Summary
Temporal expression patterns of the Bordetella pertussis alcaligin, enterobactin, and heme iron acquisition systems were examined using alcA-, bfeA-, and bhuR-tnpR recombinase fusion strains in a mouse respiratory infection model. The iron systems were differentially expressed in vivo, showing early induction of the alcaligin and enterobactin siderophore systems, and delayed induction of the heme system in a manner consistent with predicted changes in host iron source availability during infection. Previous mixed infection competition studies established the importance of alcaligin and heme utilization for B. pertussis in vivo growth and survival. In this study, the contribution of the enterobactin system to the fitness of B. pertussis was confirmed using wild-type and enterobactin receptor mutant strains in similar competition infection experiments. As a correlate to the in vivo expression studies of B. pertussis iron systems in mice, sera from uninfected and B. pertussis-infected human donors were screened for antibody reactivity with Bordetella iron-repressible cell envelope proteins. Pertussis patient sera recognized multiple iron-repressible proteins including the known outer membrane receptors for alcaligin, enterobactin, and heme, supporting the hypothesis that B. pertussis is iron-starved and responds to the presence of diverse iron sources during natural infection.
Keywords: Bordetella pertussis, iron, alcaligin, enterobactin, heme, RIVET
Introduction
It could be reasoned that the main objective of a pathogen is to survive and multiply. In order to multiply, successful pathogens must be able to scavenge growth-limiting nutrients in the host environment, yet the abundance and source of nutrients may change as an infection progresses, and these changes can decisively affect microbial growth rates and population sizes. One axiom that has emerged from in vivo expression studies of a variety of host-pathogen systems is that nutrient acquisition genes, especially those involved in iron acquisition, represent the prevalent class of in vivo expressed genes (Mahan et al., 2000).
Bordetella pertussis, the causative agent of human whooping cough or pertussis, colonizes the ciliated cells of the upper respiratory tract epithelium. The interaction of B. pertussis with its human host characteristically results in inflammation, activation of immune responses, and injury to host tissues. Over the course of infection, changes in host niche conditions are presumed to alter the array and abundance of iron sources available to B. pertussis. The alcaligin siderophore, enterobactin siderophore, and heme utilization systems of B. pertussis are genetically characterized and share key regulatory features (Brickman et al., 2001; Vanderpool and Armstrong, 2003; Anderson and Armstrong, 2004) (Table 1). All three systems are repressible by Fur and iron, and maximal expression of each system during iron starvation requires a distinct transcriptional activator that is responsive to the cognate iron source as the inducer. We hypothesize that the involvement of the substrate-inducible positive regulators AlcR (Beaumont et al., 1998; Pradel et al., 1998), BfeR (Anderson and Armstrong, 2004), and HurI (Vanderpool and Armstrong, 2001) ensures that the iron systems can be differentially expressed, allowing for effective adaption to changes in iron source availability during infection. In addition to the outer membrane receptor proteins required for transport of ferric alcaligin (FauA), ferric enterobactin (BfeA) and heme (BhuR), at least nine more genes encoding predicted TonB-dependent receptors of unknown ligand specificity are encoded in the B. pertussis genome (Parkill et al., 2003; Anderson and Armstrong, 2008).
Table 1.
Relevant features of B. pertussis iron systems analyzed using RIVET
| Iron system | Promotera | Activator | Inducer |
|---|---|---|---|
| Alcaligin siderophore biosynthesis, transport and Utilization | alcA | AlcR (AraC/XylS family) | Alcaligin |
| Enterobactin siderophore transport and utilization | bfeA | BfeR (AraC/XylS family) | enterobactin, other Catechols |
| Heme transport and utilization | bhuR | HurI (Extracytoplasmic function sigma factor) | heme compounds |
inducible promoter driving tnpR expression in a RIVET strain
Upon initial colonization of the host mucosal surface it is conceivable that B. pertussis uses available lactoferrin as an iron source by way of its native siderophore alcaligin. Other siderophores produced by transient microbes or commensals might also deliver iron to B. pertussis. As B. pertussis multiplies, inflammation and pathologic effects of Bordetella toxic factors on the host epithelial barrier could allow iron-loaded cellular and serum components to escape to the mucosal surface (Persson et al., 1991). Ultimately, host cell lysis would release yet additional iron sources such as transferrin and heme proteins that are readily utilized by B. pertussis. This postulated series of events spawned the hypothesis that iron utilization systems are differentially expressed in vivo. It was predicted that siderophore-based iron acquisition systems would be essential for B. pertussis multiplication and survival during the initial colonization stage for establishment of infection, but that heme utilization would be more important to ecological fitness later in infection once the epithelial barrier is damaged.
Previously, the importance of the alcaligin (Brickman and Armstrong, 2007) and heme (Brickman et al., 2006) systems for multiplication and survival of B. pertussis was established using mixed infection competition studies involving co-infection of mice with wild-type and fauA or bhuR receptor mutant strains. In those studies, the alcaligin system was found to be important for iron acquisition at the initial colonization stage, whereas the heme system was not essential until later in infection, after the peak of bacterial multiplication. Since the two iron systems have different substrates, the distinct phasing of alcaligin and heme system involvement implies that host iron sources changed as the infections progressed. Further investigation was required to determine whether these presumed variations in iron source availability resulted in differential expression of the B. pertussis iron systems at the transcriptional level. In this study, temporal expression patterns of key promoters of the B. pertussis alcaligin, enterobactin, and heme utilization systems were monitored using recombinase-based in vivo expression technology (RIVET) (Camilli et al., 1994) in mice. The iron systems were differentially expressed during infection in a manner consistent with variations in iron sources, showing early induction of the siderophore-based systems and late induction of the heme system. To augment the transcriptional analysis of the enterobactin utilization system in this study, and complement previous infection studies confirming that the alcaligin and heme iron systems contribute significantly to in vivo fitness of B. pertussis, the importance of the enterobactin system for B. pertussis growth and survival was established using mixed infection competition experiments in mice. As a correlate to in vivo fitness and expression studies using the mouse respiratory infection model, sera from normal human donors and pertussis patients were screened for antibody reactivity with Bordetella iron-repressible cell envelope proteins. The finding that pertussis patient sera were reactive with numerous iron-repressible cell envelope proteins including iron transport system receptor proteins suggests that B. pertussis is iron-starved in its natural human host environment and responds to the presence of diverse iron sources by expression of multiple B. pertussis iron utilization systems.
