SUMMARY
The inner ear derives from a patch of ectoderm defined by expression of the transcription factor Pax2. We recently showed this Pax2+ ectoderm gives rise not only to the otic placode but also to surrounding cranial epidermis, and that Wnt signaling mediates this placode-epidermis fate decision. We now present evidence for reciprocal interactions between the Wnt and Notch signaling pathways during inner ear induction. Activation of Notch1 in Pax2+ ectoderm expands the placodal epithelium at the expense of cranial epidermis, while loss of Notch1 leads to a reduction in the size of the otic placode. We show that Wnt signaling positively regulates Notch pathway genes such as Jag1, Notch1 and Hes1, and have used transgenic Wnt reporter mice to show that Notch signaling can modulate the canonical Wnt pathway. Gain and loss of function mutations in the Notch and Wnt pathways reveal that some aspects of otic placode development - such as Pax8 expression and the morphological thickening of the placode – can be regulated independently by either Notch or Wnt signals. Our results suggest that Wnt signaling specifies the size of the otic placode in two ways – by directly up-regulating a subset of otic genes, and by positively regulating components of the Notch signaling pathway which then act to augment Wnt signaling.
Keywords: Mouse, Otic placode, Wnt, β-catenin, Notch1, Jagged1, Inner ear
INTRODUCTION
Inner ear development is an excellent example of how a Darwinian “organ of extreme perfection and complication” can arise from simple origins. The inner ear derives from a patch of thickened ectoderm, the otic placode, lying next to the posterior hindbrain. Signals that induce the otic placode are present in the hindbrain and cranial paraxial mesoderm, although the relative contribution of these tissues to the induction process varies between species (Barald and Kelley, 2004; Groves, 2005; Riley and Phillips, 2003; Torres and Giraldez, 1998). Members of the Fibroblast Growth Factor (FGF) family play a crucial role in inducing the otic placode in all vertebrates examined (Friesel and Brown, 1992; Ladher et al., 2005; Leger and Brand, 2002; Mackereth et al., 2005; Maroon et al., 2002; Phillips et al., 2001; Vendrell et al., 2000; Wright et al., 2004; Wright and Mansour, 2003). FGF signaling induces the expression of genes such as Pax2 and Pax8 in a broad region of cranial ectoderm stretching from rhombomeres 3 to 6 (Maroon et al., 2002; Martin and Groves, 2006; Wright and Mansour, 2003). Evidence from lineage tracing in chick (Streit, 2002) and mouse (Ohyama and Groves, 2004b; Ohyama et al., 2006) suggest that this broad Pax2+ domain, which we have referred to as the “pre-otic field”, contains cells fated to become otic and epibranchial placodes, as well as cranial epidermis.
We recently showed that Wnt signaling plays an important role in defining the size of the otic placode within this Pax2+ pre-otic field. Wnt signaling is activated in a medial subset of the Pax2+ domain closest to the hindbrain (Ohyama et al., 2006). Inactivation of Wnt signaling in Pax2+ cells by conditional deletion of β-catenin leads to a large reduction in the size of the otic placode and a corresponding expansion of cranial epidermis. Conversely, activation of β-catenin in Pax2+ cells expands the otic placode at the expense of cranial epidermis (Ohyama et al., 2006). To date, however, it is not clear how Wnt signals direct cranial ectoderm towards an otic fate. It is possible that Lef/Tcf/β-catenin transcriptional complexes activated by Wnt signaling directly regulate otic genes. Alternatively, Wnt signals might act indirectly by up-regulating short-range signals that partition cranial ectoderm into otic placode and epidermis.
There is growing evidence that Wnt and Notch signaling pathways co-operate during cell fate determination in many tissues (Crosnier et al., 2006; Estrach et al., 2006; Fre et al., 2005). Notch signaling plays various roles in patterning the inner ear, ranging from specification of neurons and prosensory patches to the generation of the stereotypical pattern of mechanosensory hair cells and supporting cells (Adam et al., 1998; Brooker et al., 2006; Daudet et al., 2007; Daudet and Lewis, 2005; Haddon et al., 1998; Kiernan et al., 2005; Lanford et al., 1999; Shi et al., 2005). Both the Notch1 receptor and several of its ligands such as Jagged1 (Jag1) and Delta-like1 (Dll1) are expressed in the otic placode from very early stages (Abello et al., 2007; Adam et al., 1998; Daudet et al., 2007; Groves and Bronner-Fraser, 2000; Haddon et al., 1998). Notch signaling may therefore also have an early function during otic placode development. We now provide evidence that elements of the Notch pathway are positively regulated by Wnt signaling, and that Notch1 signaling can in turn modulate the canonical Wnt signaling pathway. We also show that while some aspects of otic placode identity are regulated only by Wnt signals, other features of placodal differentiation can be regulated independently by Wnt or Notch pathways.
MATERIALS AND METHODS
Genetically modified mice
The following lines of mice were used in this study: Pax2-Cre (Ohyama and Groves, 2004b; available from the Mutant Mouse Regional Resource Center; www.mmrrc.org/strains/10569/010569.html), conditionally activated Notch1 intracellular domain (cN1ICD; Murtaugh et al., 2003), Notch1 null mutants (Conlon et al., 1995), conditional activated β-catenin Catnblox(ex3) (cAct; Harada et al., 1999), conditional β-cateninfloxed mutants (β-cat- CKO; Brault et al., 2001) Tcf/Lef Wnt reporter (Mohamed et al., 2004); conditional Rbpj/Rbsuh mutants (Tanigaki et al., 2002) and a GFP-expressing Cre reporter (Z/EG; Novak et al., 2000). To generate cN1ICD animals, N1ICDfloxed homozygotes were crossed with Pax2-Cre animals. Age matched heterozygotes and wildtypes were used as controls for Notch1 mutant embryos. Detailed mating strategies for cAct and β-cat-CKO mice have been described previously (Ohyama et al., 2006). To generate Notch1; cAct mutants, a line that was heterozygous for Notch1; Pax2-Cre was crossed to animals that were heterozygous for Notch1; cAct. To generate cN1ICD; β-cat- CKO mutants, a line that was heterozygous for β-cat-null; Pax2-Cre was crossed to animals that were heterozygous for N1ICD and homozygous for a floxed allele of β-catenin. For each mutant genotype at least 3 embryos were analyzed except for Notch1; cAct mutants (n = 2). All animal experiments were done in accordance with the guidelines of the institution’s Animal Care and Use Committee.
Whole mount in situ hybridization, immunostaining and detection of β-galactosidase
Whole-mount in situ hybridization was performed as previously described (Ohyama et al., 2006). The following probes were used: Notch1 (Jeffrey Nye), Dll1 (Achim Gossler), Jag1 (Tim Mitsiadu), Hes1 and Hes5 (Ryoichiro Kageyama), Lunatic fringe (Lfng) (Thomas Vogt) and Wnt6 (Andrew McMahon). Probes for Pax2, Pax8, Foxi2, Dlx5, Krox20, Hoxb1, FGF3, and EphA4 have been previously described (Ohyama et al., 2006). Embryos were embedded in 15% sucrose and 7% gelatin in phosphate buffered saline (PBS) as previously described (Groves and Bronner-Fraser, 2000) and 15–30 mm thick sections cut using a Leica CM 1850 cryostat. Immunostaining and detection of β-galactosidase on cryostat sections and embryos was performed as previously described (Ohyama et al., 2006). The following primary antibodies were used: β-catenin (Zymed) at 1:200, activated Caspase-3 (R&D systems) at 1:1000, Green Fluorescent Protein (GFP) conjugated to fluorescein (Abcam) at 1: 250 to 1:500, β-galactosidase (ICN/MP Biochemicals) at 1:100, Jagged1 (Jag1; Santa Cruz) at 1:50 to 1:100, Pax2 (Zymed) at 1:500 and Phospho-Histone-H3 (PH3; Upstate/Millipore) at 1:1000. Secondary goat anti-rabbit antibody conjugated to Alexa 594 (Molecular probes) was used at 1:200. Sections were counterstained with the nuclear marker DAPI (Molecular probes). All images were captured using a Zeiss Axiocam digital camera and Axiophot2 or M2 Bio microscopes and processed using Adobe Photoshop CS software.
Quantification of thickened placode and average placode cell density in Notch1 mutants
The thickened otic placode was defined as the 2–3 cell layer of ectoderm located adjacent to rhombomere 5/6 (as identified morphologically with DAPI staining) and/or negative for Foxi2 transcripts. Quantifications of placode size were made from 15 μm serial sections from Notch1 mutants and age matched control embryos. Length measurements were made using Image J software. To allow for direct comparisons along the anterior-posterior (AP) axis of control and mutant mice, measurements were binned into 5 categories: 0–20 % (being the most anterior sections), 21–40 %, 41–60 %, 61–80 % and 81–100 % (being the most posterior sections). For a given genotype, each bin consisted of multiple sections from several embryos. The mean and standard error of the mean (SEM) were calculated for each bin. Non-parametric Mann-Whitney U-tests were performed to test for significance between genotypes. The cranial region of Notch1 mutants was comparable in size to controls. To confirm this we measured the dorsal-ventral length of the neural tube adjacent to the otic placode. The measurements were processed as described for the otic placode. We found no differences in neural tube length between Notch1 mutants and controls (data not shown). For average density measurements, serial 15 μm thick sections stained with DAPI and/or hybridized with Dlx5 or Foxi2 probes were used. Cell density for each section was calculated as follows: number of cells/μm2 × 500 and pooled for each genotype.
Quantification of cell proliferation, otic cup length and Wnt reporter domain length in cN1ICD mutants
Cell proliferation counts were performed as described previously (Ohyama et al., 2006). To account for variations in staging of embryos, the medio-lateral length of otic cup or Wnt domain was standardized against D-V neural tube length adjacent to the otic cup and expressed as a percentage. Only mid-sections from otic cups were used for quantitation and Student t-tests were performed to test for significance between genotypes.
