Abstract
Elevated activity of the eIF4F complex, which controls initiation of cap-dependent mRNA translation, has been linked to cancer progression. eIF4E recruitment to eIF4F is the rate limiting step of complex assembly and is regulated by eIF4E-Binding Proteins (4E-BPs). When stimulated, the mammalian Target of Rapamycin complex 1 (mTORC1) phosphorylates 4E-BP1, which then releases eIF4E. Hypoxia inhibits mTORC1 activity and therefore cap-dependent protein synthesis. To establish a novel genetic test of the role of eIF4F activity in regulating cell division and viability within hypoxic tumor microenvironments, we generated shRNA mediated 4E-BP1 knock-down in Rh30 rhabdomyosarcoma cells. 4E-BP1 knock-down relieved hypoxia-mediated inhibition of cycle progression in vitro and was correlated with increased expression of cyclin D1 and c-Myc. Xenograft tumors derived from these cells also displayed enhanced expression of cyclin D1 and c-Myc along with antiapoptotic genes encoding Bcl-xL, and XIAP, and failed to develop the extensive necrotic zones and edema observed in control tumors. Surprisingly, 4E-BP1 knock-down also leads to a dramatic increase in aberrant mitoses in vivo and enhanced expression of Mad2 and securin. Thus, reduced expression of the negative regulator of eIF4E has significant effects on tumor development, and is associated with enhanced cell proliferation and survival.
Introduction
Hypoxia (low O2) is a common feature of solid tumors, and cellular responses to O2 deprivation play a significant role in tumor progression 1,2. In addition to activating a large cohort of genes involved in hypoxic adaptation 3, O2 starved cells adopt numerous metabolic strategies to conserve energy. One effect of hypoxia is the inhibition of energetically demanding cellular functions. Protein synthesis, a principle ATP consumer, is suppressed at multiple levels following hypoxic exposure4,5. This adaptation conserves energy as cells shift from oxidative phosphorylation to glycolysis to produce ATP6,7.
mRNA translation initiation is regulated primarily by the assembly of two complexes: the “ternary complex” (comprised of eukaryotic initiation factor eIF2α, GTP, and methionine charged tRNA) and the “eIF4F” complex8,9. eIF4F consists of eIF4E (which binds the 5′ 7methyl-GTP cap of mRNAs), eIF4A (an RNA helicase), and the eIF4G scaffolding protein 10. Both complexes are negatively regulated by low O211,12. Severe hypoxia activates the endoplasmic reticulum kinase PERK, which phosphorylates eIF2α on serine 51 and prevents efficient ternary complex formation4. Low O2 also interferes with eIF4F assembly through sequestration of eIF4E by a family of proteins termed eIF4E-binding proteins (4E-BPs)13. 4E-BP and eIF4G interaction with eIF4E is mutually exclusive; therefore, 4E-BPs prevent productive eIF4F complex formation 14,15. Of the three 4E-BP family members (4E-BP1, 4E-BP2, and 4E-BP3) 4E-BP1 is the best characterized. eIF4E:4E-BP1 binding efficiency is negatively regulated by 4E-BP1 phosphorylation 10,16. This phosphorylation occurs hierarchically, and is initiated by the mammalian Target of Rapamycin complex 1 (mTORC1) 17–19. When mTORC1 activity is inhibited by growth factor, nutrient, or O2 deprivation, 4E-BP1 phosphorylation is decreased, allowing 4E-BP1 to bind eIF4E and inhibiting translation initiation.
We and others have shown that hypoxia inhibits protein synthesis by acting on the mTORC1 pathway5,20–22, and is dominant to other effectors such as insulin, amino acids, and phorbol esters 13. The degree of mTORC1 inhibition imposed by O2 deprivation varies considerably with cell type. Interestingly, global protein synthesis is minimally affected in some tumor cells whereas most untransformed cells are significantly affected: sensitivity to low O2 may depend on both levels of hypoxia and degree of malignancy21,23. Nevertheless, at least some hypoxic impact on mTORC1 activity has been observed in all cells tested 5,13,21.
O2 effects on mTORC1 occur via multiple mechanisms. Extended O2 starvation results in hypoxia-inducible factor (HIF) mediated induction of REDD1, which activates the tuberous sclerosis factor complex (TSC1/2), thereby inhibiting the GTPase protein Rheb and ultimately mTORC120,24. Chronic hypoxia also results in energy starvation, leading to AMP-activated protein kinase (AMPK) stimulation of TSC1/2 and mTORC1 inhibition 5. Finally, mTORC1 is regulated by the tumor suppressor PML, which disrupts mTORC1/Rheb interactions, thereby decreasing mTORC1 action25. In addition to long-term effects, decreased mTORC1 target phosphorylation is detected within 15–30 minutes of exposure to low O2 5,13. The molecular basis for this rapid mTORC1 inhibition is currently unknown.
Elevated eIF4F activity has been linked to cancer initiation and progression, both experimentally and clinically 26–29. High levels of eIF4F are not uniformly associated with increased global protein synthesis, but rather promote the more efficient translation of specific mRNAs. These mRNAs tend to have structured 5′ UTRs, and include critical growth factors and proto-oncogenes such as b-FGF, c-Myc, cyclin D1, and ornithine decarboxylase (ODC) 26,30,31. Interestingly, increased eIF-4E expression also promotes the transport of specific mRNAs from the nucleus to the cytoplasm, enhancing their expression 32. eIF4E overexpression in fibroblasts contributes to transformation 33,34, and relatively high eIF4E levels have been described in breast, head and neck, colon, bladder, prostate, cervical, and other cancers29,30.
Alternatively, decreased 4E-BP1 binding capacity can also promote eIF4F activity. 4E-BP1 is often hyperphosphorylated in human tumors, primarily due to PI3K/Akt stimulation, resulting in mTORC1 activation 35. In addition, 4E-BP1 expression levels are correlated with the severity of gastrointestinal cancer36. Decreased 4E-BP1 levels or hyperphosphorylation should result in increased eIF4F complex formation and preferential expression of specific proteins. Consistent with this idea, 4E-BP1 overexpression (or introduction of non-phosphorylatable versions of 4E-BP1) results in rapid cell death and tumor regression 37–39. These results strongly suggest that disrupted 4E-BP1 function could contribute to eIF4F-mediated tumor formation.
Postulating that tumor cells with reduced 4E-BP1 expression (and augmented eIF4F activity) would be more refractory to hypoxic stress, we performed shRNA mediated knockdown of 4E-BP1 in Rh30 rhabdomyosarcoma cells. This strategy provides an effective genetic approach to model 4E-BP1 loss or constitutive phosphorylation, without affecting other components of the mTORC1 pathway. 4E-BP1 downregulation in Rh30 cells results in significantly decreased susceptibility to hypoxia and other stresses both in vitro and in vivo. Furthermore, increased expression of specific anti-apoptotic and growth-promoting genes (Bcl-XL, XIAP, cyclin D1, and c-Myc) is associated with this phenotype. Finally, 4E-BP1 knock-down resulted in increased aberrant mitotic figures within tumors which correlated with higher Mad2 and securin levels. Of note, Mad2 overexpression has recently been reported to contribute to tumorigenesis and enhanced genomic instability40,41. Our results indicate that decreased 4E-BP1 expression promotes aspects of tumor progression by facilitating adaptation to the tumor microenvironment.
