Abstract
In the process of autophagy, a ubiquitin-like molecule, LC3/Atg8, is conjugated to phosphatidylethanolamine (PE) and associates with forming autophagosomes. In mammalian cells, the existence of multiple Atg8 homologues (referred to as LC3 paralogues) has hampered genetic analysis of the lipidation of LC3 paralogues. Here, we show that overexpression of an inactive mutant of Atg4B, a protease that processes pro-LC3 paralogues, inhibits autophagic degradation and lipidation of LC3 paralogues. Inhibition was caused by sequestration of free LC3 paralogues in stable complexes with the Atg4B mutant. In mutant overexpressing cells, Atg5- and ULK1-positive intermediate autophagic structures accumulated. The length of these membrane structures was comparable to that in control cells; however, a significant number were not closed. These results show that the lipidation of LC3 paralogues is involved in the completion of autophagosome formation in mammalian cells. This study also provides a powerful tool for a wide variety of studies of autophagy in the future.
INTRODUCTION
Macroautophagy, referred to here as autophagy, is an intracellular process in which cytosol and organelles are sequestered within double-membrane–bound structures, called autophagosomes, which deliver their contents to the lysosome/vacuole for degradation. In addition to its well understood physiological role in recycling intracellular materials as a starvation response, there is growing evidence for the participation of autophagy in other cellular processes including cellular differentiation, tissue remodeling, growth control, adaptation to adverse environments, and cellular immunity (Cuervo, 2004; Levine and Klionsky, 2004; Mizushima, 2007; Yoshimori, 2004). The mode of autophagosome formation stands apart from vesicle formation in other membrane trafficking processes, such as endocytosis and the secretory pathway (Noda et al., 2002). In autophagy, a flattened membrane sac, the so-called isolation membrane in mammals, is generated de novo, elongates, and encloses a cargo to form the autophagosome.
The LC3 (mammalian Atg8 homologue) protein is a ubiquitin-like molecule involved in autophagy. After synthesis, the C-terminal 22 residues of precursor LC3 are immediately removed by a protease, Atg4, to produce the LC3-I form. The C-terminal carboxyl base of LC3-I/Atg8 is conjugated to the head group amine of phosphatidylethanolamine (PE) through an amide bond by a sequence of ubiquitination-like reactions that involves an E1 (Atg7), an E2 (Atg3), and an E3 (protein complex including Atg5, Atg12, and Atg16L; Ichimura et al., 2000; Hanada et al., 2007; Fujita et al., 2008). The lipidated form of LC3 (LC3-II) and Atg8-PE are associated with the autophagosomal membrane (Kabeya et al., 2000; Kirisako et al., 2000). The LC3 lipidation process is reversible, because the Atg4 proteases can also catalyze the reverse modification reaction, termed delipidation, of LC3/Atg8 (Kabeya et al., 2004; Kirisako et al., 2000). In fact, most of the LC3/Atg8 is liberated from the membrane at, or before, the final stage of autophagy: fusion between autophagosomes and lysosomes (Kimura et al., 2007; Kirisako et al., 1999).
In yeast, Atg8 is proposed to function in expansion of the autophagosomal membrane (Nakatogawa et al., 2007; Xie et al., 2008). Atg8-PE causes the hemifusion of vesicles in vitro, and this property may be related to the membrane expansion step of autophagosome formation (Nakatogawa et al., 2007). In mammals, the existence of multiple Atg8 homologues (referred to as LC3 paralogues), including LC3, LC3A, LC3B, GABARAP, GATE16, and Atg8L, has been an impediment to genetic analysis of the lipidation of LC3 paralogues (Tanida et al., 2006; Wu et al., 2006).
In mammalian cells, four Atg4 homologues have been reported: Atg4A/autophagin-2, Atg4B/autophagin-1, Atg4C/autophagin-3, and autophagin-4 (Marino et al., 2003). Among these, Atg4B has a broad specificity for LC3 paralogues (Hemelaar et al., 2003; Kabeya et al., 2004; Tanida et al., 2004). Human Atg4B is a cysteine protease whose active catalytic triad consists of Cys74, His280, and Asp278 (Sugawara et al., 2005; Kumanomidou et al., 2006).
Here, we found that overexpression of a protease activity-deficient mutant of Atg4B strongly inhibits autophagosome formation. Through a mechanistic analysis, we show that excess inactive Atg4B blocks lipidation of LC3 paralogues, resulting in inhibition of autophagy. We believe this study not only demonstrates the role of the LC3 paralogues in autophagy, but also provides a powerful tool for inhibiting autophagy than will be useful in a wide variety of future studies.
MATERIALS AND METHODS
Reagents and Antibodies
Cell culture reagents were purchased from Invitrogen (Carlsbad, CA). The following antibodies were used: rabbit polyclonal anti-rat LC3 (Kabeya et al., 2000); anti-human Atg5 (Mizushima et al., 2001); anti-mouse Atg16L (Mizushima et al., 2003); anti-p62 (BIOMOL Research Laboratories, Plymouth Meeting, PA); anti-GABARAP (MBL, Nagoya, Japan); anti-GATE16 (MBL); anti-monomeric red fluorescent protein, which reacts with mStrawberry (MBL); mouse monoclonal anti-GFP (clone 7.1 and 13.1; Roche, Indianapolis, IN); anti-c-myc (clone 9E10; Gentaur Molecular Products, Kobe, Japan); anti-α-tubulin (clone B5-1-2; Sigma, St. Louis, MO). Wortmannin (Calbiochem, La Jolla, CA) was prepared as a 100 μM stock in Me2SO. All other reagents were purchased from Sigma-Aldrich.
DNA Engineering, Recombinant Adenoviruses, and Recombinant Retroviruses
The plasmid encoding monomeric red fluorescent protein (mStrawberry) was a generous gift from Dr. Roger Y. Tsien (University of California, San Diego, CA; Shaner et al., 2004). Expression vectors for green fluorescent protein (GFP)-LC3, Myc-LC3-HA (hemagglutinin), Myc-LC3G120A-HA, and mStrawberry have previously been described (Kabeya et al., 2000; Mizushima et al., 2001; Kimura et al., 2007). To construct the mStrawberry-Atg4BC74A plasmid, the Atg4B cDNA was cloned from genomic DNA isolated from mouse embryonic fibroblast (MEF) cells and was inserted into pmStrawberry-C1 using engineered BamHI and KpnI sites; the point mutation (C74A or C74S) was introduced using the QuikChange Site-Directed mutagenesis system (Stratagene, La Jolla, CA). To construct the anti-human LC3 shRNA-plasmid, two oligonucleotides, 5′-GATCCGCTGAGATCGATCAGTTCATTTCAAGAGAATGAACTGATCGATCTCAGTTTTTTGGAAA-3′ and 5′-AGCTT TTCCAAAAAACTGAGATCGATCAGTTCATTCTCTTGAAATGAACTGATCG ATCTCAGCG-3′ were synthesized and annealed, and the double-stranded fragment was subcloned into the pRNA-H1/neo vector (GenScript, Piscataway, NJ) at the BamHI/HindIII sites. To produce recombinant adenoviruses, the cDNAs corresponding to mStrawberry, mStrawberry-tagged-Atg4BWT, -Atg4BC74A, or -Atg4BC74S were subcloned into the pENTR 1A plasmid (Invitrogen). The cDNA inserts in pENTR-1A were transferred to the pAd/CMV/V5-DEST vector (Invitrogen) by means of the Gateway system using LR clonase (Invitrogen). Recombinant adenoviruses were prepared with the ViraPower Adenovirus Expression System (Invitrogen) according to the manufacturer's instructions. pMRX-IRES-puro and pMRX-IRES-bsr were donated by Dr. S. Yamaoka (Tokyo Medical and Dental University, Japan; Saitoh et al., 2003). For production of recombinant retroviruses, the cDNAs corresponding to enhanced green fluorescent protein (EGFP)-LC3, EGFP-Atg5, or mStrawberry-Atg4BC74A were transferred to pMRX-IRES-puro or pMRX-IRES-bsr. Recombinant retroviruses were prepared as described previously (Saitoh et al., 2003).