Results and Discussion
Construction of B. pertussis iron transport system promoter-resolvase fusion strains
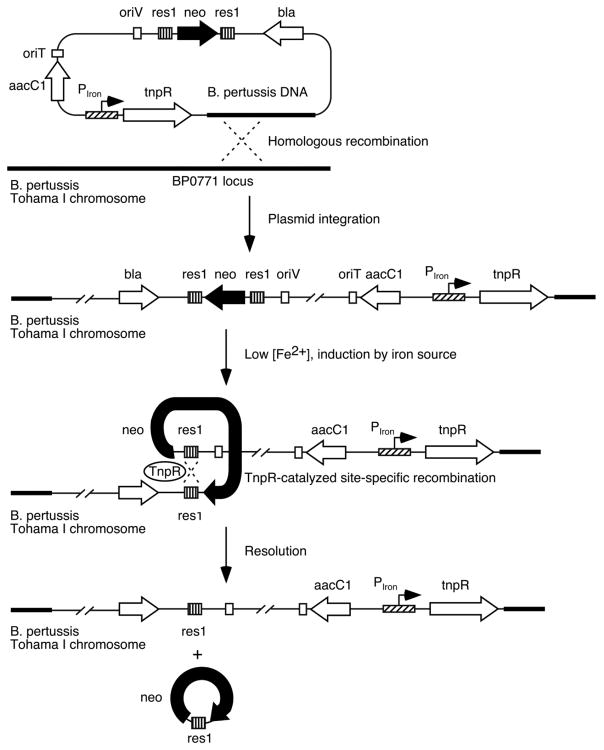
Three different B. pertussis iron transport system promoter-tnpR fusion strains were constructed for monitoring expression of the alcaligin, enterobactin, and heme utilization systems by placing tnpR expression under the control of the substrate-inducible alcA (Kang and Armstrong, 1998; Brickman et al., 2001), bfeA (Anderson and Armstrong, 2004) and bhuR (Vanderpool and Armstrong, 2004) promoters (Fig. 1). The TnpR recombinase can then catalyze the excision of the res1 site-flanked kanamycin resistance marker on the integrated fusion plasmid, resulting in a stable kanamycin-sensitive phenotype that can be scored in B. pertussis populations recovered from infected mice as an indicator of prior expression of a particular iron system. The B. pertussis RIVET system (Veal-Carr and Stibitz, 2005) can employ various combinations of the tnpR resolvase and res site variant alleles devised previously to make the reporter system amenable to use with promoters having low-to-moderate basal expression levels (Lee et al., 1999). In the current study, preliminary in vitro experiments using the attenuated tnpR135 and tnpR168 alleles to construct B. pertussis RIVET strains found that the reduced TnpR production levels from these mutant alleles were insufficient to catalyze resolution of a res1 site-flanked marker when expressed in B. pertussis from the iron system promoters (data not presented), whereas production of TnpR using the wild-type allele was associated with normally regulated expression and high resolution frequencies under culture conditions known to activate the promoters. Accordingly, all gene fusions in this study used the wild-type tnpR allele in conjunction with the res1 resolvase substrate. Resolvase substrates were stably maintained during in vitro cloning and strain construction procedures by supplementing culture media with additional iron over the standard formulations. In addition, since the bhuR heme receptor gene promoter is induced by heme under iron-limiting growth conditions (Vanderpool and Armstrong, 2001), heme-free Stainer-Scholte charcoal agar medium (SCMB) was used instead of Bordet-Gengou agar (BG) (Bordet and Gengou, 1906) in bhuR-tnpR B. pertussis strain construction procedures.
Fig. 1.
Schematic representation of RIVET analysis of B. pertussis iron transport system genes. Transcriptional activity of alcA (alcaligin), bfeA (enterobactin) and bhuR (heme) iron system promoters (designated PIron) in vivo drives production of the site-specific resolvase TnpR, which catalyzes excision of the linked kanamycin resistance marker (neo) by recombination between the res1 sites. Loss of kanamycin resistance serves as a stable heritable marker of prior gene expression.
Analysis of B. pertussis iron system promoter activities in vitro
The alcaligin, enterobactin, and heme utilization systems of B. pertussis have been studied extensively, and their genetic and transcriptional organizations, substrate and inducer specificities, and regulatory features are well characterized (Brickman et al., 2004; Brickman et al., 2007). To confirm that the iron promoter-tnpR fusion genes were transcriptionally responsive to iron nutritional status and substrate induction, resolution frequencies were determined under controlled growth conditions in vitro. It was anticipated that the resolution frequencies would be influenced primarily by the iron co-repressor concentration and its effect on the intensity of the iron starvation signal, positive regulator gene expression levels and effective inducer concentrations, and intrinsic iron system promoter strength.
After growth to post-exponential phase (48 h) under iron-replete (i.e., repressing) growth conditions, mean resolution frequencies were 7% or less for all three fusion strains. Under iron starvation growth conditions in the absence of inducer supplements, the bfeA-tnpR and bhuR-tnpR fusion strains showed moderate elevation of mean resolution frequencies to 26% and 36%, respectively, consistent with relief of Fur repression. In contrast, the alcA-tnpR strain had a mean resolution frequency of 96%, indicative of strong alcA promoter activation known to occur during iron starvation in the presence of the autogenously produced alcaligin siderophore inducer (Brickman et al., 2001). Likewise, when supplied with enterobactin or hemin inducers under iron starvation conditions, promoter activation in the bfeA-tnpR and bhuR-tnpR fusion strains resulted in mean resolution frequencies of 91% and 92%, respectively, consistent with their known inducer requirements for maximal promoter activation (Vanderpool and Armstrong, 2003; Anderson and Armstrong, 2004). These in vitro RIVET assays established that resolution frequencies for all fusion strains correlated well with previously characterized transcriptional activities associated with iron-repressed, derepressed, and induced expression levels of the alcA, bfeA, and bhuR iron system promoters (Brickman et al., 2001; Vanderpool and Armstrong, 2003; Anderson and Armstrong, 2004).
Differential expression of iron transport system promoters in vivo
The three B. pertussis RIVET strains were used individually in mouse respiratory infection experiments for temporal analysis of alcaligin, enterobactin, and heme system gene expression. Since the use of heme-free medium was necessary to prevent induction of the heme-inducible bhuR-tnpR fusion in strain TohI-res-R-bhuR, the same heme-free SCMB medium was used for the other two fusion strains as well. Plating efficiencies of B. pertussis fusion strains on SCMB were found to be similar to those on BG, with mean CFU values differing by no more than 6% when determined in parallel (data not presented).