RESULTS
Notch pathway genes are expressed during early otic placode development
The pre-otic field destined to give rise to the otic placode and surrounding epidermis is marked by Pax2 expression from the 0 somite stage (0ss; E8 in the mouse; Ohyama and Groves, 2004b; Ohyama et al., 2006). Pax2 expression later becomes restricted to the otic placode, which is morphologically visible as a thickening patch of ectoderm next to rhombomeres 5 and 6 from 8ss (E8.5) onwards (Ohyama and Groves, 2004b). To see if elements of the Notch pathway were expressed at an appropriate place and time to participate in otic placode induction, we compared the expression patterns of Notch1, Jag1, Dll1, Hes1, Lfng, and Hes5 to Pax2 (Fig. 1). At 0–1ss, no Notch pathway transcripts were detected in the pre-otic field (Fig. 1B–C). Onset of Notch1 expression was observed as early as the 4ss, becoming stronger by 5–7ss (Fig. 1C). Scattered cells in the anterior Pax2 domain adjacent to the neural tube expressed the Notch ligand Jag1 from around 5ss, although posterior cells did not express Jag1 until 8–9ss (arrowhead, Fig. 1C–D). Dll1 was also expressed adjacent to the neural tube at 4–5ss (Fig. 1B–C) and restricted to the otic placode from 9ss (Fig. 1C). Between 12–14ss Dll1 expression was gradually restricted to differentiating neuroblasts in the antero-ventral placode (data not shown; Adam et al., 1998). We were unable to detect Lfng at the pre-otic field and placode stages in whole mounts (data not shown), although at later otic vesicle stages, Lfng is expressed in the antero-ventral portion of the otic cup destined to produce the vestibuloacoustic ganglion and the utricular and saccular maculae (Morsli et al., 1998; Raft et al., 2004). Hes1 and Hes5 are effectors of the Notch pathway that function in many processes including regulation of cell fate decisions (Bray, 1998; Kageyama et al., 2007; Lai, 2004).
Fig. 1. Expression profile of Notch pathway genes during otic placode development.
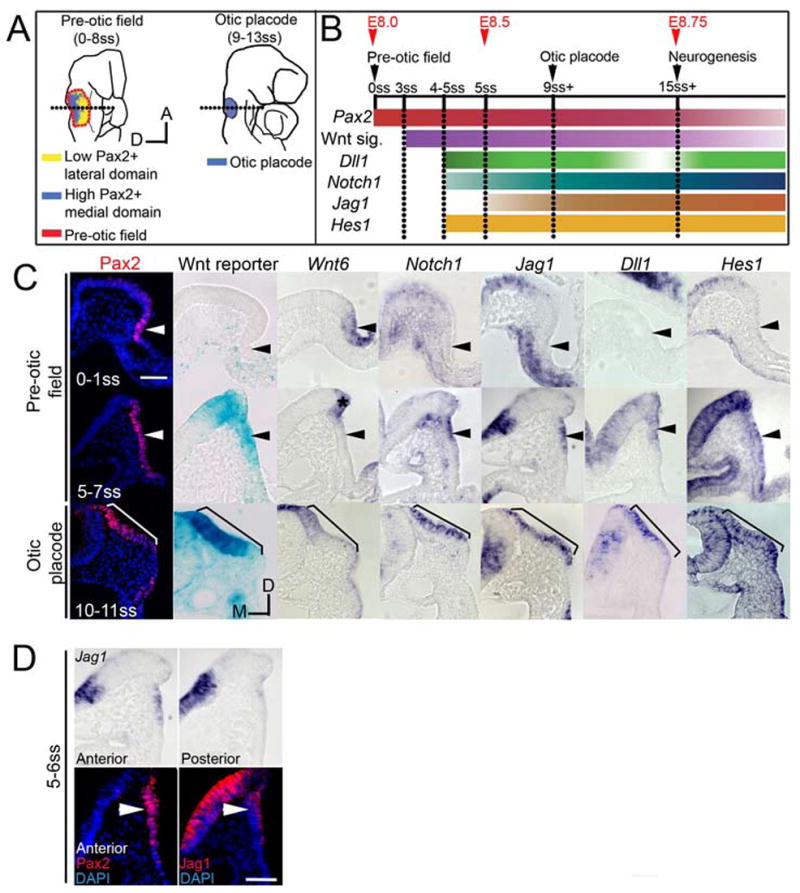
(A) Schematic views of embryos during otic placode development. The dotted line indicates the approximate level of sections in (C). (B) Summary showing the onset of Notch1 (blue), Jagged1 (Jag1; brown), Delta-like1 (Dll1; green), and Hes1 (yellow-orange) with respect to Pax2 (pink-red) and Wnt signaling (purple). Preceding neurogenesis, Dll1 expression is high in the pre-otic field and progressively becomes weaker in the placode. During neurogenesis Dll1 localizes to neuroblasts. (C) Sections comparing expression of Wnt6, Notch1, Jag1, Dll1, Hes1 with respect to Pax2 (red) and Wnt reporter (blue). Arrowheads mark the pre-otic field. Brackets mark the extent of the otic placode. (D) Top row: Jag1 expression in the anterior and posterior pre-otic field. Bottom row: consecutive serial sections through the pre-otic field immunostained with anti-Pax2 and anti-Jag1 antibodies respectively. Scale: 50 μm.
Hes1 expression was scattered throughout the pre-otic field and by 10–11ss was restricted to the otic placode (Fig. 1C). We found no evidence for Hes5 expression in the pre-otic field (data not shown). These data suggest that at least some transcriptional targets of the Notch pathway are expressed during early phases of otic placode development.
We previously used a transgenic Wnt reporter mouse line (Mohamed et al., 2004) to show that the canonical Wnt signaling pathway is activated in the pre-otic field between 3–5ss (Ohyama et al., 2006). Several Wnt family members are expressed in an appropriate location to trigger the observed Wnt reporter activity – for example, Wnt8 is expressed in rhombomere 4 (Ohyama et al., 2006). We also observed Wnt6 expression in the Pax2+ pre-otic field at 0ss. It continues to be expressed in the neural folds at 5–7ss and the dorsal-most region of the otic placode at 11ss (Fig. 1C; Lillevali et al., 2006). Since the onset of Notch pathway gene expression closely corresponded to Wnt reporter activity in the pre-otic field (Fig. 1B–C), we hypothesized that the Notch1 pathway may interact with the canonical Wnt signaling pathway in mediating the fate decision between otic placode and epidermis.
Notch pathway components are positively regulated by canonical Wnt signaling in the developing otic placode
Previous studies suggest that Notch pathway components can be regulated by β-catenin (e.g. Estrach et al., 2006; Katoh and Katoh, 2006). We therefore examined expression of Notch pathway genes in embryos carrying gain- or loss-of-function mutations of the canonical Wnt pathway in the Pax2+ pre-otic field. We crossed Pax2-Cre transgenic mice (Ohyama and Groves, 2004b) with mice in which β-catenin is constitutively activated in Cre-expressing cells (cAct; Harada et al., 1999) and examined expression of Jag1, Notch1 and Hes1. In cAct mutants, Jag1 expression was expanded ventrally to the level of the pharynx at the 9–10ss (bracket, Fig. 2A) and this ectopic expression continued until at least E9.0. Jag1 is thought to be a direct target of β-catenin, as its promoter region contains five, three and six consensus Tcf/Lef binding sites in mouse, human and rat respectively (Estrach et al., 2006; Katoh and Katoh, 2006). The domain of Notch1 and Hes1 expression also expanded, although only after a delay (from the 14–15ss; Fig. 2A, brackets). Such a delayed induction of Notch1 and Hes1 relative to Jag1 has also been observed in epidermis in which β-catenin is activated (Ambler and Watt, 2007). Other Notch pathway genes such as Dll1, Hes5 and Lfng were not expressed in cAct mutants (data not shown).
Fig. 2. The canonical Wnt pathway positively regulates components of the Notch pathway in the otic placode.
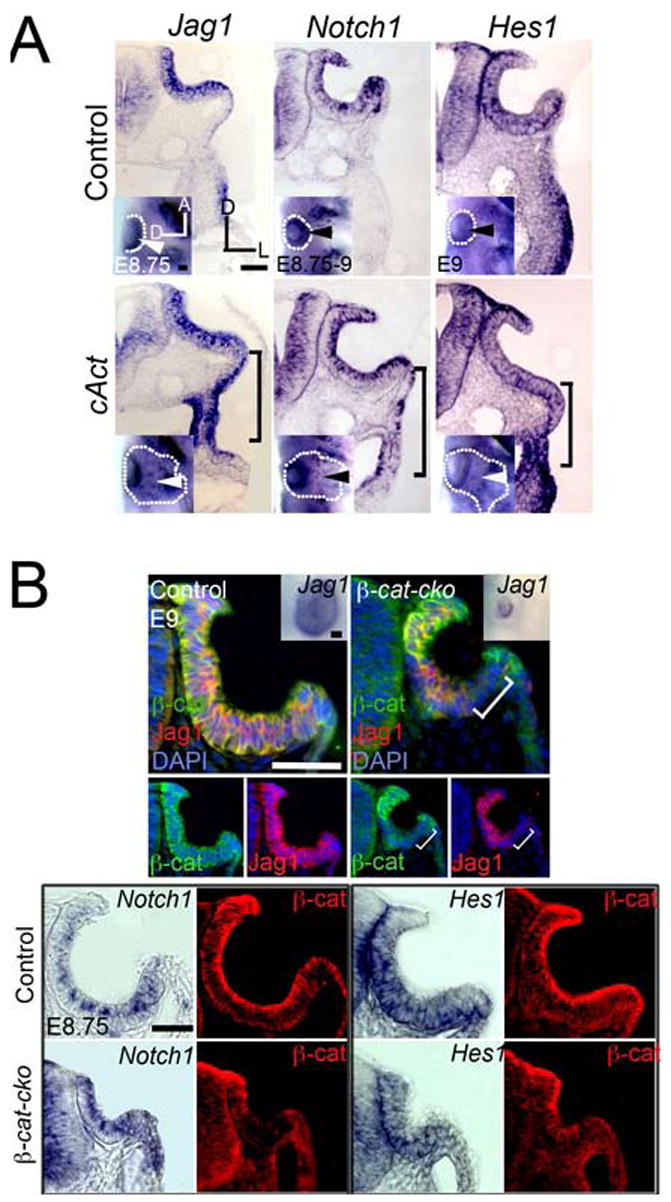
(A) Jag1, Notch1, and Hes1 domains are ectopically expanded in cAct embryos (bracket). Inset: corresponding whole-mounts. Arrowhead: normal (top row) or ectopic (bottom row) expression. Dotted outline: otic area. (B) Jag1, Notch1 and Hes1 domains are reduced in β-cat-CKO embryos. (Left panel) Anti-Jag1 (red) and β-catenin (β-cat, green) co-immunostaining. Bracket: β-catenin−: Jag1− cells. Inset: Jag1 expression at E9.5 in whole-mount. (Right panel) Whole-mount and (bottom panel) sections of Notch1 and Hes1 hybridized embryos. Scale: 50 μm.
To determine if Wnt signaling is necessary for expression of Notch pathway components, we analyzed expression of Jag1, Notch1 and Hes1 in the Pax2+ pre-otic field of mice lacking β-catenin (Brault et al., 2001; β-cat-CKO). The Jag1 domain was significantly reduced at 10–11ss, as we previously reported for Pax2 and Pax8 (Ohyama et al., 2006). Many cells within the vestigial β-cat-CKO otic vesicle were β-catenin−;Jag1−, suggesting that Wnt signaling is directly responsible for Jag1 induction in the placode (bracket; Fig. 2B). Close examination of the vestigial mutant vesicles at E9–9.5 revealed that cells expressing Jag1 were β-catenin+ and had therefore failed to undergo Cre-recombination (Fig. 2B). The domains of Notch1 and Hes1 were also significantly reduced, although the resolution of the whole mount in situ technique made it difficult to determine whether all Notch1 and Hes1 expressing cells also expressed β-catenin protein (Fig. 2B).
