Abstract
The tobacco-specific nitrosamine 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK) is a potent pulmonary carcinogen in rats and is believed to be one cause of lung cancer in smokers. NNK is metabolized to 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol (NNAL) which is also a strong lung carcinogen in rats and has a chiral center at its 1-carbon. Previous studies have demonstrated that cytochrome P450-catalyzed α-hydroxylation of NNK in the lung leading to the formation of methyl and pyridyloxobutyl (POB)-DNA adducts is critical for its carcinogenicity. α-Hydroxylation of NNAL would similarly produce pyridylhydroxybutyl (PHB)-DNA adducts, but these have not been previously investigated in vivo. POB- and PHB-DNA adduct levels can indicate the amounts of pyridyloxobutylating and pyridylhydroxybutylating agents present in tissues of NNK or NNAL treated rats at any given point. Therefore, in this study, we developed a sensitive and quantitative liquid chromatography-electrospray ionization-tandem mass spectrometry-selected reaction monitoring method to determine levels of the PHB-DNA adducts O6-[4-(3-pyridyl)-4-hydroxybut-1-yl]-2′-deoxyguanosine (O6-PHB-dGuo, 10b), O2-[4-(3-pyridyl)-4-hydroxybut-1-yl]thymidine (O2-PHB-dThd, 11b), and 7-[4-(3-pyridyl)-4-hydroxybut-1-yl]-2′-deoxyguanosine (7-PHB-dGuo, 12b), the latter as the corresponding base 7-[4-(3-pyridyl)-4-hydroxybut-1-yl]-Gua (7-PHB-Gua, 14b) in DNA isolated from liver and lung of rats treated with 10 ppm NNK, (S)-NNAL, or (R)-NNAL in the drinking water for 20 weeks, and sacrificed at 1, 2, 5, 10, 16, and 20 weeks. PHB-DNA adduct levels were higher in lung than in liver at each time point, consistent with previous studies of POB-DNA adducts in rats treated with NNK and NNAL in the drinking water. The results showed that NNK and (S)-NNAL behaved in a similar fashion while (R)-NNAL was strikingly different. In the rats treated with NNK or (S)-NNAL, levels of each adduct at each time point were remarkably similar in lung, and levels of O2-PHB-dThd were generally > than 7-PHB-Gua > O6-PHB-dGuo. The highest PHB-DNA adduct levels were found in lung and liver of rats treated with (R)-NNAL, suggesting that there are cytochrome P450s that efficiently catalyze the α-methyl hydroxylation of this compound. The results of this study provide further support for our hypothesis that (S)-NNAL is rapidly formed from NNK, sequestered at an unknown site in the lung, then released and reoxidized to NNK with consequent DNA adduct formation resulting in lung carcinogenicity.
Keywords: Pyridylhydroxybutyl DNA adducts, 4-(Methylnitrosamino)-1-(3-pyridyl)-1-butanone, 4-(Methylnitrosamino)-1-(3-pyridyl)-1-butanol
Introduction
There are over 3000 deaths per day in the world from lung cancer, the leading cause of cancer death. Ninety percent of this incredible toll results from cigarette smoking (1). There are multiple lung carcinogens in cigarette smoke, including polycyclic aromatic hydrocarbons, 1,3-butadiene, metals, and the tobacco-specific nitrosamine 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK) (2). There is little doubt that these carcinogens are collectively responsible for lung cancer in smokers. An understanding of the mechanisms by which they cause cancer can lead to rational approaches to lung cancer prevention and possibly to improved therapy.
This study focuses on NNK (1, Chart 1), an effective pulmonary carcinogen in every species tested (3,4). NNK induces lung adenoma and adenocarcinoma independent of its route of administration, and is a particularly strong pulmonary carcinogen in the rat. A total dose of only 6 mg/kg NNK, administered by s.c. injection over a period of twenty weeks, induced a significant incidence of lung tumors (5). NNK given in the drinking water to rats at a concentration of 1 ppm for 105 weeks caused a significant incidence of lung tumors, and similar treatment of rats with 5 ppm of NNK or its metabolite 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol (NNAL, 2, Chart 1) induced lung tumors in more than 85% of the rats (6). A smoker is exposed to an estimated 0.5 mg NNK per kg body weight in 30 years of smoking (7). Parallels in NNK exposure and mechanisms between rodents and humans led the International Agency for Research on Cancer to classify NNK and the related carcinogen N’-nitrosonornicotine as “carcinogenic to humans (7).”
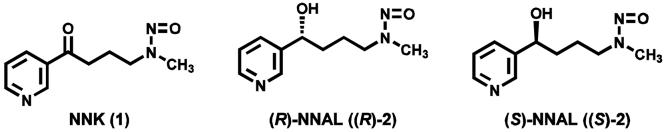
Chart 1.
Structures of NNK, (R)-NNAL and (S)-NNAL.
NNK belongs to the vast class of nitrosamine carcinogens. Volumes of literature convincingly demonstrate that virtually all nitrosamines require metabolic activation by cytochrome P450 catalyzed α-hydroxylation to exert their carcinogenic effects (8,9). The resulting diazonium ions produce DNA adducts which cause mutations in oncogenes such as K-ras, leading to uncontrolled growth, genomic instability, and cancer (4,10,11). There is no doubt that the metabolic activation of NNK to DNA adducts in the lung is a critical determinant of NNK carcinogenesis. This has been demonstrated in many studies, the most recent of which elegantly showed that NNK-induced DNA adduct formation and lung tumorigenesis was significantly reduced in mice with the NADPH-P450 reductase gene deleted in a lung-specific fashion (3,12).