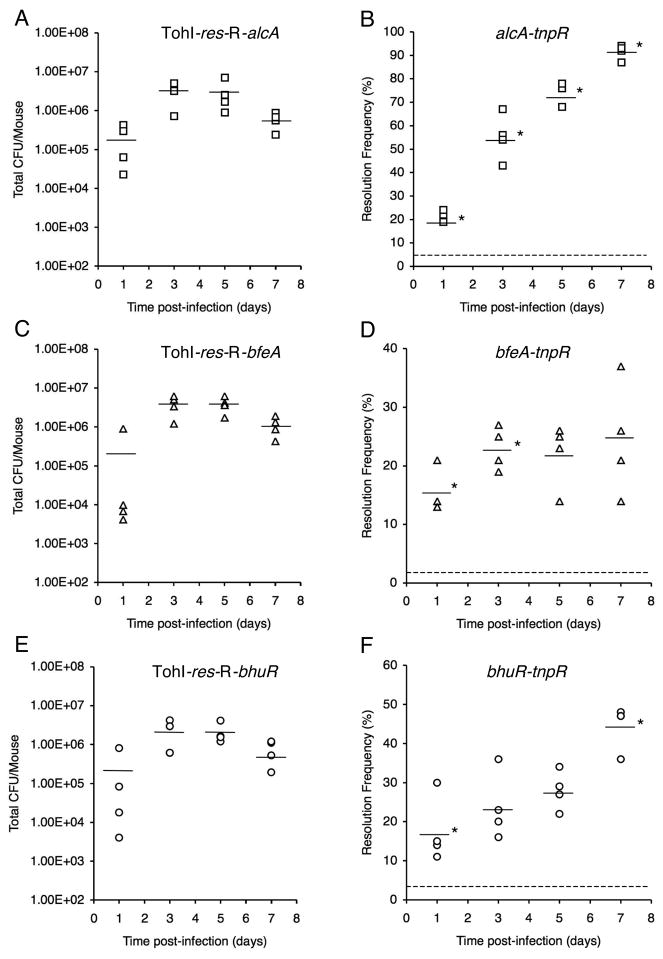
The total CFU and basal resolution frequencies for the mouse input inocula were determined to be 2.06 × 106 CFU (3.9% resolution frequency) for TohI-res-R-alcA, 2.15 × 106 CFU (1.9% resolution frequency) for TohI-res-R-bfeA, and 1.02 × 106 CFU (2.9% resolution frequency) for strain TohI-res-R-bhuR. Over the 7d course of the infection study, all three strains showed total CFU levels that are typical of B. pertussis Tohama I mouse respiratory infections (Fig. 2A, C, E). All fusion strains showed a significant increase in resolution by 1d post-infection compared with the base-line resolution frequencies of the input population; however, the temporal changes in resolution frequencies varied considerably among strains through day 7 (Fig. 2B, D, F). The alcA-tnpR alcaligin system promoter fusion strain resolved to 21% by day 1, further increased to 55% mean resolution by day 3, then displayed a progressive increase in resolution through day 5 to reach 91% by day 7. The changes in mean resolution frequencies were statistically significant for all consecutive sampling time intervals for the alcA-tnpR fusion strain (P values ranged from 0.001 to 0.047). The bhuR-tnpR heme system fusion strain resolved to 18% by day 1, displayed a limited increase to 24% and 28% resolution by days 3 and 5, but experienced a significant increase in resolution between days 5 and 7 (P = 0.022), achieving 44% resolution on day 7. The bfeA-tnpR enterobactin system fusion strain resolved to 16% by day 1, increased significantly again to 23% by day 3 (P = 0.013), held at 23% and 22% resolution at days 3 and 5, and reached only 25% by 7 days post-infection. The fold-change in resolution relative to the basal resolution frequency of the input population provides a numerical value we have termed the resolution index (Fig. 3). The resolution index is a useful measure for comparing temporal changes in iron system gene expression, normalized for differences in basal transcriptional activity and intrinsic promoter strength. The resolution index increased steadily for the alcA-tnpR strain over the entire 7d experiment, whereas the resolution index values for the bfeA-tnpR strain increased during the first day, and between days 1 and 3, but were maximal at day 3. The bhuR-tnpR strain showed a significant increase in resolution frequency during the first day, then did not increase significantly until between days 5 and 7.
Fig. 2.
Total CFU (A, C, E) and TnpR-mediated resolution frequencies (B, D, F) at various times post-infection for RIVET strains carrying the indicated fusion genes. Bars represent the mean total CFU/mouse or mean resolution frequencies (n = 4 mice per time point), and each symbol represents the total CFU or the resolution frequency for the B. pertussis population recovered from an individual mouse. An asterisk indicates that the mean resolution frequency differed significantly from the mean at the preceding time point. The dashed lines in panels B, D, and F correspond to the base-line resolution frequencies of the inocula.
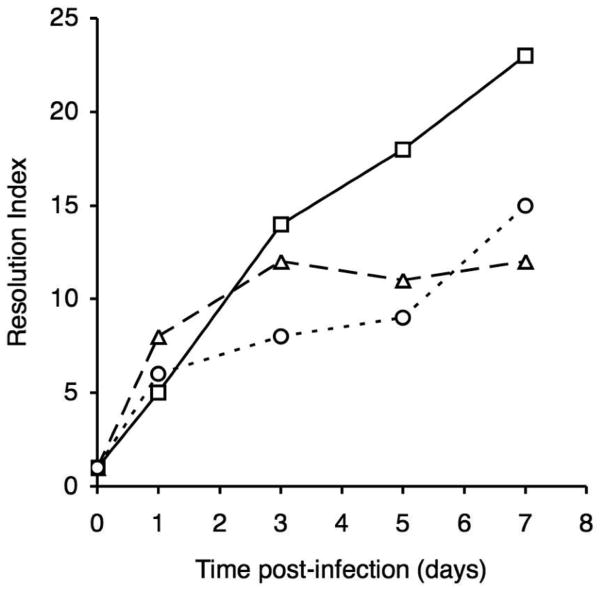
Fig. 3.
Resolution index values for RIVET strains at various times post-infection. Values represent the fold-change in mean resolution frequency compared with the base-line resolution frequency determined for the input inocula. Strains: TohI-res-R-alcA (squares), TohI-res-R-bfeA (triangles), and TohI-res-R-bhuR (circles).
Most notably, the timing of alcaligin and heme iron system induction by RIVET analysis approximated the phasing of changes in competitive indices (CIs) that were observed in previous competition infection studies in mice (Brickman et al., 2006; Brickman and Armstrong, 2007). Comparison of the relative fitness of B. pertussis wild-type and fauA alcaligin receptor mutant strains revealed the attenuating effect of the mutation early in infection. Concordantly, in the current RIVET analysis alcA transcription was significantly elevated over repressed expression levels by 1d post-infection, and was heightened to an induced level by as early as 3 days post-infection. In contrast, previous infection studies found that a bhuR heme receptor mutant showed a competitive defect only later in infection. The delay in CI reduction until after the peak of infection indicated that heme utilization contributes little to initial survival and multiplication of B. pertussis. This would imply that the heme resource or host condition responsible for the competitive exclusion of the bhuR mutant was unavailable or inactive in the host niche at the initiation of infection, but was made accessible during more advanced stages of infection, presumably by the action of Bordetella toxic factors on the host. The delayed induction of bhuR transcription seen in RIVET analyses is consistent with the hypothesis that heme is not prevalent in the host niche initially, but becomes available to B. pertussis later in infection. Early expression of the bfeA-tnpR fusion in RIVET analysis indicates that enterobactin or another catechol inducer (Anderson and Armstrong, 2006; Anderson and Armstrong, 2008) is present in the host niche early in infection.
All RIVET strains resolved to a significant degree over the input population, confirming that all three iron system promoters were active in vivo. Furthermore, the strains resolved to different levels and with different kinetics, indicating that the iron system promoters were differentially expressed in vivo. Temporal changes in iron system gene expression by RIVET could be correlated with the infection stage during which the iron systems appear to have their greatest impact on in vivo fitness. The moderate but significant increase in resolution frequencies for all fusion strains by 1d post-infection likely reflects the initial adaptation of B. pertussis to iron starvation in the host environment, causing relief of Fur repression with concomitant increases in iron system promoter activities. Since in vitro studies have established that maximal transcription from the alcA, bfeA, and bhuR promoters requires induction and activation subsequent to derepression, it is likely that the increased resolution frequencies observed after day 1 reflect the actions of the system-specific positive regulators AlcR, BfeR, and HurI in response to cognate iron source availability.
BfeA system function contributes to in vivo fitness of B. pertussis in mice
The resolution frequencies for the enterobactin system RIVET strain rose significantly during the first 3 days of infection but did not increase thereafter. The 25% resolution frequency at day 7 was relatively low compared with that of the alcA-tnpR and bhuR-tnpR strains. This lower level of resolution in the bfeA-tnpR strain may reflect differences in induction, or differences in intrinsic promoter strength under conditions that prevail in the infected mouse respiratory tract. Nonetheless, under conditions known to maximize transcriptional activity of bfeA, the same bfeA-tnpR fusion strain resolved to 91% in vitro. Since bfeA-tnpR expression was relatively low in vivo, mixed infection competition experiments were performed to evaluate the contribution of the enterobactin utilization system to multiplication and survival in the mouse respiratory infection model. In vivo fitness of B. pertussis bfeA enterobactin receptor mutant strain PM19 was determined relative to the wild-type parent strain UT25Sm1. The UT25Sm1 genetic background was used instead of the Tohama I background used in RIVET experiments to allow direct comparison with previously published mixed infection competition studies of fauA (Brickman and Armstrong, 2007) and bhuR (Brickman et al., 2006) iron receptor mutants.
