Abstract
Cystic fibrosis transmembrane conductance regulator (CFTR) is a polytopic membrane protein that functions as a Cl− channel and consists of two membrane spanning domains (MSDs), two cytosolic nucleotide binding domains (NBDs), and a cytosolic regulatory domain. Cytosolic 70-kDa heat shock protein (Hsp70), and endoplasmic reticulum-localized calnexin are chaperones that facilitate CFTR biogenesis. Hsp70 functions in both the cotranslational folding and posttranslational degradation of CFTR. Yet, the mechanism for calnexin action in folding and quality control of CFTR is not clear. Investigation of this question revealed that calnexin is not essential for CFTR or CFTRΔF508 degradation. We identified a dependence on calnexin for proper assembly of CFTR's membrane spanning domains. Interestingly, efficient folding of NBD2 was also found to be dependent upon calnexin binding to CFTR. Furthermore, we identified folding defects caused by deletion of F508 that occurred before and after the calnexin-dependent association of MSD1 and MSD2. Early folding defects are evident upon translation of the NBD1 and R-domain and are sensed by the RMA-1 ubiquitin ligase complex.
INTRODUCTION
Cystic fibrosis transmembrane conductance regulator (CFTR) is a membrane glycoprotein that is localized to the apical surface of epithelial cells that line ducts of glands and airways. CFTR functions as an ATP-gated Cl− channel that is critical for proper hydration of the mucosal layer that lines lung airways (Welsh and Smith, 1993). Individuals who inherit two mutant forms of CFTR have exceedingly viscous mucous and, due to chronic lung infections, develop cystic fibrosis and often die from lung failure. CFTR is a member of the ATP-binding cassette (ABC) transporter superfamily (Hyde et al., 1990) and is a 1480-amino acid protein that contains two membrane spanning domains (MSDs), MSD1 and MSD2; two cytosolic nucleotide binding domains (NBDs), NBD1 and NBD2; and a regulatory (R) domain (Riordan et al., 1989). The proper folding and assembly of CFTR subdomains in the endoplasmic reticulum (ER) is required for CFTR to engage the COPII machinery and be packaged into vesicles for transport to the plasma membrane (Kopito, 1999; Wang et al., 2004). The folding pathway of this complex polytopic membrane protein has been a topic of great interest, because misfolding results in premature recognition of CFTR by the ER quality control system (ERQC) and degradation by the ubiquitin proteasome system (Skach, 2000). In fact, the most common disease-causing mutation of CFTR, ΔF508CFTR, results in almost complete degradation of the protein by the ERQC system, which gives rise to a loss of function phenotype and lung disease (Ward and Kopito, 1994).
The assembly of CFTR into an ion channel is complicated because it requires the coordinated folding and assembly of its membrane and cytoplasmic domains into a functional unit (Du et al., 2005; Riordan, 2005; Cui et al., 2007). CFTR is a modular protein, and its domains can collapse to a protease-resistant conformation independently (Zhang et al., 1998). Yet, structures of related ABC transporter family members suggest that assembly of CFTR to an active conformation is a cooperative process that is dependent upon cross-contact formation between its N- and C-terminal membrane and cytosolic domains (Dawson and Locher, 2006; Mendoza and Thomas, 2007; Serohijos et al., 2008). Formation of a CFTR structure that can pass quality control occurs in cotranslational and posttranslational steps that are proposed to involve critical interactions between solvent-exposed surfaces of NBD1 and MSD2 and similar interactions between NBD2 and MSD1 (Serohijos et al., 2008). A fragment of CFTR that contains MSD1, NBD1, the R-domain, and MSD2 (CFTR 1-1172) can fold to a conformation that can escape the ER quality control system and traffic to the plasma membrane in which it exhibits ATP-gated channel activity (Cui et al., 2007). Thus, it is assumed that MSD1, NBD1, the R-domain, and MSD2 are folded and assembled cotranslationally, with the folding and subsequent assembly of NBD2 into a complex, with the remainder of the protein occurring posttranslationally.
F508 of CFTR is located on the solvent-exposed surface of NBD1 (Lewis et al., 2004; Thibodeau et al., 2005), and the crystal structures of bacterial ABC transporters suggest that it makes contacts with cytosolic surface loops on MSD2 that are critical for stabilization of CFTR structure (Dawson and Locher, 2006; Serohijos et al., 2008). This supposition is supported by the observation that the F508 mutation disrupts assembly of CFTR 1-1172 (Cui et al., 2007) and makes similar CFTR fragments susceptible to cotranslational recognition by the ER-associated ubiquitin ligase RMA1 (Younger et al., 2006). NBD2 is the last domain on CFTR that is synthesized, and its folding and assembly into a complex with amino-terminal regions of CFTR seems to be slow and thus occur posttranslationally (Du et al., 2005). Deletion of F508 hinders the folding of NBD2, but whether this occurs as a result of improper interaction of F508 NBD1 with NBD2 or global defects in CFTR assembly is not clear (Du et al., 2005; Cui et al., 2007).
The folding and assembly of CFTR is not a spontaneous process, and some of the same chaperones that are involved in the selection of misfolded CFTR for degradation are also required for CFTR folding. The 40-kDa heat shock protein (Hsp40) Hdj2 is farnesylated and localized to the cytoplasmic face of the ER in which it binds CFTR translation intermediates to facilitate aspects of its cotranslational folding (Meacham et al., 1999). Yet, severalfold more Hdj2 is found in association with CFTRΔF508, so Hdj2 may also participate in the selection of misfolded CFTR for degradation by the Hsp70/CHIP E3 ubiquitin ligase complex (Meacham et al., 2001). Hsp90 is required for CFTR biogenesis (Loo et al., 1998), and cellular depletion of the Hsp90 cochaperone Aha1 enhances CFTR folding (Wang et al., 2006); yet, the step where Hsp90 acts in the CFTR folding pathway is not clear. The ER lumenal chaperone calnexin forms transient complexes with the ER localized and immaturely glycosylated B-form of CFTR (Pind et al., 1994), but mechanistic details of calnexin function in CFTR biogenesis also remain to be elucidated.
Studies with CFTR assembly intermediates indicate that Hsp70 can bind CFTR at co- and posttranslational stages of its biogenesis (Meacham et al., 1999), but no data are available to describe the temporal relationship between Hsp70 and calnexin function in CFTR biogenesis. Calnexin binds N-linked glycans, and two such glycosylation sites are found on CFTR in extracellular loop 4 in MSD2 (Cheng et al., 1990; Farinha and Amaral, 2005). Knockdown of calnexin leads a large portion of nascent CFTR to misfold, but why calnexin is required for CFTR folding is not clear (Farinha and Amaral, 2005; Okiyoneda et al., 2008). In addition, analysis of calnexin's role in degradation of misfolded CFTR and CFTRΔF508 has provided mixed results. Overexpression of calnexin results in the accumulation of ΔF508 CFTR in the ER (Okiyoneda et al., 2004), but RNA interference (RNAi)-mediated decrease of calnexin levels or deletion of the calnexin gene does not inhibit degradation of wild-type (WT) or ΔF508 CFTR (Farinha and Amaral, 2005; Okiyoneda et al., 2008).
In this study, we sought to define the steps in CFTR folding and degradation that are catalyzed by calnexin. In addition, we examined the temporal relationship between folding defects caused by disease mutations such as ΔF508 to calnexin-dependent folding reactions. Finally, by further refining the steps in the CFTR folding pathway we hoped to clarify the events that lead to recognition of misfolded CFTR by the ERQC machinery. To accomplish these goals, we examined the influence that castanospermine (CAS), an inhibitor of the ER glucosidase I and II enzymes that process N-linked oligosaccharides to a form that can be recognized by calnexin, had on CFTR folding and degradation (Hammond et al., 1994). We found that calnexin plays a critical role in the stabilization of the MSD2 domain of CFTR. Inhibition of calnexin binding blocked CFTR folding at a stage where MSD1 and MSD2 normally associate. Disruption of this calnexin-dependent reaction also caused a downstream defect in the folding of C-terminal NBD2. Similarly, introduction of the ΔF508 mutation caused both early and late folding defects, with the early defects being monitored by the RMA-1ubiquitin ligase complex.
