Abstract
Recent reports of the slow-tight binding inhibition of bovine liver dihydrofolate reductase (bDHFR) in the presence of polyphenols isolated from green tea leaves has spurred renewed interest in the biochemical properties of bDHFR. Earlier studies were done with native bDHFR but in order to validate models of polyphenol binding to bDHFR, larger quntities of bDHFR are necessary to support structural studies. Bovine DHFR differs from its closest sequence homologue, murine DHFR, by 19 amino acids. To obtain the bDHFR cDNA, murineDHFR cDNA was transformed by a series of nested PCR reactions to reproduce the amino acid coding sequence for bovine DHFR. The bovine liver DHFR cDNA has an open reading frame of 561 base pairs encoding a protein of 187 amino acids that has a high level of conservation at the primary sequence level with other DHFR enzymes, and more so for the amino acid residues in the active site of the mammalian DHFR enzymes. Expression of the bovine DHFR cDNA in bacterial cells produced a stable recombinant protein with high enzymatic activity and kinetic properties similar to those previously reported for the native protein.
Introduction
Dihydrofolate reductase (DHFR) catalyzes the NADPH-dependent reduction of dihydrofolate to tetrahydrofolate, which is in turn converted to a key cofactor required for several one-carbon transfer reactions that are critical for the biosynthesis of DNA, RNA and certain amino acids [1–3]. Without DHFR activity, cellular metabolism is thus profoundly affected. As a result of its importance in maintaining folate pools in their active reduced state, DHFR has been studied extensively and many compounds have been synthesized and tested as potential drugs that inhibit DHFR function [4–14].
Comparison of DHFR sequences shows a high homology among vertebrate species (~75–95%) (Table 1). Structural data for several species of DHFR, both in the solid state and solution, also reveals a highly conserved topology with the differences in enzyme size primarily found in flexible loop insertions that surround a conserved active site core. The monomeric enzyme consists of an eight-stranded beta sheet structure with seven parallel strands and five helices that fold against the active site core [4].
Table 1.
Sequence alignment of mammalian DHFRs.
| CLUSTAL FORMAT for T-COFFEE Version_5.13 [http://www.tcoffee.org ],CPU=0.49 sec, SCORE=88, Nseq=7, Len=190 | |
|---|---|
| bovine | MVRPLNCIVAVSQNMGIGKNGDLPWPPLRNEFQYFQRMTTVSSVEGKQNLVIMGRKTWFS |
| hamster | MVRPLNCIVAVSQNMGIGKNGDFPWPMLRNEFKYFQRMTTTSSVEGKQNLVIMGRKTWFS |
| human | MVGSLNCIVAVSQNMGIGKNGDLPWPPLRNEFRYFQRMTTTSSVEGKQNLVIMGKKTWFS |
| murine | MVRPLNCIVAVSQNMGIGKNGDLPWPPLRNEFKYFQRMTTTSSVEGKQNLVIMGRKTWFS |
| pig | MVRPLNCIVAVSQNMGIGKNGDLPWPPLRNEYKYFQRMTTTSSVEGKQNLVIMGRKTWFS |
| rat | MVRPLNCIVAVSQNMGIGKNGDLPWPLLRNEFKYFQRMTTTSSVEGKQNLVIMGRKTWFS |
| chicken | MVRSLNSIVAVCQNMGIGKDGNLPWPPLRNEYKYFQRMTSTSHVEGKQNAVIMGKKTWFS |
| bovine | IPEKNRPLKDRINIVLSRELKEPPKGAHFLAKSLDDALELIEDPELTNKVDVVWIVGGSS |
| hamster | IPEKNRPLKDRINIVLSRELKEPPQGAHFLAKSLDDALKLIEQPELADKVDMVWIVGGSS |
| human | IPEKNRPLKGRINLVLSRELKEPPQGAHFLSRSLDDALKLTEQPELANKVDMVWIVGGSS |
| murine | IPEKNRPLKDRINIVLSRELKEPPRGAHFLAKSLDDALRLIEQPELASKVDMVWIVGGSS |
| pig | IPEKNRPLKDRINIVLSRELKEPPQGAHFLAKSLDDALKLTEQPELKDKVDMVWIVGGSS |
| rat | IPEKNRPLKDRINIVLSRELKEPPQGAHFLAKSLDDALKLIEQPELASKVDMVWVVGGSS |
| chicken | IPEKNRPLKDRINIVLSRELKEAPKGAHYLSKSLDDALALLDSPELKSKVDMVWIVGGTA |
| bovine | VYKEAMNKPGHVRLFVTRIMQEFESDAFFPEIDFEKYKLLPEYPGVPLDVQEEKGIKYKF |
| hamster | VYKEAMNQPGHLRLFVTRIMQEFESDTFFPEIDLEKYKLLPEYPGVLSEVQEEKGIKYKF |
| human | VYKEAMNHPGHLKLFVTRIMQDFESDTFFPEIDLEKYKLLPEYPGVLSDVQEEKGIKYKF |
| murine | VYQEAMNQPGHLRLFVTRIMQEFESDTFFPEIDLGKYKLLPEYPGVLSEVQEEKGIKYKF |
| pig | VYKEAMNKPGHIRLFVTRIMKEFESDTFFPEIDLEKYKLLSECSGVPSDVQEEKGIKYKF |
| rat | VYQEAMNQPGHLRLFVTRIMQEFESDTFFPEIDLEKYKLLPEYPGVLSEIQEEKGIKYKF |
| chicken | VYKAAMEKPINHRLFVTRILHEFESDTFFPEIDYKDFKLLTEYPGVPADIQEEDGIQYKF |
| bovine | EVYEKNN--- |
| hamster | EVYEKKG--- |
| human | EVYEKND--- |
| murine | EVYEKKD--- |
| pig | EVYEKNN--- |
| rat | EVYEKKD--- |
| chicken | EVYQKSVLAQ |
The pIs for these DHFRs are: 6.20, 7.76, 6.85, 8.60, 8.73, 6.77, and 7.78, respectively.
