Abstract
Clearance of misfolded proteins from the ER is central for maintenance of cellular homeostasis. This process requires coordinated recognition, ER-cytosol translocation, and finally ubiquitination-dependent proteasomal degradation. Here, we identify an ER resident seven-transmembrane protein (JAMP) that links ER chaperones, channel proteins, ubiquitin ligases, and 26S proteasome subunits, thereby optimizing degradation of misfolded proteins. Elevated JAMP expression promotes localization of proteasomes at the ER, with a concomitant effect on degradation of specific ER-resident misfolded proteins, whereas inhibiting JAMP promotes the opposite response. Correspondingly, a jamp-1 deleted Caenorhabditis elegans strain exhibits hypersensitivity to ER stress and increased UPR. Using biochemical and genetic approaches, we identify JAMP as important component for coordinated clearance of misfolded proteins from the ER.
INTRODUCTION
Degradation of misfolded proteins is part of a complex quality control system that clears diverse proteins independent of sequence or functional similarity (Hampton, 2002; Brodsky, 2007; Ron and Walter, 2007). The unfolded protein response (UPR) reduces the burden caused by unfolded protein accumulation in the endoplasmic reticulum (ER; Harding et al., 2002), in part through activation of IRE1-XBP1 and ATF6, resulting in transcriptional activation of genes important for UPR, including components of the endoplasmic reticulum-associated degradation system (ERAD; Wu and Kaufman, 2006).
ERAD is regulated by an ER quality control system that detects proteins that cannot fold or assemble into multiprotein complexes and marks them for ubiquitination-dependent degradation (Hampton, 2002; Brodsky, 2007; Ron and Walter, 2007). This quality control consists of molecular chaperones such as BiP (Brodsky, 2007; Ron and Walter, 2007), which interact with the misfolded protein enabling its transfer across the ER membrane, possibly catalyzed by the multispanning proteins Derlin-1/2/3 and Sec61 (Osborne et al., 2005) with assistance of the AAA (ATPases associated with cellular activities) ATPase p97 (also called VCP or Cdc48). Subsequent to translocation, misfolded proteins are ubiquitinated via ER-anchored ubiquitin ligases, such as the vertebrate gp78, parkin, RNF5/RMA1, or Hrd1 (Fang et al., 2001; Younger et al., 2006), followed by proteasome-mediated degradation.
The JNK-associated membrane protein (JAMP) associates with JNK and prolongs its kinase activity after stress (Kadoya et al., 2005). JAMP was originally identified in a complex with the ER-anchored ubiquitin ligase RNF5/RMA1. RNF5 was recently implicated in ERAD (Younger et al., 2006; Morito et al., 2008) through its cooperation with either CHIP or gp78. Mouse models have revealed a role for RNF5 in a condition called inclusion body myocytosis, a muscular disorder associated with extensive ER stress (Delaunay et al., 2008), consistent with its reported role in ER stress and in controlling localization and levels of LIM/LD domain-containing proteins (Didier et al., 2003; Broday et al., 2004). JAMP consists of seven predicted transmembrane domains, a zinc finger domain and a putative N-glycosylation site, is widely expressed, and is induced in response to stress, including ER stress (Kadoya et al., 2005). Here we demonstrate that JAMP is an important component in ERAD, serving as an adaptor for both ERAD and proteasome components and facilitating clearance of misfolded proteins from the ER.
MATERIALS AND METHODS
Cell Culture and Transfection
Human embryonic kidney (HEK) 293T and HeLa cells were maintained in DMEM supplemented with 10% calf serum and antibiotics (penicillin and streptomycin, 100 U/ml; Invitrogen, Carlsbad, CA) in 5% CO2 at 37°C. Transfections were performed using the calcium phosphate technique for 293T cells and Lipofectamine Plus (Invitrogen) for HeLa cells. For cycloheximide chase experiments hemagglutinin (HA)-CFTRΔ508 (2 μg) was cotransfected with Flag-JAMP (2 μg) or empty vector together with green fluorescent protein (GFP; 0.3 μg) per 10-cm dish.
Antibodies and Immunoblot Analysis
Polyclonal antibodies against the JAMP N-terminal domain (aa 1-58) were generated as described (Kadoya et al., 2005) and used for immunoblot analysis (1:500). Antibodies for proteasome subunits [Rpt6, 1:1000; and Rpt2, Rpt5, rpn12, and 20S (C2); all diluted 1:50,000] were used as described (Wójcik et al., 2006). The following antibodies were purchased from the indicated vendors: monoclonal Flag antibodies (Sigma, St. Louis, MO; 1:10,000), HA polyclonal antibodies (Santa-Cruz Biotechnology, Santa-Cruz, CA; 1:1000), monoclonal myc antibodies (Santa-Cruz; 1:5000); Antibody to Grp78 (Santa-Cruz; 1:1000), anti p97 (1:1000,) and anti Sec61α (Abcam, Cambridge, MA; 1:1000). Rpt6 monoclonal antibodies for immunohistochemical studies (Abcam; 1:50) and calnexin monoclonal antibodies for immunostaining were from BD Biosciences Pharmingen (San Diego, CA; 1:100). The following antibodies were obtained as gifts: antibodies to gp78 (1:200) from Dr. A. Weissman (National Cancer Institute, Frederick, MD); antibodies against Golgi compartment TGN38 from Dr. H. Xu (Burnham Institute for Medical Research, La Jolla, CA). Protein samples were separated on SDS-PAGE and electrotransferred onto membranes incubated with indicated primary antibodies indicated. Immunoblots were visualized using goat anti-mouse or anti-rabbit Alexa-Fluor 680 secondary antibodies (Molecular Probes, Eugene, OR) after detection with the Odyssey Infrared Imaging System (Li-Cor Biosciences, Lincoln, NE).