Cell Culture, Plasmid Transfections, and Adenovirus Infections
Plat-E cells were generously provided by Dr. T. Kitamura (The University of Tokyo; Morita et al., 2000). MCF7, 293A, NIH3T3, and Plat-E cells were grown in DMEM supplemented with 10% fetal bovine serum, 2 mM l-glutamine, and appropriate antibiotics in a 5% CO2 incubator at 37°C. For nutrient-starvation, cells were cultured in Hanks' balanced salt solution (HBSS; Invitrogen) for 1 or 2 h. Transient transfections were carried out using LipofectAMINE 2000 reagent (Invitrogen) according to the manufacturer's protocol. Stable transformants were selected in growth medium with 500 μg/ml G418, 1 μg/ml puromycin, or 10 μg/ml blastcidin. Adenovirus infections were carried out as follows: on the day before infection, ∼2 × 105 cells were plated into six-well plates and incubated at 37°C overnight in a CO2 incubator. The medium was replaced with 1.5 ml of culture medium that contained recombinant adenoviruses. After 16-h of incubation, the medium containing adenoviruses was replaced with 1.5 ml culture medium. After an additional 24-h incubation, the cells were used for experiments.
Western Blotting
Cells were rinsed with ice-cold PBS, scraped, and collected by centrifugation at 4°C. Cells were lysed in PBS containing 2% Triton X-100, 1 mM phenylmethylsulfonyl fluoride, and Protease inhibitor cocktail (Roche; Sou et al., 2006). Cell lysates were centrifuged at 15,000 × g for 15 min at 4°C, and supernatants were collected. Samples were separated by SDS-PAGE and transferred to polyvinylidene difluoride membrane. The membranes were blocked with 1% skim milk in 0.1% Tween 20/TBS and incubated with primary antibodies. Immunoreactive bands were detected using horseradish peroxidase–conjugated secondary antibodies (The Jackson Laboratory, Bar Harbor, ME) and luminol solution (1.25 mM luminol, 65 mM Tris-HCl, pH 8.0, 0.2 mM coumaric acid, and 0.01% H2O2).
Fluorescence Microscopy
Cells cultured on coverslips were fixed with 4% paraformaldehyde in PBS. Samples were examined under a fluorescence laser scanning confocal microscope, FV1000 (Olympus, Tokyo, Japan) or Olympus IX81 microscope equipped with a mercury lamp and cooled charge-coupled device camera (Cool Snap HQ; Roper Scientific, Tucson, AZ), under control of SlideBook software (Intelligent Imaging Innovations, Denver, CO).
Gel Filtration
Gel filtration analysis was performed as previously described (Mizushima et al., 2003). Briefly, 293A cells were homogenized in homogenization buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, and Protease inhibitor cocktail; Roche) by repeated passage (∼15 times) through a 1-ml syringe with a 23-gauge needle. The homogenate was centrifuged at 10,000 × g for 10 min, and the supernatant was further centrifuged at 100,000 × g for 60 min. The resulting supernatants (cytosol fraction) were separated by size exclusion chromatography on a Superose 6 column (GE Healthcare, Waukesha, WI).
Bulk Protein Degradation Assay
Cells were seeded in 24-well dishes and incubated overnight. On the following day, the cells were exchanged into labeling medium containing 14C-valine (1.5 μCi/ml) and incubated overnight. Cells were exchanged into chase medium (DMEM supplemented with 10% FBS and 10 mM unlabeled valine) and further incubated for 4 h to remove the contribution of short-lived proteins. After the chase period, cells were exchanged into growth medium containing 10 mM valine or HBSS containing 10 mM valine to induce autophagy. After a 2-h incubation, the media were collected and the trichloroacetic acid (TCA)-soluble fraction was analyzed by scintillation counting. The cells were lysed in ice-cold RIPA buffer (25 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.1% SDS, 1% Triton X-100, 1% deoxycholate, 5 mM EDTA, and Protease inhibitor cocktail; (Roche) and the TCA-insoluble fraction was isolated and analyzed by scintillation counting. To determine the rate of long-lived protein degradation, the count in the TCA-soluble fraction in the medium was divided by the equivalent TCA-insoluble count in the cell.
Electron Microscopy
Conventional electron microscopy was performed as previously described (Yoshimori et al., 2000) except that NIH 3T3 cells were postfixed with 1% OsO4 in 1% K4Fe(CN)6 and 0.1 M phosphate buffer, pH 7.4, for 1 h. Immunoelectron microscopy using the gold enhancement method was also performed, as described previously (Luo et al., 2006).
Statistics
All values shown in figures are represented with SD. Statistical significance (p value) is determined by Student's t test.
RESULTS
Atg4B Overexpression Inhibits Autophagic Flux Independent of Its Catalytic Activity
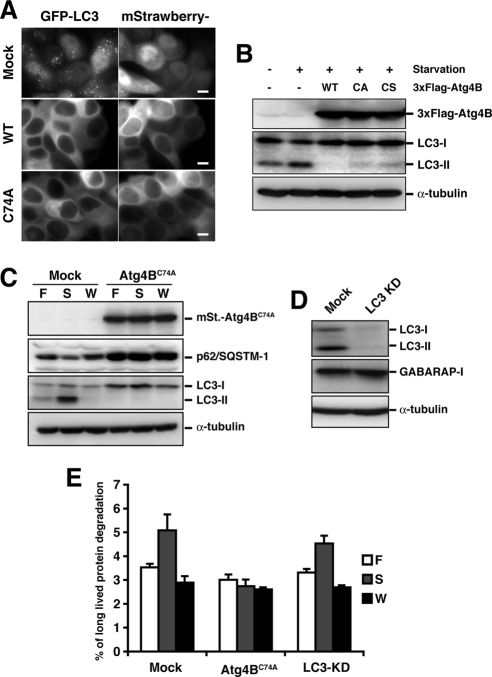
It has been reported that overexpression of Atg4B, a protease that processes LC3 paralogues, negatively affects the membrane localization and PE conjugation of LC3 (Tanida et al., 2004). In our experimental system, GFP-LC3 puncta were not observed after overexpression of Atg4B (Figure 1A). One possible mechanism underlying this negative effect is that excess Atg4B efficiently deconjugates the LC3 from PE and decreases membrane localized LC3. To test this hypothesis, inactive Atg4B mutants (Atg4BC74A or Atg4BC74S) that lack protease activity due to mutation of the catalytic cysteine residue (Cys74) were overexpressed. Unexpectedly, both mutants showed similar effects as wild-type Atg4B on membrane targeting and PE conjugation of LC3 (Figure 1, A and B), indicating that the negative effect is independent of the deconjugation activity.
Figure 1.
