [DOI] [PubMed] [Google Scholar]
- 39.Yeon SW, Jung MW, Ha MJ, Kim SU, Huh K, Savage MJ, Masliah E, Mook-Jung I. Blockade of PKC epsilon activation attenuates phorbol ester-induced increase of alpha-secretase-derived secreted form of amyloid precursor protein. Biochem Biophys Res Commun. 2001;280:782–787. doi: 10.1006/bbrc.2000.4181. [DOI] [PubMed] [Google Scholar]
- 40.Schubert D, Heineman S, Carlisle W, Tarikas H, Kimes B, Patrick J, Steinbac Jh, Culp W, Brandt BL. Clonal cell lines from rat central nervous system. Nature. 1974;249:224–227. doi: 10.1038/249224a0. [DOI] [PubMed] [Google Scholar]
- 41.Kim C, Jang CH, Bang JH, Jung MW, Joo I, Kim SU, Mook-Jung I. Amyloid precursor protein processing is separately regulated by protein kinase C and tyrosine kinase in human astrocytes. Neurosci Lett. 2002;324:185–188. doi: 10.1016/s0304-3940(02)00217-3. [DOI] [PubMed] [Google Scholar]
- 42.Sun XG, Rotenberg SA. Overexpression of protein kinase C alpha in MCF-10A human breast cells engenders dramatic alterations in morphology, proliferation, and motility. Cell Growth Differ. 1999;10:343–352. [PubMed] [Google Scholar]
- 43.Abeyweera TP, Rotenberg SA. Design and characterization of a traceable protein kinase C alpha. Biochemistry. 2007;46:2364–2370. doi: 10.1021/bi0622017. [DOI] [PubMed] [Google Scholar]
- 44.Nacro K, Bienfait B, Lee J, Han KC, Kang JH, Benzaria S, Lewin NE, Bhattacharyya DK, Blumberg PM, Marquez VE. Conformationally constrained analogues of diacylglycerol (DAG). 16. How much structural complexity is necessary for recognition and high binding affinity to protein kinase C? J Med Chem. 2000;43:921–944. doi: 10.1021/jm9904607. [DOI] [PubMed] [Google Scholar]
- 45.Angel P, Imagawa M, Chiu R, Stein B, Imbra RJ, Rahmsdorf HJ, Jonat C, Herrlich P, Karin M. Phorbol ester inducible genes contain a common cis element recognized by a TPA-modulated trans-acting factor. Cell. 1987;49:729–739. doi: 10.1016/0092-8674(87)90611-8. [DOI] [PubMed] [Google Scholar]
- 46.Young MR, Li JJ, Rincon M, Flavell RA, Sathyanarayana BK, Hunziker R, Colburn N. Transgenic mice demonstrate AP-1 (activator protein-1) transactivation is required for tumor promotion. Proc Natl Acad Sci USA. 1999;96:9827–9832. doi: 10.1073/pnas.96.17.9827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bernstein LR, Colburn NH. AP1/jun function is differentially induced in promotion-sensitive and resistant JB6 cells. Science. 1989;244:566–569. doi: 10.1126/science.2541502. [DOI] [PubMed] [Google Scholar]
- 48.Dhar A, Hu J, Reeves R, Resar LMS, Colburn NH. Dominant-negative c-Jun (TAM67) target genes: HMGA1 is required for tumor promoter-induced transformation. Oncogene. 2004;23:4466–4476. doi: 10.1038/sj.onc.1207581. [DOI] [PubMed] [Google Scholar]
- 49.Obeid LM, Linardic CM, Karolak LA, Hannun YA. Programmed Cell-Death Induced by Ceramide. Science. 1993;259:1769–1771. doi: 10.1126/science.8456305. [DOI] [PubMed] [Google Scholar]
- 50.Grant S, Jarvis WD, Swerdlow PS, Turner AJ, Traylor RS, Wallace HJ, Lin PS, Pettit GR, Gewirtz DA. Potentiation of the Activity of 1-beta-D-Arabinofuranosylcytosine by the Protein Kinase C Activator Bryostatin-1 in HL-60 Cells - Association with Enhanced Fragmentation of Mature DNA. Cancer Res. 1992;52:6270–6278. [PubMed] [Google Scholar]
- 51.Lotem J, Cragoe EJ, Sachs L. Rescue from Programmed Cell-Death in Leukemic and Normal Myeloid Cells. Blood. 1991;78:953–960. [PubMed] [Google Scholar]
- 52.Haimovitzfriedman A, Balaban N, McLoughlin M, Ehleiter D, Michaeli J, Vlodavsky I, Fuks Z. Protein-Kinase-C Mediates Basic Fibroblast Growth-Factor Protection of Endothelial Cells Against Radiation-Induced Apoptosis. Cancer Res. 1994;54:2591–2597. [PubMed] [Google Scholar]
- 53.Garcia-Bermejo ML, Leskow FC, Fujii T, Wang Q, Blumberg PM, Ohba M, Kuroki T, Han KC, Lee J, Marquez VE, Kazanietz MG. Diacylglycerol (DAG)-lactones, a new class of protein kinase C (PKC) agonists, induce apoptosis in LNCaP prostate cancer cells by selective activation of PKCalpha. J Biol Chem. 2002;277:645–655. doi: 10.1074/jbc.M107639200. [Published erratum appears in J. Biol. Chem. 2004, 279, 23846] [DOI] [PubMed] [Google Scholar]
- 54.Truman J-P, Rotenberg SA, Kang JH, Fuks Z, Kolesnick R, Marquez VE, Leibel S, Haimovitz-Friedman A. PKC-alpha activation down-regulates ATM and radio-sensitizes androgen-sensitive human prostate cancer cells in vitro and in vivo. Clin Cancer Res. 2007 doi: 10.4161/cbt.8.1.7119. submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liao WC, Haimovitz-Friedman A, Persaud RS, McLoughlin M, Ehleiter D, Zhang N, Gatei M, Lavin M, Kolesnick R, Fuks Z. Ataxia telangiectasia-mutated gene product inhibits DNA damage-induced apoptosis via ceramide synthase. J Biol Chem. 1999;274:17908–17917. doi: 10.1074/jbc.274.25.17908. [DOI] [PubMed] [Google Scholar]