DNA Constructs
Full-length (936bp) mouse JAMP cDNA was amplified by PCR and cloned into BamHI/XhoI sites of pcDNA-Flag and pEF-HA. JAMP deletion mutants (TM 1, 1–58 aa; TM 1–3, 1–157 aa; TM 4–7, 158–311 aa; and TM 6–7, 232–311 aa) were generated via PCR-based cloning of the DNA fragment into Flag-tagged pcDNA vector. Flag-Rpt6/4 constructs were a gift of K. Tanaka (Tokyo Metropolitan Institute, Tokyo, Japan). HA-CFTR Δ508 expression vector was a gift from G. Lukacs. The integrity of each construct was verified by sequencing. HA-CD3δ construct was a gift of Dr. A. Weissman (Fang et al., 2001). HA-TCRα, p97 and p97H13A constructs were a gift of Dr. R. Kopito (Stanford University, Stanford, CA; DeLaBarre et al., 2006). HA-Derlin1 construct was a gift from Dr. Y. Ye (NIH, Bethesda, MD; Ye et al., 2004).
Gene Silencing by RNA Interference
Two sequences harboring nucleotides corresponding to nucleotides 12–30 of the mouse JAMP coding sequence (TATTCAACCAGCATGCCTT and AATAGAGAACTGCTATGAT) were synthesized and cloned into BglII and HindIII sites of pSuper vector. Construct integrity was confirmed by sequencing. In transient small interfering RNA (siRNA) experiments either vector was transfected into 293T cells. Thirty-six hours after transfection, cells were harvested, and JAMP expression was assayed via immunoblot analysis. Stable clones of HeLa cells expressing siRNA of JAMP were selected after treatment with puromycin (6 μg/ml) of cultures cotransfected with pSuper containing JAMP siRNA sequences and a puromycin expression plasmid. The siJAMP clone expressing the sequence AATAGAGAACTGCTATGAT exhibited stronger inhibition of JAMP expression in immunoblots, RT-PCR, and immunohistochemical analysis and was used for all analyses shown.
Immunoprecipitation.
Co-IPs were done in all cases, unless otherwise specified, after extraction of the proteins with 1% Triton X-100 in 50 mM Tris, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1 mM PMSF, 10 μg/ml leupeptin, 10 μg/ml aprotinin, and 10 μg/ml pepstatin A. Where specified in the figure legends, immunoprecipitation (IP) was performed under mild conditions, following the protocol for immunopurification of endogenous proteasomes from cells using sonication and lysis buffer (50 mM Tris, pH 7.4, 10% glycerol, 150 mM NaCl, 5 mM MgCl2, 1 mM PMSF, 10 μg/ml leupeptin, 10 μg/ml pepstatin A, and 10 μg/ml aprotinin). In all cases, samples were centrifuged (15 min, 14,000 rpm), and supernatants were incubated (1 h) with indicated antibodies (2 μg). Immunoprecipitation was performed by incubation (40 min at 4°C) with protein G-agarose (Invitrogen). After washing three times with lysis buffer, proteins were solubilized in 3× Laemmli buffer and separated on SDS-PAGE followed by immunoblot analysis with the indicated antibodies.
Immunohistochemistry.
HeLa cells grown on coverslips (22-mm2, Chase Scientific Glass, Rockwood, TN) were fixed using freshly prepared 3% paraformaldehyde in PBS (5 min at room temperature). Cells were then washed (three times, 5 min each) in PBS, followed by permeabilization in 0.1% Triton X-100 in PBS (pH 7.4) for 1 min and an additional three 5-min washes in PBS. Cells were then incubated in PBS supplemented with 3% bovine serum albumin for 30 min. Cells were incubated with antibodies (1 h at room temperature) in a humidity chamber and then washed in PBS (three times, 5 min each) before incubation with 100-μl of Alexa-488– and Alexa-568–conjugated anti-rabbit or anti-mouse immunoglobulin G (Molecular Probes) diluted (2 μg/ml) in PBS containing 0.2% BSA (60 min at room temperature in a light-protected humidity chamber). Cells were rinsed three times in PBS. Coverslips were mounted on glass slides using Vectashield (Vector Laboratories, Burlingame, CA). For immunohistochemistry (IHC), antibodies were used at the following concentrations: Rpt6 1:100, Rpt2 (1:100), 20S (1:100), Flag (4.6 μg/ml), HA (2 μg/ml), and calnexin (1:100). Detection of JAMP by IHC requires a different method (methanol) than used for other proteins (Triton), thereby requiring us to perform parallel rather than coimmunostaining. Immunofluorescence data were obtained using Olympus TH4-100 microscope (Melville, NY) and Slidebook 4.1 digital microscopy software (Intelligent Imaging Innovations, Denver, CO). Images were deconvoluted with the aid of constrained iterative and nearest neighbor algorithms. Confocal microscopy was done with a Fluoview 1000 Olympus Laser Point Scanning Confocal Microscope. Images were processed using ImageJ software package (http://rsb.info.nih.gov/ij/).