DHFR sequences from vertebrates show high homology, but homology of these sequences to DHFR sequences from lower eukaryotes or bacteria is as low as 30%; moreover, kinetic and biochemical data show differences in the mechanism of action among the enzymes [5–14]. Structure activity correlation data from different DHFR species (bovine, murine, and E. coli DHFR) reveal variation in the hydrophobic nature of the binding site of these enzymes, which has been exploited in the design of inhibitors that show specificity for individual enzymes [9–14]. For example, structure-activity relationships of bDHFR inhibition have shown that the antifolate trimethoprim (TMP) binds about 6,000-fold better with E. coli DHFR than with bDHFR [10]. The NMR solution structure of bDHFR [15] shows that the binding of TMP is similar to that observed in the crystal structure of chicken DHFR-NADPH-TMP ternary complex [16].
Recent reports show slow-tight binding inhibition of bDHFR by the catechin (−)-epigallocatechin gallate (EGCG), a naturally occurring gallated polyphenol isolated from green tea leaves [17,18]. This unusual binding by a molecule without the signature 2,4-diaminopyrimidine ring of classical antifolates spurred renewed interest in the biochemical properties of bovine DHFR and raised the question of whether EGCG was a general inhibitor of DHFR from other species.
In order to better understand the biochemical properties of bDHFR and to validate the binding models of novel inhibitors such as the polyphenols from green tea, we needed to produce adequate quantities of bDHFR to support structural studies. The amino acid sequence of bovine DHFR is highly homologous to the DHFR sequence of mammalian enzymes, with that of murine DHFR as the closest homologue. Comparison of amino acid sequences revealed 19 substitutions between murine and bovine DHFR with the majority of these changes observed outside of the active site (Table 2). Having already successfully cloned and expressed murine DHFR from cDNA, we carried out a series of nested PCR reactions to mutate the cDNA sequence of murine DHFR to that of bDHFR and expressed the cDNA for bDHFR in bacteria. We cloned the cDNA of bDHFR into a plasmid containing a SUMO tag in order to aid in purification and enhance the solubility of the purified enzyme product. In this report we demonstrate the physical and kinetic properties of the recombinant bDHFR enzyme and illustrate that it is similar to the native enzyme previously reported in the literature. Additionally, we report preliminary results from robotic crystallization screens for the purified bDHFR enzyme complexes.
Table 2.
Sequence alignment of murine and bovine DHFR. Sequence differences are highlighted in red and underlined.
 |
MATERIALS AND METHODS
Reagents
DNA primers from Integrated DNA Technologies (IDT, Coralville, Iowa), restriction enzyme from New England Biolabs, high-fidelity PCR Kit from Invitrogen, T-easy vector kit from Promega, T4 DNA ligation kit from Roche, pETduet vector from Novagen, DNA sequencing in Roswell Park Cancer Institute, plasmid purification mini-prep kit from Qiagen, Rossetta (DE3) cells from Novagen, cells broken by microfluidizer (Microfluidics Corp., Newton, MA), Ni-NTA His beads from Qiagen.
Overlap PCR for Mutations
Site directed mutagenesis by overlap extension using the polymerase chain reaction (OE-PCR) was used to introduce a single mutation in the target DNA [19]. As illustrated (Table 3 and Table 4), 14 primers were used to incorporate the 19 residues that differ between the sequence of murine and bovine DHFR. The first step was to generate by PCR two DNA fragments having overlapping ends. These fragments were combined in a fusion reaction (5 cycles, without adding primer) in which the overlapping ends annealed, allowing the 3' overlap of each strand to serve as a primer for the extension. The resulting fusion product was amplified further by PCR (adding primers). The initial denaturation step was carried out for one cycle for 2 min at 94°C, the next denaturation, annealing and extension steps were carried out at 94°C, 50°C and 72°C for one minute each, respectively for 30 cycles, and the final extension step was carried out for 7 min at 72°C for one cycle. Specific mutations in the DNA sequence were introduced by incorporating nucleotide changes into the overlapping oligo primers.
Table 3.
Oligonucleotides used to incorporate the 19 residue changes in the protein sequence of murine and bovine DHFR as shown in Table 2
| 1. | AAG(K)33CAA(Q) |
| 2. | ACC(T)41GTG(V) |
| 3. | CGA(R)85AAA(K) |
| 4. | AGA (R)99 GAA (E) |
| 5. | CAA (Q) 103 GAC (D) |
| 6. | GCA (A) 107 ACA (T) |
| 7. | AGA (S) 108 AAT (N) |
| 8. | ATG (M) 112 GTT (V) |
| 9. | CAG (Q) 123 AAA (K) |
| 10. | CAA (Q) 128 AAA (K) |
| 11. | CTC (L) 132 GTT (V) |
| 12. | ACG (T) 147 GCA (A) |
| 13. | TTG (L) 154 TTC(F) |
| 14. | GGG (G) 155 GAG (E) |
| 15. | CTC (L) 167 CCG (P) |
| 16. | TCT (S) 168 CTC (L) |
| 17. | GAG (E) 169 GAC (D) |
| 18. | AAA (K) 186 AAC (N) |
| 19. | GAC (D) 187 AAC (N) |
Table 4.
