Abstract
Autophagy is an evolutionarily conserved bulk-protein degradation pathway in which isolation membranes engulf the cytoplasmic constituents, and the resulting autophagosomes transport them to lysosomes. Two ubiquitin-like conjugation systems, termed Atg12 and Atg8 systems, are essential for autophagosomal formation. In addition to the pathophysiological roles of autophagy in mammals, recent mouse genetic studies have shown that the Atg8 system is predominantly under the control of the Atg12 system. To clarify the roles of the Atg8 system in mammalian autophagosome formation, we generated mice deficient in Atg3 gene encoding specific E2 enzyme for Atg8. Atg3-deficient mice were born but died within 1 d after birth. Conjugate formation of mammalian Atg8 homologues was completely defective in the mutant mice. Intriguingly, Atg12–Atg5 conjugation was markedly decreased in Atg3-deficient mice, and its dissociation from isolation membranes was significantly delayed. Furthermore, loss of Atg3 was associated with defective process of autophagosome formation, including the elongation and complete closure of the isolation membranes, resulting in malformation of the autophagosomes. The results indicate the essential role of the Atg8 system in the proper development of autophagic isolation membranes in mice.
INTRODUCTION
Proteolysis in eukaryotic cells can be separated largely into two major pathways: one pathway mediated by the proteasome and the other pathway by the lysosome. The proteasome, in collaboration with the sophisticated ubiquitin system for selection of target proteins, plays crucial roles in rapid and aggressive degradation of not only a variety of short-lived regulatory proteins but also abnormal proteins with aberrant structures that should be eliminated from the cells to maintain cellular viability (Goldberg, 2003). In contrast, the lysosome is an organelle that contains a diverse array of hydrolases, which is separated from the cytosol by a limiting membrane to prevent the random destruction of cellular components. In this lysosomal pathway, degradation of plasma membrane proteins and extracellular proteins is mediated by endocytosis, whereas degradation of cytoplasmic components is achieved through autophagy, a process divided into three classes of pathways; macroautophagy, microautophagy, and chaperone-mediated autophagy (Cuervo, 2004; Klionsky, 2005).
Macroautophagy (hereafter referred to as autophagy) is the main route for sequestration of bulky portions of the cytoplasm into the lysosome. The initial steps of autophagy are the formation and subsequent elongation of the isolation membrane. The isolation membrane then enwraps cytoplasmic constituents such as organelles until its edges fuse with each other to form a double-membrane structure called the autophagosome. Finally, the outer membrane of the autophagosome fuses with the lysosome/vacuole; thereby the sequestered cytoplasmic components together with the inner membrane of the autophagosomes are completely degraded by the lysosomal/vacuolar hydrolases (Mizushima et al., 2002).
There is general agreement that the most important role of autophagy is the supply of amino acids under nutrient-poor conditions in yeast and neonate starvation period in mice (Tsukada and Ohsumi, 1993; Kuma et al., 2004; Komatsu et al., 2005). In addition, it is also noted that constitutive autophagy, which occurs even under nutrient-rich conditions, is indispensable for cell homeostasis. In fact, autophagy deficiency in the CNS and the liver leads to neurodegeneration and liver dysfunction, respectively (Komatsu et al., 2005, 2006, 2007; Hara et al., 2006). Moreover, autophagy plays a vital role in cellular remodeling during differentiation and development of multicellular organisms, such as fly, worm and slime mold (Levine and Klionsky, 2004) and cellular defense against invading streptococcus (Gutierrez et al., 2004; Nakagawa et al., 2004). Plants lacking autophagy show accelerated senescence (Hanaoka et al., 2002).
In the last decade, a set of genes, essential for autophagosome formation, were specified in yeast (Tsukada and Ohsumi, 1993; Thumm et al., 1994) and most of them are conserved in mammals. Among them, gene products involved in Atg (autophagy) conjugation systems were identified, in which the enzymatic reactions involved are, in principle, analogous to the ubiquitin conjugation system, including E1 (activating enzyme) and E2 (conjugating enzyme) (Ohsumi, 2001). Both ubiquitin-like modifiers, Atg12 and Atg8, are activated by a common E1-like enzyme, Atg7, and transferred to two different E2-like enzymes, Atg10 and Atg3, respectively (Mizushima et al., 1998a,b; Shintani et al., 1999; Tanida et al., 1999, 2002; Ichimura et al., 2000). Although Atg12 forms an isopeptide bond with Atg5, which is the only specific target reported so far, Atg8 forms an amide bond with phosphatidylethanolamine (PE) dependent on Atg12–Atg5 conjugation (Ichimura et al., 2000; Mizushima et al., 2001). The Atg12–Atg5 conjugate interacts with another ATG gene product Atg16, to ultimately form a tetrameric complex in yeast (Kuma et al., 2002). Mammalian Atg16 counterpart Atg16L also orchestrates a high molecular complex with Atg12–Atg5 conjugate (Mizushima et al., 2003). Although Atg12–Atg5 conjugation is indispensable for elongation of the isolation membrane, the PE-bound microtubule-associated protein 1 light chain 3/MAP1LC3 (LC3-II) (mammalian Atg8 homologue) is considered important for maturation of autophagosomes (Kabeya et al., 2000; Mizushima et al., 2001). Indeed, the Atg12–Atg5–Atg16L complex is localized on the isolation membranes during their elongation and disassociated from them before completion of autophagosomal formation (Mizushima et al., 2003). In comparison, the LC3-II stays on the outer and inner membranes of the isolation membranes until completion of autophagosome formation. In the final stage, LC3-II on the inner membrane is degraded by fusion of autophagosomes to lysosomes, whereas LC3-II on the outer membrane is released from autophagosomes by a specific cysteine protease, Atg4 (Kirisako et al., 2000; Tanida et al., 2004). Importantly, the attachment of PE to LC3 is almost completely impaired in Atg5-knockout cells, indicating that these two LC3/Atg8 and Atg12 conjugation systems contribute to the formation of autophagosomes in a cooperative manner (Mizushima et al., 2001). Moreover, it became clear that the Atg12–Atg5 conjugate functions as an E3-like enzyme for the Atg8 lipidation reaction (Hanada et al., 2007; Fujita et al., 2008).
In the present study, we analyzed the specific role(s) of LC3/Atg8 conjugation in mammals. To do this, we generated mice lacking Atg3, the specific E2-like enzyme for the Atg8 conjugation system, and we found that lack of Atg3 severely suppressed the formation of Atg12–Atg5 conjugate as well as completely impaired Atg8 conjugation. Surprisingly, loss of Atg3 also resulted in the accumulation of small autophagosome-like structures or disorganized isolation membranes, indicating that Atg8 conjugation plays a critical role in the proper development of isolation membranes. We also discuss the precise roles of the Atg8 conjugation system in the autophagic pathway, based on phenotypic analyses of Atg3 null-mice.
MATERIALS AND METHODS
Generation of Atg3−/− Mice
A targeting vector for Atg3 was designed to disrupt exon 10, which codes the active site cysteine, by neo-resistant gene cassette. The targeting vector was electroporated into mouse TT2 embryonic stem (ES) cells, selected with G418 (200 μg/ml; Invitrogen, Carlsbad, CA), and then screened for homologous recombinants by Southern blot analyses. Southern blot analysis was performed by digestion of genomic DNA with PstI and hybridization with the Atg3 probe. Genotyping of mice was performed by polymerase chain reaction (PCR) using the two primers sets: one set was Atg3-S1 (5′-TAGGAGCCATTTGCCACCAATCGTA-3′) and Neopr2Rv (5′-GCACTTCGCCCAATAGCAGCCA GTCCCTTC-3′), and the other set was Atg3-671-691 (5′-GGCAGCCTTTAACAGTTGAGC-3′) and Atg3S-1. To generate Atg3−/− mice with green fluorescent protein (GFP)-LC3, we crossed Atg3+/− mice with GFP-LC3 transgenic mice (Mizushima et al., 2004). Mice were housed in specific-pathogen free facilities, and the experimental protocol was approved by the Ethics Review Committee for Animal Experimentation of Tokyo Metropolitan Institute of Medical Science.
Generation of Mouse Embryonic Fibroblasts (MEFs)
MEFs were prepared as described previously (Komatsu et al., 2005). Immortalized wild-type and Atg3-deficient MEFs were established by infecting MEFs with a recombinant retrovirus carrying a temperature-sensitive simian virus 40 large T antigen (Lee et al., 1995). GFP-tagged Atg5 and FLAG-tagged Atg12 were subcloned into retrovirus vector pMXs-puro (Kitamura et al., 2003). As described recently (Ichimura et al., 2008), each GFPAtg5 and FLAGAtg12 vector was introduced into MEFs. To prepare Atg3 and Atg3C264S adenoviruses, we used the Adenovirus Expression Vector kit (Takara Bio, Otsu, Shiga, Japan). To express exogenous Atg3 and Atg3C264S proteins, the immortalized wild-type and Atg3-deficient MEFs were plated on six-well dishes in 2 ml of growth medium at 24 h before infection. The medium was replaced with fresh medium containing Atg3 or Atg3C264S adenovirus. The cells were lysed 48 h later, and then the lysates were analyzed by immunoblotting.
