Abstract
Nucleic acids are substrates for different types of damage, but little is known about the fate of damaged RNAs. We addressed the existence of an RNA-damage response in yeast. The decay kinetics of GAL1p-driven mRNAs revealed a dose-dependent mRNA stabilization upon UV-irradiation that was not observed after heat or saline shocks, or during nitrogen starvation. UV-induced mRNA stabilization did not depend on DNA repair, damage checkpoint or mRNA degradation machineries. Notably, fluorescent in situ hybridization revealed that after UV-irradiation, polyadenylated mRNA accumulated in cytoplasmic foci that increased in size with time. In situ colocalization showed that these foci are not processing-bodies, eIF4E-, eIF4G-, and Pab1-containing bodies, stress granules, autophagy vesicles, or part of the secretory or endocytic pathways. These results point to the existence of a specific eukaryotic RNA-damage response, which leads to new polyadenylated mRNA-containing granules (UV-induced mRNA granules; UVGs). We propose that potentially damaged mRNAs, which may be deleterious to the cell, are temporarily stored in UVG granules to safeguard cell viability.
INTRODUCTION
DNA repair is crucial to maintain genome integrity, but it is not the only target for nucleic acid-damaging agents, because these may affect RNA molecules as well. In the cell, RNA is more abundant than DNA, and almost all cellular RNAs either encode proteins (mRNA) or are involved in the mechanism of protein production (rRNA and tRNA). Nevertheless, only 28% of genomic DNA is transcribed into RNA, and only 5% of these transcribed sequences actually encode proteins in humans (Baltimore, 2001). Because RNA is mostly single stranded, it may be more susceptible to damaging agents than DNA, in which bases are protected by hydrogen bonding and located inside the double helix (reviewed in Bregeon and Sarasin, 2005). From this point of view, it is likely that significant RNA damage occurs when cells are exposed to genotoxic agents. In fact, alkylating agents, reactive oxygen species, and UV-irradiation have been shown to induce RNA lesions (reviewed in Bregeon and Sarasin, 2005). RNA oxidation has been implied in a variety of neurological disorders (reviewed in Nunomura et al., 2006), whereas some anticancer agents cause RNA damage that lead to cell cycle arrest and cell death as much as DNA damage does (reviewed in Bellacosa and Moss, 2003; Bregeon and Sarasin, 2005). Moreover, nitrogen mustard, cisplatin, and alkylating agents inhibit translation reactions, which indicates that a single mRNA lesion may be sufficient to block translation (Rosenberg and Sato, 1988; Masta et al., 1995; Heminger et al., 1997; Ougland et al., 2004).
The abundance of individual mRNAs in the cell is determined by the rate at which they are produced and degraded. mRNA stability can be regulated in response to a variety of stimuli, allowing for rapid alterations in gene expression. Many clinically relevant mRNAs are regulated by differential RNA stability, and the aberrant control of mRNA stability has been drawn into disease states, including cancer, chronic inflammatory responses, and coronary disease (reviewed in Hollams et al., 2002). In eukaryotes, several types of cytoplasmic particles containing dormant mRNAs have been observed under stress, during oogenesis, and in neuronal cells (reviewed in St Johnston, 2005; Anderson and Kedersha, 2006).
Components of the 5′ → 3′ mRNA decay pathway and translationally repressed messenger ribonucleoprotein (mRNPs) are enriched in granular cytoplasmic foci known as processing bodies (P-bodies; reviewed in Anderson and Kedersha, 2006; Eulalio et al., 2007; Parker and Sheth, 2007). P-body size is proportional to the flux of mRNAs undergoing the decapping step in turnover. The mRNA decay intermediates trapped in the process of degradation are localized to these structures in yeast and human cells (Sheth and Parker, 2003; Cougot et al., 2004). P-bodies are dynamic structures and are affected by a range of cellular perturbations, including glucose deprivation, osmotic stress, exposure to UV light, and the stage of cell growth (Kedersha et al., 2005; Teixeira et al., 2005; Wilczynska et al., 2005). Beyond their participation in mRNA decapping, P-bodies have been suggested to be functionally involved in nonsense-mediated decay, mRNA storage, general translation repression, microRNA-mediated repression, and viral packaging (reviewed in Anderson and Kedersha, 2006; Eulalio et al., 2007; Parker and Sheth, 2007). Recently, new cytoplasmic mRNA granules called eIF4E-, eIF4G-, and Pab1-containing bodies (EGP)-bodies have been described in yeast (Hoyle et al., 2007). EGP-bodies, which are distinct from P-bodies, arise under glucose starvation conditions and contain the translational factors eIF4E, eIF4G, as well as Pap1. EGP-bodies have been proposed to be sites where mRNAs are stored during period of translational inactivity.
In mammalian cells, different types of stress, including UV-irradiation, heat shock, and oxidative stress, inhibit translation of bulk mRNA, which aggregates in cytoplasmic structures known as stress granules (SGs; Kedersha et al., 1999; Kimball et al., 2003). SGs are dynamic and reversible structures, which assemble in response to environmental stress and disperse after recovery. SGs contain most of the 48S preinitiation complex (e.g., eIF3, eIF4, and eIF4G), PABP1, the p54/Rck helicase, the 5 → 3′ exonuclease Xrn1, many RNA-binding proteins, including the prion-like proteins TIA-1 and TIAR, and polyadenylated (poly-A+) RNA (reviewed in Anderson and Kedersha, 2008). Stress granules and P-bodies seem to interact with each other under certain circumstances (Kedersha et al., 2005; Wilczynska et al., 2005), although the diversity of different mRNP types and the mechanisms that mediate transitions between stress granules, P-bodies, and polysome-bound mRNAs remain to be established. SGs have been described in Schizosaccharomyces pombe (Dunand-Sauthier et al., 2002) but have not yet been reported in Saccharomyces cerevisiae.
Given the importance of controlled mRNA turnover and the likeliness of bulky adducts to impair translation, the existence of some kind of surveillance mechanisms is rather predictable. However, little is known about the fate of damaged mRNAs to date. We addressed the existence of an RNA-damage response in the yeast S. cerevisiae by a combination of genetic, molecular, and cellular approaches. Our results point to the existence of an RNA-damage–specific cellular response in yeast, which leads to transcript stabilization and to the accumulation of poly-A+ mRNA in a novel class of cytoplasmic granules.
MATERIALS AND METHODS
Strains and Plasmids
We used the wild-type strain W303-1A (MATa ade2-1 can1-100 his3-11,15 leu2-3112 trp1-1 ura3-1), its isogenic rad1Δ::LEU2 (W839-5D, R. Rothstein) and rad53-21 (LSY843, L. Symington), plus dcp1-2 and its isogenic wild-type DCP1 (MATa his4 leu2 lys2 ura3 trp1; Tharun and Parker, 1999). Strains BY4741 (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0), rad14Δ::KAN, rad9Δ::KAN, rrp6Δ::KAN, and lsm1Δ::KAN were purchased from EUROSCARF (Frankfurt, Germany). The pub1Δ::KAN and nam8Δ::KAN strains were obtained by replacement of the PUB1 and NAM8 gene in W303–1A. The pub1Δ::KAN nam8Δ::KAN strain was obtained by genetic cross. To analyze decay of the YLR454w gene, the GAL1 promoter fused to the 5′-most 300 base pairs of the YLR454w open-reading frame was integrated at the YLR454w locus in W303-1A (Mason and Struhl, 2003). Strains harboring green fluorescent protein (GFP)-tagged PUB1 and NAM8 genes at their respective chromosomal locus were purchased from Invitrogen (Carlsbad, CA) and backcrossed with W303. Strains harboring GFP-tagged CDC33, TIF4631, TIF4632, and PAB1 genes were kindly provided by M. Ashe (University of Manchester, United Kingdom) (yMK 885, yMK1172, yMK1214, and yMK1185). Plasmids expressing GFP-tagged proteins were provided by J. de la Cruz (Universida de Sevilla, Spain) (pRS315-Rpl25-eGFP and pRS315-Rsp2-eGFP) or constructed by inserting the open reading frame (ORF) in a pUG34 vector (pUG34-GFP-Aut7). Plasmid pRS424-GAL1 expressing the GAL1 gene under its own promoter was constructed by inserting the GAL1 ORF into a pRS424-GAL1p vector kindly provided by R. Wellinger (Universida de Sevilla, Spain).
UV-Irradiation and Recovery
Irradiation and recovery were carried out as described previously (Gaillard et al., 2007) with minor modifications. Yeast cells were grown at 30°C in YPGal (1% yeast extract, 1% peptone, and 2% galactose) to an OD600 of 0.8, harvested, and resuspended in synthetic complete (SC) medium with 0.2% galactose to an OD600 of 1.2. Four millilmeter-deep cell suspensions were irradiated with UV light using germicidal lamps (Philips T UV 15 W) at the indicated dose, as measured by a UVX radiometer (UVP, San Gabriel, CA). After irradiation, the medium was supplemented with the appropriate amino acids and glucose (2% final concentration) and incubated at room temperature in the dark in a shaker for recovery. Where indicated, cycloheximide or thiolutin (Sigma-Aldrich, St. Louis, MO) was added to UV-irradiated cells to final concentrations of 100 and 6 μg/ml, respectively. For fluorescence in situ hybridization (FISH) analysis of GAL1 RNA, cells harboring pRS424-GAL1 were grown in SCGal (SC with 2% galactose) to an OD600 of 0.3. Cells were harvested and resuspended in SC with 0.2% galactose to an OD600 of 0.5. Four-millimeter-deep cell suspensions were irradiated with 200 J/m2 UV light using germicidal lamps (Philips T UV 15 W), and the medium was supplemented with the appropriate amino acids and glucose to 2% final concentration. For RNA isolation, 10-ml samples were collected at different times and immediately frozen in liquid nitrogen. For microscopy and FISH analyses, the samples were directly processed.