Wnt and Notch signaling pathways differentially regulate expression of otic markers
The expression of Notch pathway genes in the pre-otic field and otic placode, together with the regulation of these genes by Wnt signaling suggested that Notch signaling may participate in the fate decision between otic placode and epidermis. To test this, we conditionally activated Notch1 in the pre-otic field using mice in which the active, intracellular domain of Notch1 receptor (N1ICD) was knocked into the ROSA26 locus with a transcriptional STOP cassette flanked by LoxP sites (Murtaugh et al., 2003). We drove expression of N1ICD in the Pax2+ pre-otic field using Pax2-Cre mice (Ohyama and Groves, 2004b). The Pax2-Cre mouse line expresses Cre recombinase in the midbrain and rhombomere 1 (R1) of the hindbrain (Ohyama and Groves 2004b). Conditionally activated N1ICD (cN1ICD) mutants displayed an open neural tube phenotype at the level of the midbrain-R1 region, likely resulting from over-proliferation of precursor cells induced by Notch activation. However, the patterning of the posterior hindbrain next to the ear was normal at embryonic (E) day 8.5–9.5 based on the expression of HoxB1 (rhombomere 4), FGF3 (rhombomeres 5 and 6), EphA4 and Krox20 (rhombomeres 3 and 5; Fig. S1B), suggesting that any otic placode phenotype in cN1ICD mutants is not due to changes in the adjacent hindbrain.
We examined embryos inheriting both the Cre-inducible N1ICD and Pax2-Cre transgenes for otic placode and epidermal markers. The N1ICD transgene also harbors an IRES-nGFP sequence, allowing visualization of N1ICD-expressing cells by GFP fluorescence. GFP-expressing cells were observed throughout the pre-otic field from 5–6ss (data not shown). E9.5 cN1ICD mutant embryos displayed GFP expression throughout a thickened placode-like structure which expanded to the level of the ventral pharynx (Fig. S1A). Analysis of cN1ICD embryos at 10–11ss revealed that the Pax8 domain was expanded ventrally (arrowheads and brackets, Fig. 3A, A′). We previously showed that Foxi2 is an epidermal marker expressed in a complementary manner to Pax2 and Pax8 during otic placode development (Ohyama and Groves, 2004a). By E8.75–9, Foxi2 expression was reduced dramatically in cN1ICD mutants compared to controls (dotted outline and brackets, Fig. 3C–C′), complementing the expansion of the thickened epidermis. To determine if cell proliferation was responsible for the expanded placode, we examined expression of the M-phase marker phosphohistone-3 (PH3) in cN1ICD embryos (n = 10 placodes) and control embryos produced by crossing the Pax2-Cre line with the Cre inducible Z/EG GFP-expressing line (Novak et al., 2000). We saw no significant differences in total PH3+ or PH3+;GFP+ cell counts per section (Fig. S1D).
Fig. 3. Some, but not all otic markers are expanded at the expense of epidermis in conditionally activated Notch1 (cN1ICD) embryos.
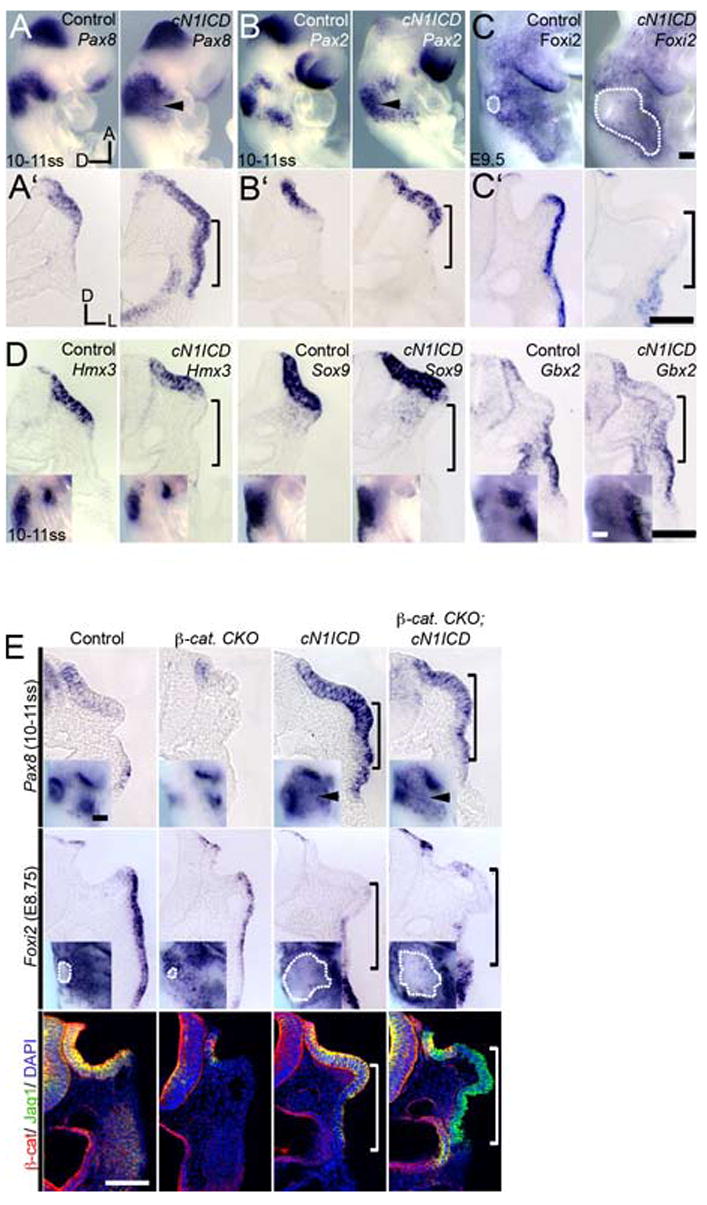
(A–C): Expanded otic placode in cN1ICD embryos hybridized with probes for Pax2 (A, A′), Pax8 (B, B′) and the epidermal marker Foxi2 (C, C′). Arrowhead: ectopic expression. (C) Dotted outline indicates the invaginating otic cup (controls) or expanded otic region (cN1ICD mutants). (A′–C′) Corresponding transverse sections. Brackets: lateral ectopic placode region. Scale: 100 μm. (D) Hmx3, Sox9 and Gbx2 otic markers are not expanded in cN1ICD mutants. (E) In β-cat-CKO; cN1ICD mutants, Pax8 (top row) and Jag1 (green; last row) are expanded at the expense of Foxi2 (middle row). Last row; β-catenin (red) and Jag1 protein expression in Foxi2 hybridized embryos. Scale: 100 μm.
The expansion of Pax8 at the expense of Foxi2 in cN1ICD embryos is strikingly similar to that seen in embryos in which the canonical Wnt pathway is activated (cAct embryos; Ohyama et al., 2006). However, in contrast to cAct embryos, we saw only a modest expansion of the Pax2 domain (bracket and arrowhead, Fig. 3B, B′; Fig. S1C) and no expansion of the otic markers Gbx2 or Sox9 (Fig. 3D). Finally, a marker of the dorso-lateral otocyst, Hmx3, which does not require either Wnt or Hedgehog signaling for its expression (Ohyama et al., 2006; Riccomagno et al., 2002) was also not expanded in cN1ICD mutants (Fig. 3D).
These results suggest that different aspects of otic placode development are differentially regulated by Wnt and Notch signaling. Placode markers such as Pax2, Gbx2, and Sox9 appear to be regulated by Wnt signaling (Ohayama et al., 1996; Saint-Germain et al., 2004), but not Notch signaling, whereas markers such as Pax8, the morphological thickening of epithelium and the repression of the epidermal marker Foxi2 can be regulated by both Notch and Wnt signals. To determine if Notch signaling can regulate these markers independently of Wnt signaling, we analyzed β-cat-CKO;cN1ICD mutant embryos in which β-catenin was inactivated and Notch1ICD was activated throughout the pre-otic field. Mutant embryos displayed greatly expanded regions of thickened placode-like epithelium that expressed both Pax8 and Jag1 (Figure 3E). This expanded region of thickened epithelium was also largely devoid of Foxi2 expression (Fig. 3E), although occasional Foxi2+ patches of cells could sometimes be detected. These results show that Notch and Wnt signals can independently regulate some aspects of otic placode development.
Inactivation of Notch1 reduces the size of the otic placode
Our results show that Notch1 activation throughout the Pax2+ pre-otic field expands some otic placode markers at the expense of epidermis. In complementary experiments, we examined Notch1 mutants, in which a substantial portion of the Notch1 gene is deleted (amino acids 1056–2049; Conlon et al., 1995). This deletion encompasses RAM and Ankyrin repeats required for RBPJk signaling (Conlon et al., 1995; Fortini and Artavanis-Tsakonas, 1994; Kurooka et al., 1998a; Kurooka et al., 1998b; Lamar et al., 2001; Nam et al., 2003; Tani et al., 2001). We confirmed that posterior hindbrain patterning was normal in Notch1 mutants by assaying for Hoxb1, FGF3 and Krox20 expression (Fig. S2A). All three genes were expressed normally, suggesting that any defects observed in otic placode development are due to deficiency in Notch1 signaling in the placode, rather than in the hindbrain.
To determine if Notch signaling was necessary for the expression of otic markers, we examined Pax2 and Pax8 expression in Notch1 mutants. By 9–11ss there was a dramatic down-regulation of Pax2 expression in mutants in both the otic region and the epibranchial placodes (Fig. 4A). While the anterior-posterior limits of Pax8 expression in the otic region was reduced, the expression in the hyoid arch was relatively unchanged (Fig. 4B). In Notch1 mutant whole mounts, the limits of Pax2/8 domains in the anterior-posterior axis were reduced (brackets, Fig. 4). Sections through Notch1 mutants also revealed a reduction in the medial-lateral extent of Pax2/8 expression (brackets, Fig. 4).
Fig. 4. Domains of Pax2 and Pax8 are reduced in Notch1 mutants.