Immunological Analysis
Lysates of MEFs were immunoblotted as described previously (Komatsu et al., 2005). The antibodies for Atg3, Atg7, Atg10, LC3, GABA(A) receptor-associated protein (GABARAP), and Golgi-associated ATPase enhancer of 16 kDa (GATE)-16 were described previously (Tanida et al., 2002; Nemoto et al., 2003; Komatsu et al., 2005). The antibody for Atg16L was described previously (Mizushima et al., 2003). The antibody against Atg12 was raised in rabbits using the recombinant protein as antigens. The antibody for actin (MAB1501R) was purchased from Millipore Bioscience Research Reagents (Temecula, CA) and that for Atg5 from Sigma (St. Louis, MO). For LC3 and Atg16L staining, MEFs were fixed and stained with anti-LC3 and anti-Atg16L antibodies, respectively, as described previously (Komatsu et al., 2005). Fluorescence images were obtained using a fluorescence microscope (model BZ-9000; Keyence, Osaka, Japan). Atg16L-positive dots per each treated cell (n = 20) were counted using software Dynamic cell count BZ-HIC (Keyence).
Time-Lapse Imaging Analysis
Each genotype of MEF expressing GFPAtg5 was cultured in Hanks' solution for 1 h, and green fluorescent protein (GFP) images were taken every 30 s by using a fluorescence microscope (model BZ-9000; Keyence). Time-lapse recordings proceeded for 30 min at 37°C with a culture dish system (Keyence) for temperature and CO2 control.
Pulse-Chase Experiments
Cells were incubated with cysteine- and methionine-free medium for 1 h, metabolically labeled with [35S]cysteine and methionine for 2 or 24 h, and then washed and chased for the indicated time. The cell lysates were immunoprecipitated with M2 agarose (Sigma) or GFP agarose (Medical & Biological Laboratories, Nagoya, Japan), fractionated by SDS-polyacrylamide gel electrophoresis (PAGE), and visualized by autoradiography.
Histological Examination
Methods for tissue fixation and subsequent procedures, including hematoxylin and eosin staining, immunofluorescence, conventional electron microscopy, and immunoelectron microscopy were described previously (Komatsu et al., 2005, 2006). For pre-embedding immunoelectron microscopy, MEFs were fixed with 4% paraformaldehyde-4% sucrose in phosphate buffer (PB; pH 7.2) followed by permeabilization with 0.25% saponin/0.1 M PB. They were immunolabeled with anti-GFP or anti-Atg16L antibody and then with nanogold-conjugated anti-rabbit Fab' (Nanoprobes, Yaphank NY). After silver enhancement with HQsilver (Nanoprobes), they were embedded in Epon 812 and sectioned for electron microscopic observation. The number of each organelle was counted and expressed as the number per cell section or per 100 μm2 of cytoplasmic area. The areas of the cytoplasm and autophagosomes were measured using MetaMorph image processing software (Molecular Devices, Sunnyvale, CA). Antibodies used for immunofluorescence and immunoelectron microscopy were as follow: rabbit polyclonal antibodies against Atg16L (Mizushima et al., 2003) and GFP (Abcam, Cambridge, MA).
Other Procedures
RNA samples were extracted from each genotyping MEFs by using TRIzol reagent (Invitrogen). For RNA blot, 10 μg of RNA per samples was loaded. Blots were probed with Atg3- and actin-cDNA as control. Cell starvation was conducted by incubating the cells in Hanks' balanced solution with 10 mM HEPES, pH 7.5, following three separate washes. Caesarean delivery, measurement of amino acids, and assay of long-lived protein degradation were performed as reported previously (Komatsu et al., 2005).
RESULTS
Generation of Atg3-Knockout Mice
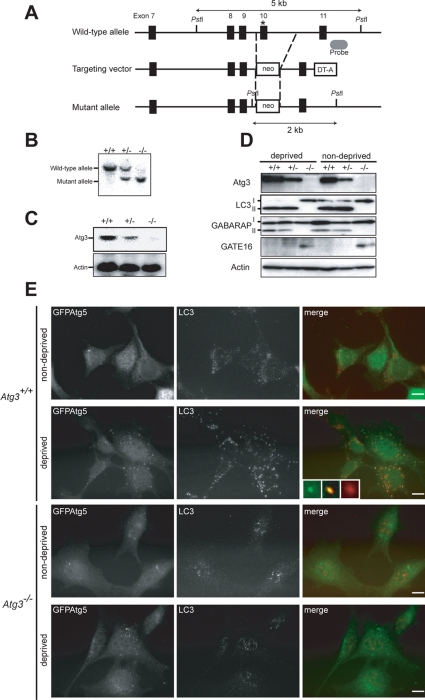
To investigate the physiological roles of the Atg8 system in vivo, we generated mice deficient in Atg3, a specific E2-like enzyme for Atg8. Mouse Atg3 gene is encoded by 12 exons that span 29.71 kb of genomic DNA. The active site cysteine residue, essential for conjugation of the substrates, is encoded by exon 10, and the targeting vector was designed to disrupt this exon by neo-resistant gene cassette (Figure 1A). Mice heterozygotes for the Atg3 allele (referred to Atg3+/− mice) were born healthy and fertile without any noticeable pathological phenotypes for 1 y. Figure 1B shows Southern blots of mice with the indicated genotypes. Neither Atg3 mRNA nor protein was detected in Atg3−/− mice (Figure 1, C and D).
Figure 1.
Generation of Atg3-knockout mice. (A) Schematic representation of the targeting vector and the targeted allele of Atg3 gene. The coding exons numbered in accordance with the initiation site as exon 1 are depicted by black boxes. The probe for Southern blot analysis is shown as gray ellipse. The asterisk denotes the essential cysteine residue on exon 10. PstI, PstI sites; neo, neomycin-resistant gene cassette; DT-A, diphtheria toxin gene. (B) Southern blot analysis of genomic DNAs extracted from mice tails. Wild-type and mutant alleles are detected as 5- and 2-kb bands, respectively. (C) Expression of Atg3 transcript. Total RNAs were extracted from each genotyping MEFs. Atg3 transcript was detected by Northern blot with mouse Atg3 cDNA. Actin cDNA was used as an internal control. (D) Immunoblot of Atg3 and Atg8 homologues in MEFs. Primary MEFs of the indicated genotypes were isolated and cultured under nutrient-rich (nondeprived) and nutrient-poor (deprived) conditions. The lysates were immunoblotted with anti-Atg3, anti-LC3, anti-GABARAP, anti-GATE-16, and anti-actin antibodies. Data shown are representative of three separate experiments. (E) Immunofluorescence analysis of MEFs using anti-LC3antibody. Each genotype immortalized MEFs with introduced GFP-tagged Atg5 were cultured in DMEM with fetal calf serum (nondeprived) or Hanks' solution (deprived). The cells were fixed and then immunostained with anti-LC3. Insets show a GFPAtg5-single- (left inset), an LC3-single- (right inset), and a double-positive (middle inset) structure, respectively. Bars; 10 μm.
Shutdown of Atg8 System in Atg3-deficient Mice
We tested the loss of Atg3 activity by measuring the conversion of LC3 to its PE-conjugated form. To do this, we isolated MEFs from each genotype, followed by culture under nutrient-rich or starvation conditions and lysis. The lysates were subjected to SDS-PAGE followed by immunoblot analyses with anti-LC3 antibody. Mammalian Atg8p homologue LC3 exists in two forms in cells, LC3-I and LC3-II. LC3-I is a cytosolic form and LC3-II is a PE-conjugated form, which localizes in autophagosomes (Kabeya et al., 2000). In Atg3+/+ and Atg3+/− MEFs, both forms were detected under nutrient-rich condition, whereas only the LC3-II form was detected under starvation condition (Figure 1D), suggesting conversion of LC3-I in response to starvation. In contrast, only the LC3-I form was recognized in Atg3−/− MEFs, regardless of the nutrient conditions. We also examined other Atg8 homologues, GABARAP and GATE-16 (Figure 1D). It is generally considered that they are also activated by Atg7, transferred to Atg3, and finally conjugated with PE for localization to autophagosomes (Tanida et al., 2001, 2002; Kabeya et al., 2004; Sou et al., 2006). Whereas GABARAP-I (free form) was converted to GABARAP-II (PE-conjugated form) in wild-type and heterozygous MEFs and the level of conversion was not affected by starvation, its conversion was completely blocked in Atg3−/− MEFs. Although GATE-16 was hardly detected in wild-type and heterozygous MEFs under both conditions, the band corresponding to GATE-16-I (free form) was clearly detected in Atg3−/− MEFs (Supplemental Figure S1). Immunofluorescence analyses with anti-LC3 antibody revealed that starvation induced the appearance of LC3-positive dots in the Atg3+/+ but not in the Atg3−/− MEFs. In contrast, Atg5-positive dots were apparently induced in wild-type MEFs (Figure 1E), but no clear increase in Atg3−/− MEFs was evident because many of those were already formed even in nutrient-rich conditions (see Figure 4). Notably, in contrast with small dots containing only GFPAtg5 (left inset), the large structures (right inset) were almost singly positive for LC3, and the rest were positive for both proteins that were considered to be intermediate structures (middle inset). Together, these results suggest impairment of the Atg8 system in Atg3-knockout mice.