Heat and Saline Shocks and Nitrogen (N) Starvation
Yeast cells were grown at 30°C in YPGal (heat and saline shocks) or SCGal (nitrogen starvation) to an OD600 of 0.5 and harvested. For saline shock, cells were resuspended in YPGal with 0.8 M NaCl, incubated at 30°C for 10 min, washed once with H2O, resuspended in YPD, and incubated in a shaker at room temperature for recovery. For heat shock, cells were resuspended in YPGal to an OD600 of 0.9. After incubation at 45°C for 10 min, the medium was supplemented with 2% glucose and incubated in a shaker at room temperature for recovery. For nitrogen starvation, cells were resuspended in SCGal without ammonium sulfate, incubated at 30°C for 3 h, harvested, washed with prewarmed H2O, resuspended in SCD (SC with 2% glucose) without ammonium sulfate, and incubated in a shaker at room temperature. All media were prewarmed to 30°C, and centrifugations were performed at 25°C. For RNA isolation, 10-ml samples were collected at different times and immediately frozen in liquid nitrogen.
Northern Analyses
RNA was extracted and Northern analyses were performed according to standard procedures. Filters were hybridized with either a 764-base pair GAL1 fragment or a 455-base pair YLR454w fragment obtained by polymerase chain reaction (PCR) using primers GAL1-A (5′-GTGCCCGAGCATAATTAAGAAAT-3′), GAL1-B (5′-TGTAGTGACTTCTACCACTCTTA-3′) and YLR3-A (5′-GATGGAAACGGAGATGACGA-3′), YLR3-B (5′-CGAAGTTCTTCAGGCTCCG-3′), respectively. Northern blots were quantified using a Fuji FLA 3000 and normalized to the rRNA levels of each sample.
Polysome Profile Analyses
Polysome preparation and analysis were performed as described previously (Kressler et al., 1997). W303-1A cells harboring pRS424-GAL1 were grown in SCGal to an OD600 of 0.8, irradiated or mock treated with 200 J/m2 UV light in SC with 0.2% galactose, and the medium was supplemented with the appropriate amino acids and glucose to 2% final concentration. Cycloheximide was added to a final concentration of 100 μg/ml immediately before harvesting. Eight A260 units of extracts were layered onto 11.2 ml of 7–50% linear sucrose gradients that were centrifuged at 39,000 rpm in an SW41 rotor (Beckman Coulter, Fullerton, CA) at 4°C for 2 h 45 min. Gradient analysis was performed using a UV-6 gradient collector (ISCO, Lincoln, NE) and continuously monitored at A254. Fractions of 0.5 ml were collected manually. For RNA analysis, fractions were adjusted to a final concentration of 10 mM Tris-HCl, pH 7.5, 10 mM EDTA, and 0.5% SDS. Total RNA was isolated by two consecutive extractions with 10 mM Tris-HCl, pH 7.5, saturated phenol:chloroform:isoamylalcohol (25:24:1) followed by a chloroform:isoamylalcohol (24:1) extraction. RNA was then precipitaded with ethanol in the presence of 0.3 M sodium acetate, pH 5.2, and the RNA pellets dissolved in 10 μl of distilled water.
Reverse Transcription (RT)-PCR Analyses
Poly-A+ RNA was isolated from total RNA to eliminate residual DNA from the samples by using oligo(dT)-coated magnetic beads (Novagen, Madison, WI) and then subjected to reverse transcription (Transcriptor First Strand cDNA Synthesis kit; Roche Applied Science, Indianapolis, IN). The sequences of the oligonucleotides used for the PCR are available upon request.
FISH
The subcellular distribution of poly-A+ RNA was examined by in situ hybridization using a digoxigenin-conjugated oligo(dT) probe and indirect immunofluorescence microscopy as described previously (Cole et al., 2002). Oligo(dT)50 was 3′ end-labeled with digoxigenin (Roche Applied Science), and rhodamine-conjugated anti-digoxigenin antibody (1:200 dilution; Roche Applied Science) was used to visualize poly-A+ RNA. To visualize the subcellular distribution of GAL1 RNA, six GAL1-specific fluorescein-conjugated probes were used. GAL1 fragments were labeled by PCR using fluorescein-conjugated nucleotides (Roche Applied Science) and purified using GFX columns (GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom). Oligonucleotides sequences are available upon request. For colocalization studies, cells were incubated overnight with a dilution of primary antibody at 4°C, washed five times (with 0.1% phosphate-buffered saline, 0.1% bovine serum albumin, and IPEGAL [Sigma-Aldrich, St. Louis, MO]), and incubated for 1 h with a dilution of secondary antibody at room temperature before performing the final washes and mounting steps of the FISH protocol.
Fluorescence Microscopy
For direct visualization of GFP-tagged proteins, cells were grown to an OD600 of 0.3 in SCGal. For UV-irradiation analyses, cells were harvested and resuspended in SC with 0.2% galactose to an OD600 of 0.5. Four-millimeter-deep cell suspensions were irradiated with 100 J/m2 UV light using germicidal lamps (Philips T UV 15 W), and the medium was supplemented with the appropriate amino acids and 2% glucose. For nitrogen starvation analyses, cells were grown in SCD, harvested, washed once with H2O and resuspended in SCD without ammonium sulfate. After incubation at room temperature in the dark for the indicated time, 2 μl of cell suspension were directly observed by fluorescence microscopy. Acquisition was done in a DM600B microscope equipped with a DFC350FX camera (both from Leica, Wetzlar, Germany) by using an HCXPL APO 100× 1.40 oil objective (Leica). Images of the colocalization experiments are a Z-series compilation of five to 10 images. Merges were generated using Adobe Photoshop 7.0 (Adobe Systems, Mountain View, CA).
Confocal Microscopy
For direct visualization of GFP-tagged proteins under glucose starvation, cells were grown to an OD600 of 0.6 in SCD, washed twice, and resuspended in either SC− (no glucose) or SCD. For UV-irradiation analyses, cells were resuspended in SC− to an OD600 of 0.5. Four-millimeter-deep cell suspensions were irradiated with 100 J/m2 UV light using germicidal lamps (Philips T UV 15 W), and the medium was supplemented with the appropriate amino acids and 2% glucose. After incubation at 30°C in the dark for 1 h, 2 μl of cell suspension was directly observed by confocal microscopy. Acquisition was done in a TCS SP5 microscope using a HCXPL APO 100× 1.40–0.70 oil objective and Application Suite Advanced Fluorescence 1.6.3 (all from Leica). Images of the colocalization experiments are a Z-series compilation of two to three images. Merges were generated using Adobe Photoshop 7.0. Quantification of colocalization was done manually in Z-series compilation of five to 12 images, making up in total 239 cells for P-bodies, and using MetaMorph 7.5.1 application software (Molecular Devices, Sunnyvale, CA) for GAL1 (>300 cells).
RESULTS
UV-Irradiation Leads to mRNA Stabilization in a Dose-dependent Manner
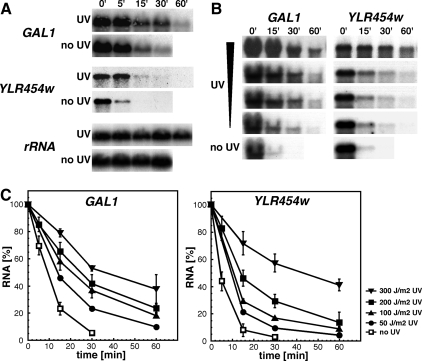
Assuming that damaged mRNA might be rapidly degraded to avoid their putative deleterious effects for the cell, we first analyzed mRNA decay of the GAL1 promoter-driven GAL1 and YLR454w transcripts after inhibition of transcription in cells that were either UV irradiated or mock treated. Wild-type cells were grown to mid-log phase in conditions of active transcription (YPGal). Immediately after irradiation with 200 J/m2 UV light, the medium (SCGal) was supplemented with glucose, leading to transcription inactivation. The amount of GAL1 and YLR454w transcripts was analyzed by Northern at different times (Figure 1A). Surprisingly, both mRNAs seemed to be significantly stabilized upon UV-irradiation compared with nonirradiated control cells. Further experiments with different UV doses indicated that the magnitude of transcript stabilization correlates with the irradiation dosage (Figure 1, B and C). Notably, UV-dependent mRNA stabilization was not significantly influenced by transcript length, because a comparable degree of stabilization was observed with the short GAL1 (1.6 kb) and the long YLR454w (7.9 kb) genes.
Figure 1.
UV-irradiation leads to mRNA stabilization in a dose-dependent manner. (A) Northern analysis of wild-type cells either irradiated with 200 J/m2 UV light (UV) or mock treated (no UV) and collected at the indicated times. The same blot was probed with GAL1-, YLR454w-, and rRNA-specific probes. (B) Northern analysis of wild-type cells after irradiation with 0, 50, 100, 200, and 300 J/m2 UV light. Results obtained with GAL1- and YLR454w-specific probes are shown. Results obtained with an rRNA-specific probe are omitted for clarity. (C) Graphical representation of the GAL1 and YLR454w decay kinetics after UV-irradiation shown in A and B. Northern blots were quantified using a Fuji FLA 3000 and normalized to the rRNA levels of each sample. Average values derived from at least two independent experiments are plotted with their standard deviations.
