Abstract
Regulation of Rho GTPase signaling is critical for cell shape determination and polarity. Here, we investigated the role of LRG1, a novel member of the GTPase-activating proteins (GAPs) of Neurospora crassa. LRG1 is essential for apical tip extension and to restrict excessive branch formation in subapical regions of the hypha and is involved in determining the size of the hyphal compartments. LRG1 localizes to hyphal tips and sites of septation via its three LIM domains. The accumulation of LRG1 as an apical cap is dependent on a functional actin cytoskeleton and active growth, and is influenced by the opposing microtubule-dependent motor proteins dynein and kinesin-1. Genetic evidence and in vitro GTPase assays identify LRG1 as a RHO1-specific GAP affecting several output pathways of RHO1, based on hyposensitivity to the glucan inhibitor caspofungin, synthetic lethality with a hyperactive β1,3-glucan synthase mutant, altered PKC/MAK1 pathway activities, and hypersensitivity to latrunculin A. The morphological defects of lrg-1 are highly reminiscent to the Ndr kinase/RAM pathway mutants cot-1 and pod-6, and genetic evidence suggests that RHO1/LRG1 function in parallel with COT1 in coordinating apical tip growth.
INTRODUCTION
Polarized growth is a multifactorial property that is coordinated by numerous signaling pathways, and monomeric GTPases of the Ras super family have been identified as key regulators of cell polarity (Etienne-Manneville and Hall, 2002; Jaffe and Hall, 2005; Bustelo et al., 2007; Park and Bi, 2007). They act as molecular switches that cycle between an active GTP–bound and an inactive GDP–bound form. Transition between these two forms is achieved through GTPase-activating proteins (GAPs) leading to the inactive form and GDP-GTP-exchange factors (GEFs) that activate the small G-protein. Originally, GTPases of the Rho subfamily were described as key regulators of the actin cytoskeleton, but now it has become obvious that they influence many cellular processes including cytoskeletal organization, vesicle transport, and transcriptional regulation. The position of small G-proteins at the bottleneck of signal transduction pathways explains the many defects seen when these GTPases are misregulated.
A challenging complication in determining the specific functions of small G-proteins is the fact that the number of GAPs and GEFs is significantly larger than the number of GTPases that they regulate. For example, the D. melanogaster genome contains only six Rho proteins, but ∼20 GEFs and more than 20 GAPs (Adams et al., 2000), indicating that understanding the specific functions of these regulators will be the key for deciphering the temporal and spatial activity of the switches. Although considerable progress has been made in understanding the activation of small GTPases through their GEFs (Gulli and Peter, 2001; Schmidt and Hall, 2002; Garcia et al., 2006), full activation of a specific GTPase requires not only the coordination of the “on” switch, but also the shuttling between both the active and inactive states of the GTPase is essential for full signaling activity of the small G-protein (Vanni et al., 2005; Barale et al., 2006). Nevertheless, little is known about how the GAPs are regulated in a spatial and temporal manner. Interestingly, the number of GAPs present in the available fungal and animal genomes is exceeding those of the GEFs (Goffeau et al., 1996; Adams et al., 2000; Borkovich et al., 2004; Jaffe and Hall, 2005), indicating that fine-tuning of the “off-switch” is of high importance for providing the necessary specificity for the Rho module.
Apical tip extension is the hallmark of filamentous fungi, and fungal hyphae share with neurons and pollen tubes the distinction of being among the most highly polarized cells in biology (Palanivelu and Preuss, 2000; Chilton, 2006; Harris, 2006), thus making them attractive models for the analysis of fundamental mechanisms underlying cellular polarity. Nevertheless, the molecular understanding of fungal morphogenesis is still a major challenge. Phylogenetic analyses and the comparison of Saccharomyces cerevisiae morphogenetic data with the limited results from various filamentous asco- and basidiomycetes have established that a core set of “polarity factors” is conserved between unicellular and filamentous fungi (reviewed in Wendland, 2001; Borkovich et al., 2004; Harris, 2006). Thus, the accumulated knowledge of baker's yeast is serving as an invaluable source for comparative morphogenetic studies, but it is becoming increasingly evident that subtle differences in the wiring of these conserved components and the presence of additional proteins that are absent in unicellular fungi result in dramatically different morphogenetic outcomes ranging from budding to filamentous growth (Boyce et al., 2003, 2005; Rottmann et al., 2003; Seiler and Plamann, 2003; Li et al., 2006; Malavazi et al., 2006).
Current fungal research is focusing on the description of the various Rho proteins and the analysis of the interplay between the different modules (Wendland and Philippsen, 2000, 2001; Boyce et al., 2003, 2005; Chen and Dickman, 2004; Leveleki et al., 2004; Rasmussen and Glass, 2005, 2007; Mahlert et al., 2006). Various studies have identified Rho1 as one key regulator of hyphal growth and polarity. Aspergillus fumigatus Rho1 has been described as part of the β1,3-glucan synthase complex that localizes to zones of active growth at the hyphal apex (Beauvais et al., 2001). A similar role in maintaining cell wall integrity was suggested for Rho1 of Ashbya gossypii, because deletion mutants showed reduced filamentous growth and high rates of cell lysis (Wendland and Philippsen, 2001). Aspergillus nidulans RhoA has been implicated in polar growth, branching, and cell wall synthesis (Guest et al., 2004). Budding yeast Rho1p as the best characterized representative of the Rho1 family has multiple functions in regulating the two main structural features of the fungal cell (reviewed in Levin, 2005; Park and Bi, 2007). The organization of the actin cytoskeleton is controlled through the interaction of the activated GTPase with the polarisome component Bni1p, whereas maintenance of the cell wall integrity is achieved via two independent mechanisms. Rho1p activates cell wall synthesis through the direct stimulation of the enzyme glucan synthase, which catalyzes the polymerization of β1,3-glucan. In addition, it activates the mitogen-activated protein (MAP) kinase pathway that monitors the cell wall integrity through its activation of protein kinase C thereby coordinating the transcription of several cell wall–specific enzymes.
To identify the critical components that contribute to polarized growth, we developed a large-scale genetic screen for the isolation of conditional mutants defective in polar and directed growth in the Neurospora crassa (Seiler and Plamann, 2003) and have identified the Ndr kinase COT1 and its upstream regulator kinase POD6 as required for hyphal tip extension. These kinases are important for normal cell differentiation and polar morphogenesis in various organisms, yet their specific functions are still elusive (summarized in Hergovich et al., 2006). The budding yeast RAM network (regulation of Ace2p activity and cellular morphogenesis) is currently the best characterized, even though still poorly understood, Ndr signaling cascade (Nelson et al., 2003; Jansen et al., 2006; and references therein). RAM-defective cells exhibit a round morphology and fail to separate after cytokinesis. The Ndr kinase Cbk1p is an essential component of this pathway, whereas the STE20-like kinase Kic1p probably functions as a direct upstream kinase of Cbk1p. In addition, Hym1p, Tao3p and Mob2p are required for Cbk1p activation and localization, The presence of these components also in higher eukaryotes have established the idea that Ndr kinases and other interacting proteins represent the core components of a conserved signaling network required for polarized morphogenesis. Here, we describe the characterization of a LIM and Rho-GAP domain–containing protein (therefore named LRG1) that acts as a RHO1–specific GAP and provide genetic evidence that LRG1 functions in parallel with COT1 in coordinating polar tip growth.
MATERIALS AND METHODS
Strains, Media, and Growth Conditions
N. crassa strains used in this study are summarized in the Table 1. General genetic procedures and media used in the handling of N. crassa have been described (Davis and DeSerres, 1970) or are available through the Fungal Genetic Stock Center (www.fgsc.net). Growth tests to determine the sensitivity of mutant strains in comparison to wild type against cytoskeletal, cell wall, and protein kinase C (PKC) inhibitors were performed on minimal medium supplemented with 0.5% sorbose to enhance colonial growth of the strains and to allow a better comparison of the different strains on a single plate. The hygromycin B and nourseothricin concentrations were adjusted to 200 and 30 μg/ml, respectively, to select for transformants. Homologous recombination events in the heterokaryotic strain HP1 containing the Δlrg-1–deleted nucleus were verified by phenotypic analysis of transformants grown on 400 μM p-fluorophenylalanine (fpa) and 200 μg/ml histidine or 5 μg/ml benomyl and 200 μg/ml pantothenic acid and by the complementation of the growth defect of potential Δlrg-1 strains with a 6-kb genomic SacII fragment containing lrg-1 (pNV16).
Table 1.