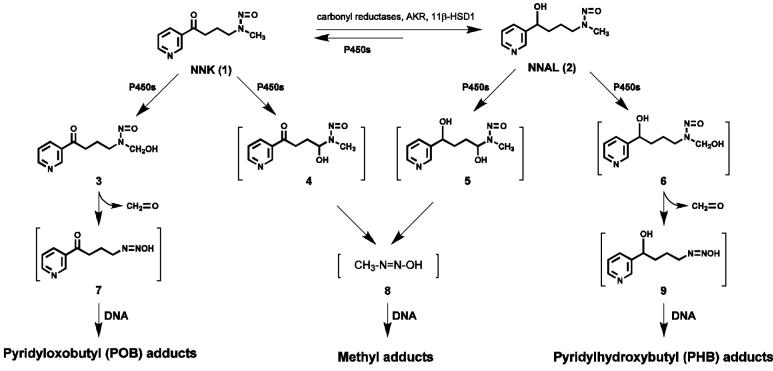
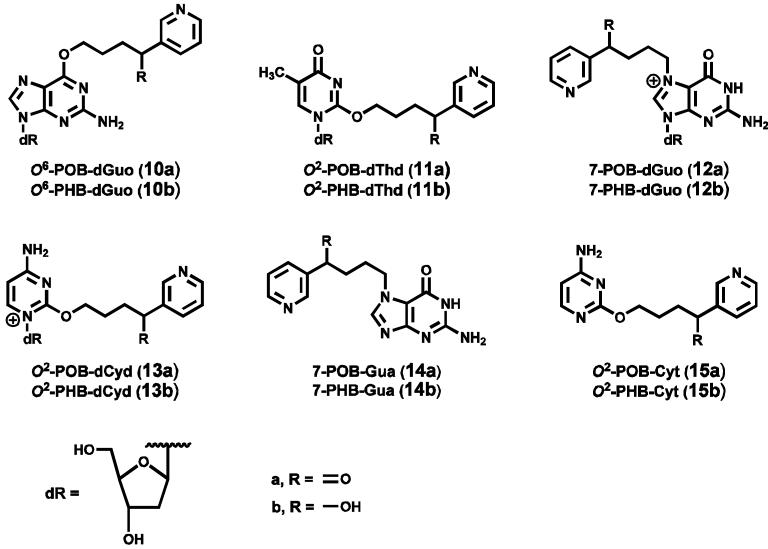
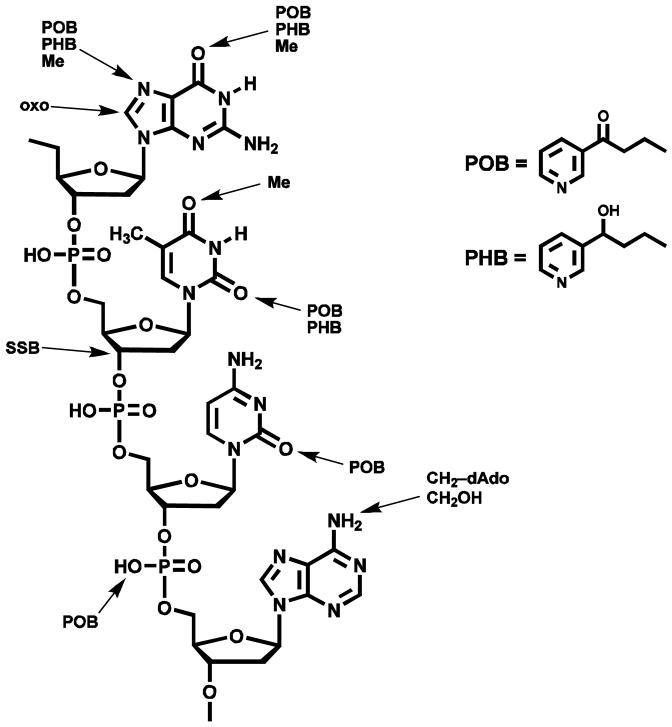
An overview of NNK and NNAL metabolic activation to DNA adducts is presented in Scheme 1. Cytochrome P450-catalyzed hydroxylation of the NNK methyl group gives 3, which spontaneously loses formaldehyde producing the pyridyloxobutanediazohydroxide 7. This intermediate or the corresponding diazonium ion alkylates DNA yielding pyridyloxobutyl (POB) adducts 10a - 13a (Chart 2)(13-17,17) as well as DNA adducts of formaldehyde (18,19). Hydroxylation of the NNK or NNAL α-methylene group produces intermediates 4 or 5, which decompose to methanediazohydroxide (8) and the methyl diazonium ion leading to the formation of methyl adducts in DNA (3). Hydroxylation of the NNAL methyl group yields intermediate 6 and diazohydroxide 9, resulting in the formation of the pyridylhydroxybutyl (PHB) adducts O6-[4-(3-pyridyl)-4-hydroxybut-1-yl]-2′-deoxyguanosine (O6-PHB-dGuo, 10b), O2-[4-(3-pyridyl)-4-hydroxybut-1-yl]thymidine (O2-PHB-dThd, 11b), 7-[4-(3-pyridyl)-4-hydroxybut-1-yl]-2′-deoxyguanosine (7-PHB-dGuo, 12b), and O2-[4-(3-pyridyl)-4-hydroxybut-1-yl]-2′-deoxycytidine (O2-PHB-dCyd, 13b) (Chart 2)(13,20).
Scheme 1.
Overview of NNK and NNAL metabolism and DNA adduct formation.
Chart 2.
Structures of POB-DNA and PHB-DNA adducts.
We have previously developed a sensitive and quantitative liquid chromatography-electrospray ionization-tandem mass spectrometry-selected reaction monitoring (LC-ESI-MS/MS-SRM) method to quantify POB-DNA adducts 10a - 13a (16). Adducts 12a and 13a, which can spontaneously lose deoxyribose, were determined as the bases 14a and 15a after neutral thermal hydrolysis. PHB adducts of NNK and NNAL have never been previously quantified. In this study, we developed and applied a method for quantitation of PHB adducts 10b - 12b, the latter as the base 7-[4-(3-pyridyl)-4-hydroxybut-1-yl]-Gua (7-PHB-Gua, 14b). Because of the low levels of O2-POB-dCyd(13a) (as the base 15a) observed in previous studies (15,16), O2-PHB-dCyd (13b) was not quantified here.
PHB adducts were measured in liver and lung DNA of rats treated chronically with NNK, (R)-NNAL, or (S)-NNAL in the drinking water under conditions which eventually produce lung tumors. PHB adduct levels were compared with those of POB adducts in the same animals, reported previously (15). The precursors to PHB and POB adducts, intermediates 7 and 9 (Scheme 1), are unstable and cannot be quantified directly. Measurement of PHB and POB adducts provides a snapshot of the relative amounts of intermediates 7 and 9 in liver and lung at any given point during NNK or NNAL treatment. This information, together with the total levels of PHB and POB adducts formed in liver and lung, can provide critical insights on mechanisms of DNA adduct formation and carcinogenicity by NNK and the enantiomers of NNAL.
Materials and Methods
Caution
NNK and NNAL are carcinogenic. They should be handled in a well-ventilated hood with extreme care and with personal protective equipment.
Chemicals
O6-PHB-dGuo (10b), O2-PHB-dThd (11b) and 7-PHB-Gua (14b) were prepared in high purity as previously described (13,14). [pyridine-D4]Ethyl nicotinate was purchased from Cambridge Isotope Laboratories (Andover, MA). NNK was obtained from Toronto Research Chemicals (Toronto, Canada). Purity was greater than 98%. (R)- and (S)-NNAL were synthesized (21). Chemical and enantiomeric purities were at least 99% (22). Micrococcal nuclease (LS004797, 15 kU) and phosphodiesterase II (LS003603, 10 U) were obtained from Worthington Biochemical Corporation (Lake- wood, NJ). Alkaline phosphatase (567752, 30 U/mL) was procured from Roche Molecular Biochemicals (Indianapolis, IN). The reagents and enzymes for DNA isolation were obtained from Gentra Systems (Minneapolis, MN). All other chemicals and solvents were acquired from Sigma-Aldrich Chemical Co. (Milwaukee, WI) or Fisher Scientific (Fairlawn, NJ).
NMR and MS
Qualitative MS was performed on an Agilent 1100LC/MSD ion trap instrument (Agilent Technologies, Inc., Palo Alto, CA) in the positive mode for characterization of deuterated standards. For the development, validation, and application of the quantitative method, we used either a Finnigan TSQ Quantum Ultra AM or a TSQ Quantum Discovery Max (Thermo Electron, San Jose, CA) instrument coupled with an Agilent 1100 series capillary HPLC system. NMR spectra were run on a Varian Inova 500 or 600 MHz instrument (Varian, Inc., Palo Alto, CA).