MATERIALS AND METHODS
Plasmids, Antibodies, and Reagents
CFTR expression plasmids of pcDNA3.1-CFTR and pcDNA3.1-ΔF508-CFTR have been described previously (Meacham et al., 1999), and CFTR constructs representing biogenic intermediates were made by use of QuikChange (Stratagene, La Jolla, CA) to introduce stop codons after the indicated amino acid. Antibodies used in this study were as follows: mouse monoclonal α-CFTR MM13-4 (N-terminal tail epitope) and α-CFTR M3A7 (NBD2 epitope) were from Millipore (Billerica, MA); rabbit polyclonal α-Hsp/c70 (SPA-757) and rabbit polyclonal α-calnexin (SPA-860) were from Nventa Biopharmaceuticals (San Diego, CA); mouse α-tubulin was purchased from Sigma (T9026); and rabbit α-Derlin-1 (PM018) was purchased from MBL International (Woburn, MA). Polyclonal α-CFTR was generated against a glutathione transferase (GST) fusion protein that contained residues 1–79 of CFTR and was a gift from Dr. K. Kirk (Department of Physiology and Biophysics, University of Alabama-Birmingham). CAS was purchased from Sigma and used at a final concentration of 5 mM to inhibit glucosidase I and II. Brefeldin A (BFA) was from Sigma and used at a final concentration of 10 μg/ml.
Cell Culture and Transfection
Human embryonic kidney (HEK)293 cells were from Stratagene and were maintained in Dulbecco's modified Eagle's medium (DMEM; Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (HyClone Laboratories, Logan, UT) and antibiotics (100 U/ml penicillin and 100 μg/ml streptomycin; Invitrogen) at 37°C in an atmosphere of 5% CO2. Cell transfections were performed using Effectene reagent (QIAGEN, Valencia, CA) with 1 μg pcDNA3.1-ΔF508-CFTR, pcDNA3.1-CFTR, pcDNA3.1-CFTR642X pcDNA3.1-CFTR653X, pcDNA3.1-CFTR673X, pcDNA3.1-CFTR837X, pcDNA3.1- CFTR1162X, pcDNA3.1CFTR1172X, or pcDNA3.1-CFTR837–1480 for one six-well for Western Blot, pulse chase, or immunoprecipitation.
Coexpression of CFTR Halves
Cells were transfected with pcDNA3.1-CFTR837X (1 μg) and pcDNA3.1-CFTR837-1480 (1 μg), individually or in combination. Reactions were balanced with pcDNA3.1 such that all transfections were performed with equal microgram quantities of DNA. Where indicated, brefeldin A (10 μg/ml) or CAS (5 mM) was added to the media 5 h after transfection or N-acetyl-L-leucinyl-L-leucinyl-L-norleucinal (ALLN) (200 μM) was added 18 h after transfection. Twenty-four hours after transfection, cells were harvested with citric saline, diluted in 2× sample buffer (100 mM Tris, pH 6.8, 4% SDS, 20% glycerol, and 0.1% bromphenol blue), sonicated for 10 s, and warmed to 37°C for 10 min before loading on 10% SDS-PAGE gels. Proteins were transferred to nitrocellulose using a mini gel wet transfer apparatus (Bio-Rad, Hercules, CA). Blots were blocked in blocking buffer containing 10% fat-free milk and 0.1% Triton-X 100 in phosphate-buffered saline (PBS) and probed with monoclonal α-CFTR N-terminal tail (MM13-4 1:1000 dilution) or α-CFTR NBD2 (M3A7 1:1000 dilution). α-Tubulin (Sigma) or α-Derlin-1 (MBL International) was used to indicate loading controls.
Analysis of CFTR Folding and Degradation
The fate of nascent CFTR and CFTRΔF508 was analyzed by pulse chase as described below. Twenty-four hours after transfection, cells were preincubated for 1 h with 5 mM CAS in DMEM where indicated, starved in methionine-free minimal essential medium (Sigma) for 20 min, pulse labeled for 30 min with [35S]methionine (100 μCi/6-well; 1200 Ci/mmol; MP Biomedicals, Irvine, CA) and then chased for the indicated amount of time. Cells were then washed twice in PBS and lysed in radioimmunoprecipitation assay (RIPA) buffer (150 mM NaCl, 1% NP-40, 0.5% deoxycholate, 0.2% SDS, and 50 mM HEPES, pH 7.4) freshly supplemented with 1 mM phenylmethylsulfonyl fluoride and Complete Protease Inhibitor cocktail (Roche Diagnostics, Indianapolis, IN). Lysates were precleared with 2% Pansorbin cells (Calbiochem, San Diego, CA) for 15 min. Radiolabeled CFTR was then immunoprecipitated by incubation with rabbit polyclonal α-CFTR antibody directed against the N terminus (provided as a kind gift from Dr. K. Kirk) and protein G beads sequentially for 1 h each at 4°C, washed three times with RIPA buffer, and eluted with 2× sample buffer at 37°C for 15 min. The samples were analyzed by SDS-PAGE and visualized by autoradiography.
Coimmunoprecipitation of CFTR with Chaperones
Cells were starved and pulse labeled as described above for the pulse-chase assay. Lysates were prepared in coimmunoprecipitation buffer with an ATP regeneration system (PBS, pH 7.4, supplemented with 1% Triton X-100, 5 mM Mg-ATP, 80 mM phosphocreatine, 500 μg/ml creatine phosphokinase and Complete Protease Inhibitor [Roche Diagnostics]) and precleared by centrifugation at 20,000 rpm for 10 min in an Allegra 64R centrifuge (Beckman Coulter, Fullerton, CA). The cleared lysates were split in to three reactions and then incubated at 4°C for 1 h with rabbit polyclonal α-CFTR antibody directed against the N terminus (provided as a kind gift from Dr. K. Kirk), rabbit polyclonal α-calnexin antibody (Nventa Biopharmaceuticals) or rabbit α-hsc/p70 antibody (Nventa Biopharmaceuticals). This was followed by the addition of 25 μl of a 70% protein G slurry, and incubations with beads were carried out for 30 min. Protein G pellets were washed twice with PBS-Tr buffer (PBS supplemented with 1% Triton X-100) and eluted in 25 μl of 2× sample buffer. For reimmunoprecipitation, the primary immunoprecipitates were washed once with PBS-Tr buffer and eluted in 25 μl of 2× sample buffer at 37°C for 15 min. Then, a 20-μl aliquot was diluted in 750 μl of PBS supplemented with 1% Triton X-100, 0.2% SDS, and 0.5% bovine serum albumin. The reimmunoprecipitation was carried out by incubation with 5 μl of polyclonal α-CFTR antibody for 1 h and subsequent incubation with 50 μl of a 50% slurry of protein A-Sepharose for 30 min at 4°C. The beads were washed twice with PBS-Tr supplemented with 0.2% SDS. Immunoprecipitated proteins were eluted in 2× sample buffer at 55°C for 15 min before loading on 10% gels, and they were visualized by autoradiography.
Limited Proteolysis
Six wells of a six-well plate containing HEK293 cells were transfected with 1 μg each of the indicated plasmids. Where indicated, 5 mM CAS was added to the media 5 h after transfection. Twenty-four hours posttransfection, the cells were harvested by citric saline and lysed in PBS-Tr (0.1%) for 1 h at 4°C. Lysates were cleared by centrifugation at 20,000 rpm for 10 min in an Allegra 64R centrifuge (Beckman Coulter). Supernatants were removed, and total microgram quantities of protein were determined by the DC protein determination assay (Bio-Rad). Cleared cell lysates were diluted in PBS-Tr (0.1%) to make 100-μl aliquots of a 2 mg/ml cell lysate solution. Twenty-five μl aliquots of trypsin stocks were then added to each aliquot to reach the indicated final trypsin concentrations. The cleavage reactions incubated on ice for 15 min, and they were quenched by addition of Complete Protease Inhibitor (Roche Diagnostics) and trypsin inhibitor. Sample buffer was added to a final 1× concentration, and samples were run on 12.5% SDS-PAGE gels. Gels were transferred to nitrocellulose and probed with CFTR antibodies as indicated above.
RNAi Analysis
Six-well samples of HEK293 cells were transfected with 100 nM total of oligonucleotides (oligos) directed at either CHIP (sequence 1, GGAGCAGGGCAAUCGUCUG; sequence 2, CCAAGCACGACAAGUACAU), RMA-1 (sequence 1, GCGCGACCUUCGAAUGUAA; sequence 2, CGGCAAGAGUGUCCAGUAU), or a nonspecific control (Dharmacon RNA Technologies, Lafayette, CO) by using Lipofectamine 2000 (Invitrogen) as a transfection reagent. Forty-eight hours after transfection cells were transfected a second time with 1 μg of plasmid DNA by using Effectene to express the indicated CFTR fragments. Cells were harvested 18 h after the second transfection. Sample buffer was added to cell pellets at a final 2× concentration, samples were sonicated, and then equal microgram amounts of cell lysate (as determined by the DC Protein Assay) were run on 12.5% SDS-PAGE gels. Gels were transferred to nitrocellulose and probed with CFTR antibodies.