Primers used to generate the nested PCR reactions to transform murine DHFR
| cDNA to bovine DHFR cDNA. |
|---|
| Primer 1: DHFR NdeI (forward primer): |
| CAT ATG GTT CGA CCA TTG AAC TGC |
| Primer2: DHFR XhoI : (for mutation 18,19, reverse primer) |
| CTC GAG TTA GTT GTT CTT CTC GTA GAC TTC AAA CTT ATA C |
| Primer 3: DHFR 002F (for mutations 1,2, forward primer) |
| tac ttc caa aga atg acc aca gtg tct tca gtg gaa ggt aaa cag |
| Primer 4: DHFR 002R (for mutations 1,2, reverse primer) |
| tgt ggt cat tct ttg gaa gta ttg gaa ctc gtt cct gag cgg |
| Primer 5: DHFR 001F: (for mutation 3, forward primer) |
| CCA CCA AAA GGA GCT CAT TTT CTT GCC |
| Primer 6: DHFR 001R (for mutation 4, reverse primer): |
| ATG AGC TCC TTT TGG TGG TCC TTT GAG TTC TCT |
| Primer 7: DHFR 003F (for mutations 4,5,6,7, forward primer): |
| GAA GAC CCG GAA TTG ACA AAT AAA GTA GAC ATG GTT TGG ATA GTC |
| Primer 8: DHFR 003R (for mutations 4,5,6,7, reverse primer): |
| ATT TGT CAA TTC CGG GTC TTC AAT AAG TTC TAA GGC ATC ATC CAAACT TTT G |
| Primer 9: DHFR008FM2V (for mutation 8, forward primer) |
| AAA GTA GAC GTT GTT TGG ATA GTC |
| Primer 10: DHFR008RM2V (for mutation 8, reverse primer) |
| GAC TAT CCA AAC AAC GTC TAC TTT |
| Primer 11: DHFR 004F(for mutations 9,10,11, forward primer): |
| GCC ATG AAT AAA CCA GGC CAC GTG AGA CTC TTT GTG ACA AGG ATC ATG |
| Primer 12: DHFR 004R(for mutations 9,10,11, reverse primer): |
| GTG GCC TGG TTT ATT CAT GGC TTC TTT GTA AAC AGA ACT GCC TCC GAC |
| Primer 13: DHFR006lastF: (for mutation 12 to 17, forward primer) |
| AAA TAT AAA CTT CTC CCA GAA TAC CCA GGC GTC CCG CTC GAC GTC CAG GAG GAA AAA GG |
| Primer 14: DHFR006lastR (for mutations 12–17, reverse primer): |
| TTC TGG GAG AAG TTT ATA TTT CTC GAA ATC AAT TTC TGG GAA AAA TGC GTC ACT TTC |
DNA sequence analysis
Multiple positive pBlueScript clones containing bovine liver DHFR cDNA inserts were sequenced using an ABI PRISM automated sequencer from Roswell Park Cancer Institute Biopolymer Facility (Buffalo, New York). T7 forward and M13 reverse primers of the vector were used to sequence and confirm the isolation of the full length bovine liver DHFR cDNA. The bovine liver DHFR cDNA sequence was compared with known DHFR sequences from different species using BLAST web services from National Center of Biotechnology Information (NCBI). Protein sequence alignment with other known DHFR species was performed using T-Coffee for multiple alignment [20] and ProtParam [21] to calculate the pI for these enzymes based on their amino acid sequences (Table 1).
Construction of Plasmid for Expression of SUMO-bovine DHFR
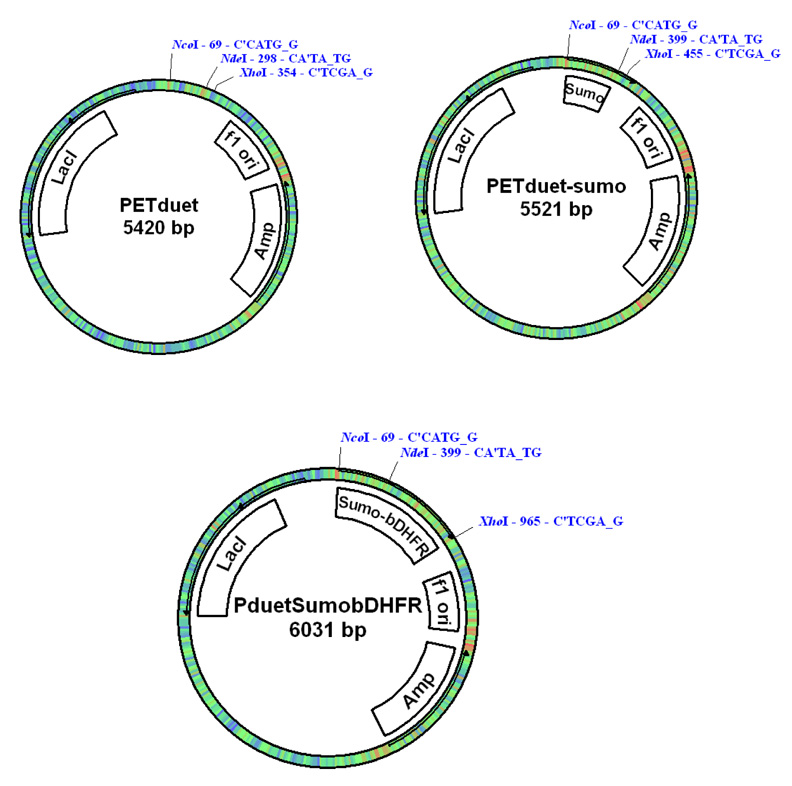
A Sumo (Small Ubiquitin-like Modifier) Tag was used to express the target protein in E. coli cells. The cDNA for the Sumo (1–99 aa, yeast) was cloned by PCR with 5’-NcoI and 3’-NdeI at the ends (Fig. 1). For the convenience of protein purification, a His6 tag (MSYSHHHHHH) was added to the N-terminal end of the Sumo cDNA that was inserted into the T-easy vector (Promega) and then subcloned into pETduet (Novagen) vector by using NcoI and NdeI. The cDNA for bDHFR was then inserted into pETduet-Sumo vector by using NdeI and XhoI. The final plasmid (pETduet-sumo-DHFR) was transformed into E. coli Rossetta competent cells for expression (Fig.1).