HPLC Analysis
HPLC was carried out with a Waters Associates (Milford, MA) system equipped with a model 440 UV-visible detector set at 254 nm. System 1, for purification of standards, used a 250 mm × 10 mm, 10 μm C-18 Vydac 201 TP column (Separations Group, Hesperia, CA) eluted at 3 mL/min with 5% CH3CN in 15 mM NH4OAc to 30% CH3CN over the course of 60 min. LC-ESI-MS for characterization of standards was carried out in System 2, with an Agilent 1100 series capillary flow HPLC ion trap MS (Agilent Technologies, Palo Alto, CA) equipped with a 150 mm × 0.5 mm Zorbax Extend-C-18, 3.5 μm column (Agilent) eluted at 15 μL/min with 5% CH3CN in 15 mM NH4OAc to 65 % CH3CN over the course of 40 min. N2 was used as drying gas (200 °C, 5L/min) and as nebulizing gas (15 psi). The system also contained an in-line UV detector. The mass spectrometer was operated in the full scan mode (m/z 100-800), with target ion abundance of 30,000, maximum accumulation time of 300 ms, and fragmentation amplitude of 0.9 V. MS/MS experiments were performed with a CID gas pressure of 1.5 mTorr and CID energy of 30 V.
Synthesis of Deuterated Standards
These were prepared based on previously published methods (13,20) and purified using HPLC system 1, as follows.
[pyridine-D4]O6-PHB-dGuo ([pyridine-D4]10b)
Retention time 15.5 min; purity 98% (HPLC-UV); isotopic purity, 98% D. 1H NMR (DMSO-d6) δ 8.1 (s, 1H, Gua C8-H), 6.2 (m, 1H, 1H’), 4.62 (br, 1H, but-4H), 4.4 (m, 2H, but-1-H), 4.23 (br, 1H, 3′H), 3.77 (br, 1H, 4′H), 3.6 (d, J=12.5, 1H, 5′Ha), 3.55 (d, J=12.5, 1H, 5′Hb), 2.16 (br, 2H, 2′H), 1.79-1.68 (m, 2H, but-3Ha,b), 1.69-1.60 (m, 2H, but-2Ha,b). UV (MeOH/pH 6.8 buffer λ max 204, 250, 282 nm. LC- positive ion ESI-MS (system 2) retention time 14.2 min, m/z 421 [M + H]+; MS/MS of m/z 421; m/z 305 [BH]+; m/z 154 [[pyridine-D4]CH(OH)C3H6]+.
[pyridine-D4]O2-PHB-dThd ([pyridine-D4]11b)
Retention time 15 min; purity 98% (HPLC-UV); isotopic purity, 98% D. 1H NMR (DMSO-d6) δ 7.8 (s, 1H, C-6H), 6.08 (t, J=6.0 Hz, 1H, 1′H), 4.6 (br, 1H, but-4-H), 4.3 (m, 2H, but-1-H), 4.2 (br, 1H, 3′H), 3.8 (br, 1H, 4′H), 3.6 (d, J=12.0 Hz, 1H, 5′Ha), 3.55 (d, J=12.5, 1H, 5′Hb), 2.15 (m, 2H, 2′H), 1.79-1.73 (m, 2H, but-3Ha,b), 1.77 (s, 3H, 5-CH3), 1.72 (m, 2H, but-2Ha,b). UV (MeOH/pH 6.8 buffer λ max 207, 258 nm. LC-positive ion ESI-MS (system 2) retention time 12 min, m/z 396 [M + H]+; MS/MS of m/z 396; m/z 280 [BH]+; m/z 154 [[pyridine-D4]CH(OH)C3H6]+.
[pyridine-D4]7-PHB-Gua ([pyridine-D4]14b)
Retention time 11.3 min; purity 98% (HPLC-UV); isotopic purity, 98% D. 1H NMR (DMSO-d6) δ 7.88 (s, 1H, GuaC8-H), 5.35 (s, 1H, but-4-OH), 4.55 (br, 1H, but-4H), 4.2 (m, 2H, but-1H), 1.8-1.7 (m, 2H, but-3Ha,b), 1.57-1.4 (m, 2H, but-2Ha,b). UV (MeOH/pH 6.8 buffer) λ max 214, 250, 285 nm. LC-positive ion ESI-MS (system 2) retention time 11.3 min, m/z 305 [M + H]+; MS/MS of m/z 305; m/z 287 [(M + H)+ - H2O]+; 154 [[pyridine-D4]CH(OH)C3H6]+.
A stock solution of [pyridine-D4]O6-PHB-dGuo (250 fmol/ μL), [pyridine-D4]O2-PHB-dThd (25 fmol/ μL), and [pyridine-D4]7-PHB-Gua (750 fmol/ μL) was prepared by NMR standardization with toluene as an internal standard, as previously described (17).
Animal Experiment
Liver and lung tissues analyzed in this study were those produced in a previous study in which rats were treated with NNK, (R)-NNAL, or (S)-NNAL (15). Briefly, the rats were randomly divided into four groups of 54 rats each: (1) control; (2) NNK; (3) (R)-NNAL; and (4) (S)-NNAL. The rats in the treatment groups received 10 ppm of the appropriate carcinogen in the drinking water, and the control rats were given tap water. Aqueous solutions of the carcinogens were prepared weekly and stored at 4 °C, conditions under which they are known to be stable. These solutions were placed in the plastic water bottles of the rat cages twice weekly. Nine rats per group were sacrificed by CO2 overdose at 1, 2, 5, 10, 16, and 20 weeks. Tissues were harvested and stored at -80 °C until DNA isolation.
Quantitation of PHB-DNA adducts by HPLC-ESI-MS/MS-SRM
DNA was isolated from the liver and lung of three rats per group as previously described (15). The analysis of PHB-DNA adducts by HPLC-ESI-MS/MS-SRM was carried out essentially as described previously (15,16). Briefly, 0.05-2 mg of each DNA sample plus 3 deuterated internal standards, was subjected to neutral thermal hydrolysis (100 °C, 30 min), then to enzymatic hydrolysis with micrococcal nuclease (75 U), phosphodiesterase II (450 mU), and alkaline phosphatase (150 U). The hydrolysate was purified on a solid-phase extraction cartridge (Strata-X-cartridge, Phenomenex, CA). The resulting sample was dissolved in 20 μL of 2% NH4OAc and 8 μL was injected on LC-MS using the same column as in System 2 with a gradient from 0 to 25% CH3CN in 15 mM NH4OAc over a period of 29 min, then 25-75% CH3CN for 5 min, then 75% CH3CN for 5 min, then returning to 0% CH3CN in 5 min, at a flow rate of 15 μL/min. The column was operated at 25 °C. The first 10 min of eluant was directed to waste, and the 10-40 min fraction was diverted to the ESI source operated in the positive mode. The retention times of the PHB-DNA adducts in this HPLC system were as follows: 7-PHB-Gua, 20.97-21.7 min; O2-PHB-dThd, 25.2-25.5 min; and O6-PHB-dGuo, 28.4-28.9 min. PHB-DNA adducts and corresponding deuterated standards were detected by monitoring the transition [M + 1]+ → [PHB]+ (Table 1). MS parameters were set as follows: spray voltage, 4 kV; sheath gas pressure, 30; capillary temperature, 200 °C; collision energy, 10 V; scan width, 0.7 amu; Q2 gas pressure, 1.0 mTorr; source CID, 10 V; and tube lens offset, 100 V. MS/MS data were acquired and processed by Xcalibur software version 1.4 (Thermo Electron). Quantitation of each PHB-DNA adduct was accomplished by comparing the MS peak area ratio of each adduct to that of its deuterated standard with a calibration curve. Calibration standards were prepared by mixing various quantities of each adduct with a constant amount of the corresponding internal standard in 2% NH4OAc. These were analyzed by LC-MS/MS without undergoing the sample preparation described above. Calibration curves were made by plotting the concentration ratio (adduct: internal standard) vs. the MS peak area ratio (adduct: internal standard). The amount of dGuo in each sample was determined by HPLC. The amount of DNA was then calculated based on 1 mg of DNA containing 3 μmol of nucleotides and rat DNA containing 22% dGuo. Adducts levels were expressed as fmol/mg DNA, mean ± S.D. of 3 liver or lung DNA samples per group, each analyzed once.