Neurospora crassa strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| Wild type | 74-OR23–1A | FGSC 987 |
| lrg-1(12–20) | lrg-1(Y926H) | Seiler and Plamann (2003) |
| LRG11–847 | lrg-1(Y926H) lrg-1(1–847)::hph(EC) | This study |
| LRG1781–1279 | lrg-1(Y926H) lrg-1(781–1279)::hph(EC) | This study |
| MYC::LRG11–847 | lrg-1(Y926H) myc9::lrg-1(1–847)::hph(EC) | This study |
| LRG1781–1279::MYC | lrg-1(Y926H) lrg-1(781–1279)::myc9::hph(EC) | This study |
| HP1 | benRhis-3+fpaS pan-2− + benS his-3−fpaR pan-2+ | Nargang et al. (1995) |
| ΔLRG1 | benRhis-3+fpaS pan-2−nat:: lrg-1Δ + benS his-3−fpaR pan-2+lrg-1+ | This study |
| LIM1* | lrg-1(Y926H) lrg-1(C121S, C124S, C98L, C101S)::hph(EC) | This study |
| LIM2* | lrg-1(Y926H) lrg-1(H185V, C188S, C162S, C165S)::hph(EC) | This study |
| LIM3* | lrg-1(Y926H) lrg-1(C492S, C495S, C469G, C472S)::hph(EC) | This study |
| LRG* | lrg-1(Y926H) lrg-1(1–1279; LIM1*, LIM2*, LIM3*)::hph(EC) | This study |
| LRG1R847L | lrg-1(Y926H) lrg-1(R847L)::hph(EC) | This study |
| LRG1K910A | lrg-1(Y926H) lrg-1(K910A)::hph(EC) | This study |
| GFP::LRG1 | benRpan-2−natR:: lrg-1Δ gfp::lrg-1::hph(EC) | This study |
| GFP::LRG1* | benRpan-2−natR:: lrg-1Δ gfp::lrg-1(1–1279; LIM1*, LIM2*, LIM3*)::hph(EC) | This study |
| gs-1(8–6) | gs-1(8–6) | Seiler and Plamann (2003) |
| gs-1(8–6);lrg-1(12–20) | gs-1(8–6) lrg-1(Y926H) | This study |
| cot-1(1) | cot-1(C102t) | FGSC 4066 |
| cot-1(1);gs-1(8–6) | cot-1(C102t) gs-1(8–6) | This study |
| ro-1 | ro-1(B15) | FGSC 146 |
| ro-10 | ro-10(AR7) | FGSC 3619 |
| ro-3 | ro-3(R2354) | FGSC 3 |
| gul-1 | gul-1(CA1) | FGSC 803 |
| gul-1;cot-1(1) | gul-1(CA1) cot-1(C102t) | FGSC 1962 |
| gul-1;lrg-1(12–20) | gul-1(CA1) lrg-1(Y926H) | This study |
| lrg-1;ro-1 | lrg-1(Y926H) ro-1(B15) | This study |
| lrg-1;ro-10 | lrg-1(Y926H) ro-10(AR7) | This study |
| lrg-1;ro-3 | lrg-1(Y926H) ro-3(R2354) | This study |
| nkin | nkin(RIP-1) | Seiler et al. (1997) |
| nkin;lrg-1 | nkin(RIP-1) | This study |
| lrg-1;Bni11029–1817 | lrg-1(Y926H) bni-11029–1817(EC) | This study |
| cot-1(1);Bni11029–1817 | cot-1(C102t) bni-11029–1817(EC) | This study |
| lrg-1; Bni11–824 | lrg-1(Y926H) bni-11–824(EC) | This study |
| cot-1(1); Bni11–824 | cot-1(C102t) bni-11–8/24(EC) | This study |
| cot-1(1);ro-1 | cot-1(C102t) ro-1(B15) | Seiler et al. (2006) |
| cot-1(1);ro-10 | cot-1(C102t) ro-10(AR7) | Seiler et al. (2006) |
| cot-1(1);ro-3 | cot-1(C102t) ro-3(R2354) | Seiler et al. (2006) |
| GFP::LRG1;ro-1 | ro-1(B15) natR:: lrg-1Δ gfp::lrg-1::hph (EC) | This study |
| GFP::LRG1*;ro-1 | ro-1(B15) natR:: lrg-1Δ gfp::lrg-1(1–1279; LIM1*, LIM2*, LIM3*)::hph (EC) | This study |
| GFP::LRG1;nkin | nkin(RIP-1) nat:: lrg-1Δ gfp::lrg-1::hph(EC) | This study |
Plasmid Construction
Primer sequences are summarized in the Supplementary Table S1. The lrg-1 deletion cassette in pNV46 was obtained by plasmid gap repair in S. cerevisiae (Orr-Weaver and Szostak, 1983). pRS416 was linearized with XbaI and XhoI, the NatR cassette was amplified by PCR with primers NV_gpd1 and NV_nat5 and pNV1 as template (Seiler et al., 2006), and the 5′ and 3′ flanking regions of lrg-1 were amplified from cosmid X8D4 using the oligonucleotides NV_KOlrg5f, NV_KOlrg5r, NV_KOlrg3f, and NV_KOlrg3r. These primers contained 25-bp homologous overhangs at their 5′ ends and allowed recombination of the four linear DNA fragments into the functional deletion cassette in yeast. To generate the GAP and LIM domain deletion constructs, a 6-kb genomic SacII fragment containing lrg-1 coding region and 1.5- and 1-kb 5′ and 3′ regions, respectively, was inserted in pBluescript SK+ (Stratagene, Amsterdam, Netherlands) to obtain pNV16. The hygromycin cassette, amplified with the primers Hyg5′Xba and Hyg3′Xba and plasmid pMP6 as template, was inserted into the unique XbaI site. LRG11–847 was generated by deleting the C-terminal domain of lrg-1 in pNV16 with ApaI and by inserting the hygromycin B resistance cassette (hph; which was amplified via ApaI-containing primers). For LRG1781–1279, the lrg-1 promoter was amplified using oligonucleotides that introduce NcoI and SacI sites and inserted together with an NcoI/SacII fragment harboring the GAP domain (aa 781-1279) into SacI/SacII-digested pBluescript SK+. The hygromycin cassette was introduced into XbaI. Point mutations in the three LIM domains were generated with the QuickChange Site-Directed Mutagenesis Kit (Stratagene) according to manufacturer's instruction using the oligonucleotides NV_LIM1mut_f, NV_LIM1mut_r, NV_LIM1mut2_f, NV_LIM1mut2_r, NV_LIM2mut_f, NV_LIM2mut_r, NV_LIM2mut2_f, NV_LIM2mut2_r, NV_LIM3mut_f, NV_LIM3mut_r, NV_LIM3mut2_f, NV_LIM3mut2_r. The zinc-finger coordinating cysteines and histidines were substituted by amino acids that lack the zinc-binding capability (LIM1: C121S, C124S, C98L, C101S; LIM2: H185V, C188S, C162S, C165A; LIM3: C492S, C495S, C469G, and C472S). The triple LIM domain mutation construct was generated by multiple mutagenesis PCRs to obtain all eight mutations of the first and second LIM domain. The third LIM domain was mutated in a parallel construct. A MscI/SacI fragment containing mutated LIM1*/2*, and a SacI/MluI fragment of LIM3* was ligated into pNV16 digested with MscI/MluI. The hygromycin cassette harboring Bsp120I restriction sites was subsequently introduced into the unique NotI site of the vector. The K910A exchange in the GAP domain was generated using the primers NV_GAPmut_f and NV_GAPmut_r, and the hygromycin resistance cassette introduced via Bsp120I/NotI. The R847L point mutation in the GAP domain was generated using the primers NV_LRG20 and NV_LRG21, and the hygromycin resistance cassette introduced via XbaI. N-terminal MYC-tagged version of LRG1 and LRG11–847 were constructed by inserting a ninefold MYC-tag into the unique MscI right after the start codon with primers NV_myc4 and NV_myc3 with pYM6 (Knop et al., 1999) as template. The C-terminal MYC9-tag in LRG1781–1279 was inserted via the EcoRI and SacII sites of pNV16 using the primers NV_myc1 and NV_myc2. Western blot analysis using a monoclonal 9E10-anti-cMYC antibody (Santa Cruz Biotechnology, Santa Cruz, CA) verified the expression of the MYC-tagged LRG1 constructs. The enhanced green fluorescent protein (eGFP) coding region was amplified from pSM1 (Poggeler et al., 2003) using oligonucleotides NV_GFP1 and NV_GFP2 and subsequently ligated via EcoRV into MscI-digested pNV16 to generate GFP::LRG1. cDNAs of the six Rho GTPases were generated by reverse transcription with RevertAid M-MuLV Reverse Transcriptase (Fermentas, Vilnius, Lithuania) from mRNA prepared with PolyATtract (Promega, Madison, WI), amplified by PCR using primers covering the genes from ATG to stop codon using oligonucleotides mentioned in Supplementary Table S1. Sequences were inserted via primer-based restriction sites (NotI-NotI for Rho1 to Rho4 or SalI-NotI for CDC42 and RAC) into modified pETM-30 (EMBL-Heidelberg, Protein Expression Facility). The GAP domain of LRG1 (aa 650-1035) was amplified with primers NV_lrg14 and NV_lrg18 and introduced via SalI/XhoI and NotI into pETM-30. The N. crassa overexpression constructs were controlled by a modified CPC promoter derived from plasmid pMP6 harboring BglII and SpeI as unique cloning sites (Seiler et al., 2006). Genomic coding regions of the six Rho proteins were amplified with primers containing the mentioned restriction sites.
Microscopy
Immunolocalization for N. crassa hyphae was conducted following a protocol adapted from Minke et al. (1999). Conidia were germinated on a small piece of GN-6 cellulose filter (Gelman Sciences,) placed on the surface of an agar plate. Filters were plunge-frozen in liquid propane and transferred to fixative (3% formaldehyde in 100% ethanol precooled to −80°C). Samples were maintained at −80°C for at least 2 d and then slowly transferred to room temperature (2 h at 20°C, 2 h at 4°C). Filters were rehydrated in a series of ethanol:buffer (100 mM phosphate, pH 7.0) solutions starting at a ratio of 90:10 and ending at 10:90. The cell wall was digested by incubating the filters for 0.5–8 min in 2 mg/ml lysing enzymes from Trichoderma harzianum (Sigma, Munich, Germany) in 100 mM potassium citrate (pH 6.0, 20 mM EGTA, 5% BSA). To block nonspecific binding of antibody, the filters were incubated for 1 h in 5% BSA. Samples were immersed in the primary antibody for at least 8 h, washed several times in phosphate buffer, incubated in the secondary antibody for ≥8 h, and visualized using standard rhodamine and FITC filter sets. For immunolocalization, samples were viewed with an ORCA ER digital camera (Hamamatsu, Hamamatsu City, Japan) mounted on an Axiovert S100 microscope (Carl Zeiss, Jena, Germany). Image acquisition was done using the Openlab 5.01 software (Improvision, Coventry, United Kingdom), and images were further processed using Photoshop CS2 (Adobe, San Jose, CA). For localization studies of GFP::LRG1 and GFP::LRG1*, an AxioObserverZ.1 microscope equipped with an ApoTome unit, an AxioCam MRm r3.0 CCD camera and X-Cite (EXFO) illumination source was used with AxioVision Software 4.6. For confocal imaging an Axiovert 100M microscope with the confocal module LSM 510 and LSM 510-Software was used (all from Zeiss). To determine the kinetics of septum formation, DIC imaging of a hyphal segment was randomly started, and the fluorescence images were manually recorded at time points when the LRG::GFP localization had visibly changed to reduce photobleaching. The absolute time points of the GFP images were used for the calculation of septation kinetics. Low magnification documentation of fungal hyphae or colonies was performed with a SZX12 stereomicroscope (Olympus, Tokyo, Japan) and a PS30 video camera (Kappa, Gleichen, Germany).
Enzymatic Assays
The RHO GTPases were purified as glutathione S-transferase (GST) fusion proteins. Transformed Rosetta2(DE3) cells (Novagen, Schwalbach, Germany) were grown at 20°C and induced for 12 h with 0.1 M IPTG. Cell extracts generated by sonification in lysing buffer (50 mM Tris, pH 7.5, 10% saccharose, 5 mM MgCl2, 1 mM PMSF, 0.008% β-mercaptoethanol, and 0.02% NP40), were bound to GSH Sepharose (Amersham, Amersham, Buckinghamshire, United Kingdom), washed (50 mM Tris, pH 7.5, 250 mM NaCl, 5 mM MgCl2, 1 mM PMSF, 0.008% β-mercaptoethanol, 0.02% NP40) and eluted (50 mM Tris, pH 8.0, 250 mM NaCl, 5 mM MgCl2, 5 mM DTT, 20 mM glutathione reduced, and 0.02% NP40). GTPase assays were performed as described (Gibbs et al., 1988). Pre-equilibration of the RHO proteins was for 15 min at 25°C in 30 μl buffer A (20 mM HEPES, pH 7.6, 25 mM sodium chloride, 2 mM EDTA, 1 mg/ml BSA, 0.5 mM DTT, and 0.005% sodium cholate) including 0.5 μM GTPase and 5 μCi (0.17 μM) [γ-32P]GTP. GTP loading was stopped on ice by adding 1 μl 0.5 M MgCl2. GTPase, 5 μl, was added to start the reaction (final concentration: 20 mM HEPES, pH 7.6, 1 mg/ml BSA, 0.1 mM DTT, 1 mM GTP, and 4 μM GST::GAP or purified GST) at 25°C. Samples of 5 μl were stopped in 1 ml ice-cold wash buffer (50 mM Tris, pH 7.5, 50 mM sodium chloride, and 5 mM MgCl2) and filtered trough BA85 nitrocellulose membranes (Whatman, Clifton, NJ). The filters were washed with 6 ml wash buffer, dried, and measured by scintillation counting in a QuantaSmart scintillation counter (Perkin Elmer-Cetus, Norwalk, CT). The phospho-p44/42 MAP kinase (Thr202/Tyr204) antibodies (Cell Signaling Technology, Beverly, MA) were used according to manufacturer's instructions.