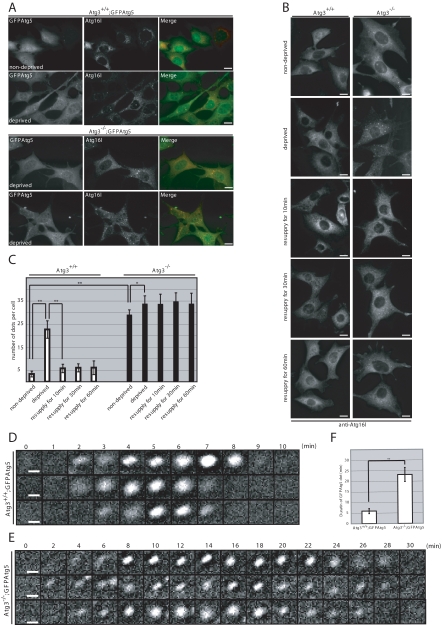
Figure 4.
Accumulation of Atg16L-positive structures in Atg3-deficient MEFs. (A) Immunofluorescence analysis of Atg16L in GFPAtg5-introduced immortalized MEFs. Each genotype MEFs were cultured in nutrient-rich (nondeprived) or Hanks' solution (deprived). The cells were fixed and then immunostained with anti-Atg16L. Bars; 10 μm. (B) Immunofluorescence analysis of Atg16L in MEFs. MEFs isolated from Atg3+/+ and Atg3−/− were cultured in nutrient-rich (nondeprived) or Hanks' solution (deprived). After deprivation of nutrients, the cells were resupplied with nutrients for 10, 30, or 60 min. The cells were fixed and then immunostained with anti-Atg16L. Bars; 10 μm. (C) The number of Atg16L-positive dots in MEF (n = 20) was determined in each genotype. Data are mean ± SD. *p < 0.05 and **p < 0.01 by Student's t test. (D and E) Time-lapse observation of starvation-induced GFPAtg5 dots. Wild-type (D) and Atg3-deficient (E) MEFs harboring GFPAtg5 were cultured in Hanks' solution for 1 h and directly observed by time-lapse video microscopy. Bars, 1 μm. (F) Duration of existence of GFPAtg5 dots in wild-type and Atg3-deficient MEFs. The time of sustained presence of GFPAtg5 dots in wild-type (n = 9) and mutant (n = 10) was measured. Data are mean ± SD. **p < 0.01 by Student's t test.
Phenotypes of Atg3-Knockout Mice
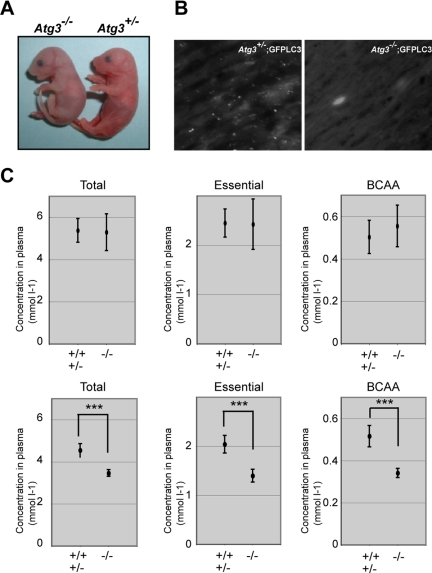
Although Atg3−/− mice were born at Mendelian frequency (+/+; +/−; −/− = 25; 38; 23), their body weight (0.99 ± 0.12 g; n = 8) was lower than that of wild-type and heterozygous mice (1.19 ± 0.105 g; n = 25; p < 0.002) (Figure 2A), and they died within 1 d after birth (n = 11), similar to Atg5 and Atg7 null-mice (Kuma et al., 2004; Komatsu et al., 2005). Autophagy is induced in newborn when the nutrients supply through the placenta terminates after birth (Kuma et al., 2004). To confirm this phenomenon, we crossed Atg3 with GFPLC3 transgenic mice (Mizushima et al., 2004) to generate Atg3+/− and Atg3−/− mice harboring GFP-LC3. Fasting for 3 h resulted in appearance of LC3-positive dots in Atg3+/−;GFPLC3 but not Atg3−/−;GFPLC3 hearts (Figure 2B). Lower production of amino acids correlates with the cause of death in autophagy-deficient mice (Kuma et al., 2004; Komatsu et al., 2005). Therefore, we next measured the survival time of Atg3−/− neonates under starvation condition and measured serum amino acids concentrations after Caesarean delivery. Atg3−/− mice died at 13.2 ± 3.5 h after Caesarean delivery, although wild-type and heterozygous mice were alive at that time. The concentrations of total, essential and branched chain amino acids (BCAAs) in sera of Atg3−/− mice were almost comparable with those of wild-type mice at 0 h after Caesarean delivery but those of Atg3−/− were significantly lower at 10 h after Caesarean delivery compared with wild-type mice (Figure 2C). These results suggest that Atg3, similar to Atg5 and Atg7 (Kuma et al., 2004; Komatsu et al., 2005), is essential for the supply of amino acids into the blood circulation and survival of newborn mice under starvation condition.
Figure 2.
Phenotypes of Atg3-deficient mice. (A) Morphology of Atg3+/- and Atg3−/− mice. (B) Deficiency of LC3-positive dots in Atg3−/− heart. Atg3+/− and Atg3−/− mice expressing GFP-LC3 were delivered by Cesarean section and analyzed by florescence microscopy. Representative results obtained from each neonatal heart at 3 h after Cesarean delivery. Bar; 50 μm. (C) The concentrations of total amino acids, essential amino acids, and BCAA in sera of mice of the indicated genotypes. Atg3+/+, Atg3+/− (n = 5) and Atg3−/− mice (n = 4) were dissected immediately (top) or at 10 h (bottom) after delivery and the plasma concentrations of amino acids were measured. Data are mean ± SD. ***p < 0.001 by Student's t test.
Atg3 Deficiency Causes Reduced Formation of Atg12–Atg5 Conjugation
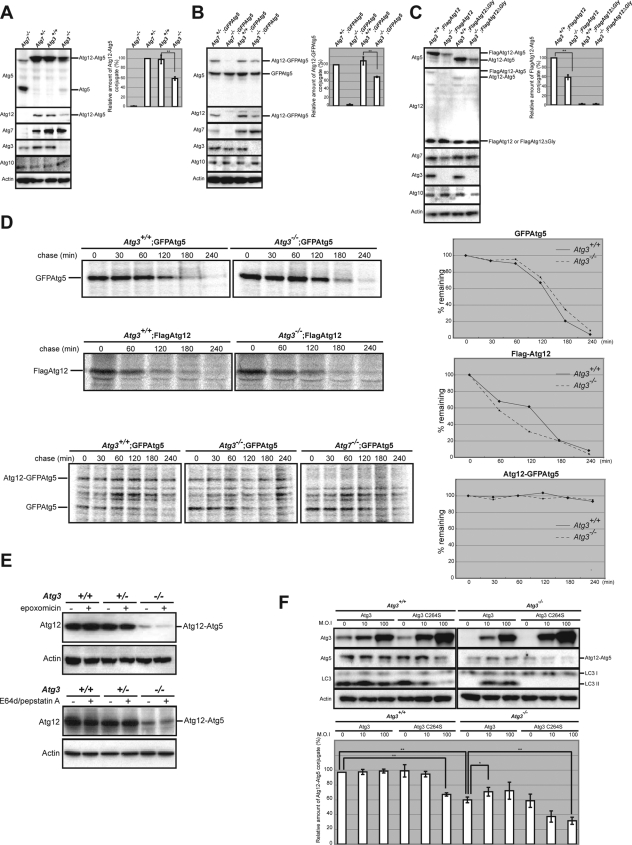
Atg5 deficiency leads to a significant reduction in LC3 conjugation to PE in yeast and mice as reported previously (Mizushima et al., 2001; Suzuki et al., 2001; Hosokawa et al., 2007), which could be explained by the E3-like activity of Atg12–Atg5 conjugate in Atg8-PE conjugation (Hanada et al., 2007; Fujita et al., 2008). Conversely, overproduction of Atg3 in human embryonic kidney 293 cells facilitates the formation of the Atg12–Atg5 conjugate (Tanida et al., 2002). These results suggest cooperation of the two conjugation systems during autophagosome formation. However, it is still puzzling how the Atg8/LC3 system affects the activity of the Atg12 system. To solve this puzzle, we conducted additional experiments in which each wild-type and Atg3-knockout MEFs were lysed and subjected to immunoblot analyses with anti-Atg12 and anti-Atg5 antibodies. Intriguingly, the amount of Atg12–Atg5 conjugate was markedly reduced in Atg3-deficient MEFs, whereas the amount of free Atg5 in the cells increased significantly (Figure 3A and Supplemental Figure S2). Even upon overexpression of either FLAG-tagged Atg12 or GFP-tagged Atg5, the efficiency of Atg12–Atg5 conjugation formation was still low in Atg3-deficient MEFs (Figure 3, B and C), suggesting the involvement of Atg3 in the conjugation reaction rather than the instability of individual components involved in the Atg12 system. In fact, the half-life of each of FLAGAtg12 and GFPAtg5 introduced into Atg3-deficient MEFs was comparable with that of wild-type MEFs (Figure 3D, top and middle). Importantly, once Atg12-GFPAtg5 conjugate was formed in Atg3-deficient cells, the conjugate was stable, similar to that in wild-type MEFs (Figure 3D, bottom). Furthermore, the amount of Atg12–Atg5 conjugates in Atg3-deficient MEFs did not change under treatment with epoxomicin, a proteasomal inhibitor (Figure 3E, top). We also confirmed that they did not change by treatment with E64d and pepstatin A, lysosomal inhibitors (Figure 3E, bottom). These results suggest inhibition of the Atg12–Atg5 conjugation reaction in Atg3-deficient MEFs.