Materials and Methods
Cell culture
Rh30 (alveolar rhabdomyosarcoma) engineered to stably express shRNA for 4E-BP1 were described previously42. Clones were cultured in RPMI medium (Gibco/Invitrogen) with 10% FCS (Hyclone), 2mM L-glutamine (Gibco/Invitrogen), and 500μg/mL G418 sulfate (Invitrogen). Hypoxia (0.5% O2, 5% CO2, 94.5% N2) was achieved using an InVivo2 hypoxic workstation (Biotrace). For serum starvation and hypoxia experiments cells were subjected to 16 hours of serum starvation prior to 24 hours of 0.5% O2 in serum-free media.
35S methionine incorporation
Following 48 hours hypoxia or control, cells were washed, and incubated for two hours with methionine free media containing dialyzed FCS. Cells were then pulsed for one hour with 0.67μCi 35S-methionine containing media. Cells were lysed in lysis buffer and spun to remove nuclei. Equal volumes of lysates were dried on filter paper and precipitated in TCA with non-radioactive methionine, followed by water, ethanol, and acetone rinses. Filters were then read in a scintillation counter. Counts were normalized to protein quantity and counts calculated as a percentage of normoxic control.
Western blotting and 7methyl-GTP affinity chromatography
Lysates for Western blots from cell culture were performed in lysis buffer containing 1% Triton-X100, 20mM Tris (pH 7.5), 150mM NaCl, 1mM EDTA, 20mM (β-glycerophosphate, 1mM Na3VO4, 10mM NaF, and complete protease inhibitor tablet (Roche). Hypoxic cells were scraped into lysis buffer in hypoxic conditions. Lysates were sonicated, spun, protein quantified by BCA assay (Pierce), and subjected to SDS-PAGE and blotting. Antibodies used for Western blots used were: eIF4E, 4E-BP1, cyclin D1, phospho-Rb, XIAP, and Bcl-xL (Cell Signaling); Rb, c-Myc, and cyclin D2 (Santa Cruz); GAPDH (Abcam); Actin (Sigma). 7methyl-GTP affinity chromatography was performed as previously described 13. All chemicals were from Sigma.
Cell cycle analysis
Cells were plated so that upon harvest they were 50% confluent. Following 48 hours hypoxia treatment, cells were pulsed with 10μM BrdU for 20 minutes, washed, trypsinized, and fixed in 70% ethanol. DMA was denatured with 2N HCI and neutralized with 0.1M Borax pH 8.5. Cells were stained with Alexa-488-labeled anti-BrdU antibodies (Invitrogen) and 5μg/mL propidium iodide. Cells were analyzed on a FACScalibur flow cytometer gated on single cells.
QRT-PCR
Total RNA was isolated from cells using Trizol reagent (Invitrogen) following manufacturer’s instructions. Tumor RNA was isolated by performing two Trizol extractions, followed by a phenol:chloroform extraction. cDNA was produced using 1μg total RNA using Superscript II (Invitrogen) with random hexamer primers (Roche). Primers against 18S (Applied Biosystems) were used for endogenous control for cell culture derived RNA in ΔΔ CT analysis. Primer sets were generated for all other targets and analysis was performed using an Applied Biosystems 7900HT Sequence Detection System, with amplification quantified using SYBR green. Primers specific for human (β-actin were used for endogenous control for tumor RNA. All sequences available upon request.
Xenograft experiments
Rh30 cells (107) were injected into the flanks of athymic nu/nu mice (Charles River) in 200μL PBS. Each mouse was injected with control and BP-KD on opposing flanks. Tumor volume was monitored throughout the experiment and determined using the formula: Volume=π/6×(large dimension)×(small dimension)2. Three independent experiments were performed and averages of volumes determined over the three experiments. Measurements over the three experiments were averaged if they occurred within one day of one another. One to two hours before sacrifice, mice were injected with 60mg/kg pimonidazole (Hypoxyprobe, Chemicon). Following removal, mass determination, and photography, tumors were either fixed in 4% paraformaldehyde for histology, or frozen in liquid nitrogen for protein and mRNA analysis. Tumor lysates for Western blots were made in RIPA buffer (50mM Tris (pH 7.4), 150mM NaCl, 1% NP-40, 0.5% Sodium deoxycholate, 0.1% SDS, and phosphatase and protease inhibitors as above), homogenized, spun twice, and protein concentrations determined by BCA assay (Pierce). Following fixation for IHC, tumors were washed and dehydrated with increasing ethanol. Tumors were then xylene perfused, paraffin embedded, and sectioned.
Histological analysis and IHC
Tumor sections were stained with hematoxylin and eosin for histological examination. IHC was performed on sections by heat for 20 minutes at 60C, clearing with xylene, and rehydration with increasing water in ethanol. Sections were boiled in antigen unmasking solution (Vector Laboratories), treated with 1% H2O2, and blocked with 4% BSA in 2% goat serum (Vector laboratories). Primary antibodies were incubated overnight at 4C in blocking buffer. Staining was revealed with biotin conjugated secondary antibodies and ABC-streptavivin-HRP kit and incubation with DAB (both Vector Laboratories). Hypoxia was revealed using Hypoxyprobe kit (HP-1 plus, Chemicon) as per manufacturer’s instructions with blocking in above buffer and primary antibody at 1:250–500 dilution. In all cases, control and BP-KD tumor sections from each individual mouse were placed on one slide to better control for IHC staining. Microscopy was performed on a Leica microscope. Sections were counted as described in figure legends (low power 10X, high power 40X). Antibodies used: eIF4E, 4E-BP1, cleaved caspase-3 (Cell Signaling); Ki-67 (Novacastra); phospho-histone H3 (Ser10) (Upstate/Millipore). Control stainings were performed with secondary antibody alone and absence of staining confirmed. All IHC sections were counterstained with hematoxylin. Determination of aberrant mitoses was performed on sections stained with phospho-histone H3 antibodies. Abnormal mitotic figures were determined by examining all positively stained cells within the field, and enumerating normal versus abnormal. Abnormal was defined by multipolar mitotic figure, anaphase bridge, or nucleus with excess DNA content relative to neighboring cells.
Statistical analyses
Statistical significance of experiments was performed using Student’s t-test based on standard deviation or standard error of the mean as indicated. Analyses were performed using Microsoft Excel.
Results
Previous studies demonstrated that eIF4E overexpression can promote tumor growth 26–29. To determine whether decreased 4E-BP1 expression, and consequent increased activity of endogenous eIF4E, would alter cellular responses to hypoxic stress, we isolated Rh30 cell clones engineered to express an shRNA transgene specific for 4E-BP1. Figure 1A shows that two cell lines (deemed BP-KD1 and BP-KD2) reveal >80% decrease in 4E-BP1 protein levels, corresponding to significantly decreased 4E-BP1 mRNA (Figure 1B). This effect is specific to 4E-BP1, as Western blot analysis of 4E-BP2 in these cells displayed no change in expression (data not shown).
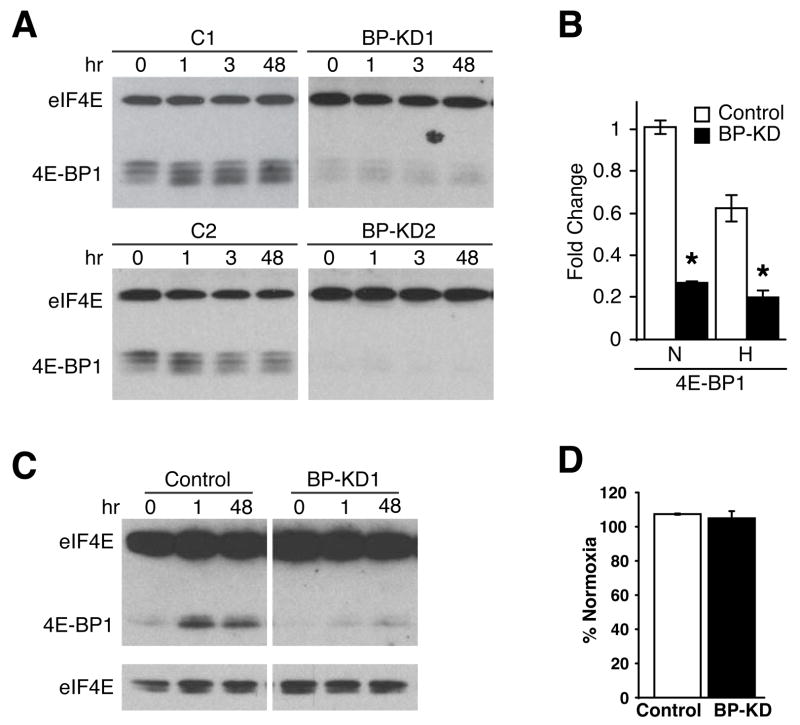
Figure 1.