RESULTS
LRG1 Is Essential for Hyphal Tip Extension
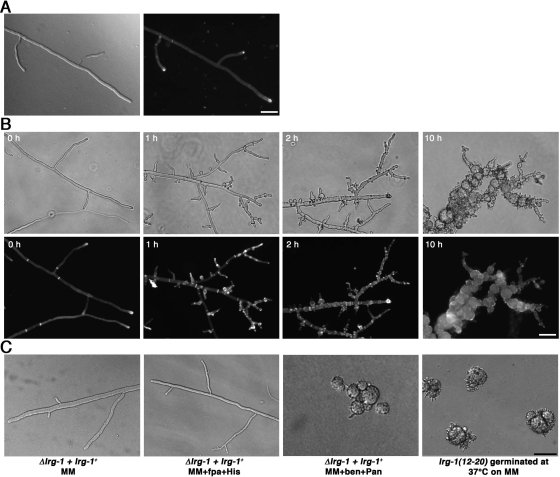
The phenotypic analysis of lrg-1(12-20) indicated no major differences in hyphal morphology or branching frequency compared with wild type when the strains were grown at permissive temperature (25°C; Figure 1, A and B). However, within 30 min of transferring lrg-1(12-20) to 37°C, we observed a rapid cessation of tip extension. In contrast to the wild-type apex, which is a dome-shaped structure, the tips of lrg-1(12-20) generated a characteristic pointed, needle-like shape. This stop of tip extension was accompanied by the appearance of numerous subapical needle-shaped branches of ≤10 μm in length that also stopped growth with a pointed tip. After prolonged incubation at restrictive temperature, the new branches as well as the primary hyphae began to swell up in a bulbous and apolar manner. Transfer of such a culture back to permissive temperature resulted in growing tips having normal growth rates, diameter, and morphology within 30 min. Germination of lrg-1(12-20) at restrictive temperature resulted in the formation of compact colonies with multiple ≤10-μm-long germ tubes with pointed tips.
Figure 1.
LRG1 is essential for hyphal tip extension. (A) wild type was grown on minimal media plates at 25°C (left, DIC image; right, Calcofluor White staining). Bar, 40 μm. (B) lrg-1(12-20) was grown on minimal media plates at 25°C and shifted for the indicated times to restrictive temperature to illustrate the cessation of tip extension with pointed, needle-like tips and the progressive hyperbranching of the mutant (phase-contrast images, top panel). Increased septation and abnormal chitin distribution was monitored by labeling with Calcofluor White (bottom panel). Bar, 40 μm. (C) Growth of the heterokaryotic lrg-1 deletion strain lrg-1+ + Δlrg-1 on minimal medium, medium containing fpa and histidine and medium containing benomyl and pantothenic acid (which forces the knockout nucleus to predominate), resulting in morphological defects identical to lrg-1(12-20) germinated at restrictive temperature. Bar, 7.5 μm.
Chitin is the primary component of fungal septae and of the inner layers of the hyphal cell wall and is therefore accessible to the Calcofluor White staining, primarily at the hyphal tips and at septae. At permissive temperature, tips and septae of lrg-1(12-20) are strongly labeled in a manner identical to wild type (Figure 1, A and B). Within 2 h at 37°C, lrg-1(12-20) showed extensive label in a patchy, subapical manner throughout the hyphae at positions of newly emerging branches, indicating excessive chitin deposition at sites of aberrant growth. In contrast, the high Calcofluor White stain at the primary tips observed in hyphae grown at the permissive temperature is nearly absent when the strains are cultured at the restrictive temperature, indicating decreased chitin synthesis at these nonelongating tips. Furthermore, we observed increased septation of lrg-1(12-20) compared with wild type grown at permissive temperature, which is further increased after shifting cells to restrictive conditions. The length of hyphal compartments at 25°C were 53.7 ± 21.0 and 148 ± 58.0 μm for lrg-1(12-20) and wild type, respectively (n = 50). After the transfer of lrg-1(12-20) to 37°C for 2.5 h the compartment length was further reduced to 11.3 ± 4.3 μm (n = 50), whereas the compartment length of wild type remained unchanged. The hyphal diameter of wild type and lrg-1(12-20) were similar and did not change during the shift experiment (8.2 ± 0.8 and 8.4 ± 1.6 μm, respectively, after 2.5 h at 37°C; n = 20).
To examine the phenotype of a lrg-1 deletion strain, we constructed a null mutant by using the sheltered disruption method (Nargang et al., 1995), which takes advantage of the fact that N. crassa is a multinucleated cell. The lrg-1 gene was deleted via homologous recombination in the heterokaryotic strain HP1, thereby allowing the generation of mutants in genes that are essential or important for growth. The resulting mutant harbors two kinds of nuclei, one with a null allele of lrg-1, and one with a wild-type copy. These nuclei contain selectable markers that allowed a shift in the nuclear ratio within the heterokaryotic cells. Growth on media containing fpa and histidine favored the propagation of the wild-type nucleus. In contrast, growth of heterokaryotic cells on media containing benomyl and pantothenic acid forced the knockout nucleus to predominate and thus led to the depletion of LRG1 and morphological defects that were indistinguishable from lrg-1(12-20) germinated at restrictive temperatures (Figure 1C). These results characterize lrg-1(12-20) as a conditional loss-of-function allele of lrg-1 and confirm an essential role of LRG1 during the apical extension of the hyphal tip and in controlling the number and position of subapical branches.
LRG1 Contains Three LIM Domains and a GAP Domain That Is Essential for Growth and Septation
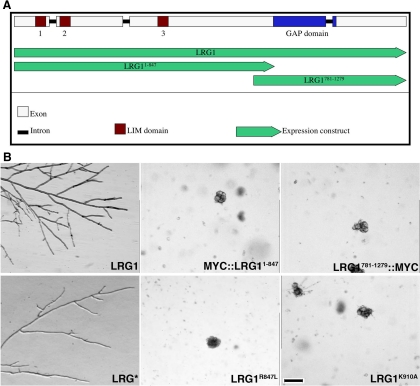
Examination of the 1279 amino acid (aa) encoding LRG1 sequence revealed two interesting features (Figure 2A). The N-terminal region from aa 1–533 is cysteine- and histidine-rich and can be arranged into three tandem zinc-finger–containing structures called LIM domains that act as versatile protein–protein interaction motifs (Schaller, 2001; Brown and Turner, 2004). Furthermore, a RHO-GAP domain was found in the C-terminal part of LRG1 (aa 791-1279). Sequence comparisons indicated that highly homologous proteins containing this domain architecture are present in all available fungal genomes. Apart from the fungal LRG1 relatives, the LIM domains of LRG1 are most closely related to the focal adhesion organizing protein paxillin (E-value of 2e−28 for Drosophila pseudoobscura paxillin vs. E = 1e−17 for mouse leupaxin, a more distantly related member of the paxillin superfamily, and E = 1e−14 for human actin-binding LIM protein 1 as the closest non-paxillin–related LIM domain-containing hit).
Figure 2.
The N-terminal region and the C-terminal Rho-GAP domain are both required for LRG1's cellular function. (A) Genomic organization of lrg-1 locus. The four exons are represented in white, the three introns in black. The lrg-1 gene encodes for a protein with three LIM domains (brown) in the N-terminal half and one Rho-GAP domain (blue) in the C-terminal part of the protein. Green arrows represent expression constructs under the control of the endogenous promoter. (B) Complementation experiments of lrg-1(12-20) transformed with the indicated constructs. Transformants were grown at 37°C to determine the complementation capability of the respective construct. Bar, 200 μm.
Sequencing of lrg-1(12-20) revealed a single amino acid substitution of the conserved tyrosine 926 (TAC) in the GAP domain by histidine (CAC), suggesting that the GAP domain is essential for the cellular function of LRG1. To analyze the function of the two LRG1 domains, we generated and expressed constructs lacking either the three LIM (LRG781–1279) or the GAP (LRG1–847) motifs in lrg-1(12-20), and determined that the two individual domains were unable to complement the tip extension defect of lrg-1(12-20) (Figure 2B). The importance of the GAP domain for hyphal growth was further supported by a construct, in which the conserved lysine 910 in the full-length protein was substituted with alanine. This mutation has been shown to result in a nonfunctional GAP due to loss of binding to the corresponding Rho (Li et al., 1997). When we expressed this construct in lrg-1(12-20) or in Δlrg-1, no complementation of the mutants defects were detected (Figure 2B). To confirm that the GAP activity of LRG1 is essential for its function, we generated an allele, in which the conserved catalytic arginine residue 847 of the GAP domain was substituted by a lysine. This construct was expressed in lrg-1(12-20), but was not sufficient for complementation (Figure 2B), thus providing strong evidence for the importance of the enzymatic activity for the function of LRG1.
An important function of the N-terminal part of LRG1 containing the three LIM domains was indicated by the failure of LRG1781–1279 to complement lrg-1(12-20). Therefore, we generated a series of lrg-1 alleles, in which four of the eight metal ion–coordinating cysteines or histidines of each of the three LIM domains were substituted with serines or alanines. These residues have been shown to mimic the behavior of cysteines, but lack their zinc-coordinating activity, which has been shown to be essential for the function of LIM domains (Schmeichel and Beckerle, 1997; Bombarda et al., 2002). Surprisingly, when we tested the three constructs that each was defective in one of the three LIM domains, we found that they did lead to full phenotypic complementation of lrg-1(12-20). Also, when we combined the three mutated LIM domains by generating LRG*, we did not observe abnormal growth of the lrg-1(12-20) (Figure 2B) or of homocaryotic Δlrg-1 transformants (data not shown) and measured overall radial growth rates on agar plates, which were comparable to wild type (LRG1: 1.1±0.1 mm/h; LRG1*: 1.0 ± 0.4 mm/h; n = 3). However, we noted a higher variability of extension rates of individual hyphal tips of LRG1* grown on microscopic slides (7.2±6.2 μm/min; n = 50) in comparison to LRG1 (6.3±4.3 μm/min; n = 50), suggesting that the dysfunctional LIM domains impair proper tip extension. Nevertheless, the placement of septae was not altered in LRG1*. The distance of the hyphal tip to the first septum was 224.0 ± 41.9 μm in LRG1 and 225.4±55.8 μm for LRG1* (n=30), whereas the distance between subapical septae was 84.9 ± 30.9 μm in LRG1 compared with 92.5 ± 32.4 μm in LRG1* (n = 100).
The LIM Domains Are Required for Localizing LRG1 to Sites of Growth
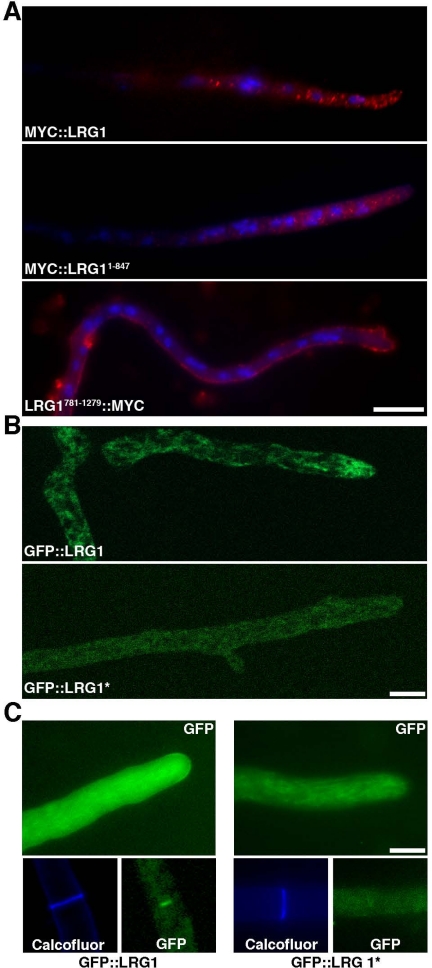
To determine the cellular distribution of LRG1, we generated an N-terminal myc9-tagged version of lrg-1, which complemented lrg-1(12-20). Immunolocalization experiments using anti-MYC antibodies revealed a punctate distribution throughout the cell, which was enriched at hyphal tips (Figure 3A). This localization was in line with a function of LRG1 at growing tips and was also observed, when we used antisera generated against LRG1 (data not shown). Next, we characterized the localization pattern of MYC9::LRG11–847 and LRG781–1279::MYC9 in the wild-type background. Although LRG781–1279::MYC9 aberrantly localized in a cortical manner throughout the whole hypha and along septae, the MYC9::LRG11–847 distribution was similar to localization of LRG1, suggesting that the N-terminal part of LRG1 is involved in the localization of the GAP domain.