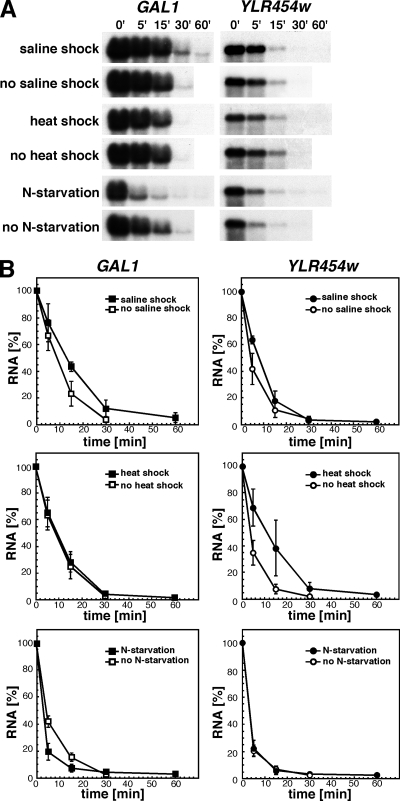
Next, we tested whether similar mRNA stabilization was observed during recovery from other types of stresses. Mid-log cells grown in YPGal were incubated for 10 min at 30°C in medium containing 0.8 M NaCl or at 45°C to induce saline and heat shock, respectively. Immediately after treatment cells were incubated in glucose-containing medium and GAL1 and YLR454w decay analyzed by Northern (Figure 2). Although heat shock lead to weak stabilization of the YLR454w transcript, neither saline nor heat shock resulted in a significant mRNA stabilization, despite cell survival of both treatments being comparable with after UV-irradiation with 200 J/m2 (60% survival). As an additional stress, we used N starvation. We incubated mid-log cells in SCGal without ammonium sulfate for 2 h, after which we changed the medium for SCD without ammonium sulfate to analyze GAL1 and YLR454w mRNA decay under N starvation conditions (Figure 2). None of the transcripts were stabilized by N starvation; in contrast, decay of the GAL1 transcript seemed to be even faster than in mock-treated cells. Thus, our results indicate that UV-dependent mRNA stabilization is not part of a general stress response.
Figure 2.
Saline and heat shocks or nitrogen starvation do not lead to mRNA stabilization. (A) Northern analysis of wild-type cells after saline shock (10-min incubation in medium containing 0.8 M NaCl), heat shock (10-min incubation at 45°C), in nitrogen (N−) starvation conditions, or mock treated. Samples were collected at the indicated times for Northern analysis. (B) Graphical representation of the GAL1 and YLR454w decay kinetics after saline and heat shocks and nitrogen starvation shown in A. Other details as in Figure 1.
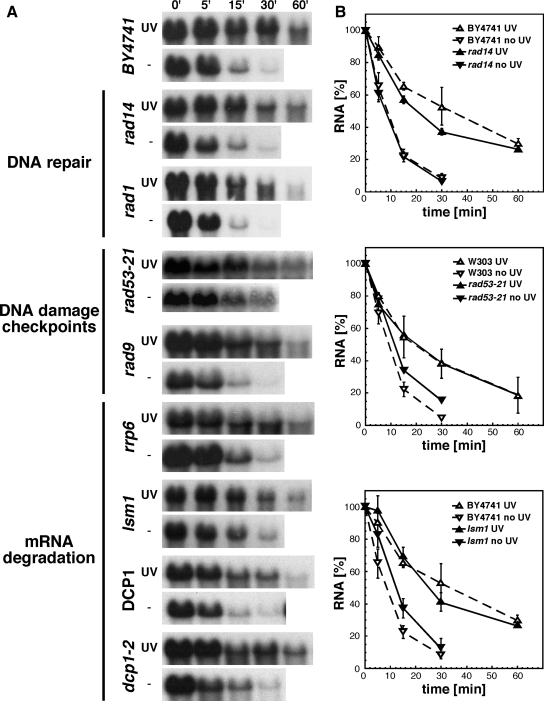
Because UV-irradiation generates DNA lesions and leads to the activation of DNA-damage checkpoints, we investigated whether UV-dependent mRNA stabilization might depend on DNA repair and checkpoint activation. Analyses of mRNA decay after UV-irradiation in nucleotide excision repair-deficient rad14Δ and rad1Δ strains showed similar mRNA stabilization as in wild-type cells (Figure 3), indicating that increased transcript stability does not depend on DNA repair. UV-dependent mRNA stabilization was also observed in the rad9Δ and rad53-21 checkpoint mutants (Figure 3), suggesting that transcript stabilization is not a consequence of classical DNA-damage checkpoint activation. To test the putative implication of mRNA degradation pathways in UV-dependent transcript stabilization, we extended our study to mutants of the nuclear (rrp6Δ) and cytoplasmic (lsm1Δ and dcp1-2) mRNA decay machineries (Figure 3). As reported previously (Tharun and Parker, 1999; Bouveret et al., 2000; Tharun et al., 2000; Kuai et al., 2005), mRNA degradation was slightly impaired compared with wild-type in rrp6Δ, lsm1Δ, and dcp1-2 cells in the absence of UV-irradiation. Importantly, UV treatment resulted in a substantial increase of transcript stabilization. Thus, our results indicate that UV-induced mRNA stabilization does not depend on functional nuclear or cytoplasmic mRNA degradation machineries.
Figure 3.
UV-dependent mRNA stabilization does not depend on DNA repair, DNA-damage checkpoint, or on functional mRNA degradation machineries. (A) Northern analysis of RNA isolated from rad14Δ, rad1Δ, rad53-21, rad9Δ, rrp6Δ, and dcp1-2 mutants and the corresponding wild-types after UV-irradiation (200 J/m2). Times of collection are indicated above the lanes. The results obtained with a GAL1-specific probe are shown, whereas the results obtained with an rRNA-specific probe are omitted for clarity. (B) Graphical representation of the GAL1 decay kinetics after UV-irradiation shown in A. for one representative mutant affected in DNA repair (rad14Δ), DNA checkpoint response (rad53-21), and mRNA degradation (lsm1Δ), respectively. Other details are as described in Figure 1.
Poly-A+ mRNA Accumulate in Discrete Cytoplasmic Foci upon UV-Irradiation
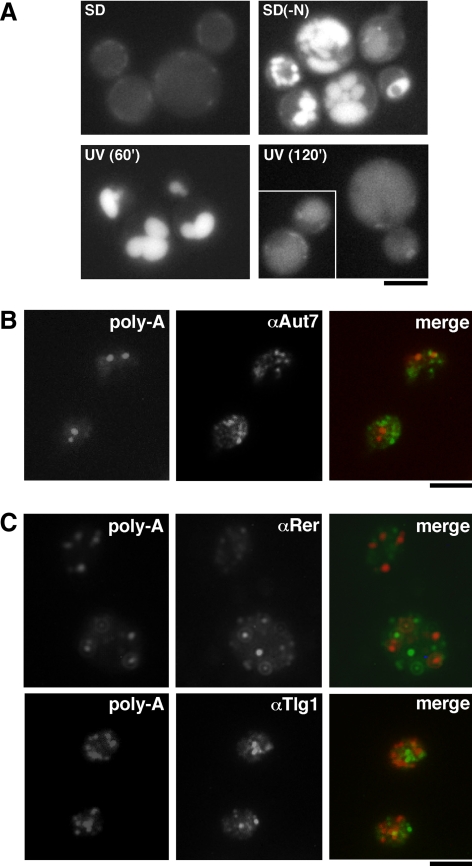
Given the strong UV-dependent stabilization of GAL1 and YLR454w transcripts, we wondered whether poly-A+ mRNAs might accumulate in the nucleus or in some other organelles upon UV-irradiation. To directly assess the cellular localization of poly-A+ mRNA after UV-irradiation, we performed FISH with oligo(dT) probe at various times. As can be seen in Figure 4, poly-A+ mRNAs started to accumulate in cytoplasmic foci 15 min after UV-irradiation. After 60 min, 80% of the cells showed bright poly-A+ foci, which tended to increase in size, whereas they decreased in number with further incubation time, as determined by the number of cells with more than two, two, one, or no foci. The results suggest that these foci identify dynamic structures. Importantly, the occurrence of UV-dependent poly-A+ foci did not correlate with cell death, because the UV dose used (200 J/m2) led to 60% survival and cells continued growing after UV treatment, as assessed by optical density measurements and cell survival at later time points (unpublished data). To test whether poly-A+ mRNA foci formation is specific to UV treatment or results from a general stress response, we checked focus formation after heat and saline shocks (Supplemental Figure S1). As described previously in yeast (Tani et al., 1995; Saavedra et al., 1996; Yoshida and Tani, 2005), poly-A+ mRNA accumulated in the nucleus after both treatments. Meaningfully, no focus formation was observed in the cytoplasm, after neither heat nor saline shocks. Thus, our results indicate that the UV-dependent poly-A+ mRNA cytoplasmic foci are not caused by a general stress response.
Figure 4.
Poly-A+ mRNA accumulate in discrete cytoplasmic foci upon UV-irradiation. Wild-type cells were irradiated with 200 J/m2 UV and collected at the indicated time points. Poly-A+ mRNA was visualized by oligo(dT) hybridization and indirect immunofluorescence. Cells with poly-A+ foci were counted for each time point and the distribution of cells without focus and with one, two, or more than two foci arising from quantification of three biological replicates (>130 cells each) are displayed on the right. Standard deviations are indicated below each value. Bar, 5 μm.