- 56.Gleave ME, Hsieh JT, Wu HC, Voneschenbach AC, Chung LWK. Serum Prostate Specific Antigen Levels in Mice Bearing Human Prostate LNCaP Tumors Are Determined by Tumor Volume and Endocrine and Growth Factors. Cancer Res. 1992;52:1598–1605. [PubMed] [Google Scholar]
- 57.Kasahara T, Djeu JY, Dougherty SF, Oppenheim JJ. Capacity of human large granular lymphocytes (LGL) to produce multiple lymphokines - interleukin-2, interferon and colony stimulating factor. J Immunol. 1983;131:2379–2385. [PubMed] [Google Scholar]
- 58.Kasahara T, Hooks JJ, Dougherty SF, Oppenheim JJ. Interleukin 2-mediated immune interferon (IFN-gamma) production by human T-cells and T-cell subsets. J Immunol. 1983;130:1784–1789. [PubMed] [Google Scholar]
- 59.Chan SH, Perussia B, Gupta JW, Kobayashi M, Pospisil M, Young HW, Wolf SF, Young D, Clark SC, Trinchieri G. Induction of interferon gamma production by natural killer cell stimulatory factor: characterization of the responder cells and synergy with other inducers. J Exp Med. 1991;173:869–879. doi: 10.1084/jem.173.4.869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bower MJ, Cohen FE, Dunbrack RL. Prediction of protein side-chain rotamers from a backbone-dependent rotamer library: a new homology modeling tool. J Mol Biol. 1997;267:1268–1282. doi: 10.1006/jmbi.1997.0926. [DOI] [PubMed] [Google Scholar]
- 61.Malolanarasimhan K, Kedei N, Sigano DM, Kelley JA, Lai CC, Lewin NE, Surawski RJ, Pavlyukovets VA, Garfield SH, Wincovitch S, Blumberg PM, Marquez VE. Conformationally constrained analogues of diacylglycerol (DAG). 27. Modulation of membrane translocation of protein kinase C (PKC) isozymes alpha and delta by diacylglycerol lactones (DAG-lactones) containing rigid-rod acyl groups. J Med Chem. 2007;50:962–978. doi: 10.1021/jm061289j. [DOI] [PubMed] [Google Scholar]
- 62.Wang QMJ, Bhattacharyya D, Garfield S, Nacro K, Marquez VE, Blumberg PM. Differential localization of protein kinase C delta by phorbol esters and related compounds using a fusion protein with green fluorescent protein. J Biol Chem. 1999;274:37233–37239. doi: 10.1074/jbc.274.52.37233. [DOI] [PubMed] [Google Scholar]
- 63.Garzotto M, White-Jones M, Jiang YW, Ehleiter D, Liao WC, Haimovitz-Friedman A, Fuks Z, Kolesnick R. 12-O-tetradecanoylphorbol-13-acetate-induced apoptosis in LNCaP cells is mediated through ceramide synthase. Cancer Res. 1998;58:2260–2264. [PubMed] [Google Scholar]
- 64.Stephenson RA, Dinney CPN, Gohji K, Ordonez NG, Killion JJ, Fidler IJ. Metastatic Model for Human Prostate Cancer Using Orthotopic Implantation in Nude Mice. J Natl Cancer Inst. 1992;84:951–957. doi: 10.1093/jnci/84.12.951. [DOI] [PubMed] [Google Scholar]
- 65.Lai CC, Duan D, Phillips LR, Marquez VE, Kelley JA. Mass spectrometric strategies for the rapid characterization of diacylglycerol-lactone combinatorial libraries. Proceedings of the 53rd ASMS Conference on Mass Spectrometry and Allied Topics; San Antonio, TX. Jun, 2005. paper MP177. [Google Scholar]
- 66.Fenselau C, Cotter RJ. Chemical aspects of fast-atom bombardment. Chem Rev. 1987;87:501–512. [Google Scholar]
- 67.Yan B, Fang LL, Irving M, Zhang S, Boldi AM, Woolard F, Johnson CR, Kshirsagar T, Figliozzi GM, Krueger CA, Collins N. Quality control in combinatorial chemistry: Determination of the quantity, purity, and quantitative purity of compounds in combinatorial libraries. J Comb Chem. 2003;5:547–559. doi: 10.1021/cc030008f. [DOI] [PubMed] [Google Scholar]
- 68.Choi Y, Kang JH, Lewin NE, Blumberg PM, Lee J, Marquez VE. Conformationally constrained analogues of diacylglycerol. 19. Synthesis and protein kinase C binding affinity of diacylglycerol lactones bearing an N-hydroxylamide side chain. J Med Chem. 2003;46:2790–2793. doi: 10.1021/jm030082c. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information Available. Crystal data and structural refinement, atomic coordinates, bond lengths, anisotropic displacement parameters, hydrogen coordinates and isotropic displacement parameters, torsion angles and hydrogen bonds for L2-F11; 1H NMR spectra, 13C NMR spectra and combustion analysis results. This material is available free of charge via the Internet at http://pubs.acs.org.