4E-BP1 knock-down in Rh30 cells. A. Western blot of eIF4E and 4E-BP1 levels in Rh30 cells stably expressing control or shRNA construct for 4E-BP1 over a time course of 0.5% O2 in hours. Two control (C1 and C2) and two knock-down (BP-KD1 and BP-KD2) clones are shown. B. QRT-PCR analyses of 4E-BP1 expression in two control and two BP-KD Rh30 clones following 16 hours serum starvation, followed by 24 hours at 21% (N) or 0.5% (H) O2. QRT-PCR is average of three independent experiments, error bars ±1 SEM. * p<0.05. C. 7m-GTP affinity chromatography performed on lysates from control and 4E-BP1 knock-down (BP-KD1) cells following exposure to 0.5% O2 for the indicated time in hours. eIF4E and 4E-BP1 protein expression is shown. Lower eIF4E panels show shorter exposure of upper panel. D. Protein synthesis evaluated by 35S-methionine incorporation following 48 hours 0.5% O2. Shown is 35S-methionine incorporation of hypoxic cells as a percentage of normoxic controls. Percentages are average of four independent experiments ±1 SEM.
When control Rh30 cells were exposed to hypoxic conditions, 4E-BP phosphorylation decreased in a time-dependent manner, as evidenced by increasing electrophoretic mobility (Figure 1A). 7-methyl-GTP affinity chromatography revealed that these changes correlated to increased 4E-BP binding to eIF-4E (Figure 1C). BP-KD cells displayed greatly reduced levels of 4E-BP1 protein associated with eIF-4E, further indicating the effectiveness of the shRNA knock-down. Interestingly, neither Rh30 control nor BP-KD cells showed perceptible changes in global translation rate under hypoxia (Fig 1D). Although hypoxia is known to decrease global protein synthesis in most cells through multiple mechanisms, including reduced 4E-BP1 phosphorylation by mTOR, this response can be suppressed in transformed cells by constitutive activation of upstream signaling pathways21. Despite the maintenance of global translation rates, it has been shown that elevated eIF4E expression is correlated with increased translation and/or nuclear-cytoplasmic transport and expression of specific mRNAs26,32.
We next investigated the effects of 4E-BP1 depletion on hypoxic inhibition of cell cycle progression (Figure 2A, B). As reported previously for other cell types43,44, control Rh30 cells accumulate in G1 when challenged with hypoxic stress and show a concomitant decrease of cells in S phase, indicating an hypoxia-induced delay in G1/S progression (Figure 2B). This effect is significantly diminished in hypoxic BP-KD cell lines, which display a reduced accumulation in G1, and a greater proportion of cells in S phase compared to control cells. These results demonstrate that 4E-BP1 can regulate the G1/S transition in hypoxic cells, presumably by altering the amount of available eIF4E. Interestingly, both normoxic and hypoxic 4E-BP1 KD cells display an increased proportion of cells accumulating in G2/M, suggesting that 4E-BP1 may have effects on multiple phases of the cell cycle in Rh30 cells.
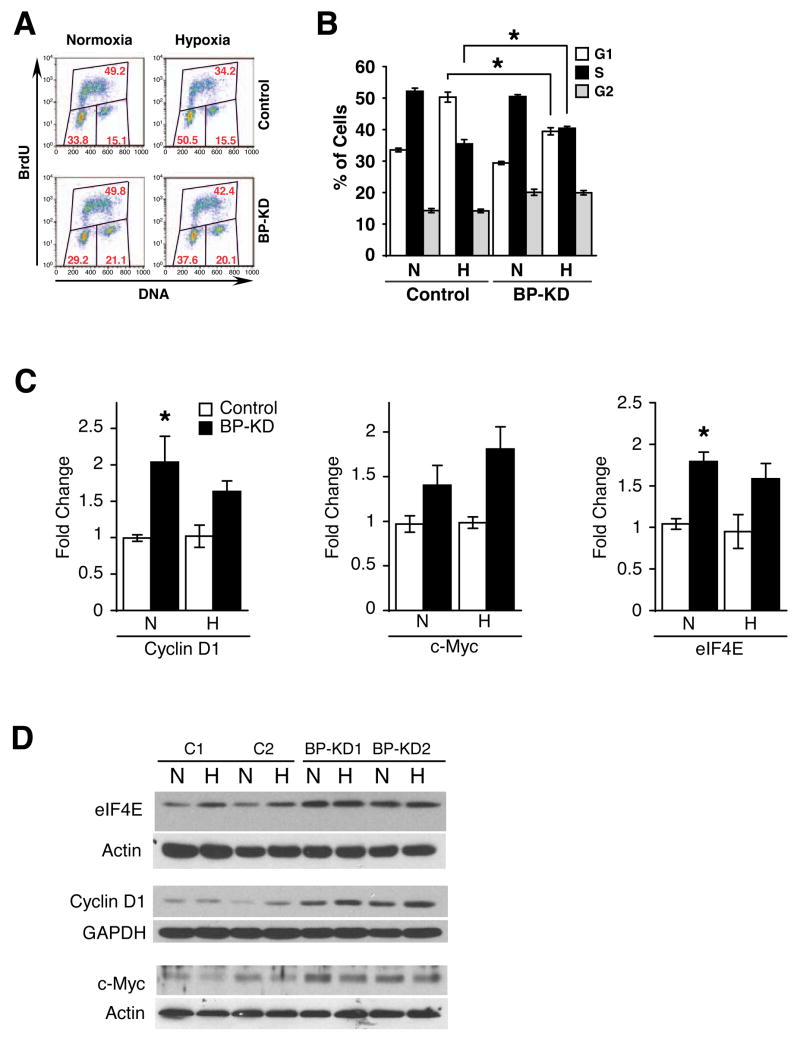
Figure 2.
Knock-down of 4E-BP1 alters cell cycle profile. A. Representative BrdU incorporation histograms from Rh30 clones grown at 21% or 0.5% O2 for 48 hours. Percentages of cells in each cell cycle phase are given in red. B. Summary of BrdU incorporation, data from two control and two BP-KD clones were combined over four independent experiments. Cells were subjected to 21% (N) or 0.5% (H) O2 for 48 hours. Averages of percentage of cells in each cell cycle stage are shown, error bars ±1 SEM. * p<0.01. C, D. QRT-PCR (C) and Western blot (D) analyses of eIF4E, cyclin D1 and c-Myc mRNA expression in two control and two BP-KD Rh30 clones following 16 hours serum starvation, followed by 24 hours at 21% (N) or 0.5% (H) O2. QRT-PCR is average of three independent experiments, error bars ±1 SEM. * p<0.01.