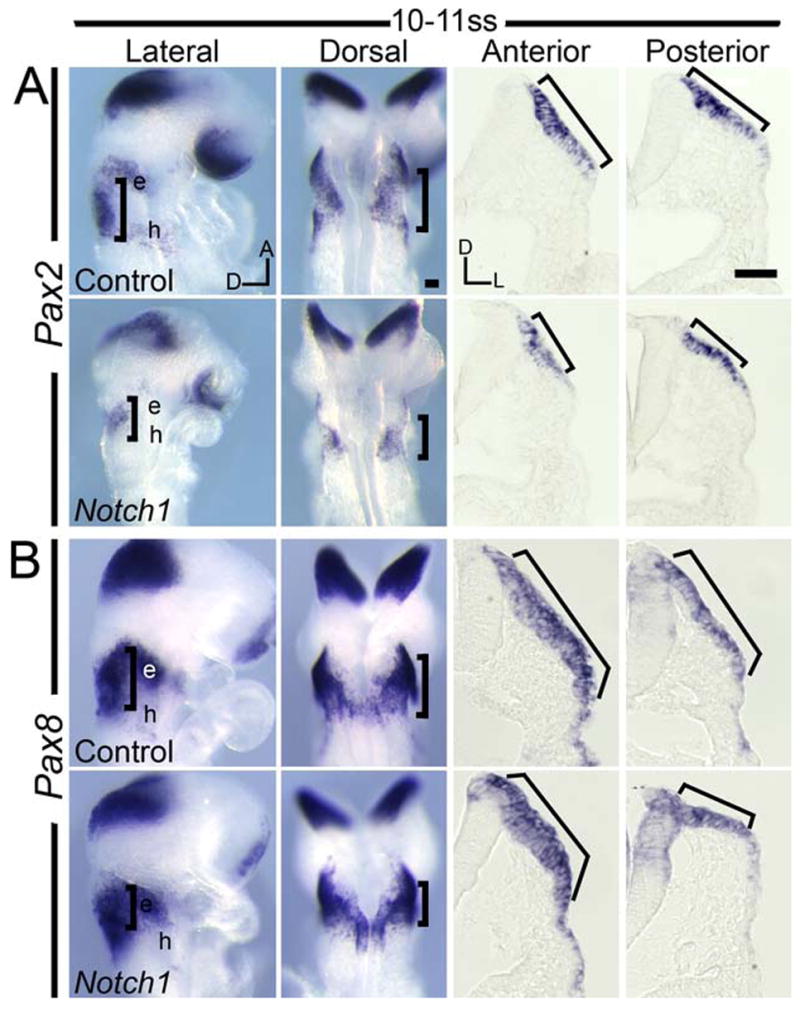
(A–B) Dorsal and lateral whole-mount views of Pax2 (A) and Pax8 (B) expression. Anterior and posterior sections through control and Notch1 mutant placodes are also shown. Brackets: otic expression. e, epibranchial placode; h, hyoid arch. Scale: 50 μm.
The reduction in the size of the otic placode in Notch1 mutants may result from increased apoptosis, increased cell density or a change in cell fate. We measured the size of the placode by examining Foxi2 expression, which is precisely excluded from the thickened placode region. The Notch1 mutant otic placode was indeed smaller at 9–13ss on the basis of Foxi2 expression (dotted outline, Fig. 5A–B). We compared the medial-lateral extent of the thickened otic placode in Notch1 mutants and controls at 9–11ss and 12–13ss (see Methods: Fig. 5C, D) Notch1 mutants (9–11ss, n = 25 placodes; 12–13ss, n = 10 placodes) had significantly smaller placodes compared to controls (9–11ss, n = 13 placodes; 12–13ss n = 6 placodes) regardless of the axial level of the section (p < 0.05–0.005; Fig. 5C–D). There were no significant changes in placode cell density at 9–11ss (n = 10 mutant placodes; n = 6 control placodes) and 12–13ss (n = 5 mutant placodes; n = 4 control placodes; p > 0.05; Fig. 5F), or in apoptosis when analyzed for activated caspase-3 expression (Fig. 5G; Conlon et al., 1995; Del Monte et al., 2007). We also confirmed that the smaller placode was not due to the precocious generation of neurons by analyzing Ngn1 expression (data not shown).
Fig. 5. The otic placode is significantly reduced in Notch1 mutants.
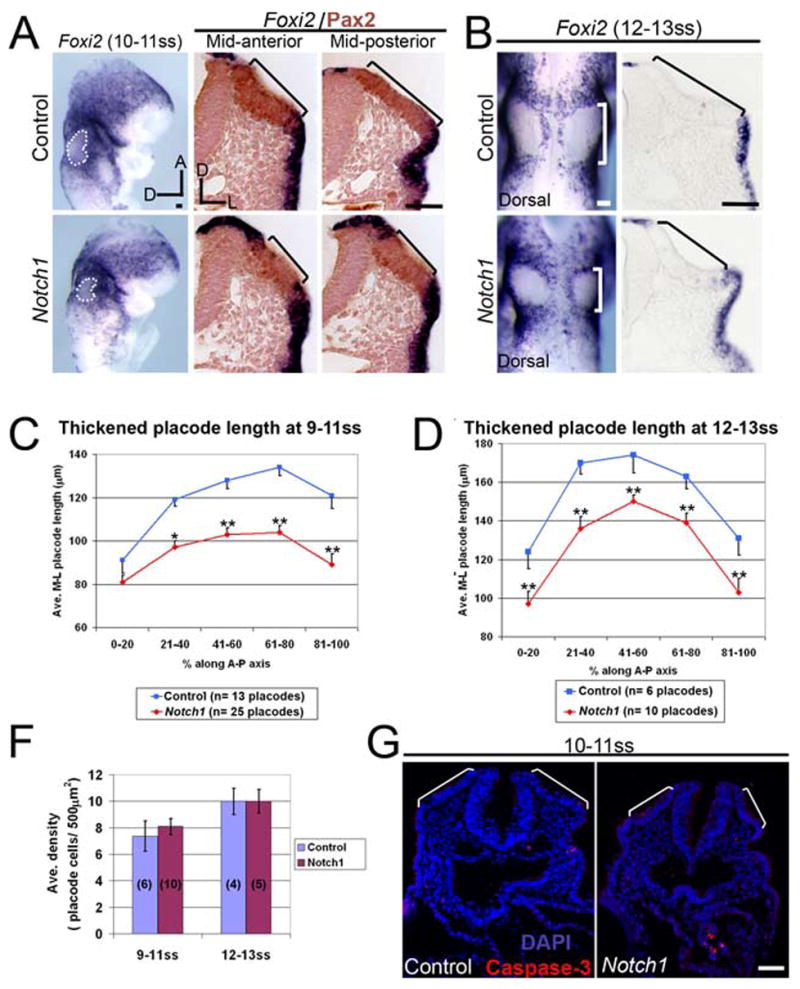
(A–B) Foxi2 expression at 9–11ss (A) and at 12–13ss (B) in whole mounts and sections. Dotted outline/brackets indicate the thickened otic placode region that does not express Foxi2. (A) Corresponding sections showing Foxi2 transcript (blue) and Pax2 protein expression (brown). (C–D) Quantitative comparison of medial-lateral placode length in control and Notch1 mutant embryos at 9–11 somite stage (ss) (C) and 12–13ss (D) (refer to Materials and Methods). (*) p < 0.05 and (**) p < 0.005. (F) Quantitation of average cell density in control and Notch1 mutant. Placode n values in parentheses. (G) Activated Caspase-3 expression (red) indicating lack of apoptotic cells within the Notch1 mutant otic placode at 10–11ss. Bracket: thickened placode. Error bars: SEM. Scale: 50 μm.
Collectively, our data show that many otic placode precursors undergo a fate change to epidermis in Notch1-deficient embryos. It is possible that the other Notch receptors are active during otic placode development in addition to Notch1. We confirmed our results by examination of conditional mutants of RBPJ/Rbsuh, a transcriptional co-factor of NICD. The otic placode still forms in these mice (Fig. S2C; see also Oka et al., 1995; de la Pompa et al., 1997), confirming that Notch signaling can modulate the size of the otic placode but is not necessary for its induction.
Daudet and colleagues recently suggested that initiation, but not maintenance of Jag1 expression in the chick otic placode is regulated independently of Notch1 signaling (Daudet et al., 2007). We confirmed this result in mice: Jag1 continued to be expressed in the placode of Notch1 mutants, but the intensity of expression was reduced compared to controls (Fig. S2B). It has been previously reported that Jag1 continues to be expressed in a morphologically distinct otic placode in mice carrying mutations of Pofut1, an O-fucosyltransferase essential for Notch signaling (Shi and Stanley, 2003). We confirmed that Jag1 and Hes1 expression can be initiated in the absence of canonical Notch signaling by examination of conditional mutants of RBPJ/Rbsuh. Both genes continue to be expressed in a morphologically visible otic cup, although Hes1 was expressed at significantly reduced levels compared to controls (Fig. S2C). This is consistent with Hes1 expression being initiated by Notch signaling, but Jag1 expression being initiated independently of Notch signaling.
Notch1 augments canonical Wnt signaling in the otic placode
Canonical Wnt signaling plays an important role in defining the size of otic placode by driving medial Pax2+ pre-otic cells towards an otic rather than cranial epidermis fate (Ohyama et al., 2006). Similarly, conditional activation of Notch1 in the Pax2+ pre-otic field expands some, but not all otic markers at the expense of epidermis (Fig. 3). Additionally, Notch pathway gene expression can be activated by canonical Wnt signaling (Fig. 2). These results suggested the possibility of reciprocal interactions between the Notch and Wnt pathways. To test whether Wnt signaling is modulated by the Notch pathway in the developing otic placode, we crossed Wnt reporter mice expressing a β-galactosidase reporter gene under the control of six Tcf/Lef DNA binding sites (Mohamed et al., 2004) to either cN1ICD or Notch1 mutant lines.
Surprisingly, although the thickened Pax8+placode was dramatically expanded to the level of the pharynx in cN1ICD embryos (Fig. 3A), Wnt reporter activity showed a much more modest expansion, extending a little beyond the lateral edge of the otic cup (Fig. 6A). We observed similar results with Dlx5, a known Wnt-responsive marker of the otic placode (bracket, Fig. 6A). To verify these results, we made use of the fact that cN1ICD mutants also express nuclear GFP after Cre recombination (Murtaugh et al., 2003). We co-immunostained cN1ICD;Wnt reporter embryos with anti-β-galactosidase and anti-GFP antibodies to mark the extent of the Wnt reporter and the expanded otic placode respectively (Fig. 6B). By E9–9.25, Wnt activity was elevated in the lateral regions of the mutant otic cup which normally demonstrate moderate or low Wnt activity (red arrowhead; Fig 6A). Furthermore, the otic cup region was larger in cN1ICD mutants compared to controls (Fig. S3, n = 13 mutant placodes, n = 14 control placodes; p < 0.005). However, the ectopic placode region lateral to the otic cup which expressed N1ICD and GFP did not express β-galactosidase (bracket, Fig. 6B). These results suggest that Notch signaling can augment Wnt signaling, but that the active Notch1 ICD does not directly regulate Wnt-responsive genes containing Tcf/Lef DNA binding sites.
Fig. 6. Notch1 signaling augments Wnt signaling in the otic placode.