The calnexin shRNAmir construct (pGIPZ calnexin; V2LHS_150212) and the nonsilencing pGipZ control (RHS4346) were purchased from Open Biosystems (Huntsville, AL), and 2 μg of plasmid was transfected per six-well by using Lipofectamine 2000 as a transfection reagent. Eighteen hours after transfection, puromycin (10 μg/ml) was added to the HEK293 cells to select for those cells that took up the pGIPZ plasmids. Cells were grown in puromycin-containing media for 5 d, and then a second transfection was performed with Effectene reagent to introduce the indicated CFTR plasmids. Eighteen hours later, the cells were harvested, and equal microgram quantities of cell lysates (as determined by the DC Protein Assay) were run on 10% SDS-PAGE gels.
RESULTS
Calnexin Acts Transiently at a Late Stage Folding Event of WT CFTR
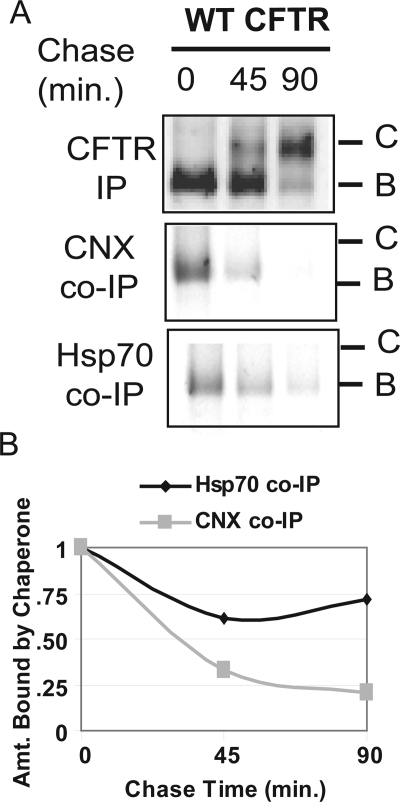
To identify the calnexin-mediated steps in CFTR folding, we first compared the timing of calnexin and Hsp70 binding and release to the newly synthesized and immaturely glycosylated B-form of [35S]CFTR in pulse-chase experiments (Figure 1). The B-form of CFTR was detected in coimmuneprecipitable complexes with calnexin immediately after the labeling period. Yet, even though a large pool of CFTR remained in the ER after a 45-min chase incubation, there was a dramatic reduction in the levels of calnexin:CFTR complexes. In contrast, the relative quantity of the B-form of CFTR that could be coimmuneprecipitated with Hsp70 did not change dramatically over the course of the 90-min chase period. Thus, complexes formed between Hsp70 and the B-form of CFTR were not as transient as those observed with calnexin. These data suggest that calnexin acts in a “hit-and-run” manner in which it binds CFTR to facilitate a specific step in the folding pathway, and then it releases the CFTR molecule before completion of its global folding.
Figure 1.
Calnexin–CFTR complexes are short lived in comparison with Hsp70–CFTR complexes. (A) HEK293 cells were transiently transfected with WT CFTR, starved for 20 min, labeled with [35S]methionine, and chased for the indicated amounts of time. Cells were lysed in PBS-Tr (1%) and immunoprecipitations were performed with α-CFTR, α-calnexin, or α-Hsp70 antibody as indicated. Results shown are representative of one experiment, but trends were identical when the experiment was repeated three individual times. (B) The amount of CFTR bound by either calnexin or Hsp70 was quantified by laser densitometry and normalized to the amount of CFTR B band remaining at that time point.
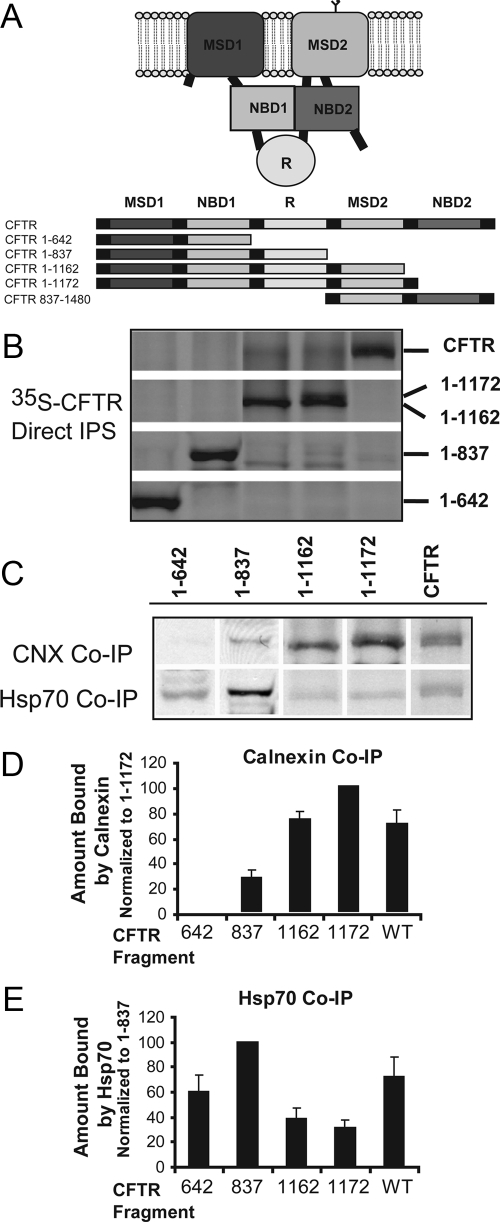
To identify the step in the CFTR folding pathway facilitated by calnexin, we determined its ability to bind different-length CFTR fragments that resemble biogenic intermediates (Figure 2A). Again for comparison, we also analyzed the binding of Hsp70 to the same CFTR fragments. Overall, this analysis performed with the CFTR fragments provides insight as to the stages of CFTR biogenesis at which chaperone action is required. However, because these fragments are overexpressed in a heterologous system, it may be that the absolute requirement for chaperones is higher than what would be seen with native protein. Direct CFTR immunoprecipitations under denaturing conditions indicate that the expression of each CFTR construct was similar, except CFTR 1-1162 accumulated to a slightly lower level (Figure 2B). Coimmunoprecipitations with chaperones under native buffer conditions were carried out from the same cell lysates as the direct immunoprecipitations. The quantity of CFTR fragment coimmunoprecipitated with the indicated chaperone was normalized to the total quantity of the respective fragment and expressed as a percentage in relation to the fragment with the highest complex formation (Figure 2, D and E). Calnexin is only detected in complex with CFTR after MSD2 has been translated, as would be expected from the glycosylation site found in MSD2. Calnexin displayed the highest affinity for CFTR 1-1172, and upon synthesis of NBD2 the ability to isolate CFTR:calnexin complexes was strongly reduced. On three repeats of the coimmunoprecipitation experiments, the percentage of increase in calnexin binding from the 837 fragment to the 1172 fragment averaged at 73% (p < 0.005), and the decrease in calnexin binding from the 1172 fragment to full-length CFTR averaged at 30% (p = 0.06). These data suggest that the point at which calnexin is required to act is after translation of the MSD2 but precedes the translation of the NBD2 domain. In addition, the presence of NBD2 seems to enhance the ability of CFTR translation intermediates to progress past the point where calnexin binding sites are exposed.
Figure 2.
Calnexin preferentially binds late folding intermediates of CFTR, whereas Hsp70 binds both early and late folding intermediates. (A) Domain structures of CFTR constructs used for coimmunoprecipitations in this figure and in later figures. Individual domains of CFTR are color-coded to indicate which domains are found in each construct. (B and C) HEK293 cells were transfected with the indicated CFTR plasmids, labeled with [35S]methionine and lysed in PBS-Tr (1%). Cell lysates were precleared and then split into three reactions and immunoprecipitated with either polyclonal α-CFTR N-terminal antibody (B) or α-Hsp70 or α-calnexin antibody (C) as described in Materials and Methods. Hsp70 and calnexin immunoprecipitations were then subjected to a secondary reimmunoprecipitation with α-CFTR antibody; these re-IPs are shown in (C). (D and E) To determine the relative amount of the CFTR fragment that interacts with calnexin (D) or Hsp70 (E), CFTR levels in the direct and reimmunoprecipitations were quantified by laser densitometry. The amount of CFTR fragment that was coimmunoprecipitated with the chaperone was normalized to the input levels of that specific fragment (as determined by the direct immunoprecipitation with CFTR antibody), and then all values were expressed as a percentage of that fragment showing the highest affinity for that specific chaperone. The data in B and C of this figure are from one representative experiment; however, average values from three independent experiments are shown in D and E.