Pulse-Chase Analysis
Cells transfected with HA-CFTRΔ508 or HA-CD3δ construct were incubated in 10% dFBS/DMEM (lacking Met and/Cys) medium for 1 h. 35S-labeled methionine/cysteine mix (0.1mCi/ml) was added for 1.5 h. Labeled medium was removed, and cells were washed in chase medium (10% FBS/DMEM supplemented with 0.5 M cold methionine and 0.5 M cysteine) and lysed at indicated time points. Cell lysates were subjected to IP with HA antibodies followed by autoradiography and phosphorimager analysis with FLA-1500 (Fujifilm, Life Science, Tokyo, Japan).
Cycloheximide Chase Analysis
Cells (293T) were transfected with the indicated plasmids, and 24 h later cells were subjected to treatment with cycloheximide (20 μg/ml). Proteins were prepared by lysis with detergent-based buffer at the indicated time points. Analysis was performed after IP of the misfolded protein (for CD3δ with HA antibodies) or straight Western blotting (for NHK). Relative changes were quantified using Li-COR scanner and are indicated under each lane.
Proteasome Purification
293T cells transfected with Flag-JAMP were sonicated (10 min) using lysis buffer (50 mM Tris, pH 7.4, 10% glycerol, 150 mM NaCl, 5 mM MgCl2, 5 mM ATP, 1 mM PMSF, 10 μg/ml leupeptin, and 10 μg/ml aprotinin). Samples were centrifuged (15 min, 8000 rpm), and supernatants were incubated for up to 1 h at 4°C with anti 20S (C2) antibody. IP was performed by incubation (40 min at 4°C) with protein G-agarose (Invitrogen). After washing (three times) with lysis buffer proteins were solubilized in 3× Laemmli buffer and separated on SDS-PAGE followed by immunoblot analysis with antibodies against proteasome subunits and Flag antibody for detection of JAMP.
Membrane Floating and Proteasome Activity
293T cells were transfected with Flag-JAMP or empty vector and 36 h later, crude microsome extract was prepared using ER extraction kit (Sigma). Three milligrams of protein were resuspended in 2.5 M sucrose solution and placed at the bottom of a ultracentrifuge tube (final concentration 2 M). A step sucrose gradient (2–0.7 M) was applied on top of the crude microsome extract and subjected to ultracentrifugation at 100,000 × g using SW41 rotor for 18 h. Fractions were collected and analyzed using Western blot with the aid of Flag and Rpt6 antibodies. Grp78, calnexin, or translocon (Sec61alpha) served as a marker for the ER fraction. Proteasome activity was measured using LLVY-AMC fluorescent substrate (Chemicon, Temecula, CA; 20S proteasome activity kit); Released fluorescence was quantified using Spectra MAX Gemini EM luminometer (380/460 filter). Samples were treated in parallel with MG132 (50 μM) for 20 min in RT, proteasome activity was measured, and results obtained in the presence of MG132 were subsequently subtracted from nontreated fractions. Purified 20S particle was used as positive control as well as control for MG132 inhibition. Specific proteasome activity was normalized to protein concentration in the respective fraction (fluorescence per microgram of protein).
Protein Identification by Mass Spectrometry
Proteins prepared from 293T cells that express exogenous Flag-JAMP were immunoprecipitated with Flag antibodies, and bound material was eluted with the aid of Flag peptide. Flag-tagged empty expression vector was used as a control. Eluted proteins were separated on 2D gels and specific spots (compared with empty construct Flag immunoprecipitation) were visualized by silver staining, digested with trypsin, batch-purified on a reversed-phase microtip, and resulting peptide pools individually analyzed by matrix-assisted laser desorption/ionization reflectron time-of-flight (MALDI-reTOF) mass spectrometry (MS; UltraFlex TOF/TOF; Bruker, Bremen, Germany) for peptide mass fingerprinting (PMF), as described (Erdjument-Bromage et al., 1998). Selected peptide ions (m/z) were taken to search a “nonredundant” human protein database (NR; 134,604 entries; National Center for Biotechnology Information, Bethesda, MD) utilizing the PeptideSearch algorithm (Matthias Mann, Max-Planck Institute for Biochemistry, Martinsried, Germany; an updated version of this program is currently available as “PepSea” from Applied Biosystems/MDS Sciex, Foster City, CA). A molecular-mass range up to twice the apparent molecular weight (as estimated from electrophoretic relative mobility) was covered, with a mass accuracy restriction of <35 ppm, and a maximum of one missed cleavage site allowed per peptide. To confirm PMF results with scores ≤40, mass spectrometric sequencing of selected peptides was done by MALDI-TOF/TOF (MS/MS) analysis on the same prepared samples, using the UltraFlex instrument in “LIFT” mode. Fragment ion spectra were taken to search NR using the MASCOT MS/MS Ion Search program, version 2.0.04 for Windows (Matrix Science, London, United Kingdom). Any tentative confirmation (Mascot score ≥30) of a PMF result thus obtained was verified by comparing the computer-generated fragment ion series of the predicted tryptic peptide with the experimental MS/MS data.