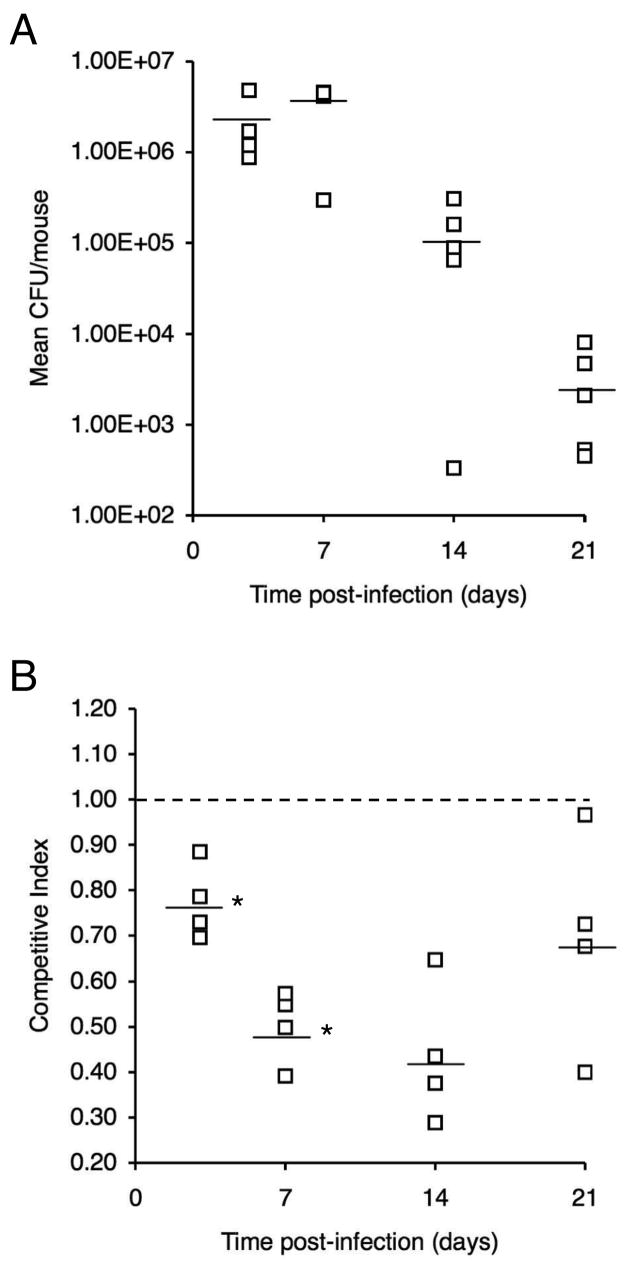
In growth competition studies in vitro, enterobactin receptor mutant PM19 and UT25Sm1 showed no significant difference in relative fitness when co-cultured for 48 h in iron-replete SS. In contrast, the wild-type strain had a distinct competitive advantage over the bfeA mutant when co-cultured in iron-depleted SS supplemented with ferric enterobactin as the sole source of iron; the mean CI value was 0.17 ± 0.06 (mean ± SD, n = 3). For in vivo competition studies, mice were infected intranasally with a 1:1 mixed strain suspension of UT25Sm1 and PM19, and total CFU (Fig. 4A) and competitive index values (CI) (Fig. 4B) were determined for the B. pertussis populations recovered in respiratory tract tissues at 3d, 7d, 14d, and 21d post-infection. At 3 days post-infection, the mean CI value had decreased significantly from the reference value of 1.00 to 0.77 (P < 0.05) indicating an early fitness cost associated with inactivation of the enterobactin utilization system. The mean CI decreased further to 0.50 by day 7, which constitutes a significant reduction from the mean CI at day 3 (P < 0.05). At subsequent time points, no additional significant change in mean CI was observed, but the mean CI remained lower than the reference value of 1.00 (mean CI = 0.44 and 0.69 at days 14 and 21, respectively; P < 0.05).
Fig. 4.
Relative in vivo fitness of a B. pertussis enterobactin receptor mutant during primary infection in mice. Mice were infected intranasally with a 1:1 mixture of wild-type strain UT25Sm1 and enterobactin receptor mutant strain PM19. (A) Total CFU over the course of the infection experiment. (B) Competitive index values were calculated as the mutant/wild-type CFU ratio in the output recovered at each time point divided by the mutant/wild-type CFU ratio in the input inoculum. CI values are shown relative to the reference CI of 1.00 (dashed line). Bars represent mean values, and each symbol represents the total CFU or CI value determined for an individual mouse. An asterisk indicates that the mean CI differed significantly from the mean at the preceding time point.
As with the alcaligin and heme utilization systems of B. pertussis, there is general agreement between the stage of infection at which the enterobactin system appears to be most important for in vivo fitness by the mixed infection competition approach, and the stage associated with a significant increase in enterobactin system transcriptional activity by RIVET. Further, the competition infection results confirm that although bfeA transcriptional activity was insufficient to drive TnpR production to levels necessary for high-frequency resolution in the RIVET strain, BfeA function significantly favors the in vivo growth and survival of B. pertussis during primary infection of the mouse respiratory tract, and is most important under conditions that exist in the host environment at the early stage of infection.
The mean CI values for the enterobactin system study (Fig. 4) and from similar, previously published infection studies of the alcaligin and heme systems were used to estimate the relative importance of the three iron systems for in vivo fitness of B. pertussis in the mouse respiratory infection model (Fig. 5). It is reasonable to hypothesize that the host niche conditions that influenced competition of the B. pertussis strains during these co-infection studies were related to limiting iron resources since co-infecting strains differed only in their production of iron receptors. Given that proposition, a value we have termed the fitness index (FI, calculated as 1 - mean CI) provides a measure of the relative in vivo importance or fitness contribution of a particular iron system for multiplication and survival of B. pertussis under the existing host niche conditions. These data support the hypothesis that the alcaligin system has a crucial role in iron assimilation both in initial colonization and during the entire infection process. The enterobactin system also contributes significantly to B. pertussis growth at the initial stage of infection but to a lesser extent than the alcaligin system, and its role is diminished after the peak of bacterial multiplication. Conversely, the heme system appears to be dispensable for growth until after B. pertussis has reached its maximum growth level, at which time it is essential for iron acquisition.
Fig. 5.