In contrast, Hsp70 bound a greater percentage of CFTR 1-837, which comprises MSD1, NBD1, and the R domain, than any of the other fragments tested. It seems that the addition of the R domain greatly contributes to this affinity because a significantly lower percentage of CFTR 1-642, which lacks the R domain, was found in complex with Hsp70 (average of 40% less 1-642 than 1-837 found in complex with Hsp70; p <0.05). On addition of the MSD2 domain, CFTR 1-1162 as well as CFTR 1-1172 are able to form a structure that is not readily recognized by Hsp70 (70% decrease for 1-1172 in comparison with 1-837; p < 0.005), but Hsp70 binding increases once again after exposure of the NBD2 domain (average 41% increase for full-length CFTR binding to Hsp70 in comparison with 1-1172; p < 0.05). These data indicate that Hsp70 is able to bind each of the cytosolic domains of CFTR but that the affinity for the NBD1 and R domains is decreased after translation of MSD2. This suggests a folding pathway for CFTR in which translation of MSD2 results in a compact folded structure in which Hsp70 is no longer necessary to stabilize the exposed NBD1 and R domains. These data suggest that Hsp70 is involved in the folding of cytosolic regions of CFTR that are localized in both the N and C terminus, whereas calnexin binds MSD2 and facilitates a folding reaction that does not require NBD2. Yet, the presence of NBD2 seems to enhance the ability of CFTR to progress past the calnexin-dependent step.
Calnexin Promotes Interactions between MSD1 and MSD2 of CFTR
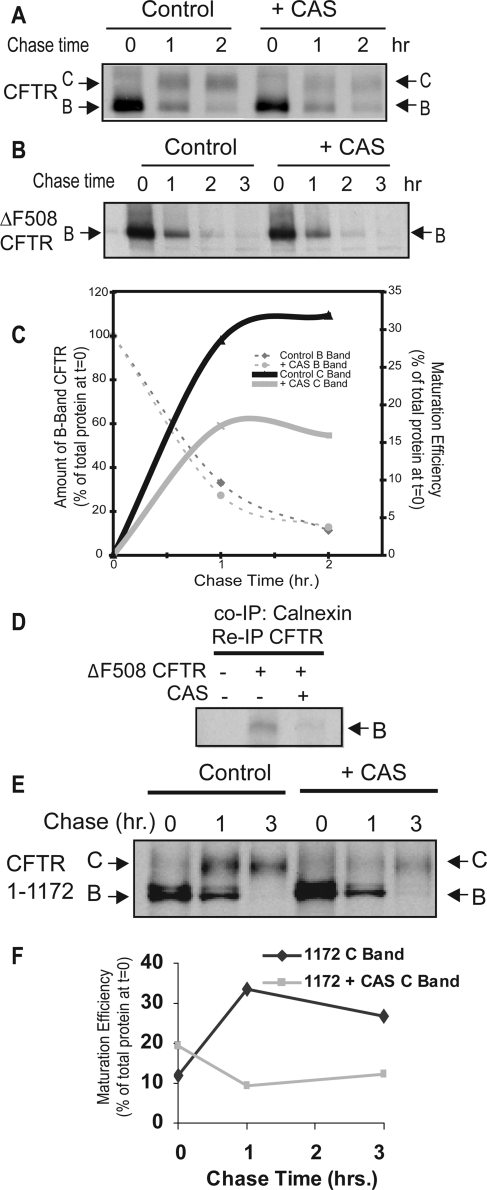
To test the concept that calnexin acts in CFTR assembly before NBD2 synthesis, we compared the effect of CAS on the biogenesis of CFTR, CFTRΔF508, and CFTR 1-1172 (Figure 3, A–F). CFTR 1-1172 lacks NBD2 but folds to a conformation that passes ERQC and functions at the cell surface as a Cl− channel (Cui et al., 2007). Treatment of cells with CAS inhibits calnexin-dependent protein folding reactions (Hammond et al., 1994; Ellgaard and Helenius, 2003) and blocks calnexin binding to CFTR (Figure 3D). We confirmed that treatment of cells with CAS decreases the folding efficiency of WT CFTR by ∼50% as indicated by the decreased maturation to C band from 32% after 2 h to 16% (Figure 3A). The inhibition in C band maturation was repeatedly observed after addition of CAS (n = 4; average 44% decrease in maturation; p < 0.05) and ranged in efficacy from 30 to 60%. CFTR folding efficiency is reduced by ∼50% in cells cultured from calnexin −/− mice, so CFTR-folding defects caused by CAS treatment of cells are nearly identical to those observed when calnexin is absent form the ER (Okiyoneda et al., 2008).
Figure 3.
Inhibition of calnexin through treatment with CAS inhibits folding of wild-type CFTR. HEK293 cells transiently transfected with pcDNA3.1-CFTR (A), pcDNA3.1-ΔF508-CFTR (B), or pcDNA3.1-CFTR 1-1172 (E) were preincubated for 1 h in the absence or presence of 5 mM CAS, and then starved, labeled with [35S]methionine, and chased for the indicated amounts of time. The cells were then lysed and incubated with α-CFTR rabbit polyclonal antibody. Results were quantified by densitometry and graphed in C and F. (D) Control samples were used to indicate the level to which CAS treatment inhibited calnexin–CFTR interactions. Samples were first subjected to a calnexin immuneprecipitation and then a secondary reimmunoprecipitation was performed with rabbit polyclonal α-CFTR. Products of immunoprecipitations were eluted in SDS sample buffer and analyzed by SDS-PAGE and autoradiography.
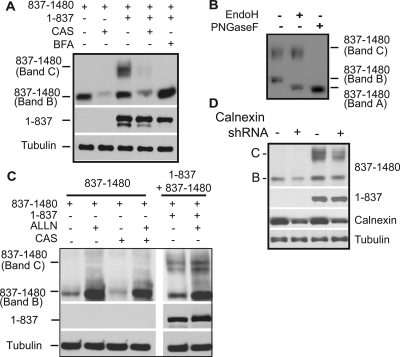
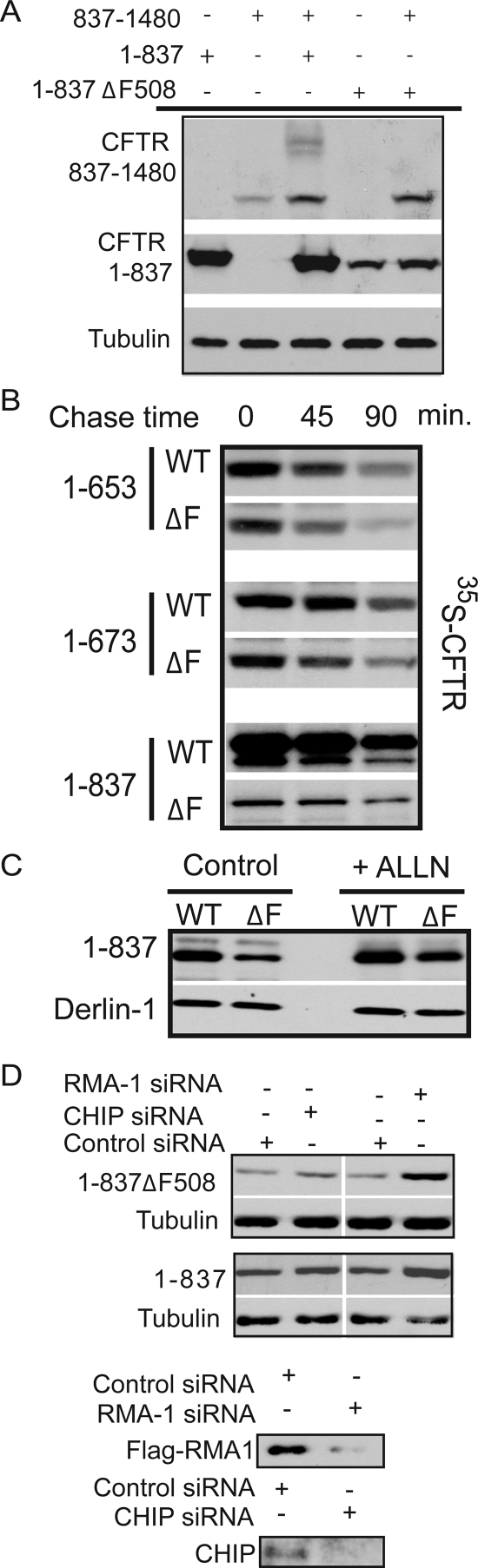
Consistent with results from studies with calnexin −/− mice (Okiyoneda et al., 2008), but in contrast to previous reports (Farinha and Amaral, 2005), CAS had little detectable effect on the degradation of WT or ΔF508 CFTR (Figure 3B). More importantly, CAS reduced the biosynthetic maturation of CFTR 1-1172 to the C-form by approximately threefold (Figure 3, E and F). Calnexin therefore seems to facilitate a folding step that involves interaction between MSD2 and amino-terminal regions of CFTR. Calnexin may facilitate CFTR folding by stabilizing MSD2 and thereby promoting formation of proper contacts between MSD1 and MSD2. To test this model, we took advantage of the observation that split CFTR fragments that individually contain the N- and C-terminal subdomains assemble into an ion channel when expressed in trans (Chan et al., 2000). N-terminal CFTR fragments containing MSD1, NBD1, and the R domain fold to a conformation that has a long half-life and accumulates at high levels when expressed alone or in trans with CFTR 837-1480 (Ostedgaard et al., 1997; Xiong et al., 1997; Meacham et al., 1999). However, when CFTR 837-1480 is expressed alone it accumulates at low levels as an immaturely glycosylated species (Figure 4A). Yet, upon coexpression with CFTR 1-837 a severalfold increase in total CFTR 837-1480 accumulation and a pool of its maturely glycosylated C-form were detected. The identity of the B-band of CFTR 837-1480 as the ER-localized immaturely glycosylated protein, and the C-Band as the post-Golgi, maturely glycosylated form was confirmed by endoglycosidase H (EndoH) and peptide N-glycosidase F (PNGaseF) digestion (Figure 4B). BFA, an inhibitor of ER-to-Golgi trafficking, also blocked the CFTR 1-837–dependent glycolytic maturation of CFTR 837-1480 and allows for better visualization of the stabilization of the 837-1480 fragment that occurs upon coexpression with CFTR 1-837 (Figure 4A). Chemical interference with calnexin binding by addition of CAS resulted in decreased levels of CFTR 837-1480 (Figure 4A). Furthermore, CAS also prevented the CFTR 1-837–dependent increase in CFTR 837-1480 levels and glycolytic maturation of CFTR 837-1480 was no longer observed (Figure 4A). In the presence of CAS, treatment with the proteasomal inhibitor ALLN restores the steady state levels of CFTR 837-1480, thereby indicating that misfolding events caused by inhibition of calnexin interactions are recognized by the ubiquitin proteasome system (Figure 4C).