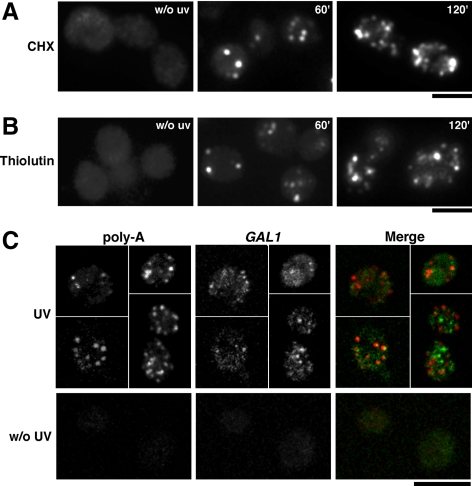
Impaired translation elongation has been shown to increase mRNA stability (Beelman and Parker, 1994). Because UV-irradiation also injures RNA molecules (Rycyna and Alderfer, 1988), it was conceivable that UV-damaged mRNA might impair translation elongation, either directly or indirectly, leading to the observed mRNA stabilization. To test this hypothesis, we checked whether poly-A+ mRNA foci are induced after cycloheximide treatment. However, we did not detect any poly-A+ foci formation in cells treated with cycloheximide, indicating that impaired translation elongation alone is not sufficient for cytoplasmic accumulation of poly-A+ mRNA in discrete foci (Figure 5A). Then again, translation might work as a sensor for damaged RNA and as such could be required for poly-A+ foci formation. To address this possibility, we tested whether cycloheximide could prevent the formation of UV-dependent poly-A+ foci (Figure 5A). Our results show that this is not the case, suggesting that poly-A+ foci formation occurs independently from translation elongation.
Figure 5.
UV-induced poly-A+ foci are formed independently from translation elongation and contain potentially damaged RNA and stabilized GAL1 transcripts. (A) FISH analyses of wild-type cells to which cycloheximide (CHX) was added to a final concentration of 100 μg/ml at the time of UV-irradiation and collected at the indicated times. The non-irradiated sample was taken 60 min after mock irradiation. (B) FISH analyses of wild-type cells to which thiolutin (Thiolutin) was added to a final concentration of 6 μg/ml at the time of UV-irradiation and collected at the indicated times. The non-irradiated sample was taken 60 min after mock irradiation. Other details are as described in Figure 4. (C) Confocal microscopic images of wild-type cells collected 30 min after irradiation with 200 J/m2 UV light. Poly-A+ and GAL1 mRNA were visualized by FISH. The merge generated by Adobe Photoshop is shown. Bar, 5 μm. Quantification of 3 biological replicates (>100 cells each) with the MetaMorph 7.5.1 application software revealed that 35 ± 7% of GAL1 colocalized with poly-A+ foci.
To assess whether poly-A+ RNAs found in the UV-dependent granules corresponded to mRNAs that were present at the time of irradiation or were transcribed a posteriori, we performed FISH analyses in cells in which transcription was blocked by the addition of thiolutin immediately after UV-irradiation (Figure 5B). We first showed that thiolutin had no effect on the formation of UV-induced poly-A+ foci, which were formed in similar amounts in thiolutin-treated and nontreated cells (Figure 4). This suggests that the foci contain transcripts that were present at the time of UV-irradiation; therefore, they are potentially damaged RNAs. Next, we performed colocalization studies using a GAL1-specific probe in addition to the poly(dT) probe, to assess whether the stabilized GAL1 reporter mRNA actually resided in the UV-induced poly-A+ foci. The results (Figure 5C) were obtained 30 min after UV-irradiation (and transcription switch-off), a time point at which intact GAL1 transcripts are found in UV-irradiated cells but not in nonirradiated cells (Figure 1) and at which UV-induced poly-A+ foci are detected (Figure 4). A relevant fraction (35%) of the GAL1-specific foci colocalized with poly-A+ foci, indicating that the UVGs contain stabilized reporter transcripts. In other cases, GAL1 foci were found aside from poly-A+ foci. We believe that these foci could correspond to transcripts being degraded within processing bodies (P-bodies).
As an additional approach to study the effect of UV-irradiation on translation and the distribution of our reporter GAL1 mRNA in nontranslated and translated fractions, we performed polysome profile analyses 30 min after UV-irradiation and transcription switch-off. Extracts were fractionated on sucrose gradient and absorbance at 254 nm continuously monitored (Figure 6A). Our results show that polysomes are not disrupted in response to UV-irradiation, even if a decrease in the overall amount of polysomes is observed, probably due to the previously described UV-dependent down-regulation of transcriptional activity (Smerdon et al., 1990; Reagan and Friedberg, 1997; Gaillard et al., 2007). To assess whether stabilized GAL1 transcript accumulates in polysome-free fractions, as predicted by our FISH results, poly-A+ RNAs were purified from polysomal and polysome-free fractions, and the presence of GAL1 transcript monitored by RT-PCR (Figure 6B). The highly transcribed housekeeping RPB4 mRNA was used as internal control. We observed an enrichment in GAL1 transcript in the polysome-free fraction of UV-irradiated cells, indicating that the stabilized transcript are not associated with polysomes and thus not in the process of being translated.
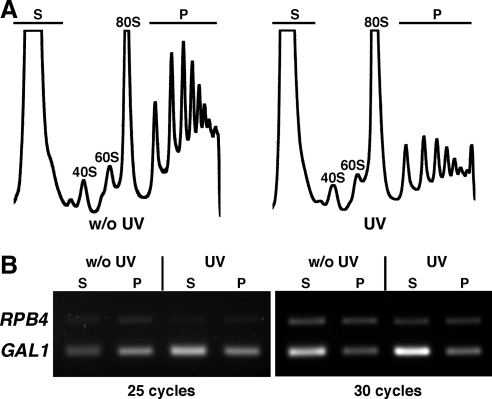
Figure 6.
Distribution of GAL1 mRNA between polysome-free and polysomal fractions. (A) Wild-type cells harboring plasmid pRS424-GAL1 were irradiated with 200 J/m2 UV and collected 30 min after UV-irradiation and GAL1p shutoff. Cell extracts were resolved in sucrose gradients and the A254 was continuously measured. The peaks of polysome-free molecules (S), free 40S and 60S subunits, 80S free couples/monosomes, and polysomes (P) are indicated. Representative polysome profiles for mock- or UV-irradiated cells are shown (w/o UV and UV, respectively). (B) Poly-A+ RNA was prepared from polysomal and polysome-free fractions. RT-PCR products for the GAL1 and the RPB4 mRNA, which was used as internal control, are shown after 25 and 30 cycles of amplification.
UV-dependent Poly-A+ Foci Represent New RNA Granules
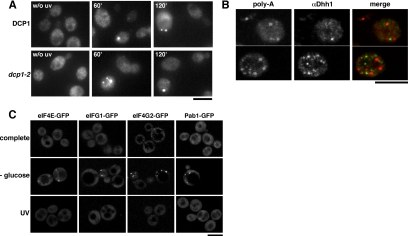
The major pathway of mRNA degradation has been shown to occur in defined cytoplasmic bodies (P-bodies) in yeast (Sheth and Parker, 2003). P-bodies have been shown to increase in number and size in mutants in which mRNA turnover is inhibited at, or after decapping as well as in stress conditions, including UV-irradiation and saline shock (Teixeira et al., 2005). To check whether the UV-dependent cytoplasmic mRNA foci correspond to P-bodies, we analyzed poly-A+ mRNA localization in the dcp1-2 decapping mutant by FISH (Figure 7A). We did not observe any significant difference between dcp1-2 and wild-type cells, indicating that UV-dependent poly-A+ foci formation occurs independently of the Dcp1 protein and that, in contrast to P-bodies, impairment of the major mRNA degradation pathway does not enhance UV-dependent poly-A+ foci. To investigate the possible connection between P-bodies and UV-dependent poly-A+ foci more directly, we performed colocalization studies using an antibody against the P-body marker Dhh1 (Figure 7B). Our results show that most UV-dependent poly-A+ foci do not colocalize with P-bodies; therefore, they represent distinct cellular structures. However, a small fraction (11%) of poly-A+ foci partially colocalized with P-bodies, whereas another fraction (10%) was found just aside from P-bodies, suggesting that UV-induced poly-A+ foci and P-bodies might interact functionally. Given the recent discovery of EGP-bodies (Hoyle et al., 2007), which are distinct from P-bodies and are caused by glucose starvation-induced translation repression, we assessed whether the UV-induced poly-A+ foci might correspond to EGP-bodies. First, we analyzed whether EGP-bodies might be induced upon UV-irradiation by looking at the cellular localization of GFP-tagged eIF4E, eIF4G1, eIFG2, and Pab1 proteins (Figure 7C). Although all four proteins accumulated in EGP-bodies upon glucose starvation, UV-irradiation failed to induce a similar response. Second, to investigate whether poly-A+ foci might be induced in glucose starvation conditions and whether UV-induced poly-A+ foci and glucose starvation-induced EGP-bodies might colocalize in cells subjected to both stresses simultaneously, we performed colocalization studies using antibodies again eIF4E and eIF4G1 (Supplemental Figure S3). Our results show that UV-dependent poly-A+ foci and EGP are induced only upon UV-irradiation or glucose starvation, respectively, and that they do not colocalize in cells subjected to both stresses. Therefore, we conclude that UV-induced poly-A+ foci are distinct from EGP-bodies.
Figure 7.
UV-dependent poly-A+ foci are different from P- and EGP-bodies. (A) FISH analysis of wild-type (DCP1) and dcp1-2 cells irradiated with 200 J/m2 UV or mock treated and collected at the indicated times. Other details as in Figure 4. (B) Confocal microscopic images of wild-type cells collected 60 min after irradiation with 200 J/m2 UV light. Poly-A+ mRNA was visualized by FISH (left). Dhh1 was visualized by immunofluorescence (middle). The merge generated by Adobe Photoshop is shown (right). (C) Direct confocal analysis of wild-type cells expressing a GFP-tagged version of the eIF4E, eIF4G1, eIF4G2, or Pap1 proteins collected after 60-min incubation in SCD (complete), SC− (−glucose), or SCD after irradiation with 100 J/m2 UV light (UV). Bar, 5 μm.