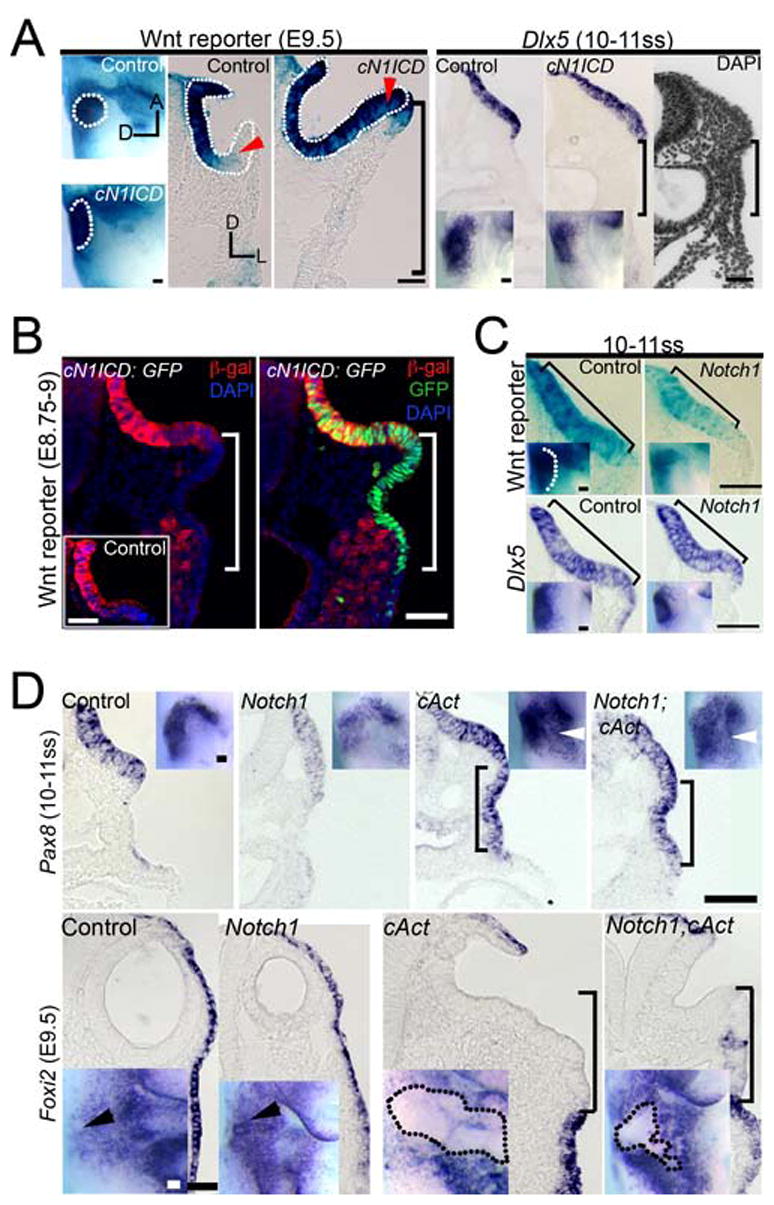
(A) Wnt reporter and Dlx5 mRNA expression is increased in conditionally over-expressing Notch-ICD (cN1ICD) embryos in whole-mount (top row) and corresponding mid-placode transverse sections (bottom row). Dotted outline: otic cup. Red arrowhead: medio-lateral otic region. (B) Wnt reporter expression in transverse sections of control and cN1ICD; Wnt reporter embryos; Wnt reporter mice co-immunostained with anti-β-galactosidase (β-gal; red) and anti-GFP (green) antibodies. Note that only the medial part of the expanded placode expresses the Wnt reporter. The inset shows anti-β-galactosidase staining in a normal Wnt reporter mouse. (A–B) Bracket: ectopic lateral placode region is negative for Wnt reporter and Dlx5. (C) Wnt reporter and Dlx5 expression is diminished in Notch1 mutants relative to controls. Brackets: otic placode. (D) A comparison of Pax8 and Foxi2 expression in Notch1 mutant, cAct and Notch1; cAct double mutant littermates. (A, C, D) Arrowhead: otic expression. Scale: 50 μm.
To test if Wnt signaling can also be modulated by loss of Notch1 activity, we examined Notch1 mutant mice crossed to a Wnt reporter mouse background. As expected, Wnt reporter activity was detected in Notch1 mutant placodes (Fig. 6C). However, the intensity of Wnt activity, as measured by time-matched β– galactosidase reactions was weaker compared to controls. Additionally, the medio-lateral extent of the Wnt reporter and expression of the Wnt-responsive gene Dlx5 was slightly reduced (Fig. 6C), reflecting the observed reduction in the placode size due to Notch1 deficiency (Fig. 4). Taken together with our data showing that Wnt signaling can up-regulate Notch pathway components, our results are consistent with a model in which the Wnt pathway can positively regulate components of the Notch pathway, and can in turn be augmented by Notch signaling. One prediction of this model is that maximal activation of Wnt signaling by a constitutively activated β–catenin mutation will be unaffected by a Notch1 mutation. To test this, we analyzed Pax8 and Foxi2 expression in Notch1 mutant embryos that also carried the activated β-catenin (cAct) mutation. As expected, the size of the expanded Pax8 domain seen in cAct embryos was not significantly different from Notch1; cAct mutants (Fig. 6D). Similarly, the reduced domain of epidermal Foxi2 expression seen in cAct mutants was not significantly different in Notch1; cAct mutants (Fig. 6D).
DISCUSSION
Notch signaling plays multiple roles in inner ear patterning, from specification of neurons and prosensory patches to the generation of the stereotypical pattern of hair cells (Adam et al., 1998; Brooker et al., 2006; Daudet et al., 2007; Daudet and Lewis, 2005; Haddon et al., 1998; Kiernan et al., 2005; Lanford et al., 1999; Shi et al., 2005). Here, we have uncovered new roles for Notch and Wnt signaling in the early development of the ear. Conditional activation of Notch1 in the Pax2+ pre-otic field causes the expansion of some, but not all otic markers at the expense of epidermis. Conversely, in the absence of Notch1 signaling, the otic placode is significantly smaller. We have also shown that Wnt signaling regulates components of the Notch pathway, such as Jag1, and that Notch signaling positively regulates Wnt signaling. Our results suggest that Notch augments the Wnt signaling pathway to help define the size of the otic placode
The expression of Notch signaling pathway components in the otic placode – a role for Wnt signaling
Our expression data shows that several components of the Notch signaling pathway – Notch1, Jag1, Dll1 and Hes1 – are expressed in a medial subset of the mouse Pax2+ pre-otic field from the 5ss onwards, and that Wnt signaling initiates expression of at least some Notch pathway components. Notch1, Jag1, Dll1 and Hes1 are all expressed in the pre-otic field after Wnt6, Wnt8 and the first signs of Wnt reporter activity (Fig. 1C and Ohyama et al., 2006). Expression of these Notch pathway genes occurs only within the region of the medial Pax2+ pre-otic field that responds to Wnt signaling (Fig. 1C). Consistent with previous reports (Duncan et al., 2005; Espinosa et al., 2003; Estrach et al., 2006), we found that ectopic activation of the canonical Wnt pathway induced expression of Jag1, Notch1 and Hes1 (Fig. 2A), while conditional deletion of β-catenin greatly reduced their expression (Fig. 2B). Wnt signaling can control transcription of Notch pathway genes by directly acting on elements located in their promoters (Duncan et al., 2005; Espinosa et al., 2003; Estrach et al., 2006; Katoh and Katoh, 2006). In the case of Jag1 promoter, there are multiple Tcf/Lef binding sites that are conserved between mouse and human (Estrach et al., 2006; Katoh and Katoh, 2006). Putative Tcf/Lef binding sites have also been identified in the Notch1 promoter (Galceran et al., 2004), however, although the Dll1 promoter also has Tcf/Lef binding sites (Galceran et al., 2004), its expression was not expanded in embryos expressing activated β-catenin. Recent evidence suggests that factors distinct from Notch signaling are required to initiate Jag1 expression in the chick otocyst (Daudet et al., 2007), although maintenance of Jag1 is Notch-dependent. Our results suggest that Jag1 initiation in the developing ear may be directly regulated by Wnt signaling (Fig. 2B, 7) while Notch1 and Hes1 expression may be initiated by Wnt signaling and possibly also by FGFs (Norgaard et al., 2003; Zhou and Armstrong, 2007), in addition to Notch signaling itself.
Fig. 7. Model of how Wnt and Notch pathways interact to regulate the size of the otic placode.
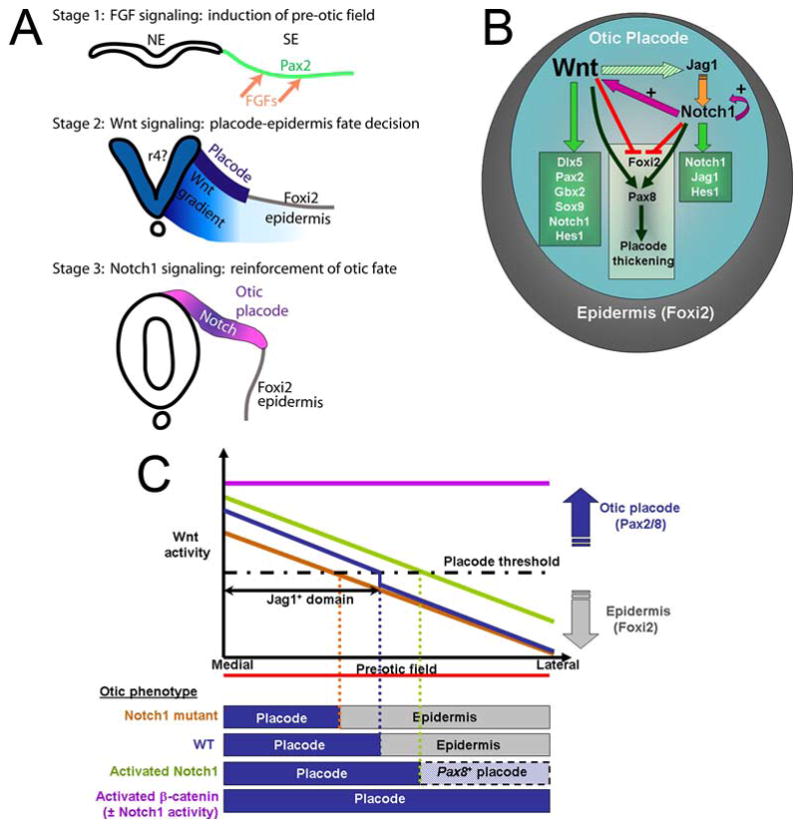
(A) The generation of the otic placode can be divided into three stages. Pax2 in the pre-otic field is induced by FGFs (arrows). A gradient of Wnts (light blue) determines the size of the otic field; above a certain threshold, Wnts drive cells towards an otic fate (dark blue) and below the threshold, cranial epidermis is formed (Foxi2) (Ohyama and Groves, 2003). Notch1 signaling is superimposed on the Wnt gradient (pink-blue) and acts to augment otic fate imposed by Wnts. NE; Neuroectoderm, SE; Surface ectoderm (this paper). (B) The Wnt pathway is the primary signal (denoted by bold lettering) that controls otic fate (blue region) by positively regulating (green arrows) the expression of Dlx5, Sox9, Gbx2, Pax2/8 and components of the Notch1 pathway such as Notch1 and Hes1 (Figs. 1 and 2). Jag1 expression is initiated by Wnts (dashed green arrow) (Fig. 2). Notch1 acts to: (1) augment Wnt and Notch1 activity within otic cells (pink arrow; plus sign) and (2) co-operate with Wnt to negatively regulate Foxi2 (red) and positively regulate Pax8 (dark green) and maintain a thickened otic placode. (C) A model summarizing the various otic placode phenotypes observed in this study. A gradient of Wnt activity emanating from the midline is established across the medio-lateral axis of the pre-otic field. Cells exposed to a certain threshold of Wnt signals express Jag1 and differentiate as otic placode (blue). Below this threshold, cells differentiate as epidermis (grey). Jag1-Notch1 signaling augments Wnt signals in the medial region of the otic placode, whereas more lateral regions are not exposed to Notch1 signals and Wnt signaling is not augmented. In the absence of Notch1 (yellow line), the gradient of Wnt signaling becomes weaker, resulting in a smaller placode and more epidermis. When Notch1 is activated in the pre-otic field (green line), the Wnt gradient is augmented further. Some Wnt-dependent markers (Dlx5) are expressed only in the expanded Wnt domain, whereas markers such as Pax8 are expressed throughout the pre-otic field (marked as lateral placode). When β-catenin is activated in the entire pre-otic field (purple line), all cells differentiate as otic placode (Ohyama et al., 2006).
