Abstract
The majority of eukaryotic secretory and membrane proteins contain disulfide bonds, which are strongly conserved within protein families because of their crucial role in folding or function. The exact role of these disulfide bonds during folding is unclear. Using virus-driven evolution we generated a viral glycoprotein variant, which is functional despite the lack of an absolutely conserved disulfide bond that links two antiparallel β-strands in a six-stranded β-barrel. Molecular dynamics simulations revealed that improved hydrogen bonding and side chain packing led to stabilization of the β-barrel fold, implying that β-sheet preference codirects glycoprotein folding in vivo. Our results show that the interactions between two β-strands that are important for the formation and/or integrity of the β-barrel can be supported by either a disulfide bond or β-sheet favoring residues.
INTRODUCTION
Protein folding is studied at several levels. Formation of secondary structure elements such as α-helices and β-sheets, for example, is investigated in vitro in model peptides or small proteins. Acquisition of tertiary structure is studied either in vitro or in vivo; formation of disulfide bonds is followed, for instance, during oxidative folding in the endoplasmic reticulum (ER) of intact cells. The results of a completed folding process are analyzed as a protein is expressed at the cell surface or secreted, for example, by NMR or x-ray crystallography. The ultimate test is the functional analysis of a completely folded protein. Despite the availability of a variety of assay systems to study protein folding, the exact role of disulfide bonds and other peptide motifs remains obscure. Viruses with a high mutation rate provide excellent tools to study protein evolution. We therefore used virus-driven evolution of a folding defective human immunodeficiency virus (HIV)-1 gp120 variant to obtain better insight into glycoprotein folding and the role of a disulfide bond in particular.
The HIV-1 envelope glycoprotein complex (Env) mediates viral attachment and entry in susceptible cells (Wyatt and Sodroski, 1998; Eckert and Kim, 2001; Poignard et al., 2001; Gallo et al., 2003). Env is synthesized as a gp160 precursor protein, which is cotranslationally translocated into the ER. Here, Env acquires carbohydrate chains and disulfide bonds, folds, trimerizes, and looses its leader peptide (Land et al., 2003). Like other glycoproteins, gp160 folds with assistance of molecular chaperones, such as BiP, calnexin, and calreticulin (Earl et al., 1991; Otteken et al., 1996; Knarr et al., 1999). Subsequently, gp160 is transported to the Golgi complex in which it is cleaved into gp120 and gp41, which stay associated noncovalently (Stein and Engleman, 1990; Moulard and Decroly, 2000), and in which about half of the ∼30 carbohydrates are modified (Leonard et al., 1990). In contrast to the folding of many model single domain proteins that exhibit a rapid two-state folding behavior, folding of Env is complex and slow, taking >4 h to complete (Land et al., 2003).
We have studied previously the importance of all individual disulfide bonds in the oxidative folding of HIV-1 Env (van Anken et al., 2008). Five of 10 disulfide bonds were dispensable for folding. Surprisingly, two were also largely dispensable for Env function and viral replication. The remaining five disulfide bonds were required for proper oxidative folding of Env in the ER. We generated a mutant virus lacking the conserved disulfide bond at the base of the V4 domain of gp120. This disulfide bond is located in a β-barrel in the outer domain of gp120, and it is required for proper oxidative folding of wild-type (wt) Env and virus replication. After prolonged virus culture and selection for function, we identified an evolved variant, in which improved local hydrogen bonding and side chain packing within the β-barrel structure compensated for the absence of the disulfide bond.
MATERIALS AND METHODS
Cloning
The pRS1, pcDNA3-Env-gp120, and pLAI plasmids containing the appropriate mutations in the env gene were generated as described in van Anken et al. (2008). Polymerase chain reaction (PCR)-generated gp120 sequences from evolved viruses (see below) were cloned into the pRS1 shuttle vector (Sanders et al., 2004a) by using the BsaB1 and Nhe1 sites and subsequently cloned into the pLAI infectious molecular clone (Peden et al., 1991) as SalI-BamHI fragments. NotI-XhoI fragments were subcloned into the pcDNA3 expression vector for use in folding experiments. Numbering of individual amino acids is based on the sequence of HXB2 gp160. Residues 385, 415, and 418 correspond to 390, 420, and 423 in LAI gp160.
Cells and Transfections
HeLa cells (American Type Culture Collection, Manassas, VA), and HT1080 cells were cultured in minimal essential medium (MEM) (Invitrogen, Groningen, The Netherlands) supplemented with 10% fetal calf serum (FCS; Hybond), 100 U/ml penicillin, and 100 μg/ml streptomycin. SupT1 cells were cultured in RPMI 1640 medium supplemented with 10% FCS, penicillin, and streptomycin. C33A cervix carcinoma cells were maintained in DMEM (Invitrogen), supplemented with 10% FCS, penicillin, and streptomycin, as described previously (Sanders et al., 2002a). SupT1 and C33A cells were transfected with pLAI by electroporation and Ca3(PO4)2 precipitation, respectively, as described previously (Das et al., 1999).
Viruses and Infections
Virus stocks were produced by transfecting C33A cells with the appropriate pLAI constructs. The virus containing supernatant was harvested 3 d posttransfection, filtered, and stored at −80°C, and the virus concentration was quantitated by capsid CA-p24 enzyme-linked immunosorbent assay (ELISA) as described previously (Jeeninga et al., 2000). These values were used to normalize the amount of virus in subsequent infection experiments. Infection experiments were performed as follows. We infected 50 × 103 SupT1 T-cells with 2.5 ng of CA-p24 of C33A-produced HIV-1LAI per well in a 96-well plate, and virus spread was measured for 14 d by using CA-p24 ELISA.
Virus Evolution
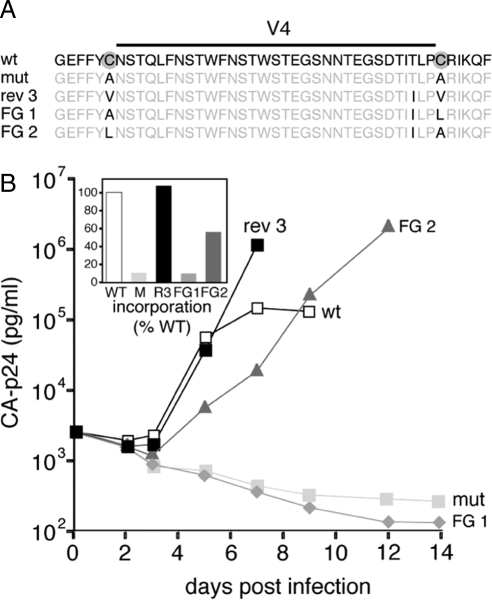
Evolution experiments were essentially performed as described previously (Sanders et al., 2004a,b). SupT1 cells (5 × 106) were transfected with 10 μg of the pLAI C385A/C418A construct by electroporation. The culture was inspected regularly for the emergence of revertant viruses, by using CA-p24 ELISA and/or the appearance of syncytia as indicators of virus replication. Cells were passaged twice a week. Decreasing amounts (1.0 ml, 100 μl, 10 μl, and 1.0 μl) of virus were passaged cell free onto fresh cells when the cells were (almost) wasted due to infection. The intervals and volumes of cell free passage depended on the replication efficiency and cytopathogenicity of the evolving virus. After prolonged culturing (∼2 mo) on SupT1 T-cells, we identified a faster replicating virus in a culture of the C385A/C418A mutant virus. DNA was extracted from infected cells (Das et al., 1997), and proviral gp120 sequences were PCR amplified with primers A (5′-GCTCCATGGCTTAGGGCAACATATATCTATG-3′) and B (5′-GTCTCGAGATGCTGCTCC-3′) and sequenced. Population sequencing revealed two reversions: a first-site pseudoreversion A418V and a second-site reversion at a nearby residue: T415I (Figure 1). Sequencing of individual env clones revealed several with the individual T415I reversion, implying that this mutation occurred first during the course of evolution (Figure 1). For simplicity, we hereafter refer to the respective variants as mutant (mut; C385A/C418A), revertant 1 (R1; C385A/C418A/T415I), and revertant 2 (R2; C385A/A418V/T415I). The R2 variant was recloned into pLAI for a subsequent round of evolution. After an additional period of culturing (∼6 mo), we identified an improved variant with a first-site pseudoreversion A385V. This variant A385V/A418V/T415I was designated revertant 3 (R3).