As eIF4E overexpression has been shown to augment the expression of many key cell cycle regulators, notably cyclin D1 and c-Myc32,45, we investigated the expression of these proteins in normoxic and hypoxic Rh30 and BP-KD cells. BP-KD cells displayed increased cyclin D1 and c-Myc protein and mRNA expression levels (Figure 2C, D). eIF4E mRNA expression is also increased in these cells, possibly a result of direct transcriptional activation by c-Myc, a known regulator of eIF4E transcription46 (Figure 2D). Interestingly, hypoxic effects on cyclin D1 and c-Myc mRNA and protein expression were subtle, suggesting that elevated baseline expression of cyclin D1, c-Myc and other targets in BP-KD cells may confer the altered cell cycle response to hypoxic stress. These data also argue against significant changes in translational efficiency of cyclin D1 and c-Myc mRNAs under hypoxia. The mechanisms by which 4E-BP1 knock-down results in increased mRNA levels for cyclin D1 and c-Myc are not known, but is consistent with a previous report45 and may be a result of enhanced nuclear-cytoplasmic transport32.
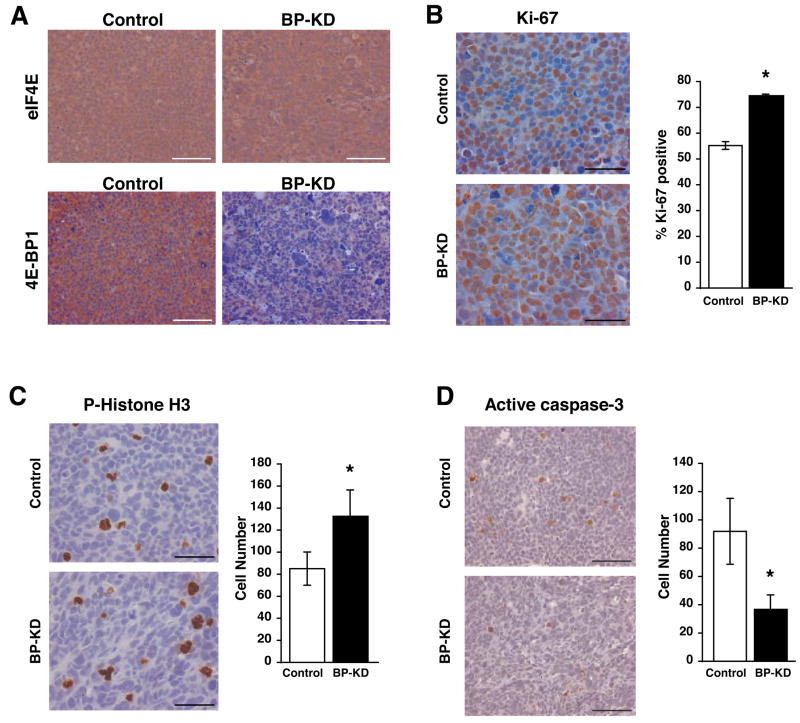
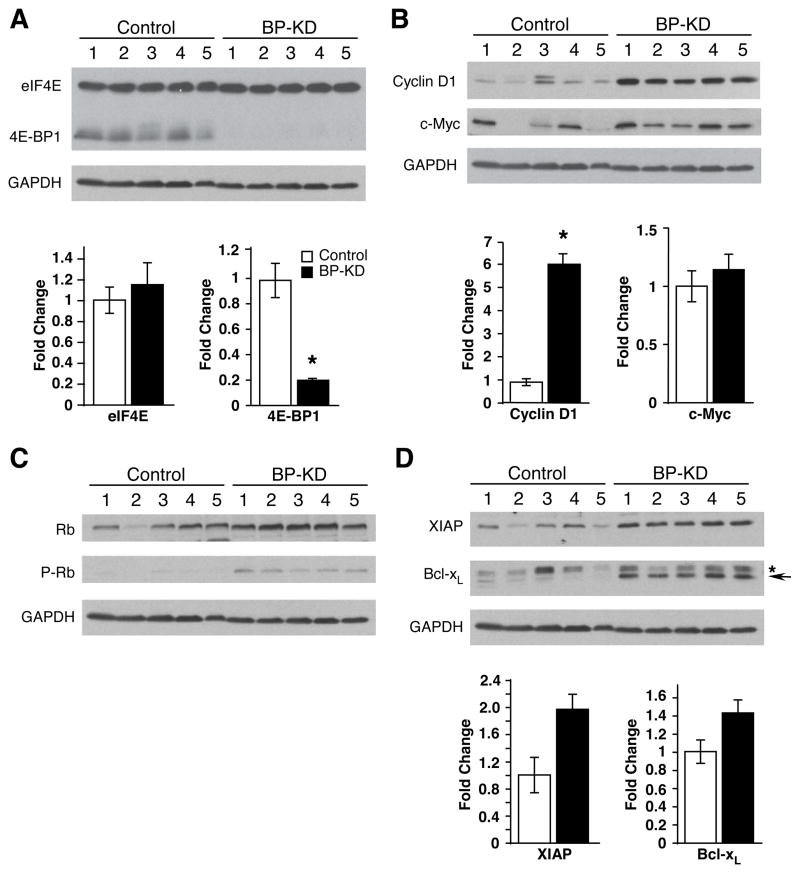
The aberrant vascular development and rapid growth of solid tumors often produce areas characterized by moderate to severe hypoxia, acidosis, and growth factor and nutrient deprivation47. To compare the relative responses of Rh30 and BP-KD cells to tumor microenvironments, we injected control Rh30 cells (line C1 or C2) and BP-KD cells (line BP-KD1 or BP-KD2) subcutaneously into opposing flanks of nude mice. The data shown in Figures 3–6 were obtained from C1 and BP-KD2 derived tumors, but similar results were obtained from C2 and BP-KD1 tumors. Immunohistochemical (IHC) analysis of morphologically similar tumor sections revealed that 4E-BP1 protein expression was stably reduced in BP-KD tumors over the time course of the experiment, whereas eIF4E expression was equivalent in tumors from both genotypes (Figure 3A). Relative cellular proliferation was determined by staining tumor sections for Ki-67 and phospho-histone H3 expression: elevated levels of nuclei staining positive for each marker were observed in BP-KD tumors relative to controls (Figure 3B and 3C). Similarly, the BP-KD tumors had lower levels of cleaved caspase 3 staining, consistent with fewer apoptotic cells (Figure 3D). These results were reflected by changes observed in Western blot and mRNA analyses of individual excised tumors (Figure 4). Interestingly, BP-KD tumors displayed an even greater increase in cyclin D1 mRNA and protein levels than that observed for these cells grown in culture (Figure 4B). This increased cyclin D1 expression correlated to higher levels of phosphorylated pRb protein in BP-KD tumors, consistent with increased cell cycle progression (Figure 3B, C). In contrast to cells in culture, expression levels of c-Myc mRNA in BP-KD tumors were approximately equivalent to control Rh30 tumors, whereas c-Myc protein levels were variable, but generally higher in BP-KD tumors. Finally, BP-KD tumors also displayed increased mRNA and protein expression of anti-apoptotic genes Bcl-xL and XIAP (Figure 4D), which correlated with decreased cleaved caspase-3 staining in tumor sections (Figure 3D).
Figure 3.