Figure 4.
Calnexin interactions are required for efficient assembly of MSD1 and MSD2. (A and C) Assembly of CFTR membrane spanning domains. HEK293 cells were transfected with CFTR 1-837 or CFTR 837-1480, and cells were either treated 5 h posttransfection with 10 μg/ml BFA or 5 mM CAS, or 18 h posttransfection with 200 μM ALLN where indicated. Cells were harvested 24 h after transfection, and cell lysates subjected to Western analysis with CFTR N-terminal or NBD2 specific antibodies. Maturely glycosylated 837-1480 is indicated as C band, and immaturely glycosylated 837-1480 is indicated as B band. Tubulin is used as a loading control. (B) HEK293 cells were transfected with pcDNA CFTR 1-837 and pcDNA CFTR 837-1480, and harvested 24 h after transfection. Cells were then lysed in 1% SDS with 0.1 mM B-mercaptoethanol, sonicated, and incubated at 37°C for 10 min. Samples were then diluted fivefold into reaction buffer (5% Triton, 100 mM sodium phosphate, pH 5.5, for Endo H or 100 mM sodium phosphate, pH 7.6, for PNGaseF), the indicated enzyme (PNGaseF or EndoH) was added, and samples were incubated overnight. Sample buffer was added to a final 1× concentration, and samples were run on 10% SDS-PAGE gels. Western blots were performed with the α-NBD2 CFTR antibody. Bands B and C represent the immature and maturely glycosylated forms, respectively, and band A represents the nonglycosylated form of CFTR 837-1480. (D) HEK 293 cells were transfected with the calnexin shRNAmir construct or the nonsilencing control as indicated in Materials and Methods. Cell lysates were subjected to Western Blot analysis with the indicated antibodies.
Because CAS is a chemical inhibitor of the glucosidase I and II enzymes, we also used RNAi techniques to confirm that the above-observed effects were directly due to inhibition of calnexin binding. A vector encoding a short hairpin RNA (shRNA) against calnexin was used to decrease the endogenous calnexin levels, and then the stability of CFTR 837-1480 as well as CFTR fragment assembly was monitored by expressing CFTR 837-1480 alone or in trans with CFTR 1-837 (Figure 4D). In agreement with the CAS data, upon calnexin knockdown we observed a severalfold decrease in the stability of 837-1480, and in the 1-837–dependent formation of 837-1480 C band. The level of destabilization and inhibition of fragment assembly seemed to correlate well with the level of calnexin knockdown achieved. Overall, these data suggest that calnexin plays an important role in CFTR folding, and these studies show that one of its functions is to stabilize MSD2, which is required for proper assembly of the MSD1/MSD2 complex.
Mechanism of ΔF508-induced Misfolding of CFTR
The common ΔF508 mutation in CFTR causes defective association of MSD1 and MSD2 (Chen et al., 2004). Data obtained with CAS show that inhibition of MSD assembly leads to instability of the second half of the CFTR molecule. Thus, we investigated the possibility that the ΔF508 mutation causes similar defects in MSD2 stability. This is an important question because contact formation between F508 and intracellular loops exposed by MSD2 seems to be critical for CFTR assembly (Mendoza and Thomas, 2007; Serohijos et al., 2008). To determine whether the misfolding events caused by the deletion of F508 were similar to those caused by CAS-induced instability of CFTR, we coexpressed CFTR 1-837ΔF508 with CFTR 837-1480. Normally, coexpression of the wild-type forms of the two CFTR halves results in both the stabilization of 837-1480 as well as the maturation of CFTR 837-1480 to a maturely glycosylated C form (Figures 4A and 5A). However, expression of CFTR 1-837ΔF508 in trans with CFTR 837-1480 results in the stabilization of CFTR 837-1480, but not in glycolytic maturation (Figure 5A). These results indicate that the F508 residue is not required for CFTR 1-837 to stabilize CFTR 837-1480. Yet, F508 seems essential for downstream folding events that enable CFTR 837-1480 to fold and escape the ER.
Figure 5.
Folding defects caused by introduction of the ΔF508 mutation. (A) HEK293 cells were transfected with CFTR 1-837, CFTR 1-837ΔF508, or CFTR 837-1480 where indicated and cultured for 24 h. Steady-state levels were then determined by Western blot. (B) HEK293 cells were transfected with the indicated CFTR plasmids, and 24 h after transfection, the cells were pulse labeled with [35S]methionine, chased for the indicated periods, and lysed in PBS-Tr (1%). Cell lysates were then immunoprecipitated with a α-CFTR N-terminal antibody and visualized by SDS-PAGE analysis and autoradiography. (C) HEK293 cells were transfected with CFTR 1-837 or CFTR 1-837 ΔF508, and 18 h after transfection they were treated with 200 μM ALLN where indicated. Western blot analysis was performed with the CFTR N-terminal antibody (MM1-34). Derlin-1 blots were performed to indicate loading controls. (D) Transfections with RNAi oligos and CFTR plasmids were performed as indicated in the Materials and Methods, cells were lysed in 2× sample buffer, and equal microgram quantities of protein were run on SDS-PAGE gels. Western blot analysis was used to determine steady-state levels of indicated proteins. Tubulin blots were used as loading controls. Control experiments were performed with overexpressed FLAG-RMA1 to indicate the efficacy of the RMA1 RNAi oligos, and endogenous CHIP levels were monitored in response to RNAi addition through the use of a polyclonal CHIP antibody.
How deletion of F508 causes defects in CFTR folding is not clear and during the course of this experiment, we observed that the deletion of F508 resulted in a dramatic decrease in the steady-state levels of CFTR 1-837 (Figure 5A). Therefore, the F508 residue seems to be important for the proper folding of CFTR 1-837, and defects in this process seem to prevent proper folding of C-terminal regions in CFTR. Based on these data, we sought to pinpoint the first step at which the ΔF508 mutation exerts its effect on CFTR folding, and we analyzed the stability of wild-type and mutant CFTR fragments by pulse chase (Figure 5B). We first looked at the effect of deletion of F508 from a CFTR fragment consisting of amino acids 1–653, which stops at the NBD1 boundary before inclusion of the regulatory extension (Lewis et al., 2004; Baker et al., 2007). We found that CFTR 1-653 ΔF508 accumulated to slightly lower levels than the wild-type fragment immediately after the labeling period (i.e., there was 27% less of 1-653 ΔF508 in comparison with WT 1-653 at t = 0). Furthermore, deletion of F508 resulted in a slight increase in the rate of degradation of this fragment over the chase period such that there was 37% of total wild-type protein remaining after the 90-min chase in comparison to 19% of the 1-653 ΔF508 protein remaining. However, the defect observed with CFTR 1-653ΔF508 did not seem to match the severity of the defect observed with the full-length protein (Ward and Kopito, 1994; Ward et al., 1995; Meacham et al., 2001) or with the steady-state levels of CFTR 1-837 ΔF508 (Figure 5A). Therefore, to identify other regions of CFTR that are affected by deletion of F508, we performed pulse-chase analysis on fragments containing the NBD1 plus the regulatory extension (RE) (CFTR 1-673) as well as a fragment containing the complete R domain (1-837) (Baker et al., 2007). First of all, we observed that as we included more of the R domain, the protein became more stable over the chase period (37% of 1-653 remained after 90 min, 53% of 1-673, and 71% of 1-837). Second, the pattern observed with CFTR 1-673 was very similar to that observed with 1-653 in which deletion of F508 results in slightly decreased protein levels at t = 0 of the chase (a 24% decrease in total levels of CFTR 1-653 at t = 0), as well as a slight increase in degradation rates (53% WT 1-653 protein remaining after 90-min chase vs. 40% of mutant 1-653ΔF508 protein remaining). Yet, consistent with what we observed in CFTR fragment assembly assays, there was a drastic decrease in the accumulation of 35S-labeled CFTR 1-837ΔF508 (a 80% decrease in total levels at t = 0). The pool of 1-837ΔF508 protein that did accumulate during the labeling period also seemed to have an increased rate of degradation such that 71% of the wild-type 1-837 protein remained after the 90-min chase, in comparison with 53% of the mutant 1-837ΔF508. The simplest interpretation of these data are that the F508 deletion causes a folding defect that only modestly enhances that ability of ERQC factors to select CFTR fragments that contain MSD1 and NBD1 for degradation. Yet, the F508 deletion dramatically disturbs the folding of MSD1, NBD1, and the R-domain into a stable complex, which causes a large pool of CFTRΔF508 to be selected for proteasomal degradation.