Table 1.
Ion transitions for SRM of PHB-DNA adducts and corresponding internal standards
| Adducts | Parent Ion ([M+1]+ , m/z) | Daughter Ion (m/z) |
|---|---|---|
| O6-PHB-dGuo (10b) | 417.1 | 150.1 [PHB]+ |
| [pyridine-D4]O6-PHB-dGuo | 421.1 | 154.1 ([pyridine-D4]PHB)+ |
| O2-PHB-dThd (11b) | 392.1 | 150.2 [PHB]+ |
| [pyridine-D4]O2-PHB-dThd | 396.1 | 154.2 ([pyridine-D4]PHB)+ |
| 7-PHB-Gua (14b) | 301.1 | 150.1 [PHB]+ |
| [pyridine-D4]7-PHB-Gua | 305.1 | 154.1 ([pyridine-D4]PHB)+ |
Statistical Analyses
Repeated measures ANOVA was chosen to compare the three PHB-DNA adducts within each group. Time was treated as a fixed effect. Due to a highly skewed distribution, the PHB-DNA adducts were analyzed on the natural log scale. Adjustments for multiple comparisons among PHB-DNA adducts were made according to the Tukey method. A p-value < 0.05 was considered to be statistically significant, except for comparisons of individual PHB-DNA adducts within each group at each time point, for which a more restrictive p-value cutoff of less than 0.01 was used to judge statistical significance.
Results
The accuracy of the method for analysis of PHB-DNA adducts was tested by adding various amounts of each adduct standard to 1 mg of calf-thymus DNA, and carrying out the analysis. The results, which are summarized in Table 2, demonstrate excellent agreement between measured and added amounts. Precision was determined by analyzing 6 aliquots of a sample containing 500 fmol/mg calf thymus DNA of O6-PHB-dGuo, 50 fmol/mg DNA of O2-PHB-dThd, and 1500 fmol/mg DNA of 7-PHB-Gua. Coefficients of variation were 1.8, 0.9, and 4.3%, respectively. Limits of detection (fmol/mg DNA) were: O6-PHB-dGuo (1.5), O2-PHB-dThd (3), and 7-PHB-Gua (18). Recoveries (% ± S.D.) were: O6-PHB-dGuo (90 ± 10), O2-PHB-dThd (60 ± 12), and 7-PHB-Gua (90 ± 10).
Table 2.
Accuracy of quantitation of PHB-DNA adductsa
| PHB-DNA Adduct (fmol) | |||||
|---|---|---|---|---|---|
| O6-PHB-dGuo | O2-PHB-dThd | 7-PHB-Gua | |||
| Added | Found | Added | Found | Added | Found |
| 50 | 51 | 5 | 8 | 150 | 159 |
| 100 | 100 | 10 | 12 | 300 | 344 |
| 150 | 149 | 15 | 13 | 450 | 471 |
| 200 | 195 | 20 | 16 | 600 | 584 |
| 250 | 248 | 25 | 28 | 750 | 752 |
| 300 | 294 | 30 | 34 | 900 | 812 |
| 350 | 358 | 35 | 35 | 1050 | 990 |
| 400 | 387 | 40 | 35 | 1200 | 1180 |
| 450 | 441 | 45 | 39 | 1350 | 1433 |
| 500 | 501 | 50 | 54 | 1500 | 1488 |
| R2 = 1.0 | R2 = 0.93 | R2 = 0.98 | |||
Standards were added to 1 mg of calf thymus DNA and the analysis performed as described in “Materials and Methods”
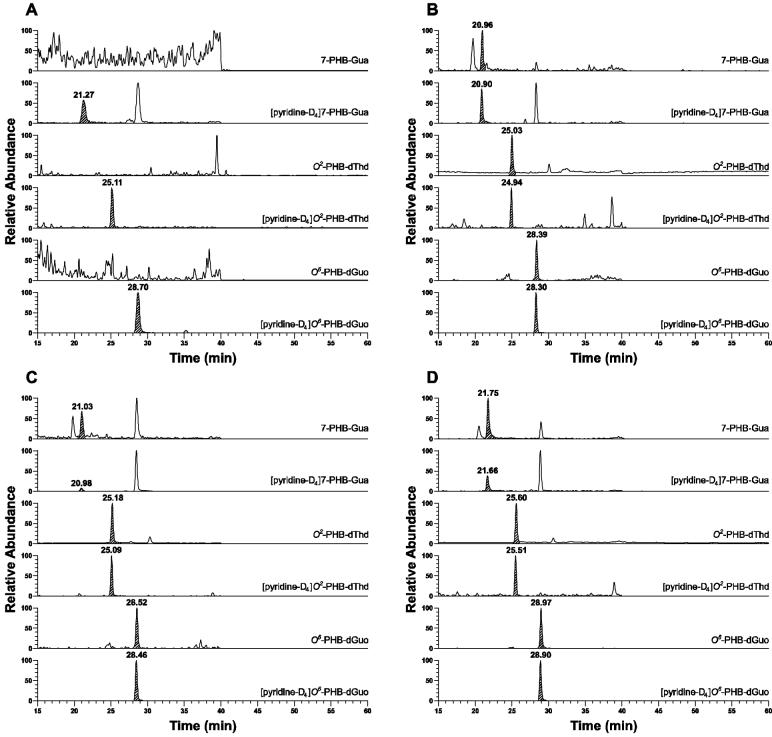
Representative chromatograms of PHB-DNA adducts in lung DNA of control rats and rats treated with NNK, (S)-NNAL, or (R)-NNAL for 16 weeks are shown in Figure 1A-D. Clear peaks co-eluting with the internal standards were observed for each adduct in the DNA of the treated rats and these were not present in lung DNA from control rats. Similar chromatograms were obtained from the liver samples in which PHB-DNA adducts were detected. Under our conditions, diastereomers of O6-PHB-dGuo and O2-PHB-dThd were not separated.
1.
Chromatograms obtained upon LC-ESI-MS/MS-SRM analysis of lung DNA from rats treated with (A) nothing; or with 10 ppm carcinogens in the drinking water for 16 weeks; (B) NNK; (C) (S)-NNAL; (D) (R)-NNAL. Individual PHB-DNA adducts or internal standards were monitored as indicated on each channel, using the transitions shown in Table 1. The DNA samples were obtained from a previous experiment (15). The peaks eluting at 28.5 min in the 7-PHB-Gua and [pyridine-D4]7-PHB-Gua channels are derived from transitions in the spectra of O6-PHB-dGuo and [pyridine-D4]O6-PHB-dGuo.
Levels of PHB-DNA adducts at each time point in lung and liver are summarized in Table 3. In the rats treated with NNK, all three adducts were detected in lung DNA at each time point. Amounts of O2-PHB-dThd were significantly (P<0.01) lower than those of 7-PHB-Gua at weeks 1 and 2, but significantly (P<0.01) higher at weeks 16 and 20. Both of these adducts were present in amounts significantly (P<0.01) greater than those of O6-PHB-dGuo at all time points. Adduct levels in liver were generally less than in lung, with O2-PHB-dThd predominating at most time points. O6-PHB-dGuo was not detected in liver DNA.