Figure 1.
Local reversions in gp120 improve viral replication. (A) Schematic of gp120 with the five conserved domains (C1–C5) and five variable domains (V1–V5). The location of the V4 base disulfide bond is indicated (gray sphere). Disulfide bonds and sites for N-linked glycosylation are indicated based on Leonard et al. (1990). Sites for N-linked glycosylation are shown. (B) Time scale of gp120 evolution. The presence of the revertants R1 (C385A/C418A/T415I), R2 (C385A/C418V/T415I), and R3 (C385V/C418V/T415I) as confirmed by sequencing is indicated on the timeline. (C) Sequences of the V4 loop and flanking regions of wt, mutant, and revertant viruses. No mutations were found outside this region. (D) The SupT1 T-cells were infected with wt, mutant, and revertant viruses and viral spread was measured for 14 d. () Inset, gp120 contents of virus. The gp120 amounts were standardized for CA-p24 input, and the gp120 contents of mutants in the respective fractions are given as percentages of the wt gp120 contents (arbitrarily set at 1). The results are representative for results from at least three independent experiments.
Single Cycle Infection and Virus Neutralization
The TZM-bl reporter cell line (Derdeyn et al., 2000; Wei et al., 2002) stably expresses high levels of CD4 and HIV-1 coreceptors CCR5 and CXCR4 and contains the luciferase and β-galactosidase genes under control of the HIV-1 long terminal repeat (LTR) promoter. The TZM-bl cell line was obtained through the National Institutes of Health AIDS Research and Reference Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases, National Institutes of Health: TZM-bl from Drs. J. C. Kappes, X. Wu (both of the University of Alabama at Birmingham, Birmingham, AL), and Tranzyme (Durham, NC). One day before infection, TZM-bl cells were plated on a 96-well plate in DMEM (Invitrogen) containing 10% fetal bovine serum, 1× MEM (Invitrogen), and penicillin/streptomycin (both at 100 U/ml), and then the cells were incubated at 37°C with 5% CO2. A fixed amount of virus produced in C33A cells (1 ng of CA-p24) was preincubated for 30 min at room temperature with escalating concentrations of monoclonal antibodies (2G12; obtained from H. Katinger through the National Institutes of Health AIDS Research and Reference Reagent Program; b12, a gift from D. Burton, The Scripps Research Institute, La Jolla, CA) or CD4 (CD4-IgG2; a gift from W. Olson, Progenics Pharmaceuticals). This mixture was added to the cells in the presence of 400 nM saquinavir (Roche Diagnostics, Indianapolis, IN) and 40 μg/ml DEAE in a total volume of 200 μl. Two days after infection, the medium was removed, and cells were washed once with phosphate-buffered saline and lysed in reporter lysis buffer (Promega, Madison, WI). Luciferase activity was measured using the luciferase assay kit (Promega) and a Glomax luminometer according to the manufacturer's instructions (Turner Designs, Sunnyvale, CA). All infections were performed in duplicate, and luciferase measurements were also performed in duplicate. Uninfected cells were used to correct for background luciferase activity. The infectivity of each mutant without inhibitor was set at 100%. Nonlinear regression curves were determined, and IC50 values were calculated using Prism software version 4.0c (GraphPad Software, San Diego, CA).
Quantitation of gp120 in Cell, Virion, and Supernatant Fractions
C33A cells were transfected with 40 μg of pLAI per T-75 flask. Medium was refreshed at day 1 after transfection. The culture supernatant was harvested at 3 d after transfection, centrifuged, and passed through a 0.45-μm filter to remove residual cells and debris. Cells were resuspended in 1.0 ml of lysis buffer (50 mM Tris, pH 7.4, 10 mM EDTA, 100 mM NaCl, and 1% SDS). Virus particles were pelleted by ultracentrifugation (100,000 × g for 45 min at 4°C) and resuspended in 0.5 ml of lysis buffer. The virus free supernatant, containing gp120 shed from the cell and virion surface, was concentrated using Amicon centrifugal filter units (Millipore, Billerica, MA), and SDS was added to a 1% final concentration.
Gp120 in cell, virion, and supernatant fractions was measured as described previously (Moore and Ho, 1993; Sanders et al., 2002b), with minor modifications. ELISA plates were coated overnight with sheep antibody D7324 (10 μg/ml; Aalto Bioreagents, Ratharnham, Dublin, Ireland), directed to the gp120 C5 region, in 0.1 M NaHCO3. After blocking with 2% milk powder in Tris-buffered saline (TBS) for 30 min, gp120 was captured by incubation for 2 h at room temperature. Recombinant HIV-1LAI gp120 (Progenics Pharmaceuticals, Tarrytown, NY) was used as a reference. Unbound gp120 was washed away with TBS and purified serum immunoglobulin (Ig) from an HIV-1–positive individual (HIVIg) was added for 1.5 h in 2% milk, 20% sheep serum, and 0.5% Tween 20. HIVIg binding was detected with alkaline phosphatase-conjugated goat anti-human Fc (1:10,000; Jackson ImmunoResearch Laboratories, West Grove, PA) in 2% milk, 20% sheep serum, and 0.5% Tween 20. Detection of alkaline phosphatase activity was performed using AMPAK reagents (Dako North America, Carpinteria, CA). The measured gp120 contents in cells, virus, and supernatant were normalized for CA-p24.
Env Folding
For folding assays, mutant gp120 was expressed using a recombinant Vaccinia virus vector system (Kieny et al., 1986). Folding of gp120 mutants was analyzed by pulse-chase labeling and immunoprecipitation with anti-Env sera, as described previously (Land et al., 2003). wt, mutant, and revertant gp120 were expressed in HeLa cells under control of the T7 promoter by using a recombinant Vaccinia virus vector system (Fuerst et al., 1986). Cells were placed on ice directly after the pulse or after various chase times. Culture supernatants were collected, and cells were washed, incubated with iodoacetamide to block free sulfhydryl groups, and lysed. Gp120 from cell lysates and culture supernatants was immunoprecipitated and treated with endoglycosidase H to remove oligomannose glycans. Formation of disulfide bonds was assayed by SDS-polyacrylamide gel electrophoresis (PAGE) mobility changes of deglycosylated, alkylated, nonreduced samples. Reduced samples were used to follow signal sequence cleavage.
Molecular Dynamics Simulations
The starting structures for molecular dynamics (MD) simulations were generated by SWISS-MODEL (Guex and Peitsch, 1997) by using the primary sequence of the LAI isolate as input and the crystal structures of gp120 of the HXB2 isolate (Protein Data Bank [PDB] entries 1G9M, 1GC1; Kwong et al., 1998, 2000) and that of the YU2 isolate (PDB entry 1G9N; Kwong et al., 2000) as templates. The flexible N and C termini and the hypervariable V1, V2, and V3 loops, which are missing in the crystal structures, were not included to limit the size of the simulations. The missing V4 loop was modeled by SWISS-MODEL by using default settings. In addition to the wt sequence of LAI, point mutations, which correspond to the mutations and reversions described in this study, were introduced in silico by modifying the input primary sequences for model generations.