Figure 3.
LRG1 localization to sites of growth at the hyphal tip and the septum is dependent on functional LIM domains. (A) MYC::LRG1 and MYC::LRG11–847 immunolocalized in vesicular-reticulate structures that are enriched at hyphal tips, whereas LRG1781–1279::MYC mis-localized to the hyphal cortex. Bar, 10 μm. (B) Localization data generated in the ApoTome modus indicate an apical vesicular–reticulate localization of GFP::LRG1 and a more diffuse localization of GFP::LRG1*. Bar, 5 μm. (C) GFP::LRG1 localized as a crescent at growing hyphal tips and at septae (left), whereas GFP::LRG1* containing the three mutated LIM domains did localize in a diffuse manner throughout the cell (right). Bar, 5 μm.
For a better resolution of the dynamics of LRG1, we generated N-terminal GFP fusion constructs with LRG1 and with LRG1* (containing the three mutated LIM domains). Both constructs were introduced into homocaryotic Δlrg-1 and complemented the growth defect of the deletion strain. ApoTome-based fluorescence microscopy confirmed the apical vesicular-reticulate localization of GFP::LRG1, but resulted in a more diffuse localization of GFP::LRG1* lacking the prominent streaks (Figure 3B). In addition to this tip-enriched vesicular–reticulate localization, we frequently observed a GFP::LRG1 cap along the apical cortex and strong staining around the septal pore (Figure 3C). In contrast, GFP::LRG1* was distributed in a more diffuse manner throughout the hypha with only a weak accumulation at the hyphal apex and no localization at septae. Thus, both the immunolocalization data and the GFP-fusion proteins indicated a function of the LIM domains in localizing the GAP domain to sites of active growth in the apical region and along septae.
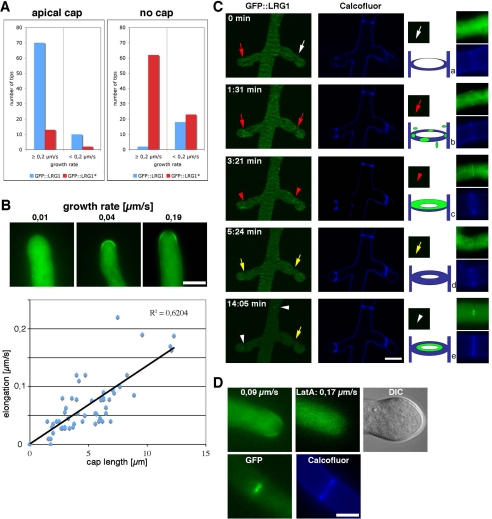
Confocal microscopy was used to characterize the dynamics of GFP::LRG1 in more detail. We first asked, if the localization of the apical cap is growth-dependent and analyzed 100 randomly chosen tips of the GFP::LRG1 expressing strain (Figure 4A). At a growth rate of ≥0.2 μm/s, 70% displayed an apical cap, whereas 2% did not. In contrast, only 10% of the hyphae growing at rates of <0.2 μm/s and displayed an apical cap, whereas 18% did not. Thus, the localization of GFP::LRG1 as a cap-like structure at the hyphal tip was dependent on active growth. When we quantified the localization of GFP::LRG1*, we observed the opposite behavior. Only 13% fast-growing and 2% slow-growing tips displayed a cap, whereas 62 and 23%, respectively, did not show an apical cap of GFP::LRG1, confirming that the LIM domains are responsible for localizing LRG1 to the growing hyphal apex in addition to their importance in targeting LRG1 to sites of septation.
Figure 4.
The localization of GFP::LRG1 is dependent on active growth. (A) One hundred randomly chosen hyphal tips were assayed for the presence of an apical GFP::LRG1 or GFP::LRG1* cap. In fast growing hyphae, an apical GFP::LRG1 cap was detected in 70% hyphal tips, whereas only 13% of GFP::LRG1* hyphae displayed a visible cap. (B) The size of the apical GFP::LRG1 cap correlated with the growth rate of the tip (determined by the distance between the tip in two images taken at 20-s intervals). Examples of slow, medium, and fast growing tips are shown in the top panel. Bar, 5 μm. (C) The GFP::LRG1 localization at the forming septum occurred in two phases. Approximately 1.5 min after the appearance of Calcofluor White–positive septae (a; white arrow) GFP::LRG1 began to localize along the septal plate initially as punctate specs (b; red arrow) that soon merged into a diffuse plate (c; red arrowheads). This septal GFP::LRG1 localization disappeared then for ca. 10 min (d; yellow arrow), before it strongly accumulated at the central part of the septum along the septal pore (e; white arrowheads). Bar, 5 μm. (D) Treatment of the GFP::LRG1 expressing strain with 1 μM latrunculin A affected the localization of GFP::LRG1 at the hyphal apex within 30 s (top left, and middle images) and before any morphological change, which occurred within 5 min (DIC image; top right image), but had no influence on the terminal localization of GFP::LRG1 along as bright spot at the septal pore (bottom images). Bar, 5 μm.
During this analysis, we noted that the size of the apical cap varied with the growth rate. GFP::LRG1 accumulated in slow growing hyphae primarily along the central apical cortex, whereas in fast growing tips, an extended cap structure was visible, with the highest GFP intensity frequently accumulating as a subapical ring ∼2 μm behind the apex (Figure 4B). When we quantified the length of the GFP::LRG1 marked apical cortex relative to the growth rate of the tip, we observed a positive correlation (R2 = 0.6204) between growth rate and size of the apical cap.
Septum formation in filamentous fungi requires the formation of actin rings as an initial step before cross wall formation. The myosin-dependent constriction of these rings then serves as track for the delivery of glucan- and chitin-rich vesicles (visualized by Calcofluor White staining) that then form the septal plate (Harris, 2001; Rasmussen and Glass, 2005, 2007). To investigate LRG1's function during septum formation, we analyzed the dynamics of GFP::LRG1 in subapical regions (Figure 4C). Calcofluor White positive cross walls appeared before the accumulation of weak GFP::LRG1 dots along the cortical region of a forming septum. These dots merged into a septal plate, resulting in weak labeling of the whole septum after ∼3 min. This diffuse septal localization disappeared then for ca. 10 min, before GFP::LRG1 strongly accumulated in the central region around the septal pore. This final localization remained permanent and was visible at most older septae. When we incubated growing hyphae with 1 μM latrunculin A, we found that the apical cap was disappearing within 30 s (Figure 4D), before the cessation of growth. The actin-depolymerizing agent induced a morphological change of the hyphal tip after ∼5 min. Nevertheless, the septal pore-associated localization of GFP::LRG1 remained unaffected by this treatment, indicating that this terminal association of LRG1 with the central region of the septum is independent of a functional actin cytoskeleton.
The Apical Localization of LRG1 Is Influenced by a Functional Microtubule System and Opposing Motor Proteins
We also tested the dependence of the localization of LRG1 on a functional microtubule cytoskeleton by growing GFP::LRG1 on 4 μg/ml nocodazole or 5 μg/ml benomyl. For both treatments, the apical LRG1 cap persisted as long as growth was observed, indicating that a functional microtubule cytoskeleton is dispensable for LRG1's localization (data not shown). However, the size of the apical GFP::LRG1 cap was reduced by ca. 50% in inhibitor-treated cells when compared with untreated wild-type tips growing at comparable rates (Figure 5, suggesting that a functional microtubule cytoskeleton is necessary for a stable accumulation of LRG1 at the hyphal tip.
Figure 5.
The localization of GFP::LRG1 is influenced by opposing microtubule-dependent motor proteins. (A) Growth rate and size of the GFP::LRG1 cap was determined in hyphae treated with 4 μM nocodazole for 2 min and compared with untreated control cells. Because the inhibitor treatment resulted in reduced growth rates, only slow-growing control cells with growth rates of up to 0.15 μm/s were included in the comparison. (B) lrg-1(12-20), cot-1(1) and the respective double mutants with components defective in the dynein/dynactin subunits ro-1, ro-3, and ro-10 or in the phosphatase-associated factor gul-1 were cultivated for 3 d at 37°C. The increased colony diameter of lrg-1(12-20);ro-1/-3/-10 mutants compared with lrg-1(12-20) indicate suppression of the GAP defect by loss of dynein/dynactin function. (C) Apical GFP::LRG1 caps were detected in 78% of wild-type tips with growth rates of >0.05 μm/s. This rate of GFP::LRG1 caps increased in mutants defective in the anterograde directed motor protein nkin to 91% and decreased to 19% in the mutants defective in the retrograde directed motor protein ro-1. (D) GFP::LRG1* in a ro-1 background localized along the cortex and the hyphal morphology of the double mutant closely resembled the deletion phenotype.
The high rate of fungal tip extension requires efficient microtubule-based and motor protein-dependent transport systems for supplying the growing tip with all necessary growth supplies and regulatory factors (Steinberg, 2007). This transport toward the growing tip could be the reason for the observed microtubule-dependence of apical LRG1 localization. Thus we analyzed the impact of mutants defective in anterograde- (kinesin-1) and retrograde-directed (dynein) on apical GFP::LRG1 localization (Seiler et al., 1999; Zhang et al., 2003). We compared the growth rates of various ropy (= dynein/dynactin) strains in single as well as double mutants in combination with lrg-1(12-20) and found that ropy mutations partially suppressed lrg-1(12-20) (Figure 5B). In contrast to this common phenotype of lrg-1(12-20) and the Ndr pathway mutants cot-1(1) and pod-6(31–21), gul-1 that had been implicated in phosphatase-associated, COT1/POD6-antagonizing functions (Terenzi and Reissig, 1967; Seiler et al., 2006), only suppressed the growth defects of cot-1(1) and pod-6(31–21), but not those of lrg-1(12-20). Next, we tested for a genetic connection between lrg-1(12-20) and nkin, which had been shown to counteract dynein's retrograde transport activity (Seiler et al., 1999; Zhang et al., 2003; Seiler et al., 2006). The growth rate of a lrg-1(12-20);nkin double mutant at semirestrictive temperature was reduced by 42% when compared with lrg-1(12-20) as the slower growing of the two parental strains, indicating a synthetic interaction between lrg-1(12-20) and nkin.
These data suggested that both opposing microtubule-dependent motor proteins are involved in localizing LRG1. Therefore, we determined the localization of GFP-tagged LRG1 in these motor mutants. As determined previously (Figure 4A), the majority of slow-growing hyphal tips of wild type displayed no GFP::LRG1 cap. The growth rates of the two motor mutants is ca. 20% of wild type (Seiler et al., 1997, 1999), and thus, a visible cap should be rarely visible, if the GFP::LRG1 localization is independent of the two motors. We observed apical caps in only 22% of wild-type tips at growth rates <0.05 μm/s (Figure 5C). This contrasted sharply with 81% of ro-1 tips growing at rates of <0.05 μm/s that displayed an apical GFP::LRG1 cap, while the number of GFP::LRG1 caps at growth rates >0.05 μm/s was reduced to 8% in the nkin background (Figure 5C). Thus the apical localization of LRG is influenced by opposing microtubule-dependent motor proteins. This localization dependence of LRG1 on both motor proteins and functional LIM domains was further supported when we introduced GFP::LRG1* in the ro-1 background. The resulting progeny closely resembled the lrg-1 deletion phenotype, with poorly growing and highly hyperbranched cells (Figure 5D). Interestingly, GFP::LRG1* localized in an aberrant cortical manner similar to what we have determined for the GAP domain alone (Figure 3 A), suggesting that either of the two localization mechanism alone is sufficient for apical localization of LRG1, but disturbing of both targeting mechanisms results in abnormal localization of LRG1.
