Abstract
Ubiquitination regulates many cellular functions, including protein localization and degradation. Each function is specified by unique determinants in the conjugate. Ubiquitinated Jun is localized to lysosomes for degradation. Here, we characterized determinants of Jun ubiquitination and lysosomal localization by using ubiquitin-mediated fluorescence complementation (UbFC) in living cells and analysis of the stoichiometry of ubiquitin linked to Jun extracted from cells. The δ region of Jun and isoleucine-44 in ubiquitin were required for lysosomal localization of the conjugate. Ubiquitin containing only lysine-27, but no other single-lysine ubiquitin, mediated Jun ubiquitination, albeit at lower stoichiometry than wild-type ubiquitin. These conjugates were predominantly nuclear, but coexpression of lysine-27 and lysine-less ubiquitins enhanced the mean stoichiometry of Jun ubiquitination and lysosomal localization of the conjugate. Hepatocyte growth factor-regulated tyrosine kinase substrate (HRS) and tumor susceptibility gene 101 (TSG101) colocalized with ubiquitinated Jun. Knockdown of HRS or TSG101 inhibited lysosomal localization of ubiquitinated Jun and reduced Jun turnover. Ubiquitination of other Fos and Jun family proteins had distinct effects on their localization. Our results indicate that Jun is polyubiquitinated by E3 ligases that produce lysine-27–linked chains. Lysosomal localization of the conjugate requires determinants in Jun and in ubiquitin that are recognized in part by TSG101 and HRS, facilitating selective translocation and degradation of ubiquitinated Jun.
INTRODUCTION
Ubiquitination regulates cellular proteins through a variety of mechanisms, including control of their subcellular localization and degradation. The diverse roles of ubiquitination require each function to be defined by a distinct combination of determinants in the conjugate (Hershko and Ciechanover, 1998; Hicke, 2001; Pickart and Fushman, 2004). Many characteristics, including the site(s) of ubiquitination, the number of ubiquitins added, the isopeptide linkages between ubiquitins as well as determinants intrinsic to the substrate protein can distinguish different conjugates.
Ubiquitinated proteins have been difficult to visualize, particularly in living cells, because the subpopulation of any one protein that is modified by ubiquitin is generally very small. We developed a method for the visualization of specific ubiquitinated proteins in living cells designated ubiquitin-mediated fluorescence complementation (UbFC) analysis (Fang and Kerppola, 2004). This approach is based on the formation of a fluorescent conjugate when ubiquitin fused to a nonfluorescent fragment of a fluorescent protein is conjugated to a substrate fused to a complementary fluorescent protein fragment. The UbFC assay allows visualization of the subcellular distributions of specific ubiquitinated proteins in living cells. UbFC analysis has been used to visualize the ubiquitination of proteins in different subcellular compartments (Fang and Kerppola, 2004; van der Horst et al., 2006; Hinman et al., 2008).
Jun is a transcription regulatory protein with a short half-life regulated by ubiquitination. The protein encoded by the v-jun oncogene contains a deletion of the δ region, resulting in a longer half-life that is thought to contribute to cell transformation (Treier et al., 1994). Many different E3 ligases can ubiquitinate Jun in different cell types and in response to different stimuli (Fang and Kerppola, 2004; Gao et al., 2004; Nateri et al., 2004; Wertz et al., 2004; Wei et al., 2005). Whereas Jun is predominantly nuclear, ubiquitinated Jun colocalizes with lysosomal proteins, cofractionates with lysosomal enzyme activity, and is stabilized in cells treated with inhibitors of lysosomal proteases (Fang and Kerppola, 2004). The molecular determinants that direct ubiquitinated Jun to lysosomes have not been elucidated.
Several pathways for protein degradation in lysosomes have been described (Klionsky and Emr, 2000; Hurley and Emr, 2006). Membrane receptors for extracellular growth factors are translocated to lysosomes by an endosomal trafficking pathway (Hurley and Emr, 2006). The ubiquitinated receptors are recognized by a series of proteins containing ubiquitin-binding domains (Strous et al., 1996; Katzmann et al., 2001; Hicke et al., 2005; Huang et al., 2006; Hurley and Emr, 2006). The proteins required for lysosomal localization of ubiquitinated Jun are unknown.
Here, we have investigated the determinants for lysosomal localization of ubiquitinated Jun. We identified amino acid residues in Jun and ubiquitin that were required for the lysosomal localization of the conjugate. We characterized the isopeptide linkages that define the topology of the polyubiquitin chain linked to Jun, and we examined the effect of the stoichiometry of ubiquitination on conjugate localization. We determined the roles of ubiquitin-binding proteins in the localization and degradation of ubiquitinated Jun. The results define several unique characteristics of ubiquitinated Jun that are required for its lysosomal localization and degradation.
MATERIALS AND METHODS
Plasmids and Antibodies
Plasmids encoding amino acid residues 1-155 of Venus (VN) fused to the amino terminus of ubiquitin and amino acid residues 156-238 of cyan fluorescent protein (CFP) (CC) fused to the C termini of Jun, JunB, JunD, Fos, FosB, Fra1, and Fra2 were constructed. Mutations in Jun and ubiquitin were generated by polymerase chain reaction. The detailed construction strategy for each plasmid, and the sources of antibodies are listed in supporting materials and methods.
UbFC Analysis of the Distribution of Ubiquitinated Jun
The UbFC assay described originally (Fang and Kerppola, 2004) required incubation of the cells at 30°C to promote fluorophore maturation. Here, we modified the UbFC assay by using residues 1-155 of VN fused to ubiquitin and residues 156-238 of CFP fused to Jun and other putative substrates of ubiquitination. The modified UbFC assay enabled visualization of the conjugate at 37°C, avoiding potential changes in the formation, distribution or degradation of ubiquitin conjugates caused by incubation at lower temperature. The UbFC conjugates formed by these fusions had at least fivefold higher fluorescence intensities when imaged in living cells at 37°C compared with UbFC conjugates formed by the fluorescent protein fragments used originally (Fang and Kerppola, 2004). The modified assay enabled visualization of many ubiquitin conjugates that could not be detected using the original assay.
To visualize UbFC conjugates, cells transfected with plasmids encoding the fusion proteins indicated in each experiment were imaged 24 h after transfection by using 500-nm excitation and 535-nm emission filters. For studies of subcellular localization, the cells were incubated with fluorescent labels (LysoTracker and epidermal growth factor [EGF]-rhodamine) and imaged directly or fixed and immunostained using the antibodies indicated. The distributions of UbFC conjugates were compared with LysoTracker Red, hepatocyte growth factor-regulated tyrosine kinase substrate (HRS), and tumor susceptibility gene 101 (TSG101) by using both epifluorescence and confocal microscopy.
Quantification of the Ratio of Cytoplasmic to Nuclear Localization of UbFC Conjugates
All images were acquired using identical acquisition parameters and the raw images were used for quantitation. Images were segmented by semiautomated image analysis by using SimplePCI software. The nucleus was labeled using Hoechst, imaged using 436-nm excitation and 470-nm emission filters, and used as a mask to determine localization of UbFC fluorescence. The nuclear area was defined after three applications of the “erode” function to the area corresponding to Hoechst signal to avoid perinuclear signal. The cytoplasm was defined as the difference between three and 50 applications of the “dilate” function to the area corresponding to Hoechst signal. The background signal in an area without a cell was subtracted from all pixels. The cytoplasmic and nuclear UbFC and yellow fluorescent protein (YFP) fluorescence were quantified and plotted as a scatterplot. At least 30 cells were analyzed for each combination of proteins examined in each experiment. The best linear fit to the data was determined to define the cytoplasm to nuclear (C/N) ratio and SD for each UbFC conjugate by using linear regression in SigmaPlot software. The C/N slope was not materially affected by removal of 10% of the data from any experiment. Experiments were performed using double-blind design to avoid bias.
Analysis of Stoichiometry of Jun Ubiquitination, Rate of Jun Degradation, and Transcription Activation
COS-1 cells transfected with plasmids that expressed the Xpress-Jun and hemagglutinin (HA)-ubiquitin variants indicated were harvested 36 h after transfection. Cell extracts were immunoprecipitated using anti-Xpress antibody and analyzed by immunoblotting by using anti-HA antibody as described previously (Fang and Kerppola, 2004). To measure the rate of Jun degradation, cells were treated with 50 μg/ml cycloheximide, harvested at the indicated times, and analyzed by immunoblotting. The intensities of bands were imaged using nonsaturated exposures and quantified using ImageJ (http://rsb.info.nih.gov/ij/). Transcription activation by Jun was measured by transient transfection of Xpress-Jun or the Xpress vector plasmid together with the pAP1-TA-Luc reporter plasmid (Clontech, Mountain View, CA) and the pCMV-Renilla internal control plasmid. Luciferase activities were measured 24 h after transfection using dual luciferase assay reagents (Promega, Madison, WI).
Derivation of Knockdown Cell Lines
COS-7 cells were transfected with plasmids that contained sequences encoding short hairpin RNA (shRNA) directed against HRS, TSG101, or a control sequence in pSUPER.puro (Oligoengine, Seattle, WA) vectors. Stable clones were selected in the presence of 1 μg/ml puromycin and screened for TSG101 or HRS protein expression by immunoblotting. To restore TSG101 or HRS expression, plasmids encoding the corresponding mouse proteins that differ in the sequences targeted by the shRNAs were transfected into the cells. For more detailed descriptions of the materials and methods used, please see Supplemental Material.