C. elegans Strains and Tunicamycin Treatment
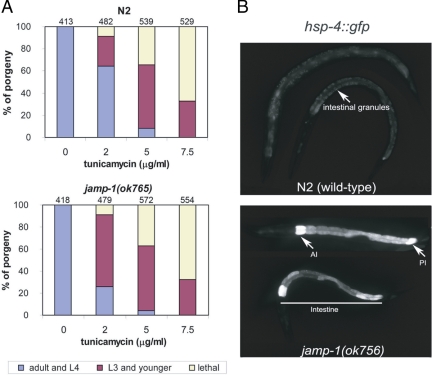
The putative seven-transmembrane protein orthologue to JAMP in C. elegans is encoded by the gene R01B10.5, designated jamp-1. The jamp-1(ok765) mutant worms (isolated by the C. elegans Gene Knockout Consortium, Oklahoma) harbor a 851-bp deletion and are viable. The 3′ breakpoint of the deletion causes the formation of a premature stop codon, and therefore the predicted protein includes only the first 41aa of the N-terminus of JAMP-1. Because mRNAs that contain premature stop codons are usually degraded by the nonsense-mediated mRNA decay system (Pulak and Anderson, 1993), this allele probably represents a loss-of-function allele. The jamp-1(ok765) mutant was outcrossed three times before analysis. The hsp-4::gfp integrated strain zcIs4[hsp-4::GFP] was received from the CGC and crossed to the jamp-1(ok765) strain to construct the jamp-1(ok765); zcIs4[hsp-4::GFP] strain. Homozygous jamp-1(ok765) offspring were identified by PCR genotyping. C. elegans strains were cultivated at 20°C. For analysis of tunicamycin sensitivity, gravid adults were allowed to lay eggs on plates containing tunicamycin. Eggs were counted and analyzed 72 h later. At least four independent experiments were performed for each tunicamycin concentration (0–7.5 μg/ml), and two or three plates for each treatment were analyzed in each experiment; n values above the bars in Figure 6 are the total embryos of all four experiments.
Figure 6.
A jamp-1 deleted C. elegans strain exhibits basal stress and is hypersensitive to ER stress. (A) Increased sensitivity to ER stress exposure of a jamp-1(ok765) mutant. N2 (wild-type) and jamp-1(ok765) embryos were treated with indicated concentrations of tunicamycin, and developmental stages were assessed after a 72-h incubation on a bacterial lawn at 20°C. Animals were categorized as follows: adults and L4, arrested L3 and younger, and dead larvae. Each category is represented by the bar graph as the percentage of the total number of embryos (indicated above each bar). (B) Elevated basal expression of the hsp-4::gfp transcriptional reporter in jamp-1(ok765) worms. GFP fluorescence of control WT larvae and jamp-1(ok765) mutant carrying the hsp-4::gfp reporter. Both WT and mutant worms were grown in normal medium at 20°C. Wild-type (N2) larvae (top) exhibit low GFP levels. Autofluorescent granules in the cytoplasm of intestinal cells are shown (arrow). The jamp-1(ok765) larvae (bottom) show elevated fluorescence in intestine and exhibit high-GFP expression in anterior (AI) and posterior (PI) intestinal cells (arrows).
RESULTS
JAMP Is Localized in the ER and Associates with AAA-ATPase Components of the 19S Proteasomal Regulatory Complex
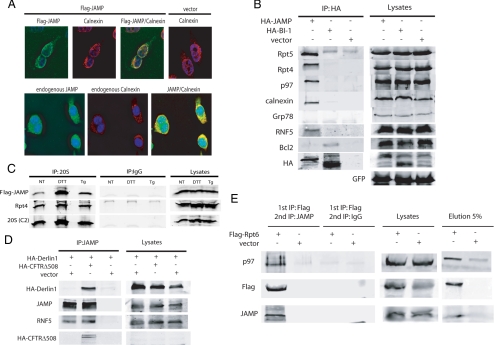
Immunostaining of exogenous as well as endogenous JAMP was consistent with ER localization, as shown by its colocalization with calnexin (Figure 1A). Consistent with these findings, membrane-floating experiments using sucrose density gradient ultracentrifugation also identified JAMP within the ER-enriched fractions (see data below). Two-dimensional PAGE of JAMP-associated proteins followed by mass spectrometry analysis identified several polypeptides as AAA-ATPase components of the proteasome (data not shown): among them were Sug1/Rpt6/S8 (which we will refer to as Rpt6) and Sug2/Rpt4/S10b (which we will refer to as Rpt4), both subunits of PA700, the 19S regulatory complex of the 26S proteasome (Rubin et al., 1996; Russell et al., 1996). Not all 19S cap components were found in this analysis as JAMP-associated protein either because of limited sensitivity or because only select set of components are required for JAMP association–dependent function on the proteasomes. Rpt4 and Rpt5 association with JAMP, but not with the six-transmembrane, ER-anchored protein BI-1 (Chae et al., 2004), was confirmed by IP (Figure 1B). Similarly, IP reactions performed following a protein extraction protocol that preserves the proteasome complex, revealed that JAMP associates with other AAA-ATPase components of the proteasome, including S4/Rpt2 and TBP1/Rpt5, and with the non-ATPase subunit p31/Rpn12 (Supplemental Figure S1). These data suggest that JAMP is an ER-localized protein associating with multiple components of the proteasome. Immunopurification of intact proteasome particles from cells confirmed association with JAMP under physiological conditions before and after ER stress (Figure 1C). These data suggest that JAMP associates with proteasomes under nonstress and after ER stress conditions.
Figure 1.