LRG1 Is a RHO1-specific GAP Affecting Several Output Pathways of RHO1
Given LRG1's homology with Rho-GAP proteins, we determined, which of the six Rho-GTPases present in the N. crassa genome are regulated by LRG1. When we overexpressed the wild-type forms of all RHO proteins in lrg-1(12-20), only RHO1 was capable of partially complementing the lrg-1(12-20) growth defect. Also dominant active (Rho1G15V) as well as dominant negative (Rho1E41I) RHO1 alleles were able to overcome the lrg-1(12-20) dependent growth defect initially and generated hyphal tips that are capable of apical extension (Figure 6A). Nevertheless, all growing transformants died within 2 d with swollen and lysed hyphae (data not shown). This death phenotype was also observed when we overexpressed the three RHO1 alleles in wild type and is thus likely the result of interfering with endogenous RHO1 function (Figure 6B). A deletion strain of rho-1 was available through the Neurospora Genome Project (Dunlap et al., 2007). Its heterocaryotic nature already suggested the essentiality of RHO1. When we plated asexually derived conidiospores on hygromcyin-containing media, we detected a mixture of germinating and isotropically growing spores, indicating that RHO1 is essential for establishing polarity (Figure 6C). Similarly, we were unable to obtain viable hygromycin-resistant ascospores from wild type × Δrho-1::hph + rho-1+ crosses (Figure 6C). The majority of the hygR ascospores did germinate apolarly, indicating that RHO1 is required for polarity establishment in both types spores. Rarely, we observed the formation of hygR colonies, which then showed a high rate of cell lysis, suggesting that suppressor mutation can overcome the rho-1 deletion, but cell wall integrity remains affected.
Figure 6.
LRG1 is a RHO1–specific GTPase activating protein. (A) Overexpression of wild type as well as dominant-active (rho-1G15V) and dominant-negative (rho-1E41I) rho-1 alleles, but not of any of the other Rho proteins was sufficient to overcome the morphological defect of lrg-1(12-20) at restrictive temperature. Bar, 50 μm. (B) Death phenotypes of wild-type cells transformed with the overexpression constructs of the indicated RHO1 alleles. (C) Conidia (left side) of a heterocaryotic Δrho-1::hphR + rho-1+ strain were germinated on minimal media supplemented with hygromycin. Spores harboring only the deletion nucleus germinated isotropically (red arrows), whereas heterokaryotic cells were able to produce polar germ tubes (blue arrowheads). Bar, 20 μm. Ascospore progeny (right side) of Δrho-1::hphR + rho-1+× wild-type crosses predominatly germinated apolarly on medium supplemented with hygromycin (top image). Bar, 20 μm. Rarely (<1 of 1000 ascospores) we observed growing hygR colonies, which then displayed a high rate of cell lysis (bottom image). Bar, 200 μm. (D) Intrinsic and LRG1650–1035 stimulated GTPase activities of the six Rho proteins. Bacterially expressed and affinity-purified Rho proteins were preloaded with [γ-32-P]GTP, and the reactions started by the addition of LRG1650–1035 or buffer. GTPase activities were determined by measuring remaining RHO-bound [γ-32P]GTP after 8 min at 25°C (see Materials and Methods). (E) Kinetics of in vitro RHO1 GTPase activity in the presence and absence on LRG1650–1035.
Next, we performed in vitro GTPase assays with the six bacterially expressed and affinity-purified His6::GST::Rho fusion proteins (Figure 6, D and E). All G-proteins had significant intrinsic GTP-GDP turnover rates, indicating that the purified enzymes were active. The addition of the GST::GAP domain of LRG1 stimulated only the GTPase activity of RHO1 (ca. 10-fold). None of the other GTPases exhibited any significant change in activity. Thus, both the genetic and the in vitro experiments identify LRG1 as a RHO1–specific GAP.
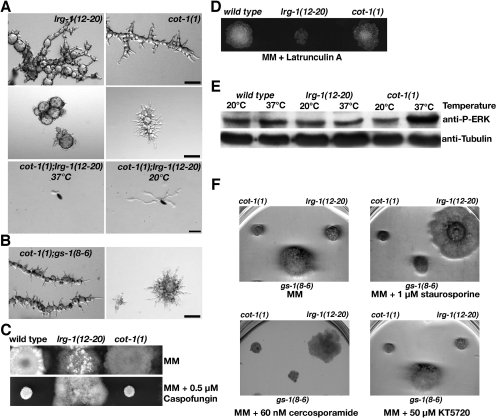
RHO1 is a bona fide regulatory subunit of the cell wall enzyme β1,3-glucan synthase in yeasts and filamentous fungi. Furthermore, it is activating the cell wall integrity MAP kinase pathway (Beauvais et al., 2001; Levin, 2005). Therefore, we tested the impact of defective LRG1 function on these signaling routes. We have recently described conditional mutants in gs-1, a regulator of β1,3-glucan synthase (Seiler and Plamann, 2003), and when we generated a lrg-1(12-20);gs-1(8-6) double mutant, it did display strong synthetic defects (Figure 7A). Its growth rate at permissive or semirestrictive temperature was reduced by 25 and 41%, respectively, when compared with lrg-1(12-20) as the slower growing of the two parental strains. At restrictive conditions, gs-1(8-6) generated growth-inhibited hyperbranched hyphae that were still capable of slow apical growth. In contrast, lrg-1(12-20);gs-1(8-6) formed apolarly growing spheres when germinated at 37°C. When germinated hyphae were transferred to restrictive conditions, chains of spherical-growing cells were generated within 8 h. Furthermore, these strains were less sensitive against caspofungin at semipermissive conditions (Figure 7B), a specific inhibitor of fungal β1,3-glucan synthesis (Denning, 2003), indicating hyperactive β1,3-glucan synthase in lrg-1(12-20) and gs-1(8-6). Although the growth rate of lrg-1(12-20) increased in the presence of caspofungin, hyphal morphology, and branching frequency was not different in treated versus untreated cells (Supplementary Figure S1).
Figure 7.
LRG1 is affecting several RHO1-dependent effector pathways. (A) The indicated strains were grown at permissive temperature and shifted to 37°C for 10 h (top) or germinated at restrictive temperature for 15 h (bottom) to illustrate the synthetic nature of the two mutations. Bar, 40 μm. (B) lrg-1(12-20), gs-1(8-6), and lrg-1(12-20);gs-1(8-6) grown at permissive temperature are hyposensitive against the β1,3-glucan synthase inhibitor caspofungin. (C) Growth of the indicated strains for 48 h at 32°C on plates supplemented with the kinase inhibitors staurosporine, KT5720, or cercosporamide. The growth defects of lrg-1(12-20) were suppressed by staurosporine and cercosporamide, but not by KT5720. The growth of wild-type cells was not affected by the mentioned conditions. One example of three consistent experiments is shown. (D) Total soluble protein was extracted from wild type and lrg-1(12-20) shifted to 37°C for the indicated times. The blot was probed with anti-phospho-ERK (α-P-ERK) antibody to detect activated MAK1 (top panel) induced by the temperature stress. A replicate was probed with anti-ERK to confirm equal loading (bottom panel). Wild type but not lrg-1(12-20) showed stress-induced MAK1 activation after 20 min at 37°C. (E) MAK1 phosphorylation induced by the addition of 7 mM H2O2 for 30 min to wild type and lrg-1(12-20) indicate that the response capacity of the MAK1 pathway is not affected in lrg-1 mutants. One example of five consistent experiments is shown in D and E (originating from individually treated biological cultures and independent protein preparations). (F) lrg-1(12-20) grown at permissive conditions is hypersensitive against the actin-depolymerizing drug latrunculin A.
The abnormal chitin distribution observed in lrg-1(12-20) (Figure 1A) could be a result of increased activity of the N. crassa PKC/MAK1 cell integrity pathway and subsequently the altered expression of additional cell wall enzymes. Therefore, we tested for altered activity levels of PKC in the GAP mutant. We observed a partial suppression of the lrg-1(12-20) growth defect on media supplemented with 1 μM staurosporine, a protein kinase inhibitor with highest specificity toward PKC (Figure 7C). Nevertheless, staurosporine is also inhibiting PKA (although with a ca. 10-fold lower affinity). Therefore, we tested the impact of 50 μM KT5720, a PKA-specific inhibitor, on the growth behavior of lrg-1(12-20), but observed no morphological changes (Figure 7C), indicating that PKC is hyperactive in lrg-1(12-20). This was further supported by growth tests on media supplemented with 60 nM cercosporamide, a highly specific inhibitor for PKC (Sussman et al., 2004).
Next, we tested the activity of the cell integrity MAP kinase MAK1 by the use of phospho-specific antibodies against activated ERK-type MAPKs. No obvious differences were found in MAK1 phosphorylation pattern in wild-type extracts from cultures grown at 25 and 37°C. Stressing these cells by a temperature shift from 25 to 37°C for 20 min resulted in activation of cell integrity signaling and increased MAK1 activity (Figure 7D). In contrast, the MAK1 phosphorylation pattern in lrg-1(12-20) shifted to restrictive temperature for 20 min did not result in heat stress-induced phosphorylation of MAK1, suggesting that MAK1 regulation may be affected in lrg-1(12-20). However, we did not detect increased MAK1 activity levels in lrg-1(12-20) after prolonged temperature shift (Figure 7D) or in Δlrg-1 germinated on benomyl and panthotenic acid (data not shown) as one would predict for increased RHO1–MAK1 signaling. To test, whether the capacity of the MAK1 pathway to respond to stress signals is affected in lrg-1(12-20) in a general manner, wild type and lrg-1(12-20) were grown at 37°C and stressed by the addition of 7 mM H2O2 for 20 min (Figure 7E). Both strains displayed identical activation patterns as detected by MAK1 phosphorylation, indicating that the response capacity of the MAK1 pathway is not affected in lrg-1(12-20). Taken together, these data indicate that RHO1–PKC signaling is increased in lrg-1(12-20), but also suggest that the downstream MAK1 MAP kinase pathway is only affected to a minor extent.
Furthermore, we determined that lrg-1(12-20) was hypersensitive against the actin depolymerizing drug latrunculin A, suggesting an altered actin cytoskeleton in the GAP mutant (Figure 7F). In summary, we suggest that LRG1 acts as a general GAP for RHO1 and regulates several RHO1-dependent signaling pathways in N. crassa.