RESULTS
To identify the determinants required for Jun ubiquitination and lysosomal localization of the conjugate in living cells, we used a modified version of the UbFC assay with enhanced sensitivity (see Materials and Methods). We compared the distributions of UbFC conjugates formed by wild-type and mutated Jun and ubiquitin to identify amino acid residues that affected the localization of ubiquitinated Jun. To quantify differences in distribution, we measured the fluorescence intensities in the cytoplasm and nucleus of at least 30 cells for each combination of proteins in each of three independent experiments, and we used the best linear fit to the data to determine the ratio of C/N UbFC conjugates. To determine whether the fluorescent protein fragments fused to Jun and ubiquitin affected Jun ubiquitination, we compared the effects of the same mutations on conjugate formation by proteins lacking the fragments by using immunoprecipitation and immunoblotting to detect the conjugates.
The majority of UbFC conjugates formed by wild-type Jun were cytoplasmic (Figure 1A; C/N = 2.2 ± 0.07), whereas the total pool of mostly nonubiquitinated Jun fused to full-length YFP was primarily nuclear (Supplemental Figure 1B; C/N = 0.4 ± 0.02). Chimeric proteins in which ubiquitin is fused in tandem to protein substrates often mimic ubiquitin conjugates (Haglund et al., 2003; Mosesson et al., 2003). In contrast, fusion of ubiquitin to the N terminus of Jun had no detectable effect on its localization (Supplemental Figure 1C; C/N = 0.5 ± 0.02), suggesting that a specific ubiquitination site, multiple ubiquitins, or both were required for the cytoplasmic localization of Jun conjugates.
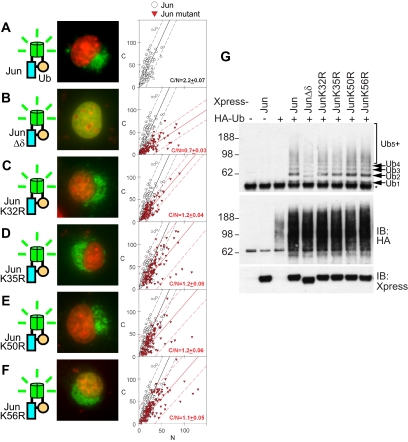
Figure 1.
Localization and stoichiometry of ubiquitin conjugates formed by Jun mutants. (A–F) Distributions of conjugates formed by Jun mutants indicated in the diagrams to the left of each image with wild-type ubiquitin were visualized using UbFC analysis in living COS-7 cells. Cells transfected with plasmids encoding the Jun mutants are indicated, and wild-type ubiquitin fused to complementary fluorescent protein fragments were imaged 24 h after transfection. The UbFC fluorescence (green) was superimposed with Hoechst staining of DNA (red). The diagrams to the left of the images depict the proteins expressed and the mutations in Jun (Jun, blue; ubiquitin chain, orange; fluorescent protein fragments, green). For each combination of proteins, the UbFC fluorescence intensities of the cytoplasm (C) and nucleus (N) were measured in at least 30 cells as described in Materials and Methods. The fluorescence intensities are shown as a scatterplot (graphs to the right of images), with data for each mutant plotted as red triangles and data for wild-type Jun plotted as open circles for reference. The best linear fit to the data is shown as a solid line, and the 95% confidence interval is shown as dashed lines. The slope and SD of the fit are shown in each graph by using red characters for the mutants and black characters for wild-type Jun. Each graph is representative of three independent experiments. (G) The ubiquitination stoichiometries of conjugates formed by the Jun mutants were analyzed by immunoprecipitation followed by immunoblotting. Cells transfected with plasmids encoding the Jun mutants indicated above the lanes fused to the Xpress tag and wild-type ubiquitin fused to the HA tag were lysed, the extracts were immunoprecipitated using anti-Xpress antibodies, and then they were analyzed by immunoblotting using anti-HA antibodies (top). The mobilities of Jun conjugates containing different numbers of ubiquitins (Ubn) as well as cross-reactive bands (*) are indicated to the right of the immunoblot. The same membrane was probed using anti-Xpress antibodies (bottom) to determine the levels of the Jun mutants. Aliquots of the extracts were analyzed by immunoblotting using anti-HA antibodies (middle) to determine the effects of Jun expression on the overall level of HA-ubiquitin conjugation to all cellular proteins.
The δ Region Is Required for the Cytoplasmic Localization of Ubiquitinated Jun
We examined the localization of UbFC conjugates formed by Jun mutants to determine whether the localization of ubiquitinated Jun correlated with its degradation. We focused on the δ region because of its known role in Jun degradation (Treier et al., 1994). Jun lacking the δ region produced predominantly nuclear UbFC conjugates (Figure 1B; C/N = 0.7 ± 0.03). This deletion reduced the amount of conjugates formed, but it did not alter the stoichiometry of ubiquitin in each conjugate (Figure 1G). The discrete ladder of bands detected in the immunoprecipitated samples was shifted by the 3-kDa difference in size between wild-type Jun and Jun lacking the δ region, demonstrating that these bands corresponded to ubiquitinated Jun rather than interaction partners that could be coimmunoprecipitated with Jun. The overall level of ubiquitin conjugates increased in cells that expressed exogenous Jun (Figure 1G, middle), indicating that the ubiquitination of cellular proteins increased in response to Jun expression. Consequently, deletion of the δ region affected both the efficiency of Jun ubiquitination as well as the localization of the conjugates that were formed.
We examined the localization of UbFC conjugates formed by Jun mutants in which individual lysine residues in the δ region were replaced by arginines. These substitutions had intermediate effects on the ratio of cytoplasmic-to-nuclear UbFC conjugates (Figure 1, C–F; C/N = 1.2 ± 0.04, 1.2 ± 0.06, 1.2 ± 0.06, and 1.1 ± 0.05 for K32R, K35R, K50R, and K56R mutants, respectively), and they had no detectable effect on the level or stoichiometry of Jun ubiquitination (Figure 1G). These results suggest that the lysine residues in the δ region had partially redundant effects on the cytoplasmic localization of ubiquitinated Jun.
Specific Recognition of Ubiquitin Is Required for the Cytoplasmic Localization of Ubiquitinated Jun
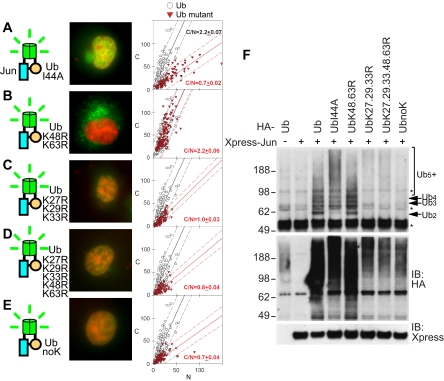
To investigate whether specific recognition of ubiquitin was required for the cytoplasmic localization of UbFC conjugates formed by Jun, we substituted isoleucine-44 in ubiquitin by alanine. This mutation eliminates binding by most ubiquitin recognition motifs that have been characterized previously (Beal et al., 1996; Hicke et al., 2005). The I44A mutation in ubiquitin (UbI44A) markedly reduced the cytoplasmic localization of UbFC conjugates formed by Jun (Figure 2A; C/N = 0.7 ± 0.02). Cells that expressed ubiquitin containing the I44A mutation produced more ubiquitin conjugates, and they had a higher mean stoichiometry of Jun ubiquitination than cells that expressed wild-type ubiquitin (Figure 2F). A low level of ubiquitinated Jun was detected in the cytoplasm of cells that expressed UbI44A (Figure 2A). These conjugates could be localized to the cytoplasm independently of I44 recognition, but they could also represent mixed conjugates that contained endogenous ubiquitin. In either case, the low efficiency of cytoplasmic localization of ubiquitinated Jun in cells that expressed UbI44A indicates that endogenous ubiquitin did not prevent detection of the effects of mutations in ubiquitin on the localization of ubiquitinated Jun. Likewise, the higher level and mean stoichiometry of Jun conjugates in cells that expressed ubiquitin containing the I44A mutation indicates that endogenous ubiquitin did not prevent detection of the effects of mutations in ubiquitin on the stoichiometry of Jun ubiquitination. Although the cells have a high level of endogenous ubiquitin, the majority of the ubiquitin is linked in conjugates, and it does not compete with conjugation by the exogenously expressed ubiquitin mutants. Endogenous ubiquitin is therefore unlikely to affect interpretation of the results (see Supplemental Material).
Figure 2.
Effects of mutations in ubiquitin on the localization and stoichiometry of ubiquitinated Jun. (A–E) Distributions of conjugates formed by the ubiquitin mutants indicated in the diagrams to the left of each image with wild-type Jun were visualized using UbFC analysis in living COS-7 cells as described in Figure 1, A–F. (F) The stoichiometries of conjugates formed by the ubiquitin mutants indicated above the lanes with wild type Jun were analyzed by immunoprecipitation followed by immunoblotting as described in Figure 1G (top). The levels of Jun expression and HA-ubiquitin mutant conjugation to all cellular proteins were determined by immunoblotting by using anti-Xpress and anti-HA antibodies, respectively (bottom and middle).