To confirm that the dramatically reduced levels of the CFTR1-837 fragment observed upon deletion of F508 are resultant from its premature proteasomal degradation, we first examined the steady-state levels of wild type and mutant ΔF508 forms of this protein in the absence and presence of the proteasome inhibitor ALLN (Figure 5C). In the presence of ALLN, steady-state levels of CFTR 1-837ΔF508 were increased by 50%. Second, small interfering RNA knockdown of the E3 ubiquitin ligase RMA1, which is proposed to detect defects in CFTR folding cotransationally (Younger et al., 2006), led to an average 135% increase in accumulation of CFTR 1-837ΔF508 in comparison with an average 33% increase for the wild-type 1-837 protein (values are averages from 3 trials). The drastic increase seen with 1-837ΔF508 in comparison with 1-837 was statistically significant, with a p value <0.05, and it is in agreement with data previously published showing that CFTR was more sensitive to RMA1 after deletion of the F508 residue (Younger et al., 2006). In contrast, knockdown of the E3 ubiquitin ligase CHIP, which seems to primarily recognize later folding defects (Younger et al., 2006), only had minimal effects on the accumulation of WT 1-837 or 1-837 ΔF508 (14 and 11% average increases, respectively). Thus, a CFTRΔF508 folding defect that occurs before calnexin action is related to misassembly of N-terminal regions of CFTR, and these defects are primarily detected by the RMA1 E3 ubiquitin ligase complex.
Global Misfolding of CFTR as Assayed by Limited Proteolysis
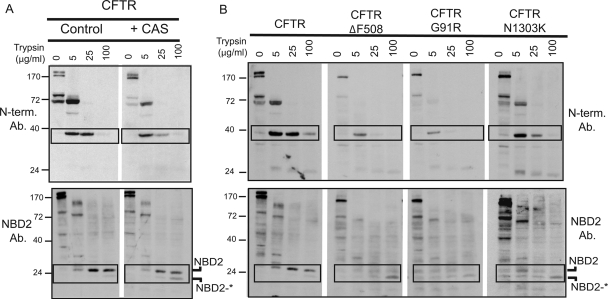
Next, limited proteolysis was utilized to probe the global structure of CFTR when calnexin-dependent folding steps were inhibited (Zhang et al., 1998; Cui et al., 2007). Then, we compared the conformation of CFTR whose folding was arrested by loss of calnexin binding with the conformation of disease-related CFTR mutant proteins that contain point mutations in different subdomains. Proteolytic fragments of CFTR generated by adding increasing concentrations of trypsin to detergent-solublized cell extracts were detected by Western blot with an antibody directed against the N-terminal tail or NBD2 (Figure 6). The N-terminal tail antibody was able to detect two major trypsin cleavage products of CFTR. The first major cleavage product is a band of ∼72 kDa, which runs just below the 1-837 fragment composed of the MSD1, NBD1, and R domain. The next cleavage product is a band of ∼40 kDa, which corresponds to a cleavage immediately preceding NBD1. Inhibition of calnexin binding to CFTR did not significantly affect the cleavage pattern detected by the N-terminal antibody, but there was a noticeable increase in the sensitivity of both the 72- and 40-kDa fragments to digestion. For example, in wild type CFTR, the 40-kDa fragment is stable when trypsin concentrations are increased from 5 to 25 μg/ml (see boxed fragments in Figure 6A, top). However, upon treatment with CAS, this fragment increases in sensitivity to trypsin. These data are consistent with the notion that calnexin-dependent formation of interdomain contacts between helices in MSD1 and MSD2 is required for proper folding of both membrane-inserted and cytosolic domains of full-length CFTR.
Figure 6.
Limited Proteolysis of WT CFTR and CFTR mutants. (A and B) HEK293 cells were transiently transfected with the indicated plasmids, and 5 h after transfection, they were treated with 5 mM CAS where indicated. Twenty-four hours after transfection cells were harvested and lysed in PBS-Tr (0.1%). Equal microgram quantities of cleared cell lysates were exposed to the indicated concentrations of trypsin for 15 min on ice. Cell lysates were then run on 12.5% gels and subjected to Western Blot analysis with the indicated antibody (either α-N-terminal CFTR or α-NBD2 CFTR).
The NBD2 antibody was able to detect three major cleavage products in WT CFTR; an ∼80-kDa band, a 35-kDa band, and a 26-kDa band. Based on molecular weight, these bands likely correspond to cleavage events toward the end of the R domain, near the end of the MSD2 domain, and right at the beginning of the NBD2 domain, respectively. We noticed that the band corresponding to the NBD2 domain existed as a doublet separated by ∼3 kDa (indicated as NBD2 and NBD2*), with the top band being the dominant one for WT CFTR. However, upon addition of CAS, we noticed a shift in the ratio of these bands, such that NBD2* increased in prominence (see boxed fragments in Figure 6A, bottom). These data suggest that improper MSD assembly leads NBD2 folding to become arrested. In the absence of proper calnexin function, it seems that NBD2 still collapses to a protease-resistant state, but it fails to bury a small loop that can now be cleaved by trypsin and this gives rise to NBD2*.
We next compared the trypsin proteolysis patterns of misfolded CFTR from CAS-treated cells with the patterns observed from CFTRΔF508, CFTRG91R, and CFTR N1303K, which contain mutations localized to the NBD1, MSD1, and NBD2 domains, respectively (Osborne et al., 1992; Xiong et al., 1997). Similar to what we observed upon CAS treatment, the sizes of the N-terminal fragments produced by trypsin digestion of the CFTRΔF508 and CFTRG91R mutants did not change significantly, but a significant increase in the sensitivity of the 40-kDa fragment to digestion by 25 μg/ml trypsin was observed (Figure 6B). In contrast, mutation of N1303K in NBD2 did not drastically affect the stability of CFTR N-terminal fragments. Yet, the protease resistance of NBD2 was reduced to a greater extent with the three mutant CFTR proteins than what we observed with CAS-treated CFTR. In each mutant, there was a decrease in the protease resistance of the NBD2 band, and the smaller NBD2* band became the more prevalent band of the two (see boxed fragments in Figure 6B, bottom). This is in agreement with results published by Zhang et al. (1998) in which they demonstrate that the ΔF508 mutation results in disruption of the NBD2 domain. Yet, the fact that the G91R mutation, and to a lesser extent, inhibition of calnexin function also hinder NBD2 folding suggests that general disruption of MSD assembly prevents proper folding of NBD2. These data suggest the calnexin helps facilitate the cooperative folding of CFTR through promoting interdomain contacts that facilitate folding of both N- and C-terminal domains.
DISCUSSION
The data presented herein define the point at which calnexin acts in the CFTR folding pathway and help to delineate the mechanism of misfolding events that result in CFTR's recognition by the ERQC pathway. We have found that calnexin action is essential for both stabilizing the C-terminal half of CFTR as well as for promoting proper association between MSD1 and MSD2. Calnexin-dependent association of CFTR's membrane regions is important for proper folding of CFTR's N-terminal domains and complete collapse of the NBD2 domain. Interestingly, calnexin action juxtaposes a critical point in the CFTR folding pathway, as disease causing mutations such as ΔF508 disrupt CFTR folding in similar ways. For example, deletion of F508 has also been shown to block membrane spanning domain assembly (Chen et al., 2004) and NBD2 folding (Du et al., 2005). Furthermore, we have found that these late folding defects caused by the ΔF508 mutation are accompanied by an early folding defect that becomes evident after translation of the R domain and is sensed by the RMA1 ubiquitin ligase.