Genetic Interactions of lrg-1 with the Ndr Kinase Mutant cot-1 Indicate Two Parallel Pathways for Polar Tip Growth
The morphological defects of lrg-1(12-20) and of Δlrg-1 were strikingly similar to those of the Ndr kinase mutant cot-1 and its upstream regulating kinase pod-6 (Yarden et al., 1992; Seiler et al., 2006). The common defects of these mutants are a block in apical tip extension and the generation pointed, needle-like tips, the induction of multiple subapical branches, excessive and mislocalized chitin distribution, and a highly thickened cell wall. Furthermore, several recessive alleles of cot-1, pod-6, and lrg-1 showed unlinked noncomplementation (Seiler and Plamann, 2003) and suggest LRG1 and COT1/POD6 either as components of two independent pathways that act in parallel to support apical tip growth or hint for a physical interaction of the three proteins. In contrast to a cot-1(1);pod-6(31–21) double mutant, which displayed defects identical to the two parental strains (Seiler et al., 2006), cot-1(1);lrg-1(12-20) or pod-6(31–21);lrg-1(12-20) double mutants were synthetically lethal (Figure 8A). Double mutant ascospores germinated at permissive conditions generated polar germ tubes, but grew in a compact and highly vacuolated manner with very slow tip extension rates and frequent lysis of hyphal tips or subapical regions. Germination at 37°C resulted in apolar germination and death of the double mutants. In combination with the observation that lrg-1 and cot-1/pod-6 share the same suppressors (Figure 5B), these genetic data indicate two parallel, COT1/POD6- and LRG1-dependent, pathways for apical tip extension.
Figure 8.
LRG1 acts in a pathway parallel to the Ndr kinase COT1 (A) lrg-1(12-20) and cot-1(1) displayed nearly identical defects with tip extension terminating with 2–10- and 10–30-μm-long pointed tips, respectively, when shifted from permissive to restrictive conditions for 12 h (top panel) or when germinated at 37°C for 15 h (middle panel). Double mutant ascospores germinated at 25°C produced compact, highly vacuolated and slow growing hyphae. Germination at 37°C resulted in apolar germination and death of lrg-1(12-20);cot-1(1) ascospores. (bottom panel). Bars, 40 μm. (B) Double mutants of cot-1(1) and gs-1(8-6) did not display synthetic defects after shift to restrictive temperatures (left image) or when germinated under restrictive conditions (right image). Bar, 40 μm. (C) The growth behavior of cot-1(1) is identical to wild type on medium supplemented with 0.5 μM caspofungin. (D) cot-1(1) is not hypersensitive to the addition of 0.5 μM latrunculin A. (E) Total soluble protein of the indicated strains grown at 25 or shifted to 37°C for 10 h was extracted. The Blot was probed with an anti-phospho-ERK antibody to detect increased MAK1 activity levels in cot-1(1) grown at restrictive conditions (top panel). To confirm equal loading the blot was reprobed with anti-tubulin antibody (bottom panel). One example of five consistent experiments is shown. (F) The PKC-specific inhibitors staurosporine and cercosporamide partially suppressed the tip extension defects of lrg-1(12-20), but not those of cot-1(1). The growth of wild-type cells was not affected by the mentioned conditions. One example of three consistent experiments is shown.
Intrigued by these results, we tested for a potential connection between COT1 and RHO1 signaling and analyzed the RHO1 effector pathways that are affected by lrg-1. A highly thickened cell wall of cot-1(1) grown at 37°C described in electron microscopic sections (Gorovits et al., 2000) and the excessive chitin distribution at sites of aberrant growth (Seiler et al., 2006) already indicated a defect in cell wall organization. However, we did not observe any synthetic interaction in a cot-1(1);gs-1(8-6) double mutant (Figure 8B) and detected growth characteristics of cot-1(1) identical to wild type on media supplemented with caspofungin (Figure 8C), indicating no direct regulation of β1,3 glucan synthase by COT1. Furthermore, in contrast to the hypersensitivity of lrg-1(12-20) to latrunculin A, we did not observe any difference of cot-1(1) compared with wild type when the strains were grown on media supplemented with 0.5 μM latrunculin A (Figure 8D). However, when we analyzed the activity of the cell wall integrity pathway in cot-1(1), we observed increased MAK1 activity levels in cot-1(1) cultures grown at restrictive temperature (Figure 8E), implying COT1 as a potential negative regulator of the MAK1 pathway. Interestingly, the cot-1(1) defect was not suppressed by the PKC inhibitors straurosporine and cercosporamide (Figure 8F), indicating that MAK1 MAP kinase activation is not induced via RHO1–PKC signaling. Taken together, these results establish distinct effects of LRG1 and of COT1 on the analyzed RHO1-dependent effector pathways.
DISCUSSION
Cellular polarity is a fundamental property of every cell, and fungal hyphae are among the most highly polarized cells known in nature. In a visual screen designed to identify factors critical for hyphal morphogenesis, we isolated lrg-1 as a component that is essential for apical tip extension and to restrict excessive branch formation in subapical regions of the hypha (Seiler and Plamann, 2003). The data presented here characterize LRG1 as a RHO1–specific GAP. In vitro, only the GTPase rate of RHO1 is stimulated significantly by LRG1, and the in vivo results indicate several misregulated RHO1 effector pathways in mutants lacking a functional GAP. Interesting, but difficult to explain is that overexpression of wild type and of both dominant rho-1 versions is partially compensating the lrg-1 growth defects. Similar results were obtained for mammalian Cdc42 during cell proliferation, where the stable overexpression of the constitutively active G12V mutant did result in a dominant negative phenotype after prolonged growth (Vanni et al., 2005). Furthermore, the fact that lrg-1(12-20) cells, which are deficient for Rho-GAP activity, are defective in polarized growth, suggests that cycling of RHO1, rather than its GTP-bound form alone is important for its function. Thus, our results indicate that polar tip extension and the regulation of branch formation may involve cycling of N. crassa RHO1 to gain full activation, rather than its functioning as simple on/off switch.
Rho1 is a key regulator of hyphal growth and polarity and has been described as integral part of the β1,3-glucan synthase complex in yeasts and A. fumigatus (Arellano et al., 1996; Beauvais et al., 2001; Levin, 2005). The observed caspofungin hyposensitivity and the genetic interaction of lrg-1(12-20) with gs-1(8-6) are consistent with increased β1,3-glucan synthase activity in lrg-1(12-20) and gs-1(8-6). Furthermore, the latrunculin A sensitivity suggest altered RHO1–actin signaling in lrg-1(12-20). Difficult to interpret are the data presented for the PKC/MAK1 pathway. The suppression of lrg-1(12-20) by the PKC specific inhibitors staurosporine and cercosporamide indicate hyperactivity of PKC in the GAP mutant. Nevertheless, downstream MAK1 activity is not enhanced, suggesting that the MAP kinase cascade is not regulated by LRG1. However, the MAK1 activation pattern in heat-stressed lrg-1(12-20) is different from wild type, supporting a regulatory function of the cell integrity pathway by LRG1. Thus, additional feedback mechanisms may regulate the activation of the PKC–MAK1 part of the cell integrity pathway in N. crassa.
Which of these RHO1 effector pathways may be responsible for the block in tip extension and the subapical hyperbranching observed in lrg-1? On the basis of data presented here, we do not believe that the altered activities of the PKC–MAK1 pathway are the primary defect. Nevertheless, the unregulated deposition of excessive chitin with in the cell wall of lrg-1 may be a result transmitted through PKC. As demonstrated in budding yeast, cell wall stress can result in up to a 10-fold increase of the cell wall's chitin content by stress-induced mobilization of Chs3p from chitosome vesicles to the plasma membrane. This stress signal is transmitted via Rho1p and Pkc1p, involving components of the cell integrity pathway, but does not include the downstream MAP kinase components (Valdivia and Schekman, 2003).
Loss of LRG1 results in increased glucan formation, which may interfere with the plasticity of the hyphal cell wall and could thus lead to the cessation of tip extension and the induction of branch formation in subapical regions of the hypha. This hypothesis is supported by a common hyperbranching phenotype of lrg-1(12–10) and gs-1(8-6) as a result of increased glucan deposition in the cell walls of both strains as monitored by the hyposensitivity to the glucan synthase inhibitor caspofungin. Interesting is that the lrg-1(12–10);gs-1(8-6) double mutant loose cellular polarity when hyphae are shifted to restrictive conditions and are unable to establish polarity when conidia are germinated at high temperature. This phenotype is reminiscent to conditional mutants of key regulators of the actin cytoskeleton such as bem-1 and cdc-24 (Seiler and Plamann, 2003) and suggests a tight connection between cell wall function and the actin cytoskeleton for apical tip extension and regulation of branch formation. Thus, we speculate that increased GS1 activity and altered actin organization are the major cause for the observed lrg-1 defects.
A clear homolog of LRG1 is also found in S. cerevisiae (E = 1e−111), but conflicting evidence about the Rho protein(s) and the Rho effector pathway(s) that are regulated by Lrg1p was reported. In vitro and yeast two hybrid analyses identified Lrg1p as a GAP for Rho1p, Rho2p, or Cdc42p or for combinations of these GTPases (Lorberg et al., 2001; Roumanie et al., 2001; Watanabe et al., 2001; Fitch et al., 2004). However, most genetic data link Lrg1p with Rho1p functions (Lorberg et al., 2001; Watanabe et al., 2001; Fitch et al., 2004; Varelas et al., 2006; Stewart et al., 2007), suggesting that Lrg1p is a Rho1p specific GAP in budding yeast. Furthermore, the effector pathways that are regulated by Lrg1p are discussed in a controversial manner. Two studies report enhanced MAP kinase activity (Lorberg et al., 2001; Stewart et al., 2007) in contrast to a report describing no effect on the MAP kinase (Watanabe et al., 2001). In addition, increased glucan synthase activity was reported for Δlrg-1 (Lorberg et al., 2001; Watanabe et al., 2001; Fitch et al., 2004), but no effect of Lrg1p on the Bni1p/actin branch of Rho1p signaling could be identified in any of these studies. In contrast, Nakano et al. (2001) reported delocalized actin patches in a mutant defective in rga-1, the fission yeast homolog of LRG1.
The domain analysis of LRG1 revealed that both parts of the protein are essential for its cellular function. Whereas deletion of the GAP domain and point mutations in residues that are required for either the interaction of LRG1 with RHO1 or for the GAP activity of LRG1 strongly support the importance of the enzymatic function of LRG1, several lines of evidence indicate that the N-terminal part of the protein is required for the localization of the GAP domain to sites of active growth. A construct lacking the first 780 aa (LRG781–1279) is not capable of complementing the mutant defects, but this N-terminal domain (LRG1–847) is sufficient for correct localization. The morphological defects observed in the mutant and the localization pattern of LRG1 indicate a function of the GAP at sites of active growth at the hyphal tip and during septation. This is further supported by its growth-dependent localization of LRG1 at the hyphal apex and along septae and by the correlation between the growth rate and size of the GFP::LRG1 cap at the hyphal tip. A clear indication for the involvement of the three LIM domains within the N-terminus of LRG1 in the localization process is the altered localization pattern in GFP::LRG1*, in which the three LIM domains have been mutated. However, despite its localization defect, GFP::LRG1* was capable of rescuing the growth defect of Δlrg-1, indicating additional functional motifs within the N-terminus. Interesting in this respect is the high sequence similarity of these domains with paxillin (E = 1e−28, whereas other LIM domain proteins aside from LRG1 homologues have much weaker similarities of E = 1e−14), suggesting that LRG1 might act as a cytoskeletal organizer similar to paxillin in focal adhesion complexes (Schaller, 2001; Brown and Turner, 2004). Another possibility could be the interaction of microtubule-dependent motor proteins with additional sites within the N-terminus of LRG1. We showed that treatment of cells with anti-microtubule drugs reduced the apical LRG1 localization and that motor-driven transport affected the localization of LRG1. The reduced retrograde transport of dynein/dynactin mutations resulted in enriched apical localization of LRG1, whereas its abundance is reduced in nkin, which is defective for apical-directed transport. Thus we suggest that the opposing transport rates of NKIN and dynein result in a balanced distribution of LRG1 and thus allow growth that is independent of the functionality of the LIM domains. The abolishment of both—the microtubule- and LIM domain-dependent—localization mechanisms by deleting the whole N-terminus is resulting in the loss of function of LRG1 as observed in LRG1781–1279 and by the introduction of LRG1* into the ro-1 background. Both conditions resulted cells resembling the lrg-1 loss-of-function phenotype and in an aberrant cortical localization of the GAP. A similar dependence for correct localization on opposing motor proteins was also observed for cot-1 and pod-6 and suggest a general mechanism to supply all regions of fast growing, highly polar cells with the necessary components. In contrast to this microtubule-dependent distribution of LRG1 throughout the hypha, its localization as an apical cap during active growth is clearly F-actin–dependent, as determined in the latrunculin A inhibitor experiments.