The multiple ubiquitins conjugated to Jun could be linked to different lysine residues or joined as a chain. We examined whether the replacement of lysines in ubiquitin with arginines affected the localization of UbFC conjugates formed by Jun. K48R and K63R substitutions individually or in combination had little effect on the fluorescence intensity or localization of UbFC conjugates formed by Jun (Figure 2B, data not shown; C/N = 1.7 ± 0.16, 2.2 ± 0.19, and 2.2 ± 0.06 for UbK48R, UbK63R, and UbK48.63R, respectively). In contrast, the K27R, K29R, and K33R substitutions together as well as in combination with other substitutions reduced the fluorescence intensity of UbFC conjugates formed by Jun and inhibited their cytoplasmic localization (Figure 2, C–E; C/N = 1.0 ± 0.03, 0.8 ± 0.04, and 0.7 ± 0.04 for UbK27.29.33R, UbK27.29.33.48.63R, and UbnoK, respectively). Substitution of lysine residues in ubiquitin caused changes in the levels of ubiquitinated Jun detected by immunoprecipitation and immunoblot analysis that corresponded closely to the changes in the fluorescence intensities of UbFC conjugates formed by ubiquitin fusions containing the same substitutions (Figure 2F).
Ubiquitin Containing Only Lysine-27 Can Support Polyubiquitination of Jun
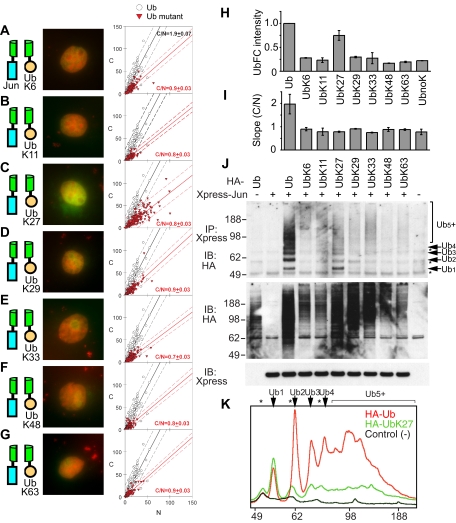
To determine whether individual lysine residues in ubiquitin were sufficient for Jun ubiquitination and to establish the localization of these conjugates, we performed UbFC analysis by using ubiquitin variants in which all lysines but one were replaced by arginines. Cells that expressed ubiquitin containing only lysine-27 (UbK27) had 75% of the fluorescence intensity of cells that expressed wild-type ubiquitin (Figure 3, C and H). The fluorescence intensities produced by other single-lysine variants were similar to that produced by lysine-less ubiquitin (Figure 3, A and B, D–H). The UbFC conjugates produced by all single-lysine ubiquitins were predominantly nuclear (Figure 3I, C/N <1 for all variants). These results show that ubiquitin containing only K27 was sufficient for UbFC conjugate formation but that UbK27 alone was not sufficient for efficient lysosomal localization of the conjugates. The stabilities of conjugates formed by different ubiquitin variants could affect their steady-state levels, but it is likely that K27 in ubiquitin affected Jun ubiquitination rather than conjugate stability alone (see Discussion).
Figure 3.
Ubiquitination of Jun by single-lysine ubiquitin variants and their effects on conjugate localization and stoichiometry. (A–G) Distributions of conjugates formed by the single-lysine ubiquitin variants indicated in the diagrams to the left of each image with wild-type Jun were visualized using UbFC analysis in living COS-7 cells as described in Figure 1, A–F. (H) Mean UbFC intensities of cells that expressed the single-lysine ubiquitin variants indicated below the bars and wild-type Jun. The data represent the mean and SD of results from three independent experiments, normalized by the mean fluorescence intensity of cells that expressed wild-type ubiquitin. (I) Slopes of best fits to the cytoplasmic and nuclear fluorescence intensities of cells that expressed the ubiquitin variants indicated above the bars and wild-type Jun. (J) Stoichiometries of Jun ubiquitination by the single-lysine ubiquitin variants indicated above the lanes were analyzed by immunoprecipitation and immunoblot analysis as described for Figure 1G (top). The levels of Jun expression and HA-ubiquitin mutant conjugation to all cellular proteins were determined by immunoblotting using anti-Xpress and anti-HA antibodies (bottom and middle, respectively). (K) Quantitation of the stoichiometries of Jun ubiquitination by wild-type and K27-only ubiquitin. The lanes corresponding to the ubiquitin variants indicated in the top right corner of the graph were analyzed to determine the relative amounts of conjugates with different ubiquitination stoichiometries. The mobilities of Jun conjugates containing different numbers of ubiquitins (Ubn) as well as cross-reactive bands (*) are indicated above the graph.
We examined the stoichiometries of Jun ubiquitination by the single-lysine ubiquitin variants. Cells that expressed ubiquitin containing only K27 produced 50–100% more monoubiquitinated Jun (Ub1) than cells that expressed the corresponding wild-type ubiquitin (Figure 3, J and K, and Supplemental Table 1). In the same cells, diubiquitinated Jun (Ub2) was reduced by >90%, oligoubiquitinated Jun (Ub3-7) by 85–90%, and polyubiquitinated Jun (Ub8+) by 75–85%. The maximum size of conjugates produced in cells that expressed ubiquitin containing only K27 was approximately the same as that produced in cells that expressed wild-type ubiquitin, indicating that the processivity of ubiquitination was comparable for K27-only and wild type ubiquitin. Although we cannot strictly exclude the possibility of multiple monoubiquitination, many lines of evidence suggest that Jun is polyubiquitinated by a ubiquitin chain (see Supplemental Material). Low levels of ubiquitinated Jun were produced in cells that expressed ubiquitins containing only K6, K29, or K33. Other proteins whose ubiquitination was enhanced in cells that expressed exogenous Jun were also preferentially ubiquitinated by UbK27, UbK29, and UbK33, suggesting that some of these proteins were ubiquitinated by related mechanisms (Figure 3J, middle). Single-lysine variants of ubiquitin other than UbK27 did not support monoubiquitination of Jun, indicating that K27 in ubiquitin affected both mono- and polyubiquitination of Jun. Thus, UbFC analysis and conjugates detected in cell extracts corroborate the unique role of K27 of ubiquitin in Jun ubiquitination.
The synthesis of polyubiquitin conjugates proceeds through a series of cycles of ubiquitin addition. We compared the relative amounts of conjugates that differ by one ubiquitin between cells that expressed wild-type and K27-only ubiquitin. At steady state, the ratio between oligomers that differ by one ubiquitin reflects the interconversion between different oligomeric states. Cells that expressed ubiquitin containing only K27 had a markedly reduced ratio of mono- to diubiquitinated Jun compared with cells that expressed wild-type ubiquitin, but there was little difference for other cycles of ubiquitin addition/removal (Figures 3K and 4I and Supplemental Table 1). The small increase in the ratio of tri- to diubiquitinated Jun in cells that expressed ubiquitin containing only K27 could be due to the depletion of diubiquitinated Jun. The reduced efficiency of Jun diubiquitination in cells that expressed K27-only ubiquitin was propagated through the chain of ubiquitin addition cycles, reducing the amount of polyubiquitinated Jun. The shift in the stoichiometry of Jun ubiquitination in cells that expressed K27-only compared with wild-type ubiquitin was specific to Jun, because there was little change in the size distribution of ubiquitin conjugates formed on other proteins in the same cells (Figures 3J and 4G, middle). The stoichiometry of Jun ubiquitination could be affected by either ubiquitination or deubiquitination, but we favor the interpretation that the changes observed in these experiments reflect changes in ubiquitination (see Discussion).
Figure 4.
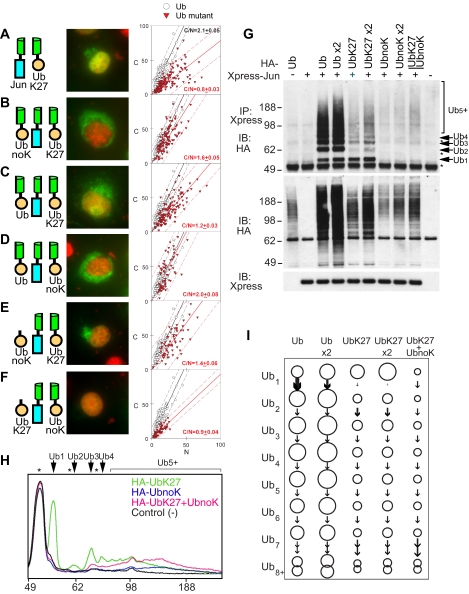
Effects of coexpression of ubiquitin variants on the localization and stoichiometry of ubiquitinated Jun. (A–F) Distributions of conjugates formed by the combinations of ubiquitin variants indicated in the diagrams to the left of each image with wild-type Jun were visualized using UbFC analysis in living COS-7 cells as described in Figure 1, A–F. Cells that were cotransfected with plasmids encoding the ubiquitin variants indicated fused to either a fluorescent protein fragment (UbK27 and UbnoK) or the HA tag (HA-UbK27 and HA-UbnoK) together with wild-type Jun fused to a complementary fragment were imaged 24 h after transfection. (G) Stoichiometries of Jun ubiquitination by different amounts and combinations of the ubiquitin variants indicated above the lanes were analyzed by immunoprecipitation and immunoblot analysis as described for Figure 1G (top). The levels of Jun expression and HA-ubiquitin mutant conjugation to other cellular proteins were determined by immunoblotting using anti-Xpress and anti-HA antibodies, respectively (bottom and middle). In lanes indicated x2, twice the standard amount of plasmid encoding the ubiquitin fusion was transfected. The bar above lane 9 indicates that plasmids encoding the two ubiquitin variants were cotransfected into the cell. (H) Quantitation of the stoichiometries of Jun ubiquitination by combinations of ubiquitin variants. The lanes corresponding to the ubiquitin variants indicated in the top right corner of the graph were analyzed as described for Figure 3K to determine the relative amounts of conjugates with different ubiquitination stoichiometries. (I) Comparison of the relative amounts of Jun conjugates with different stoichiometries of ubiquitination in cells transfected with different amounts and combinations of plasmids encoding different ubiquitin variants. The area of each circle indicates the relative amount of Jun conjugates formed by the ubiquitin variant(s) indicated above each column with the number of ubiquitins indicated to the left of each row. The arrows between the circles indicate the net efficiency of conversion of the lower stoichiometry conjugate to the higher stoichiometry conjugate. For numerical values and more details, see Supplemental Table S1.