Cells expressing Atg4BC74A exhibit a block in autophagic degradation. (A) MCF7 cells stably expressing GFP-LC3 were infected with adenovirus bearing mStrawberry (Mock), mStrawberry-Atg4BWT (WT), or mStrawberry-Atg4BC74A (C74A) and incubated for 40 h. The cells were then cultured in HBSS for 2 h, fixed, and observed using fluorescence microscopy. Bar, 10 μm. (B) PC12 cells were infected with adenovirus bearing GFP (−), 3xFlag-tagged wild-type Atg4B (WT), Atg4BC74A (CA), or Atg4BC74S (CS). Cells were cultured either in growth or starvation medium, and lysates were examined by Western blotting using each antibody. Top panel, anti-Flag; middle panel, anti-LC3; bottom panel, anti-α-tubulin. (C) 293A cells stably expressing empty vector (Mock) or mStrawberry-Atg4BC74A (Atg4BC74A) were grown in growth medium (F), HBSS (S), or HBSS with 100 nM wortmannin (W) for 2 h. Cell lysates were examined by Western blotting using each antibody. From top panel, anti-RFP, anti-p62, anti-LC3, and anti-α-tubulin. (D) 293A cells stably expressing empty vector (Mock) or shRNA against LC3 (LC3 KD) were cultured in HBSS for 2 h and collected. Cell lysates were examined by Western blotting using each antibody. From top panel, anti-LC3, anti-GABARAP, and anti-α-tubulin. (E) Mock, mStrawberry-Atg4BC74A-expressing (Atg4BC74A), or LC3-knockdown (LC3-KD) 293A cells were grown in growth medium (F), HBSS (S), or HBSS with 100 nM wortmannin (W) for 2 h. Long-lived protein degradation was scored as described in Materials and Methods.
Next, we examined the effect of Atg4BC74A overexpression on autophagic flux in 293A cells. As it has been reported that p62/SQSTM1 is a selective substrate of autophagy (Bjorkoy et al., 2005; Mizushima and Yoshimori, 2007), we investigated autophagic degradation by monitoring endogenous p62 protein levels. At steady state, the p62 level in Atg4BC74A overexpressing cells was much higher than in mock cells, suggesting a defect in constitutive autophagic clearance (Figure 1C). In mock cells, nutrient starvation, which induces autophagy, resulted in a slight decrease of p62 protein level; this was blocked by 100 nM wortmannin, an inhibitor of phosphatidylinositol 3-kinase that blocks autophagic activity (Blommaart et al., 1997). A similar reduction was not observed in Atg4BC74A-overexpressing cells (Figure 1C). We then monitored long-lived protein degradation upon nutrient starvation in mock cells, cells stably expressing mStrawberry-Atg4BC74A, and cells stably expressing shRNA against LC3 (Figure 1D). In mock and LC3-knockdown cells, long-lived protein degradation was elevated by nutrient starvation, and the elevation was blocked by wortmannin (Figure 1E). This suggests that LC3 deficiency has little effect on autophagy flux. It is possible that this is due to the presence of LC3 paralogues, such as GABARAP, that may have redundant function. On the other hand, in cells stably expressing mStrawberry-Atg4BC74A, the degradation of long-lived proteins was significantly inhibited (Figure 1E), indicating a defect in autophagy.
Overexpression of Atg4BC74A Inhibits PE Conjugation by Sequestering LC3
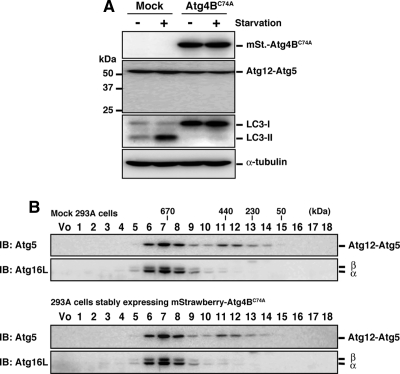
We next assessed which step in the PE conjugation pathway was affected by the Atg4BC74A mutant. In the LC3/Atg8 conjugation reaction, the Atg12-Atg5 conjugate and Atg16L form an 800-kDa super complex (referred as to the Atg16L complex; Mizushima et al., 2003) that plays an E3-like role by recruiting an E2 (Atg3)-LC3 intermediate to the site of conjugation (Hanada et al., 2007; Fujita et al., 2008). In 293A cells stably expressing mStrawberry-Atg4BC74A, the formation of the Atg12-Atg5 conjugate was not affected (Figure 2A). To examine the size of the Atg16L complex, cytosolic fractions of mock 293A or 293A cells stably expressing mStrawberry-Atg4BC74A were separated by size exclusion chromatography and immunoblotted with anti-Atg5 or anti-Atg16L antibody. As shown in Figure 2B, the molecular mass of the Atg16L complex was not shifted by mStrawberry-Atg4BC74A overexpression. From these results, we conclude that the Atg16L complex formation is not affected by overexpression of Atg4BC74A.
Figure 2.
Effect of Atg4BC74A overexpression on the Atg16L complex. (A) 293A cells stably expressing empty vector (Mock) or mStrawberry-Atg4BC74A (Atg4BC74A) were cultured in growth medium or HBSS for 2 h, and Western blotting was performed using each antibody. From top panel, anti-RFP, anti-Atg5, anti-LC3, anti-α-tubulin. Anti-RFP antibody reacts with mStrawberry. (B) Cytosolic fractions of 293A cells stably expressing empty vector or mStrawberry-Atg4BC74A were separated by size exclusion chromatography. Fractions were subjected to Western blotting using the indicated antibodies. The positions of the molecular-mass standards are shown. Vo, void fraction.
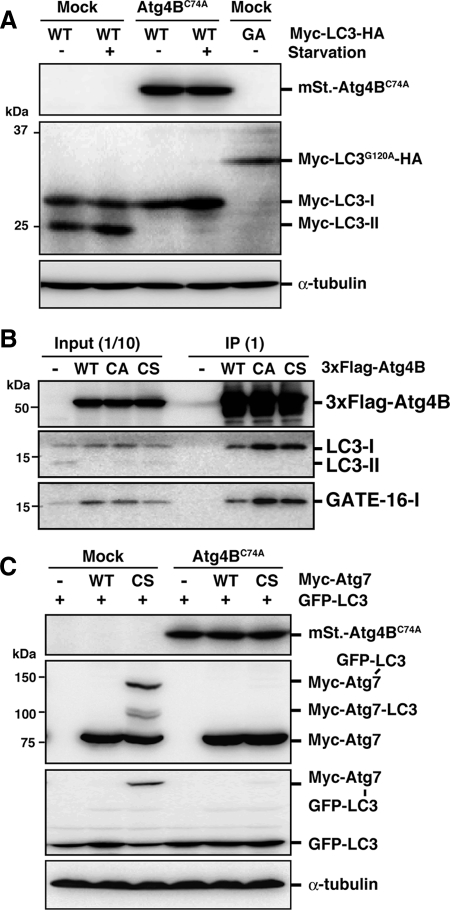
LC3 paralogues are synthesized as a proform that must be cleaved by Atg4 to expose a glycine residue that is engaged in conjugation. It is possible that this initial processing step is competitively inhibited by excess inactive Atg4B, resulting in failure to expose the glycine residue. To determine whether cleavage was inhibited, we used a Myc-LC3-HA construct, in which the HA tag is fused at the C-terminus of LC3. This construct enables us to easily distinguish between pro-LC3 and LC3-I by monitoring size (Kabeya et al., 2000). As shown in Figure 3A, the negative control Myc-LC3G120A-HA protein was not processed. However, Myc-LC3-I was detected both in 293A cells stably expressing mStrawberry-Atg4BC74A and in mock cells. This result shows that pro-LC3 processing is not affected by overexpression of Atg4BC74A.