The GROMACS 3.0 MD package (Lindahl et al., 2001) with the GROMOS 43A1 force field (Daura et al., 1998) was used with a protocol described previously (Hsu and Bonvin, 2004). In short, starting structures were individually solvated using the simple point charge water model (Berendsen et al., 1981) in periodic cubic boxes with a 1.4-nm solute-wall minimum distance. Chloride ions were introduced in all systems to obtain an electroneutralized system after a first steepest descent energy minimization. The resulting systems consisted of 317 amino acids and ∼30,500 water molecules that amount to a total number of ∼94,000 atoms. A second energy minimization was then performed, followed by five successive 20-ps MD runs with decreasing positional restraint force constants on the solutes (Kposres = 1000, 1000, 100, 10, and 0 kJ mol−1 nm−2) before the production runs.
All simulations were run for a period of 10 ns at 300K and 1 atmosphere. Solute, solvent, and counterions were independently coupled to a reference temperature bath, and the pressure was maintained by weakly coupling the system to an external pressure bath at one atmosphere (Berendsen et al., 1984). Nonbonded interactions were calculated using twin range cutoffs of 0.8 and 1.4 nm. Long-range electrostatic interactions beyond the cut-off were treated with the generalized reaction field model (Tironi et al., 1995) by using a dielectric constant of 54. A 4-fs integration time step was used for the integration of the equations of motions. The LINCS algorithm (Hess et al., 1997) was used for bond length constraining in conjunction with dummy atoms for the aromatic rings and amino group in side chains (Feenstra et al., 1999).
RESULTS
Rescue of a Folding Defective Glycoprotein by Evolution
To examine the role of a single conserved structure element in folding, we generated a mutant virus lacking the conserved disulfide bond at the base of the V4 domain of HIV-1 gp120 (Figure 1A). We found that removal of this disulfide bond by replacing the cysteines by alanines (C385A/C418A) led to Env misfolding (van Anken et al., 2008). Whereas the defective phenotype is no surprise considering the conserved nature of the disulfide, the folding-defective mutant did mediate some cell entry when placed in the context of the complete virus, albeit insufficient to cause a spreading virus infection (van Anken et al., 2008). A minority of the mutant Env molecules apparently folded correctly, exited the ER, and reached the virion surface to mediate attachment and membrane fusion.
Because of its residual function, we used this mutant for protein evolution studies, to identify and characterize escape routes that result in restoration of Env folding and function in the absence of the conserved disulfide bond. For an 8-mo evolution experiment of mutant virus, SupT1 T-cells were transfected with 10 μg of the molecular clone of the HIV-1LAI isolate (pLAI) containing the C385A and C418A substitutions (mut), and cells were passaged until they were wasted due to viral infection. Virus was then passaged cell free onto fresh cells, and the process was repeated for 73 d. At day 73, the env gene was PCR amplified and sequenced. The sequences revealed two reversions: A418V and T415I (Figure 1, B and C). The PCR fragment was recloned into pLAI and individual pLAI clones were sequenced, revealing two clones with the A418V and T415I reversions (R2), whereas one clone only contained the T415I reversion (R1), indicating that the latter reversion occurred first and that the R2 virus did not constitute the entire virus population yet at day 73. Fresh SupT1 cells then were transfected with 10 μg of the molecular clone containing R2 Env, and evolution was continued for an additional 180 d. The env gene was PCR amplified and sequenced at the time points indicated by an arrow in Figure 1B. At week 24, a third reversion occurred: A385V (R3). Both A385V and A418V were first-site reversions, but the wt cysteines were not restored, which is consistent with our design of the mutants. Conversion of alanine codons back into a cysteine codon required at least two nucleotide changes, providing a high mutational threshold and favoring alternative repair pathways. The change from alanine to valine represents a relatively simple evolutionary event (GCC to GTC). Several other simple evolution routes could have been chosen starting from the alanine codons, but we only found the reversions to valine. Moreover, we did not observe further evolution of the valine codons (Figure 1B; data not shown). The A385V reversion emerged again in an independent evolution experiment (data not shown), confirming that the evolution to valine is preferred. Inspection of gp120 sequences in the Los Alamos sequence database showed that an isoleucine at position 415 is found in ∼6% of the virus isolates, whereas threonine is found in ∼80%; the cysteines at positions 385 and 418 are absolutely conserved (http://www.hiv.lanl.gov/). In summary, we observed the sequential appearance of three amino acid substitutions: T415I, A418V, and A385V, respectively (R1, R2, and R3; Figure 1, B and C).
To establish that the identified substitutions accounted for the revertant phenotype, the relevant env fragments were recloned into the molecular clone HIV-1LAI to produce virus stocks. SupT1 T cells were infected with wt, mutant, and revertant viruses, and subsequent virus spread was monitored (Figure 1D). The mutant virus did not cause a spreading infection (van Anken et al., 2008), but viral replication improved increasingly for revertants R1, R2, and R3. These results indicate that a three-step evolution process took place upon removal of the V4-base disulfide bond, with all three reversions contributing to the final revertant phenotype.
To further study the contribution of each reversion to the improvement in virus replication we performed quantitative single cycle infection experiments by using reporter cells carrying the luciferase gene under control of the HIV-1 LTR (Wei et al., 2002). This assay provides a quantitative measure of Env-mediated viral entry, because upon entry the viral Tat protein transactivates the LTR and promotes luciferase expression. We used the assay to compare the different evolved variants and included a number of variants that did not emerge during the evolution experiment to provide a better understanding of each reversion.
In line with the replication data, viral infectivity and hence Env function gradually improved in R1-R3 compared with mut, with R3 displaying infectivity as high as 68% of wt (Figure 2). To examine the individual and pairwise contribution of each reversion to the revertant phenotype of R3, we constructed viruses lacking the first reversion, T415I, but containing the second or third reversion or both (AV, VA, and VV). Infectivity of both AV and VV was lower than that of R2 and R3, indicating that T415I did contribute to the revertant phenotype of R2 and R3 (Figure 2). In contrast, the VV variant was not more infectious than the VA variant, suggesting that the second reversion (A418V), although obviously beneficial in R2, did not contribute to the improved phenotype of R3 (Figure 2).
Figure 2.
Reversions restore virus infectivity. TZM-bl reporter cells (∼17 × 103; confluence of 70–80%) were infected with 5.0 ng of mutant or wt virus in the presence of SQV in a 96-well plate, and luciferase activity was measured after 48 h. The amino acids at positions 385, 418, and 415, respectively, are given below the graph. The relative infectivities were as follows: AA (mut), 7.5%; AAI (R1), 10.1%; AVI (R2), 25.6%; VVI (R3), 68.1%; AV, 20.2%; VA, 58.2%; VV, 54.8%; ALI (FG1), 1.9%; and LAI (FG2), 19.1% (*p < 0.05, **p < 0.01, ***p < 0.005).
Restoration of Env Folding and Incorporation into Virions
Because folding-defective Env mutants such as C385A/C418A are retained in the ER, they produce virions virtually devoid of Env molecules (van Anken et al., 2008). We therefore determined the content of revertant Env molecules on virions, expressed as gp120/p24 ratio, relative to wt (Figure 1D, inset). Only ∼10% of C385A/C418A Env was found on virus particles relative to wt, consistent with the severe folding defect measured for this mutant (van Anken et al., 2008). Revertants R1, R2, and R3 displayed gp120 virion contents of 17, 62, and 109%, suggesting that protein folding had improved with each successive substitution.