Enhancement of the Cytoplasmic Localization of Ubiquitinated Jun by Coexpression of Ubiquitin Variants
To investigate whether a combination of ubiquitin variants containing different lysine residues facilitated the cytoplasmic localization of ubiquitinated Jun, we coexpressed UbK27 together with other single-lysine ubiquitins. Coexpression of UbK27 with each of the other single-lysine ubiquitins did not facilitate the cytoplasmic localization of UbFC conjugates formed by Jun (Supplemental Figure 2, A–F; C/N <1 for all combinations). Surprisingly, coexpression of UbK27 with lysine-less ubiquitin (UbnoK) enhanced the cytoplasmic localization of UbFC conjugates (Figure 4B; C/N = 1.6 ± 0.05) compared with expression of these ubiquitin variants separately (Figure 4A, C/N = 0.8 ± 0.03 and Figure 2E, C/N = 0.7 ± 0.04). Transfection of two equivalents of the plasmid encoding either ubiquitin variant separately did not enhance the cytoplasmic localization of ubiquitinated Jun (data not shown). Coexpression of either UbK27 or UbnoK with wild-type ubiquitin did not enhance the cytoplasmic localization of the conjugates (Figure 4C, C/N = 1.2 ± 0.03 and Figure 4D, C/N = 2.0 ± 0.08). Thus, UbK27 and lysine-less ubiquitin had a combinatorial effect on the localization of ubiquitinated Jun.
We examined the effect of coexpression of ubiquitin containing only K27 and lysine-less ubiquitin on the stoichiometry of Jun ubiquitination. Coexpression of these ubiquitin variants reduced the total amount of ubiquitinated Jun, but it increased the mean stoichiometry of ubiquitination due to selective reduction of the levels of low-stoichiometry conjugates (Figure 4, G–I). The level of Ub1 was reduced by >90%, but there was no significant difference in the amount of Ub8+ whether ubiquitin containing only K27 and lysine-less ubiquitin were expressed together or separately (Figure 4, H and G, and Supplemental Table 1). The ratio of mono- to diubiquitinated Jun could not be determined accurately because of the low levels of these conjugates. However, subsequent cycles of ubiquitin addition/removal were comparable in cells that coexpressed UbK27 with lysine-less ubiquitin and in cells that expressed UbK27 alone. The coexpression of lysine-less ubiquitin therefore preferentially suppressed the synthesis of low-stoichiometry ubiquitin conjugates of Jun. There was no detectable change in the ubiquitination of other proteins observed by immunoblotting of extracts from cells that expressed these ubiquitin variants together and separately (Figure 4G, middle). The increased mean stoichiometry of Jun ubiquitination in cells that coexpressed UbK27 with lysine-less ubiquitin correlated with cytoplasmic localization of the conjugates.
To determine whether the level of ubiquitin expression affected the stoichiometry of Jun ubiquitination, we compared cells transfected with one and two equivalents of the plasmids that expressed wild-type, K27-only, and lysine-less ubiquitin. Doubling the amount of plasmid transfected nearly doubled the amount of ubiquitinated Jun detected in cells that expressed either wild-type or K27-only ubiquitin. There was a slightly larger increase in the level of monoubiquitinated Jun and progressively smaller changes in the levels of higher stoichiometry ubiquitin conjugates (Figure 4, G and I, and Supplemental Table 1). The level of ubiquitin expression therefore primarily affected the efficiency of monoubiquitination, and it had little effect on the relative amounts of other ubiquitin conjugates. The low level of conjugates produced by lysine-less ubiquitin alone was not significantly affected by the amount of plasmid transfected. These could represent endogenous ubiquitin chains capped by lysine-less ubiquitin. The amount of ubiquitin expressed therefore did not account for the effect of coexpression of lysine-less ubiquitin with K27-only ubiquitin on the stoichiometry of Jun ubiquitination. We hypothesize that lysine-less ubiquitin preferentially inhibited monoubiquitination of Jun (see Supplemental Material).
To investigate whether fusion of both UbK27 and UbnoK to fluorescent protein fragments was required to enhance of the cytoplasmic localization of UbFC conjugates, we coexpressed the two variants, but we fused only one of them to a fluorescent protein fragment. Coexpression of lysine-less ubiquitin lacking a fusion (HA-UbnoK) with UbK27 fused to a fluorescent protein fragment enhanced the cytoplasmic localization of UbFC conjugates (Figure 4E; C/N = 1.4 ± 0.06). In contrast, coexpression of UbK27 lacking a fusion (HA-UbK27) with lysine-less ubiquitin fused to a fluorescent protein fragment (UbnoK) had little effect on the localization of the low level of UbFC conjugates formed (Figure 4F; C/N = 0.9 ± 0.04). In the former case, the fluorescent conjugates contain UbK27, but they may not contain HA-UbnoK, whereas in the latter case, the fluorescent conjugates contain UbnoK, but they may not contain HA-UbK27. The nonequivalent roles of UbK27 and UbnoK are consistent with the model that UbnoK expression enhances the cytoplasmic localization of UbFC conjugates by increasing the stoichiometry of UbK27 conjugated to Jun.
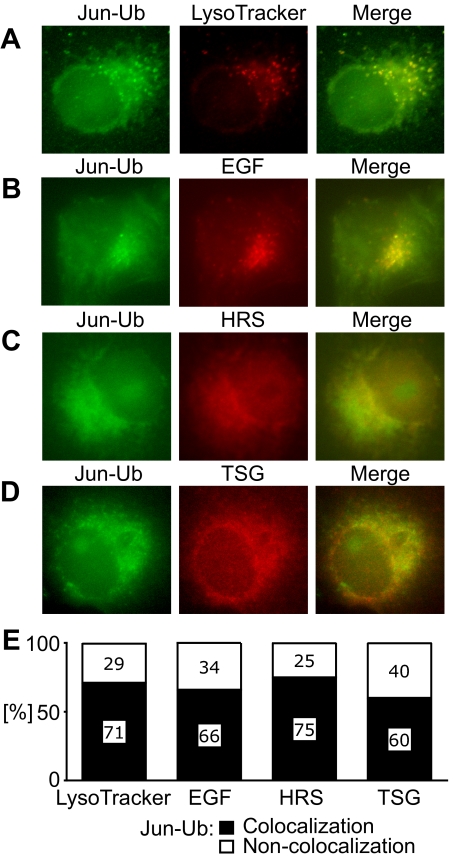
Ubiquitinated Jun Colocalizes with Internalized EGF Receptor, HRS, and TSG101
To identify proteins that could affect the localization of ubiquitinated Jun, we compared the distribution of UbFC conjugates formed by Jun with markers of various subcellular compartments. The majority of UbFC conjugates colocalized with LysoTracker Red in small cytoplasmic foci in living cells (Figure 5A). The UbFC conjugates were also found in a diffuse distribution that frequently formed a perinuclear halo. Similar distributions of UbFC conjugates were observed in HEK293T, COS-1, and COS-7 cells. The foci of UbFC conjugates and part of the diffuse fluorescence colocalized with Texas Red-labeled EGF bound to internalized EGF receptor (Figure 5B). We observed no colocalization with markers of early or late endosomes, proteasomes, or autophagosomes (Supplemental Figure 3, A–E).
Figure 5.
Colocalization of ubiquitinated Jun with ubiquitin-binding proteins. The distribution of ubiquitinated Jun, visualized using UbFC analysis (green), was compared with LysoTracker Red (A) and EGF-rhodamine labeling (B) as well as anti-HRS (C) and anti-TSG101 (D) immunofluorescence (red). Cells that were transfected with plasmids encoding Jun and ubiquitin fused to the fluorescent protein fragments were either incubated with LysoTracker Red or EGF-rhodamine and imaged live or the cells were fixed and anti-HRS or anti-TSG101 antibodies were used to image their distributions. Images were acquired by fluorescence microscopy by using filters selective for UbFC fluorescence (left) and the respective labels (center). The images were superimposed (right). (E) Percentage of cells where more than half of the UbFC fluorescence overlapped with the fluorescence of the markers. At least 30 cells that had UbFC fluorescence intensities higher than the background signal in nontransfected cells but lower than the 10% of cells with the highest fluorescence intensities were analyzed.