Fig. 1.
Plasmid map for the pET-duet Sumo-bDHFR contstruct.
Expression and Purification of SUMO-bDHFR and bDHFR
One single clone was picked to inoculate an overnight starter culture (100 ml) of LB media supplemented with 100 µg/ml ampicillin and 35 µg/ml chloramphenicol. Subsequently, 20 ml of culture was added to 1 L of LB media (supplemented with 100 µg/ml ampicillin and 35 µg/ml chloramphenicol) in a 2L shaker flask. The cells were cultured at 37°C for 3–4 hours. The temperature for the shaker was then changed to 20°C when the OD600 reached to 0.6–0.8. A preliminary test of protein expression induced by the addition of 1.0 mM IPTG (isopropyl β-D-thiogalactopyranoside) for 4 hours at 30°C revealed reduced expression so that the final expression was carried out overnight at 20°C. The cells were harvested by centrifugation at 6000 g for 15 minutes, and the cell paste was frozen and stored at −80°C.
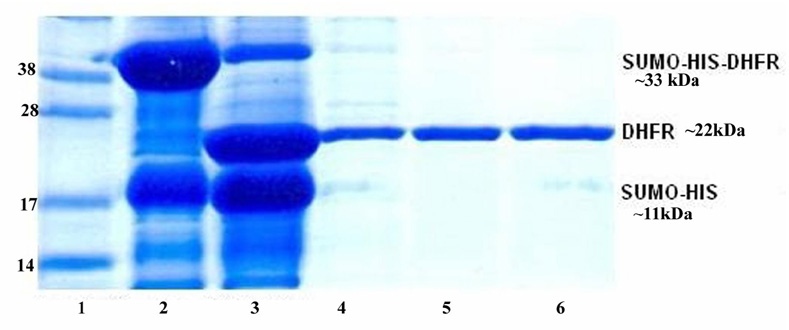
For protein purification, the cell pellets from 2L cell cultures was re-suspended in 150 ml His Tag binding buffer (25mM NaH2PO4, pH 7.0, 500 mM NaCl, 10 mM imidazole). The cells were broken by using a microfluidizer M-110EH (Microfluidics Corp., Newton, MA). The whole cell lysate was centrifuged at 35000× g for 1 hr. The supernatant was mixed with 10 ml Ni-NTA His beads equilibrated with His Tag binding buffer. The Ni-NTA His beads were harvested by centrifugation at 700 rpm for 5 minutes. The harvested His beads were loaded into an empty column and then rinsed by 10 column volumes (CV) of the His binding buffer. The Sumo-bDHFR was eluted from the Ni-NTA His beads by using 2 CV of His Tag elution buffer (25 mM NaH2PO4, pH 7.0, 500 mM NaCl, 500 mM imidazole). The eluted protein was dialyzed in the buffer (50 mM Tris, pH8.0, 500 mM NaCl, 1mM DTT) to remove the imidazole. In order to remove the Sumo Tag from the fusion protein, sumo protease (Ulp1) [22] was then added to the protein solution at the ratio (1:100) and incubated at 4°C for 2 hours. SDS-PAGE was used to examine the completeness of digestion. The proteins His6-Ulp1 and His-Sumo were removed by passing through Ni-NTA His column (Fig. 2). The flow through solution containing the target protein was collected and concentrated to ~18 mg/ml. Based on the pellet from 1L of cell growth, lane 2 represents about 30–40 mg of His-SUMO-bDHFR, lane 3 shows about 15–18 mg of bDHFR and lanes 4–6 show 13 mg of purified bDHFR. A 5% glycerol stock was added to the protein buffer. The final bDHFR sample was divided into small aliquots (100 µl/tube) and then stored at −80°C.
Figure 2.
SDS-PAGE for the purification of bovine DHFR. Lane 1: Protein Marker (Invitrogen, SeeBlue Plus2); lane 2, whole cell lysate, lane 3: digest with Sumo protease (Ulp1) at 4°C for 1 hour, lane 4: elution from Ni-NTA 5ml HisTrap, lanes 5 and 6: flow through by using Ni-NTA His tag second time.