Table 3.
PHB-DNA adducts in lung and liver of rats treated with NNK, (S)-NNAL, or (R)-NNAL
| Compound/Adducts | Adduct levels (fmol/mg DNA) | |||||
|---|---|---|---|---|---|---|
| Weeks of treatmenta | ||||||
| A. Lung | 1 | 2 | 5 | 10 | 16 | 20 |
| NNK | ||||||
| O6-PHB-dGuo | 22 ± 6.4 | 35 ± 10.3 | 28 ± 9.7 | 40 ± 18 | 25 ± 7.5 | 15 ± 1.9 |
| O2-PHB-dThd | 71 ± 13 | 163 ± 69 | 277 ± 60 | 516 ± 255 | 789 ± 303 | 322 ± 95 |
| 7-PHB-Gua | 142 ± 57 | 255 ± 96 | 261 ± 98 | 354 ± 181 | 312 ± 65 | 147 ± 33 |
| (S)-NNAL | ||||||
| O6-PHB-dGuo | 21 ± 11 | 41 ± 18 | 24 ± 3.6 | 31 ± 17 | 35 ± 15 | 16 ± 3.8 |
| O2-PHB-dThd | 91 ± 32 | 258 ± 94 | 291 ± 82 | 540 ± 150 | 921 ± 420 | 394 ± 86 |
| 7-PHB-Gua | 148 ± 88 | 335 ± 157 | 218 ± 48 | 271 ± 112 | 334 ± 56 | 159 ± 56 |
| (R)-NNAL | ||||||
| O6-PHB-dGuo | 219 ± 54 | 223 ± 98 | 368 ± 98 | 425 ± 120 | 516 ± 37 | 455 ± 123 |
| O2-PHB-dThd | 721 ± 183 | 1310 ± 1240 | 3460 ± 1560 | 4970 ± 1360 | 9610 ± 58 | 8080 ± 1750 |
| 7-PHB-Gua | 1300 ± 330 | 1620 ± 1320 | 2260 ± 915 | 3670 ± 564 | 4390 ± 373 | 3770 ± 1020 |
| B. Liver | 1 | 2 | 5 | 10 | 16 | 20 |
|---|---|---|---|---|---|---|
| NNK | ||||||
| O6-PHB-dGuo | NDb | ND | ND | ND | ND | ND |
| O2-PHB-dThd | 33 ± 5 | 92 ± 71 | 38 ± 14 | 407 ± 104 | 431 ± 371 | 454 ± 554 |
| 7-PHB-Gua | 28 ± 3.6 | 53 ± 23 | 26 ± 12 | 241 ± 337 | ND | ND |
| (S)-NNAL | ||||||
| O6-PHB-dGuo | ND | ND | ND | ND | ND | ND |
| O2-PHB-dThd | 54.5 ± 20 | 403 ± 175 | 366 ± 275 | 213 ± 123 | 1960 ± 480 | 140 ± 13 |
| 7-PHB-Gua | ND | ND | ND | ND | ND | ND |
| (R)-NNAL | ||||||
| O6-PHB-dGuo | 14.6 ± 2.1 | 10.2 ± 6.1 | 7.3 ± 6.8 | ND | ND | ND |
| O2-PHB-dThd | 230 ± 76.4 | 471 ± 123 | 861 ± 613 | 1450 ± 929 | 1380 ± 1170 | 1570 ± 846 |
| 7-PHB-Gua | 437 ± 31 | 552 ± 202 | 571 ± 205 | 476 ± 117 | 551 ± 326 | 757 ± 423 |
Male F-344 rats were treated with NNK, (S)-NNAL, or (R)-NNAL at a concentration of 10 ppm in the drinking water for 20 weeks. Groups of 3 rats were sacrificed at the intervals shown. Further details are in reference (15). Tissue samples are from that study.
ND = not detected
Levels of each adduct in lung DNA of rats treated with (S)-NNAL were remarkably similar to those observed in the rats treated with NNK. Levels of O2-PHB-dThd were significantly greater than those of the other adducts at weeks 10-20, similar to the pattern seen in the NNK-treated animals. Levels of O2-PHB-dThd and 7-PHB-Gua in lung were significantly greater (P<0.01) than those of O6-PHB-dGuo at all time points, as in the NNK treated animals. In liver, O2-PHB-dThd was the only adduct detected.
The highest PHB-DNA adduct levels were clearly seen in the rats treated with (R)-NNAL. Total adduct levels were significantly higher (P<0.001) in lung and liver DNA of these rats compared to the DNA from rats treated with either NNK or (S)-NNAL, and they were higher in lung than in liver DNA, as seen in the NNK and (S)-NNAL-treated rats.
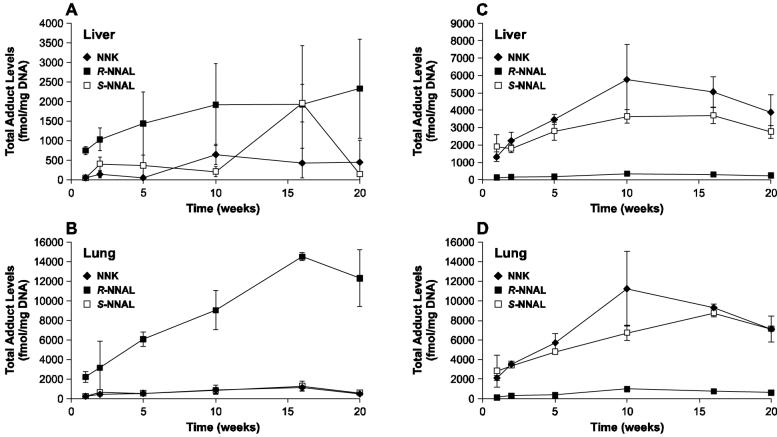
Comparative levels of total PHB-DNA and POB-DNA adducts (from our previous study (15)) in lung and liver of NNK, (R)-NNAL, and (S)-NNAL-treated rats are summarized in Figure 2A-D.
2.
Levels of total adducts (fmol/mg DNA) ± S.D. vs time (weeks): A) PHB-DNA adducts in liver; B) PHB-DNA adducts in lung; C) POB-DNA adducts in liver; D) POB-DNA adducts in lung. Panels C and D are included from a previous publication (15) for comparison.
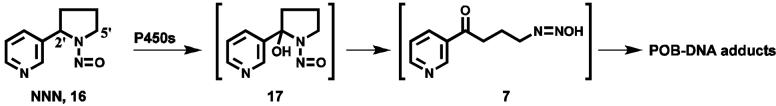
As a control for the possible interconversion of pyridyloxobutylating and pyridylhydroxybutylating agents or the corresponding DNA adducts, two DNA samples, one each from the livers of rats treated with either enantiomer of N’-nitrosonornicotine (NNN, 16, Scheme 2) in a previous study (23), were analyzed for PHB-DNA adducts. None were detected.
Scheme 2.
Formation of POB-DNA adducts from NNN.
Discussion
There were three major notable results of this study. First, we detected and quantified, for the first time in vivo, PHB-DNA adducts of NNK and NNAL. Second, there was a remarkable kinship between NNK and (S)-NNAL in levels of PHB-DNA adducts, particularly in lung, and similar to our previous observations regarding POB-DNA adducts of these two compounds, while the pattern of adduct levels from (R)-NNAL was very different. Third, (R)-NNAL produced significantly higher levels of PHB-DNA adducts than either NNK or (S)-NNAL.