Figure 3.
Formation of Atg12–Atg5 conjugates is partially inhibited by Atg3 deficiency. (A) Immunoblot analyses of the Atg12–Atg5 conjugate. Immortalized MEFs of the indicated genotype were lysed, and the lysates were subjected to SDS-PAGE and analyzed by immunoblotting with the indicated antibodies. As a negative control for detection of Atg12–Atg5 conjugate, we used the lysate of Atg7-deficient MEFs. Note the low production of the Atg12–Atg5 conjugate in Atg3-deficient cells. (B) Each genotype immortalized MEFs with introduced GFP-tagged Atg5 were lysed, and then the lysates were subjected to SDS-PAGE followed by immunoblotting with the indicated antibodies. As a negative control for detection of Atg12–GFPAtg5 conjugate, we used the lysate of GFPAtg5-introduced Atg7-deficient MEFs. (C) Immortalized wild-type and Atg3-deficient MEFs-introduced FLAG-tagged Atg12 or Atg12ΔGly were lysed, and then the lysates were subjected to SDS-PAGE followed by immunoblotting with the indicated antibodies. As a negative control for detection of FLAGAtg12-Atg5 conjugate, we used the lysate of FLAGAtg12ΔGly-expressing MEFs. Graphs shown in A–C indicate the ratios of Atg12–Atg5, Atg12–GFPAtg5, and FLAGAtg12–Atg5 relative to actin, respectively. Data are mean ± SD of triplicate experiments. **p < 0.01 by Student's t test. (D) Half-lives of GFPAtg5, FLAGAtg12, and GFPAtg5-Atg12 conjugation in Atg3-deficient MEFs. Each immortalized wild-type and Atg3-deficient MEFs harboring GFPAtg5 or FLAGAtg12 were labeled with [35S]cysteine and methionine for 2 h and chased for the indicated time. For labeling of GFPAtg5-12 conjugate, the cells were labeled with [35S]cysteine and methionine for 24 h. Subsequently, each lysate was immunoprecipitated with anti-GFP and FLAG antibodies and then subjected to SDS-PAGE and visualized by autoradiography. As a negative control for detection of Atg12–GFPAtg5 conjugate, we used the lysate of GFPAtg5-introduced Atg7-deficient MEFs. The decay curves of GFPAtg5 (top, right), FLAGAtg12 (middle, right) and Atg12-GFPAtg5 (bottom, right) were generated from quantification of the band shown in the left panels. (E) The MEFs of the indicated genotypes were cultured in the presence or absence of a proteasome inhibitor (epoxomicin) for 16 h (top) or lysosomal inhibitors (E64d and pepstatin A: E/P) for 24 h (bottom). The cell lysates were subjected to SDS-PAGE and analyzed by immunoblotting with anti-Atg12 and anti-actin antibodies. (F) Wild-type and Atg3-deficient immortalized MEFs were infected with Atg3 and Atg3C264S at 10, or 100 multiplicity of infection (M.O.I.). After 48-h infection, the cell lysates were subjected to SDS-PAGE and analyzed by immunoblotting with the indicated antibodies. Bottom graph shows the ratios of Atg12–Atg5 relative to actin, respectively. Data are mean ± SD. **p < 0.01 by Student's t test. Data are representative of three separate experiments.
Next, to determine whether the loss of E2 activity of Atg3 is involved in the reduced Atg12–Atg5 conjugate in Atg3-deficient MEFs, we used the mutant Atg3C264S. Atg3C264S has a mutation at the active site cysteine; thus, it is unable to form Atg8/LC3-PE conjugate. When Atg3 or Atg3C264S was expressed exogenously into Atg3-deficient MEFs by using adenovirus system, they were detected in sufficient amounts in both MEF genotypes (Figure 3F). Predictably, LC3 lipidation was complemented by exogenous expression of Atg3 but not Atg3C264S mutant (Figure 3F). Similarly, expression of Atg3, but not Atg3C264S mutant, recovered partial defect in the Atg12–Atg5 conjugate in Atg3-deficient cells (Figure 3F). Moreover, expression of Atg3C264S into wild-type MEFs led to not only partial inhibition of LC3-PE but also suppression of the Atg12–Atg5 conjugate formation (Figure 3F). These results suggest that the Atg8/LC3 system participates in the reaction of Atg12–Atg5 conjugation through the E2 activity of Atg3.
Accumulation of Atg16L-positive Structures in Atg3-deficient MEFs
Next, we examined the localization of Atg16L and Atg5 in Atg3-deficient MEFs stably expressing GFPAtg5. It has been shown that Atg16L is localized in isolation membranes in Atg5-dependent manner and disassociated from the membrane before completion of autophagosome formation (Mizushima et al., 2003). We also observed the induction of dot structures that were double positive for GFPAtg5 and Atg16L after nutrient deprivation in wild-type MEFs (Figure 4A). Although the Atg16L/GFPAtg5-positive dots were only rarely detected in wild-type MEFs in nutrient-rich conditions, these dots were distinctly observed in Atg3-deficient MEFs (Figure 4A). Moreover, their number was slightly but significantly increased upon nutrient deprivation (Figure 4, A and C), suggesting that the formation of isolation membranes is induced in response to starvation even in mutant cells. Wortmannin, an inhibitor of phosphatidylinositol 3-kinases, which are known to inhibit autophagy, suppressed the appearance of the GFPAtg5/Atg16L-positive dots during nutrient deprivation in both wild-type and mutant MEFs (Supplemental Figure S3). These results suggest that the appearance of GFPAtg5/Atg16L-positive dots in Atg3−/− MEFs is due to a standard autophagy mechanism requiring functional phosphatidylinositol 3-kinases. A similar Atg16L-staining pattern was also observed in Atg3-knockout MEFs without exogenous GFPAtg5, where almost all Atg5 existed as Atg12–Atg5 conjugates (Figures 3A and 4B), implying colocalization of Atg12–Atg5 conjugate and Atg16L in Atg3-deficient MEFs.
If elongation of isolation membrane and/or completion of autophagosome formation were impaired in Atg3-deficient cells, starvation-induced Atg16L-positive structures in mutant cells should remain even after the resupply of nutrients. As expected, although the starvation-induced Atg16L-positive structures disappeared within 10 min upon resupply of nutrients in wild-type MEFs, Atg16L-positive dots in mutant MEFs remained for 60 min after the resupply of nutrients (Figure 4, B and C). To directly determine the life span of these isolation membranes in Atg3-deficient MEFs, we carried out time-lapse video microscopic analysis. Consistent with the previous report (Mizushima et al., 2001), upon nutrient deprivation, the GFPAtg5 dots occurred at random in the cytosol of wild-type MEFs, dwelled, and disappeared within ∼5 min (Figure 4D and Supplemental Video 1). In contrast, although new GFPAtg5-dots in addition to the preexisting dots occurred in response to nutrient deprivation in the cytosol of mutant MEFs, the disappearance was significantly delayed; and in most cases, their presence was noted over >20 min (Figure 4E and Supplemental Video 2). Moreover, the numbers of de novo GFPAtg5-dots in wild-type and mutant MEFs under 10-min starvation conditions were 12.4 ± 2.2 and 8.7 ± 2.1, respectively (n = 5, p < 0.05). These results suggest impaired dissociation of the Atg12-Atg5-Atg16L complex from the isolation membranes in Atg3-deficient mice.