Overlapping and distinct functions of Notch and Wnt signaling in the otic placode
We recently showed that the mouse pre-otic field defined by expression of Pax2 undergoes a fate decision to give rise to cranial epidermis and otic placode (Ohyama et al., 2006). The placode-epidermis fate decision is mediated by the canonical Wnt pathway, such that conditional deletion of β-catenin in Pax2+ cells drastically reduces the otic placode and expands epidermis, whereas conditional activation of β-catenin in Pax2+ cells expands the otic placode at the expense of epidermis (Ohyama et al., 2006). In the light of the expression of many components of the Notch signaling pathway in the developing otic placode (Fig. 1), we hypothesized that Notch signaling may act with the canonical Wnt pathway to specify otic placode identity.
Activation of Notch1 signaling in the pre-otic field leads to a massive expansion of thickened, placode-like epithelium expressing Pax8 at the expense of Foxi2+epidermis in a manner very similar to activation of β-catenin (Fig. 3). In contrast, although Pax2 expression can be expanded by activation of Wnt signaling (Ohyama et al., 2006) it showed only modest expansion in cN1ICD mutants compared to Pax8 (compare Fig. 3A and B). Pax2 and Pax8 are known to be differentially regulated by FGF signaling and the foxi1 transcription factor during induction of the zebrafish ear (Hans et al., 2004; Nissen et al., 2003, Solomon et al., 2003; 2004), and our results suggests these genes may also be differentially regulated by Notch signaling. In particular, Pax8 can be regulated by either canonical Wnt signaling, or by Notch signaling. However, it is not clear whether the two pathways regulate Pax8 in entirely different ways or whether they converge on a nodal point, such as the binding of Lef/Tcf complexes to the Pax8 promoter (Schmidt-Ott et al., 2007). Pax8 expression correlates with epithelial thickening in all experiments in our study. However, further experiments are required to determine whether Pax8 is directly responsible for regulating this morphological change in the otic placode.
Examination of Notch1 mutants consistently showed a significant reduction in the size of the otic placode (Fig. 4B–C; 5). This small reduction is unlikely to be due to redundancy with other Notch genes, as there is no detectable expression of Notch2–4 in the otic placode (Lewis et al., 1998; Williams et al., 1995). A similar persistence of the otic placode is seen after treating chick otic ectoderm with DAPT, a γ-secretase inhibitor that abolishes Notch signaling (Abello et al., 2007; Daudet et al., 2007), in mice carrying mutations in Pofut1, an O-fucosyltransferase that is an essential component of the Notch pathway (Shi and Stanley, 2003; C.J., unpublished), and in mice lacking RBPJ/Rbsuh/CSL (Oka et al., 1995; de la Pompa et al., 1997; Fig. S2C). In all these experiments, any reduction in placode size in the absence of Notch signaling is much more modest than that seen in mice in which Wnt signaling is blocked by conditional deletion of β-catenin (Ohyama et al., 2006; Fig. 2B).
Our results suggest a model (Fig. 7C) in which both Notch and Wnt signaling can specify the size of the epithelium destined to form the otic placode by virtue of their regulation of Pax8, Foxi2, Jag1 and the induction of a thickened epithelial morphology. Our data from mice in which Wnt signaling is activated in the absence of Notch1 (Fig. 6D), or Notch1 is activated in the absence of β-catenin (Fig. 3C) show that the two pathways can regulate these genes independently of each other. However, unlike the Wnt pathway, Notch signaling does not regulate the expression of otic placode-specific genes such as Gbx2, Sox9 and Hmx3, as these are unchanged in cN1ICD mutants (Fig. 3D). In addition, our results, taken together with previously published studies, suggest that Notch signaling also acts to augment Wnt signaling during otic placode induction, rather than being absolutely necessary for placode induction
Notch signaling acts to augment Wnt signaling during otic placode induction
To integrate our gain- and loss-of-function experiments with the Notch and Wnt pathways, we propose a model in which some Notch pathway components such as Jag1 are induced by Wnt signaling. Subsequently, activation of Notch1 by Jag1 feeds back to augment the Wnt response (Fig. 7B). This feedback activity has no effect on the most medial regions of the pre-otic field - which receive the highest levels of Wnt signaling - but acts to increase Wnt signaling in medio-lateral regions of ectoderm that receive modest to low levels of Wnt signaling. Thus, Notch-mediated feedback serves to sharpen and refine the initial medio-lateral gradient of Wnt activity during the pre-otic field (Ohyama et al., 2006) into a more binary pattern at the otic placode stage, where Wnt signaling is either active (giving rise to otic placode) or silenced (giving rise to epidermis; Fig. 7C).
Our data support this model in four ways. First, Notch1 deficiency causes a reduction in the area and intensity of β-galactosidase activity in Wnt reporter mice and a reduction of the domain of the Wnt-responsive gene Dlx5 (Fig. 6C). However, loss of Notch1 does not abolish the expression of either marker, consistent with the notion that Notch1 signaling augments the Wnt response but does not initiate it. Second, the reduction in Wnt signaling resulting from loss of Notch1 (Fig. 4; 5) causes a consistent reduction in the size of the otic placode, but does not eliminate it entirely. The otic placode also forms in mice lacking other crucial components of the Notch pathway, such as Pofut1 or Rbpj/Rbsuh/CSL (Oka et al., 1995; de la Pompa et al., 1997; Shi and Stanley, 2003). Third, mutation of Notch1 has no effect on the size of the otic placode in embryos also expressing constitutively active β-catenin in the entire pre-otic field (Fig. 6D), presumably because cells expressing artificially high levels of activated β-catenin are not dependent on Notch1 function for stabilization of otic fate. Finally, artificial N1ICD activation throughout the pre-otic field greatly expands Pax8 to the ventral pharynx, but this is not the case for Dlx5 or Wnt activity (Fig. 6A). This suggests that ectopic activation of N1ICD in regions of the pre-otic field that receive no Wnt signals is insufficient to augment or initiate the Wnt response (Fig. 7). Furthermore, Wnt reporter expression is enhanced in regions receiving moderate levels of Wnt activity in cN1ICD mutants (Fig. 6A). Although, the mechanism of how Notch signaling augments Wnt activity is not clear, this result suggests that it is unlikely that N1ICD can directly activate transcription of Wnt-responsive genes by itself. A growing body of evidence suggests that Wnt and Notch pathways interact during cell fate determination (Aoyama et al., 2007; Arias and Hayward, 2006; Crosnier et al., 2006; Estrach et al., 2006; Fre et al., 2005). Notch signaling can act upstream of the Wnt pathway (Balint et al., 2005; Johnston and Edgar, 1998; Neumann and Cohen, 1996), or downstream (Estrach et al., 2006). Stimulation of the Wnt pathway can either antagonize or activate the Notch pathway in different contexts – for example, dishevelled can antagonize Notch signaling (Axelrod et al., 1996), whereas down-regulation of GSK3 activity by Wnt signaling stimulates the Notch pathway (Espinosa et al., 2003). The Notch receptor is also able to antagonize β-catenin activity (Nicolas et al., 2003), sometimes in an NICD-independent manner (Hayward et al., 2006; Hayward et al., 2005).
Taken together, our current and previously published data suggest a model of otic placode induction where FGF signaling initially establishes a Pax2+ pre-otic field that is then patterned by a gradient of Wnt signaling arising from the midline. Wnt signaling up-regulates components of the Notch pathway, which then act locally to augment the Wnt response and to mediate the placode-epidermis fate decision in the pre-otic field.
Acknowledgments
We thank the following colleagues for generously providing transgenic mice for this study: Charles Murtaugh (N1ICD mice), Daniel Dufort (Wnt reporter mice), Ron Conlon (Notch1 mutant mice), Tasuku Honjo (conditional Rbpj mice) and Mark Taketo (cAct β-catenin mice). We also thank Jeffrey Nye, Achim Gossler, Tim Mitsiadu, Ryoichiro Kageyama, Thomas Vogt and Andy McMahon for gifts of probes, Juan Llamas, Welly Makmura, Sheri Juntilla, Francesca Della Ripa and Juemei Wang for excellent technical support, and for Groves and Segil lab members for help and advice during this project. This work was funded by the House Ear Institute, by a March of Dimes Research Grant (A.K.G.), and by RO1DC04675 (A.K.G.), and RO1DC06185 (A.K.G. and N.S.).