In addition to this motor-dependent localization of LRG1 and COT1/POD6, mutants defective in the two kinases of the Ndr signaling network and in lrg-1 displayed nearly identical phenotypes. The major difference of the kinase mutants versus lrg-1 is the reduced length of the subapical branches that are induced by the temperature shift in lrg-1. Our double mutant analysis, nonalleleic noncomplementing alleles and common suppressors provide strong evidence that the COT1/POD6 complex is acting in parallel to LRG1/RHO1 signaling. Genetic data in S. cerevisiae suggest that the COT1 homolog Cbk1p may negatively regulate the small GTPase Rho1p, although the mechanism of interaction remained open (Jorgensen et al., 2002; Schneper et al., 2004). A similar negative effect of COT1 on RHO1 signaling in N. crassa would explain the observed synthetic lethality of the lrg-1(12-20);cot-1(1) double mutant. However, loss of COT1 function did not affect GS1 activity or the actin cytoskeleton, and the only RHO1 effector pathway that is altered in cot-1(1) is the MAK1 MAP kinase pathway (but not the upstream acting PKC). Thus, the molecular basis for the observed genetic interactions remains open. Interesting in this context is that a physical interaction between the Ndr kinase ORB6 and a different Rho-GAP has been proposed to exist in fission yeast based on two hybrid studies (Das et al., 2007). An indication that the connection between Rho GTPases and Ndr signaling is conserved between fungi and animals has been provided by studies in D. melanogaster and Caenorhabditis elegans (Zallen et al., 2000; Emoto et al., 2004). Thus, the genetic connection established, but also the distinctions between COT1/POD6 and LRG1/RHO1 signaling during hyphal growth, and the similarities of LRG1 to the focal adhesion organizer paxillin may provide insights in the regulation of morphogenesis in other highly polar cells such as neurons.
Figure 9.
Model summarizing components and potential connections between the LRG1/RHO1 and COT1/POD6 pathways in the regulation of hyphal tip extension in N. crassa. Details are discussed in the text.
Supplementary Material
ACKNOWLEDGMENTS
The authors acknowledge the help of Inga von Behrens and Seema Singh in the initial phase of this work and thank Angela Hoffman (University of Portland, Portland, OR) for a gift of cercosporamide. Caspofungin was kindly provided by MSD Sharp & Dohme, GMBH. This research project was financially supported by the Deutsche Forschungsgemeinschaft through the DFG Schwerpunkt Zellpolarität (SPP1111) and the DFG Research Center of Molecular Physiology of the Brain (CMPB).
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E07-12-1266) on August 20, 2008.
REFERENCES
- Adams M. D., et al. The genome sequence of. Drosophila melanogaster. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- Alberts A. S. Identification of a carboxyl-terminal diaphanous-related formin homology protein autoregulatory domain. J. Biol. Chem. 2001;276:2824–2830. doi: 10.1074/jbc.M006205200. [DOI] [PubMed] [Google Scholar]
- Arellano M., Duran A., Perez P. Rho 1 GTPase activates the (1–3)beta-d-glucan synthase and is involved in Schizosaccharomyces pombe morphogenesis. EMBO J. 1996;15:4584–4591. [PMC free article] [PubMed] [Google Scholar]
- Barale S., McCusker D., Arkowitz R. A. Cdc42p GDP/GTP cycling is necessary for efficient cell fusion during yeast mating. Mol. Biol. Cell. 2006;17:2824–2838. doi: 10.1091/mbc.E05-11-1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beauvais A., Bruneau J. M., Mol P. C., Buitrago M. J., Legrand R., Latge J. P. Glucan synthase complex of. Aspergillus fumigatus. J. Bacteriol. 2001;183:2273–2279. doi: 10.1128/JB.183.7.2273-2279.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bombarda E., Cherradi H., Morellet N., Roques B. P., Mely Y. Zn(2+) binding properties of single-point mutants of the C-terminal zinc finger of the HIV-1 nucleocapsid protein: evidence of a critical role of cysteine 49 in Zn(2+) dissociation. Biochemistry. 2002;41:4312–4320. doi: 10.1021/bi015956g. [DOI] [PubMed] [Google Scholar]
- Borkovich K. A., et al. Lessons from the genome sequence of Neurospora crassa: tracing the path from genomic blueprint to multicellular organism. Microbiol. Mol. Biol. Rev. 2004;68:1–108. doi: 10.1128/MMBR.68.1.1-108.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyce K. J., Hynes M. J., Andrianopoulos A. Control of morphogenesis and actin localization by the Penicillium marneffei RAC homolog. J. Cell Sci. 2003;116:1249–1260. doi: 10.1242/jcs.00319. [DOI] [PubMed] [Google Scholar]
- Boyce K. J., Hynes M. J., Andrianopoulos A. The Ras and Rho GTPases genetically interact to co-ordinately regulate cell polarity during development in Penicillium marneffei. Mol. Microbiol. 2005;55:1487–1501. doi: 10.1111/j.1365-2958.2005.04485.x. [DOI] [PubMed] [Google Scholar]
- Brown M. C., Turner C. E. Paxillin: adapting to change. Physiol. Rev. 2004;84:1315–1339. doi: 10.1152/physrev.00002.2004. [DOI] [PubMed] [Google Scholar]
- Bustelo X. R., Sauzeau V., Berenjeno I. M. GTP-binding proteins of the Rho/Rac family: regulation, effectors and functions. in vivo. Bioessays. 2007;29:356–370. doi: 10.1002/bies.20558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C., Dickman M. B. Dominant active Rac and dominant negative Rac revert the dominant active Ras phenotype in Colletotrichum trifolii by distinct signalling pathways. Mol. Microbiol. 2004;51:1493–1507. doi: 10.1111/j.1365-2958.2003.03932.x. [DOI] [PubMed] [Google Scholar]
- Chilton J. K. Molecular mechanisms of axon guidance. Dev. Biol. 2006;292:13–24. doi: 10.1016/j.ydbio.2005.12.048. [DOI] [PubMed] [Google Scholar]
- Das M., Wiley D. J., Medina S., Vincent H. A., Larrea M., Oriolo A., Verde F. Regulation of cell diameter, For3p localization, and cell symmetry by fission yeast Rho-GAP Rga4p. Mol. Biol. Cell. 2007;18:2090–2101. doi: 10.1091/mbc.E06-09-0883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis R. D., DeSerres F. J. Genetic and microbiological research techniques for Neurospora crassa. Methods Enzymol. 1970;17:79–143. [Google Scholar]
- Denning D. W. Echinocandin antifungal drugs. Lancet. 2003;362:1142–1151. doi: 10.1016/S0140-6736(03)14472-8. [DOI] [PubMed] [Google Scholar]
- Dunlap J. C., et al. Enabling a community to dissect an organism: overview of the Neurospora functional genomics project. Adv. Genet. 2007;57:49–96. doi: 10.1016/S0065-2660(06)57002-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emoto K., He Y., Ye B., Grueber W. B., Adler P. N., Jan L. Y., Jan Y. N. Control of dendritic branching and tiling by the Tricornered-kinase/Furry signaling pathway in Drosophila sensory neurons. Cell. 2004;119:245–256. doi: 10.1016/j.cell.2004.09.036. [DOI] [PubMed] [Google Scholar]
- Etienne-Manneville S., Hall A. Rho GTPases in cell biology. Nature. 2002;420:629–635. doi: 10.1038/nature01148. [DOI] [PubMed] [Google Scholar]
- Fitch P. G., Gammie A. E., Lee D. J., de Candal V. B., Rose M. D. Lrg1p Is a Rho1 GTPase-activating protein required for efficient cell fusion in yeast. Genetics. 2004;168:733–746. doi: 10.1534/genetics.104.028027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia P., Tajadura V., Garcia I., Sanchez Y. Role of Rho GTPases and Rho-GEFs in the regulation of cell shape and integrity in fission yeast. Yeast. 2006;23:1031–1043. doi: 10.1002/yea.1409. [DOI] [PubMed] [Google Scholar]
- Gibbs J. B., Schaber M. D., Allard W. J., Sigal I. S., Scolnick E. M. Purification of ras GTPase activating protein from bovine brain. Proc. Natl. Acad. Sci. USA. 1988;85:5026–5030. doi: 10.1073/pnas.85.14.5026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goffeau A., et al. Life with 6000 genes. Science. 1996;274:546. doi: 10.1126/science.274.5287.546. [DOI] [PubMed] [Google Scholar]
- Gorovits R., Sjollema K. A., Sietsma J. H., Yarden O. Cellular distribution of COT1 kinase in Neurospora crassa. Fungal Genet. Biol. 2000;30:63–70. doi: 10.1006/fgbi.2000.1198. [DOI] [PubMed] [Google Scholar]
- Guest G. M., Lin X., Momany M. Aspergillus nidulans RhoA is involved in polar growth, branching, and cell wall synthesis. Fungal Genet. Biol. 2004;41:13–22. doi: 10.1016/j.fgb.2003.08.006. [DOI] [PubMed] [Google Scholar]
- Gulli M. P., Peter M. Temporal and spatial regulation of Rho-type guanine-nucleotide exchange factors: the yeast perspective. Genes Dev. 2001;15:365–379. doi: 10.1101/gad.876901. [DOI] [PubMed] [Google Scholar]
- Harris S. D. Septum formation in Aspergillus nidulans. Curr. Opin. Microbiol. 2001;4:736–739. doi: 10.1016/s1369-5274(01)00276-4. [DOI] [PubMed] [Google Scholar]
- Harris S. D. Cell polarity in filamentous fungi: shaping the mold. Int. Rev. Cytol. 2006;251:41–77. doi: 10.1016/S0074-7696(06)51002-2. [DOI] [PubMed] [Google Scholar]
- Hergovich A., Stegert M. R., Schmitz D., Hemmings B. A. NDR kinases regulate essential cell processes from yeast to humans. Nat. Rev. Mol. Cell Biol. 2006;7:253–264. doi: 10.1038/nrm1891. [DOI] [PubMed] [Google Scholar]
- Jaffe A. B., Hall A. Rho GTPases: biochemistry and biology. Annu. Rev. Cell Dev. Biol. 2005;21:247–269. doi: 10.1146/annurev.cellbio.21.020604.150721. [DOI] [PubMed] [Google Scholar]
- Jansen J. M., Barry M. F., Yoo C. K., Weiss E. L. Phosphoregulation of Cbk1 is critical for RAM network control of transcription and morphogenesis. J. Cell Biol. 2006;175:755–766. doi: 10.1083/jcb.200604107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen P., Nelson B., Robinson M. D., Chen Y., Andrews B., Tyers M., Boone C. High-resolution genetic mapping with ordered arrays of Saccharomyces cerevisiae deletion mutants. Genetics. 2002;162:1091–1099. doi: 10.1093/genetics/162.3.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop M., Siegers K., Pereira G., Zachariae W., Winsor B., Nasmyth K., Schiebel E. Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast. 1999;15:963–972. doi: 10.1002/(SICI)1097-0061(199907)15:10B<963::AID-YEA399>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Leveleki L., Mahlert M., Sandrock B., Bölker M. The PAK family kinase Cla4 is required for budding and morphogenesis in Ustilago maydis. Mol. Microbiol. 2004;54:396–406. doi: 10.1111/j.1365-2958.2004.04296.x. [DOI] [PubMed] [Google Scholar]