Analysis of Rh30 xenograft growth. A. IHC staining for eIF4E and 4E-BP1 in control and BP-KD tumor sections. Bar = 100μm. B. IHC performed for Ki-67 positive cells. Ten high power sections were counted from three tumors for each tumor type and average percentage of Ki-67 positive tumors was determined with standard deviation on right. Left, representative IHC stainings, bar = 50μm. * p<0.01. C. IHC with phospho-specific histone H3 (serine 10) performed on control and BP-KD tumors. Six low power sections from three tumors were counted and averages with standard deviation determined, right. Left, representative staining at high power, bar = 50μm. * p<0.05. D. IHC performed for active caspase-3 on control and BP-KD tumors. Ten low power sections were counted from three tumors for each tumor type and the average number of cells with standard deviation is shown on right. Left, high power representative images of stained cells, bar = 100μm. * p<0.05.
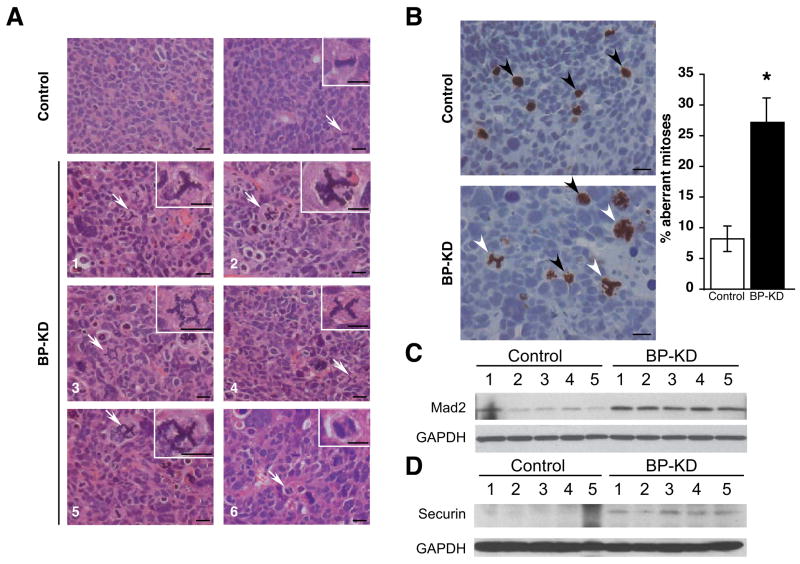
Figure 6.
BP-KD tumors display aberrant mitoses. A. Examples (H&E) of mitotic figures observed in tumors from control and BP-KD tumors. Top panels: sections from control tumors. Left, section showing typical nuclei; right, typical mitosis. Bottom six panels show examples of aberrant mitoses in sections from BP-KD tumors. In all cases, arrows point to cells in insets. All bars = 20μm. B. IHC performed with phospho-specific histone H3 (serine 10) antibody. Six low power sections from three tumors of each type were counted and percentage of aberrant mitoses determined, error bars ± 1 SEM. Left panels show high power examples of tumor stainings. Bar = 20μm. * p<0.05. C, D. Western blot analysis for MAD2 (C) and securin (D) levels from extracts of control and BP-KD tumors.
Figure 4.
Key cell cycle and apoptotic proteins are differentially regulated in control and BP-KD tumors. Extracts of control and BP-KD tumors were analyzed by Western and/or QT-PCR for expression of: A. eIF4E and 4E-BP1, B. Cyclin D1 and c-Myc, C. Retinblastoma (Rb) protein and phospho-Rb, and D. XIAP and Bcl-XL. For Bcl-XL panel, * designates a non-specific band, whereas the arrow indicates Bcl-XL protein. For each panel, lysates from five tumors of each type are shown, mRNA expression levels are averaged over the five tumors, with each set of RNA assayed three times with error bars, ± 1 SEM. * p<0.05. GAPDH protein levels in tumor lysates are shown as a loading control for each panel.
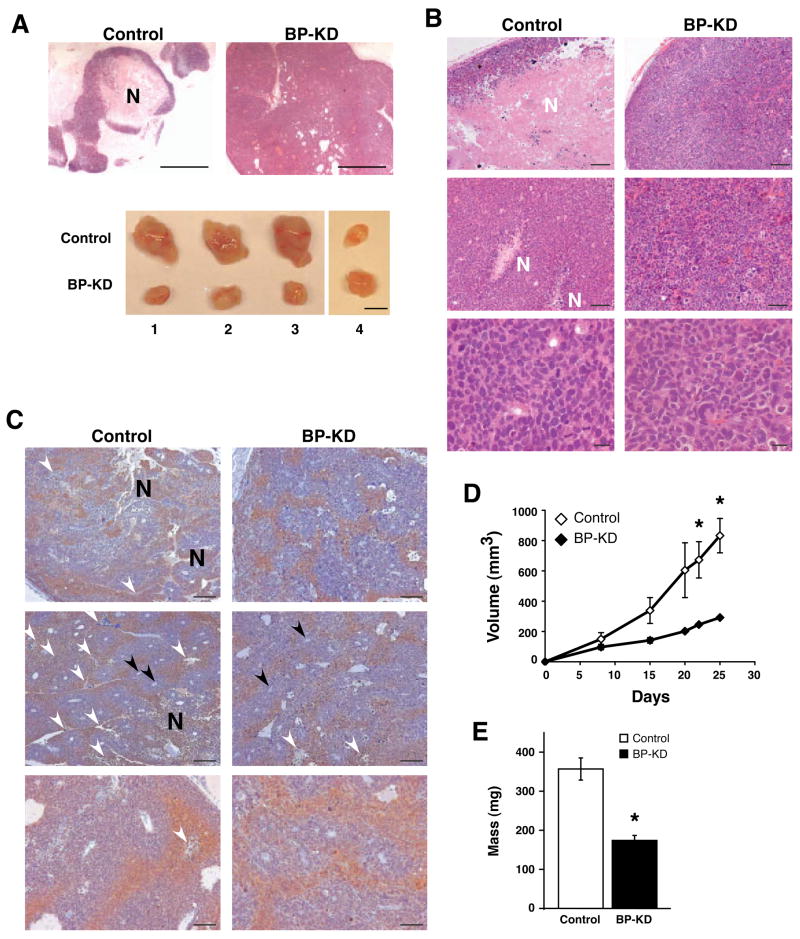
The most striking difference between control Rh30 and BP-KD tumors was their internal architecture. All Rh30 tumors displayed large necrotic regions and fluid-filled spaces, often comprising more than 50% of the total tumor volume, surrounded by a penumbra of viable cells. In contrast, the BP-KD tumors were essentially devoid of necrotic tissue (Figure 5A, B), and contained densely packed cells. Upon excision, the Rh30 tumors were soft, pliable and easily disrupted by minor physical manipulation. In contrast, BP-KD tumors were inelastic and refractory to physical disruption. The remarkable disparity in morphological phenotype accounts, at least in part, for the counterintuitive tumor growth rates of Rh30 and BP-KD tumors (Figure 5D, E). Although both tumor types grew at an equivalent rate over the first week following subcutaneous injection, the overall size of the Rh30 tumors increased at an accelerated rate relative to the BP-KD tumors (Figure 5D). Intriguingly, this difference in tumor size was not reflected in total tumor cellularity: we recovered significantly less total genomic DNA per gram of tumor tissue from Rh30 tumors (9+/−2 μg/mg tissue) than from BP-KD tumors (15+/−3 μg/mg tissue). It is important to emphasize that the comparisons of Ki-67, phospho-histone H3, and cleaved caspase 3 between Rh30 and BP-KD tumors shown in Figure 3 were made using regions of comparable cellularity, and did not contain the necrotic regions of the Rh30 tumors. The level and pattern of tumor hypoxia, as revealed by piminidazole staining (Figure 5C) appear similar in the control and BP-KD tumors, although other stresses are difficult to assess in tumor sections. Our data suggest that cells in the control Rh30 tumors succumb to local microenvironmental stresses (hypoxia, low pH, nutrient and growth factor deprivation), whereas cells in the BP-KD tumors survive these stresses better and are more proliferative, generating smaller, denser tumors. These data are consistent with the idea that elevated levels of free eIF4E, and consequent expression of specific mRNAs, can protect cells from environmental stresses.