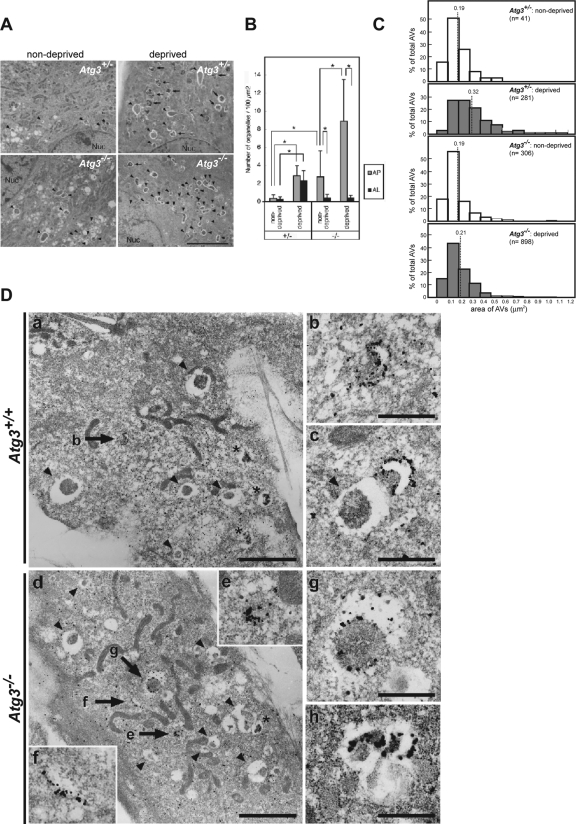
Appearance of Small Autophagosome-like Structures but Not Autolysosomes in Atg3-deficient Cells
Surprisingly, electron microscopic analyses revealed the presence of some autophagosome-like structures in Atg3-deficient MEFs cultured in normal media (Figure 5A), and their number was higher than in wild-type MEFs (Figure 5B). Nutrient deprivation induced a marked increase in the autophagosome-like structures even in Atg3-deficient MEFs (Figure 5, A and B), although the rate of increase was lower than that in wild-type MEFs (Figure 5B; see Discussion). However, quantitative analysis showed that the average size of the structures (represented as the encircled area) was smaller than that in wild-type MEFs cultured under nutrient-poor conditions (Figure 5C). The induction of autophagosome-like structures in mutant MEFs was suppressed by pretreatment with wortmannin (Supplemental Figure S4), suggesting that similar to the GFPAtg5/Atg16L-positive dots, these autophagosome-like structures are also derived from autophagic mechanisms involving the function of phosphatidylinositol 3-kinases.
Figure 5.
Induction of autophagosomes under starvation condition in Atg3-knockout cells. (A) Electron micrographs of primary MEFs from the indicated genotype mice under nutrient-rich (nondeprived) or -poor (deprived) condition. Bar, 5 μm. Arrowheads, autophagosomes; arrows, autolysosomes. (B) Numbers of autophagosomes (AP) and autolysosomes (AL) in each genotype were counted (n = 20; for details, see Materials and Methods). Data are mean ± SD. *p < 0.01, by Student's t test. (C) Area of autophagosome-like vacuoles (AVs) in MEFs from the indicated genotype under nutrient-rich (nondeprived) and -poor (deprived) conditions. Frequencies (percentages) of the given range of area are expressed in the histogram. The dotted line represents the mean value of the area in each genotype. (D) Immunoelectron micrograph showing labeling of GFP (a and b) and Atg16L (c–h) in wild-type (a–c) and Atg3-deficient MEFs (d–h) harboring GFPAtg5 under nutrient deprivation conditions. b is a higher magnification view of the GFPAtg5-positive structure indicated by the arrow in a. Higher magnification views of the Atg16L-positive structures indicated by arrows in d are shown in insets e and f. Arrowheads indicate autophagosome-like structures without GFPAtg5 or Atg16L signal. Asterisks indicate autolysosomes. Bars, 0.5 μm.
To examine the fine structure of Atg5/Atg16L-positive dots, we carried out immunoelectron microscopic analysis in wild-type and Atg3-deficient MEFs expressing GFPAtg5. Two hours after starvation, silver-enhanced gold particles, representing GFPAtg5 or Atg16L, accumulated extensively on the small crescent-shaped membrane compartments in both wild-type and mutant MEFs (Figure 5D, b, c, e, and f), which are reported to represent the early developmental structures of the isolation membrane (Mizushima et al., 2001; Mizushima et al., 2003). Meanwhile, GFPAtg5 or Atg16L signal was hardly detected on the autophagosomal and autolysosomal membranes (Figure 5D, a and c). They were occasionally localized on the cup-shaped isolation membranes (Figure 5D, a and c) but with less intense signal compared with those on the crescent-shaped isolation membranes in both wild-type and mutant MEFs. It is of note that the atypical autophagosome-like structures described below (see Figure 7) in mutant MEFs also contained the signal for Atg16L (Figure 5Dh) and such Atg16L-positive structures were hardly observed in wild-type MEFs. These results indicate that most of the accumulated Atg5/Atg16L-positive structures observed by epifluorescent microscopy in the Atg3-deficient MEFs represent immature isolation membranes rather than autophagosome-like structures detected by electron microscopy.
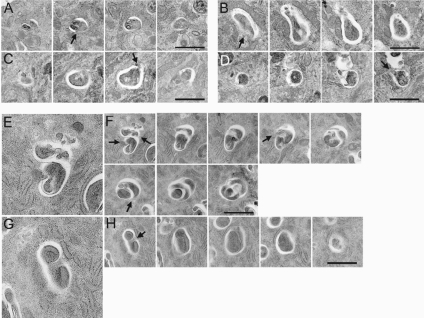
Figure 7.
Impaired elongation and closure in autophagosome formation in Atg3-deficient MEFs. Electron microscopic analyses of serial sections of wild-type (A–D) and Atg3-deficient MEFs (E–H). Arrows indicate open regions of autophagosome-like structures. Note that the isolation membranes seem to elongate in random directions, thus these membranes sequestrate certain area of the cytoplasm in some parts. Therefore, isolation membranes in mutant cells did not close to form complete autophagosomes. Bars, 1 μm.
Failure of Autolysosome Formation in Atg3-deficient MEFs
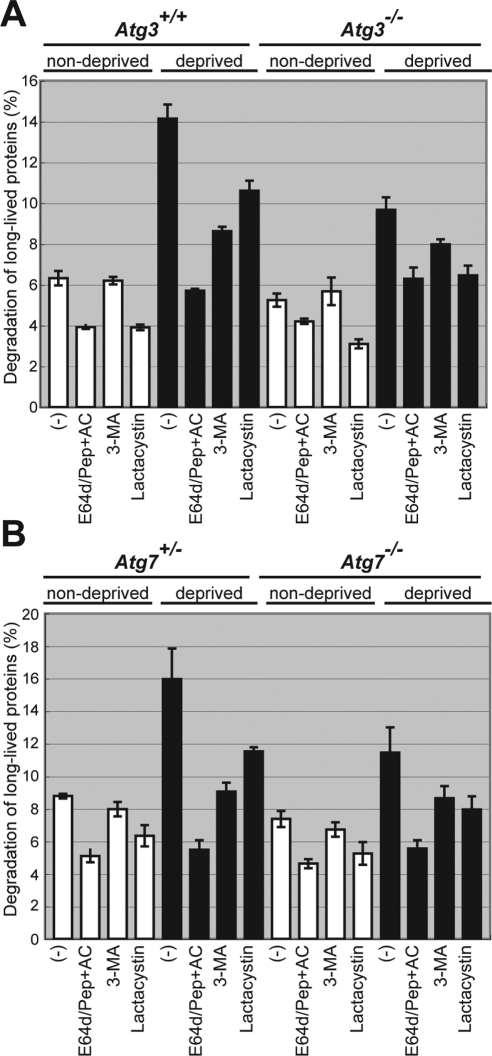
We also noticed that Atg3-deficient cells hardly contained autolysosomal structures, which contain a high electron-dense material with a hollow beneath the limiting membrane, which were otherwise usually detected in wild-type MEFs (Figure 5, A and B). To investigate whether this represents impaired autolysosome formation in Atg3 deficiency, we examined protein degradation in Atg3-deficient cells under poor nutrient condition. In wild-type MEFs, nutrient deprivation induced a significant protein degradation, which was suppressed by the addition of lysosomal inhibitors, 3-methyladenine (3-MA), E64d, pepstatin, and ammonium chloride (E64d/PepA + AC) (Figure 6A). The induced degradation was also inhibited by treatment with lactacystin, a proteasome inhibitor, although the inhibition was less than that observed with lysosomal inhibitors (Figure 6A). These results suggest that starvation-induced protein degradation is mainly mediated through the lysosomal pathway in MEFs. Although degradation of long-lived protein was induced by nutrient deprivation even in Atg3−/− MEFs, the level was significantly lower than in wild-type MEFs (Figure 6A) and comparable with that in Atg7-deficient MEFs (Figure 6B). However, it is likely that other pathways such as chaperone-mediated autophagy and microautophagy also mediate starvation-induced protein degradation in Atg3- and Atg7-deficient MEFs, although to a lesser extent, consistent with recent reports (Kiffin et al., 2006; Massey et al., 2006). Together, we concluded that nutrient-deprivation induced the formation of small autophagosome-like structures but not autolysosomes in Atg3-knockout cells.
Figure 6.
(A and B) Impaired long-lived protein degradation in Atg3-deficient MEFs. Primary MEFs from wild-type, Atg3- (A) and Atg7-knockout (B) mice were isolated and labeled with [14C]leucine for 24 h, and degradation of long-lived protein in deprived and nondeprived conditions was measured. 3-MA and/or E64d, pepstatin A and ammonium chloride (E64d/Pep + AC), or lactacystin was added as indicated. Data are mean ± SD of triplicate experiments.