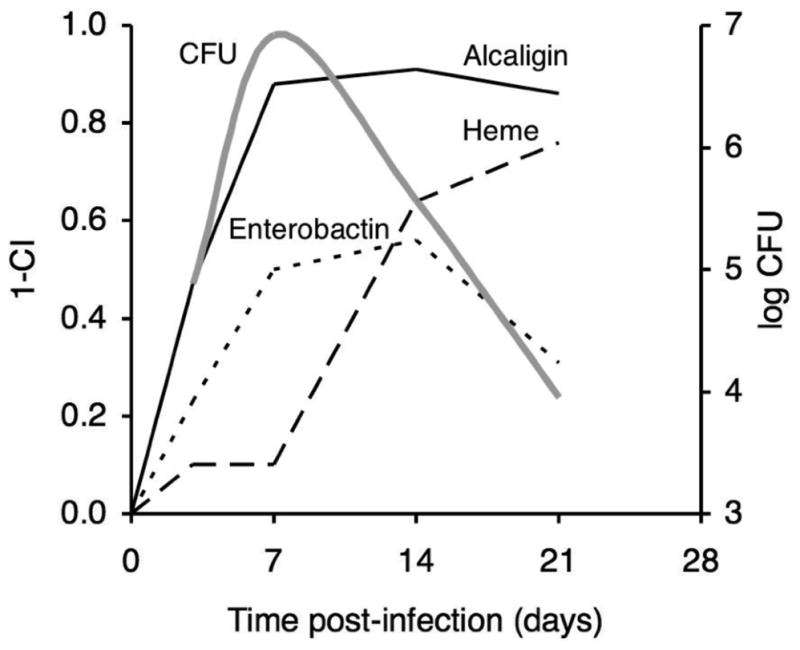
Relative importance of iron systems for in vivo fitness of B. pertussis at various phases of infection. Mean competitive index values from mixed infection competition experiments involving co-infection of mice with wild-type and alcaligin (Brickman and Armstrong, 2007), enterobactin (see Fig. 4), or heme (Brickman et al., 2006) receptor mutant strains were used to calculate a fitness index value (FI, calculated as 1 − mean CI), at various times post-infection. The FI provides a measure of the relative in vivo importance or fitness contribution an iron system for multiplication and survival of B. pertussis under the existing host niche conditions. Solid line, alcaligin system; dotted line, enterobactin system; dashed line, heme system. A plot (grey line) showing CFU levels that are typical of B. pertussis mouse respiratory infections is overlaid for reference.
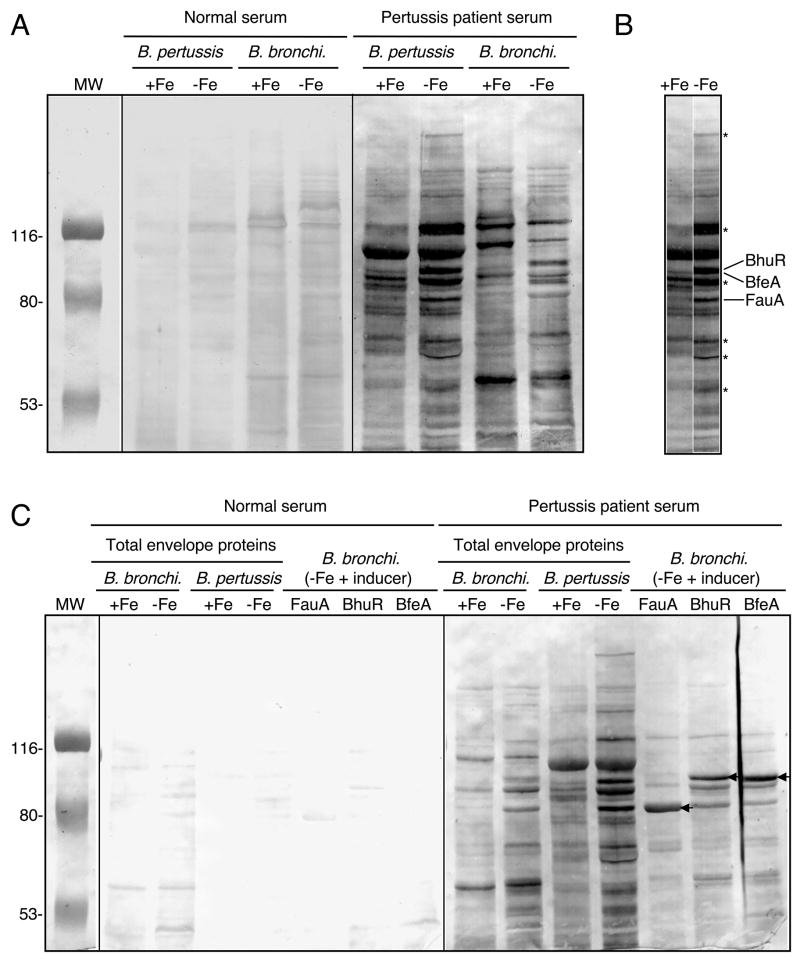
Pertussis patient sera recognize B. pertussis iron transport system receptor proteins
The RIVET experiments have determined that the B. pertussis alcaligin, enterobactin, and heme utilization systems are expressed in vivo, and competition studies established the importance of these iron systems individually for B. pertussis growth and survival during primary infection in mice. Since the mouse is not a natural host for B. pertussis and thus, has its limitations as an infection model, we investigated the expression of B. pertussis iron systems in the natural human host by screening human sera for antibody reactivity with the three iron source receptors. Representative immunoblot results are shown in Figure 6A and C. Cell envelope preparations from iron replete, iron-starved and iron source-induced Bordetella cells were probed blindly using a panel of human sera obtained from normal adult donors and from B. pertussis culture-positive donors (Mobberley-Schuman et al., 2003). Use of B. bronchiseptica mutant strains lacking fauA, bfeA, or bhuR (data not presented) and strains over-expressing those genes (Fig. 6C) aided identification of the three receptors in immunoblot analyses. Although this limited survey found no consistent pattern of serum reactivity with Bordetella envelope proteins, most sera from B. pertussis culture-positive donors (8 out of 11) reacted with multiple iron-repressible proteins from both Bordetella species, including FauA, BfeA, and BhuR (Fig. 6C), whereas 3 out of 10 serum samples from uninfected donors reacted strongly with these receptor proteins. It is possible that this serum reactivity could be due to antibodies against other TonB-dependent receptors that happen to crossreact with FauA, BfeA, and BhuR; however, this is not likely since it is the surface-exposed, substrate-specific receptor domains that are the most antigenic regions of these outer membrane receptors. In addition to the three known receptors, other iron-repressible proteins of unknown identity also reacted strongly with B. pertussis culture-positive donor sera (Fig. 6B).
Fig. 6.
Immunoblot reactivity of human sera with Bordetella iron-repressible envelope proteins. (A) Total envelope proteins prepared from B. pertussis UT25 and B. bronchiseptica B013N cultured under iron-replete (+Fe) and iron-depleted (−Fe) conditions were probed in parallel using sera from an uninfected human donor (normal serum) and a B. pertussis culture-positive human donor (pertussis patient serum). (B) Juxtaposition of B. pertussis sample lanes from panel A. Known positions of BhuR, BfeA and FauA are indicated, and asterisks designate positions of select iron-repressible envelope proteins. (C) The same antigens analyzed in panel A, and total envelope proteins from FauA, BhuR or BfeA receptor-overproducing B. bronchiseptica (B. bronchi.) strains were probed using serum from different donors than those in panel A. Positions of FauA, BhuR and BfeA are indicated by the arrows.
The presence of iron receptor-specific antibodies in pertussis patient sera is presumptive evidence that the proteins are produced by B. pertussis in response to the cognate iron source inducer during natural infection. It is unknown whether antibodies specific for known outer membrane iron receptors or other iron-repressible envelope proteins have any functional importance in protective immunity against B. pertussis infection. Since the alcaligin and enterobactin siderophore-dependent iron systems are important in the early stages of infection, and B. pertussis-infected humans appear to respond immunologically to the FauA and BfeA siderophore receptors, immunity to those receptors might inhibit colonization. Furthermore, BhuR-specific immunity could interfere with heme utilization and hasten clearance of B. pertussis from the host.