JAMP is a seven-transmembrane ER-resident protein that associates with the proteasome and ERAD components. (A) JAMP is localized in the ER. Exogenously expressed JAMP (Flag-JAMP) and endogenous JAMP were subjected to coimmunostaining in HeLa cells using anti-Flag (top panels) or anti-JAMP antibody (bottom panels). Endogenous calnexin was used as a marker for the ER. (B) JAMP interacts with endogenous proteasome subunits and different components of the ERAD. Cells were transfected with HA-JAMP (3 μg) or HA-BI1 (3 μg), and proteins prepared 36 h later using detergent-based extraction protocols to isolate membrane proteins (lysis buffer supplemented with 1% Triton X-100) were subjected to IP with antibodies to HA followed by immunoblotting using antibodies to endogenous Rpt5, Rpt4, p97, RNF5, calnexin, and Grp78. Bcl2 was used as a positive control for BI-1 interaction. (C) Association of JAMP with proteasome subunits. 293T cells transfected with Flag-JAMP (3 μg) were treated with DTT (10 mM) or thapsigargin (Tg; 0.5 μM). Nontreated cells (NT) were used as control. Proteins were prepared using extraction protocol without detergent to preserve proteasome complex (see Materials and Methods for details). Endogenous proteasomes were immunopurified with antibodies against the 20S C2 subunit and blotted for presence of Flag-JAMP. Association of Rpt4 with the 20S was used as a control for 19S and 20S binding and intact structure of proteasomes. (D) JAMP interaction with Derlin-1 increases in the presence of misfolded protein CFTRΔ508. HA-Derlin-1 was overexpressed with and without CFTRΔ508. After 36 h cells were lysed and subjected to immunoprecipitation with antibodies to JAMP (left). Immunoblots were carried out using the indicated antibodies. Right, expression of relevant proteins in lysates. GFP expression served as control for transfection efficiency. (E) JAMP forms a complex with the p97 chaperone and the 19S proteasome subunit Rpt6. Flag-Rpt6 or empty vector were transfected, and after 36 h cells were lysed and subjected to IP (1st IP) with Flag antibody in the presence of protein G beads. Immunoprecipitated protein complexes were eluted with Flag peptide (2 μg/μl at 4°C overnight), and eluates were subjected to IP (2nd IP) with JAMP antibodies followed by immunoblot analysis with the indicated antibodies.
JAMP Is a Component in a Multiprotein Complex with Regulatory Components of the ERAD
IPs also revealed that JAMP associates with important components of the ERAD machinery, including p97, Rpt4, Rpt5 and its cofactor Ufd1, and the E3 ubiquitin ligases RNF5 and gp78, but not with Grp78 (Figure 1B and Supplemental Figure S2 and data not shown). JAMP can also be coprecipitated with Sec61α and Derlin1, although the presence of misfolded protein CFTRΔ508, was required for JAMP-Derlin1 binding (Figure 1D and Supplemental Figure S2). These findings indicate that JAMP associates with a complex of ERAD regulatory components and that such association can be enhanced by the presence of misfolded proteins.
To determine whether JAMP and select ERAD and proteasome components are in a same complex, we performed sequential IP reactions. Immunoprecipitation of the proteasome subunit Rpt6 was followed by elution of the complex using Flag peptide, and eluted material was subjected to a second IP with antibodies to JAMP. Analysis of proteins co-IPed with JAMP (after initial IP by Rpt6) identified p97, suggesting that both Rpt6 and p97 are in a JAMP-associated protein complex (Figure 1E). Similarly, IP of exogenously expressed JAMP followed by elution of IPed material enabled the identification of Rpt6 and p97 among its associated components (data not shown). These findings suggest that JAMP is part of the ERAD system.
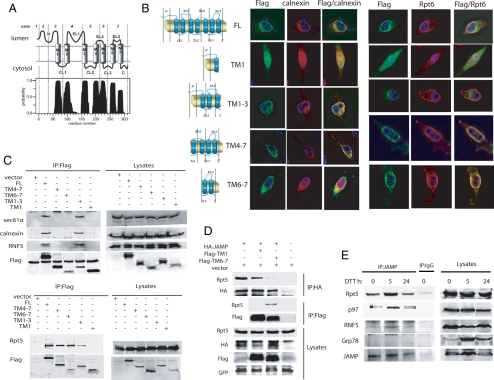
Based on topology predictions generated by the TMHMM software (http://www.cbs.dtu.dk/services/TMHMM; Krogh et al., 2001), the JAMP C-terminal domain should be localized in the cytosol, whereas the N-terminus should be in the ER lumen (Figure 2A). Consistent with this prediction, the JAMP N-terminus contains a glycosylation site (Kadoya et al., 2005). Accordingly, we generated JAMP fragments encompassing the N-terminus (TM 1), an extended N-terminus (including TM 1–3), the C-terminus (including TM 6–7), and an extended C-terminus (including TM domains 4–7), as illustrated in Figure 2B. Immunostaining revealed colocalization of full-length, TM 1–3, TM 4–7, and TM 6–7 with calnexin, indicating that N- and C-terminal fragments of JAMP are localized at the ER. However, TM 6–7 and TM 4–7 but not TM 1 or TM 1–3 colocalized with the proteasome subunit Rpt6 (Figure 2B). These data suggest that although JAMP C-terminal domains are localized primarily at the ER, they are required for colocalization with the proteasome subunits. Conversely, JAMP N-terminal domains do not associate with proteasome subunits or affect their localization, despite their localization at the ER. In agreement, the N-terminal fragment encompassing TM 1–3 could be coprecipitated with the ER-residing protein calnexin, the channel protein Sec61α, and the RNF5 ubiquitin ligase (Figure 2C). In contrast, C-terminal fragments encompassing TM 4–7 and TM 6–7 and, to a lesser degree the N-terminal fragment TM 1–3, but not TM 1, interacted with proteasome components, represented by Rpt5 (Figure 2C). Further, IP reactions confirmed that JAMP associates with Rpt5 through TM 6–7, because TM 6–7 overexpression, but not that of TM 1, inhibited association of JAMP with Rpt5 (Figure 2D). These data suggest that JAMP associates with proteasome components through its C-terminal domain, consistent with the prediction that its C-terminus is cytoplasmic.