Figure 3.
Stable complex formation between excess Atg4BC74A and LC3 prevents access to Atg7. (A) 293A cells stably expressing empty vector (Mock) or mStrawberry-Atg4BC74A (Atg4BC74A) were transfected with Myc-LC3-HA (WT) or Myc-LC3G120A-HA (GA). Thirty-six hours after transfection, cells were cultured in growth medium or HBSS for 2 h, and Western blotting were performed. From top panel, anti-RFP, anti-Myc, and anti-α-tubulin. (B) PC12 cells were infected with adenovirus bearing 3xFlag-tagged wild-type Atg4B (WT), Atg4BC74A (CA), or Atg4BC74S (CS). After 40-h incubation, cell lysates were subjected to immunoprecipitation with anti-Flag M2-conjugated agarose beads. Coimmunoprecipitated molecules were examined by Western blotting using each antibody. From top panel, anti-Flag, anti-LC3, and anti-GATE16. Total cell lysate (Input) and immunoprecipitated proteins (IP) are shown. (C) 293A cells stably expressing empty vector (Mock) or mStrawberry-Atg4BC74A (Atg4BC74A) were transfected with GFP-LC3 and Myc-Atg7 as indicated. Thirty-six hours after transfection, cell lysates were examined by Western blotting using each antibody. From top panel, anti-RFP, anti-myc, anti-GFP, and anti-α-tubulin.
GFP-LC3 is usually detected throughout the cytoplasm and within the nucleus when it does not form puncta. Nuclear localization of GFP-LC3 likely depends on the nature of GFP, because indirect immunofluorescence analysis of endogenous LC3 using an anti-LC3 antibody does not show a nuclear localization pattern (Komatsu et al., 2005). Interestingly, in Atg4B-overexpressing cells, GFP-LC3 dot formation was suppressed and GFP-LC3 signal was detected only in the cytoplasm (Figure 1A). The absence of nuclear GFP-LC3 suggests that it is trapped by excess Atg4B that localizes only in the cytoplasm. In support of this hypothesis, we ascertained that Atg4B and its inactive mutants form stable complexes with LC3 paralogues. 3xFlag-Atg4B mutants efficiently pulled down LC3-I and GATE16-I (Figure 3B). Accordingly, the formation of the LC3-Atg7 intermediate should be inhibited by overexpression of Atg4BC74A. To confirm this, we used a mutant E1 (Atg7C567S) enzyme whose active-site cysteine residue is replaced by serine. This mutant stabilizes the high-molecular-mass LC3-Atg7C567S intermediate, because a stable ester bond is formed between the enzyme and substrate instead of a labile thioester bond (Tanida et al., 2001). Although the LC3-Atg7 intermediate was detected in mock 293A cells, it was not detected in 293A cells stably expressing mStrawberry-Atg4BC74A (Figure 3C). We also observed that the LC3-Atg7 intermediate was not detected in wild-type Atg4B, as well as Atg4BC74A mutant, overexpressing cells (Supplemental Figure S1). These results indicate that the formation of Atg7-LC3 intermediate is the step that is inhibited by overexpression of Atg4B.
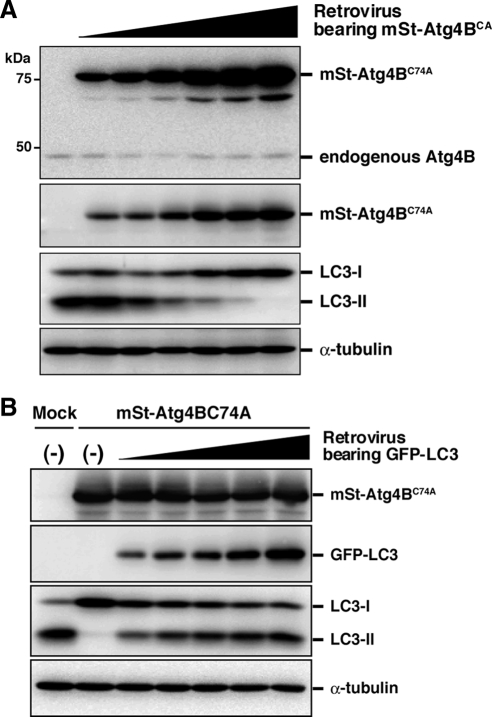
Atg4BC74A mutant inhibits LC3 lipidation in a dose-dependent manner (Figure 4A). Accumulating data set suggest that the cause of the inhibitory effect is sequestration of LC3 paralogues by excess Atg4B mutant. If so, the inhibitory effect should be dependent on the molecular ratio of Atg4B to LC3 paralogues. To test this model, we expressed GFP-tagged LC3 paralogues in NIH3T3 cells stably expressing mStrawberry-Atg4BC74A. As expected, the inhibitory effect of Atg4B mutant on LC3 lipidation was suppressed by exogenous LC3 or other LC3 paralogues in a dose-dependent manner (Figure 4B and Supplemental Figure S2). Collectively, we conclude that sequestration of LC3 paralogues by excess Atg4BC74A prevents access of LC3 paralogues to Atg7 and leads to a defect in autophagic degradation.
Figure 4.
The inhibitory effect of Atg4B mutant on LC3 lipidation is suppressed by exogenous LC3 in a dose-dependent manner. (A) NIH3T3 cells were infected with different amounts of retroviruses bearing mStrawberry-Atg4BC74A, and stable transformants were selected. The stable cells were cultured in HBSS (Starved) for 1 h, and cell lysates were examined by Western blotting using each antibody. From top panel, anti-Atg4B, anti-RFP, anti-LC3, and anti-α-tubulin. (B) NIH3T3 cells stably expressing mStrawberry-Atg4BC74A were infected with different amounts of retroviruses bearing GFP-LC3 and then double stable transformants were selected. Parent NIH3T3 cells and the stable transformants were cultured in HBSS (Starved) for 1 h, and cell lysates were examined by Western blotting using each antibody. From top panel, anti-RFP, anti-GFP, anti-LC3, and anti-α-tubulin.
The LC3 Paralogues Are Involved in Closing the Isolation Membrane
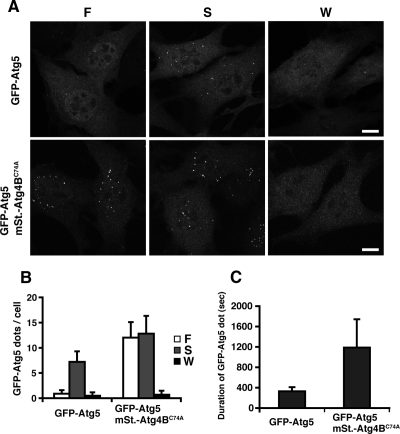
As the Atg4B mutant sequesters LC3 paralogues, cells overexpressing this protein provide a useful system for analysis of the role of the LC3 paralogues in autophagosome formation. To this end, we utilized NIH3T3 cells, which are suited to morphological analysis of the autophagic membrane, because of well-spread cytoplasm. The inhibitory effect of Atg4B mutant overexpression on PE conjugation of LC3 paralogues and membrane targeting of LC3 were also observed in this cell line (Figure 4, Supplemental Figure S3 and S4A). Moreover, the number of p62 bodies was significantly increased in mStrawberry-Atg4BC74A–expressing cells (Supplemental Figure S4C). In wild-type cells, isolation membranes were detected as punctate GFP-Atg5 signals. When the isolation membrane elongates and fuses to form the autophagosome, Atg5 detaches from the membrane. Therefore, GFP-Atg5 can be detected only on nascent autophagosomes, but not on completely formed ones (Mizushima et al., 2001). In a cell line in which GFP-Atg5 and mStrawberry-Atg4BC74A were stably coexpressed, the number of punctate GFP-Atg5 signals was increased in both nutrient-rich and starvation conditions (Figure 5, A and B). We obtained similar results by immunostaining with anti-Atg16L antibody (Supplemental Figure S4B). The membrane localization of the Atg16L complex is dependent on the phosphatidylinositol 3-kinase (Fujita et al., 2008; Mizushima et al., 2001). The punctate GFP-Atg5 signals in mStrawberry-Atg4BC74A–expressing cells were also dispersed by wortmannin treatment, as in control cells (Figure 5, A and B). The average lifetime of the GFP-Atg5 structure, as measured by time-lapse videomicroscopy, was ∼5 min in control cells, but was prolonged to ∼20 min in Atg4BC74A-overexpressing cells (Supplemental Movies S1 and S2, and Figure 5C).