In mammalian cells, poly-A+ mRNA has been shown to accumulate in discrete cytoplasmic foci under stress condition (SGs). SGs formation depends on the prion-like proteins TIA-1 and TIAR, which accumulate in the granules together with poly-A+ mRNA and most of the 48S preinitiation complex including the small but excluding the large ribosomal subunits (reviewed in Anderson and Kedersha, 2008). SGs have not yet been reported in S. cerevisiae, although they have been described in S. pombe (Dunand-Sauthier et al., 2002). To approach the putative existence of SGs in S. cerevisiae and whether the UV-dependent mRNA foci might correspond to SGs, we first checked the localization of both the large and the small ribosomal subunits after UV damage by using GFP-tagged fusion of the Rpl25 and Rsp2 proteins, respectively (Figure 8A). Both proteins show diffuse cytoplasmic staining that was not affected by UV-irradiation, indicating that neither the small nor the large ribosomal subunit accumulates in discrete foci upon UV-irradiation. Next, we looked for yeast homologues of the TIA-1 and TIAR proteins by BLAST analysis. Two putative homologues were found, yPub1 and yNam8 (35 and 30% identity with TIA-1, respectively). We analyzed the localization of Pub1 and Nam8 after UV-irradiation by using GFP-tagged proteins (Figure 8B), and we did not observe any cytoplasmic accumulation of either protein after UV-irradiation. In addition, we generated pub1Δ, nam8Δ and pub1Δnam8Δ deletion strains and analyzed UV-dependent poly-A+ mRNA foci formation by FISH (Figure 8C). We did not detect any significant difference among pub1Δ, nam8Δ, pub1nam8Δ, and wild-type cells. Thus, our results do not support the existence of SGs in the yeast S. cerevisiae and indicate that the putative TIA-1 and TIAR homologues Pub1 and Nam8 are not involved in UV-dependent poly-A+ mRNA foci formation.
Figure 8.
UV-dependent poly-A+ foci do not correspond to mammalian stress granules. (A) Direct fluorescence analysis of wild-type cells expressing a GFP-tagged version of the Rpl25 or Rsp2 ribosomal proteins collected 60 min after irradiation with 0 or 100 J/m2 UV light. (B) Direct fluorescence analysis of wild-type cells expressing a GFP-tagged version of the Pub1 or Nam8 proteins collected 60 min after irradiation with 0 or 100 J/m2 UV light. (C) FISH analysis of pub1Δ, nam8Δ, or pub1Δnam8Δ cells irradiated with 200 J/m2 UV or mock treated and then collected at the indicated times. (D) Distribution of cells with no, one, two, and more than two foci arising from quantification of three biological replicates (>50 cells each). Standard deviations are indicated below each value. Other details as in Figure 4. Bar, 5 μm.
Autophagy is a process in which cytosol and organelles are sequestered within double-membrane vesicles that deliver the contents to the lysosome/vacuole for degradation and recycling of the resulting macromolecules. It plays an important role in the cellular response to stress, is involved in various developmental pathways, and functions in tumor suppression, resistance to pathogens and extension of lifespan (reviewed in Klionsky, 2005a,b). Because many of the UV-dependent poly-A+ mRNA foci were observed around the vacuole, we decided to check whether autophagy might be involved in their formation. Using a GFP-fusion of the Aut7 autophagy marker protein, we found that the autophagy pathway is indeed induced by UV-irradiation, although to a lesser extent than in starvation conditions (Figure 9A). However, the kinetics of UV-induced autophagy and poly-A+ foci formation were different, because the autophagy response disappeared 120 min after UV-irradiation, whereas poly-A+ foci could be observed as long as 5 h after UV-irradiation (Figure 4). Nevertheless, to be sure that UV-induced poly-A+ foci did not arise from autophagy vesicle, we performed colocalization studies 30 and 60 min after UV-irradiation—time points in which both responses are active—using an antibody against Aut7 (Figure 9B; unpublished data). We did not detect any colocalization, indicating that Aut7 is absent from UV-induced poly-A+ foci.
Figure 9.
UV-dependent poly-A+ foci are not autophagy vesicles nor do they colocalize with the secretory and endocytic pathways. (A) Direct fluorescence analysis of wild-type cells expressing a GFP-tagged version of the Aut7 protein incubated for 2 h in minimal medium lacking a nitrogen source [SD(−N)] or mock treated (SD), or collected at the indicated times after irradiation with 100 J/m2 UV light. (B) Colocalization analysis of wild-type cells collected 60 min after irradiation with 200 J/m2 UV light. Poly-A+ mRNA was visualized by FISH (left). Aut7 was visualized by immunofluorescence (middle). The merge generated by Adobe Photoshop is shown (right). (C) Colocalization analysis of wild-type cells collected 60 min after irradiation with 200 J/m2 UV light. Poly-A+ mRNA was visualized by FISH (left). Rer1 or Tlg1 was visualized by immunofluorescence (middle). The merges generated by Adobe Photoshop are shown (right). Other details are as described in Figure 4. Bar, 5 μm.
Recently, mRNA and the poly-A+–binding protein Pab1 have been shown to interact with coat protein complex I (COPI)-coated vesicles generated from Golgi membranes (Trautwein et al., 2004). Because the poly-A+ mRNA foci formed upon UV-irradiation reminded of the punctuate composite of the Golgi network and endocytic pathway, we performed colocalization studies using antibodies against the cis-Golgi marker Rer1 and the early endosome marker Tlg1 (Figure 9C). None of them colocalized with UV-induced poly-A+ foci, suggesting that they are not part of the secretory pathway.
Together, our results point to the existence of a RNA-damage specific cellular response in yeast, which leads to the accumulation of poly-A+ mRNA in cytoplasmic foci. The fact that these poly-A+ foci do not relate to P-bodies, nor to EGP-bodies, nor to stress granules, nor to autophagy vesicles, nor to the secretory network suggests that they might represent a novel kind of RNA granule (UVGs).
DISCUSSION
UV light produces cyclobutane pyrimidine dimers and pyrimidine-pyrimidone (6-4) photoproducts, which considerably distort the DNA double-helical structure. These products can be found in RNA as well as in DNA (Rycyna and Alderfer, 1988). However, the fate of UV-damaged RNA has been poorly documented to date. The first assumption would be that damaged RNAs are useless or even deleterious for living cells and might therefore be rapidly degraded. Indeed, exposure to UV radiation after treatment with the mRNA-synthesis inhibitor cordycepin has been shown to cause mRNA degradation in Arabidopsis thaliana (Revenkova et al., 1999). In yeast, mRNA degradation profiling after UV-irradiation was performed using thiolutin, a drug inhibiting RNA and protein synthesis. In wild-type cells, some transcripts were stabilized (26%), others destabilized (12%), whereas the majority remained unchanged (62%) 45 min after UV-irradiation (Hieronymus et al., 2004). We took advantage of the regulable GAL1 promoter to analyze mRNA decay after UV-irradiation, thus avoiding the use of general inhibitors of RNA synthesis, which might induce a stress response and activate checkpoints on their own. Surprisingly, our results show that substantial and dose-dependent mRNA stabilization occurs upon UV-irradiation in yeast (Figure 1). In human cells, a general and dose-dependent stabilization of mRNA has been observed after UV-irradiation by using the regulable tet off promoter (Bollig et al., 2002; Gowrishankar et al., 2005), indicating that the phenomenon is not restricted to yeast. The observation that UV-dependent mRNA stabilization occurred independently of DNA repair and of the DNA-damage checkpoint response (Figure 3) suggests that it results from damaged RNA. Other stresses that do not induce physical damage in nucleic acids, such as heat and saline shocks or nitrogen starvation, did not lead to a similar mRNA stabilization in our conditions (Figure 2). It is worth noticing that a previous report showed an inhibition of mRNA deadenylation, leading to transcript stabilization during strong hyperosmolarity and heat shock in yeast (Hilgers et al., 2006). However, the way experiments were performed differs between the two studies. We assessed the stability of transcripts during recovery from mild saline and heat shocks that led to cell viability levels similar to those obtained after UV-irradiation at 200 J/m2, whereas the previous report used strong continuous stresses. Indeed, the differences between both results indicate that the response to stresses strongly depends on the experimental conditions used. Despite the transcript stabilization observed after UV-irradiation (this study) and during strong hyperosmolarity and heat shock (Hilgers et al., 2006) both being independent of the translational state of the mRNAs, a major difference between both responses is the localization of the mRNA pool. This is found in cytoplasmic UVGs after UV (Figure 4), whereas mRNAs are retained in the nucleus during/after saline or heat shocks (Supplemental Figure S1; Tani et al., 1995; Saavedra et al., 1996; Yoshida and Tani, 2005).
When present in RNA, UV lesions may surely inhibit translation as cisplatin cross-links do (Rosenberg and Sato, 1988; Heminger et al., 1997). We did not find a transcript length dependency of the mRNA stabilization response, in agreement with the assumption that a single damage on an RNA molecule would be enough to block translation and possibly trigger the response. Because impaired translation elongation has been shown to increase mRNA stability (Beelman and Parker, 1994), it was conceivable that trapping of translation at a UV lesion in the template mRNA could trigger the observed mRNA stabilization. Yet, the observation that cycloheximide, a drug impairing translation elongation and known to reduce mRNA decay rates (Stimac et al., 1984; Herrick et al., 1990), did not lead to poly-A+ foci formation (Figure 5) argues against the idea that a translational machinery stalled at RNA lesions might be sufficient to trigger an RNA-damage response. Then again, translation might work as a sensor for damaged RNA and be required for poly-A+ foci formation. However, our results show that addition of cycloheximide does not prevent the formation of UVG (Figure 5) and that polysomes do not get disassembled upon UV-irradiation (Figure 6). Together, these results indicate that UVG formation is independent from translation elongation. Interestingly, our results and previous work indicating that stalled ribosomes trigger a rapid RNA decay response termed no-go decay (Doma and Parker, 2006) suggest that potentially damaged mRNA might be trapped in UVG before their engagement in translation. This hypothesis is further supported by the observation that the stabilized GAL1-reporter transcript accumulates in polysome-free fractions in UV-irradiated cells (Figure 6).