Impairment of Closing Step of Autophagosome Formation Followed by Appearance of Aberrant Membranous Structures in Atg3-deficient MEFs
We further examined in more details the autophagosome-like structures in serial sections. A conventional autophagosome in wild-type MEF consists of a single mass of engulfed material surrounded by a single isolation membrane, and the whole structure looked round or elliptical (Figure 7, A–D), as has been described extensively. We also found that more than half of the autophagosomes were not closed and the engulfed contents were continuous with the cytoplasm. In Atg3-deficient MEF, however, the engulfed portion was often divided by an isolation membrane that seemed to elongate in a random manner (Figure 7, F and H). Therefore, in some cases, the single set of autophagosome-like structures looked like a cluster of small autophagosomes, whereas in others, they looked just like an abnormally elongated structure (Figure 5A). The frequency of such aberrant structures in Atg3-deficient MEFs (22.0 ± 10.2% of AVi) was significantly (p < 0.01 by Student's t test) higher than those in wild-type MEFs (5.5 ± 5.9% of AVi). We could not compare the proportion of unclosed autophagosome-like structures between wild-type and Atg3-deficient MEFs because of the complex membranous extensions in the mutant MEFs and the limitation of using serial sections, in which we can observe objects along only a given direction. Nevertheless, these results strongly suggest that Atg8 system is indispensable for the proper elongation step of isolation membranes, and probably their closure.
Suppression of Development of Early Isolation Membranes and Appearance of Aberrant Membranous Structures in Atg3-deficient Hepatocytes
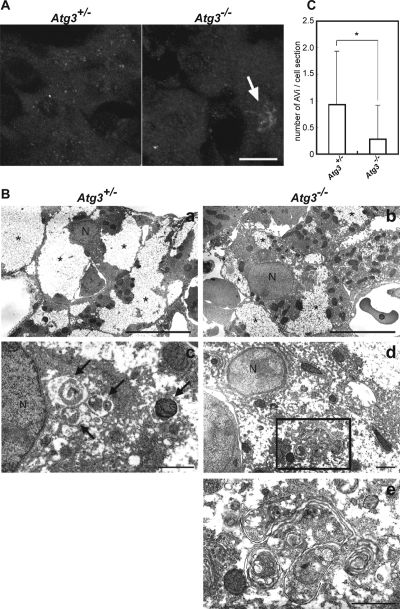
Finally, to investigate whether the autophagosome-like structures are found in other Atg3-deficient mouse tissues, we examined the liver of heterozygous and Atg3-knockout neonates under starvation condition after Caesarean delivery. Immunofluorescence microscopy using anti-Atg16L antibody showed that numerous Atg16L-positive structures occurred in heterozygous liver, but such structures were hardly observed in the mutant liver (Figure 8A). Instead, some pleomorphic structures weakly positive for Atg16L were often observed in Atg3-deficient hepatocytes (Figure 8A, arrow). Subsequent electron microscopic analyses revealed a tendency for a smaller glycogen area in mutant hepatocytes than heterozygous hepatocytes (Figure 8B, a and b), similar to Atg7-deficient adult hepatocytes (Komatsu et al., 2005). Although the nutrient-deprived heterozygous hepatocytes contained typical autophagosomes (Figure 8Bc), the number of such structures was significantly lower in nutrient-deprived mutant hepatocytes (Figure 8Bb). Instead, the Atg3-knockout hepatocytes occasionally contained aberrant membranous complexes that sometimes resembled the autophagosomes. However, the membranes were often elongated and wrapped more than two cytoplasmic portions and/or wrapped a portion several times, forming a multilamellar structure. These results strengthen the hypothesis that the Atg8 system is indispensable for the regulated elongation of isolation membranes, and its defect leads to the formation of autophagosome-like structures, such as multilamellar structures.
Figure 8.
Suppression of formation of early isolation membranes and appearance of aberrant membranous structures in Atg3-deficient hepatocytes. (A) Immunofluorescence analysis of Atg16L in hepatocytes from Atg3+/− (left) and Atg3−/− (right) newborn mice. Each newborn mouse was fixed at 6 h after Cesarean delivery. Atg16L-positive structures were observed in Atg3+/−, but only few in Atg3−/− neonate hepatocytes. Arrow indicates a pleomorphic structure positive for Atg16L. Bar, 10 μm. (B) Electron microscopic analysis of hepatocytes from Atg3+/− (a and c) and Atg3−/− (b, d, and e) newborn mice. Each newborn mouse was fixed for electron microscopy at 6 h after Cesarean delivery. Typical autophagosomes (arrows) were observed in Atg3+/−, but fewer in Atg3−/− neonatal hepatocytes. Aberrant membranous structures occurred in Atg3−/− neonate hepatocytes (d and e). e is a magnification view of the boxed region in d. Note that the glycogen area (asterisks) is smaller in mutant hepatocytes than heterozygous hepatocytes. N, nucleus. Bars, 10 μm (a and b) and 1 μm (c–e). (C) The number of autophagosomes in a section of hepatocyte in each genotype (mean ± SD; n = 60). *p < 0.001.
DISCUSSION
In this study, we showed the roles of Atg3, a specific E2-like enzyme for Atg8, in the autophagic pathway by generating Atg3 null-mice. Although Atg3-deficient mice displayed no apparent developmental defects based on our histological analyses (Supplemental Figure S5), they died within 1 d after birth, similar to Atg5- and Atg7-deficient mice (Kuma et al., 2004; Komatsu et al., 2005). The short survival time after Caesarean delivery of Atg3-deficient mice was comparable with those of other autophagy-deficient mice such as Atg5 and Atg7 mice (Kuma et al., 2004; Komatsu et al., 2005). Furthermore, the low concentration of amino acids in Atg3-deficient mice after starvation was similar to those in Atg5- and Atg7-deficient mice (Figure 2) (Kuma et al., 2004; Komatsu et al., 2005). We reported recently that autophagy is responsible for constitutive protein turnover in quiescent hepatocytes and neurons even under nutrient-rich conditions and that defects in autophagy lead to accumulation of ubiquitin-containing large inclusions (Komatsu et al., 2005; 2006; Hara et al., 2006). Thus, we expected that ubiquitin-positive inclusions would also be present in Atg3-deficient mice. Such inclusions were observed in the pons and hepatocytes of Atg3-deficient mice at P0 (data not shown). All examined phenotypes of Atg3-knockout mice resembled those of other Atg knockout mice. All results are consistent with impaired autophagy in Atg3-deficient cells.
Our results showed that Atg3 is essential for Atg8-conjugation in mammals (Figure 1). Immunofluorescent analyses revealed that LC3 did not form cup-shaped or ring-like structure in Atg3-deficient MEFs and myocardium (Figures 1 and 2). In mammals, LC3 has at least two other homologues, GABARAP and GATE-16, which share common biochemical characteristics and localize to autophagosomes (Tanida et al., 2001, 2002; Kabeya et al., 2004). The modification and levels of these molecules were also affected in the mutant MEFs. Similar to LC3 states, mutant cells showed impairment of modification of GABARAP, whereas the PE-conjugated form was observed in the wild-type cells under both nutrient-rich and -deprived conditions. Because the modified forms of LC3 and GABARAP were formed even in nutrient-rich conditions, they might play a role in constitutive autophagy, which occurs irrelevant of the nutrient state, in addition to their roles in starvation-induced autophagy. Although neither GATE-16-I nor GATE-16-II was detected in wild-type cells under both nutrient-available and -deprived conditions, GATE-16-I was clearly detected in mutant cells, suggesting that GATE-16 might be rapidly degraded depending on the autophagic pathway. Actually, treatment with lysosomal inhibitor led to acute accumulation of GATE-16-II (Supplemental Figure S1). These findings are similar to those observed in Atg7-deficient mice (Komatsu et al., 2005). The difference of action among Atg8 homologues in their response to nutrient conditions might reflect their distinct roles in constitutive or starvation-induced autophagy. We sought to determine their localizations under nutrient-rich and deprived conditions, but our antibodies for these molecules were not applicable for immunofluorescent analyses. Further analysis is required to unravel the roles of GABARAP and GATE-16 in autophagy, and such analysis is currently underway by generating transgenic mice harboring each GFP-GABARAP and GFP-GATE-16 (Tanida and Kominami, unpublished data).
Intriguingly, Atg5 deficiency affects another line of conjugation system that causes significant reduction in the conversion of LC3 to PE-conjugated form in yeast and mice (Mizushima et al., 2001; Suzuki et al., 2001; Hosokawa et al., 2007). Moreover, Atg10, a specific E2 enzyme for Atg12, interacts with LC3, and overproduction of Atg10 accelerates conversion into the LC3-II form (Nemoto et al., 2003). Furthermore, overproduction of Atg3 promotes the formation of Atg12–Atg5 conjugates (Tanida et al., 2002). These results strongly suggest functional cooperation between the two conjugation systems. However, the role of Atg8 system in Atg12 system based on the loss of function has not yet been examined so far. In this study, we showed that Atg3 deficiency markedly inhibits the formation of the Atg12–Atg5 conjugate (Figure 3). Why is Atg12–Atg5 conjugation suppressed by loss of Atg3? Comprehensive analyses of yeast two-hybrid screening revealed interaction between Atg3 and Atg12 in yeast (Uetz et al., 2000). This physical interaction was also confirmed in mammalian cells (Tanida et al., 2002). This interaction might be critical for effective formation of the Atg12–Atg5 conjugate. However, in in vitro reconstitution assay for the Atg12–Atg5 conjugate in the presence or absence of Atg3 protein, we could not detect enhanced formation of the Atg12–Atg5 conjugate (Supplemental Figure S6), suggesting the involvement of other cellular factor(s) in the accelerated formation of the Atg12–Atg5 conjugate by Atg3. Indeed, unlike the in vitro results, inhibition of Atg12–Atg5 conjugate formation was complemented by exogenous expression of wild-type but not the active site mutant of Atg3 into Atg3-deficient MEFs (Figure 3), implying that the Atg8/LC3-conjugation system might assist Atg12–Atg5 conjugation reaction. Another possibility is that the retarded dissociation of Atg12–Atg5 conjugate from the isolation membrane (Figures 4 and 5) might suppress any new conjugation between Atg12 and Atg5. Further analysis is required to unravel the role of Atg3 in the formation of the Atg12–Atg5 conjugate.