Figure 5.
Effect of Atg4BC74A overexpression on GFP-Atg5–positive membrane structures. (A and B) NIH3T3 cells stably expressing GFP-Atg5 or both GFP-Atg5 and mStrawberry-Atg4BC74A were grown in growth medium (F), HBSS (S), or HBSS with 100 nM wortmannin (W) for 1 h and then fixed. Three-dimensional image stacks were obtained from sequential optical sections acquired 0.3 μm apart by confocal laser scanning microscopy (FV1000, Olympus) (A). Bar, 10 μm. The number of GFP-Atg5 puncta was counted in more than 100 cells. The value indicated is the mean ± SD (B). (C) NIH3T3 cells stably expressing GFP-Atg5 or both GFP-Atg5 and mStrawberry-Atg4BC74A were grown in HBSS for 1 h and directly observed by time-lapse video microscopy. The duration of each GFP-Atg5 puncta was measured for more than 50 cases. The value indicated is the mean ± SD.
To further characterize the Atg5-positive membrane structure, we examined the localization of Atg9L1, which is a mammalian homolog of yeast Atg9, a membrane protein engaged in autophagosome formation (Yamada et al., 2005). In mammalian cells, Atg9L is reported to cycle between the trans-Golgi network and the Golgi (Young et al., 2006). As shown in Supplemental Figure S5A, Atg9L1 did not show high colocalization with GFP-Atg5 in the presence or absence of Atg4BC74A, suggesting that the defect seen in Atg4BC74A-overexpressing cells is not related to Atg9L localization. We then analyzed the localization of the uncoordinated 51-like kinase1 (ULK1), the putative mammalian orthologue of yeast Atg1, a protein kinase also engaged in autophagosome formation (Chan et al., 2007; Hara et al., 2008). In control cells, myc-TEV-Flag–tagged ULK1 colocalized with GFP-Atg5 (Supplemental Figure S5B). In Atg5 knockout cells expressing the conjugation-deficient mutant of Atg5K130R, small crescent-like Atg5-positive membrane compartments accumulated (Mizushima et al., 2001). In Atg5 knockout MEF cells expressing GFP-Atg5K130R, ULK1 is also recruited to the structure (Supplemental Figure S6). It was reported that ULK1 and GABARAP, an LC3 paralogue, physically interact (Okazaki et al., 2000); therefore, it is possible that Atg4BC74A overexpression leads to a defect in ULK1 localization. However, colocalization of ULK1 and Atg5 was observed in Atg4BC74A-overexpressing cells (Supplemental Figure S5B), showing that the GFP-Atg5–positive structures that lack LC3-PE contain ULK1.
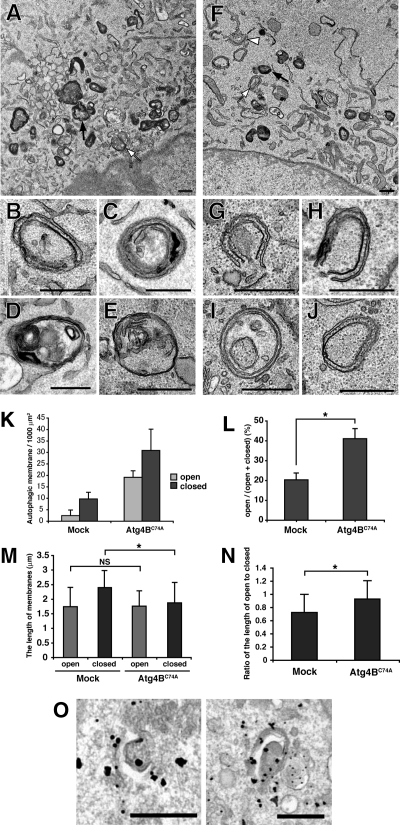
Next, we examined the Atg5-positive structures at the ultrastructural level. In mock cells, isolation membranes (Figure 6B), autophagosomes (Figure 6, C–E), and many autolysosome-like structures, which are characterized by highly electron-dense signals, were observed by electron microscopy (Figure 6A). In addition to autolysosomes, there are other electron-dense structures within cells, and therefore certain structures, such as amphisomes or autolysosomes, cannot be absolutely distinguished without specific markers (Eskelinen, 2008). Because membrane localization of LC3, the sole specific marker for autophagosomes and autolysosomes, was severely suppressed by Atg4BC74A overexpression (Supplemental Figure S4A), we cannot interpret the electron-dense structures further. Therefore, we counted the isolation membranes and double-membraned autophagic membranes. In contrast to mock cells, many isolation membranes (Figure 6, G and H) and autophagosome-like structures (Figure 6, I and J) were observed in cells stably overexpressing Atg4BC74A (Figure 6, F and K). Because we observed cross-sections of the cells, it was sometimes difficult to determine whether the autophagosome-like structures were really closed. However, the ratio of completely open structures to total autophagic structures was significantly higher in Atg4BC74A-expressing cells than in mock cells (Figure 6L). We also observed that the elevation in the number of autophagic structures and the ratio of open structures to total autophagic structures by excess Atg4B was diminished by overexpression of GFP-LC3 (Supplemental Figure S7). Although there was no significant difference in the length of open-autophagic membranes between mock and mStrawberry-Atg4BC74A–overexpressing cell, the length of the closed-autophagic membranes in Atg4BC74A-overexpressing cells was slightly shorter than the length in mock cells (Figure 6M). The ratio of the length of open to closed autophagic membrane in Atg4BC74A-overexpressing cells was significantly higher than in mock cells (Figure 6N), suggesting that the defect exists at a late stage in the autophagosome formation. Finally, to correlate these structures with fluorescence microscopy, we performed immunoelectron microscopy. NIH3T3 cells expressing GFP-Atg5 and mStrawberry-Atg4BC74A were grown in HBSS for 1 h, and the localization of GFP-Atg5 was examined by gold-enhanced immunogold electron microscopy using an anti-GFP antibody. As shown in Figure 6O, the isolation membranes, which elongate relatively well, were positive for GFP-Atg5 in Atg4BC74A-overexpressing cells. These lines of evidence indicate that the LC3 paralogues are involved in the completion of autophagosome formation in mammalian cells.
Figure 6.