The data presented here provide convincing evidence for the presence of PHB-DNA adducts in rats treated with NNK or the enantiomers of NNAL. Clean chromatograms, such as those illustrated in Figure 1, were obtained, and in each case where adducts were detected, peaks having the correct fragmentation pattern, as detected by SRM, co-eluted with the appropriate internal standard. These results leave no doubt that PHB-DNA adducts are formed in vivo as a consequence of methyl hydroxylation of NNAL, as illustrated in Scheme 1. As shown in Chart 3, 15 different types of DNA damage have now been identified in lung or liver of NNK treated rats or mice, resulting from pyridyloxobutylation of dGuo, dThd, dCyd, and phosphate, pyridylhydroxybutylation of dGuo and dThd, methylation of dGuo and dThd, and formaldehyde alkylation of dAdo (3,15,16,18,24). Single strand breaks (SSB) and 8-oxo-dGuo have also been reported (3). The formation of these multiple DNA adducts of NNK is certainly consistent with its potent carcinogenicity.
Chart 3.
DNA adducts formed in rats or mice treated with NNK.
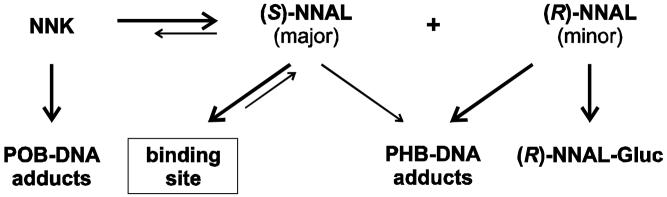
Total quantified PHB-DNA adduct levels in the lung of rats treated with NNK and (S)-NNAL were strikingly similar and very different from the results obtained with (R)-NNAL, echoing the results which we have previously obtained upon analysis of POB-DNA adducts in the same rats (Figure 2) (15). There were also similarities in liver. These data can be understood more clearly based on the results of previous studies. NNK is rapidly converted to NNAL in vivo in the rat, with a plasma half-life of 25 min (25). Studies carried out in vitro demonstrate that rat liver and lung microsomes and cytosol, as well as red blood cells, readily convert NNK to NNAL, which is >95% (S)-NNAL (26). In agreement with these findings, (S)-NNAL predominates in plasma upon administration of NNK to rats (27). Therefore, since NNK is efficiently converted to (S)-NNAL in rats, it is reasonable that after administration of (S)-NNAL to rats, as in the present study, one observes properties that are quite similar to those of NNK.
Our results are also consistent with those obtained by Zimmerman et al who compared the metabolism, pharmacokinetics, and tissue distribution of (S)-NNAL and (R)-NNAL in rats (28). (S)-NNAL had a much larger volume of distribution (Vss = 1792 ± 570 ml) than did (R)-NNAL (Vss = 645 ± 230 ml). These large Vss values suggest extensive tissue distribution of both compounds, but particularly of (S)-NNAL, which was also selectively retained in the lung. There were major differences in metabolism, with almost 40% of the (R)-NNAL dose being eliminated as (R)-NNAL-glucuronide, while only small amounts of (S)-NNAL-glucuronide were formed. Furthermore, (S)-NNAL was converted to NNK more extensively than (R)-NNAL, based on analysis of urinary metabolites [4-oxo-4-(3-pyridyl)butyric acid from (S)-NNAL via NNK, and 4-hydroxy-4-(3-pyridyl)butyric acid from (R)-NNAL] (28). Overall, NNK and (S)-NNAL behave as closely related siblings, while (R)-NNAL is a distant cousin.
Since the pyridyloxobutylating and pyridylhydroxybutylating intermediates 7 and 9 of Scheme 1 are unstable and are apparently not interconvertible (see below), POB-DNA adducts and PHB-DNA adducts provide a snapshot of their tissue concentrations at a given time. Thus, our results (Figure 2) indicate that the flux of these intermediates from NNK and (S)-NNAL are similar, and very different from (R)-NNAL, although adduct levels from all three compounds increased through 16 weeks of treatment. The finding of similar levels of POB-DNA adducts from NNK and (S)-NNAL thus clearly requires reconversion of (S)-NNAL to NNK. Yet, the biological equilibrium between NNK and (S)-NNAL is far to the side of (S)-NNAL in the rat (25,27). Furthermore, there appears to be relatively little α-methyl hydroxylation of (S)-NNAL in the rat in vivo (28), in spite of the fact that rat liver and lung microsomes catalyze this process in vitro (26). How do we resolve these apparent contradictions? We hypothesize that (S)-NNAL, after its rapid formation from NNK, is sequestered at receptor sites in rat lung and perhaps also in liver (see Scheme 3). While sequestered, it is protected from α-methyl hydroxylation. It may be released from this protected site by oxidation to NNK within the site (which could be an oxidoreductase or cytochrome P450), or it may be released from the site and then efficiently reconverted to NNK, a reaction that is catalyzed more effectively by cytochromes P450 and rat liver and lung microsomes for (S)-NNAL than for (R)-NNAL (26). This NNK then undergoes α-hydroxylation to POB-DNA adducts. These relationships are summarized in Scheme 3. This hypothesis is consistent with the previously observed selective retention of (S)-NNAL in the rat lung (29), and could explain in part the high pulmonary carcinogenicity of NNK in rats as well as the high levels of POB-DNA adducts in rat lung after administration of NNK. It is also consistent with previous studies which have demonstrated binding of NNK to nicotinic acetylcholine and β-adrenergic receptors, although there are no data reported for NNAL (30-32).
Scheme 3.
Proposed relationships of NNK, (S)-NNAL, (R)-NNAL, and their DNA adducts in vivo in the rat.
This hypothesis depends on the veracity of the assumption that pyridyloxobutylating intermediates 7 and 9, as well as POB-DNA and PHB-DNA adducts, are not interconvertible, so that the latter do really provide a snapshot of the former. Intermediates 7 and 9 and the consequent diazonium ions are expected to be short-lived, thus their interconversion seems unlikely, as does interconversion of the resulting DNA adducts. We examined this by analyzing PHB-DNA adducts in the liver of rats treated with NNN (16, Scheme 2) (23). NNN undergoes conversion to intermediate 7, but not 9, following metabolic 2′-hydroxylation, and POB-DNA adducts are present in tissues of rats treated with NNN. We did not detect PHB-DNA adducts in these samples, demonstrating that intermediate 7 is not converted to 9 in vivo, nor are POB-DNA adducts converted to PHB-DNA adducts. Thus, it is safe to assume that POB-DNA adducts result from pyridyloxobutylating species, while PHB-DNA adducts result from pyridylhydroxybutylating species, and these pathways are well insulated in vivo once the reactive intermediate is formed.
The extensive formation of PHB-DNA adducts from (R)-NNAL was unexpected. Levels of these adducts in lung were similar to those of the POB-DNA adducts produced from NNK and (S)-NNAL, and all adducts persisted throughout treatment. Rat lung microsomes were less able to catalyze the α-methyl hydroxylation of (R)-NNAL than (S)-NNAL, while liver microsomes catalyzed these reactions equally (26). Rat P450 2A3 was a less efficient catalyst of α-methyl hydroxylation of racemic NNAL than of NNK (33). In addition, (R)-NNAL is more readily detoxified by glucuronidation than is (S)-NNAL (28). These results suggested that PHB-DNA adduct formation from (R)-NNAL would be less than from (S)-NNAL or NNK in rat lung, but this was not observed. The higher levels of PHB-DNA adducts observed from (R)-NNAL than (S)-NNAL can be explained by the sequestration hypothesis discussed above, as α-methyl hydroxylation of (S)-NNAL would be inhibited. But the comparable levels in rat lung of PHB-DNA adducts from (R)-NNAL and POB-DNA adducts from NNK are not so readily explained in view of the documented differences in the rates of α-methyl hydroxylation of NNAL and NNK by P450 2A3, found in rat lung (33), and the detoxification of (R)-NNAL by glucuronidation. Apparently, there are P450s in rat lung and liver that efficiently catalyze the α-methyl hydroxylation of (R)-NNAL.