Size Exclusion of bDHFR
A superdex 75 high resolution gel filtration column was used to examine the size of bDHFR. The buffer used for gel filtration is 50mM Tris, pH8.0, 500mM NaCl, 1mM DTT, 1mM EDTA. 100 µl of bDHFR (1 mg/ml) was loaded to the column and eluted with 1.5 column volumes of buffer. The flow rate was set at 0.5 ml/min and the pressure limit was 1.5 Mpa.
Enzyme and inhibitor assay
The activities of the recombinant bovine liver DHFR was measured spectrophotometrically for the decrease in absorbance at 340 nm that occurs when NADPH and dihydrofolate are converted to NADP+ and tetrahydrofolate; the assay also contained saturating concentrations of substrate (18–90 µM dihydrofolic acid (DHFA)) and cofactor (117 µM NADPH), with 150 mM KCl, as previously described [23,25]. Assays contained sufficient DHFR protein to produce a change in substrate concentration of 2.5 µM/min. Assays were performed either at 37°C or at 30°C, as noted in the tables.
Km values were calculated based upon the Michaelis-Menton equation, using Prism 4.0 (GraphPad. Inc.). Catalytic efficiency is the ratio of the rate constant kcat divided by the Km of the enzyme for DHFA, and thus has the units of s−1 µM−1. IC50 curves were performed for each inhibitor at several different concentrations of the substrate DHFA, using concentrations of inhibitor to produce rates ranging from 5 to 95% of control uninhibited rates. Ki values were calculated from these data, using the model for competitive inhibition for TMP (this inhibitor affects only Km and not Vmax); for methotrexate (MTX), the model for a tight binding inhibitor was used [26].
Crystallization Trials
Crystallization trials were submitted to the high throughput HWI robot facility in which 1536 different conditions were screened [27]. Crystallization experiments were set up using the microbatch-under-oil technique [28] with a Robbins Scientific Tango liquid handling system. Each of the 1536 experiments contained 200 nL of crystallization cocktail solution combined with 200 nL of protein solution un USP grade mineral oil contained in a 1536-well plate. The experimental plate was stored and imaged at 23°C. Images were recorded 24 hours after set up and thereafter weekly for 4 weeks. Images were manually reviewed for crystals.
RESULTS AND DISCUSSION
Conformation of the cDNA sequence for bDHFR
To carry out the 19 specific amino acid substitutions needed to convert murine DHFR to bovine DHFR, we designed 14 primers that were predicted to insert the required codon changes to convert the cDNA for murine DHFR to cDNA that would encode bovine DHFR (Table 2). A series of nested PCR reactions (Table 3,Table 4) accomplished the conversion and produced a cDNA product that was the correct size. The sequence of this cDNA product was compared with known DHFR sequences from different species using BLAST web services from National Center of Biotechnology Information (NCBI). The translated sequence perfectly matched the reported sequence of bovine DHFR, thus confirming that the mutations were successfully inserted.
Conformation of physical properties for bDHFR
Native bovine DHFR has a molecular weight of 22.1 kDa based on equilibrium sedimentation [29]. Expressed recombinant protein contains the 187 amino acids found in the native protein. In size exclusion chromatography bDHFR eluted as a single peak that verified the expected molecular weight of the monomeric enzyme (~22 kDa) which agrees with the predicted molecular weight for the recombinant protein of 21,604 based on amino acid sequence.
Prior work has shown two forms of bDHFR in a ratio of 20:1, with pIs of 7.15 and 5.94 and identical specific activities with dihydrofolate and folate [30]. There is an indication that bDHFR can form a dimer with molecular weights 23 and 46 kDa. We saw no evidence of dimer formation in either size exclusion chromatography or PAGE (Fig. 2). Fluorescence data shows a slow conformational change in bDHFR induced by NADPH binding [23,24]. This interconversion between two protein conformers shows an increase in the apparent rate of enzymatic reaction catalyzed by DHFR. Our assay conditions include preincubation of the DHFR with NADPH in KCl buffer before addition of substrate or inhibitor and substrate; thus, pre-activation by NADPH is built into the standard assay and is not visible as a separate process.
Expression of catalytically active bDHFR
Bovine DHFR prepared and purified as described here was catalytically active and could be characterized kinetically. Using enzyme concentrations determined by titration with MTX [30,31], kcat and catalytic efficiency could also be determined and the values compared to those calculated by the same methods for related recombinant DHFR (Table 5). The results show that bovine and murine DHFR had similar Km values for substrate and cofactor, as well as kcat and catalytic efficiency. Human DHFR showed a slightly higher Km for DHFA and a resultant lower catalytic efficiency.
Table 5.
Kinetic characterization of bDHFR
| Enzyme | Km DHFA µM | Km NADPH µM | kcat s−1 | Catalytic efficiency s−1 µM−1 |
|---|---|---|---|---|
| bDHFR | 0.35 ± 0.13 | 3.2 ± 0.6 | 24 ± 7 | 68 |
| mDHFR | 0.32 ± 0. 05 | 2.4 ± 0.5 | 29 ± 5 | 92 |
| hDHFR | 2.7 ± 0.5 | 5.7 ± 0.9 | 40 ± 2 | 15 |
All assays were conducted at 37°C with KCl in the assay, as described in methods.
b = bovine DHFR; m = murine DHFR; h = human DHFR.