Figure 5.
Phenotypic comparison of control and BP-KD tumors. A. Top panel: low power image of control and BP-KD tumors demonstrating level of necrosis. Bar = 500μm. Bottom panel: matched control and BP-KD tumors excised from opposing flanks of nude mice. Bar = 1 cm. B. H&E stains of tumor sections from control and BP-KD tumors. Necrotic regions are marked with “N”. Top panels bar = 200μm, middle panels bar = 100μm, bottom panels bar = 20μm. C. Analysis of hypoxia in tumors using pimonidazole staining. Mice were injected with pimonidazole prior to sacrifice and IHC performed on sections from control and BP-KD tumors using anti-pimonidazole antibodies. Large necrotic areas are marked by “N”, smaller areas of necrosis by white arrowheads. Black arrowheads mark vessels. Top and middle panels, bar = 200μm. Bottom panels, bar = 100μm. D. Volume measurements of control and BP-KD tumors grown in nude mice. Shown are averages for indicated days of three independent experiments (total n=26) with two different sets of clones. Error bars ±1 SEM. * p<0.05. E. Tumor mass average shown from three independent experiments (total n=26) with two sets of clones. Error bars±1 SEM, *p<0.01.
Finally, we observed a large number of aberrant mitotic figures in BP-KD tumors, largely absent in controls (Figure 6A). These figures included tripolar and multipolar spindles, anaphase bridges and other highly aberrant nuclei. In addition to aberrant mitoses, BP-KD tumors also displayed extensive nuclear atypia characterized by increased size, which may reflect increased ploidy (Figure 6B). To quantify the extent of abnormal mitoses we further examined tumor sections stained for phospho-histone H3, and determined the percentage of mitotic cells that were grossly abnormal (e.g., multipolar spindles or nuclei with disordered or excessive DMA content). BP-KD tumors had a significantly higher percentage (27%) of aberrant figures than control Rh30 tumors (8%) (p<0.05). To further characterize the mechanism behind these observations we examined the expression of Mad2 and securin in control and BP-KD tumors. Mad2 overexpression has recently been linked to tumorigenesis and genomic instability and has been reported to be elevated in human tumors, leading to worse prognosis48. Strikingly, Mad2 and securin expression levels were higher in BP-KD tumors compared to controls (Figure 6C, D). These observations suggest that 4E-BP1 downregulation disrupts one or more aspects of mitotic spindle function in Rh30 cells, possibly accounting for the relative preponderance of cells in G2/M when grown in vitro (Figure 2B), although the precise mechanisms underlying this phenotype are not yet clear.
Discussion
eIF4E overexpression has been shown to enhance tumor growth in transgenic mice27, and many human tumors exhibit increased eIF4E expression26. Mounting evidence indicates that elevated eIF4E activity can also modulate the translation and nuclear-cytoplasmic transport of mRNAs encoding factors associated with increased malignancy (VEGF, b-FGF, cyclin D1, and c-Myc)32. The clinical outcome of patients with dysregulated 4E-BP1 has been examined 36,49, and results indicate that lower 4E-BP1 expression or increased 4E-BP1 phosphorylation correlate with poorer prognosis. Therefore eIF4E regulation by 4E-BP1 is likely to be important in malignant disease.
Here we examined the physiological consequences of decreasing 4E-BP1 expression in cells exposed to hypoxia in vitro or hypoxic tumor microenvironments in vivo. Hypoxia elicits numerous cellular responses, including suppression of cap-dependent protein synthesis via mTORC1 inhibition 11,12. Since mTORC1 promotes cap-dependent translation by phosphorylating 4E-BP1, decreased 4E-BP1 expression should bypass some of the translational effects of mTORC1 inhibition. We observed that4E-BP1 knock-down in Rh30 cells resulted in a reversal of hypoxic inhibition of cell cycle progression in vitro, and a higher proliferative index and decreased apoptosis in tumors grown in vivo (comparing regions of similar cellularity). These phenotypes correlated to increased expression of cyclin D1 and c-Myc in 4E-BP1 cells relative to Rh30 cells, both in vitro and in vivo.
Control Rh30 tumors displayed extensive necrotic regions whereas the BP-KD tumors did not, presumably due to increased cell viability. The control tumors also exhibited extensive edema and cellular debris, which appears to account for their counterintuitive greater tumor volume and mass relative to BP-KD tumors. Interestingly, tumors derived from both cell types displayed similar patterns and extent of tumor hypoxia, but our data suggest that decreased 4E-BP1 expression, and commensurate increases in eIF4E activity, provide a proliferative and survival benefit to Rh30 cells under these conditions.
Reduced 4E-BP1 expression was also associated with increased cellular proliferation and elevated cyclin D1 mRNA levels. It has been observed that eIF4E overexpression leads to increased cyclin D1 mRNA levels, perhaps due to translation of unknown transcription factor(s) and/or altered nuclear-cytoplasmic mRNA transport32, 45, 50. More cyclin D1 and c-Myc protein was detected in vivo and in Rh30 cells cultured in serum-free conditions. Cyclin D1 and c-Myc expression was also maintained under combined conditions of hypoxia and growth factor withdrawal, which inhibit mTORC1 signaling 35. Continued cyclin D1 and c-Myc expression under stress conditions indicates that 4E-BP1 downregulation bypasses some aspects of mTORC1 inhibition, most likely by increasing eIF4E activity.
It is interesting to note that hypoxia inhibits mRNA translation by modulating the activity of at least two different regulatory complexes, eIF2α and eIF4E. Specifically, hypoxia can promote PERK-dependent phosphorylation of eIF2α, thereby disrupting formation of the ternary complex. In addition, hypoxia inhibits mTORC1 activity, and thereby 4E-BP1 phosphorylation, through multiple mechanisms. Ablating eIF2α phosphorylation by targeted mutagenesis of the critical serine residue (S51) results in decreased tumor formation by transformed MEFs51, a phenotype associated with increased cell death. Thus, the inappropriate maintenance of mRNA translation in cells that cannot phosphorylate eIF2α compromises their survival under conditions of environmental stress. In contrast, our results indicate that reduced 4E-BP1 expression, the major negative regulator of eIF4E, resulted in enhanced cell cycle progression and survival associated with elevated expression of cyclin D1, c-Myc, XIAP and Bcl-XL. These apparently disparate results likely reflect important cell type-specific differences between MEFs and rhabdomyosarcoma cells, but may also highlight the fact that PERK activation and mTORC1 (and hence 4E-BP1) inhibition occur at different levels of hypoxia. For example, extremely low levels of O2 (0.2%) are necessary to activate PERK, whereas mTORC1 activity is inhibited at significantly higher levels of O2 (1.5%). These results suggest a graded hierarchy of stress responses that integrate multiple cellular pathways, including translation, to modulate cell survival and proliferation under varying levels of tumor hypoxia. As global mRNA translation in 4E-BP1 knock-down cells is not detectably affected by hypoxia (presumably due to elevated signal pathway activation in Rh30 cells), it is all the more striking that cell cycle progression and survival are significantly effected in vitro and in vivo.