Consistent with their kinship, (S)-NNAL was equally as tumorigenic in A/J mouse lung as NNK, both having induced about 25 lung tumors per mouse, while (R)-NNAL was less tumorigenic but still produced 8.2 lung tumors per mouse, compared to 0.1 in the vehicle controls (21). Human exposure to NNAL is primarily through metabolic reduction of NNK, as there is relatively little NNAL in tobacco products (7). While (S)-NNAL was the major enantiomer formed in human liver cytosol, both enantiomers have been observed in substantial quantities in experiments with human liver microsomes, and (R)-NNAL predominated in human lung microsomes (26,34). The enantiomeric distribution of free NNAL in smokers’ urine was 54% (S)- and 46% (R)-NNAL, while that of NNAL glucuronides was 68% (S)- and 32% (R)-NNAL (22). Collectively, these results indicate that there may be significant exposure to both tumorigenic enantiomers of NNAL in humans, suggesting that PHB-DNA adducts may be formed in tandem with the POB-DNA adducts that have already been observed (35,36).
In summary, the results of this study demonstrate for the first time the formation of PHB-DNA adducts in lung and liver of rats treated with NNK and the enantiomers of NNAL. Adduct formation was similar from NNK and (S)-NNAL, supporting a mechanism of NNK carcinogenesis in which (S)-NNAL is sequestered, then re-oxidized to NNK, resulting in the formation of POB-DNA adducts. Surprisingly high levels of PHB-DNA adducts were found in rats treated with (R)-NNAL.
Acknowledgments
We thank Bruce Lindgren of the University of Minnesota Cancer Center Bioastatistics Core Facility for statistical analysis. Mass spectrometry was carried out in the Analytical Biochemistry Core Facility. The core facilities are supported in part by grant CA-77598 from the National Cancer Institute. Stephen S. Hecht is an American Cancer Society Research Professor, supported in part by ACS grant RP-00-138. This work was supported by grant CA-81301 from the National Cancer Institute. We thank Bob Carlson for editorial assistance.
References
- 1.International Agency for Research on Cancer . IARC Monographs on the Evaluation of Carcinogenic Risks to Humans. Vol. 83. IARC; Lyon, FR: 2004. Tobacco Smoke and Involuntary Smoking; pp. 1179–1187. [PMC free article] [PubMed] [Google Scholar]
- 2.International Agency for Research on Cancer . IARC Monographs on the Evaluation of Carcinogenic Risks to Humans. Vol. 83. IARC; Lyon, FR: 2004. Tobacco Smoke and Involuntary Smoking; pp. 53–119. [PMC free article] [PubMed] [Google Scholar]
- 3.Hecht SS. Biochemistry, biology, and carcinogenicity of tobacco-specific N-nitrosamines. Chem. Res. Toxicol. 1998;11:559–603. doi: 10.1021/tx980005y. [DOI] [PubMed] [Google Scholar]
- 4.Hecht SS. Progress and challenges in selected areas of tobacco carcinogenesis. Chem. Res. Toxicol. 2008;21:160–171. doi: 10.1021/tx7002068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Belinsky SA, Foley JF, White CM, Anderson MW, Maronpot RR. Dose-response relationship between O6-methylguanine formation in Clara cells and induction of pulmonary neoplasia in the rat by 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone. Cancer Res. 1990;50:3772–3780. [PubMed] [Google Scholar]
- 6.Rivenson A, Hoffmann D, Prokopczyk B, Amin S, Hecht SS. Induction of lung and exocrine pancreas tumors in F344 rats by tobacco-specific and Areca-derived N-nitrosamines. Cancer Res. 1988;48:6912–6917. [PubMed] [Google Scholar]
- 7.International Agency for Research on Cancer . IARC Monographs on the Evaluation of Carcinogenic Risks to Humans. Vol. 89. IARC; Lyon, FR: 2007. Smokeless tobacco and tobacco-specific nitrosamines. [PMC free article] [PubMed] [Google Scholar]
- 8.Preussmann R, Stewart BW. N-Nitroso Carcinogens. In: Searle CE, editor. Chemical Carcinogens, Second Edition, ACS Monograph 182. Vol. 2. American Chemical Society; Washington, DC: 1984. pp. 643–828. [Google Scholar]
- 9.Mirvish SS. Role of N-nitroso compounds (NOC) and N-nitrosation in etiology of gastric, esophageal, nasopharyngeal and bladder cancer and contribution to cancer of known exposures to NOC. Cancer Lett. 1995;93:17–48. doi: 10.1016/0304-3835(95)03786-V. [DOI] [PubMed] [Google Scholar]
- 10.Ronai Z, Gradia S, Peterson LA, Hecht SS. G to A transitions and G to T transversions in codon 12 of the Ki-ras oncogene isolated from mouse lung tumors induced by 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK) and related DNA methylating and pyridyloxobutylating agents. Carcinogenesis. 1993;14:2419–2422. doi: 10.1093/carcin/14.11.2419. [DOI] [PubMed] [Google Scholar]
- 11.Johnson L, Mercer K, Greenbaum D, Bronson RT, Crowley D, Tuveson DA, Jacks T. Somatic activation of the K-ras oncogene causes early onset lung cancer in mice. Nature. 2001;410:1111–1116. doi: 10.1038/35074129. [DOI] [PubMed] [Google Scholar]
- 12.Weng Y, Fang C, Turesky RJ, Behr M, Kaminsky LS, Ding X. Determination of the role of target tissue metabolism in lung carcinogenesis using conditional cytochrome P450 reductase-null mice. Cancer Res. 2007;67:7825–7832. doi: 10.1158/0008-5472.CAN-07-1006. [DOI] [PubMed] [Google Scholar]
- 13.Hecht SS, Villalta PW, Sturla SJ, Cheng G, Yu N, Upadhyaya P, Wang M. Identification of O2-substituted pyrimidine adducts formed in reactions of 4-(acetoxymethylnitrosamino)-1-(3-pyridyl)-1-1butanone and 4-(acetoxymethylnitrosamino)-1-(3-pyridyl)-1-butanol with DNA. Chem. Res. Toxicol. 2004;17:588–597. doi: 10.1021/tx034263t. [DOI] [PubMed] [Google Scholar]
- 14.Wang M, Cheng G, Sturla SJ, Shi Y, McIntee EJ, Villalta PW, Upadhyaya P, Hecht SS. Identification of adducts formed by pyridyloxobutylation of deoxyguanosine and DNA by 4-(acetoxymethylnitrosamino)-1-(3-pyridyl)-1-butanone, a chemically activated form of tobacco-specific carcinogens. Chem. Res. Toxicol. 2003;16:616–626. doi: 10.1021/tx034003b. [DOI] [PubMed] [Google Scholar]