Lysosomal localization of the EGF receptor is thought to involve HRS (also called HGS) (Komada and Kitamura, 1995) as well TSG101 (Lu et al., 2003). Both of these proteins require I44 in ubiquitin for binding (Bishop et al., 2002; Hicke et al., 2005; Hurley and Emr, 2006), suggesting their potential involvement in the lysosomal localization of ubiquitinated Jun. We compared the distributions of HRS and TSG101 with UbFC conjugates formed by Jun. HRS and TSG101 were distributed in spotted patterns that colocalized with UbFC conjugates formed by Jun in 75 and 60% of cells, respectively (Figure 5, C and D).
The level of exogenous Jun expression in the transfected cells was, on average, 10-fold higher than the level of endogenous Jun in resting cells (Supplemental Figure 4C). This difference is comparable with the increase in the level of Jun expression in response to growth factor stimulation. The linear relationship between the fluorescence intensities of the cytoplasm and the nucleus of cells that had overall fluorescence intensities that varied by more than an order of magnitude indicates that the level of ubiquitinated Jun in this range did not affect the efficiency of cytoplasmic localization of the conjugates (see graphs in Figures 1–4). However, we noticed that cells with higher fluorescence intensities exhibited a lower colocalization of UbFC conjugates with HRS, TSG101, and LysoTracker dyes (Supplemental Figure 4A). Overexpression of Jun at 100-fold higher levels than endogenous Jun, on average, resulted in nuclear accumulation of the UbFC conjugates in a majority of cells, suggesting that the pathway for lysosomal localization was saturated (Supplemental Figure 4B).
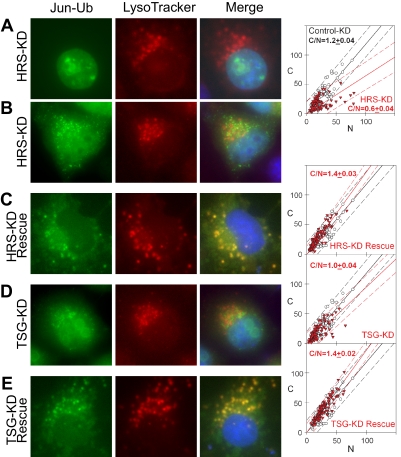
Effects of HRS and TSG101 Knockdown on the Localization of Ubiquitinated Jun
We used cell lines that stably expressed shRNAs directed against HRS and TSG101 to test their roles in the localization of ubiquitinated Jun (Supplemental Figure 5). To analyze the distributions of UbFC conjugates in HRS and TSG101 knockdown cell lines, we compared their localization with LysoTracker Red in these cells (Figure 6). UbFC conjugates formed by Jun in HRS knockdown cells were localized to the nucleus and cytoplasmic foci that did not colocalize with LysoTracker Red (Figure 6, A and B). The ratio of cytoplasmic to nuclear UbFC conjugates in HRS knockdown cells was lower than that observed in control cells (Figure 6, A and B; C/N = 0.6 ± 0.04 vs. 1.2 ± 0.04). Reexpression of HRS in these cells restored the colocalization of UbFC conjugates with LysoTracker Red and increased the cytoplasmic localization of the conjugates (Figure 6C; C/N = 1.4 ± 0.03).
Figure 6.
Localization of ubiquitinated Jun in HRS and TSG101 knockdown cells. (A–E) Distributions of ubiquitinated Jun visualized using UbFC analysis (left) were compared with the distribution of LysoTracker Red (middle) in HRS (A and B) and TSG101 (D) knockdown cells as well as in knockdown cells where HRS (C) or TSG101 (E) was reexpressed. The UbFC (green) and LysoTracker Red (red) fluorescence were superimposed together with Hoechst staining of DNA (blue) to produce merged images (right). The fluorescence intensities in the cytoplasm and nucleus of individual cells were plotted as described for Figure 1, A–F. Data for each knockdown and rescued cell line were plotted using red triangles and data for the control cell line that expressed an unrelated shRNA were plotted using open circles. Each plot is representative of at least three independent experiments.
The UbFC conjugates formed by Jun in TSG101 knockdown cells were also localized to the nucleus and cytoplasmic foci that did not colocalize with LysoTracker Red (Figure 6D). The cytoplasmic to nuclear ratio in these cells was marginally lower than that observed in control cells (Figure 6D; C/N = 1.0 ± 0.04 vs. 1.2 ± 0.04). Reexpression of TSG101 restored the colocalization of UbFC conjugates with LysoTracker Red (Figure 6E; C/N = 1.4 ± 0.02). Together, these results indicate that both HRS and TSG101 contributed to the lysosomal localization of ubiquitinated Jun.
Roles of HRS and TSG101 in Jun Degradation and Transcription Activation
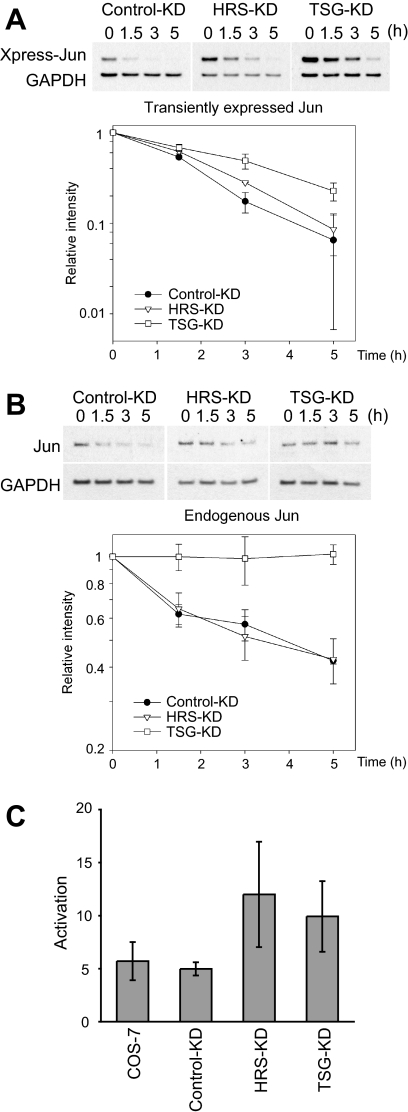
We examined the turnover rates of both transiently expressed and endogenous Jun in HRS and TSG101 knockdown cells. Transiently expressed Jun had a half-life of 1.5 h in the parental cells as well as in cells that expressed the control shRNA (Figure 7A). The half-life increased to ∼2 h in HRS knockdown cells and 3 h in TSG101 knockdown cells. Transiently expressed Jun accumulated at higher levels in TSG101 knockdown cells than in HRS knockdown cells, which in turn had higher levels than control cells (Figure 7A). Reexpression of either TSG101 or HRS reduced the rate of Jun turnover in both control and knockdown cells, possibly because of secondary effects of overexpression (data not shown).
Figure 7.
Turnover rates of transiently expressed and endogenous Jun in HRS and TSG101 knockdown cells and transcription activation. (A) Turnover rates of transiently expressed Jun in control, HRS, and TSG101 knockdown cells. Xpress-tagged Jun was transfected into cells that expressed control shRNA (Control-KD), HRS shRNA (HRS-KD), or TSG101 shRNA (TSG-KD). The cells were treated with cycloheximide and incubated for the times indicated above the lanes before analysis of the cell extracts by immunoblotting using a mixture of anti-Xpress and anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) antibodies. (B) Turover rates of endogenous Jun in control, HRS, and TSG101 knockdown cells. Cells that expressed control shRNA (Control-KD), HRS shRNA (HRS-KD), or TSG101 shRNA (TSG-KD) were treated with cycloheximide and incubated for the times indicated above the lanes before analysis of the cell extracts by immunoblotting using anti-Jun and anti-GAPDH antibodies. The optical densities of the bands corresponding to Jun and GAPDH were measured from multiple nonsaturated exposures, and they were quantified using ImageJ. The ratio was plotted as a function of time after cycloheximide addition. The data represent the mean and SD from three independent experiments for both transiently expressed and endogenous Jun. (C) Activation of reporter gene expression by transiently expressed Jun in COS-7, control, HRS, and TSG101 knockdown cells. Transcription activation by Jun was quantified by measuring the ratio between the normalized luciferase activities produced in cells transfected with Xpress-Jun versus the Xpress vector plasmid together with the pAP1-TA-Luc reporter plasmid and the pCMV-Renilla internal control plasmid. The data represent the mean and SD of three replicates from two independent experiments.
Endogenous Jun had a half-life of ∼3 h in control cells (Figure 7B). There was no significant change in the half-life of endogenous Jun in HRS knockdown cells. In contrast, endogenous Jun was markedly stabilized in TSG101 knockdown cells where no reduction in Jun levels was observed over 5 h. The levels of endogenous Jun in the HRS and TSG101 knockdown cells were comparable with the level observed in the control cells, suggesting feedback regulation of Jun expression.