References
- Abello G, Khatri S, Giraldez F, Alsina B. Early regionalization of the otic placode and its regulation by the Notch signaling pathway. Mech Dev. 2007;124:631–45. doi: 10.1016/j.mod.2007.04.002. [DOI] [PubMed] [Google Scholar]
- Adam J, Myat A, Le Roux I, Eddison M, Henrique D, Ish-Horowicz D, Lewis J. Cell fate choices and the expression of Notch, Delta and Serrate homologues in the chick inner ear: parallels with Drosophila sense-organ development. Development. 1998;125:4645–54. doi: 10.1242/dev.125.23.4645. [DOI] [PubMed] [Google Scholar]
- Ambler CA, Watt FM. Expression of Notch pathway genes in mammalian epidermis and modulation by beta-catenin. Dev Dyn. 2007;236:1595–601. doi: 10.1002/dvdy.21151. [DOI] [PubMed] [Google Scholar]
- Aoyama K, Delaney C, Varnum-Finney B, Kohn AD, Moon RT, Bernstein ID. The Interaction of The Wnt and Notch Pathways Modulates Nk vs. T Cell Differentiation. Stem Cells. 2007 doi: 10.1634/stemcells.2007-0102. [DOI] [PubMed] [Google Scholar]
- Arias AM, Hayward P. Filtering transcriptional noise during development: concepts and mechanisms. Nat Rev Genet. 2006;7:34–44. doi: 10.1038/nrg1750. [DOI] [PubMed] [Google Scholar]
- Axelrod JD, Matsuno K, Artavanis-Tsakonas S, Perrimon N. Interaction between Wingless and Notch signaling pathways mediated by dishevelled. Science. 1996;271:1826–32. doi: 10.1126/science.271.5257.1826. [DOI] [PubMed] [Google Scholar]
- Balint K, Xiao M, Pinnix CC, Soma A, Veres I, Juhasz I, Brown EJ, Capobianco AJ, Herlyn M, Liu ZJ. Activation of Notch1 signaling is required for beta-catenin-mediated human primary melanoma progression. J Clin Invest. 2005;115:3166–76. doi: 10.1172/JCI25001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barald KF, Kelley MW. From placode to polarization: new tunes in inner ear development. Development. 2004;131:4119–30. doi: 10.1242/dev.01339. [DOI] [PubMed] [Google Scholar]
- Brault V, Moore R, Kutsch S, Ishibashi M, Rowitch DH, McMahon AP, Sommer L, Boussadia O, Kemler R. Inactivation of the beta-catenin gene by Wnt1-Cre-mediated deletion results in dramatic brain malformation and failure of craniofacial development. Development. 2001;128:1253–64. doi: 10.1242/dev.128.8.1253. [DOI] [PubMed] [Google Scholar]
- Bray S. Notch signaling in Drosophila: three ways to use a pathway. Seminars in Cell and Developmental Biology. 1998;9:591. doi: 10.1006/scdb.1998.0262. [DOI] [PubMed] [Google Scholar]
- Brooker R, Hozumi K, Lewis J. Notch ligands with contrasting functions: Jagged1 and Delta1 in the mouse inner ear. Development. 2006:dev.02284. doi: 10.1242/dev.02284. [DOI] [PubMed] [Google Scholar]
- Conlon RA, Reaume AG, Rossant J. Notch1 is required for the coordinate segmentation of somites. Development. 1995;121:1533–45. doi: 10.1242/dev.121.5.1533. [DOI] [PubMed] [Google Scholar]
- Crosnier C, Stamataki D, Lewis J. Organizing cell renewal in the intestine: stem cells, signals and combinatorial control. Nat Rev Genet. 2006;7:349–59. doi: 10.1038/nrg1840. [DOI] [PubMed] [Google Scholar]
- Daudet N, Ariza-McNaughton L, Lewis J. Notch signalling is needed to maintain, but not to initiate, the formation of prosensory patches in the chick inner ear. Development. 2007;134:2369–2378. doi: 10.1242/dev.001842. [DOI] [PubMed] [Google Scholar]
- Daudet N, Lewis J. Two contrasting roles for Notch activity in chick inner ear development: specification of prosensory patches and lateral inhibition of hair-cell differentiation. Development. 2005;132:541–51. doi: 10.1242/dev.01589. [DOI] [PubMed] [Google Scholar]
- De la Pompa JL, Wakeham A, Correia KM, Samper E, Brown S, Aguilera RJ, Nakano T, Honjo T, Mak TW, Rossant J, Conlon RA. Conservation of the Notch signalling pathway in mammalian neurogenesis. Development. 1997;124:1139–48. doi: 10.1242/dev.124.6.1139. [DOI] [PubMed] [Google Scholar]
- Del Monte G, Grego-Bessa J, Gonzalez-Rajal A, Bolos V, De La Pompa JL. Monitoring Notch1 activity in development: Evidence for a feedback regulatory loop. Dev Dyn. 2007 doi: 10.1002/dvdy.21246. [DOI] [PubMed] [Google Scholar]
- Duncan AW, Rattis FM, DiMascio LN, Congdon KL, Pazianos G, Zhao C, Yoon K, Cook JM, Willert K, Gaiano N, et al. Integration of Notch and Wnt signaling in hematopoietic stem cell maintenance. Nat Immunol. 2005;6:314–22. doi: 10.1038/ni1164. [DOI] [PubMed] [Google Scholar]
- Espinosa L, Ingles-Esteve J, Aguilera C, Bigas A. Phosphorylation by Glycogen Synthase Kinase-3{beta} Down-regulates Notch Activity, a Link for Notch and Wnt Pathways. J Biol Chem. 2003;278:32227–32235. doi: 10.1074/jbc.M304001200. [DOI] [PubMed] [Google Scholar]
- Estrach S, Ambler CA, Lo Celso C, Hozumi K, Watt FM. Jagged 1 is a {beta}-catenin target gene required for ectopic hair follicle formation in adult epidermis. Development. 2006:dev.02644. doi: 10.1242/dev.02644. [DOI] [PubMed] [Google Scholar]
- Fortini ME, Artavanis-Tsakonas S. The suppressor of hairless protein participates in notch receptor signaling. Cell. 1994;79:273–82. doi: 10.1016/0092-8674(94)90196-1. [DOI] [PubMed] [Google Scholar]
- Fre S, Huyghe M, Mourikis P, Robine S, Louvard D, Artavanis-Tsakonas S. Notch signals control the fate of immature progenitor cells in the intestine. Nature. 2005;435:964–8. doi: 10.1038/nature03589. [DOI] [PubMed] [Google Scholar]
- Friesel R, Brown SA. Spatially restricted expression of fibroblast growth factor receptor-2 during Xenopus development. Development. 1992;116:1051–8. doi: 10.1242/dev.116.4.1051. [DOI] [PubMed] [Google Scholar]
- Galceran J, Sustmann C, Hsu S-C, Folberth S, Grosschedl R. LEF1-mediated regulation of Delta-like1 links Wnt and Notch signaling in somitogenesis. Genes Dev. 2004;18:2718–2723. doi: 10.1101/gad.1249504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groves AK, Bronner-Fraser M. Competence, specification and commitment in otic placode induction. Development. 2000;127:3489–99. doi: 10.1242/dev.127.16.3489. [DOI] [PubMed] [Google Scholar]
- Groves AK. The induction of the otic placode. In: Popper AN, Kelley MW, Wu DK, editors. Development of the Inner Ear, (Springer Handbook of Auditory Research. Vol. 26. New York: Springer Verlag; 2005. [Google Scholar]
- Haddon C, Jiang YJ, Smithers L, Lewis J. Delta-Notch signalling and the patterning of sensory cell differentiation in the zebrafish ear: evidence from the mind bomb mutant. Development. 1998;125:4637–44. doi: 10.1242/dev.125.23.4637. [DOI] [PubMed] [Google Scholar]
- Hans S, Liu D, Westerfield M. Pax8 and Pax2a function synergistically in otic specification, downstream of the Foxi1 and Dlx3b transcription factors. Development. 2004;131:5091–102. doi: 10.1242/dev.01346. [DOI] [PubMed] [Google Scholar]
- Harada N, Tamai Y, Ishikawa T, Sauer B, Takaku K, Oshima M, Taketo MM. Intestinal polyposis in mice with a dominant stable mutation of the beta-catenin gene. Embo J. 1999;18:5931–42. doi: 10.1093/emboj/18.21.5931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward P, Balayo T, Martinez Arias A. Notch synergizes with axin to regulate the activity of armadillo in Drosophila. Dev Dyn. 2006;235:2656–66. doi: 10.1002/dvdy.20902. [DOI] [PubMed] [Google Scholar]
- Hayward P, Brennan K, Sanders P, Balayo T, DasGupta R, Perrimon N, Arias AM. Notch modulates Wnt signalling by associating with Armadillo/{beta}-catenin and regulating its transcriptional activity. Development. 2005;132:1819–1830. doi: 10.1242/dev.01724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirata H, Yoshiura S, Ohtsuka T, Bessho Y, Harada T, Yoshikawa K, Kageyama R. Oscillatory expression of the bHLH factor Hes1 regulated by a negative feedback loop. Science. 2002;298:840–3. doi: 10.1126/science.1074560. [DOI] [PubMed] [Google Scholar]
- Johnston LA, Edgar BA. Wingless and Notch regulate cell-cycle arrest in the developing Drosophila wing. Nature. 1998;394:82–4. doi: 10.1038/27925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kageyama R, Ohtsuka T, Kobayashi T. The Hes gene family: repressors and oscillators that orchestrate embryogenesis. Development. 2007;134:1243–1251. doi: 10.1242/dev.000786. [DOI] [PubMed] [Google Scholar]
- Katoh M, Katoh M. Notch ligand, JAG1, is evolutionarily conserved target of canonical WNT signaling pathway in progenitor cells. Int J Mol Med. 2006;17:681–5. [PubMed] [Google Scholar]
- Kiernan AE, Cordes R, Kopan R, Gossler A, Gridley T. The Notch ligands DLL1 and JAG2 act synergistically to regulate hair cell development in the mammalian inner ear. Development. 2005;132:4353–62. doi: 10.1242/dev.02002. [DOI] [PubMed] [Google Scholar]
- Kurooka H, Kuroda K, Honjo T. Roles of the ankyrin repeats and C-terminal region of the mouse notch1 intracellular region. Nucleic Acids Res. 1998a;26:5448–55. doi: 10.1093/nar/26.23.5448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurooka H, Kuroda K, Honjo T, Tani S, Kurooka H, Aoki T, Hashimoto N, Honjo T, Nam Y, Weng AP, et al. Roles of the ankyrin repeats and C-terminal region of the mouse notch1 intracellular region. Nucleic Acids Res. 1998b;26:5448–55. doi: 10.1093/nar/26.23.5448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladher RK, Wright TJ, Moon AM, Mansour SL, Schoenwolf GC. FGF8 initiates inner ear induction in chick and mouse. Genes Dev. 2005;19:603–613. doi: 10.1101/gad.1273605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai EC. Notch signaling: control of cell communication and cell fate. Development. 2004;131:965–73. doi: 10.1242/dev.01074. [DOI] [PubMed] [Google Scholar]