BP-KD Rh30 tumors were also more prone to aberrant mitoses than controls. We observed numerous examples of multipolar mitotic figures, anaphase bridges, and cells with excess DNA content. The reasons for this are not yet clear, but results from several reports suggest a possible mechanism. Rh30 cells carry multiple chromosomal abnormalities52, which may be exacerbated by increased survival in BP-KD cells. In addition, hypoxia or ischemia (O2 deprivation coupled with glucose deprivation) result in a polyploid phenotype if apoptotic pathways are blocked 53. Cells with reduced 4E-BP1 may be protected from cell death triggered by mitotic catastrophe and therefore accumulate more chromosomal aberrations. We also observed an increase in Mad2 levels in BP-KD cells in vitro and in vivo. A component of the spindle checkpoint, Mad2 overexpression has recently been reported to drive tumorigenesis and increase genetic instability in numerous systems40, 41. Consistent with this observation, securin levels were also higher in BP-KD tumors, indicating that enhanced Mad2 levels could result in the persistence of securin, which may be indicative of genomic instability.
Data presented here have important implications for anti-tumor therapy. eIF4F activation is a common event in human tumors (either through eIF4E overexpression, activation of the PI3K/Akt pathway, or 4E-BP1 reduction) and has been observed in breast, colon, lung, head and neck, cervical, ovarian, hematological, and others29, 30. Our results also have ramifications for treatment with rapamycin analogues54, as tumors experimentally induced by eIF4E overexpression are resistant to rapamycin-based therapy28. Presumably, tumors with reduced 4E-BP1 will also be resistant to mTORC1 inhibition. Alternatively, targeting eIF4F itself should lower the apoptotic threshold and act as an anti-proliferative treatment. Our data suggest that 4E-BP1 expression level or its phosphorylation status should be considered in addition to assessment of eIF4E expression levels within human tumors.
References
- 1.Brown JM, Wilson WR. Exploiting tumour hypoxia in cancer treatment. Nat Rev Cancer. 2004;4:437–47. doi: 10.1038/nrc1367. [DOI] [PubMed] [Google Scholar]
- 2.Hockel M, Vaupel P. Tumor hypoxia: definitions and current clinical, biologic, and molecular aspects. J Natl Cancer Inst. 2001;93:266–76. doi: 10.1093/jnci/93.4.266. [DOI] [PubMed] [Google Scholar]
- 3.Semenza GL. Regulation of mammalian O2 homeostasis by hypoxia-inducible factor 1. Annu Rev Cell Dev Biol. 1999;15:551–78. doi: 10.1146/annurev.cellbio.15.1.551. [DOI] [PubMed] [Google Scholar]
- 4.Koumenis C, et al. Regulation of protein synthesis by hypoxia via activation of the endoplasmic reticulum kinase PERK and phosphorylation of the translation initiation factor eIF2alpha. Mol Cell Biol. 2002;22:7405–16. doi: 10.1128/MCB.22.21.7405-7416.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu L, et al. Hypoxia-induced energy stress regulates mRNA translation and cell growth. Mol Cell. 2006;21:521–31. doi: 10.1016/j.molcel.2006.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim JW, Tchernyshyov I, Semenza GL, Dang CV. HIF-1-mediated expression of pyruvate dehydrogenase kinase: a metabolic switch required for cellular adaptation to hypoxia. Cell Metab. 2006;3:177–85. doi: 10.1016/j.cmet.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 7.Papandreou I, Cairns RA, Fontana L, Lim AL, Denko NC. HIF-1 mediates adaptation to hypoxia by actively downregulating mitochondrial oxygen consumption. Cell Metab. 2006;3:187–97. doi: 10.1016/j.cmet.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 8.Kapp LD, Lorsch JR. The molecular mechanics of eukaryotic translation. Annu Rev Biochem. 2004;73:657–704. doi: 10.1146/annurev.biochem.73.030403.080419. [DOI] [PubMed] [Google Scholar]
- 9.Richter JD, Sonenberg N. Regulation of cap-dependent translation by eIF4E inhibitory proteins. Nature. 2005;433:477–80. doi: 10.1038/nature03205. [DOI] [PubMed] [Google Scholar]
- 10.Gingras AC, Raught B, Sonenberg N. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu Rev Biochem. 1999;68:913–63. doi: 10.1146/annurev.biochem.68.1.913. [DOI] [PubMed] [Google Scholar]
- 11.Liu L, Simon MC. Regulation of transcription and translation by hypoxia. Cancer Biol Ther. 2004;3:492–7. doi: 10.4161/cbt.3.6.1010. [DOI] [PubMed] [Google Scholar]
- 12.Wouters BG, et al. Control of the hypoxic response through regulation of mRNA translation. Semin Cell Dev Biol. 2005;16:487–501. doi: 10.1016/j.semcdb.2005.03.009. [DOI] [PubMed] [Google Scholar]
- 13.Arsham AM, Howell JJ, Simon MC. A novel hypoxia-inducible factor-independent hypoxic response regulating mammalian target of rapamycin and its targets. J Biol Chem. 2003;278:29655–60. doi: 10.1074/jbc.M212770200. [DOI] [PubMed] [Google Scholar]
- 14.Haghighat A, Mader S, Pause A, Sonenberg N. Repression of cap-dependent translation by 4E-binding protein 1: competition with p220 for binding to eukaryotic initiation factor-4E. Embo J. 1995;14:5701–9. doi: 10.1002/j.1460-2075.1995.tb00257.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pause A, et al. Insulin-dependent stimulation of protein synthesis by phosphorylation of a regulator of 5′-cap function. Nature. 1994;371:762–7. doi: 10.1038/371762a0. [DOI] [PubMed] [Google Scholar]
- 16.Gingras AC, et al. Hierarchical phosphorylation of the translation inhibitor 4E-BP1. Genes Dev. 2001;15:2852–64. doi: 10.1101/gad.912401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brunn GJ, et al. Phosphorylation of the translational repressor PHAS-I by the mammalian target of rapamycin. Science. 1997;277:99–101. doi: 10.1126/science.277.5322.99. [DOI] [PubMed] [Google Scholar]
- 18.Burnett PE, Barrow RK, Cohen NA, Snyder SH, Sabatini DM. RAFT1 phosphorylation of the translational regulators p70 S6 kinase and 4E-BP1. Proc Natl Acad Sci U S A. 1998;95:1432–7. doi: 10.1073/pnas.95.4.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mothe-Satney I, Yang D, Fadden P, Haystead TA, Lawrence JC., Jr Multiple mechanisms control phosphorylation of PHAS-I in five (S/T)P sites that govern translational repression. Mol Cell Biol. 2000;20:3558–67. doi: 10.1128/mcb.20.10.3558-3567.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brugarolas J, et al. Regulation of mTOR function in response to hypoxia by REDD1 and the TSC1/TSC2 tumor suppressor complex. Genes Dev. 2004;18:2893–904. doi: 10.1101/gad.1256804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Connolly E, Braunstein S, Formenti S, Schneider RJ. Hypoxia inhibits protein synthesis through a 4E-BP1 and elongation factor 2 kinase pathway controlled by mTOR and uncoupled in breast cancer cells. Mol Cell Biol. 2006;26:3955–65. doi: 10.1128/MCB.26.10.3955-3965.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koritzinsky M, et al. Gene expression during acute and prolonged hypoxia is regulated by distinct mechanisms of translational control. Embo J. 2006;25:1114–25. doi: 10.1038/sj.emboj.7600998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koritzinsky M, et al. Phosphorylation of eIF2alpha is required for mRNA translation inhibition and survival during moderate hypoxia. Radiother Oncol. 2007 doi: 10.1016/j.radonc.2007.04.031. [DOI] [PubMed] [Google Scholar]