We examined transcription activation by Jun in HRS and TSG101 knockdown cells by using a reporter gene assay. Transiently expressed Jun increased reporter gene activity to a greater extent in HRS and TSG101 knockdown cells compared with either control knockdown cells or the parental cell line (Figure 7C). It was not possible to determine whether this increase in transcription activation potential was due to altered localization or degradation of Jun, because the overexpression of HRS and TSG101 enhanced transcription activation by Jun in all of the cell lines, reminiscent of their effects on the turnover rates of Jun.
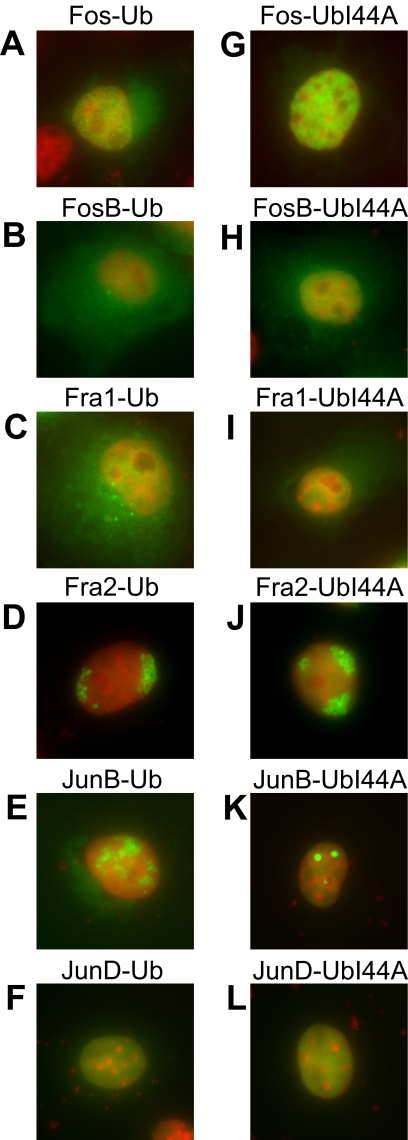
Effects of Ubiquitination on the Localization of Fos and Jun Family Proteins
We investigated the effects of ubiquitination on the localization of other Fos and Jun family proteins by using UbFC analysis. The total population of each Fos and Jun family protein fused to YFP or CFP was predominantly nuclear (data not shown). UbFC conjugates formed by Fos had a diffuse distribution in part of the cytoplasm as well as in the nucleoplasm (Figure 8A). UbFC conjugates formed by FosB were distributed throughout the cell, except for the nucleoli (Figure 8B). UbFC conjugates formed by Fra1 were enriched in small foci near the nucleus in addition to a uniform distribution throughout the cell except for the nucleoli (Figure 8C). The I44A mutation in ubiquitin eliminated the cytoplasmic localization of these conjugates, indicating that the cytoplasmic localization of UbFC conjugates formed by Fos, FosB, and Fra1 required specific recognition by ubiquitin-binding proteins (Figure 8, G–I). The distinct distributions of these UbFC conjugates suggest that they do not reflect the ubiquitination of a shared interaction partner such as Jun.
Figure 8.
Localization of ubiquitinated Fos and Jun family members and effects of the I44A mutation in ubiquitin on their localization. The distributions of ubiquitinated Fos and Jun family proteins were visualized using UbFC analysis (green). The distributions of conjugates formed by wild-type ubiquitin (A–F) and the I44A mutant (G–L) were compared. The UbFC fluorescence (green) was superimposed with Hoechst staining of DNA (red). Each image is representative of >60% of the cells in the population.
UbFC conjugates formed by Fra2 were enriched near the nuclear periphery (Figure 8D). UbFC conjugates formed by JunB were enriched in irregular areas within the nucleus (Figure 8E). UbFC conjugates formed by JunD were uniformly distributed in the nucleoplasm (Figure 8F). The I44A mutation in ubiquitin altered the subnuclear distribution of the UbFC conjugates formed by JunB, but it did not appreciably affect the distributions of the UbFC conjugates formed by Fra2 or JunD (Figure 8, J–L). Thus, ubiquitin conjugates formed by different Fos and Jun family members had distinct distributions, and these distributions were differentially affected by the I44A mutation in ubiquitin. The distinct distributions of the UbFC conjugates formed by all Fos and Jun family proteins except for JunD compared with the distributions of the total populations of these proteins support the interpretation that these conjugates represented specific complexes rather than spontaneous association of the fluorescent protein fragments. Additional studies of these ubiquitin conjugates are needed to elucidate the mechanisms that control their localization and functions.
DISCUSSION
Many nuclear proteins are translocated to the cytoplasm upon ubiquitination, often as a prelude to their degradation (Tomoda et al., 1999; Fukuchi et al., 2001; Li et al., 2003; Fang and Kerppola, 2004). Most such proteins are thought to be degraded by proteasomes. Jun is the first nuclear protein found to be localized to lysosomes for degradation. The novelty of this degradation pathway is reflected by the unique structure of the polyubiquitin chain conjugated to Jun. The specificity of ubiquitinated Jun translocation to lysosomes is corroborated by the distinct distributions of UbFC conjugates formed by other Fos and Jun family proteins. These differences are also consistent with the dissimilar rates of degradation of different family members
A Specific Conjugate Structure Is Required for Lysosomal Localization of Ubiquitinated Jun
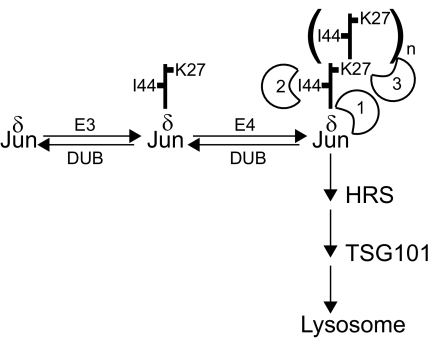
Three distinct determinants for the localization of ubiquitinated Jun to lysosomes were identified: 1) the δ region of Jun, 2) I44 in ubiquitin, and 3) a polyubiquitin chain (Figure 9). In contrast to many other substrates of ubiquitination, the translational fusion of ubiquitin to Jun had no detectable effect on its localization, consistent with the role of a specific conjugate structure in the lysosomal localization of ubiquitinated Jun.
Figure 9.
Model for Jun ubiquitination and recognition of determinants for lysosomal localization. Jun ubiquitination is mediated by at least two distinct mechanisms, here represented by separate E3 and E4 enzyme complexes. Lysosomal localization of ubiquitinated Jun requires recognition of the δ region of Jun (1), I44 on ubiquitin (2), and a polyubiquitin chain (3). These determinants could be recognized by one or more proteins either simultaneously or sequentially. The thick vertical line represents ubiquitin with the N terminus pointing up.
The δ region was required for localization of ubiquitinated Jun to lysosomes, and it enhances the rate of Jun degradation (Treier et al., 1994). Deletion of the δ region reduced the level of ubiquitinated Jun, but it did not affect the stoichiometry of ubiquitin in each conjugate. Individual lysine residues within the delta region had partially redundant roles in the lysosomal localization of ubiquitin conjugates, consistent with their redundant effects on Jun degradation (Treier et al., 1994). The correspondence between the effects of mutations in Jun on lysosomal localization and degradation together with the stabilization of Jun by inhibitors of lysosomal proteases (Fang and Kerppola, 2004) indicates that lysosomes are an important site for Jun degradation.
A Unique Role for K27 of Ubiquitin in Jun Ubiquitination
Isopeptide linkages involving all lysine residues in ubiquitin have been detected by mass spectrometry of total cellular protein (Peng et al., 2003). Only a subset of the linkages has been associated with specific protein substrates, and even fewer have known biological functions (Pickart and Fushman, 2004). K27 in ubiquitin was the only lysine residue that supported both mono- and polyubiquitination of Jun, suggesting that it was either recognized by the E3 ligase or stabilized the conjugates. Jun is the first protein found to be ubiquitinated by K27-only ubiquitin to the exclusion of other single-lysine ubiquitins.
Cells that expressed K27-only versus wild-type ubiquitin had different stoichiometries of Jun ubiquitination. K27-only ubiquitin selectively reduced the conversion of mono- to diubiquitinated Jun. We hypothesize that monoubiquitination and subsequent chain extension are mediated by different mechanisms and that the transition between these mechanisms is affected by lysine residues other than K27 in ubiquitin. One model consistent with the data is that separate E3 and processive E4 ubiquitin ligases catalyze conjugation of the first and subsequent ubiquitins to Jun (Figure 9). The reduced efficiency of chain extension for mono-K27-only ubiquitinated Jun could be due to a difference in conformation, localization, interactions, or E4 ligase recognition between Jun monoubiquitinated by wild-type versus K27-only ubiquitin. Discrimination among these and other mechanisms requires comparison of the characteristics of these monoubiquitinated Jun conjugates.
The effects of the level of ubiquitin expression as well as the coexpression of lysine-less ubiquitin on Jun ubiquitination are consistent with the model that the first and subsequent ubiquitins are conjugated by different mechanisms (Figure 9). Increased levels of ubiquitin preferentially enhanced monoubiquitination, whereas coexpression of lysine-less ubiquitin inhibited monoubiquitination. Neither condition affected the efficiencies of subsequent cycles of ubiquitin addition/removal. The mechanisms whereby these conditions selectively affect monoubiquitination remain unknown. A simulation of ubiquitin conjugation based on this model can predict the relative amounts of conjugates with different stoichiometries of Jun ubiquitination with a high degree of accuracy (Supplemental Figure 6).