- Lamar E, Deblandre G, Wettstein D, Gawantka V, Pollet N, Niehrs C, Kintner C. Nrarp is a novel intracellular component of the Notch signaling pathway. Genes Dev. 2001;15:1885–1899. doi: 10.1101/gad.908101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanford PJ, Lan Y, Jiang R, Lindsell C, Weinmaster G, Gridley T, Kelley MW. Notch signalling pathway mediates hair cell development in mammalian cochlea. Nat Genet. 1999;21:289–92. doi: 10.1038/6804. [DOI] [PubMed] [Google Scholar]
- Leger S, Brand M. Fgf8 and Fgf3 are required for zebrafish ear placode induction, maintenance and inner ear patterning. Mech Dev. 2002;119:91–108. doi: 10.1016/s0925-4773(02)00343-x. [DOI] [PubMed] [Google Scholar]
- Lewis AK, Frantz GD, Carpenter DA, de Sauvage FJ, Gao WQ. Distinct expression patterns of notch family receptors and ligands during development of the mammalian inner ear. Mechanisms of Development. 1998;78:159. doi: 10.1016/s0925-4773(98)00165-8. [DOI] [PubMed] [Google Scholar]
- Lillevali K, Haugas M, Matilainen T, Pussinen C, Karis A, Salminen M. Gata3 is required for early morphogenesis and Fgf10 expression during otic development. Mech Dev. 2006;123:415–29. doi: 10.1016/j.mod.2006.04.007. [DOI] [PubMed] [Google Scholar]
- Mackereth MD, Kwak SJ, Fritz A, Riley BB. Zebrafish pax8 is required for otic placode induction and plays a redundant role with Pax2 genes in the maintenance of the otic placode. Development. 2005;132:371–82. doi: 10.1242/dev.01587. [DOI] [PubMed] [Google Scholar]
- Maroon H, Walshe J, Mahmood R, Kiefer P, Dickson C, Mason I. Fgf3 and Fgf8 are required together for formation of the otic placode and vesicle. Development. 2002;129:2099–108. doi: 10.1242/dev.129.9.2099. [DOI] [PubMed] [Google Scholar]
- Martin K, Groves AK. Competence of cranial ectoderm to respond to Fgf signaling suggests a two-step model of otic placode induction. Development. 2006;133:877–887. doi: 10.1242/dev.02267. [DOI] [PubMed] [Google Scholar]
- Mohamed OA, Clarke HJ, Dufort D. Beta-catenin signaling marks the prospective site of primitive streak formation in the mouse embryo. Dev Dyn. 2004;231:416–24. doi: 10.1002/dvdy.20135. [DOI] [PubMed] [Google Scholar]
- Morsli H, Choo D, Ryan A, Johnson R, Wu DK. Development of the mouse inner ear and origin of its sensory organs. J Neurosci. 1998;18:3327–35. doi: 10.1523/JNEUROSCI.18-09-03327.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murtaugh LC, Stanger BZ, Kwan KM, Melton DA. Notch signaling controls multiple steps of pancreatic differentiation. Proc Natl Acad Sci U S A. 2003;100:14920–5. doi: 10.1073/pnas.2436557100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam Y, Weng AP, Aster JC, Blacklow SC. Structural requirements for assembly of the CSL.intracellular Notch1.Mastermind-like 1 transcriptional activation complex. J Biol Chem. 2003;278:21232–9. doi: 10.1074/jbc.M301567200. [DOI] [PubMed] [Google Scholar]
- Neumann CJ, Cohen SM. A hierarchy of cross-regulation involving Notch, wingless, vestigial and cut organizes the dorsal/ventral axis of the Drosophila wing. Development. 1996;122:3477–85. doi: 10.1242/dev.122.11.3477. [DOI] [PubMed] [Google Scholar]
- Nicolas M, Wolfer A, Raj K, Kummer JA, Mill P, van Noort M, Hui CC, Clevers H, Dotto GP, Radtke F. Notch1 functions as a tumor suppressor in mouse skin. Nat Genet. 2003;33:416–21. doi: 10.1038/ng1099. [DOI] [PubMed] [Google Scholar]
- Nissen RM, Yan J, Amsterdam A, Hopkins N, Burgess SM. Zebrafish foxi one modulates cellular responses to Fgf signaling required for the integrity of ear and jaw patterning. Development. 2003;130:2543–54. doi: 10.1242/dev.00455. [DOI] [PubMed] [Google Scholar]
- Norgaard GA, Jensen JN, Jensen J. FGF10 signaling maintains the pancreatic progenitor cell state revealing a novel role of Notch in organ development. Dev Biol. 2003;264:323–38. doi: 10.1016/j.ydbio.2003.08.013. [DOI] [PubMed] [Google Scholar]
- Novak A, Guo C, Yang W, Nagy A, Lobe CG. Z/EG, a double reporter mouse line that expresses enhanced green fluorescent protein upon Cre-mediated excision. genesis. 2000;28:147–55. [PubMed] [Google Scholar]
- Ohyama T, Groves AK. Expression of mouse Foxi class genes in early craniofacial development. Dev Dyn. 2004a;231:640–6. doi: 10.1002/dvdy.20160. [DOI] [PubMed] [Google Scholar]
- Ohyama T, Groves AK. Generation of Pax2-Cre mice by modification of a Pax2 bacterial artificial chromosome. genesis. 2004b;38:195–9. doi: 10.1002/gene.20017. [DOI] [PubMed] [Google Scholar]
- Ohyama T, Mohamed OA, Taketo MM, Dufort D, Groves AK. Wnt signals mediate a fate decision between otic placode and epidermis. Development. 2006;133:865–875. doi: 10.1242/dev.02271. [DOI] [PubMed] [Google Scholar]
- Oka C, Nakano T, Wakeham A, de la Pompa JL, Mori C, Sakai T, Okazaki S, Kawaichi M, Shiota K, Mak TW, Honjo T. Disruption of the mouse RBP-J kappa gene results in early embryonic death. Development. 1995;121:3291–301. doi: 10.1242/dev.121.10.3291. [DOI] [PubMed] [Google Scholar]
- Phillips BT, Bolding K, Riley BB. Zebrafish fgf3 and fgf8 encode redundant functions required for otic placode induction. Dev Biol. 2001;235:351–65. doi: 10.1006/dbio.2001.0297. [DOI] [PubMed] [Google Scholar]
- Raft S, Nowotschin S, Liao J, Morrow BE. Suppression of neural fate and control of inner ear morphogenesis by Tbx1. Development. 2004;131:1801–12. doi: 10.1242/dev.01067. [DOI] [PubMed] [Google Scholar]
- Riccomagno MM, Martinu L, Mulheisen M, Wu DK, Epstein DJ. Specification of the mammalian cochlea is dependent on Sonic hedgehog. Genes Dev. 2002;16:2365–2378. doi: 10.1101/gad.1013302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley BB, Phillips BT. Ringing in the new ear: resolution of cell interactions in otic development. Dev Biol. 2003;261:289–312. doi: 10.1016/s0012-1606(03)00245-8. [DOI] [PubMed] [Google Scholar]
- Saint-Germain N, Lee YH, Zhang Y, Sargent TD, Saint-Jeannet JP. Specification of the otic placode depends on Sox9 function in Xenopus. Development. 2004;131:755–63. doi: 10.1242/dev.01066. [DOI] [PubMed] [Google Scholar]
- Schmidt-Ott KM, Masckauchan TN, Chen X, Hirsh BJ, Sarkar A, Yang J, Paragas N, Wallace VA, Dufort D, Pavlidis P, et al. beta-catenin/TCF/Lef controls a differentiation-associated transcriptional program in renal epithelial progenitors. Development. 2007;134:3177–90. doi: 10.1242/dev.006544. [DOI] [PubMed] [Google Scholar]
- Shi S, Stahl M, Lu L, Stanley P. Canonical Notch Signaling Is Dispensable for Early Cell Fate Specifications in Mammals. Mol Cell Biol. 2005;25:9503–9508. doi: 10.1128/MCB.25.21.9503-9508.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi S, Stanley P. Protein O-fucosyltransferase 1 is an essential component of Notch signaling pathways. Proc Natl Acad Sci U S A. 2003;100:5234–9. doi: 10.1073/pnas.0831126100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon KS, Kudoh T, Dawid IB, Fritz A. Zebrafish foxi1 mediates otic placode formation and jaw development. Development. 2003;130:929–40. doi: 10.1242/dev.00308. [DOI] [PubMed] [Google Scholar]
- Solomon KS, Kwak SJ, Fritz A. Genetic interactions underlying otic placode induction and formation. Dev Dyn. 2004;230:419–33. doi: 10.1002/dvdy.20067. [DOI] [PubMed] [Google Scholar]
- Streit A. Extensive Cell Movements Accompany Formation of the Otic Placode. Developmental Biology. 2002;249:237. doi: 10.1006/dbio.2002.0739. [DOI] [PubMed] [Google Scholar]
- Tani S, Kurooka H, Aoki T, Hashimoto N, Honjo T. The N- and C-terminal regions of RBP-J interact with the ankyrin repeats of Notch1 RAMIC to activate transcription. Nucleic Acids Res. 2001;29:1373–80. doi: 10.1093/nar/29.6.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanigaki K, Han H, Yamamoto N, Tashiro K, Ikegawa M, Kuroda K, Suzuki A, Nakano T, Honjo T. Notch-RBP-J signaling is involved in cell fate determination of marginal zone B cells. Nat Immunol. 2002;3:443–50. doi: 10.1038/ni793. [DOI] [PubMed] [Google Scholar]
- Torres M, Giraldez F. The development of the vertebrate inner ear. Mech Dev. 1998;71:5–21. doi: 10.1016/s0925-4773(97)00155-x. [DOI] [PubMed] [Google Scholar]
- Vendrell V, Carnicero E, Giraldez F, Alonso MT, Schimmang T. Induction of inner ear fate by FGF3. Development. 2000;127:2011–9. doi: 10.1242/dev.127.10.2011. [DOI] [PubMed] [Google Scholar]
- Williams R, Lendahl U, Lardelli M. Complementary and combinatorial patterns of Notch gene family expression during early mouse development. Mechanisms of Development. 1995;53:357. doi: 10.1016/0925-4773(95)00451-3. [DOI] [PubMed] [Google Scholar]
- Wright TJ, Ladher R, McWhirter J, Murre C, Schoenwolf GC, Mansour SL. Mouse FGF15 is the ortholog of human and chick FGF19, but is not uniquely required for otic induction. Dev Biol. 2004;269:264–75. doi: 10.1016/j.ydbio.2004.02.003. [DOI] [PubMed] [Google Scholar]
- Wright TJ, Mansour SL. Fgf3 and Fgf10 are required for mouse otic placode induction. Development. 2003;130:3379–90. doi: 10.1242/dev.00555. [DOI] [PubMed] [Google Scholar]
- Zhou YX, Armstrong RC. Interaction of fibroblast growth factor 2 (FGF2) and notch signaling components in inhibition of oligodendrocyte progenitor (OP) differentiation. Neurosci Lett. 2007;421:27–32. doi: 10.1016/j.neulet.2007.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]