- 24.Reiling JH, Hafen E. The hypoxia-induced paralogs Scylla and Charybdis inhibit growth by down-regulating S6K activity upstream of TSC in Drosophila. Genes Dev. 2004;18:2879–92. doi: 10.1101/gad.322704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bernardi R, et al. PML inhibits HIF-1 alpha translation and neoangiogenesis through repression of mTOR. Nature. 2006;442:779–85. doi: 10.1038/nature05029. [DOI] [PubMed] [Google Scholar]
- 26.Mamane Y, et al. eIF4E--from translation to transformation. Oncogene. 2004;23:3172–9. doi: 10.1038/sj.onc.1207549. [DOI] [PubMed] [Google Scholar]
- 27.Ruggero D, et al. The translation factor eIF-4E promotes tumor formation and cooperates with c-Myc in lymphomagenesis. Nat Med. 2004;10:484–6. doi: 10.1038/nm1042. [DOI] [PubMed] [Google Scholar]
- 28.Wendel HG, et al. Survival signalling by Akt and eIF4E in oncogenesis and cancer therapy. Nature. 2004;428:332–7. doi: 10.1038/nature02369. [DOI] [PubMed] [Google Scholar]
- 29.Bjornsti MA, Houghton PJ. The TOR pathway: a target for cancer therapy. Nat Rev Cancer. 2004;4:335–48. doi: 10.1038/nrc1362. [DOI] [PubMed] [Google Scholar]
- 30.De Benedetti A, Graff JR. eIF-4E expression and its role in malignancies and metastases. Oncogene. 2004;23:3189–99. doi: 10.1038/sj.onc.1207545. [DOI] [PubMed] [Google Scholar]
- 31.Mamane Y, et al. Epigenetic Activation of a Subset of mRNAs by eIF4E Explains Its Effects on Cell Proliferation. PLoS ONE. 2007;2:e242. doi: 10.1371/journal.pone.0000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Culjkovic B, Topisirovic I, Skrabanek L, Ruiz-Gutierrez M, Borden KL. eIF4E is a central node of an RNA regulon that governs cellular proliferation. J Cell Biol. 2006;175:415–26. doi: 10.1083/jcb.200607020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lazaris-Karatzas A, Sonenberg N. The mRNA 5′ cap-binding protein, eIF-4E, cooperates with v-myc or E1A in the transformation of primary rodent fibroblasts. Mol Cell Biol. 1992;12:1234–8. doi: 10.1128/mcb.12.3.1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lazaris-Karatzas A, Montine KS, Sonenberg N. Malignant transformation by a eukaryotic initiation factor subunit that binds to mRNA 5′ cap. Nature. 1990;345:544–7. doi: 10.1038/345544a0. [DOI] [PubMed] [Google Scholar]
- 35.Hay N, Sonenberg N. Upstream and downstream of mTOR. Genes Dev. 2004;18:1926–45. doi: 10.1101/gad.1212704. [DOI] [PubMed] [Google Scholar]
- 36.Martin ME, et al. 4E binding protein 1 expression is inversely correlated to the progression of gastrointestinal cancers. Int J Biochem Cell Biol. 2000;32:633–42. doi: 10.1016/s1357-2725(00)00007-8. [DOI] [PubMed] [Google Scholar]
- 37.Avdulov S, et al. Activation of translation complex eIF4F is essential for the genesis and maintenance of the malignant phenotype in human mammary epithelial cells. Cancer Cell. 2004;5:553–63. doi: 10.1016/j.ccr.2004.05.024. [DOI] [PubMed] [Google Scholar]
- 38.Jacobson BA, et al. Repression of cap-dependent translation attenuates the transformed phenotype in non-small cell lung cancer both in vitro and in vivo. Cancer Res. 2006;66:4256–62. doi: 10.1158/0008-5472.CAN-05-2879. [DOI] [PubMed] [Google Scholar]
- 39.Li S, et al. Translational control of cell fate: availability of phosphorylation sites on translational represser 4E-BP1 governs its proapoptotic potency. Mol Cell Biol. 2002;22:2853–61. doi: 10.1128/MCB.22.8.2853-2861.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hernando E, et al. Rb inactivation promotes genomic instability by uncoupling cell cycle progression from mitotic control. Nature. 2004;430:797–802. doi: 10.1038/nature02820. [DOI] [PubMed] [Google Scholar]
- 41.Sotillo R, et al. Mad2 overexpression promotes aneuploidy and tumorigenesis in mice. Cancer Cell. 2007;11:9–23. doi: 10.1016/j.ccr.2006.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang S, et al. Sustained activation of the JNK cascade and rapamycin-induced apoptosis are suppressed by p53/p21(Cip1) Mol Cell. 2003;11:1491–501. doi: 10.1016/s1097-2765(03)00180-1. [DOI] [PubMed] [Google Scholar]
- 43.Gardner LB, et al. Hypoxia inhibits G1/S transition through regulation of p27 expression. J Biol Chem. 2001;276:7919–26. doi: 10.1074/jbc.M010189200. [DOI] [PubMed] [Google Scholar]
- 44.Goda N, et al. Hypoxia-inducible factor 1 alpha is essential for cell cycle arrest during hypoxia. Mol Cell Biol. 2003;23:359–69. doi: 10.1128/MCB.23.1.359-369.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rosenwald IB, et al. Eukaryotic translation initiation factor 4E regulates expression of cyclin D1 at transcriptional and post-transcriptional levels. J Biol Chem. 1995;270:21176–80. doi: 10.1074/jbc.270.36.21176. [DOI] [PubMed] [Google Scholar]
- 46.Clemens MJ, Bommer UA. Translational control: the cancer connection. Int J Biochem Cell Biol. 1999;31:1–23. doi: 10.1016/s1357-2725(98)00127-7. [DOI] [PubMed] [Google Scholar]
- 47.Fukumura D, Jain RK. Tumor microenvironment abnormalities: causes, consequences, and strategies to normalize. J Cell Biochem. 2007;101:937–49. doi: 10.1002/jcb.21187. [DOI] [PubMed] [Google Scholar]
- 48.Kops GJ, Weaver BA, Cleveland DW. On the road to cancer: aneuploidy and the mitotic checkpoint. Nat Rev Cancer. 2005;5:773–85. doi: 10.1038/nrc1714. [DOI] [PubMed] [Google Scholar]
- 49.Castellvi J, et al. Phosphorylated 4E binding protein 1: a hallmark of cell signaling that correlates with survival in ovarian cancer. Cancer. 2006;107:1801–11. doi: 10.1002/cncr.22195. [DOI] [PubMed] [Google Scholar]
- 50.Rosenwald IB, Lazaris-Karatzas A, Sonenberg N, Schmidt EV. Elevated levels of cyclin D1 protein in response to increased expression of eukaryotic initiation factor 4E. Mol Cell Biol. 1993;13:7358–63. doi: 10.1128/mcb.13.12.7358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bi M, et al. ER stress-regulated translation increases tolerance to extreme hypoxia and promotes tumor growth. Embo J. 2005;24:3470–81. doi: 10.1038/sj.emboj.7600777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rodriguez-Perales S, et al. Molecular cytogenetic characterization of rhabdomyosarcoma cell lines. Cancer Genet Cytogenet. 2004;148:35–43. doi: 10.1016/s0165-4608(03)00216-4. [DOI] [PubMed] [Google Scholar]
- 53.Nelson DA, et al. Hypoxia and defective apoptosis drive genomic instability and tumorigenesis. Genes Dev. 2004;18:2095–107. doi: 10.1101/gad.1204904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Easton JB, Houghton PJ. mTOR and cancer therapy. Oncogene. 2006;25:6436–46. doi: 10.1038/sj.onc.1209886. [DOI] [PubMed] [Google Scholar]