Experimental procedures
Bacterial strains and in vitro growth conditions
Bordetella pertussis strains Tohama I (Kasuga et al., 1954), UT25 (Field and Parker, 1979), UT25Sm1 (Brickman and Armstrong, 1996), and PM19 were grown on BG. Bordetella bronchiseptica B013N (Armstrong and Clements, 1993) and its derivatives were cultured on LB agar (Sambrook et al., 1989) or blood agar plates with selective antibiotics. SCMB was used as solid agar growth medium in B. pertussis RIVET experiments. To prepare SCMB, SS was supplemented with 66 μM iron, 0.5% powdered activated charcoal, 0.1% methylated β-cyclodextrin, 1% casamino acids, 0.15% bovine serum albumin, and 1.5% agar. Modified Stainer-Scholte medium (SS) (Schneider and Parker, 1982; Stainer and Scholte, 1970) was used as chemically defined broth culture medium for Bordetella cultures.
B. pertussis and B. bronchiseptica growth in SS was monitored as optical density using a spectrophotometer or a Klett-Summerson colorimeter equipped with a #54 filter (Klett Mfg. Co.). Escherichia coli DH5α (Bethesda Research Laboratories) was used as the host for routine plasmid propagation and DNA cloning procedures, and as the donor strain in tri-parental matings to transfer plasmids to Bordetella strains. E. coli strains were grown in Luria-Bertani (LB) broth or on LB agar plates. Antibiotics were used at the indicated final concentrations: ampicillin, 100 μg/ml; gentamicin, 5 μg/ml; kanamycin, 10 μg/ml; chloramphenicol, 30 μg/ml; and tetracycline, 15 μg/ml.
General genetic methods
Conjugal transfer of plasmids to Bordetella strains was performed using previously described methods (Brickman and Armstrong, 1996), and transformation of E. coli strains by electroporation used standard methods. General genetic techniques were performed essentially as described previously (Sambrook et al., 1989). DNA and protein sequence analysis used the Lasergene sequence analysis software system for the Macintosh PowerPC computer (DNASTAR, Inc.). Synthetic DNA oligonucleotides were purchased from Integrated DNA Technologies, and nucleotide sequencing was performed by the University of Minnesota BioMedical Genomics Center.
Recombinase fusion plasmid construction
TnpR fusion plasmids were constructed as described previously using plasmid vector pSS3110-TnpR (Veal-Carr and Stibitz, 2005). The alcA, bfeA and bhuR promoter regions were PCR-amplified from Tohama I genomic DNA templates using primer-adapters (Table 2) to add EcoRI and SalI restriction sites to the products’ termini. The EcoRI- and SalI-digested PCR products were ligated with EcoRI-SalI-digested pSS3110-TnpR plasmid DNA, and then the 3.2-kb NotI-PspOM1 res1-neo-res1 cassette of pRES1 was inserted at the unique NotI site of the resulting plasmids to produce the pSS3110-TnpR-res-alcA, -bfeA, and -bhuR promoter fusion plasmids. Cloned Bordetella promoter DNA fragments were confirmed by nucleotide sequencing. Nucleotide coordinates of Tohama I (The Wellcome Trust Sanger Institute, ftp://ftp.sanger.ac.uk/pub/pathogens/bp/) iron transport system promoter fragments cloned into pSS3110-TnpR for RIVET were alcA: 2597374 -> 2597813 (440 bp), bfeA: 3076704 -> 3077139 (436 bp), and bhuR: 351402 <- 350898 (505 bp).
Table 2.
Oligonucleotide primers used for PCR cloning of B. pertussis promoter regions and for genomic mapping of integrated tnpR fusion plasmids
| Name | Nucleotide sequence | Base positionsa |
|---|---|---|
| alcA-EcoRI | GGCCgaattcAGACATTGCAAGCCCCTGATb | 2597374..2597393 |
| alcA-SalI | GGCCgtcgacGTCCGATACCGATAGCCACGAA | 2597813..2597792 |
| bfeA-EcoRI | GGCCgaattcGCGCAGGCCGGGCTTGAGTTC | 3076704..3076724 |
| bfeA-SalI | GGCCgtcgacGGGGGTGGACATGCGGCTTCT | 3077139..3077119 |
| bhuR-EcoRI | GGCCgaattcCGCCGACAACCGCACCCACTC | 351402..351382 |
| bhuR-SalI | GGCCgtcgacGCCCTGATCGCAAGCGTAAACCAT | 350898..350921 |
| BP0771c | CCGCCCGCGCCGTAGTTCACCAC | 792956..792978 |
| 3110junc | CGCGGACCACAGCCCCATCACCAG | NAd |
Nucleotide coordinates of the B. pertussis Tohama I genome sequence (The Wellcome Trust Sanger Institute, ftp://ftp.sanger.ac.uk/pub/pathogens/bp/).
Nucleotides in lower case represent restriction enzyme recognition site adapters incorporated for cloning to pSS3110-TnpR. Underlined nucleotides are complementary to B. pertussis chromosomal DNA sequences.
complementary to the B. pertussis chromosomal site of pSS3110-TnpR integration
Not applicable, primer is complementary to the pSS3110-TnpR plasmid vector
RIVET strain construction
The pSS3110-TnpR-res-promoter plasmids were delivered to B. pertussis Tohama I by triparental mating using E. coli DH5α as the donor, with helper functions supplied by pRK2013. Exconjugants were selected on BG supplemented with selective antibiotics, 30 μM FeSO4, and crude colicin B, except for the bhuR-tnpR fusion strain, which was selected on SCMB instead of BG. Integration of recombinase fusion plasmids at the appropriate B. pertussis Tohama I chromosomal site was confirmed by PCR analysis of genomic DNA using primers BP0771 and 3110junc (Table 2), yielding the B. pertussis Tohama I derivative strains TohI-res-R-alcA, TohI-res-R-bfeA, and TohI-res-R-bhuR.
In vitro resolution assays
B. pertussis Tohama I derivative strains TohI-res-R-alcA and TohI-res-R-bfeA, carrying the integrated alcA-tnpR and bfeA-tnpR promoter fusion plasmids with their resolvase substrates, were cultured for 2 days on BG supplemented with selective antibiotics and 30 μM FeSO4. The bhuR-tnpR strain TohI-res-R-bhuR was cultured similarly, except using SCMB instead of BG to prevent induction of bhuR transcription by the heme present in BG, which contains defibrinated sheep blood. Plate growth was used to inoculate 10-ml SS cultures supplemented with selective antibiotics and 66 μM FeSO4 at an initial optical density of approximately 0.1 OD600. SS cultures of the bhuR-tnpR strain from SCMB growth were also supplemented with 1% casamino acids. After 18 to 24 h growth at 37°C, bacteria were washed using iron-free SS, and subcultured to 10-ml volumes of iron-replete and iron-depleted SS. In resolution assays with TohI-res-R-bfeA, 6-μM apo-enterobactin inducer was added to the iron-depleted culture at 15 h, and for TohI-res-R-bhuR, 1.25-μM bovine hemin chloride inducer was added to the iron-depleted culture at 8 h. The alcA promoter of the TohI-res-R-alcA strain responds to the autogenously produced inducer alcaligin, thus requires no inducer supplement. At various times, cultures were sampled for differential CFU determinations in triplicate on BG with kanamycin and gentamicin versus BG with gentamicin to determine resolution frequencies, except that TohI-res-R-bhuR CFU counts used heme-free SCMB instead of BG. Resolution frequencies (% KanS CFU) were calculated as .