Figure 2.
JAMP is an ER stress–inducible protein that associates with ERAD and proteasome components using N- and C-terminal domains, respectively. (A) TMHMM-based prediction for the membrane organization of JAMP. Possible position of JAMP C- and N-terminal domains within the cytosol and lumen, respectively, is outlined based on the TMHMM program. (B) Colocalization of the C-ter and N-Ter fragments of JAMP with ER and proteasome components. Truncated fragments representing different domains of JAMP (schematic outline on the left) were generated based on structural prediction and assessed for colocalization with endogenous calnexin or Rpt6. Flag-tagged JAMP domains were transfected into HeLa cells, and immunostaining was performed 24 h later with the indicated antibodies. (C) The C-terminal of JAMP binds proteasome subunits, whereas the N-terminal binds ERAD components. Deletion mutants of JAMP (Flag-tagged) were transfected into 293T cells, and 24 h later proteins were prepared and IPed with Flag antibodies. Immunoblots were carried out using the indicated antibodies against endogenous Rpt5, Sec61α, calnexin, and RNF5. (D) Overexpression of the JAMP C-terminal disrupts interaction between JAMP and proteasome subunits. HA-JAMP was cotransfected with Flag-TM 1, Flag-TM 6–7, or vector as control. After 36 h cells were lysed and subjected to IP with HA antibody after immunoblot analysis with Rpt5 and HA antibodies. Lysates were also subjected to parallel IP with Flag antibodies to compare interaction of Rpt5 with C-terminal (TM 6–7) versus N-terminal (TM1) JAMP. GFP expression served as control for transfection efficiency. (E) ER stress increases endogenous JAMP expression and its association with Rpt5, RNF5, and p97. 293T cells subjected to ER stress (10 mM DTT for 30 min), and proteins were prepared (lysis buffer supplemented with 1% Triton X-100) at the indicated time points. Left, IP with anti-JAMP antibody was followed by immunoblot analyses using indicated antibodies; right, total lysates.
Although JAMP protein is expressed under nonstress conditions, its expression increases transiently after exposure of cells to ER stress (Figure 2E, Supplemental Figure S3), with kinetics similar to grp78 (Figure 2E) and other ERAD regulatory components including EDEM and Derlin1, 2, and 3 (Oda et al., 2006). Corresponding with an increase in JAMP expression after ER stress is its association with p97, Rpt5, and RNF5 (Figure 2E). These data suggest that basal expression of JAMP is further induced by ER stress and that such an increase coincides with elevated JAMP association with ERAD components.
JAMP Expression Promotes Localization of Proteasome Subunits at the ER
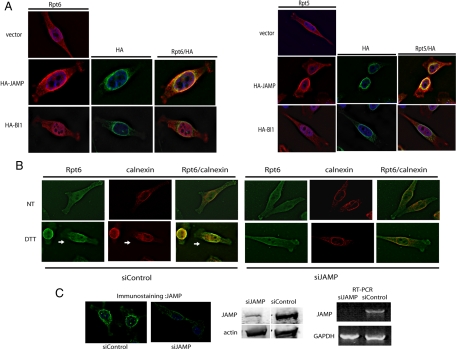
Immunostaining confirmed colocalization of JAMP with endogenous Rpt5 and Rpt6 (Figure 3A). Ectopically expressed JAMP also colocalized with other 26S proteasome components, including Rpt2 (PA700) and C-2 (20S proteasome subunit alpha type 1; data not shown). Significantly, although 19S proteasome AAA-ATPases exhibit a generally diffuse staining pattern, ectopic expression of JAMP promoted their colocalization with JAMP and the ER (Figure 3A; Supplemental Figures S4 and S5) but not with Golgi proteins (Supplemental Figure S6). Unlike the effect of JAMP, overexpression of the ER-residing six-transmembrane protein, BI-1, did not alter localization of proteasome components (Figure 3A). These observations suggest that JAMP expression promotes colocalization of proteasomes at the ER.
Figure 3.
JAMP localizes proteasome components to the proximity of the ER. (A) JAMP recruits proteasome components to the ER vicinity. HA-JAMP or HA-BI1 or empty vector construct (0.5 μg per well, in six-well plate) were transfected into HeLa cells, which were subjected to immunostaining 24 h later using antibodies to HA, Rpt6, or Rpt5. (B) siRNA of JAMP reduces localization of Rpt6 to the ER. Cell clones stably expressing siRNA-JAMP or control siRNA (see C for siRNA data) were assessed for changes in Rpt6 localization before and after ER stress. Colocalization of endogenous Rpt6 and calnexin was monitored before and 3 h after treatment with DTT (10 mM). Shown are representative figures from multiple fields in more than three experiments. (C) Stable inhibition of JAMP expression by siRNA. HeLa cells that stably express pRS-JAMP (siJAMP) or control scrambled siRNA sequence were selected and characterized for the degree of JAMP inhibition. Analysis was performed on clones transfected with different siRNA sequences against JAMP to eliminate off target effects (not shown). Selected clones were assessed for changes in JAMP expression compared with the control clone using immunostaining of endogenous JAMP (top panel), immunoblot analysis of endogenous JAMP protein levels (bottom left panel), and RT-PCR to assay JAMP mRNA levels (bottom right panel). At least one additional siRNA sequence was identified capable of eliciting similar inhibition of JAMP expression (not shown).