Although both Atg8 and its homologue LC3 are considered suitable markers for autophagosomes (Kabeya et al., 2000; Kirisako et al., 2000; Mizushima et al., 2004), their functions in autophagosome formation are poorly understood. Herein, we showed that loss of Atg3 was associated with autophagosome malformation with defects in multiple steps of autophagosome formation after generation of early isolation membranes. The first defect was failure of localization of LC3 to the isolation membranes due to the loss of its conjugation to PE (Figure 1). Second, the absence of LC3 on the isolation membrane led to delay of dissociation of Atg12–Atg5–Atg16L complex from the membrane (Figure 4). Third, the loss of LC3-PE and/or prolonged association of Ag12–Atg5–Atg16L complex with the membranes caused a dysregulated elongation of isolation membranes in response to nutrient deprivation (Figure 5). Fourth, loss of LC3-PE on the isolation membranes caused impairment of fusion of each membrane sac, resulting in the accumulation of unclosed autophagosome-like structures, some of which could be seen as multilamellar structures in hepatocytes (Figures 7 and 8). The fourth criterion is in agreement with the data showing accumulation of intermediate autophagic structures (isolation membranes) in cells with almost complete impairment of LC3 conjugation by overexpression of Atg4B (Fujita, Noda, and Yoshimori, personal communication). These findings emphasize the indispensable role of the Atg8/LC3 system in the development of isolation membranes, consistent with a recent report in yeasts (Nakatogawa et al., 2007), although we could not exclude the involvement of Atg8 system in the fusion of autophagosomes with lysosomes.
Although numerous early isolation membranes double-positive for Atg16L- and Atg5 were observed in Atg3-deficient MEFs, we think that incidence of these isolation membranes in Atg3-deficient mice are probably lower than that in wild-type mice. Because the half-life of autophagosomes is ∼10 min (Mizushima et al., 2001), if the isolation membranes in mutant mice were normally developed and only their elongation and closing steps were impaired, numerous autophagosome-like structures should accumulate in mutant mice. However, their number after 2-h starvation in the mutant remained twofold that in wild-type MEFs (Figure 5), suggesting a low production of the isolation membranes in Atg3 deficiency. This conclusion is also supported by the reduced production of Atg16L-positive structures in Atg3-deficient hepatocytes (Figure 8). Moreover, time-lapse analysis revealed fewer de novo GFPAtg5 dots in mutant MEFs. Therefore, we infer that Atg3 deficiency affects the generation of early isolation membranes, as well as their development. We stress that almost all phenotypes observed in Atg3-deficient mice were due to impaired Atg8-system rather than low production of Atg12–Atg5 conjugate because the presence of a very low level of Atg12–Atg5 is sufficient for autophagy (Hosokawa et al., 2007).
Importantly, small autophagosome-like structures observed in Atg3-deficient MEFs were also recognized in Atg7-deficient MEFs (our unpublished data). In addition, several elongated isolation membranes and numerous double membrane structures were found even in Atg5-deficient ES cells and Purkinje cells, respectively (Mizushima et al., 2001; Nishiyama et al., 2007). Moreover, membrane complex structures recognized in Atg3-deficient neonate hepatocytes resemble the multilamellar structures observed in Atg7-deficient adult hepatocytes (Komatsu et al., 2005). Furthermore, such multilamellar structures were also observed in Atg5- and Atg7-deficient Purkinje cells (Komatsu et al., 2007; Nishiyama et al., 2007). Therefore, the Atg8 system and perhaps the Atg12 system may be essential for proper membrane development and closure for the completion of autophagosome formation. Our Atg3 mutant mice, together with Atg5−/− and Atg7−/− mice, should be useful for examining and delineating autophagosome formation in mammalian cells.
Supplementary Material
ACKNOWLEDGMENTS
We thank T. Kouno for the excellent technical assistance; A. Yabashi, K. Kanno, and Y. Sekikawa for help in electron microscopic analyses; Drs. J. Ezaki, I. Tanida, Y. Ichimura, and N. Furuya for the helpful discussion; Dr. M. Hagiwara (Tokyo Medical and Dental University) for providing the pLRT-X retrovirus vectors; and Dr. T. Kitamura (Tokyo University) for providing the pMXs-puro retrovirus vectors. This work was supported by grants from the Japan Science and Technology Agency (to M. K.); the Ministry of Education, Science and Culture of Japan (to M. K. and K. T.); and Takeda Science Foundation (to K. T.).
Abbreviations used:
- BCAA
branched chain amino acids
- ES
embryonic stem
- GABARAP
GABA(A) receptor-associated protein
- GATE-16
Golgi-associated ATPase enhancer of 16 kDa
- GFP
green fluorescent protein
- LC3
microtubule-associated protein 1 light chain 3/MAP1LC3
- MEF
mouse embryonic fibroblasts
- PE
phosphatidylethanolamine.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-03-0309) on September 3, 2008.
REFERENCES
- Cuervo A. M. Autophagy: in sickness and in health. Trends Cell Biol. 2004;14:70–77. doi: 10.1016/j.tcb.2003.12.002. [DOI] [PubMed] [Google Scholar]
- Fujita N., Itoh T., Omori H., Fukuda M., Noda T., Yoshimori T. The Atg16L complex specifies the site of LC3 lipidation for membrane biogenesis in autophagy. Mol. Biol. Cell. 2008;19:2092–2100. doi: 10.1091/mbc.E07-12-1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg A. L. Protein degradation and protection against misfolded or damaged proteins. Nature. 2003;426:895–899. doi: 10.1038/nature02263. [DOI] [PubMed] [Google Scholar]
- Gutierrez M. G., Master S. S., Singh S. B., Taylor G. A., Colombo M. I., Deretic V. Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell. 2004;119:753–766. doi: 10.1016/j.cell.2004.11.038. [DOI] [PubMed] [Google Scholar]
- Hanada T., Noda N. N., Satomi Y., Ichimura Y., Fujioka Y., Takao T., Inagaki F., Ohsumi Y. The Atg12-Atg5 conjugate has a novel E3-like activity for protein lipidation in autophagy. J. Biol. Chem. 2007;282:37298–37302. doi: 10.1074/jbc.C700195200. [DOI] [PubMed] [Google Scholar]
- Hanaoka H., Noda T., Shirano Y., Kato T., Hayashi H., Shibata D., Tabata S., Ohsumi Y. Leaf senescence and starvation-induced chlorosis are accelerated by the disruption of an Arabidopsis autophagy gene. Plant Physiol. 2002;129:1181–1193. doi: 10.1104/pp.011024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara T., et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–889. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- Hosokawa N., Hara Y., Mizushima N. Generation of cell lines with tetracycline-regulated autophagy and a role for autophagy in controlling cell size. FEBS Lett. 2007;581:2623–2629. doi: 10.1016/j.febslet.2007.05.061. [DOI] [PubMed] [Google Scholar]
- Ichimura Y., et al. A ubiquitin-like system mediates protein lipidation. Nature. 2000;408:488–492. doi: 10.1038/35044114. [DOI] [PubMed] [Google Scholar]
- Ichimura Y., Kumanomido T., Sou Y. S., Mizuchima T., Ezaki J., Ueno T., Kominami E., Yamane T., Tanaka K., Komatsu M. Structural basis for sorting mechanism of p62 in selective autophagy. J. Biol. Chem. 2008;283:22847–22857. doi: 10.1074/jbc.M802182200. [DOI] [PubMed] [Google Scholar]
- Kabeya Y., Mizushima N., Ueno T., Yamamoto A., Kirisako T., Noda T., Kominami E., Ohsumi Y., Yoshimori T. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeya Y., Mizushima N., Yamamoto A., Oshitani-Okamoto S., Ohsumi Y., Yoshimori T. LC3, GABARAP and GATE16 localize to autophagosomal membrane depending on form-II formation. J. Cell Sci. 2004;117:2805–2812. doi: 10.1242/jcs.01131. [DOI] [PubMed] [Google Scholar]
- Kiffin R., Bandyopadhyay U., Cuervo A. M. Oxidative stress and autophagy. Antioxid. Redox Signal. 2006;8:152–162. doi: 10.1089/ars.2006.8.152. [DOI] [PubMed] [Google Scholar]