At first glance, the UVG shared many of the properties of P-bodies, which are currently thought to be cellular sites of mRNA turnover (reviewed in Anderson and Kedersha, 2006; Eulalio et al., 2007; Parker and Sheth, 2007). For example, P-bodies have been shown to function in RNA sorting and sequestration (Brengues et al., 2005; Bhattacharyya et al., 2006). In addition, P-bodies are dynamic and are affected by a range of cellular perturbations, including glucose deprivation, osmotic stress, and exposure to UV light, and by the stage of cell growth (Kedersha et al., 2005; Teixeira et al., 2005; Wilczynska et al., 2005). However, not only do we show that UVGs do not colocalize with P-bodies (Figure 7) but also several lines of evidence indicate that UVGs and P-bodies represent different structures. Thus, UVGs have been observed exclusively during recovery from UV-irradiation, in contrast to P-bodies, which are present in the absence of exogenous stress (Sheth and Parker, 2003). P-bodies were shown to increase in size and number after osmotic stress and during glucose starvation (Teixeira et al., 2005) as well as in a mutant impaired in RNA 5′-end decapping (Sheth and Parker, 2003). However, UVGs are not formed in response to osmotic stress (Supplemental Figure S1) nor during glucose starvation (Supplemental Figure S3; Brengues and Parker, 2007), and the amount of UVGs does not increase in dcp1-2 cells (Figure 5). Importantly, P-body formation depends on translation elongation (Brengues et al., 2005), whereas UVGs arise in UV-irradiated cells independently of translation elongation (Figures 5 and 6).
Recently, new cytoplasmic bodies distinct from P-bodies, called EGP-bodies, have been described in yeast (Hoyle et al., 2007). EGP-bodies are caused by glucose starvation-induced translation repression, and they contain the translational factors eIF4E, eIF4G, as well as Pap1. Our results indicate that UVGs and EGP-bodies are distinct structures (Figures 7 and Supplemental Figure S3). Moreover, several observations further support this finding. First, only 11% of UVG colocalize with P-bodies, whereas half of EGP-bodies do so. Second, UVG formation is independent of translation elongation (Figure 5), whereas EGP-bodies arise from key translation factors segregating away from ribosomal subunits. Finally, UV-irradiation does not lead to polysome disruption (Figure 6), in contrast to the EGP-inducing stresses upon which polysomes are completely disassembled.
Evidence exists indicating that components of the translation repression complex found in P-bodies are functionally and physically associated with the ER, which might play a role in targeting some transcripts for localized translation (reviewed in Decker and Parker, 2006). In the same line, COPI-coated vesicles have been proposed to act as short-range mRNA transport and localization vehicles, because they interact with mRNA and the poly-A+–binding protein Pab1, which is required to restrict ASH1 mRNA to the bud tip in yeast (Trautwein et al., 2004). The recent finding that Pab1 and poly-A+ mRNA can be found in P-bodies during glucose depletion and the stationary phase (Brengues and Parker, 2007) further support the implication of P-bodies in mRNA storage, transport, or both. From this point of view, it was plausible that storage and transport of damaged mRNA, possibly associated with damaged proteins, might occur via the secretory pathway and lead to the accumulation of UVG. However, neither Rer1 (cis-Golgi) nor Tlg1 (early endosome) colocalized with UV-induced poly-A+ foci (Figure 9), suggesting that UVG are not part of the secretory pathway.
In addition to P-bodies, which are conserved from yeast to humans, several kinds of RNA granules have been observed in higher eukaryotic cells under stress, during oogenesis, and in neuronal cells (reviewed in St Johnston, 2005; Anderson and Kedersha, 2008). Strikingly, these particles share certain components with P-bodies and the common function of storing nontranslating mRNAs. SGs are formed in mammalian cells in response to different types of stress, including UV-irradiation, heat shock, and oxidative stress (Kedersha et al., 1999; Kimball et al., 2003). Furthermore, recent work showed that a range of stresses lead to mRNA stabilization by inhibiting deadenylation in human cells (Gowrishankar et al., 2006). The yeast RNA-binding proteins Pub1 and Nam8 are the putative homologues of the prion-like proteins TIA-1 and TIAR (this study), which are necessary for SGs formation in mammalian cells (Kedersha et al., 1999). Pub1 is a major poly-A–binding protein (Anderson et al., 1993; Matunis et al., 1993), which has been recently involved as a regulator of cellular mRNA decay (Ruiz-Echevarria and Peltz, 2000; Vasudevan and Peltz, 2001; Grigull et al., 2004; Duttagupta et al., 2005). Nam8 has been shown to be a component of yeast U1 snRNP, to facilitate 5′ splice site (ss) recognition by interacting with nonconserved sequences downstream from the 5′ ss (Puig et al., 1999) and to be required for meiosis-specific splicing (Ogawa et al., 1995; Nakagawa and Ogawa, 1999). Analogous functions between Pub1 and its mammalian homologues TIA-1 and HuR, which both localize at SGs (Kedersha and Anderson, 2002), as well as the findings that TIA-1 is a splicing regulator acting through intron sequences adjacent to the 5′ ss (Del Gatto-Konczak et al., 2000; Forch et al., 2000), suggest at least a partial conservation of functions from yeast to humans. However, we could not find any involvement of Pub1 or Nam8 in UV-dependent poly-A+ foci formation or evidence for the existence of SGs in S. cerevisiae (Figure 8 and Supplemental Figures S1 and S3). Thus, although SGs exist in S. pombe (Dunand-Sauthier et al., 2002), similar structures seem to be absent in S. cerevisiae.
Another described stress response is autophagy, a process in which cytosol and organelles are sequestered within double-membrane vesicles that deliver the contents to the lysosome/vacuole for degradation and recycling of the resulting macromolecules (reviewed in Klionsky, 2005a,b). Although autophagy is primarily induced by nutrient starvation, other kinds of stress might be able to activate the response as well. Indeed, our results indicate that the autophagy pathway is induced by UV-irradiation in yeast, although to a lesser extent than in starvation conditions. However, the kinetics of UV-induced autophagy and UVG formation was different and the autophagy vesicle protein Aut7 absent from UV-induced poly-A+ foci (Figure 9). Thus, the activation of autophagy by UV-irradiation likely represents a response to DNA-damage checkpoint activation rather than to damaged RNA.
We have shown that UVGs contain poly-A+ RNAs that were present at the time of UV-irradiation and therefore are potentially damaged (Figure 5). The accumulation of putatively damaged poly-A+ RNA into novel cytoplasmic structures suggests that those transcripts are not designated for degradation, because removal of the poly-A+ tail is required before mRNAs can be decapped (Decker and Parker, 1993; Muhlrad et al., 1994). This is consistent with simultaneous transcript stabilization, localization of 35% of the stabilized GAL1 reporter transcripts in UVGs, accumulation of stabilized GAL1 mRNA in polysome-free fractions, and with the formation of granules distinct from P-bodies, thus avoiding direct contact with the mRNA degradation machinery. The biological significance of UVG might be as simple as minimizing the deleterious, or even toxic, effects that damaged mRNAs are likely to exert on basic cellular processes (reviewed in Bregeon and Sarasin, 2005; Falnes et al., 2007). Furthermore, sequestering the pool of mRNAs that have been subjected to UV radiation into a nontranslating state would allow for preferential translation of newly transcribed mRNAs. The clustering of potentially damaged mRNAs into UVG might also serve to sort the unspoiled transcripts that can return to the translating pool from the damaged transcripts, the latter probably being then targeted to degradation. This idea is further supported by the observation that a small fraction of UVG localizes at or near to P-bodies (Figure 7).
Alternatively, because transcripts exist whose half-lives span several life cycles (Herrick et al., 1990), it might be worth repairing damaged transcripts rather than discarding them. Studies from the 1970s suggested that photoreactivation might repair UV-damaged viral RNA in plants (Merriam and Gordon, 1965; Murphy and Gordon, 1971; Hurter et al., 1974). More recent work showed that alkylation damage to RNA is repaired in vivo both in bacteria and in human cells (Aas et al., 2003; Ougland et al., 2004). Thus, we cannot exclude that RNA repair mechanisms might exist in yeast.
Together, our results point to the existence of a RNA-damage specific cellular response in yeast, which leads to transcript stabilization and to the accumulation of potentially damaged poly-A+ mRNA in a novel class of cytoplasmic granules (UVGs). Further work will be required to identify the proteins associated with these granules and whether they represent a transient accumulation of bulk mRNA, regardless of their integrity, or exclusively contain damaged mRNAs or mRNA–protein aggregates. Nonetheless, it is tempting to speculate that damaged mRNAs, which are predicted to be deleterious or even toxic for the cell, might be stored until the cell recovers, being either repaired, degraded, or recycled to a later time.
Supplementary Material
ACKNOWLEDGMENTS
We thank R. Rothstein, L. Symington, R. Parker, and M. Ashe for strains; J. de la Cruz and K. Struhl for plasmids; K. Weis, D. Klionsky, M. Muñiz, H. Pelham, M. Ashe, and M. Altmann for antibodies; P. Domínguez for confocal microscopy assistance; J. de la Cruz and laboratory for assistance in the polysome profile analyses; R. Wellinger and M. Muñiz for discussions; and D. Haun and R. Stuckey for style supervision. This research was funded by grants from the Spanish Ministry of Science and Education (BFU2006-05260 and BFU2007-28647) and from the Junta de Andalucia (CVI102 and CVI2549). H. G. was granted by the Swiss National Science Foundation (PBEZA-100700 and PA00A-105027) and the Novartis Foundation.