The recombinant bDHFR was also assayed at 30°C to facilitate comparison of the recombinant protein to the native protein that had been studied previously. Under these conditions, the recombinant bDHFR showed a specific activity of 65 ± 5 µmoles DHFA/min/mg protein, which compares favorably to the value of 29.6 reported by Bauman and Wilson [29] for the native enzyme. Kaufman and Kemerer [31] reported a turnover number of 578 moles DHFA/min/mole enzyme for the native enzyme assayed at 28°C, which also compares well with the value of 1350 ± 107 moles DHFA/min/mole enzyme we determined for recombinant bDHFR assayed at 30°C.
Inhibition of bDHFR enzymatic activity
Recombinant bovine liver DHFR was tested for inhibition by several compounds that had previously been tested with native bovine DHFR. Methotrexate represents a classical ptertidine analogue, and trimethoprim is a lipophilic 2,4-diaminopyrimidine analogue. Gulli et al [32] determined Ki values by calorimetry for MTX and TMP, using native bovine DHFR purified by affinity chromatography (Table 6). Both Ki values determined for recombinant bDHFR were similar to those reported by Gilli et al [32].
Table 6.
Ki values for mammalian DHFR
| Enzyme | Source | Ki TMP µM | Ki MTX pM |
|---|---|---|---|
| bDHFR | This study | 0.19 ± 0.01 | 8.9 ± 0.6 |
| bDHFR | Gilli (1990) (32) | 0.33 | 33 |
| mDHFR | This study | 0.5 | 2.9 ± 0.1 |
| hDHFR | This study | 5.2 ± 0.7 | 93.3 ± 21 |
All assays were conducted at 37°C.
The tricyclic antidepressants represent an unusual class of compounds reported to be inhibitory to native bDHFR, although the potency was low [33]. We tested the inhibitory properties of the most potent compound, nortriptyline, from the series tested by Blake et al [33] (Table 7). Our results with recombinant protein confirmed the ability of the tricyclic antidepressant nortriptyline to inhibit bDHFR, but the Ki values for the recombinant protein were nearly two orders of magnitude lower than those reported for the native protein. These differences may be related to differences in assay conditions as well as methods to measure Ki.
Table 7.
Ki values for Nortriptyline with bDHFR
| Enzyme | Source | Ki Nortriptline µM |
|---|---|---|
| bDHFR | This study | 0.84 ± 0.25 |
| bDHFR | Blake & Gould (1985) (33) | 45 ± 15, 70 ± 17 |
All assays were conducted at 30°C.
The ability of EGCG to inhibit bDHFR was also measured with the recombinant bovine DHFR. In our hands a 70 minute preincubation with 0.68 mM EGCG in 1 mM ascorbate, followed by dilution and assay at 37°C with 117 µM NADPH and 10 µM DHFA and 100 µM NaCl, produced 26% inhibition compared to a sample preincubated in the same way but without EGCG. This inhibition is less than the 82% inhibition reported by Navarro-Peran [18] following a 70 minute preincubation at 25°C with 40 µM EGCG in 1 mM ascorbate and subsequent assay at 25°C. Thus, our conditions differed only in temperature and in having a higher concentration of EGCG in the preincubation.
Preincubation and assay of native rat liver DHFR under the same conditions showed this enzyme to be unstable; assay without preincubation failed to show inhibition by EGCG. Mouse DHFR assayed in the presence of 122 µM EGCG showed less than 10% inhibition when the DHFA concentration was 90 µM: inhibition increased to 18% when the DHFA concentration was reduced to 45 µM. When these assays were followed for 30 minutes, no change in inhibition over time was noted. Subsequent experiments showed no additional inhibition was gained at lower substrate concentrations. Experiments using E. coli DHFR, recombinant Pneumocystis carinii DHFR, native Toxoplasma gondii DHFR, and native Mycobacterium avium DHFR failed to show inhibition by EGCG over a 3 or 30 minute time course. We conclude that in our hands EGCG is a weak inhibitor with activity under our conditions only demonstrable for murine DHFR and bovine DHFR.
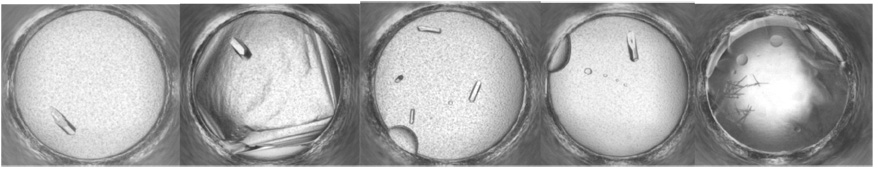
Proof of the mechanism of action of EGCG will come from structural studies showing how the compounds bind to bDHFR. As illustrated in Fig. 3, five crystallization conditions produced crystals from solutions of bDHFR incubated with NADPH and MTX. Further studies are aimed at producing crystals with EGCG with or without NADPH.
Fig. 3.
Examples of hits from the HWI crystal screening robot for bDHFR in complex with NADPH and methotrexate.
SUMMARY
Recombinant bovine liver DHFR shows predicted physical properties when expressed in E. coli and purified by affinity chromatography. The kinetic characterization showed that the recombinant protein is generally more catalytically active than the native preparations. The recombinant bDHFR resembles the murine DHFR in kinetic properties but both differ from human DHFR in having a lower Km for DHFA. The recombinant bDHFR, like the native bDHFR, is sensitive to inhibition by trimethoprim, methotrexate, nortriptyline, and to a lesser extent, EGCG. Ki values for the first three of these compounds were lower for recombinant bDHFR than the values previously reported for native bDHFR.