Confocal microscopy analysis confirmed colocalization of endogenous Rpt5 with Flag-JAMP (Supplemental Figure S5; compare nontransfected (arrow) with transfected (arrowhead) cells). Using the Manders coefficient algorithm (Manders et al., 1992), which allows quantification of the extent of signal overlap between green (Rpt5) and red (Flag-JAMP), the value calculated for JAMP-Rpt5 colocalization was significant (value = 0.7796, where 1.0 is the maximal value). Colocalization of JAMP with proteasome components was confirmed in microscopy analysis of multiple cells in >10 independent analyses. Collectively, these analyses establish colocalization of Flag-JAMP with proteasome subunits at the ER.
Significantly, ER stress promoted localization of Rpt6 (Figure 3B) and Rpt4 (data not shown) to the ER. However, such localization was impaired upon inhibition of JAMP expression by siRNA (Figure 3, B and C). These data suggest that JAMP expression, which is induced by ER stress, is important for localization of proteasome subunits at the ER.
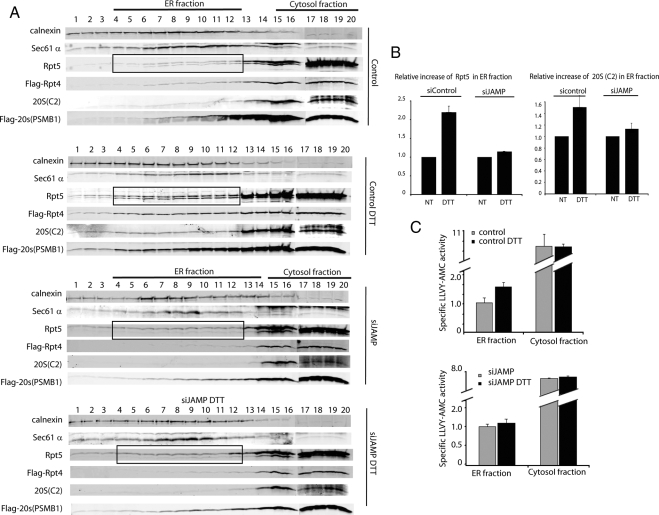
Biochemical analyses provided independent support for the effect of JAMP on localization of proteasome components at the ER. To this end, membrane floating (sucrose gradient-based fractionation) analysis was carried out on proteins prepared from cells subjected to ER stress in the presence of endogenous JAMP or siRNA to inhibit JAMP expression. Similar to effects observed after immunostaining, ER stress increased levels of Rpt4, Rpt5, and 20S (C2 and PSMB1) subunits in ER-enriched fractions (Figure 4A, quantification based on relative amounts of proteasome subunits to the ER-resident protein calnexin is shown in Figure 4B). Significantly, such increases were no longer observed after siRNA-mediated inhibition of JAMP expression (Figure 4, A and B). Analysis of proteasome activity revealed an increase in ER-enriched fractions (Figure 4C). These data suggest that JAMP facilitates the localization and activity of proteasomes at the ER in response to ER stress.
Figure 4.
JAMP is required for proteasome localization to the ER after ER stress. (A) ER stress induces JAMP-dependent recruitment of proteasome subunits to the ER compartment. HeLa cell clones expressing control scrambled siRNA or siRNA-JAMP were transfected with Flag-Rpt4 or Flag-20S (PSMB2) constructs. Cells were subjected to mock or DTT treatment, and 3 h later proteins were prepared and subjected to subcellular fractionation by membrane floatation technique. Fractions were collected and subjected to immunoblot analysis with antibodies to ER markers calnexin and translocon (Sec61α). Recruitment of endogenous as well as ectopically expressed proteasome subunits was monitored using antibodies to Tbp1/Rpt5, 20S (C2 subunit) and Flag antibodies (to 20S subunit PSMB1). Marked box highlights Rpt5 as a proteasome subunit affected by JAMP expression. (B) Quantification of proteins within ER-enriched fractions. Band intensity of endogenous proteasome subunits found within ER-enriched fractions before and after DTT treatment in JAMP-expressing or knockdown cells (shown in A) was quantified using the LICOR (Odyssey) scanner, which enables linear analysis of band intensity. Intensity was first adjusted to the background area of the blot and was normalized against the relative expression of the ER resident protein calnexin. The average intensity from fractions 4–12 (ER fraction) is shown for ER fraction. The average band intensity from nontreated cells is presented as 1.0 (100%) against average band intensity from ER fraction of DTT-treated cells. Error bars, SEs. (C) ER stress-induced proteasome activity is JAMP-dependent. ER-enriched fractions from experiments shown in A were assayed for proteasome activity using the fluorescence LLVY-AMC peptide (see Materials and Methods) in the presence and absence of MG132. Range of data shown was calculated based on activity assessed in the absence of MG132 minus the activity seen in the presence of MG132, as detailed in Materials and Methods. Data represents two independent analyses.