Abbreviations used:
- EGP-bodies, eIF4E-,eIF4G-
and Pab1-containing bodies
- N starvation
nitrogen starvation
- P-body
processing body
- poly-A+
polyadenylated
- SG
stress granule
- UVG
UV-induced granule.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-02-0193) on September 3, 2008.
REFERENCES
- Aas P. A., Otterlei M., Falnes P. O., Vagbo C. B., Skorpen F., Akbari M., Sundheim O., Bjoras M., Slupphaug G., Seeberg E., Krokan H. E. Human and bacterial oxidative demethylases repair alkylation damage in both RNA and DNA. Nature. 2003;421:859–863. doi: 10.1038/nature01363. [DOI] [PubMed] [Google Scholar]
- Anderson J. T., Paddy M. R., Swanson M. S. PUB1 is a major nuclear and cytoplasmic polyadenylated RNA-binding protein in Saccharomyces cerevisiae. Mol. Cell. Biol. 1993;13:6102–6113. doi: 10.1128/mcb.13.10.6102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson P., Kedersha N. RNA granules. J. Cell Biol. 2006;172:803–808. doi: 10.1083/jcb.200512082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson P., Kedersha N. Stress granules: the Tao of RNA triage. Trends Biochem. Sci. 2008;33:141–150. doi: 10.1016/j.tibs.2007.12.003. [DOI] [PubMed] [Google Scholar]
- Baltimore D. Our genome unveiled. Nature. 2001;409:814–816. doi: 10.1038/35057267. [DOI] [PubMed] [Google Scholar]
- Beelman C. A., Parker R. Differential effects of translational inhibition in cis and in trans on the decay of the unstable yeast MFA2 mRNA. J. Biol. Chem. 1994;269:9687–9692. [PubMed] [Google Scholar]
- Bellacosa A., Moss E. G. RNA repair: damage control. Curr. Biol. 2003;13:R482–R484. doi: 10.1016/s0960-9822(03)00408-1. [DOI] [PubMed] [Google Scholar]
- Bhattacharyya S. N., Habermacher R., Martine U., Closs E. I., Filipowicz W. Stress-induced reversal of microRNA repression and mRNA P-body localization in human cells. Cold Spring Harb. Symp. Quant. Biol. 2006;71:513–521. doi: 10.1101/sqb.2006.71.038. [DOI] [PubMed] [Google Scholar]
- Bollig F., Winzen R., Kracht M., Ghebremedhin B., Ritter B., Wilhelm A., Resch K., Holtmann H. Evidence for general stabilization of mRNAs in response to UV light. Eur. J. Biochem. 2002;269:5830–5839. doi: 10.1046/j.1432-1033.2002.03300.x. [DOI] [PubMed] [Google Scholar]
- Bouveret E., Rigaut G., Shevchenko A., Wilm M., Seraphin B. A Sm-like protein complex that participates in mRNA degradation. EMBO J. 2000;19:1661–1671. doi: 10.1093/emboj/19.7.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bregeon D., Sarasin A. Hypothetical role of RNA damage avoidance in preventing human disease. Mutat. Res. 2005;577:293–302. doi: 10.1016/j.mrfmmm.2005.04.002. [DOI] [PubMed] [Google Scholar]
- Brengues M., Parker R. Accumulation of polyadenylated mRNA, Pab1p, eIF4E, and eIF4G with P-bodies in Saccharomyces cerevisiae. Mol. Biol. Cell. 2007;18:2592–2602. doi: 10.1091/mbc.E06-12-1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brengues M., Teixeira D., Parker R. Movement of eukaryotic mRNAs between polysomes and cytoplasmic processing bodies. Science. 2005;310:486–489. doi: 10.1126/science.1115791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole C. N., Heath C. V., Hodge C. A., Hammell C. M., Amberg D. C. Analysis of RNA export. Methods Enzymol. 2002;351:568–587. doi: 10.1016/s0076-6879(02)51869-3. [DOI] [PubMed] [Google Scholar]
- Cougot N., Babajko S., Seraphin B. Cytoplasmic foci are sites of mRNA decay in human cells. J. Cell Biol. 2004;165:31–40. doi: 10.1083/jcb.200309008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker C. J., Parker R. A turnover pathway for both stable and unstable mRNAs in yeast: evidence for a requirement for deadenylation. Genes Dev. 1993;7:1632–1643. doi: 10.1101/gad.7.8.1632. [DOI] [PubMed] [Google Scholar]
- Decker C. J., Parker R. CAR-1 and trailer hitch: driving mRNP granule function at the ER? J. Cell Biol. 2006;173:159–163. doi: 10.1083/jcb.200601153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Gatto-Konczak F., Bourgeois C. F., Le Guiner C., Kister L., Gesnel M. C., Stevenin J., Breathnach R. The RNA-binding protein TIA-1 is a novel mammalian splicing regulator acting through intron sequences adjacent to a 5′ splice site. Mol. Cell. Biol. 2000;20:6287–6299. doi: 10.1128/mcb.20.17.6287-6299.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doma M. K., Parker R. Endonucleolytic cleavage of eukaryotic mRNAs with stalls in translation elongation. Nature. 2006;440:561–564. doi: 10.1038/nature04530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunand-Sauthier I., Walker C., Wilkinson C., Gordon C., Crane R., Norbury C., Humphrey T. Sum1, a component of the fission yeast eIF3 translation initiation complex, is rapidly relocalized during environmental stress and interacts with components of the 26S proteasome. Mol. Biol. Cell. 2002;13:1626–1640. doi: 10.1091/mbc.01-06-0301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duttagupta R., Tian B., Wilusz C. J., Khounh D. T., Soteropoulos P., Ouyang M., Dougherty J. P., Peltz S. W. Global analysis of Pub1p targets reveals a coordinate control of gene expression through modulation of binding and stability. Mol. Cell. Biol. 2005;25:5499–5513. doi: 10.1128/MCB.25.13.5499-5513.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulalio A., Behm-Ansmant I., Izaurralde E. P bodies: at the crossroads of post-transcriptional pathways. Nat. Rev. Mol. Cell Biol. 2007;8:9–22. doi: 10.1038/nrm2080. [DOI] [PubMed] [Google Scholar]
- Falnes P. O., Klungland A., Alseth I. Repair of methyl lesions in DNA and RNA by oxidative demethylation. Neuroscience. 2007;145:1222–1232. doi: 10.1016/j.neuroscience.2006.11.018. [DOI] [PubMed] [Google Scholar]
- Forch P., Puig O., Kedersha N., Martinez C., Granneman S., Seraphin B., Anderson P., Valcarcel J. The apoptosis-promoting factor TIA-1 is a regulator of alternative pre-mRNA splicing. Mol. Cell. 2000;6:1089–1098. doi: 10.1016/s1097-2765(00)00107-6. [DOI] [PubMed] [Google Scholar]
- Gaillard H., Wellinger R. E., Aguilera A. A new connection of mRNP biogenesis and export with transcription-coupled repair. Nucleic Acids Res. 2007;35:3893–3906. doi: 10.1093/nar/gkm373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowrishankar G., Winzen R., Bollig F., Ghebremedhin B., Redich N., Ritter B., Resch K., Kracht M., Holtmann H. Inhibition of mRNA deadenylation and degradation by ultraviolet light. Biol. Chem. 2005;386:1287–1293. doi: 10.1515/BC.2005.146. [DOI] [PubMed] [Google Scholar]
- Gowrishankar G., Winzen R., Dittrich-Breiholz O., Redich N., Kracht M., Holtmann H. Inhibition of mRNA deadenylation and degradation by different types of cell stress. Biol. Chem. 2006;387:323–327. doi: 10.1515/BC.2006.043. [DOI] [PubMed] [Google Scholar]
- Grigull J., Mnaimneh S., Pootoolal J., Robinson M. D., Hughes T. R. Genome-wide analysis of mRNA stability using transcription inhibitors and microarrays reveals posttranscriptional control of ribosome biogenesis factors. Mol. Cell. Biol. 2004;24:5534–5547. doi: 10.1128/MCB.24.12.5534-5547.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heminger K. A., Hartson S. D., Rogers J., Matts R. L. Cisplatin inhibits protein synthesis in rabbit reticulocyte lysate by causing an arrest in elongation. Arch. Biochem. Biophys. 1997;344:200–207. doi: 10.1006/abbi.1997.0198. [DOI] [PubMed] [Google Scholar]
- Herrick D., Parker R., Jacobson A. Identification and comparison of stable and unstable mRNAs in Saccharomyces cerevisiae. Mol. Cell. Biol. 1990;10:2269–2284. doi: 10.1128/mcb.10.5.2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hieronymus H., Yu M. C., Silver P. A. Genome-wide mRNA surveillance is coupled to mRNA export. Genes Dev. 2004;18:2652–2662. doi: 10.1101/gad.1241204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilgers V., Teixeira D., Parker R. Translation-independent inhibition of mRNA deadenylation during stress in Saccharomyces cerevisiae. RNA. 2006;12:1835–1845. doi: 10.1261/rna.241006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollams E. M., Giles K. M., Thomson A. M., Leedman P. J. MRNA stability and the control of gene expression: implications for human disease. Neurochem. Res. 2002;27:957–980. doi: 10.1023/a:1020992418511. [DOI] [PubMed] [Google Scholar]
- Hoyle N. P., Castelli L. M., Campbell S. G., Holmes L. E., Ashe M. P. Stress-dependent relocalization of translationally primed mRNPs to cytoplasmic granules that are kinetically and spatially distinct from P-bodies. J. Cell Biol. 2007;179:65–74. doi: 10.1083/jcb.200707010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurter J., Gordon M. P., Kirwan J. P., McLaren A. D. In vitro photoreactivation of ultraviolet-inactivated ribonucleic acid from tobacco mosaic virus. Photochem. Photobiol. 1974;19:185–190. doi: 10.1111/j.1751-1097.1974.tb06497.x. [DOI] [PubMed] [Google Scholar]