- Kirisako T., Ichimura Y., Okada H., Kabeya Y., Mizushima N., Yoshimori T., Ohsumi M., Takao T., Noda T., Ohsumi Y. The reversible modification regulates the membrane-binding state of Apg8/Aut7 essential for autophagy and the cytoplasm to vacuole targeting pathway. J. Cell Biol. 2000;151:263–276. doi: 10.1083/jcb.151.2.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura T., Koshino Y., Shibata F., Oki T., Nakajima H., Nosaka T., Kumagai H. Retrovirus-mediated gene transfer and expression cloning: powerful tools in functional genomics. Exp. Hematol. 2003;31:1007–1014. [PubMed] [Google Scholar]
- Klionsky D. J. The molecular machinery of autophagy: unanswered questions. J. Cell Sci. 2005;118:7–18. doi: 10.1242/jcs.01620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M., et al. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J. Cell Biol. 2005;169:425–434. doi: 10.1083/jcb.200412022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M., et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–884. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- Komatsu M., Wang Q. J., Holstein G. R., Friedrich V. L., Jr, Iwata J., Kominami E., Chait B. T., Tanaka K., Yue Z. Essential role for autophagy protein Atg7 in the maintenance of axonal homeostasis and the prevention of axonal degeneration. Proc. Natl. Acad. Sci. USA. 2007;104:14489–14494. doi: 10.1073/pnas.0701311104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuma A., Hatano M., Matsui M., Yamamoto A., Nakaya H., Yoshimori T., Ohsumi Y., Tokuhisa T., Mizushima N. The role of autophagy during the early neonatal starvation period. Nature. 2004;432:1032–1036. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- Kuma A., Mizushima N., Ishihara N., Ohsumi Y. Formation of the approximately 350-kDa Apg12-Apg5.Apg16 multimeric complex, mediated by Apg16 oligomerization, is essential for autophagy in yeast. J. Biol. Chem. 2002;277:18619–18625. doi: 10.1074/jbc.M111889200. [DOI] [PubMed] [Google Scholar]
- Lee G. H., Ogawa K., Drinkwater N. R. Conditional transformation of mouse liver epithelial cells. An in vitro model for analysis of genetic events in hepatocarcinogenesis. Am. J. Pathol. 1995;147:1811–1822. [PMC free article] [PubMed] [Google Scholar]
- Levine B., Klionsky D. J. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev. Cell. 2004;6:463–477. doi: 10.1016/s1534-5807(04)00099-1. [DOI] [PubMed] [Google Scholar]
- Massey A. C., Kaushik S., Sovak G., Kiffin R., Cuervo A. M. Consequences of the selective blockage of chaperone-mediated autophagy. Proc. Natl. Acad. Sci. USA. 2006;103:5805–5810. doi: 10.1073/pnas.0507436103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N., Kuma A., Kobayashi Y., Yamamoto A., Matsubae M., Takao T., Natsume T., Ohsumi Y., Yoshimori T. Mouse Apg16L, a novel WD-repeat protein, targets to the autophagic isolation membrane with the Apg12-Apg5 conjugate. J. Cell Sci. 2003;116:1679–1688. doi: 10.1242/jcs.00381. [DOI] [PubMed] [Google Scholar]
- Mizushima N., Noda T., Yoshimori T., Tanaka Y., Ishii T., George M. D., Klionsky D. J., Ohsumi M., Ohsumi Y. A protein conjugation system essential for autophagy. Nature. 1998a;395:395–398. doi: 10.1038/26506. [DOI] [PubMed] [Google Scholar]
- Mizushima N., Ohsumi Y., Yoshimori T. Autophagosome formation in mammalian cells. Cell Struct. Funct. 2002;27:421–429. doi: 10.1247/csf.27.421. [DOI] [PubMed] [Google Scholar]
- Mizushima N., Sugita H., Yoshimori T., Ohsumi Y. A new protein conjugation system in human. The counterpart of the yeast Apg12p conjugation system essential for autophagy. J. Biol. Chem. 1998b;273:33889–33892. doi: 10.1074/jbc.273.51.33889. [DOI] [PubMed] [Google Scholar]
- Mizushima N., Yamamoto A., Hatano M., Kobayashi Y., Kabeya Y., Suzuki K., Tokuhisa T., Ohsumi Y., Yoshimori T. Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells. J. Cell Biol. 2001;152:657–668. doi: 10.1083/jcb.152.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N., Yamamoto A., Matsui M., Yoshimori T., Ohsumi Y. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol. Biol. Cell. 2004;15:1101–1111. doi: 10.1091/mbc.E03-09-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa I., et al. Autophagy defends cells against invading group A Streptococcus. Science. 2004;306:1037–1040. doi: 10.1126/science.1103966. [DOI] [PubMed] [Google Scholar]
- Nakatogawa H., Ichimura Y., Ohsumi Y. Atg8, a ubiquitin-like protein required for autophagosome formation, mediates membrane tethering and hemifusion. Cell. 2007;130:165–178. doi: 10.1016/j.cell.2007.05.021. [DOI] [PubMed] [Google Scholar]
- Nemoto T., Tanida I., Tanida-Miyake E., Minematsu-Ikeguchi N., Yokota M., Ohsumi M., Ueno T., Kominami E. The mouse APG10 homologue, an E2-like enzyme for Apg12p conjugation, facilitates MAP-LC3 modification. J. Biol. Chem. 2003;278:39517–39526. doi: 10.1074/jbc.m300550200. [DOI] [PubMed] [Google Scholar]
- Nishiyama J., Miura E., Mizushima N., Watanabe M., Yuzaki M. Aberrant membranes and double-membrane structures accumulate in the axons of Atg5-null Purkinje cells before neuronal death. Autophagy. 2007;3:591–596. doi: 10.4161/auto.4964. [DOI] [PubMed] [Google Scholar]
- Ohsumi Y. Molecular dissection of autophagy: two ubiquitin-like systems. Nat. Rev. Mol. Cell Biol. 2001;2:211–216. doi: 10.1038/35056522. [DOI] [PubMed] [Google Scholar]
- Shintani T., Mizushima N., Ogawa Y., Matsuura A., Noda T., Ohsumi Y. Apg10p, a novel protein-conjugating enzyme essential for autophagy in yeast. EMBO J. 1999;18:5234–5241. doi: 10.1093/emboj/18.19.5234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sou Y. S., Tanida I., Komatsu M., Ueno T., Kominami E. Phosphatidylserine in addition to phosphatidylethanolamine is an in vitro target of the mammalian Atg8 modifiers, LC3, GABARAP, and GATE-16. J. Biol. Chem. 2006;281:3017–3024. doi: 10.1074/jbc.M505888200. [DOI] [PubMed] [Google Scholar]
- Suzuki K., Kirisako T., Kamada Y., Mizushima N., Noda T., Ohsumi Y. The pre-autophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation. EMBO J. 2001;20:5971–5981. doi: 10.1093/emboj/20.21.5971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanida I., Mizushima N., Kiyooka M., Ohsumi M., Ueno T., Ohsumi Y., Kominami E. Apg7p/Cvt2p: a novel protein-activating enzyme essential for autophagy. Mol. Biol. Cell. 1999;10:1367–1379. doi: 10.1091/mbc.10.5.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanida I., Sou Y. S., Ezaki J., Minematsu-Ikeguchi N., Ueno T., Kominami E. HsAtg4B/HsApg4B/autophagin-1 cleaves the carboxyl termini of three human Atg8 homologues and delipidates microtubule-associated protein light chain 3- and GABAA receptor-associated protein-phospholipid conjugates. J. Biol. Chem. 2004;279:36268–36276. doi: 10.1074/jbc.M401461200. [DOI] [PubMed] [Google Scholar]
- Tanida I., Tanida-Miyake E., Komatsu M., Ueno T., Kominami E. Human Apg3p/Aut1p homologue is an authentic E2 enzyme for multiple substrates, GATE-16, GABARAP, and MAP-LC3, and facilitates the conjugation of hApg12p to hApg5p. J. Biol. Chem. 2002;277:13739–13744. doi: 10.1074/jbc.M200385200. [DOI] [PubMed] [Google Scholar]
- Tanida I., Tanida-Miyake E., Ueno T., Kominami E. The human homolog of Saccharomyces cerevisiae Apg7p is a protein-activating enzyme for multiple substrates including human Apg12p, GATE-16, GABARAP, and MAP-LC3. J. Biol. Chem. 2001;276:1701–1706. doi: 10.1074/jbc.C000752200. [DOI] [PubMed] [Google Scholar]
- Thumm M., Egner R., Koch B., Schlumpberger M., Straub M., Veenhuis M., Wolf D. H. Isolation of autophagocytosis mutants of Saccharomyces cerevisiae. FEBS Lett. 1994;349:275–280. doi: 10.1016/0014-5793(94)00672-5. [DOI] [PubMed] [Google Scholar]
- Tsukada M., Ohsumi Y., et al. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 1993;333:169–174. doi: 10.1016/0014-5793(93)80398-e. [DOI] [PubMed] [Google Scholar]
- Uetz P., et al. A comprehensive analysis of protein-protein interactions in. Saccharomyces cerevisiae. Nature. 2000;403:623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.