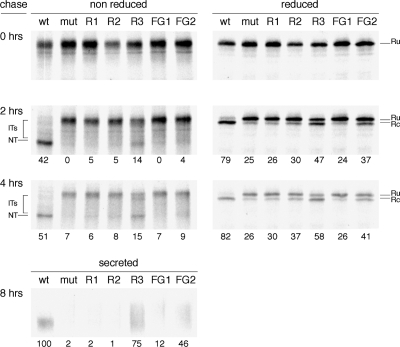
To confirm that improved folding of the revertants accounted for the gain in Env incorporation into virions, we monitored gp120 maturation in HeLa cells by pulse-chase analysis. We compared maturation kinetics of wt, mutant, and revertants by using three readouts we have developed previously (Land et al., 2003): 1) formation of disulfide bonds detectable through mobility changes of cellular gp120 in nonreducing SDS-PAGE, 2) signal peptide cleavage visible via reducing SDS-PAGE, and 3) secretion of gp120.
Immediately after the pulse, all gp120 variants displayed the same mobility in reducing and nonreducing gels (Figure 3), indicating that only few disulfide bonds had formed. After 2 h of chase, wt gp120 migrated faster in the nonreducing gel, as the fully oxidized native state. In contrast, most gp120 molecules of the mutant and revertants remained in the original, relatively unfolded, state, whereas a minority was present as faster migrating “smear,” especially in R3, representing partially oxidized folding intermediates. The mutant did not display detectable levels of the native state even after 4 h, but the revertants did reach a native-like state. Whereas for R1 and R2 the native band was faint, a considerable fraction of R3 reached native after 4 h of chase. Approximately 50% of R3 Env was still immature after 4 h, perhaps because only ∼50% folds correctly. If so, this 50% would be incorporated into the virion very efficiently (Figure 1D, inset). Alternatively, the R3 protein folds slower than wt and will continue to fold successfully after 4 h. We have not studied later time points because the pulse-chase data in this expression system become less reliable after 4 h. Regardless of the scenario, the reversions did not completely compensate for the lack of a disulfide bond.
Figure 3.
Reversions restore gp120 folding. HeLa cells were infected with VVT7 and transfected with plasmids encoding wt gp120, mut, revertant (R1, R2, or R3) or two additional mutants FG1 and FG2. Cells were pulse labeled for 5 min and chased for the indicated times. Cells were lysed and gp120 was immunoprecipitated from lysates. Immunoprecipitates were deglycosylated with endoglycosidase H and analyzed by either reducing or nonreducing 7.5% SDS-PAGE. Folding intermediates (IT), the native form (NT), the reduced state from which the signal peptide was cleaved off (Rc) or not (Ru) are indicated. In addition, secreted gp120 was immunoprecipitated from the culture supernatant after 8 h of chase and directly analyzed by reducing SDS-PAGE (bottom). Percentages of NT = native (nonreduced), cleaved = Rc (reduced), and secreted gp120 are given below lanes of chase samples. Percentage of secretion is relative to wt.
An unusual property of Env is that it must undergo some oxidative folding before its signal peptide can be removed (Land et al., 2003). We therefore used signal peptide cleavage as an additional measure for successful folding. In reducing gels, the single band present at the end of the pulse, corresponding with the preprotein form of gp120 (Ru; reduced uncleaved), changed into two bands, the lower one corresponding to cleaved gp120 (Rc; reduced cleaved). After 4 h of chase, uncleaved species were no longer detectable for wt. Signal peptide cleavage was significantly reduced for the mutant and R1, but R2 and in particular R3 displayed restored cleavage.
The third readout confirmed restoration of productive R3 folding: unlike the mutant, R1, and R2, a substantial fraction of R3 gp120 molecules had been secreted after 8 h (Figure 3, bottom). Secreted wt occurred as a compact band, but secreted R3 smeared out, most likely because of changed glycan modifications (Trombetta and Parodi, 2003). These results are consistent with the Env incorporation and virus replication experiments, and they confirm that virus-driven evolution resulted in a gradual repair of gp120 folding.
To analyze the outcome of the R3 folding process, we performed neutralization experiments with the wt and R3 viruses by using reagents that are dependent on gp120 conformation. The viruses were preincubated with the respective reagents and subsequently added to target cells containing a luciferase reporter gene under control of the HIV-1 LTR (Wei et al., 2003). First, we tested the ability of CD4 to inhibit these viruses. CD4 binding is highly dependent on conformation making contacts with both the inner and outer domain of gp120 (Kwong et al., 1998). wt and R3 were equally sensitive to inhibition by CD4-IgG2, indicating that the structure of the CD4 binding site on these viruses was similar (Figure 4A). b12 is a potent broadly neutralizing antibody with a conformational epitope that overlaps with the CD4 binding site, although most contacts are made with the outer domain of gp120 only (Pantophlet et al., 2003; Zhou et al., 2007). The b12 epitope is close to the 385–418 disulfide bond and residues in this area, e.g., N386, are known contact sites for b12 (Pantophlet et al., 2003; Zhou et al., 2007). Overall, the inhibition curves of wt and R3 were similar, although R3 was slightly less sensitive to inhibition by b12 (Figure 4B). We next performed neutralization experiments with 2G12, which binds to a conformation-dependent mannose epitope on N-linked carbohydrates (Trkola et al., 1996; Scanlan et al., 2002; Sanders et al., 2002b). The carbohydrate at N386, located next to the 385–418 disulfide bridge, is involved in 2G12 binding (Trkola et al., 1996; Sanders et al., 2002b; Scanlan et al., 2002) and local structural perturbations can affect the binding of 2G12 (Sanders et al., 2008). We did not observe major differences in inhibition by 2G12 (Figure 4C). Combined these results indicated that the R3 gp120 folded into a structure that is similar to the wt gp120.
Figure 4.
wt and R3 viruses show similar sensitivities for conformational antibodies. Single cycle infection experiments in TZM-bl cells containing a luciferase reporter construct under control of the HIV-1 LTR were carried out in the presence of escalating concentrations of CD4-IgG2 (A) and monoclonal antibodies b12 (B) and 2G12 (C). The luciferase activity in the absence of inhibiting reagents was set at 100%.
Molecular Dynamics Simulations Reveal Restoration of Local β-Sheet Structure
To understand improved folding and function of the revertants at the atomic level, we performed MD simulations of gp120 variants. Complete sampling of complete protein (un)folding by MD simulation would require enormous amounts of computational capacity, which cannot be mustered for a protein the size of gp120. We therefore carried out a comparative study of individual variants in their folded state at a residue-specific level. Changes in dynamics and stability in the gp120 native state have implications for its folding as the quality control recognition system is closely associated with structural integrity. Events of local unfolding, which are reflected in the loss of intramolecular hydrogen bonding and increased structural fluctuation, are indicative of the tendency toward global unfolding.
We performed simulations with wt gp120 and all variants (mut, R1, R2, and R3, and two additional mutants FG1 and FG2; see below). Starting from identical conformations, simulations of gp120 variants were conducted in explicit solvent (water) for an extended period (10 ns at 300 K). Despite the small differences in primary sequence (2 or 3 of 317 amino acids), the time to reach equilibrium required for each variant varied significantly as judged from the backbone (N, Cα, and C′) positional root-mean-square deviation (RMSD) evolution profiles (Supplemental Figure 1). Apparently, point mutations can cause subtle structural perturbations that require a longer time scale to reach equilibrium. We therefore focused subsequent analyses on the 5- to 10-ns segments, for which all variants were stable with little variation in secondary structure and nonbonded energies (Supplemental Table 1). Still, overall protein structure seemed very similar throughout the simulations for wt, mut, and revertants. We also carried out several simulations for wt, mut, R1, R2, and R3 at elevated temperatures (400K). Overall dynamics were similar as judged from the overall secondary structure content (with slightly higher RMSD and relative root-mean-square fluctuation [RMSF]). We concluded that neither the lack of disulfide bond 385–418 nor the reversions caused major structural changes in the native structure of gp120. We therefore focused on local perturbations of the structure in the vicinity of the mutated residues.