- Kedersha N., Anderson P. Stress granules: sites of mRNA triage that regulate mRNA stability and translatability. Biochem. Soc. Trans. 2002;30:963–969. doi: 10.1042/bst0300963. [DOI] [PubMed] [Google Scholar]
- Kedersha N., Stoecklin G., Ayodele M., Yacono P., Lykke-Andersen J., Fritzler M. J., Scheuner D., Kaufman R. J., Golan D. E., Anderson P. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J. Cell Biol. 2005;169:871–884. doi: 10.1083/jcb.200502088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N. L., Gupta M., Li W., Miller I., Anderson P. RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2 alpha to the assembly of mammalian stress granules. J. Cell Biol. 1999;147:1431–1442. doi: 10.1083/jcb.147.7.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimball S. R., Horetsky R. L., Ron D., Jefferson L. S., Harding H. P. Mammalian stress granules represent sites of accumulation of stalled translation initiation complexes. Am. J. Physiol. Cell Physiol. 2003;284:C273–C284. doi: 10.1152/ajpcell.00314.2002. [DOI] [PubMed] [Google Scholar]
- Klionsky D. J. Autophagy. Curr. Biol. 2005a;15:R282–R283. doi: 10.1016/j.cub.2005.04.013. [DOI] [PubMed] [Google Scholar]
- Klionsky D. J. The molecular machinery of autophagy: unanswered questions. J. Cell Sci. 2005b;118:7–18. doi: 10.1242/jcs.01620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kressler D., de la Cruz J., Rojo M., Linder P. Fal1p is an essential DEAD-box protein involved in 40S-ribosomal-subunit biogenesis in Saccharomyces cerevisiae. Mol. Cell. Biol. 1997;17:7283–7294. doi: 10.1128/mcb.17.12.7283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuai L., Das B., Sherman F. A nuclear degradation pathway controls the abundance of normal mRNAs in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA. 2005;102:13962–13967. doi: 10.1073/pnas.0506518102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason P. B., Struhl K. The FACT complex travels with elongating RNA polymerase II and is important for the fidelity of transcriptional initiation in vivo. Mol. Cell. Biol. 2003;23:8323–8333. doi: 10.1128/MCB.23.22.8323-8333.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masta A., Gray P. J., Phillips D. R. Nitrogen mustard inhibits transcription and translation in a cell free system. Nucleic Acids Res. 1995;23:3508–3515. doi: 10.1093/nar/23.17.3508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matunis M. J., Matunis E. L., Dreyfuss G. PUB 1, a major yeast poly(A)+ RNA-binding protein. Mol. Cell. Biol. 1993;13:6114–6123. doi: 10.1128/mcb.13.10.6114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merriam V., Gordon M. P. Photoreactivation of tobacco mosaic virus ribonucleic acid following near ultraviolet irradiation. Proc. Natl. Acad. Sci. USA. 1965;54:1261–1268. doi: 10.1073/pnas.54.4.1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlrad D., Decker C. J., Parker R. Deadenylation of the unstable mRNA encoded by the yeast MFA2 gene leads to decapping followed by 5′ – >3′ digestion of the transcript. Genes Dev. 1994;8:855–866. doi: 10.1101/gad.8.7.855. [DOI] [PubMed] [Google Scholar]
- Murphy T. M., Gordon M. P. Light-mediated regulation of TMV-RNA photoreactivation. Photochem. Photobiol. 1971;13:45–55. doi: 10.1111/j.1751-1097.1971.tb06090.x. [DOI] [PubMed] [Google Scholar]
- Nakagawa T., Ogawa H. The Saccharomyces cerevisiae MER3 gene, encoding a novel helicase-like protein, is required for crossover control in meiosis. EMBO J. 1999;18:5714–5723. doi: 10.1093/emboj/18.20.5714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunomura A., Honda K., Takeda A., Hirai K., Zhu X., Smith M. A., Perry G. Oxidative damage to RNA in neurodegenerative diseases. J. Biomed. Biotechnol. 2006;2006:82323. doi: 10.1155/JBB/2006/82323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa H., Johzuka K., Nakagawa T., Leem S. H., Hagihara A. H. Functions of the yeast meiotic recombination genes, MRE11 and MRE2. Adv. Biophys. 1995;31:67–76. doi: 10.1016/0065-227x(95)99383-z. [DOI] [PubMed] [Google Scholar]
- Ougland R., Zhang C. M., Liiv A., Johansen R. F., Seeberg E., Hou Y. M., Remme J., Falnes P. O. AlkB restores the biological function of mRNA and tRNA inactivated by chemical methylation. Mol. Cell. 2004;16:107–116. doi: 10.1016/j.molcel.2004.09.002. [DOI] [PubMed] [Google Scholar]
- Parker R., Sheth U. P bodies and the control of mRNA translation and degradation. Mol. Cell. 2007;25:635–646. doi: 10.1016/j.molcel.2007.02.011. [DOI] [PubMed] [Google Scholar]
- Puig O., Gottschalk A., Fabrizio P., Seraphin B. Interaction of the U1 snRNP with nonconserved intronic sequences affects 5′ splice site selection. Genes Dev. 1999;13:569–580. doi: 10.1101/gad.13.5.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reagan M. S., Friedberg E. C. Recovery of RNA polymerase II synthesis following DNA damage in mutants of Saccharomyces cerevisiae defective in nucleotide excision repair. Nucleic Acids Res. 1997;25:4257–4263. doi: 10.1093/nar/25.21.4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revenkova E., Masson J., Koncz C., Afsar K., Jakovleva L., Paszkowski J. Involvement of Arabidopsis thaliana ribosomal protein S27 in mRNA degradation triggered by genotoxic stress. EMBO J. 1999;18:490–499. doi: 10.1093/emboj/18.2.490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg J., Sato P. Messenger RNA loses the ability to direct in vitro peptide synthesis following incubation with cisplatin. Mol. Pharmacol. 1988;33:611–616. [PubMed] [Google Scholar]
- Ruiz-Echevarria M. J., Peltz S. W. The RNA binding protein Pub1 modulates the stability of transcripts containing upstream open reading frames. Cell. 2000;101:741–751. doi: 10.1016/s0092-8674(00)80886-7. [DOI] [PubMed] [Google Scholar]
- Rycyna R. E., Alderfer J. L. Ultraviolet irradiation of nucleic acids: formation, purification, and solution conformational analyses of the cis-syn and trans-syn photodimers of UpU. Biochemistry. 1988;27:3142–3151. doi: 10.1021/bi00409a005. [DOI] [PubMed] [Google Scholar]
- Saavedra C., Tung K. S., Amberg D. C., Hopper A. K., Cole C. N. Regulation of mRNA export in response to stress in Saccharomyces cerevisiae. Genes Dev. 1996;10:1608–1620. doi: 10.1101/gad.10.13.1608. [DOI] [PubMed] [Google Scholar]
- Sheth U., Parker R. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science. 2003;300:805–808. doi: 10.1126/science.1082320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smerdon M. J., Bedoyan J., Thoma F. DNA repair in a small yeast plasmid folded into chromatin. Nucleic Acids Res. 1990;18:2045–2051. doi: 10.1093/nar/18.8.2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Johnston D. Moving messages: the intracellular localization of mRNAs. Nat. Rev. Mol. Cell Biol. 2005;6:363–375. doi: 10.1038/nrm1643. [DOI] [PubMed] [Google Scholar]
- Stimac E., Groppi V. E., Jr, Coffino P. Inhibition of protein synthesis stabilizes histone mRNA. Mol. Cell. Biol. 1984;4:2082–2090. doi: 10.1128/mcb.4.10.2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tani T., Derby R. J., Hiraoka Y., Spector D. L. Nucleolar accumulation of poly (A)+ RNA in heat-shocked yeast cells: implication of nucleolar involvement in mRNA transport. Mol. Biol. Cell. 1995;6:1515–1534. doi: 10.1091/mbc.6.11.1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira D., Sheth U., Valencia-Sanchez M. A., Brengues M., Parker R. Processing bodies require RNA for assembly and contain nontranslating mRNAs. RNA. 2005;11:371–382. doi: 10.1261/rna.7258505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tharun S., He W., Mayes A. E., Lennertz P., Beggs J. D., Parker R. Yeast Sm-like proteins function in mRNA decapping and decay. Nature. 2000;404:515–518. doi: 10.1038/35006676. [DOI] [PubMed] [Google Scholar]
- Tharun S., Parker R. Analysis of mutations in the yeast mRNA decapping enzyme. Genetics. 1999;151:1273–1285. doi: 10.1093/genetics/151.4.1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trautwein M., Dengjel J., Schirle M., Spang A. Arf1p provides an unexpected link between COPI vesicles and mRNA in Saccharomyces cerevisiae. Mol. Biol. Cell. 2004;15:5021–5037. doi: 10.1091/mbc.E04-05-0411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasudevan S., Peltz S. W. Regulated ARE-mediated mRNA decay in Saccharomyces cerevisiae. Mol. Cell. 2001;7:1191–1200. doi: 10.1016/s1097-2765(01)00279-9. [DOI] [PubMed] [Google Scholar]
- Wilczynska A., Aigueperse C., Kress M., Dautry F., Weil D. The translational regulator CPEB1 provides a link between dcp1 bodies and stress granules. J. Cell Sci. 2005;118:981–992. doi: 10.1242/jcs.01692. [DOI] [PubMed] [Google Scholar]
- Yoshida J., Tani T. Hsp16p is required for thermotolerance in nuclear mRNA export in fission yeast Schizosaccharomyces pombe. Cell Struct. Funct. 2005;29:125–138. doi: 10.1247/csf.29.125. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.