ACKNOWLEDGEMENTS
This work was supported in part by funds from GM51670 (VC). The authors thank Jim Pace for crystal screening, Christopher D. Lima, Sloan Kettering Institute for the use of his SUMO protease construct, and Dr. John McGuire for helpful discussions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Sirotnak FM, Burchall JJ, Ensminger WD, Montgomery JA. Folate Antagonists as Therapeutic Agents. Vol 1. New York: Academic Press; 1984. [Google Scholar]
- 2.Blakley RL, Benkovic SJ. Folates and Pteridines. Vol. 1. New York: John Wiley & Sons; 1984. [Google Scholar]
- 3.Blakley RL. Eukaryotic Dihydrofolate Reductase. Adv Enzymol. & Rel. Areas of Mol. Biol. 1995;70:23–102. doi: 10.1002/9780470123164.ch2. [DOI] [PubMed] [Google Scholar]
- 4.Cody V, Schwalbe CH. Structural Characteristics of Antifolate Dihydrofolate Reductase Enzyme Interactions. Crystallography Reviews. 2006;12:301–333. [Google Scholar]
- 5.Chan DCM, Anderson AC. Towards Species-specific Antifolates. Curr. Med. Chem. 2006;13:377–398. doi: 10.2174/092986706775527938. [DOI] [PubMed] [Google Scholar]
- 6.Gangjee A, Jain H, Kurup S. Recent Advances in Classical and Non-classical Antifolates as Antitumor and Antiopportunistic Infection Agents, Part I. Anticancer Agents Med. Chem. 2007;7:523–542. doi: 10.2174/187152007781668724. [DOI] [PubMed] [Google Scholar]
- 7.Gangjee A, Jain H, Kurup S. Recent Advances in Classical and Non-classical Antifolates as Antitumor and Antiopportunistic Infection Agents, Part II. Anticancer Agents Med. Chem. 2008;8:205–231. doi: 10.2174/187152008783497064. [DOI] [PubMed] [Google Scholar]
- 8.Gangjee A, Kurup S, Namjoshi O. Dihydrofolate Reductase as a Target for Chemotherapy in Parasites. Curr. Pharm. Des. 2007;13:609–639. doi: 10.2174/138161207780162827. [DOI] [PubMed] [Google Scholar]
- 9.Queener SF. New Drug Developments for Opportunistic Infections in Immunosuppressed Patients: Pneumocystis carinii. J. Med. Chem. 1995;38:4739–4759. doi: 10.1021/jm00024a001. [DOI] [PubMed] [Google Scholar]
- 10.Dietrich SW, Blaney JM, Reynolds MA, Jow PYC, Hansch C. Quantitative Structure-Selectivity Relationships. Comparison of the Inhibition of Escherichia coli and Bovine Liver Dihydrofolate Reductases by 5-(Substituted-benzyl)-2,4-diaminopyrimidines. J. Med. Chem. 1980;23:1205–1212. doi: 10.1021/jm00185a011. [DOI] [PubMed] [Google Scholar]
- 11.Blaney JM, Dietrich SW, Reynolds MA, Hansch C. Quantitative Structure-Activity Relationship of 5-(X-benzyl)-2,4-Diaminopyrimidines Inhibiting Bovine Liver Dihydrofolate Reductase. J. Med. Chem. 1979;22:614–617. doi: 10.1021/jm00192a003. [DOI] [PubMed] [Google Scholar]
- 12.Kim KH, Dietrich SW, Hansch C, Dolnick BJ, Bertino JR. Inhibition of Dihydrofolate Reductase. 3. 4,6-Diamino-1,2-dihydro-2,2-dimethyl-1-(2-substitutedphenyl)-s-triazine Inhibition of Bovine Liver and Mouse Tumor Enzymes. J. Med. Chem. 1980;23:1248–1251. doi: 10.1021/jm00185a022. [DOI] [PubMed] [Google Scholar]
- 13.Guo Z-R, Dietrich SW, Hansch C, Dolnick BJ, Bertino JR. A Comparison of the Inhibition of Bovine and Murine Leukemia Dihydrofolate Reductase by 4,6-Diamino-1,2-dihydro-2,2-dimethyl-1-(3-X-phenyl)-s-Triazines. Molecul. Pharmacol. 1981;20:649–656. [PubMed] [Google Scholar]
- 14.Li R-L, Hansch C, Kaufman BT. A Comparison of the Inhibitory Action of 5-(Substituted-benzyl)-2,4-diaminopyrimidines on Dihydrofolate Reductase from Chicken Liver with That from Bovine Liver. J. Med. Chem. 1982;25:435–440. doi: 10.1021/jm00346a020. [DOI] [PubMed] [Google Scholar]
- 15.Jayalakshmi V, Krishna NR. Determination of the Conformation of Trimethoprim in the Binding Pocket of Bovine Dihydrofolate Reductase from a STD-NMR Intensity-Restrained CORCEMA-ST Optimization. J. Amer. Chem. Soc. 2005;127:14080–14084. doi: 10.1021/ja054192f. [DOI] [PubMed] [Google Scholar]
- 16.Matthews DA, Bolin JT, Burridge JM, Filman DJ, Volz KW, Kaufman BT, Beddell CR, Champness JN, Stammers DK, Kraut J. Refined Crystal Structures of E. coli and Chicken Liver Dihydrofolate Reductase Containing Bound Trimethoprim. J. Biol. Chem. 1985;260:381–391. [PubMed] [Google Scholar]