An alternative interpretation for the unique effect of K27-only ubiquitin on the conversion of mono- to diubiquitinated Jun is that a non-K27 isopeptide linkage is favored only at this step in the pathway for Jun ubiquitination. This interpretation does not explain why coexpression of UbK27 with each of the other single-lysine variants did not enhance the cytoplasmic localization of ubiquitinated Jun. It also does not explain the changes in the ratio of mono- to oligoubiquitinated Jun in response to the level of ubiquitin expression or the coexpression of lysine-less ubiquitin. In either case, all subsequent cycles of ubiquitin addition were equally efficient for K27-only and wild-type ubiquitin, indicating that K27-isopeptide linkages are likely to be prevalent in ubiquitinated Jun.
Potential Effects of Conjugate Degradation and Deubiquitination on the Detection of Ubiquitinated Jun
Jun ubiquitinated by a K27-linked polyubiquitin chain could be more stable than Jun ubiquitinated by wild-type ubiquitin. However, it is unlikely that K27-linked ubiquitin conjugates were the only single-lysine ubiquitin conjugates detected solely because of conjugate stability. The UbFC signal intensity of cells that expressed UbK27 was lower than that of cells that produced wild-type ubiquitin. Coexpression of UbK27 with wild-type ubiquitin did not enhance the intensity of the UbFC signal compared with the expression of wild-type ubiquitin alone. Monoubiquitination of Jun was detected only in cells that expressed UbK27, although the structures of monoubiquitin conjugates formed by other single-lysine ubiquitin variants would differ only at the individual lysine residues. The detection of only K27-linked Jun ubiquitination was therefore likely due to the selective synthesis and not to the stabilization of these conjugates.
The changes in the stoichiometry of Jun ubiquitination in cells that expressed different ubiquitin mutants could be due to altered ubiquitination or deubiquitination. We favor the interpretation that the changes were caused by altered ubiquitination. Although it is possible that Jun diubiquitinated by K27-only ubiquitin is more susceptible to deubiquitination, it is not obvious why this effect would only affect the diubiquitin conjugate and not higher stoichiometry conjugates. In addition, the increase in the level of monoubiquitinated Jun in cells that expressed higher levels of ubiquitin as well as the decrease in the level of monoubiquitinated Jun in cells that coexpressed lysine-less ubiquitin was consistent with their predicted effects on ubiquitin conjugation but are difficult to explain based on changes in deubiquitination. The association between the fluorescent protein fragments could also stabilize ubiquitin conjugates by preventing deubiquitination. However, because the results of UbFC analysis were consistent with those obtained using immunoprecipitation and immunoblotting of epitope-tagged proteins, the association of the fluorescent protein fragments did not affect the selectivity of ubiquitination by the proteins examined. Although deubiquitination could regulate the extent of Jun ubiquitination, it seems that the differences in ubiquitination observed in the present experiments were primarily caused by changes in ubiquitin conjugation.
Relationship between the Stoichiometry of Ubiquitination and the Localization of Ubiquitinated Jun
The correlation between the lower stoichiometry and reduced lysosomal localization of ubiquitinated Jun in cells that expressed UbK27 and the higher stoichiometry and increased lysosomal localization in cells that coexpressed UbK27 and lysine-less Jun suggested that the stoichiometry of Jun ubiquitination affected the efficiency of lysosomal localization. The exact relationship between the number of ubiquitins added and the efficiency of lysosomal localization cannot be determined based on these data, but it seems that monoubiquitinated Jun was not efficiently localized to lysosomes. We found previously that ubiquitinated Jun, including conjugates with low stoichiometries of ubiquitination, copurify with lysosomes during subcellular fractionation (Fang and Kerppola, 2004). It is possible that the ubiquitin conjugate is edited during or subsequent to lysosomal localization as observed in the case of ubiquitinated EGF receptor trafficking (Hicke et al., 2005; Hurley and Emr, 2006). Alternatively, the stoichiometry of Jun ubiquitination could be altered by ubiquitin hydrolases or nonspecific proteases during cell fractionation. In the present experiments, the subcellular localization was visualized in living cells and the stoichiometry of ubiquitination was determined under conditions designed to minimize the possibility of conjugate remodeling subsequent to cell lysis. We therefore interpret the results to indicate that polyubiquitination was required for lysosomal localization, but we cannot exclude the possibility of conjugate remodeling during or after translocation.
Roles of Ubiquitin-binding Proteins in Lysosomal Localization and Degradation of Ubiquitinated Jun
The requirement for specific sequences in Jun and in ubiquitin and the effects of HRS and TSG101 knockdown on the localization of ubiquitinated Jun are consistent with an active mechanism for the lysosomal localization of ubiquitinated Jun. The accumulation of conjugates with a high stoichiometry of ubiquitin containing the I44A substitution is consistent with a role for I44 of ubiquitin in the degradation of the conjugate. Both HRS and TSG101 can bind the surface of ubiquitin surrounding I44 (Bishop et al., 2002; Hicke et al., 2005; Hurley and Emr, 2006), and they were required for the lysosomal localization of ubiquitinated Jun. Depletion of either protein caused mislocalization of ubiquitinated Jun to sites similar to those observed in cells that overexpressed Jun. Depletion of TSG101 stabilized both endogenous as well as transiently expressed Jun, but the depletion of HRS had no detectable effect on the stability of endogenous Jun. It is possible that residual HRS or related proteins sustained the degradation of Jun in lysosomes or that alternative degradation pathways compensated for the lack of HRS.
It may be significant that the pathways for the degradation of Jun and of several growth factor receptors share the requirement for HRS and TSG101 ubiquitin-binding proteins. This provides a potential mechanism for coordinate regulation of the receptors that initially respond to extracellular stimuli and at least one of the ultimate targets of the signaling cascade that is initiated by receptor activation. This also presents a challenge for analysis of the functions that lysosomal degradation of Jun serves, because alterations in this pathway are likely to have many consequences unrelated to Jun degradation. Further studies of the mechanisms whereby HRS and TSG101 mediate the translocation and degradation of ubiquitinated Jun as well as of other components of this trafficking pathway are necessary to address this issue.
Proteins That Modulate Jun Ubiquitination
The correlation between the stoichiometry of Jun ubiquitination and lysosomal localization suggested that polyubiquitination facilitated lysosomal localization of the conjugate. Several E3 ligases have been proposed to ubiquitinate Jun and to contribute to its degradation (Fang and Kerppola, 2004; Gao et al., 2004; Nateri et al., 2004; Wertz et al., 2004; Wei et al., 2005). Jun ubiquitination by many of these E3 ligases can be modulated by signaling (Gao et al., 2004; Nateri et al., 2004; Wei et al., 2005; Gao et al., 2006). The stoichiometries of Jun ubiquitination and the isopeptide linkages formed by most of these E3 ligases are unknown. Itch expression enhances polyubiquitination of Deltex in cells that express ubiquitin containing only K29 (Chastagner et al., 2006).
Many proteins that can interact with the δ region of Jun regulate the ubiquitination, cytoplasmic localization and degradation of other nuclear proteins (Claret et al., 1996; Tomoda et al., 1999; Bech-Otschir et al., 2002; Wan et al., 2002; Grossman et al., 2003; Kim et al., 2004). Several of these proteins have E4 ligase activity or the ability to activate E3 ligases (Bech-Otschir et al., 2002; Grossman et al., 2003). These proteins could modulate the stoichiometry of Jun ubiquitination and control its lysosomal localization and degradation. The multiple determinants required for the lysosomal localization of ubiquitinated Jun provide numerous potential targets for control of Jun localization and stability.
Supplementary Material
ACKNOWLEDGMENTS
We thank Deyu Fang for sharing reagents and valuable suggestions, Yurii Chinenov for construction of plasmids expressing Fos and Jun family proteins fused to fluorescent protein fragments, Stanley Cohen for generously sharing plasmids encoding HRS and TSG101, and members of the Kerppola laboratory for constructive criticism. This research was supported by a grant from the Dana Foundation and by the Howard Hughes Medical Institute.
Abbreviations used:
- UbFC
ubiquitin-mediated fluorescence complementation
- YFP
yellow fluorescent protein
- HRS
hepatocyte growth factor-regulated tyrosine kinase substrate
- TSG101
tumor susceptibility gene 101
- Ubn
ubiquitin chain composed of n protomers
- VN
residues 1-155 of Venus fluorescent protein
- CC
residues 156-238 of cyan fluorescent protein.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-05-0496) on August 20, 2008.