Atomic structures are available of the core of gp120 in different states: unliganded SIV gp120, CD4-bound HIV-1 gp120, and HIV-1 in complex with the neutralizing antibody b12 (Kwong et al., 1998; Kwong et al., 2000; Chen et al., 2005; Zhou et al., 2007). These structures reveal remarkable structural rearrangements of gp120 upon binding to CD4: the inner and outer domains of gp120 undergo large reorientation upon CD4 binding, whereby a four-stranded bridging sheet is formed. Despite the large differences in the free, CD4-bound and b12-bound states, the six-stranded β-barrel structure in the outer domain of gp120, which includes (the reverted) residues 385, 415, and 418, is particularly unaffected by these conformational rearrangements (Figure 5A). The backbone Cα atoms of the β-barrel exhibited a marginal deviation between the states with a positional RMSD of 0.17 nm (data not shown). The six β-strands of the barrel (β16, β17, β19, β13, β12, and β22) are bridged by three disulfide bonds, one of which is formed by the C385/C418 pair (Figure 5A). The first and last strands of the β-barrel (β16 and β22) are not linked through classical β-sheet interactions (i.e., interstrand backbone–backbone hydrogen bonds are absent), but only through the disulfide bond between C378 and C445. We turned to detailed analysis of the β-barrel for parameters that afford sufficient statistics, namely, RMSF and hydrogen bond occurrence. The local motions were sampled reliably over the equilibrated 5-ns trajectories. The RMSF and hydrogen bond occurrence analyses can provide evidence of local unfolding events, i.e., loss of native structures, which have implications for the folding process.
Figure 5.
Reversions change dynamics within a six-stranded β-barrel in the outer domain of HIV gp120. (A) Structure of HIV gp120 core and the β-barrel structure in the outer domain. The four-stranded bridging sheet, which joins the inner and outer domains, is indicated. The β-barrel structure in the outer domain is boxed and shown in detail on the right (see Supplemental Table 3 for definitions). The individual β-strands are labeled and colored in red, yellow, and green, corresponding to the pairs that are bridged by disulfides C296/C331, C385/C418, and C378/C445, respectively. Positions of mutations studied here (C385, C415, and T418) are labeled. The β-barrel of gp120 was defined as follows based on the structural alignment (Chen et al., 2005) (residue numbers in the structure of CD4-bound HIV-1 gp120, PDB code 1G9M, and residue numbers in unliganded SIV gp120, PDB code 2BF1 in parentheses): β12, residues 291-297 (305-311); β13, 330-334 (342-346); β16-β17, 374-387 (342-346); β19, 413-422 (426-435); and β22, 443-450 (456-463). The β-barrel structures in the unliganded SIV gp120 and the CD4-bound HIV-1 gp120 deviate marginally with a backbone Cα RMSD (44 residues, excluding the GAG linker between β12 and β13) of 0.17 nm, as calculated by ProFit (http://acrmwww.biochem.ucl.ac.uk/software/profit/). (B) Relative sums of interstrand backbone–backbone hydrogen bond occurrence among various variants τTotal (variant). The occurrence of interstrand backbone–backbone hydrogen bonds across a β-strand pair j and k of individual variants τj−k (variant) is defined as a normalized sum of the occurrence of individual interstrand backbone–backbone hydrogen bonds i within the β-strand pair j-k of the β-barrel with respect to that of wt, τj−k(variant) ≡ Σiτij−k(variant)/Σiτij−k(wt) × 100% (data not shown). The sum of all interstrand backbone–backbone hydrogen bonds within the β-barrel τTotal (variant)—those between strands 22-12, 12-13, 13-19, 19-17, and 17-16 (see Figure 5A)—is used for comparison in the text. The exact percentages relative to wt are mut, 81.1; R1, 90.7; R2, 88.4; R3, 90.9; FG1, 86.4; and FG2, 83.8. (C) Relative atomic positional RMSF of individual residues (filled gray circles) and overall average of those in the β-barrel structure (open circles). A reference line of wt (100%) is drawn (dashed line) for comparison. (D) Snapshots of selected hydrophilic residues around the mutation sites that are involved in interstrand side chain–side chain hydrogen bonding. The structures are sampled every 100 ps during the 5- to 10-ns trajectories. All structures were fit onto the backbone atoms of these residues. Oxygen, nitrogen, and carbon atoms are colored in red, blue, and green, respectively.
As a first measure for structural stability (or integrity) of the β-barrel, we evaluated the interstrand backbone–backbone hydrogen bond occurrence (percentage of time) between the individual strands of the barrel [τ (variant), defined in the legend of Figure 5; Kabsch and Sander, 1983]. The different side chains introduced by the mutations, and reversions resulted in large fluctuations of interstrand hydrogen bond occurrence within the 6-stranded β-barrel (Figure 5B). The folding deficient mutant showed a substantial loss of total interstrand hydrogen bonding with a τTotal (mut) of 81.1% of wt, whereas in the revertants the τTotal was largely restored: (R1) 90.7%, (R2) 88.4%, and (R3) 90.9% (Figure 5B). This indicates that the backbone structure of the β-barrel is sensitive to elimination of the disulfide bond. Yet, only the first reversion contributed to a substantial gain in interstrand backbone–backbone hydrogen bonding.
Next, we explored whether the differences between wt, mut, and revertants correlated to alterations in side chain packing. We assessed the degree of flexibility of individual residues within the barrel by calculating their relative RMSF (i.e., the sum of atomic positional RMSF per residue normalized to the corresponding wt values). A lower relative RMSF value is indicative for improved ordering of a particular side chain. Taking wt as 100%, mut had the broadest distribution of per residue RMSF values with a maximum as large as 275% (Figure 5C). The distributions of per residue RMSF as well as the overall averages of R1, R2, and R3 reduced progressively, reflecting improved side chain packing. To visualize this, we compared selected side chain structure ensembles of wt, mut, and R3 throughout the simulations (Figure 5D). The conformations of several hydrophilic side chains were more heterogeneous in mut than in R3 and wt (e.g., Y384 and S375), implying more flexibility in the mutant and increased order in the revertant. In addition, the relative side chain orientations of R3 were more similar to those in wt than in mut (e.g., S375, N386, and R419), thereby restoring side chain–side chain hydrogen bond formation. For example, the hydrogen bond between the hydroxyl group of S375 (OHγ) and the tyrosyl group of Y384 (Oη) (highlighted by an orange arrow in Figure 5D) was highly stable in wt with an occurrence of 95%, and it was destabilized in mut with an occurrence of only 16% (14% of which was bridged by a water molecule). In R3, this hydrogen bond was recovered (84% occurrence, without any additional water bridge), implying a higher degree of compactness. We found similar effects for hydrophobic side chain contacts in the interior of the barrel (data not shown).