Ultrastructual analysis of autophagic membranes in Atg4BC74A-overexpressing cells. NIH3T3 cells stably expressing empty vector (Mock) (A–E) or mStrawberry-Atg4BC74A (F–J) were cultured in HBSS for 1 h, fixed, and subjected to conventional electron microscopic analysis. (B–E) Typical autophagic structures in mock cells; isolation membrane (B), autophagosomes (C–E). (G–J) Typical autophagic structures in mStrawberry-Atg4BC74A-expressing cells; isolation membranes (G and H), closed double-membrane structures (I and J). Examples of electron-dense structures (black arrows), closed autophagic membranes (white arrows), and open autophagic membranes (white arrowheads) are indicated. Bar, 500 nm. (K) The number of autophagic structures in mock and mStrawberry-Atg4BC74A-expressing cells. ▩, open autophagic structures; ■, closed autophagic structures. Data are the means ± SD of triplicates from representative experiments. (L) The ratio of open structures to total autophagic structures. Data are the means ± SD of triplicates from representative experiments. *p < 0.05. (M) The length of autophagic membranes in mock and mStrawberry-Atg4BC74A-expressing cells. ▩, open autophagic structures; ■, closed autophagic structures. For the length of autophagic membranes determination, ImageJ version 1.40 was used (http://rsb.info.nih.gov/ij/). The value indicated is the mean ± SD. At least 20 samples were examined for each structure. *p < 0.05; NS, not significant. (N) The ratio of the length of open to closed autophagic structures in mock and mStrawberry-Atg4BC74A-expressing cells. *p < 0.05. (O) NIH3T3 cells expressing both GFP-Atg5 and mStrawberry-Atg4BC74A were grown in HBSS for 1 h and fixed. The localization of GFP-Atg5 was examined by gold-enhanced immunogold electron microscopy using an anti-GFP antibody. Bar, 500 nm.
DISCUSSION
We found that overexpression of Atg4B has an inhibitory effect on autophagy independent of its delipidation activity in mammalian cells. The inactive Atg4BC74A mutant specifically blocks the lipidation of LC3 paralogues by sequestering LC3 paralogues in stable complexes, resulting in blockade of the Atg7-LC3 reaction. It is interesting that the Atg4B enzyme has high affinity for its product, processed LC3 paralogues. This is in sharp contrast to overexpression of inactive ubiquitin-deconjugating enzymes, which act in a dominant-negative manner, elevating levels of their substrate, ubiquitin-conjugated proteins (Hang and Dasso, 2002; Li et al., 2002). In the course of the enzymatic reaction, Atg4 and LC3 paralogues should associate, but once cleavage has occurred, the proteins should dissociate for the next step, the Atg7-LC3 reaction. It is possible that an unknown factor is necessary for dissociating LC3 from Atg4, and Atg4 overexpression might result in limitation of this factor.
In cells overexpressing Atg4BC74A, a large number of the autophagic structures were not closed, although the length of these membranes was comparable to the length of autophagosomal membranes in control cells (Figure 6M). This observation fits with the results that Atg5-positive membrane structures accumulated and had prolonged lifetimes (Figure 5), based on the previous report that the Atg16L complex detach once the autophagosome formation is completed (Mizushima et al., 2001). In Atg5 knockout cells expressing the GFP-Atg5K130R mutant, in which Atg12-Atg5 conjugation does not occur, GFP-Atg5K130R signals also remain longer in membranous structures. These cells have incomplete Atg16L complexes, which lack Atg12, whereas in Atg4BC74A overexpressing cells, the Atg16L complex is intact. Therefore, it seems that the trigger that liberates the Atg16L complex from the membrane upon completion of autophagosome formation is not within the Atg16L complex.
It is interesting that autophagosome formation proceeded to a relatively late stage in Atg4BC74A-overexpressing cells; formation of autophagosome-like structures takes place. In addition, ULK1 is recruited to the structure in a manner similar to that observed in the isolation membranes of control cells (Supplemental Figure S5). The apparent defect in autophagosome completion is closure of the end of each elongating membrane. This does not necessarily exclude the proposal that Atg8 functions in expansion of autophagosomal membranes in yeast (Xie et al., 2008), because we observe only terminal phenotype and cannot exclude the possibility that elongation speed is slower. Atg8-PE can cause hemifusion of vesicles in vitro (Nakatogawa et al., 2007). One possibility is that LC3 paralogues function to complete autophagosome formation by fusing membranes in mammalian cells. Interestingly, in atg8Δ yeast, autophagosome-like structures were also detected by electron microscopy, however, at low frequency (Kirisako et al., 1999). There remains a possibility that expansion of autophagosomal membrane is not completely hampered by the deletion of Atg8.
We do not believe the phenotypes we observed are due to incomplete inhibition of LC3 paralogues function, because autophagosome-like structures were also observed in Atg3 (a specific E2 enzyme for Atg8 homologues) knockout MEF cells, where PE conjugation of LC3 paralogues is defective (Dr. Keiji Tanaka and Dr. Masaaki Komatsu, personal communication). By utilizing an inactive mutant of Atg4B, we could exclude secondary effects that might be brought about by overexpression of wild-type Atg4B, such as hyperdelipidation. Therefore, the phenotypes we observed likely reflect the physical sequestration and deficiency of the LC3 paralogues.
One strategy that may provide important information is artificial inhibition of autophagy. Indeed, treatment with drugs such as wortmannin or 3-methyladenine is widely used in studies of autophagy, but these drugs have side effects. RNA interference–mediated gene knockdown is a potential approach; however, nearly complete suppression of the ATG genes is needed to fully inhibit autophagy, and this is often difficult to achieve (Hosokawa et al., 2006; Yoshimura et al., 2006). The use of genetic knockouts of ATG genes is an alternative option for complete inhibition, but available cell types are restricted. In the case of Atg4BC74A overexpression, it seems possible to fully inhibit autophagy in any type of cell. Several studies have suggested that the Atg12-Atg5 conjugate has other roles in addition to autophagy (Pyo et al., 2005; Yousefi et al., 2006; Takeshita et al., 2007). However, currently reported Atg5- or Atg7-deficient cells do not distinguish between autophagic and nonautophagic function of the Atg12-Atg5 conjugate, as it is lacking in both cell types. As overexpression of the Atg4B mutant inhibits formation of autophagosomes, but not generation of the Atg12-Atg5 conjugate, such problems can be avoided. We believe that the inactive Atg4B mutant will provide a useful tool for a broad range of studies analyzing autophagy.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank Dr. Kouichi Matsunaga (Yoshimori lab) for the gift of MCF7 cells stably expressing GFP-LC3; Dr. Roger Y. Tsien (University of California, San Diego, CA) for the gift of mStrawberry cDNA; Dr. Shoji Yamaoka for the gifts of pMRX-IRES-puro and pMRX-IRES-bsr; Dr. Toshio Kitamura (The University of Tokyo, Japan) for the gift of Plat-E cells; Dr. Noboru Mizushima (Tokyo Medical and Dental University, Japan) for the gift of anti-Atg16L antibody; Mr. Takuya Hayashi (Yoshimori lab) for the gift of myc-TEV-Flag-tagged Atg9L1 construct; and Dr. Kyouhei Umebayashi (Yoshimori lab) for helpful discussion. The work described in this report was supported in part by Special Coordination Funds for Promoting Science and Technology of the Ministry of Education, Culture, Sports, Science and Technology (MEXT).