REFERENCES
- Beal R., Deveraux Q., Xia G., Rechsteiner M., Pickart C. Surface hydrophobic residues of multiubiquitin chains essential for proteolytic targeting. Proc. Natl. Acad. Sci. USA. 1996;93:861–866. doi: 10.1073/pnas.93.2.861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bech-Otschir D., Seeger M., Dubiel W. The COP9 signalosome: at the interface between signal transduction and ubiquitin-dependent proteolysis. J. Cell Sci. 2002;115:467–473. doi: 10.1242/jcs.115.3.467. [DOI] [PubMed] [Google Scholar]
- Bishop N., Horman A., Woodman P. Mammalian class E vps proteins recognize ubiquitin and act in the removal of endosomal protein-ubiquitin conjugates. J. Cell Biol. 2002;157:91–101. doi: 10.1083/jcb.200112080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chastagner P., Israel A., Brou C. Itch/AIP4 mediates Deltex degradation through the formation of K29-linked polyubiquitin chains. EMBO Rep. 2006;7:1147–1153. doi: 10.1038/sj.embor.7400822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claret F. X., Hibi M., Dhut S., Toda T., Karin M. A new group of conserved coactivators that increase the specificity of AP-1 transcription factors. Nature. 1996;383:453–457. doi: 10.1038/383453a0. [DOI] [PubMed] [Google Scholar]
- Fang D., Kerppola T. K. Ubiquitin-mediated fluorescence complementation reveals that Jun ubiquitinated by Itch/AIP4 is localized to lysosomes. Proc. Natl. Acad. Sci. USA. 2004;101:14782–14787. doi: 10.1073/pnas.0404445101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuchi M., Imamura T., Chiba T., Ebisawa T., Kawabata M., Tanaka K., Miyazono K. Ligand-dependent degradation of Smad3 by a ubiquitin ligase complex of ROC1 and associated proteins. Mol. Biol. Cell. 2001;12:1431–1443. doi: 10.1091/mbc.12.5.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao M., Labuda T., Xia Y., Gallagher E., Fang D., Liu Y. C., Karin M. Jun turnover is controlled through JNK-dependent phosphorylation of the E3 ligase Itch. Science. 2004;306:271–275. doi: 10.1126/science.1099414. [DOI] [PubMed] [Google Scholar]
- Gao B., Lee S. M., Fang D. The tyrosine kinase c-Abl protects c-Jun from ubiquitination-mediated degradation in T cells. J. Biol. Chem. 2006;281:29711–29718. doi: 10.1074/jbc.M604596200. [DOI] [PubMed] [Google Scholar]
- Grossman S. R., Deato M. E., Brignone C., Chan H. M., Kung A. L., Tagami H., Nakatani Y., Livingston D. M. Polyubiquitination of p53 by a ubiquitin ligase activity of p300. Science. 2003;300:342–344. doi: 10.1126/science.1080386. [DOI] [PubMed] [Google Scholar]
- Haglund K., Sigismund S., Polo S., Szymkiewicz I., Di Fiore P. P., Dikic I. Multiple monoubiquitination of RTKs is sufficient for their endocytosis and degradation. Nat. Cell Biol. 2003;5:461–466. doi: 10.1038/ncb983. [DOI] [PubMed] [Google Scholar]
- Hershko A., Ciechanover A. The ubiquitin system. Annu. Rev. Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- Hicke L. Protein regulation by monoubiquitin. Nat. Rev. 2001;2:195–201. doi: 10.1038/35056583. [DOI] [PubMed] [Google Scholar]
- Hicke L., Schubert H. L., Hill C. P. Ubiquitin-binding domains. Nat. Rev. 2005;6:610–621. doi: 10.1038/nrm1701. [DOI] [PubMed] [Google Scholar]
- Hinman J. D., Chen C. D., Oh S. Y., Hollander W., Abraham C. R. Age-dependent accumulation of ubiquitinated 2′,3′-cyclic nucleotide 3′-phosphodiesterase in myelin lipid rafts. Glia. 2008;56:118–133. doi: 10.1002/glia.20595. [DOI] [PubMed] [Google Scholar]
- Huang F., Kirkpatrick D., Jiang X., Gygi S., Sorkin A. Differential regulation of EGF receptor internalization and degradation by multiubiquitination within the kinase domain. Mol. Cell. 2006;21:737–748. doi: 10.1016/j.molcel.2006.02.018. [DOI] [PubMed] [Google Scholar]
- Hurley J. H., Emr S. D. The ESCRT complexes: structure and mechanism of a membrane-trafficking network. Annu. Rev. Biophys. Biomol. Struct. 2006;35:277–298. doi: 10.1146/annurev.biophys.35.040405.102126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzmann D. J., Babst M., Emr S. D. Ubiquitin-dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex, ESCRT-I. Cell. 2001;106:145–155. doi: 10.1016/s0092-8674(01)00434-2. [DOI] [PubMed] [Google Scholar]
- Kim B. C., Lee H. J., Park S. H., Lee S. R., Karpova T. S., McNally J. G., Felici A., Lee D. K., Kim S. J. Jab1/CSN5, a component of the COP9 signalosome, regulates transforming growth factor beta signaling by binding to Smad7 and promoting its degradation. Mol. Cell. Biol. 2004;24:2251–2262. doi: 10.1128/MCB.24.6.2251-2262.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky D. J., Emr S. D. Autophagy as a regulated pathway of cellular degradation. Science. 2000;290:1717–1721. doi: 10.1126/science.290.5497.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komada M., Kitamura N. Growth factor-induced tyrosine phosphorylation of Hrs, a novel 115-kilodalton protein with a structurally conserved putative zinc finger domain. Mol. Cell. Biol. 1995;15:6213–6221. doi: 10.1128/mcb.15.11.6213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Brooks C. L., Wu-Baer F., Chen D., Baer R., Gu W. Mono- versus polyubiquitination: differential control of p53 fate by Mdm2. Science. 2003;302:1972–1975. doi: 10.1126/science.1091362. [DOI] [PubMed] [Google Scholar]
- Lu Q., Hope L. W., Brasch M., Reinhard C., Cohen S. N. TSG101 interaction with HRS mediates endosomal trafficking and receptor down-regulation. Proc. Natl. Acad. Sci. USA. 2003;100:7626–7631. doi: 10.1073/pnas.0932599100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosesson Y., Shtiegman K., Katz M., Zwang Y., Vereb G., Szollosi J., Yarden Y. Endocytosis of receptor tyrosine kinases is driven by monoubiquitylation, not polyubiquitylation. J. Biol. Chem. 2003;278:21323–21326. doi: 10.1074/jbc.C300096200. [DOI] [PubMed] [Google Scholar]
- Nateri A. S., Riera-Sans L., Da Costa C., Behrens A. The ubiquitin ligase SCFFbw7 antagonizes apoptotic JNK signaling. Science. 2004;303:1374–1378. doi: 10.1126/science.1092880. [DOI] [PubMed] [Google Scholar]
- Peng J., Schwartz D., Elias J. E., Thoreen C. C., Cheng D., Marsischky G., Roelofs J., Finley D., Gygi S. P. A proteomics approach to understanding protein ubiquitination. Nat. Biotechnol. 2003;21:921–926. doi: 10.1038/nbt849. [DOI] [PubMed] [Google Scholar]
- Pickart C. M., Fushman D. Polyubiquitin chains: polymeric protein signals. Curr. Opin. Chem. Biol. 2004;8:610–616. doi: 10.1016/j.cbpa.2004.09.009. [DOI] [PubMed] [Google Scholar]
- Strous G. J., van Kerkhof P., Govers R., Ciechanover A., Schwartz A. L. The ubiquitin conjugation system is required for ligand-induced endocytosis and degradation of the growth hormone receptor. EMBO J. 1996;15:3806–3812. [PMC free article] [PubMed] [Google Scholar]
- Tomoda K., Kubota Y., Kato J. Degradation of the cyclin-dependent-kinase inhibitor p27Kip1 is instigated by Jab1. Nature. 1999;398:160–165. doi: 10.1038/18230. [DOI] [PubMed] [Google Scholar]
- Treier M., Staszewski L. M., Bohmann D. Ubiquitin-dependent c-Jun degradation in vivo is mediated by the delta domain. Cell. 1994;78:787–798. doi: 10.1016/s0092-8674(94)90502-9. [DOI] [PubMed] [Google Scholar]
- van der Horst A., de Vries-Smits A. M., Brenkman A. B., van Triest M. H., van den Broek N., Colland F., Maurice M. M., Burgering B. M. FOXO4 transcriptional activity is regulated by monoubiquitination and USP7/HAUSP. Nat. Cell Biol. 2006;8:1064–1073. doi: 10.1038/ncb1469. [DOI] [PubMed] [Google Scholar]
- Wan M., Cao X., Wu Y., Bai S., Wu L., Shi X., Wang N., Cao X. Jab1 antagonizes TGF-beta signaling by inducing Smad4 degradation. EMBO Rep. 2002;3:171–176. doi: 10.1093/embo-reports/kvf024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei W., Jin J., Schlisio S., Harper J. W., Kaelin W. G., Jr The v-Jun point mutation allows c-Jun to escape GSK3-dependent recognition and destruction by the Fbw7 ubiquitin ligase. Cancer Cell. 2005;8:25–33. doi: 10.1016/j.ccr.2005.06.005. [DOI] [PubMed] [Google Scholar]
- Wertz I. E., O'Rourke K. M., Zhang Z., Dornan D., Arnott D., Deshaies R. J., Dixit V. M. Human de-etiolated-1 regulates c-Jun by assembling a CUL4A ubiquitin ligase. Science. 2004;303:1371–1374. doi: 10.1126/science.1093549. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.