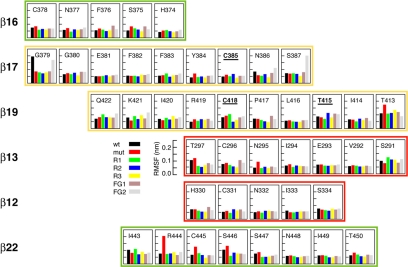
When examining each residue within the β-barrel, we did not find clear evidence of changes in relative RMSF between wt, mut, and R1–R3 in the strands in which the mutations were introduced (β17 and β19) (Figure 6). Other interesting correlations did emerge, however, for residues that have a high RMSF in mut but show progressive RMSF reduction in R1–R3 (Figure 6): 1) many residues in β16 (all residues except H374) and β22 (all residues except I443); 2) strand-end residues (C378, T413, T297, H330, and T450); 3) cysteines (or substituted residues; C378, C/A/V418, C296, and C445); and 4) residues flanking cysteines (N377, Y384, T297, N295, H330, P417, R440, R444, and S446). Because the barrel is buckled up by a number of disulfide bonds, the loss of one of these may lead to pronounced long-range perturbations, propagated through the barrel. This may explain the RMSF changes in cysteines and flanking residues distant from the mutated sites. The RMSF evolution of β16 and β22 seems somewhat surprising, but these strands are only connected by the 378–445 disulfide bond and not by sheet interactions. Hence, structural perturbations in the barrel may have a more pronounced impact on these two strands.
Figure 6.
Absolute positional RMSF for individual residues within the β-barrel. Residues 385, 418, and 415 are labeled in bold and underlined.
The MD analysis hence revealed that the functional and biochemical data can be explained at the atomic level by improved backbone–backbone hydrogen bonding and local side chain packing within the β-barrel. Our results suggest that the disulfide bond, despite its conservation, can be functionally replaced by alternative local structural features within the β-barrel.
Strengthening β-Sheets or Filling the Gap?
During our evolution experiments, we consistently encountered introduction of β-branched amino acids (T415I, A418V, and A385V) to restore folding efficiency of gp120 (Figure 1). The improvements we found in backbone hydrogen bonding and side chain packing within the β-barrel could be attributed to two factors. Knowledge-based predictions (Chou and Fasman, 1974; Levitt, 1978) and theoretical calculations (Rossmeisl et al., 2003) have shown that β-branched amino acids such as valine and isoleucine exhibit high β-sheet propensities: they promote β-sheet formation. In contrast, introduction of a methyl group (when valine replaces alanine) may fill the gap introduced by replacement of cysteine by alanine. Both β-sheet propensity and “gap filling” could therefore be responsible for the improved folding of revertant gp120. To discriminate between these two scenarios, we constructed two additional hydrophobic mutants where one of the alanines was replaced by a non–β-branched leucine rather than valine, in the context of the R1 variant (i.e., including the T415I reversion; Figure 7A): fill-gap mutant 1 (FG1: C385A/C418L/T415I; ALI) and FG2 (C385L/C418A/T415I; LAI). Valine and leucine have similar biochemical properties (both are hydrophobic and the size difference is one methyl group), but valine is β-branched and is accommodated much better in β-sheets than leucine is. FG1 displayed minimal infectivity in single cycle infection experiments (Figure 2), mirrored by minimal replication (Figure 7B). The folding assays revealed that FG1 did not fold properly: negligible FG1 gp120 reached the native state and signal peptide cleavage was also minimal (Figure 3). The folding, infectivity and replication of FG2 was better than of FG1 and R1, but less than wt, R2, and R3 (Figures 2, 3, and 7B), indicating that replacing A385 with leucine did improve protein folding to some extent. Both FG variants showed decreased in silico stability compared with R1, and R2 and R3, as indicated by decreased hydrogen bonding and increased per residue RMSF (Figures 5 and 6).
Figure 7.
Virus replication and folding of the non–β-branched fill-gap mutants FG1 (C385A/C418L/T415I) and FG2 (C385L/C418A/T415I). (A) Sequences of the V4 loop and flanking regions of wt, mutant, and revertant viruses. The original mutations are indicated with gray circles, the mutations and reversions with black letters. (B) SupT1 T-cells (50 × 103) were infected with 2.5 ng CA-p24, and viral spread was measured for 14 d. Inset, Gp120 and CA-p24 contents in virus were measured by ELISA. The gp120 amounts were standardized for CA-p24 input and the gp120 contents of mutants in the respective fractions are given as percentages of the wt gp120 contents (arbitrarily set at 1).
Thus, at position 418 a valine can improve folding and function, whereas a leucine cannot. In fact, leucine was inferior to alanine. At position 385, leucine conferred improved folding and function compared with alanine, but it was inferior to valine. The results imply that β-branching improves stability of the β-barrel and hence gp120 folding. Gap filling does contribute somewhat to improved gp120 folding, but only at position 385 and not 418.
DISCUSSION
We used a rapidly mutating virus for an evolution screen in which we identified a glycoprotein variant (gp120) that was functional despite the lack of a completely conserved and otherwise essential disulfide bond. MD simulations explained the restoration of folding and function through improvement of local hydrogen bonding and side chain packing. Our results suggest that this particular disulfide bond, despite its conservation, can be functionally replaced by alternative local structural features within the β-barrel.
The outer domain of gp120 contains three disulfide bonds, all located in this central β-barrel domain (Figure 5A). One of these (C296/C331 between strands β12 and β13) is absolutely required for folding (van Anken et al., 2008). Accordingly, parallel evolution experiments starting from a mutant lacking this disulfide bond failed to yield any functional gp120 variants. As we show in this study, the second disulfide bond (C385/C418; between β17 and β19) is also required for folding, but it can be replaced by acquisition of multiple β-sheet favoring residues. The third disulfide bond (C378/C445; between β16 and β22) is not absolutely required for protein folding and virus replication, although it does accelerate folding (van Anken et al., 2008). A clear hierarchy hence exists of disulfide bond importance within the β-barrel structure, with C296/C331 being the most important, followed by C385/C418, and finally C378/C445. Together, we showed these disulfide bonds play an important role in directing the formation or in maintaining the structure of this β-barrel during gp120 folding.
During our evolution experiments, we consistently observed reversion to β-branched amino acids (T415I, A418V, and A385V) (Figure 1). Our findings nicely agree with knowledge-based predictions (Chou and Fasman, 1974; Levitt, 1978) and theoretical calculations (Rossmeisl et al., 2003) that β-branched amino acids convey high β-sheet propensities and thus promote β-sheet formation. Consequently, the results are in accordance with β-sheet preferences as established in small model proteins (Kim and Berg, 1993; Smith et al., 1994; Minor and Kim, 1994b). The generalized rules on β-sheet preference apply to β17 and β19, because they are central strands and not edge strands (Minor and Kim, 1994a). Alternatively, the introduction of the methyl group caused by the replacement of alanine by valine fills the space that was occupied by the sulfur atom from the wt cysteine. Engineered Env variants in which the gap was filled by nonbranching side chains (A385L and A418L) performed worse than the variants containing valine (Figures 2, 3, 5, 6, and 7). Thus, although “gap filling” may play a role, β-branching contributes to the improved integrity of the β-barrel and hence to gp120 folding.
Using virus-driven protein evolution, we showed that a conserved disulfide bond facilitates formation or maintenance of a β-barrel structure during protein folding. Increasing the local β-sheet propensity can compensate for the lack of this disulfide bond, implying that β-sheet propensity is a major determinant in directing protein folding. Our data indicate that the simple β-sheet rules deduced from experiments with small model proteins also hold for the intricate folding of a complicated glycoprotein such as gp120 in living cells. These findings have implications for our molecular understanding of protein folding in vivo, as well as for protein engineering and de novo protein design.