- 17.Navarro-Peran E, Cabezas-Herrera J, Garcia-Canovas F, Durrant MC, Thorneley RNF, Rogriguez-Lopez JN. The Antifolate Activity of Tea Catechins. Cancer Res. 2005;65:2059–2064. doi: 10.1158/0008-5472.CAN-04-3469. [DOI] [PubMed] [Google Scholar]
- 18.Navarro-Peran E, Cabezas-Herrera J, Hiner ANP, Sadunishvili T, Garcia-Canovas F, Rodriguez-Lopez JN. Kinetics of the Inhibition of Bovine Liver Dihydrofolate Reductase by Tea Catechins: Origin of Slow-Binding Inhibition and pH Studies. Biochemistry. 2005;44:7512–7525. doi: 10.1021/bi050160t. [DOI] [PubMed] [Google Scholar]
- 19.Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. Site-Directed Mutagenesis by Overlap Extension Using the Polymerase Chain Reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 20.Notredame C, Higgins D, Heringa J. T-Coffee: A Novel Method for Multiple Sequence Alignments. J. Mol. Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- 21.Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins MR, Appel RD, Bairoch A. Protein Identification and Analysis Tools on the ExPASy Server. In: Walker John M., editor. The Proteomics Protocols Handbook. Humana Press; 2005. pp. 571–607. [Google Scholar]
- 22.Mossessova E, Lima CD. Ulp1-SUMO Crystal Structure and Genetic Analysis Reveal Conserved Interactions and a Regulatory Element Essential for Cell Growth in Yeast. Molecular Cell. 2000;5:865–876. doi: 10.1016/s1097-2765(00)80326-3. [DOI] [PubMed] [Google Scholar]
- 23.Rimet O, Chauvet M, Sarrazin M, Bourdeaux R. Conformational Change Induced by Coenzyme Binding to Bovine Liver Dihydrofolate Reductase: A Spectrofluorimetric Study. Biochimica et Biophysica Acta-Protein Structure and Molecular Enzymology. 1991;1076:435–438. doi: 10.1016/0167-4838(91)90488-l. [DOI] [PubMed] [Google Scholar]
- 24.Rimet O, Chauvet M, Dell’Amico M, Noat G, Bourdeaux M. Variations in Fluorescence and Enzymic Properties of Bovine Dihydrofolate Reductase-NADPH Complex During the Slow Conformational Change Induced by Coenzyme Binding. Eur. J. Biochem. 1995;228:55–59. doi: 10.1111/j.1432-1033.1995.0055o.x. [DOI] [PubMed] [Google Scholar]
- 25.Kaufman BT, Jumar AA, Blankenship DT, Freisheim JH. Activation of Bovine and Chicken Liver Dihydrofolate Reductases and Its Relationship to a Specific Cysteine Residue in Their NH2-Terminal Amino Acid Sequences. J. Biol. Chem. 1980;255:6542–6545. [PubMed] [Google Scholar]
- 26.Appleman JR, Prendergast N, Delcamp TJ, Freisheim JH, Blakley RL. Kinetics of the Formation and Isomerization of Methotrexate Complexes of Recombinant Human Dihydrofolate Reductase. J. Biol. Chem. 1988;263:10304–10313. [PubMed] [Google Scholar]
- 27.Luft JR, Collins RJ, Fehrman NA, Lauricella AM, Veatch CK, DeTitta GT. A Deliberate Approach to Screening for Initial Crystallization Conditions of Biological Macromolecules. J. Struct. Biol. 2003;142:170–179. doi: 10.1016/s1047-8477(03)00048-0. [DOI] [PubMed] [Google Scholar]
- 28.Chayen NE, Shaw SPD, Blow DM. Microbatch Crystallization under Oil - a New Technique Allowing Many Small-volume Crystallization Trials. J. Crystal Growth. 1992;122:176–180. [Google Scholar]
- 29.Baumann H, Wilson KJ. Dihydrofolate Reductase from Bovine Liver: Enzymatic and Structural Properties. Eur. J. Biochem. 1975;60:9–15. doi: 10.1111/j.1432-1033.1975.tb20969.x. [DOI] [PubMed] [Google Scholar]
- 30.Peterson DL, Gleisner JM, Blakley RL. Bovine Liver Dihydrofolate Reductase: Purification and Properties of the Enzyme. Biochemistry. 1975;14:5261–5267. doi: 10.1021/bi00695a005. [DOI] [PubMed] [Google Scholar]
- 31.Kaufman RT, Kemerer VF. Purification and Characterization of Beef Liver Dihydrofolate Reductase. Archives of Biochem, & Biophys. 1976;172:289–300. doi: 10.1016/0003-9861(76)90078-3. [DOI] [PubMed] [Google Scholar]
- 32.Gilli RM, Sari JC, Lopez CL, Rimet OS, Briand CM. Comparative Thermodynamic Study of the Interaction of Some Antifolates with Dihydrofolate Reductase. Biochimica et Biophysica Acta. 1990;1040:245–250. doi: 10.1016/0167-4838(90)90083-r. [DOI] [PubMed] [Google Scholar]
- 33.Blake C, Gould BJ, Bridges JW. The Inhibition of Bovine Liver Dihydrofolate Reductase by Triclyclic Antidepressant Drugs. Biochem. Pharmacol. 1985;12:2212–2214. doi: 10.1016/0006-2952(85)90422-8. [DOI] [PubMed] [Google Scholar]