RIVET analysis in mice
All research involving experimental animals was performed in accordance with federal guidelines and institutional policies. B. pertussis RIVET strains were cultured for 48 h at 37°C on SCMB with selective antibiotics. Bacterial growth was washed using SS supplemented with 1% casamino acids and 66 μM iron, and used to inoculate SS supplemented with 66 μM iron, 1% casamino acids, and gentamicin (5 μg/ml) plus kanamycin (10 μg/ml) at an initial cell density corresponding to an OD600 of 0.1. Cultures were grown at 37°C for 18 h, washed using cold phosphate-buffered saline (PBS), and resuspended in cold PBS to an OD600 of 0.1, which corresponds to approximately 2 × 108 CFU/ml. The total CFU in the inocula were enumerated by standard plate counting on SCMB agar (gentamicin, 5 μg/ml), and in parallel on BG agar to determine the relative plating efficiencies of B. pertussis on SCMB versus BG. Resolution frequencies in the inoculum suspensions were calculated as described for the in vitro resolution assays after differential CFU counting on SCMB.
Female BALB/cAnNHsd mice (10–20 g) (Harlan Sprague Dawley, Inc.) were mildly sedated by isoflurane inhalation and infected intranasally with ~2 × 106 CFU of a single strain in a 10-μl volume. At 1d, 3d, 5d, and 7d post-infection, 4 mice from each strain infection group were euthanized, respiratory tissues (lungs and trachea) were aseptically collected, and homogenates were immediately prepared in ice-cold SS liquid medium supplemented with 66 μM iron and 1% casamino acids. Homogenates were diluted in the same medium, and 100-μl volumes of undiluted homogenates and 10−1 to 10−4 10-fold serial dilutions were plated in parallel on SCMB medium with gentamicin versus SCMB medium with gentamicin plus kanamycin for total CFU, differential CFU counting, and determination of resolution frequencies as described for the in vitro resolution assays. For each strain infection group, Student’s t-test was used to determine whether any changes in mean resolution frequencies between consecutive time points were significant; probabilities (P) of ≤ 0.05 were considered significant.
Mixed infection competition experiments
To construct the B. pertussis enterobactin receptor mutant strain PM19, two non-contiguous segments of the B. pertussis UT25 bfeA region were PCR-amplified and joined by cloning into the allelic exchange vector pSS1129, generating a bfeA allele with a 920-nt deletion that was replaced by a HindIII restriction site. A kanamycin resistance cassette from plasmid pBSL86 (Alexeyev, 1995) was inserted at the HindIII site, and the ΔbfeA::kan allele was crossed to the chromosome of B. pertussis strain UT25Sm1 by allelic exchange.
Inocula for mixed infection competition experiments in mice were prepared as described previously (Brickman et al., 2006; Brickman and Armstrong, 2007). Wild-type B. pertussis strain UT25Sm1 and the isogenic ΔbfeA::kan enterobactin siderophore receptor mutant strain PM19 were subcultured from BG to separate SS cultures. After 36 h, bacteria were subcultured to SS medium containing 18 μM FeSO4 and grown for an additional 32 hours. Bacteria were washed, resuspended in PBS, and the two strain suspensions were combined at a 1:1 strain ratio to prepare a mixed strain suspension determined by CFU counting to have 7 × 107 CFU/ml.
Female BALB/cAnNHsd mice (10–20 g) (Harlan Sprague Dawley, Inc.) were mildly sedated by isoflurane inhalation, and infected with ~7 × 105 total CFU of a 1:1 mixture of UT25Sm1 and PM19 (10-μl volume instilled intranasally). At 3d, 7d, 14d, and 21d post-infection, 5 mice were euthanized, and respiratory tissue (lungs and trachea) homogenates were plated in the presence of appropriate antibiotics for differential enumeration of bacterial strains. The CI was calculated as the mutant/wild-type CFU ratio in the output recovered at each time point divided by the mutant/wild-type CFU ratio in the input inoculum. Each mean CI value is the mean of 4 to 5 independent mouse infections. Student’s t-test was used to determine whether the mean CI at each time point differed significantly from the hypothesized mean value of 1.00 (the predicted mean CI if there was no difference in fitness between the two strains), and from the mean CI at the preceding time point. Probabilities (P) of ≤ 0.05 were considered significant.
Immunoreactivity of pertussis patient sera with iron-regulated envelope proteins of B. pertussis
All research involving human subjects was performed in accordance with federal guidelines and institutional policies. Sera from B. pertussis culture-positive donors (aged 6 to 17 years old, previously vaccinated) and a control group of normal adult donors not known to have been infected with B. pertussis were analyzed by immunoblotting (Towbin et al., 1979) for antibody reactivity with Bordetella cell envelope proteins. Serum samples were provided by Dr. Alison Weiss, University of Cincinnati (Mobberley-Schuman et al., 2003), or were from our laboratory collection.
Total cell envelope proteins for immunoblot analysis were prepared from B. pertussis strain UT25 (Field and Parker, 1979) and B. bronchiseptica strain B013N (Armstrong and Clements, 1993) bacterial cells cultured in iron-replete and iron-depleted SS. Total cell envelope fractions were also prepared from B. bronchiseptica strains BRM18(pBB24) (Brickman and Armstrong, 1999), B013N(pBB32) (S. K. Armstrong, unpublished results), and BRM23(pRK37) (Vanderpool and Armstrong, 2003), which overproduce the iron receptor proteins FauA, BfeA, and BhuR, respectively. For BhuR and BfeA overproduction by B. bronchiseptica B013N(pBB32) and BRM23(pRK37), iron-depleted SS cultures were supplemented with the inducers, 1.25 μM hemin chloride or 3.25 μM enterobactin, respectively. FauA overproduction by BRM18(pBB24) was responsive to the autogenously produced inducer, alcaligin; thus, it needed no inducer supplement.
Bordetella cell fractionation for isolation of total cell envelopes used a modification of the Schnaitman method (Schnaitman, 1971). Bordetella cells were suspended in 50 mM N-2-hydroxyethylpiperazine- N9-2-ethanesulfonic acid (HEPES; pH 7.4), frozen and thawed, then disrupted using a French pressure cell (American Instrument Company). The pressates were centrifuged at 5000 × g at 4°C for 5 min to sediment unbroken cells, and the resulting supernatant fluids were centrifuged at 100,000 × g at 4°C for 1 h to obtain the insoluble cell fractions. The insoluble residue was washed twice using 10 mM Tris (pH 8)/1 mM EDTA buffer, then suspended in 50 mM HEPES buffer (pH 7.4) to yield the total cell envelope fraction.
For immunoblots, protein samples (adjusted to approximately 40 μg/lane) were separated by 0.1% sodium dodecyl sulfate-7.5% polyacrylamide gel electrophoresis and transferred to nitrocellulose membranes. Human sera were used at 1:400 dilution, and detection used goat anti-human IgGAM F(ab′)2 conjugated to horseradish peroxidase (1:2000) (Cappel) with 4-chloronapthol as the substrate.
Acknowledgments
We are grateful to Scott Stibitz and Wendy Veal-Carr for sharing RIVET plasmids pSS3110-TnpR, pSS3110-TnpR135, and pSS3110-TnpR168, for pRES1 originally obtained from Andrew Camilli, and for helpful discussions about RIVET studies in B. pertussis. We are thankful to Alison Weiss for providing samples of normal human sera and pertussis patient sera, and to Carin Vanderpool for help with interpretation of immunoblot results. This work was supported by Public Health Service grant AI-31088 from the National Institute of Allergy and Infectious Diseases.