Supplementary Material
ACKNOWLEDGMENTS
We thank Els Busser, Sonja Tillemans, and Stephan Heynen for excellent technical assistance. We are grateful to Dennis Burton and William Olson for reagents. This work was sponsored in part by the Dutch AIDS fund 5003 (to B. B.) and 1028 (to I. Braakman) and by grants from the Netherlands Organization for Scientific Research (NWO-Medical Sciences; to E.v.A., I. Braakman, and B. B.) and NWO-Chemical Sciences (to M. L. and I. Braakman). R.W.S. is a recipient of an amfAR Mathilde Krim research fellowship, an Anton Meelmeijer fellowship, and a VENI fellowship from NWO-Chemical Sciences. S.-T.D.H. is a recipient of a Netherlands Ramsay Fellowship from the Royal Netherlands Academy of Arts and Sciences and a long-term fellowship from the Human Frontier Science Program. A.M.J.J.B. is a recipient of NWO Jonge Chemici and VICI grants. The molecular dynamics simulations were supported by the National Computing Facilities, with financial support from NWO.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-07-0670) on September, 17, 2008.
REFERENCES
- Berendsen H.J.C., Postma J.P.M., Van Gunsteren W. F., Di-Nola A., Haak J. R. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 1984;81:3684–3690. [Google Scholar]
- Berendsen H J.C., Postma J.P.M., Van Gunsteren W. F., Hermans J. Interaction Models for Water in Relation to Protein Hydration. Dordrecht, The Netherlands: Reidel Publishing; 1981. [Google Scholar]
- Chen B., Vogan E. M., Gong H., Skehel J. J., Wiley D. C., Harrison S. C. Structure of an unliganded simian immunodeficiency virus gp120 core. Nature. 2005;433:834–841. doi: 10.1038/nature03327. [DOI] [PubMed] [Google Scholar]
- Chou P. Y., Fasman G. D. Conformational parameters for amino acids in helical, beta-sheet, and random coil regions calculated from proteins. Biochemistry. 1974;13:211–222. doi: 10.1021/bi00699a001. [DOI] [PubMed] [Google Scholar]
- Das A. T., Klaver B., Berkhout B. A hairpin structure in the R region of the human immunodeficiency virus type 1 RNA genome is instrumental in polyadenylation site selection. J. Virol. 1999;73:81–91. doi: 10.1128/jvi.73.1.81-91.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das A. T., Klaver B., Klasens B.I.F., van Wamel J.L.B., Berkhout B. A conserved hairpin motif in the R-U5 region of the human immunodeficiency virus type 1 RNA genome is essential for replication. J. Virol. 1997;71:2346–2356. doi: 10.1128/jvi.71.3.2346-2356.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daura X., Mark A. E., Van Gunsteren W. F. Parametrization of aliphatic CHn united atoms of GROMOS96 force field. J. Comput. Chem. 1998;19:535–547. [Google Scholar]
- Derdeyn C. A., Decker J. M., Sfakianos J. N., Wu X., O'Brien W. A., Ratner L., Kappes J. C., Shaw G. M., Hunter E. Sensitivity of human immunodeficiency virus type 1 to the fusion inhibitor T-20 is modulated by coreceptor specificity defined by the V3 loop of gp120. J. Virol. 2000;74:8358–8367. doi: 10.1128/jvi.74.18.8358-8367.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earl P. L., Moss B., Doms R. W. Folding, interaction with GRP78-BiP, assembly, and transport of the human immunodeficiency virus type 1 envelope protein. J. Virol. 1991;65:2047–2055. doi: 10.1128/jvi.65.4.2047-2055.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckert D. M., Kim P. S. Mechanisms of viral membrane fusion and its inhibition. Annu. Rev. Biochem. 2001;70:777–810. doi: 10.1146/annurev.biochem.70.1.777. [DOI] [PubMed] [Google Scholar]
- Feenstra K. A., Hess B., Berendsen H. J. C. Improving efficiency of large time-scale molecular dynamics of hydrogen-rich systems. J. Comput. Chem. 1999;20:786–798. doi: 10.1002/(SICI)1096-987X(199906)20:8<786::AID-JCC5>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- Fuerst T. R., Niles E. G., Studier F. W., Moss B. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc. Natl. Acad. Sci. USA. 1986;83:8122–8126. doi: 10.1073/pnas.83.21.8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallo S. A., Finnegan C. M., Viard M., Raviv Y., Dimitrov A., Rawat S. S., Puri A., Durell S., Blumenthal R. The HIV Env-mediated fusion reaction. Biochim. Biophys. Acta. 2003;1614:36–50. doi: 10.1016/s0005-2736(03)00161-5. [DOI] [PubMed] [Google Scholar]
- Guex N., Peitsch M. C. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- Hess B., Bekker H., Berendsen H.J.C., Fraaije J. LINCS: a linear constraint solver for comparative protein modeling. J. Comput. Chem. 1997;18:1463–1472. [Google Scholar]
- Hsu S. T., Bonvin A. M. Atomic insight into the CD4 binding-induced conformational changes in HIV-1 gp120. Proteins. 2004;55:582–593. doi: 10.1002/prot.20061. [DOI] [PubMed] [Google Scholar]
- Jeeninga R. E., Hoogenkamp M., Armand-Ugon M., de Baar M., Verhoef K., Berkhout B. Functional differences between the long terminal repeat transcriptional promoters of HIV-1 subtypes A through G. J. Virol. 2000;74:3740–3751. doi: 10.1128/jvi.74.8.3740-3751.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W., Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- Kieny M. P., et al. AIDS virus ENV protein expressed from a recombitant Vaccinia virus. Biotech. 1986;4:790–795. [Google Scholar]
- Kim C. A., Berg J. M. Thermodynamic beta-sheet propensities measured using a zinc-finger host peptide. Nature. 1993;362:267–270. doi: 10.1038/362267a0. [DOI] [PubMed] [Google Scholar]
- Knarr G., Modrow S., Todd A., Gething M. J., Buchner J. BiP-binding sequences in HIV gp160. Implications for the binding specificity of bip. J. Biol. Chem. 1999;274:29850–29857. doi: 10.1074/jbc.274.42.29850. [DOI] [PubMed] [Google Scholar]
- Kwong P. D., Wyatt R., Majeed S., Robinson J., Sweet R. W., Sodroski J., Hendrickson W. A. Structures of HIV-1 gp120 envelope glycoproteins from laboratory-adapted and primary isolates. Struct. Fold. Des. 2000;8:1329–1339. doi: 10.1016/s0969-2126(00)00547-5. [DOI] [PubMed] [Google Scholar]
- Kwong P. D., Wyatt R., Robinson J., Sweet R. W., Sodroski J., Hendrickson W. A. Structure of an HIV gp120 envelope glycoprotein in a complex with the CD4 receptor and a neutralizing human antibody. Nature. 1998;393:648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Land A., Zonneveld D., Braakman I. Folding of HIV-1 envelope glycoprotein involves extensive isomerization of disulfide bonds and conformation-dependent leader peptide cleavage. FASEB J. 2003;17:1058–1067. doi: 10.1096/fj.02-0811com. [DOI] [PubMed] [Google Scholar]
- Leonard C. K., Spellman M. W., Riddle L., Harris R. J., Thomas J. N., Gregory T. J. Assignment of intrachain disulfide bonds and characterization of potential glycosylation sites of the type 1 recombinant human immunodeficiency virus envelope glycoprotein (gp120) expressed in Chinese hamster ovary cells. J. Biol. Chem. 1990;265:10373–10382. [PubMed] [Google Scholar]