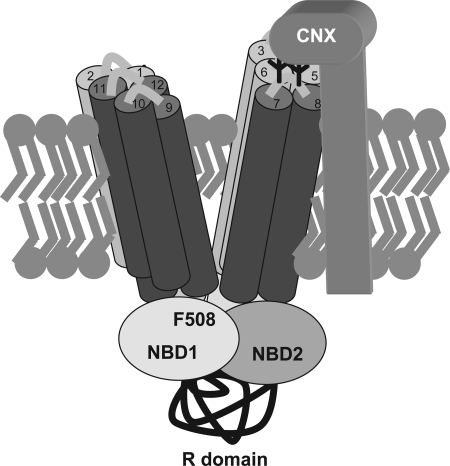
The point at which calnexin acts in the CFTR folding pathway was previously unknown, and it was not immediately obvious, considering that the lectin domain of calnexin is localized to the ER lumen, where little of the CFTR protein is found. However, based on our data we now propose a model in which the binding of calnexin to the sugars attached to extracellular loop 4 of MSD2 acts to stabilize or orient this domain in such a manner as to promote productive interactions with MSD1 (Figure 7). Structural studies of the related ABC transporter protein Sav1866 show that the transmembrane domains adopt a complex structure in which transmembrane domains from MSD1 cross-interact with transmembrane domains of MSD2 to form a two-winged pore structure (Dawson and Locher, 2006). A three-dimensional structural model of CFTR, which is based on the Sav1866 structure, predicts that transmembrane (TM) helices 1 and 2 of CFTR pack next to TM helices 9, 10, 11, and 12 to make one wing of the pore, whereas TM helices 7 and 8 pack next to TM helices 3, 4, 5, and 6 to form the other wing of the pore (Serohijos et al., 2008). Because the CFTR glycosylation sites are found in the extracellular loop connecting TM 7 and 8, we propose that calnexin binds this segment of CFTR to assemble TM 7 and 8 into the proper wing of the pore (Figure 7). The need for proper folding and assembly of the CFTR transmembrane domains is highlighted by the large number of disease-causing mutations found in TM domains. There are ∼625 missense cystic fibrosis-causing mutations identified in CFTR, with ∼300 of these localized to transmembrane domains or their connecting extracellular loops (Cheung and Deber, 2008).
Figure 7.
Model of calnexin action in CFTR folding. Calnexin is shown binding to CFTR through the glycosylation sites found in the fourth extracellular loop between TM helices 7 and 8. This binding is proposed to orient TM 7/8 into the proper wing of the CFTR two-winged pore structure. CFTR TM helices are represented by cylinders with MSD1 TM helices colored light gray and MSD2 helices colored dark gray. The number of the TM helix is indicated at the top of each cylinder.
A major defect in CFTRΔF508 folding predicted by the Sav1866 structure is defective interaction of NBD1 with a hydrophobic surface exposed on intracellular loop 4 on MSD2. Yet, how this defect leads to misfolding and premature degradation of CFTR is not clear. Data obtained with split CFTR fragments suggest that regions of MSD2 are not stably inserted into the ER membrane and that calnexin binding and association with MSD2 are required to stabilize MSD1. The F508 deletion leads a fragment of CFTR 1-1162 to be rapidly degraded and prevents the glycolytic maturation of a slightly longer fragment CFTR 1-1172 (Younger et al., 2006; Cui et al., 2007). However, the deletion of F508 from the CFTR 1-837 fragment does not affect the ability of this N-terminal fragment to stabilize a C-terminal fragment (CFTR 837-1480), Yet, it does prevent CFTR 1-837 from promoting the glycolytic maturation of CFTR 837-1480. Thus, in the context of experiments with CFTR fragment assembly, the F508 deletion seems to hinder a step in CFTR folding that occurs after the calnexin-dependent stabilization of MSD2, which might involve NBD2 (Du et al., 2005).
During the course of our study with split CFTR molecules, we also observed that compared with CFTR 1-837, the accumulation of CFTR 1-837ΔF508 was dramatically reduced. Reductions in CFTR 1-837ΔF508 seemed to result from its misfolding and selection for proteasomal degradation by the RMA1 E3 ubiquitin ligase. The F508 deletion caused a modest decrease in the stability of CFTR 1-653, an MSD1-NBD1 fragment, but the presence of the complete R-domain resulted in a much more dramatic defect. It may be that the 1-653ΔF508 fragment misfolds but is lacking a recognition motif for ER QC factors, which then prevents the severe destabilization effect observed with 1-837ΔF508. Another likely possibility is that the deletion of the F508 residue affects the interaction of the R domain with other regions of CFTR. The R domain is thought to be largely disordered (Ostedgaard et al., 2000), but it contains ordered segments of helical structure (Baker et al., 2007), and it has been shown to interact directly with both the NBD1 domain (Baker et al., 2007), and with the N-terminal tail of CFTR (Naren et al., 1999). Translation of the R-domain is also important in ensuring that N-terminal regions of CFTR achieve a compact folded structure that can no longer be recognized by the Hsp40 Hdj-2 (Meacham et al., 1999). Because the F508 residue is localized to NBD1, it is possible that the reduced stability of 1-837ΔF508 is due to a disruption of MSD1–NBD1–R domain interactions, which then results in recognition by the RMA1 E3 machinery.
The conglomeration of current data suggest a model in which interdomain contacts are essential for the folding of CFTR, and when disrupted result in specific misfolding events that are then recognized by the ERQC machinery. There seems to be an interplay between the ER lumenal chaperone system and the cytosolic chaperone system that allows for the proper folding and assembly of membrane spanning domains and cytosolic domains of CFTR. We have also found that MSD assembly is a critical aspect of CFTR's folding pathway, because it is disrupted by a variety of mechanisms, including inhibition of calnexin interactions, or introduction of disease-causing mutations such as ΔF508. Because deletion of F508 causes defects in CFTR folding that occur at early and late stages of CFTR assembly, drugs that correct CFTR folding defects might need to act at multiple steps in CFTR biogenesis.
ACKNOWLEDGMENTS
We thank Dr. K. Kirk for providing reagents. Work in the laboratory of D.M.C. is supported by the National Institutes of Health and the Cystic Fibrosis Foundation.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-04-0357) on August 20, 2008.