References
- Anderson MT, Armstrong SK. The BfeR regulator mediates enterobactin-inducible expression of Bordetella enterobactin utilization genes. J Bacteriol. 2004;186:7302–7311. doi: 10.1128/JB.186.21.7302-7311.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson MT, Armstrong SK. The Bordetella bfe system: growth and transcriptional response to siderophores, catechols, and neuroendocrine catecholamines. J Bacteriol. 2006;188:5731–5740. doi: 10.1128/JB.00495-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson MT, Armstrong SK. Norepinephrine mediates acquisition of transferrin-iron in Bordetella bronchiseptica. J Bacteriol. 2008;190:3940–3947. doi: 10.1128/JB.00086-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong SK, Clements MO. Isolation and characterization of Bordetella bronchiseptica mutants deficient in siderophore activity. J Bacteriol. 1993;175:1144–1152. doi: 10.1128/jb.175.4.1144-1152.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaumont FC, Kang HY, Brickman TJ, Armstrong SK. Identification and characterization of alcR, a gene encoding an AraC-like regulator of alcaligin siderophore biosynthesis and transport in Bordetella pertussis and Bordetella bronchiseptica. J Bacteriol. 1998;180:862–870. doi: 10.1128/jb.180.4.862-870.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordet J, Gengou O. Le microbe de la coqueluche. Ann Inst Pasteur (Paris) 1906;20:731–741. [Google Scholar]
- Brickman TJ, Armstrong SK. The ornithine decarboxylase gene odc is required for alcaligin siderophore biosynthesis in Bordetella spp.: putrescine is a precursor of alcaligin. J Bacteriol. 1996;178:54–60. doi: 10.1128/jb.178.1.54-60.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brickman TJ, Kang HY, Armstrong SK. Transcriptional activation of Bordetella alcaligin siderophore genes requires the AlcR regulator with alcaligin as inducer. J Bacteriol. 2001;183:483–489. doi: 10.1128/JB.183.2.483-489.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brickman TJ, Vanderpool CK, Armstrong SK. Bordetella. Iron Transport in Bacteria. 2004:311–328. [Google Scholar]
- Brickman TJ, Vanderpool CK, Armstrong SK. Heme transport contributes to in vivo fitness of Bordetella pertussis during primary infection in mice. Infect Immun. 2006;74:1741–1744. doi: 10.1128/IAI.74.3.1741-1744.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brickman TJ, Armstrong SK. Impact of alcaligin siderophore utilization on in vivo growth of Bordetella pertussis. Infect Immun. 2007;75:5305–5312. doi: 10.1128/IAI.00849-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brickman TJ, Anderson MT, Armstrong SK. Bordetella iron transport and virulence. Biometals. 2007;20:303–322. doi: 10.1007/s10534-006-9031-1. [DOI] [PubMed] [Google Scholar]
- Camilli A, Beattie DT, Mekalanos JJ. Use of genetic recombination as a reporter of gene expression. Proc Natl Acad Sci U S A. 1994;91:2634–2638. doi: 10.1073/pnas.91.7.2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field LH, Parker CD. Differences observed between fresh isolates of Bordetella pertussis and their laboratory-passaged derivatives. In: Manclark CR, Hill JC, editors. International Symposium on Pertussis. Washington, D.C.: U.S. Dept. of Health, Education and Welfare, Public Health Service; 1979. pp. 124–132. [Google Scholar]
- Kang HY, Armstrong SK. Transcriptional analysis of the Bordetella alcaligin siderophore biosynthesis operon. J Bacteriol. 1998;180:855–861. doi: 10.1128/jb.180.4.855-861.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasuga T, Nakase Y, Ukishima K, Takatsu K. Studies on Haemophilus pertussis. V. Relation between the phase of bacilli and the progress of the whooping-cough. Kitasato Arch Exp Med. 1954;27:57–62. [PubMed] [Google Scholar]
- Lee SH, Hava DL, Waldor MK, Camilli A. Regulation and temporal expression patterns of Vibrio cholerae virulence genes during infection. Cell. 1999;99:625–634. doi: 10.1016/s0092-8674(00)81551-2. [DOI] [PubMed] [Google Scholar]
- Mahan MJ, Heithoff DM, Sinsheimer RL, Low DA. Assessment of bacterial pathogenesis by analysis of gene expression in the host. Annu Rev Genet. 2000;34:139–164. doi: 10.1146/annurev.genet.34.1.139. [DOI] [PubMed] [Google Scholar]
- Mobberley-Schuman PS, Connelly B, Weiss AA. Phagocytosis of Bordetella pertussis incubated with convalescent serum. J Infect Dis. 2003;187:1646–1653. doi: 10.1086/374741. [DOI] [PubMed] [Google Scholar]
- Parkhill J, Sebaihia M, Preston A, Murphy LD, Thomson N, Harris DE, et al. Comparative analysis of the genome sequences of Bordetella pertussis, Bordetella parapertussis and Bordetella bronchiseptica. Nat Genet. 2003;35:32–40. doi: 10.1038/ng1227. [DOI] [PubMed] [Google Scholar]
- Persson CG, Erjefalt I, Alkner U, Baumgarten C, Greiff L, Gustafsson B, Luts A, Pipkorn U, Sundler F, Svensson C, et al. Plasma exudation as a first line respiratory mucosal defence. Clin Exp Allergy. 1991;21:17–24. doi: 10.1111/j.1365-2222.1991.tb00799.x. [DOI] [PubMed] [Google Scholar]
- Pradel E, Guiso N, Locht C. Identification of AlcR, an AraC-type regulator of alcaligin siderophore synthesis in Bordetella bronchiseptica and Bordetella pertussis. J Bacteriol. 1998;180:871–880. doi: 10.1128/jb.180.4.871-880.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning : a laboratory manual. Cold Spring Harbor, N.Y.: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- Schnaitman CA. Solubilization of the cytoplasmic membrane of Escherichia coli by Triton X-100. J Bacteriol. 1971;108:545–552. doi: 10.1128/jb.108.1.545-552.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider DR, Parker CD. Effect of pyridines on phenotypic properties of Bordetella pertussis. Infect Immun. 1982;38:548–553. doi: 10.1128/iai.38.2.548-553.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stainer DW, Scholte MJ. A simple chemically defined medium for the production of phase I Bordetella pertussis. J Gen Microbiol. 1970;63:211–220. doi: 10.1099/00221287-63-2-211. [DOI] [PubMed] [Google Scholar]
- Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderpool CK, Armstrong SK. The Bordetella bhu locus is required for heme iron utilization. J Bacteriol. 2001;183:4278–4287. doi: 10.1128/JB.183.14.4278-4287.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderpool CK, Armstrong SK. Heme-responsive transcriptional activation of Bordetella bhu genes. J Bacteriol. 2003;185:909–917. doi: 10.1128/JB.185.3.909-917.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderpool CK, Armstrong SK. Integration of environmental signals controls expression of Bordetella heme utilization genes. J Bacteriol. 2004;186:938–948. doi: 10.1128/JB.186.4.938-948.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veal-Carr WL, Stibitz S. Demonstration of differential virulence gene promoter activation in vivo in Bordetella pertussis using RIVET. Mol Microbiol. 2005;55:788–798. doi: 10.1111/j.1365-2958.2004.04418.x. [DOI] [PubMed] [Google Scholar]