- 15.Lao Y, Yu N, Kassie F, Villalta PW, Hecht SS. Formation and accumulation of pyridyloxobutyl DNA adducts in F344 rats chronically treated with 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone and enantiomers of its metabolite, 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol. Chem. Res. Toxicol. 2007;20:235–245. doi: 10.1021/tx060207r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lao Y, Villalta PW, Sturla SJ, Wang M, Hecht SS. Quantitation of pyridyloxobutyl DNA adducts of tobacco-specific nitrosamines in rat tissue DNA by high performance liquid chromatography-electrospray ionization-tandem mass spectrometry. Chem. Res. Toxicol. 2006;19:674–682. doi: 10.1021/tx050351x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sturla SJ, Scott J, Lao Y, Hecht SS, Villalta PW. Mass spectrometric analysis of relative levels of pyridyloxobutylation adducts formed in the reaction of DNA with a chemically activated form of the tobacco-specific carcinogen 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone. Chem. Res. Toxicol. 2005;18:1048–1055. doi: 10.1021/tx050028u. [DOI] [PubMed] [Google Scholar]
- 18.Wang M, Cheng G, Villalta PW, Hecht SS. Development of liquid chromatograpy electrospray ionization tandem mass spectrometry methods for analysis of DNA adducts of formaldehyde and their application to rats treated with N-nitrosodimethylamine or 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone. Chem. Res. Toxicol. 2008;20:1141–1148. doi: 10.1021/tx700189c. [DOI] [PubMed] [Google Scholar]
- 19.Cheng G, Wang M, Upadhyaya P, Villalta PW, Hecht SS. Formation of formaldehyde adducts in the reactions of DNA and deoxyribonucleosides with α-acetates of 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK), 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol (NNAL), and N-nitrosodimethylamine (NDMA) Chem. Res. Toxicol. 2008 doi: 10.1021/tx7003823. epub ahead of print Jan 18, 2008. [DOI] [PubMed] [Google Scholar]
- 20.Upadhyaya P, Sturla S, Tretyakova N, Ziegel R, Villalta PW, Wang M, Hecht SS. Identification of adducts produced by the reaction of 4-(acetoxymethynitrosamino)-1-(3-pyridyl)-1-butanol with deoxyguanosine and DNA. Chem. Res. Toxicol. 2003;16:180–190. doi: 10.1021/tx0256376. [DOI] [PubMed] [Google Scholar]
- 21.Upadhyaya P, Kenney PMJ, Hochalter JB, Wang M, Hecht SS. Tumorigenicity and metabolism of 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol (NNAL) enantiomers and metabolites in the A/J mouse. Carcinogenesis. 1999;20:1577–1582. doi: 10.1093/carcin/20.8.1577. [DOI] [PubMed] [Google Scholar]
- 22.Carmella SG, Ye M, Upadhyaya P, Hecht SS. Stereochemistry of metabolites of a tobacco-specific lung carcinogen in smokers’ urine. Cancer Res. 1999;59:3602–3605. [PubMed] [Google Scholar]
- 23.Lao Y, Yu N, Kassie F, Villalta PW, Hecht SS. Analysis of pyridyloxobutyl DNA adducts in F344 rats chronically treated with (R)- and (S)-N’-nitrosonornicotine. Chem. Res. Toxicol. 2007;20:246–256. doi: 10.1021/tx060208j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haglund J, Henderson AP, Golding BT, Tornqvist M. Evidence for phosphate adducts in DNA from mice treated with 4-(N-methyl-N-nitrosamino)-1-(3-pyridyl)-1-butanone (NNK) Chem. Res. Toxicol. 2002;15:773–779. doi: 10.1021/tx015542o. [DOI] [PubMed] [Google Scholar]
- 25.Adams JD, LaVoie EJ, Hoffmann D. On the pharmacokinetics of tobacco-specific N-nitrosamines in Fischer rats. Carcinogenesis. 1985;6:509–511. doi: 10.1093/carcin/6.4.509. [DOI] [PubMed] [Google Scholar]
- 26.Upadhyaya P, Carmella SG, Guengerich FP, Hecht SS. Formation and metabolism of 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol enantiomers in vitro in mouse, rat and human tissues. Carcinogenesis. 2000;21:1233–1238. [PubMed] [Google Scholar]
- 27.Wu Z, Upadhyaya P, Carmella SG, Hecht SS, Zimmerman CL. Disposition of 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK) and 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol (NNAL) in bile duct-cannulated rats: stereoselective metabolism and tissue distribution. Carcinogenesis. 2002;23:171–179. doi: 10.1093/carcin/23.1.171. [DOI] [PubMed] [Google Scholar]
- 28.Zimmerman CL, Wu Z, Upadhyaya P, Hecht SS. Stereoselective metabolism and tissue retention in rats of the individual enantiomers of 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol (NNAL), metabolites of the tobacco-specific nitrosamine, 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK) Carcinogenesis. 2004;25:1237–1242. doi: 10.1093/carcin/bgh120. [DOI] [PubMed] [Google Scholar]
- 29.Hecht SS, Kenney PMJ, Wang W, Trushin N, Upadhyaya P. Effects of phenethyl isothiocyanate and benzyl isothiocyanate, individually and in combination, on lung tumorigenesis induced in A/J mice by benzo[a]pyrene and 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone. Cancer Lett. 2000;150:49–56. doi: 10.1016/s0304-3835(99)00373-0. [DOI] [PubMed] [Google Scholar]
- 30.West KA, Brognard J, Clark AS, Linnoila IR, Yang X, Swain SM, Harris C, Belinsky S, Dennis PA. Rapid Akt activation by nicotine and a tobacco carcinogen modulates the phenotype of normal human airway epithelial cells. J. Clin. Invest. 2003;111:81–90. doi: 10.1172/JCI16147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schuller HM. Nitrosamines as nicotinic receptor ligands. Life Sci. 2007;80:2274–2280. doi: 10.1016/j.lfs.2007.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schuller HM. Mechanisms of smoking-related lung and pancreatic adenocarcinoma development. Nat. Rev. Cancer. 2002;2:455–463. doi: 10.1038/nrc824. [DOI] [PubMed] [Google Scholar]
- 33.Jalas J, Hecht SS, Murphy SE. Cytochrome P450 2A enzymes as catalysts of metabolism of 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK), a tobacco-specific carcinogen. Chem. Res. Toxicol. 2005;18:95–110. doi: 10.1021/tx049847p. [DOI] [PubMed] [Google Scholar]
- 34.Breyer-Pfaff U, Martin HJ, Ernst M, Maser E. Enantioselectivity of carbonyl reduction of 4-methylnitrosamino-1-(3-pyridyl)-1-butanone by tissue fractions from human and rat and by enzymes isolated from human liver. Drug Metab. Dispos. 2004;32:915–922. [PubMed] [Google Scholar]
- 35.Foiles PG, Akerkar SA, Carmella SG, Kagan M, Stoner GD, Resau JH, Hecht SS. Mass spectrometric analysis of tobacco-specific nitrosamine-DNA adducts in smokers and nonsmokers. Chem. Res. Toxicol. 1991;4:364–368. doi: 10.1021/tx00021a017. [DOI] [PubMed] [Google Scholar]
- 36.Hölzle D, Schlöbe D, Tricker AR, Richter E. Mass spectrometric analysis of 4-hydroxy-1-(3-pyridyl)-1-butanone-releasing DNA adducts in human lung. Toxicology. 2007;232:277–285. doi: 10.1016/j.tox.2007.01.016. [DOI] [PubMed] [Google Scholar]