- Levitt M. Conformational preferences of amino acids in globular proteins. Biochemistry. 1978;17:4277–4285. doi: 10.1021/bi00613a026. [DOI] [PubMed] [Google Scholar]
- Lindahl E., Hess B., Van der Spoel D. GROMACS 3.0, a package for molecular simulation and trajectory analysis. J. Mol. Model. 2001;7:306–317. [Google Scholar]
- Minor D. L., Jr, Kim P. S. Context is a major determinant of beta-sheet propensity. Nature. 1994a;371:264–267. doi: 10.1038/371264a0. [DOI] [PubMed] [Google Scholar]
- Minor D. L., Jr, Kim P. S. Measurement of the beta-sheet-forming propensities of amino acids. Nature. 1994b;367:660–663. doi: 10.1038/367660a0. [DOI] [PubMed] [Google Scholar]
- Moore J. P., Ho D. D. Antibodies to discontinuous or conformationally sensitive epitopes on the gp120 glycoprotein of human immunodeficiency virus type 1 are highly prevalent in sera of infected humans. J. Virol. 1993;67:863–875. doi: 10.1128/jvi.67.2.863-875.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moulard M., Decroly E. Maturation of HIV envelope glycoprotein precursors by cellular endoproteases. Biochim. Biophys. Acta. 2000;1469:121–132. doi: 10.1016/s0304-4157(00)00014-9. [DOI] [PubMed] [Google Scholar]
- Otteken A., Earl P. L., Moss B. Folding, assembly, and intracellular trafficking of the human immunodeficiency virus type 1 envelope glycoprotein analyzed with monoclonal antibodies recognizing maturational intermediates. J. Virol. 1996;70:3407–3415. doi: 10.1128/jvi.70.6.3407-3415.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantophlet R., Ollmann S. E., Poignard P., Parren P. W., Wilson I. A., Burton D. R. Fine mapping of the interaction of neutralizing and nonneutralizing monoclonal antibodies with the CD4 binding site of human immunodeficiency virus type 1 gp120. J. Virol. 2003;77:642–658. doi: 10.1128/JVI.77.1.642-658.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peden K., Emerman M., Montagnier L. Changes in growth properties on passage in tissue culture of viruses derived from infectious molecular clones of HIV-1LAI, HIV-1MAL, and HIV-1ELI. Virology. 1991;185:661–672. doi: 10.1016/0042-6822(91)90537-l. [DOI] [PubMed] [Google Scholar]
- Poignard P., Saphire E. O., Parren P. W., Burton D. R. gp 120, biologic aspects of structural features. Annu. Rev. Immunol. 2001;19:253–274. doi: 10.1146/annurev.immunol.19.1.253. [DOI] [PubMed] [Google Scholar]
- Rossmeisl J., Kristensen I., Gregersen M., Jacobsen K. W., Norskov J. K. Beta-sheet preferences from first principles. J. Am. Chem. Soc. 2003;125:16383–16386. doi: 10.1021/ja0359658. [DOI] [PubMed] [Google Scholar]
- Sanders R. W., Busser E., Moore J. P., Lu M., Berkhout B. Evolutionary repair of HIV type 1 gp41 with a kink in the N-terminal helix leads to restoration of the six-helix bundle structure. AIDS Res. Hum. Retroviruses. 2004a;20:742–749. doi: 10.1089/0889222041524544. [DOI] [PubMed] [Google Scholar]
- Sanders R. W., Dankers M. M., Busser E., Caffrey M., Moore J. P., Berkhout B. Evolution of the HIV-1 envelope glycoproteins with a disulfide bond between gp120 and gp41. Retrovirology. 2004b;1:3. doi: 10.1186/1742-4690-1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders R. W., de Jong E. C., Baldwin C. E., Schuitemaker J. H., Kapsenberg M. L., Berkhout B. Differential transmission of human immunodeficiency virus type 1 by distinct subsets of effector dendritic cells. J. Virol. 2002a;76:7812–7821. doi: 10.1128/JVI.76.15.7812-7821.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders R. W., et al. The carbohydrate at asparagine 386 on HIV-1 gp120 is not essential for protein folding and function but is involved in immune evasion. Retrovirology. 2008;5:10. doi: 10.1186/1742-4690-5-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders R. W., Venturi M., Schiffner L., Kalyanaraman R., Katinger H., Lloyd K. O., Kwong P. D., Moore J. P. The mannose-dependent epitope for neutralizing antibody 2G12 on human immunodeficiency virus type 1 glycoprotein gp120. J. Virol. 2002b;76:7293–7305. doi: 10.1128/JVI.76.14.7293-7305.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scanlan C. N., Pantophlet R., Wormald M. R., Ollmann S. E., Stanfield R., Wilson I. A., Katinger H., Dwek R. A., Rudd P. M., Burton D. R. The broadly neutralizing anti-human immunodeficiency virus type 1 antibody 2G12 recognizes a cluster of alpha1->2 mannose residues on the outer face of gp120. J. Virol. 2002;76:7306–7321. doi: 10.1128/JVI.76.14.7306-7321.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C. K., Withka J. M., Regan L. A thermodynamic scale for the beta-sheet forming tendencies of the amino acids. Biochemistry. 1994;33:5510–5517. doi: 10.1021/bi00184a020. [DOI] [PubMed] [Google Scholar]
- Stein B. S., Engleman E. G. Intracellular processing of the gp160 HIV-1 envelope precursor. J. Biol. Chem. 1990;265:2640–2649. [PubMed] [Google Scholar]
- Tironi I. G., Sperb R., Smith P. E., Van Gunsteren W. F. A generalized reaction field method for molecular dynamics simulations. J. Chem. Phys. 1995;102:5451–5459. [Google Scholar]
- Trkola A., Purtscher M., Muster T., Ballaun C., Buchacher A., Sullivan N., Srinivasan K., Sodroski J., Moore J. P., Katinger H. Human monoclonal antibody 2G12 defines a distinctive neutralization epitope on the gp120 glycoprotein of human immunodeficiency virus type 1. J. Virol. 1996;70:1100–1108. doi: 10.1128/jvi.70.2.1100-1108.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trombetta E. S., Parodi A. J. Quality control and protein folding in the secretory pathway. Annu. Rev. Cell Dev. Biol. 2003;19:649–676. doi: 10.1146/annurev.cellbio.19.110701.153949. [DOI] [PubMed] [Google Scholar]
- van Anken E., Sanders R. W., Liscaljet M., Land A., Bontjer I., Tillemans S., Nabatov A. A., Paxton W. A., Berkhout B., Braakman I. Only five out of ten strictly conserved disulfide bonds are essential for folding and eight for function of the HIV-1 envelope glycoprotein. Mol. Biol. Cell. 2008;19:4298–4309. doi: 10.1091/mbc.E07-12-1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X., Decker J. M., Liu H., Zhang Z., Arani R. B., Kilby J. M., Saag M. S., Wu X., Shaw G. M., Kappes J. C. Emergence of resistant human immunodeficiency virus type 1 in patients receiving fusion inhibitor (T-20) monotherapy. Antimicrob. Agents Chemother. 2002;46:1896–1905. doi: 10.1128/AAC.46.6.1896-1905.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X., et al. Antibody neutralization and escape by HIV-1. Nature. 2003;422:307–312. doi: 10.1038/nature01470. [DOI] [PubMed] [Google Scholar]
- Wyatt R., Sodroski J. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science. 1998;280:1884–1888. doi: 10.1126/science.280.5371.1884. [DOI] [PubMed] [Google Scholar]
- Zhou T., et al. Structural definition of a conserved neutralization epitope on HIV-1 gp120. Nature. 2007;445:732–737. doi: 10.1038/nature05580. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.