REFERENCES
- Baker J. M., Hudson R. P., Kanelis V., Choy W. Y., Thibodeau P. H., Thomas P. J., Forman-Kay J. D. CFTR regulatory region interacts with NBD1 predominantly via multiple transient helices. Nat. Struct. Mol. Biol. 2007;14:738–745. doi: 10.1038/nsmb1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan K. W., Csanady L., Seto-Young D., Nairn A. C., Gadsby D. C. Severed molecules functionally define the boundaries of the cystic fibrosis transmembrane conductance regulator's NH(2)-terminal nucleotide binding domain. J. Gen. Physiol. 2000;116:163–180. doi: 10.1085/jgp.116.2.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen E. Y., Bartlett M. C., Loo T. W., Clarke D. M. The DeltaF508 mutation disrupts packing of the transmembrane segments of the cystic fibrosis transmembrane conductance regulator. J. Biol. Chem. 2004;279:39620–39627. doi: 10.1074/jbc.M407887200. [DOI] [PubMed] [Google Scholar]
- Cheng S. H., Gregory R. J., Marshall J., Paul S., Souza D. W., White G. A., O'Riordan C. R., Smith A. E. Defective intracellular transport and processing of CFTR is the molecular basis of most cystic fibrosis. Cell. 1990;63:827–834. doi: 10.1016/0092-8674(90)90148-8. [DOI] [PubMed] [Google Scholar]
- Cheung J. C., Deber C. M. Misfolding of the cystic fibrosis transmembrane conductance regulator and disease. Biochemistry. 2008;47:1465–1473. doi: 10.1021/bi702209s. [DOI] [PubMed] [Google Scholar]
- Cui L., Aleksandrov L., Chang X. B., Hou Y. X., He L., Hegedus T., Gentzsch M., Aleksandrov A., Balch W. E., Riordan J. R. Domain interdependence in the biosynthetic assembly of CFTR. J. Mol. Biol. 2007;365:981–994. doi: 10.1016/j.jmb.2006.10.086. [DOI] [PubMed] [Google Scholar]
- Dawson R. J., Locher K. P. Structure of a bacterial multidrug ABC transporter. Nature. 2006;443:180–185. doi: 10.1038/nature05155. [DOI] [PubMed] [Google Scholar]
- Du K., Sharma M., Lukacs G. L. The DeltaF508 cystic fibrosis mutation impairs domain-domain interactions and arrests post-translational folding of CFTR. Nat. Struct. Mol. Biol. 2005;12:17–25. doi: 10.1038/nsmb882. [DOI] [PubMed] [Google Scholar]
- Ellgaard L., Helenius A. Quality control in the endoplasmic reticulum. Nat. Rev. Mol. Cell Biol. 2003;4:181–191. doi: 10.1038/nrm1052. [DOI] [PubMed] [Google Scholar]
- Farinha C. M., Amaral M. D. Most F508del-CFTR is targeted to degradation at an early folding checkpoint and independently of calnexin. Mol. Cell. Biol. 2005;25:5242–5252. doi: 10.1128/MCB.25.12.5242-5252.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond C., Braakman I., Helenius A. Role of N-linked oligosaccharide recognition, glucose trimming, and calnexin in glycoprotein folding and quality control. Proc. Natl. Acad. Sci. USA. 1994;91:913–917. doi: 10.1073/pnas.91.3.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyde S. C., Emsley P., Hartshorn M. J., Mimmack M. M., Gileadi U., Pearce S. R., Gallagher M. P., Gill D. R., Hubbard R. E., Higgins C. F. Structural model of ATP-binding proteins associated with cystic fibrosis, multidrug resistance and bacterial transport. Nature. 1990;346:362–365. doi: 10.1038/346362a0. [DOI] [PubMed] [Google Scholar]
- Kopito R. R. Biosynthesis and degradation of CFTR. Physiol. Rev. 1999;79:S167–173. doi: 10.1152/physrev.1999.79.1.S167. [DOI] [PubMed] [Google Scholar]
- Lewis H. A., et al. Structure of nucleotide-binding domain 1 of the cystic fibrosis transmembrane conductance regulator. EMBO J. 2004;23:282–293. doi: 10.1038/sj.emboj.7600040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo M. A., Jensen T. J., Cui L., Hou Y., Chang X. B., Riordan J. R. Perturbation of Hsp90 interaction with nascent CFTR prevents its maturation and accelerates its degradation by the proteasome. EMBO J. 1998;17:6879–6887. doi: 10.1093/emboj/17.23.6879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meacham G. C., Lu Z., King S., Sorscher E., Tousson A., Cyr D. M. The Hdj-2/Hsc70 chaperone pair facilitates early steps in CFTR biogenesis. EMBO J. 1999;18:1492–1505. doi: 10.1093/emboj/18.6.1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meacham G. C., Patterson C., Zhang W., Younger J. M., Cyr D. M. The Hsc70 co-chaperone CHIP targets immature CFTR for proteasomal degradation. Nat. Cell Biol. 2001;3:100–105. doi: 10.1038/35050509. [DOI] [PubMed] [Google Scholar]
- Mendoza J. L., Thomas P. J. Building an understanding of cystic fibrosis on the foundation of ABC transporter structures. J. Bioenerg. Biomembr. 2007;39:499–505. doi: 10.1007/s10863-007-9117-7. [DOI] [PubMed] [Google Scholar]
- Naren A. P., Cormet-Boyaka E., Fu J., Villain M., Blalock J. E., Quick M. W., Kirk K. L. CFTR chloride channel regulation by an interdomain interaction. Science. 1999;286:544–548. doi: 10.1126/science.286.5439.544. [DOI] [PubMed] [Google Scholar]
- Okiyoneda T., Harada K., Takeya M., Yamahira K., Wada I., Shuto T., Suico M. A., Hashimoto Y., Kai H. Delta F508 CFTR pool in the endoplasmic reticulum is increased by calnexin overexpression. Mol. Biol. Cell. 2004;15:563–574. doi: 10.1091/mbc.E03-06-0379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okiyoneda T., Niibori A., Harada K., Kohno T., Michalak M., Duszyk M., Wada I., Ikawa M., Shuto T., Suico M. A., Kai H. Role of calnexin in the ER quality control and productive folding of CFTR; differential effect of calnexin knockout on wild-type and DeltaF508 CFTR. Biochim. Biophys. Acta. 2008;1783:1585–1594. doi: 10.1016/j.bbamcr.2008.04.002. [DOI] [PubMed] [Google Scholar]
- Osborne, et al. Incidence and expression of the N1303K mutation of the cystic fibrosis (CFTR) gene. Hum. Genet. 1992;89:653–658. doi: 10.1007/BF00221957. [DOI] [PubMed] [Google Scholar]
- Ostedgaard L. S., Baldursson O., Vermeer D. W., Welsh M. J., Robertson A. D. A functional R domain from cystic fibrosis transmembrane conductance regulator is predominantly unstructured in solution. Proc. Natl. Acad. Sci. USA. 2000;97:5657–5662. doi: 10.1073/pnas.100588797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostedgaard L. S., Rich D. P., DeBerg L. G., Welsh M. J. Association of domains within the cystic fibrosis transmembrane conductance regulator. Biochemistry. 1997;36:1287–1294. doi: 10.1021/bi962174s. [DOI] [PubMed] [Google Scholar]
- Pind S., Riordan J. R., Williams D. B. Participation of the endoplasmic reticulum chaperone calnexin (p88, IP90) in the biogenesis of the cystic fibrosis transmembrane conductance regulator. J. Biol. Chem. 1994;269:12784–12788. [PubMed] [Google Scholar]
- Riordan J. R. Assembly of functional CFTR chloride channels. Annu. Rev. Physiol. 2005;67:701–718. doi: 10.1146/annurev.physiol.67.032003.154107. [DOI] [PubMed] [Google Scholar]
- Riordan J. R., et al. Identification of the cystic fibrosis gene: cloning and characterization of complementary DNA. Science. 1989;245:1066–1073. doi: 10.1126/science.2475911. [DOI] [PubMed] [Google Scholar]
- Serohijos A. W., Hegedus T., Aleksandrov A. A., He L., Cui L., Dokholyan N. V., Riordan J. R. Phenylalanine-508 mediates a cytoplasmic-membrane domain contact in the CFTR 3D structure crucial to assembly and channel function. Proc. Natl. Acad. Sci. USA. 2008;105:3256–3261. doi: 10.1073/pnas.0800254105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skach W. R. Defects in processing and trafficking of the cystic fibrosis transmembrane conductance regulator. Kidney Int. 2000;57:825–831. doi: 10.1046/j.1523-1755.2000.00921.x. [DOI] [PubMed] [Google Scholar]
- Thibodeau P. H., Brautigam C. A., Machius M., Thomas P. J. Side chain and backbone contributions of Phe508 to CFTR folding. Nat. Struct. Mol. Biol. 2005;12:10–16. doi: 10.1038/nsmb881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Matteson J., An Y., Moyer B., Yoo J. S., Bannykh S., Wilson I. A., Riordan J. R., Balch W. E. COPII-dependent export of cystic fibrosis transmembrane conductance regulator from the ER uses a di-acidic exit code. J. Cell Biol. 2004;167:65–74. doi: 10.1083/jcb.200401035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., et al. Hsp90 cochaperone Aha1 downregulation rescues misfolding of CFTR in cystic fibrosis. Cell. 2006;127:803–815. doi: 10.1016/j.cell.2006.09.043. [DOI] [PubMed] [Google Scholar]
- Ward C. L., Kopito R. R. Intracellular turnover of cystic fibrosis transmembrane conductance regulator. Inefficient processing and rapid degradation of wild-type and mutant proteins. J. Biol. Chem. 1994;269:25710–25718. [PubMed] [Google Scholar]
- Ward C. L., Omura S., Kopito R. R. Degradation of CFTR by the ubiquitin-proteasome pathway. Cell. 1995;83:121–127. doi: 10.1016/0092-8674(95)90240-6. [DOI] [PubMed] [Google Scholar]
- Welsh M. J., Smith A. E. Molecular mechanisms of CFTR chloride channel dysfunction in cystic fibrosis. Cell. 1993;73:1251–1254. doi: 10.1016/0092-8674(93)90353-r. [DOI] [PubMed] [Google Scholar]
- Xiong X., Bragin A., Widdicombe J. H., Cohn J., Skach W. R. Structural cues involved in endoplasmic reticulum degradation of G85E and G91R mutant cystic fibrosis transmembrane conductance regulator. J. Clin. Invest. 1997;100:1079–1088. doi: 10.1172/JCI119618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Younger J. M., Chen L., Ren H. Y., Rosser M. F., Turnbull E. L., Fan C. Y., Patterson C., Cyr D. M. Sequential quality-control checkpoints triage misfolded cystic fibrosis transmembrane conductance regulator. Cell. 2006;126:571–582. doi: 10.1016/j.cell.2006.06.041. [DOI] [PubMed] [Google Scholar]
- Zhang F., Kartner N., Lukacs G. L. Limited proteolysis as a probe for arrested conformational maturation of delta F508 CFTR. Nat. Struct. Biol. 1998;5:180–183. doi: 10.1038/nsb0398-180. [DOI] [PubMed] [Google Scholar]