Abbreviations used:
- HBSS
Hanks' balanced salt solution
- MEF
mouse embryonic fibroblast
- PE
phosphatidylethanolamine
- TCA
trichloroacetic acid
- ULK
uncoordinated 51-like kinase.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-03-0312) on September 3, 2008.
REFERENCES
- Bjorkoy G., Lamark T., Brech A., Outzen H., Perander M., Overvatn A., Stenmark H., Johansen T. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J. Cell Biol. 2005;171:603–614. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blommaart E. F., Krause U., Schellens J. P., Vreeling-Sindelarova H., Meijer A. J. The phosphatidylinositol 3-kinase inhibitors wortmannin and LY294002 inhibit autophagy in isolated rat hepatocytes. Eur. J. Biochem. 1997;243:240–246. doi: 10.1111/j.1432-1033.1997.0240a.x. [DOI] [PubMed] [Google Scholar]
- Chan E. Y., Kir S., Tooze S. A. siRNA screening of the kinome identifies ULK1 as a multidomain modulator of autophagy. J. Biol. Chem. 2007;282:25464–25474. doi: 10.1074/jbc.M703663200. [DOI] [PubMed] [Google Scholar]
- Cuervo A. M. Autophagy: in sickness and in health. Trends Cell Biol. 2004;14:70–77. doi: 10.1016/j.tcb.2003.12.002. [DOI] [PubMed] [Google Scholar]
- Eskelinen E. L. To be or not to be? Examples of incorrect identification of autophagic compartments in conventional transmission electron microscopy of mammalian cells. Autophagy. 2008;4:257–260. doi: 10.4161/auto.5179. [DOI] [PubMed] [Google Scholar]
- Fujita N., Itoh T., Omori H., Fukuda M., Noda T., Yoshimori T. The Atg16L complex specifies the site of LC3 lipidation for membrane biogenesis in autophagy. Mol. Biol. Cell. 2008;19:2092–2100. doi: 10.1091/mbc.E07-12-1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanada T., Noda N. N., Satomi Y., Ichimura Y., Fujioka Y., Takao T., Inagaki F., Ohsumi Y. The Atg12-Atg5 conjugate has a novel E3-like activity for protein lipidation in autophagy. J. Biol. Chem. 2007;282:37298–37302. doi: 10.1074/jbc.C700195200. [DOI] [PubMed] [Google Scholar]
- Hang J., Dasso M. Association of the human SUMO-1 protease SENP2 with the nuclear pore. J. Biol. Chem. 2002;277:19961–19966. doi: 10.1074/jbc.M201799200. [DOI] [PubMed] [Google Scholar]
- Hara T., Takamura A., Kishi C., Iemura S., Natsume T., Guan J. L., Mizushima N. FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells. J. Cell Biol. 2008;181:497–510. doi: 10.1083/jcb.200712064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemelaar J., Lelyveld V. S., Kessler B. M., Ploegh H. L. A single protease, Apg4B, is specific for the autophagy-related ubiquitin-like proteins GATE-16, MAP1-LC3, GABARAP, and Apg8L. J. Biol. Chem. 2003;278:51841–51850. doi: 10.1074/jbc.M308762200. [DOI] [PubMed] [Google Scholar]
- Hosokawa N., Hara Y., Mizushima N. Generation of cell lines with tetracycline-regulated autophagy and a role for autophagy in controlling cell size. FEBS Lett. 2006;580:2623–2629. doi: 10.1016/j.febslet.2006.04.008. [DOI] [PubMed] [Google Scholar]
- Ichimura Y., et al. A ubiquitin-like system mediates protein lipidation. Nature. 2000;408:488–492. doi: 10.1038/35044114. [DOI] [PubMed] [Google Scholar]
- Kabeya Y., Mizushima N., Ueno T., Yamamoto A., Kirisako T., Noda T., Kominami E., Ohsumi Y., Yoshimori T. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeya Y., Mizushima N., Yamamoto A., Oshitani-Okamoto S., Ohsumi Y., Yoshimori T. LC3, GABARAP and GATE16 localize to autophagosomal membrane depending on form-II formation. J. Cell Sci. 2004;117:2805–2812. doi: 10.1242/jcs.01131. [DOI] [PubMed] [Google Scholar]
- Kimura S., Noda T., Yoshimori T. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy. 2007;3:452–460. doi: 10.4161/auto.4451. [DOI] [PubMed] [Google Scholar]
- Kirisako T., Baba M., Ishihara N., Miyazawa K., Ohsumi M., Yoshimori T., Noda T., Ohsumi Y. Formation process of autophagosome is traced with Apg8/Aut7p in yeast. J. Cell Biol. 1999;147:435–446. doi: 10.1083/jcb.147.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirisako T., Ichimura Y., Okada H., Kabeya Y., Mizushima N., Yoshimori T., Ohsumi M., Takao T., Noda T., Ohsumi Y. The reversible modification regulates the membrane-binding state of Apg8/Aut7 essential for autophagy and the cytoplasm to vacuole targeting pathway. J. Cell Biol. 2000;151:263–276. doi: 10.1083/jcb.151.2.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M., et al. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J. Cell Biol. 2005;169:425–434. doi: 10.1083/jcb.200412022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumanomidou T., Mizushima T., Komatsu M., Suzuki A., Tanida I., Sou Y. S., Ueno T., Kominami E., Tanaka K., Yamane T. The crystal structure of human Atg4b, a processing and de-conjugating enzyme for autophagosome-forming modifiers. J. Mol. Biol. 2006;355:612–618. doi: 10.1016/j.jmb.2005.11.018. [DOI] [PubMed] [Google Scholar]
- Levine B., Klionsky D. J. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev. Cell. 2004;6:463–477. doi: 10.1016/s1534-5807(04)00099-1. [DOI] [PubMed] [Google Scholar]
- Li M., Chen D., Shiloh A., Luo J., Nikolaev A. Y., Qin J., Gu W. Deubiquitination of p53 by HAUSP is an important pathway for p53 stabilization. Nature. 2002;416:648–653. doi: 10.1038/nature737. [DOI] [PubMed] [Google Scholar]
- Luo H., Nakatsu F., Furuno A., Kato H., Yamamoto A., Ohno H. Visualization of the post-Golgi trafficking of multiphoton photoactivated transferrin receptors. Cell Struct. Funct. 2006;31:63–75. doi: 10.1247/csf.31.63. [DOI] [PubMed] [Google Scholar]
- Marino G., Uria J. A., Puente X. S., Quesada V., Bordallo J., Lopez-Otin C. Human autophagins, a family of cysteine proteinases potentially implicated in cell degradation by autophagy. J. Biol. Chem. 2003;278:3671–3678. doi: 10.1074/jbc.M208247200. [DOI] [PubMed] [Google Scholar]
- Mizushima N. Autophagy: process and function. Genes Dev. 2007;21:2861–2873. doi: 10.1101/gad.1599207. [DOI] [PubMed] [Google Scholar]
- Mizushima N., Kuma A., Kobayashi Y., Yamamoto A., Matsubae M., Takao T., Natsume T., Ohsumi Y., Yoshimori T. Mouse Apg16L, a novel WD-repeat protein, targets to the autophagic isolation membrane with the Apg12-Apg5 conjugate. J. Cell Sci. 2003;116:1679–1688. doi: 10.1242/jcs.00381. [DOI] [PubMed] [Google Scholar]