- Levin D. E. Cell wall integrity signaling in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 2005;69:262–291. doi: 10.1128/MMBR.69.2.262-291.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R., Zhang B., Zheng Y. Structural determinants required for the interaction between Rho GTPase and the GTPase-activating domain of p190. J. Biol. Chem. 1997;272:32830–32835. doi: 10.1074/jbc.272.52.32830. [DOI] [PubMed] [Google Scholar]
- Li S., Du L., Yuen G., Harris S. D. Distinct ceramide synthases regulate polarized growth in the filamentous fungus Aspergillus nidulans. Mol. Biol. Cell. 2006;17:1218–1227. doi: 10.1091/mbc.E05-06-0533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorberg A., Schmitz H. P., Jacoby J. J., Heinisch J. J. Lrg1p functions as a putative GTPase-activating protein in the Pkc1p-mediated cell integrity pathway in Saccharomyces cerevisiae. Mol. Genet. Genomics. 2001;266:514–526. doi: 10.1007/s004380100580. [DOI] [PubMed] [Google Scholar]
- Mahlert M., Leveleki L., Hlubek A., Sandrock B., Bolker M. Rac1 and Cdc42 regulate hyphal growth and cytokinesis in the dimorphic fungus Ustilago maydis. Mol. Microbiol. 2006;59:567–578. doi: 10.1111/j.1365-2958.2005.04952.x. [DOI] [PubMed] [Google Scholar]
- Malavazi I., Semighini C. P., Kress M. R., Harris S. D., Goldman G. H. Regulation of hyphal morphogenesis and the DNA damage response by the Aspergillus nidulans ATM homolog AtmA. Genetics. 2006;173:99–109. doi: 10.1534/genetics.105.052704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minke P. F., Lee I. H., Plamann M. Microscopic analysis of Neurospora ropy mutants defective in nuclear distribution. Fungal Genet. Biol. 1999;28:55–67. doi: 10.1006/fgbi.1999.1160. [DOI] [PubMed] [Google Scholar]
- Nakano K., Mutoh T., Mabuchi I. Characterization of GTPase-activating proteins for the function of the Rho-family small GTPases in the fission yeast. Schizosaccharomyces pombe. Genes Cells. 2001;6:1031–1042. doi: 10.1046/j.1365-2443.2001.00485.x. [DOI] [PubMed] [Google Scholar]
- Nargang F. E., Kunkele K. P., Mayer A., Ritzel R. G., Neupert W., Lill R. ‘Sheltered disruption’ of Neurospora crassa MOM22, an essential component of the mitochondrial protein import complex. EMBO J. 1995;14:1099–1108. doi: 10.1002/j.1460-2075.1995.tb07093.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson B., et al. RAM: a conserved signaling network that regulates Ace2p transcriptional activity and polarized morphogenesis. Mol. Biol. Cell. 2003;14:3782–3803. doi: 10.1091/mbc.E03-01-0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr-Weaver T. L., Szostak J. W. Yeast recombination: the association between double-strand gap repair and crossing-over. Proc. Natl. Acad. Sci. USA. 1983;80:4417–4421. doi: 10.1073/pnas.80.14.4417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palanivelu R., Preuss D. Pollen tube targeting and axon guidance: parallels in tip growth mechanisms. Trends Cell Biol. 2000;10:517–524. doi: 10.1016/s0962-8924(00)01849-3. [DOI] [PubMed] [Google Scholar]
- Park H. O., Bi E. Central roles of small GTPases in the development of cell polarity in yeast and beyond. Microbiol. Mol. Biol. Rev. 2007;71:48–96. doi: 10.1128/MMBR.00028-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poggeler S., Masloff S., Hoff B., Mayrhofer S., Kuck U. Versatile EGFP reporter plasmids for cellular localization of recombinant gene products in filamentous fungi. Curr. Genet. 2003;43:54–61. doi: 10.1007/s00294-003-0370-y. [DOI] [PubMed] [Google Scholar]
- Rasmussen C. G., Glass N. L. A Rho-type GTPase, rho-4, is required for septation in Neurospora crassa. Eukaryot. Cell. 2005;4:1913–1925. doi: 10.1128/EC.4.11.1913-1925.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen C. G., Glass N. L. Localization of RHO-4 indicates differential regulation of conidial versus vegetative septation in the filamentous fungus Neurospora crassa. Eukaryot. Cell. 2007;6:1097–1107. doi: 10.1128/EC.00050-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rottmann M., Dieter S., Brunner H., Rupp S. A screen in Saccharomyces cerevisiae identified CaMCM1, an essential gene in Candida albicans crucial for morphogenesis. Mol. Microbiol. 2003;47:943–959. doi: 10.1046/j.1365-2958.2003.03358.x. [DOI] [PubMed] [Google Scholar]
- Roumanie O., Weinachter C., Larrieu I., Crouzet M., Doignon F. Functional characterization of the Bag7, Lrg1 and Rgd2 RhoGAP proteins from Saccharomyces cerevisiae. FEBS Lett. 2001;506:149–156. doi: 10.1016/s0014-5793(01)02906-4. [DOI] [PubMed] [Google Scholar]
- Schaller M. D. Paxillin: a focal adhesion-associated adaptor protein. Oncogene. 2001;20:6459–6472. doi: 10.1038/sj.onc.1204786. [DOI] [PubMed] [Google Scholar]
- Schmeichel K. L., Beckerle M. C. Molecular dissection of a LIM domain. Mol. Biol. Cell. 1997;8:219–230. doi: 10.1091/mbc.8.2.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt A., Hall A. Guanine nucleotide exchange factors for Rho GTPases: turning on the switch. Genes Dev. 2002;16:1587–1609. doi: 10.1101/gad.1003302. [DOI] [PubMed] [Google Scholar]
- Schneper L., Krauss A., Miyamoto R., Fang S., Broach J. R. The Ras/protein kinase A pathway acts in parallel with the Mob2/Cbk1 pathway to effect cell cycle progression and proper bud site selection. Eukaryot. Cell. 2004;3:108–120. doi: 10.1128/EC.3.1.108-120.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seiler S., Nargang F. E., Steinberg G., Schliwa M. Kinesin is essential for cell morphogenesis and polarized secretion in Neurospora crassa. EMBO J. 1997;16:3025–3034. doi: 10.1093/emboj/16.11.3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seiler S., Plamann M. The genetic basis of cellular morphogenesis in the filamentous fungus Neurospora crassa. Mol. Biol. Cell. 2003;14:4352–4364. doi: 10.1091/mbc.E02-07-0433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seiler S., Plamann M., Schliwa M. Kinesin and dynein mutants provide novel insights into the roles of vesicle traffic during cell morphogenesis in Neurospora. Curr. Biol. 1999;9:779–785. doi: 10.1016/s0960-9822(99)80360-1. [DOI] [PubMed] [Google Scholar]
- Seiler S., Vogt N., Ziv C., Gorovits R., Yarden O. The STE20/germinal center kinase POD6 interacts with the NDR kinase COT1 and is involved in polar tip extension in Neurospora crassa. Mol. Biol. Cell. 2006;17:4080–4092. doi: 10.1091/mbc.E06-01-0072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinberg G. Preparing the way: fungal motors in microtubule organization. Trends Microbiol. 2007;15:14–21. doi: 10.1016/j.tim.2006.11.007. [DOI] [PubMed] [Google Scholar]
- Stewart M. S., Krause S. A., McGhie J., Gray J. V. Mpt5p, a stress tolerance- and lifespan-promoting PUF protein in Saccharomyces cerevisiae, acts upstream of the cell wall integrity pathway. Eukaryot. Cell. 2007;6:262–270. doi: 10.1128/EC.00188-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sussman A., et al. Discovery of cercosporamide, a known antifungal natural product, as a selective Pkc1 kinase inhibitor through high-throughput screening. Eukaryot. Cell. 2004;3:932–943. doi: 10.1128/EC.3.4.932-943.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terenzi H. F., Reissig J. L. Modifiers of the cot gene in Neurospora: the gulliver mutants. Genetics. 1967;56:321–329. doi: 10.1093/genetics/56.2.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdivia R. H., Schekman R. The yeasts Rho1p and Pkc1p regulate the transport of chitin synthase III (Chs3p) from internal stores to the plasma membrane. Proc. Natl. Acad. Sci. USA. 2003;100:10287–10292. doi: 10.1073/pnas.1834246100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanni C., Ottaviano C., Guo F., Puppo M., Varesio L., Zheng Y., Eva A. Constitutively active Cdc42 mutant confers growth disadvantage in cell transformation. Cell Cycle. 2005;4:1675–1682. doi: 10.4161/cc.4.11.2170. [DOI] [PubMed] [Google Scholar]
- Varelas X., Stuart D., Ellison M. J., Ptak C. The Cdc34/SCF ubiquitination complex mediates Saccharomyces cerevisiae cell wall integrity. Genetics. 2006;174:1825–1839. doi: 10.1534/genetics.106.059154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe D., Abe M., Ohya Y. Yeast Lrg1p acts as a specialized RhoGAP regulating 1,3-beta-glucan synthesis. Yeast. 2001;18:943–951. doi: 10.1002/yea.742. [DOI] [PubMed] [Google Scholar]
- Wendland J. Comparison of morphogenetic networks of filamentous fungi and yeast. Fungal Genet. Biol. 2001;34:63–82. doi: 10.1006/fgbi.2001.1290. [DOI] [PubMed] [Google Scholar]
- Wendland J., Philippsen P. Determination of cell polarity in germinated spores and hyphal tips of the filamentous ascomycete Ashbya gossypii requires a rhoGAP homolog. J. Cell Sci. 2000;113:1611–1621. doi: 10.1242/jcs.113.9.1611. [DOI] [PubMed] [Google Scholar]
- Wendland J., Philippsen P. Cell polarity and hyphal morphogenesis are controlled by multiple rho-protein modules in the filamentous ascomycete Ashbya gossypii. Genetics. 2001;157:601–610. doi: 10.1093/genetics/157.2.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarden O., Plamann M., Ebbole D. J., Yanofsky C. cot-1, a gene required for hyphal elongation in Neurospora crassa, encodes a protein kinase. EMBO J. 1992;11:2159–2166. doi: 10.1002/j.1460-2075.1992.tb05275.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zallen J. A., Peckol E. L., Tobin D. M., Bargmann C. I. Neuronal cell shape and neurite initiation are regulated by the Ndr kinase SAX-1, a member of the Orb6/COT-1/warts serine/threonine kinase family. Mol. Biol. Cell. 2000;11:3177–3190. doi: 10.1091/mbc.11.9.3177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Li S., Fischer R., Xiang X. Accumulation of cytoplasmic dynein and dynactin at microtubule plus ends in Aspergillus nidulans is kinesin dependent. Mol. Biol. Cell. 2003;14:1479–1488. doi: 10.1091/mbc.E02-08-0516. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.