- Mizushima N., Yamamoto A., Hatano M., Kobayashi Y., Kabeya Y., Suzuki K., Tokuhisa T., Ohsumi Y., Yoshimori T. Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells. J. Cell Biol. 2001;152:657–668. doi: 10.1083/jcb.152.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N., Yoshimori T. How to interpret LC3 immunoblotting. Autophagy. 2007;3:542–545. doi: 10.4161/auto.4600. [DOI] [PubMed] [Google Scholar]
- Morita S., Kojima T., Kitamura T. Plat-E: an efficient and stable system for transient packaging of retroviruses. Gene Ther. 2000;7:1063–1066. doi: 10.1038/sj.gt.3301206. [DOI] [PubMed] [Google Scholar]
- Nakatogawa H., Ichimura Y., Ohsumi Y. Atg8, a ubiquitin-like protein required for autophagosome formation, mediates membrane tethering and hemifusion. Cell. 2007;130:165–178. doi: 10.1016/j.cell.2007.05.021. [DOI] [PubMed] [Google Scholar]
- Noda T., Suzuki K., Ohsumi Y. Yeast autophagosomes: de novo formation of a membrane structure. Trends Cell Biol. 2002;12:231–235. doi: 10.1016/s0962-8924(02)02278-x. [DOI] [PubMed] [Google Scholar]
- Okazaki N., Yan J., Yuasa S., Ueno T., Kominami E., Masuho Y., Koga H., Muramatsu M. Interaction of the Unc-51-like kinase and microtubule-associated protein light chain 3 related proteins in the brain: possible role of vesicular transport in axonal elongation. Brain Res. Mol. Brain Res. 2000;85:1–12. doi: 10.1016/s0169-328x(00)00218-7. [DOI] [PubMed] [Google Scholar]
- Pyo J. O., et al. Essential roles of Atg5 and FADD in autophagic cell death: dissection of autophagic cell death into vacuole formation and cell death. J. Biol. Chem. 2005;280:20722–20729. doi: 10.1074/jbc.M413934200. [DOI] [PubMed] [Google Scholar]
- Saitoh T., Nakayama M., Nakano H., Yagita H., Yamamoto N., Yamaoka S. TWEAK induces NF-kappaB2 p100 processing and long lasting NF-kappaB activation. J. Biol. Chem. 2003;278:36005–36012. doi: 10.1074/jbc.M304266200. [DOI] [PubMed] [Google Scholar]
- Shaner N. C., Campbell R. E., Steinbach P. A., Giepmans B. N., Palmer A. E., Tsien R. Y. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 2004;22:1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- Sou Y. S., Tanida I., Komatsu M., Ueno T., Kominami E. Phosphatidylserine in addition to phosphatidylethanolamine is an in vitro target of the mammalian Atg8 modifiers, LC3, GABARAP, and GATE-16. J. Biol. Chem. 2006;281:3017–3024. doi: 10.1074/jbc.M505888200. [DOI] [PubMed] [Google Scholar]
- Sugawara K., Suzuki N. N., Fujioka Y., Mizushima N., Ohsumi Y., Inagaki F. Structural basis for the specificity and catalysis of human Atg4B responsible for mammalian autophagy. J. Biol. Chem. 2005;280:40058–40065. doi: 10.1074/jbc.M509158200. [DOI] [PubMed] [Google Scholar]
- Takeshita F., Kobiyama K., Miyawaki A., Jounai N., Okuda K. The non-canonical role of Atg family members as suppressors of innate antiviral immune signaling. Autophagy. 2007;4:67–69. doi: 10.4161/auto.5055. [DOI] [PubMed] [Google Scholar]
- Tanida I., Sou Y. S., Ezaki J., Minematsu-Ikeguchi N., Ueno T., Kominami E. HsAtg4B/HsApg4B/autophagin-1 cleaves the carboxyl termini of three human Atg8 homologues and delipidates microtubule-associated protein light chain 3- and GABAA receptor-associated protein-phospholipid conjugates. J. Biol. Chem. 2004;279:36268–36276. doi: 10.1074/jbc.M401461200. [DOI] [PubMed] [Google Scholar]
- Tanida I., Sou Y. S., Minematsu-Ikeguchi N., Ueno T., Kominami E. Atg8L/Apg8L is the fourth mammalian modifier of mammalian Atg8 conjugation mediated by human Atg4B, Atg7 and Atg3. FEBS J. 2006;273:2553–2562. doi: 10.1111/j.1742-4658.2006.05260.x. [DOI] [PubMed] [Google Scholar]
- Tanida I., Tanida-Miyake E., Ueno T., Kominami E. The human homolog of Saccharomyces cerevisiae Apg7p is a protein-activating enzyme for multiple substrates including human Apg12p, GATE-16, GABARAP, and MAP-LC3. J. Biol. Chem. 2001;276:1701–1706. doi: 10.1074/jbc.C000752200. [DOI] [PubMed] [Google Scholar]
- Wu J., Dang Y., Su W., Liu C., Ma H., Shan Y., Pei Y., Wan B., Guo J., Yu L. Molecular cloning and characterization of rat LC3A and LC3B–two novel markers of autophagosome. Biochem. Biophys. Res. Commun. 2006;339:437–442. doi: 10.1016/j.bbrc.2005.10.211. [DOI] [PubMed] [Google Scholar]
- Xie Z., Nair U., Klionsky D. J. Atg8 controls phagophore expansion during autophagosome formation. Mol. Biol. Cell. 2008;19:3290–3298. doi: 10.1091/mbc.E07-12-1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada T., Carson A. R., Caniggia I., Umebayashi K., Yoshimori T., Nakabayashi K., Scherer S. W. Endothelial nitric-oxide synthase antisense (NOS3AS) gene encodes an autophagy-related protein (APG9-like2) highly expressed in trophoblast. J. Biol. Chem. 2005;280:18283–18290. doi: 10.1074/jbc.M413957200. [DOI] [PubMed] [Google Scholar]
- Yoshimori T. Autophagy: a regulated bulk degradation process inside cells. Biochem. Biophys. Res. Commun. 2004;313:453–458. doi: 10.1016/j.bbrc.2003.07.023. [DOI] [PubMed] [Google Scholar]
- Yoshimori T., Yamagata F., Yamamoto A., Mizushima N., Kabeya Y., Nara A., Miwako I., Ohashi M., Ohsumi M., Ohsumi Y. The mouse SKD1, a homologue of yeast Vps4p, is required for normal endosomal trafficking and morphology in mammalian cells. Mol. Biol. Cell. 2000;11:747–763. doi: 10.1091/mbc.11.2.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura K., Shibata M., Koike M., Gotoh K., Fukaya M., Watanabe M., Uchiyama Y. Effects of RNA interference of Atg4B on the limited proteolysis of LC3 in PC12 cells and expression of Atg4B in various rat tissues. Autophagy. 2006;2:200–208. doi: 10.4161/auto.2744. [DOI] [PubMed] [Google Scholar]
- Young A. R., Chan E. Y., Hu X. W., Kochl R., Crawshaw S. G., High S., Hailey D. W., Lippincott-Schwartz J., Tooze S. A. Starvation and ULK1-dependent cycling of mammalian Atg9 between the TGN and endosomes. J. Cell Sci. 2006;119:3888–3900. doi: 10.1242/jcs.03172. [DOI] [PubMed] [Google Scholar]
- Yousefi S., Perozzo R., Schmid I., Ziemiecki A., Schaffner T., Scapozza L., Brunner T., Simon H. U. Calpain-mediated cleavage of Atg5 switches autophagy to apoptosis. Nat. Cell Biol. 2006;8:1124–1132. doi: 10.1038/ncb1482. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.