JAMP Facilitates Clearance of ER-bound Misfolded Proteins
JAMP-dependent increases in localization and activity of proteasomes at the ER after ER stress led us to ask whether JAMP also contributes to ERAD. To do so, we monitored changes in the stability of HA-TCRα and HA-CD3δ, as well as of the cystic fibrosis mutant protein CFTRΔ508, which are cleared by the ERAD machinery. Coexpression of JAMP with CD3δ or TCRα confirmed their association and markedly decreased the steady-state levels of CD3δ and TCRα (Supplemental Figure S7), The effect of JAMP on steady-state level of CD3δ was proteasome-dependent (Figure 5A). Comparing the effect of JAMP or its TM 1 or TM 6–7 fragments on CD3δ or NHK, using cycloheximide chase analysis, revealed that although full-length JAMP reduced the stability of CD3δ, it did not alter stability of the luminal misfolded protein NHK (Supplemental Figure S8). Interestingly, TM 6–7 expression delayed changes in the stability of CD3δ and NHK, probably due to competition for recruitment of proteasomes to the ER (Supplemental Figure S8). This finding implies that JAMP is effective toward membranal but not luminal misfolded proteins. Consistent with these observations, JAMP did not affect the steady-state levels of a cytosolic misfolded protein, CL-1 (Supplemental Figure S9).
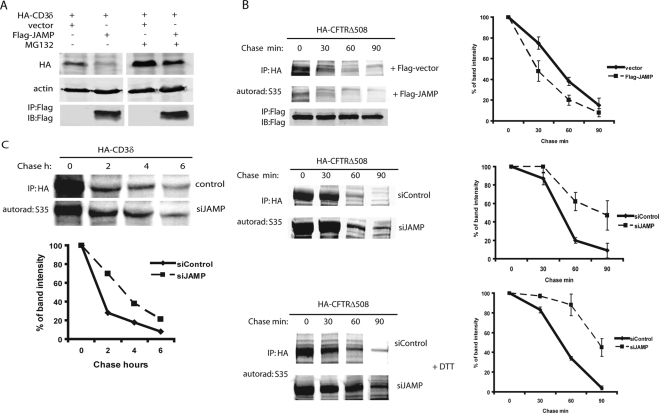
Figure 5.
JAMP promotes degradation of ERAD substrates and malfolded proteins. (A) JAMP promotes degradation of CD3δ in a proteasome-dependent manner. 293T cells were cotransfected with HA-CD3δ and Flag-JAMP (3:1 ratio) or empty vector. Cells were treated with MG132 (5 h; 40 μM) and subjected to immunoblot analysis as indicated. β-actin served as loading control. (B) JAMP expression facilitates degradation of misfolded proteins. Top, Overexpression of Flag-JAMP promotes degradation of the ERAD substrate CFTRΔ508. HeLa cells were cotransfected with HA-CFTRΔ508/vector, or CFTRΔ508/Flag-JAMP (ratio 1:1), and 24 h later cells were subjected to pulse-chase analysis using 35S-labeling (see Materials and Methods for details). Proteins collected at the indicated time points were subjected to IP with antibodies to HA (CFTRΔ508) followed by exposure to autoradiography and quantification by phosphorimager. Image shown represent example of autorad, and graphs the quantification of three experiments. Middle panel, siRNA of JAMP prolongs the half-life of CFTRΔ508. Experiment was performed as indicated above, except that cells that stably express siRNA JAMP (and control siRNA) were transfected with HA-CFTRΔ508 and subjected to pulse-chase analysis. Autorad depicts a representative analysis and graph represents three analyses which were quantified using phosphorimager. Bottom panel, siRNA of JAMP prolongs half-life of HA-CFTRΔ508 after ER stress. Experiment was performed as indicated above, except that cells were subjected to DTT treatment 3 h before the addition of 35S-labeled methionine/cysteine mix for pulse-chase analysis. Right panel depicts results of three analyses which were quantified using phosphorimager. (C) siRNA of JAMP prolongs the half-life of CD3δ. Experiment was performed as indicated in B, except that cells that stably express siRNA JAMP were transfected with CD3δ before subjected to pulse-chase analysis using 35S-labeled methionine/cysteine mix. Bottom panel depicts results of three analyses that were quantified using the phosphorimager.
Pulse-chase analysis using 35S labeling revealed that ectopic expression of JAMP effectively reduced the half-life of CFTRΔ508 from 50 to 30 min (Figure 5B, top). Conversely, cells stably expressing JAMP siRNA (Figure 3D) exhibited increased CFTRΔ508 half-life from 45 to 90 min (Figure 5B, middle). CFTRΔ508 stability also increased after ER stress of cells expressing JAMP siRNA (from 50 to 85 min; Figure 5B, bottom). Similarly, inhibition of JAMP expression also prolonged the half-life of CD3δ (Figure 5C). Independent analyses using cycloheximide chase confirmed that siRNA of JAMP prolonged the half-life of CFTRΔ508 (Supplemental Figure S10) and CD3δ (Supplemental Figure S11). These findings suggest that through its ability to accelerate degradation of misfolded proteins, JAMP serves to facilitate clearance of misfolded proteins.
Because JAMP can be found in a complex with ERAD components and contributes to accelerated degradation of misfolded proteins, we asked whether JAMP expression might affect ER stress or UPR, which may explain some changes noted above. JAMP overexpression did not appear to affect ER stress and UPR markers, including Grp78, CHOP, and p58IPK (Supplemental Figure S12). Further, the half-life of endogenous JAMP, estimated around 3 h (Supplemental Figure S13) is not affected by ERAD regulatory components, such as p97 (Supplemental Figure S14) or RNF5 (data not shown). Further, because JAMP degradation is not affected by dominant negative form of p97, we suggest that JAMP is not an ERAD substrate, although it is a regulatory component in the ERAD that is subject to degradation upon completion of its role in ERAD. Similarly, mannosidase, a component required for glycoprotein ERAD, is rapidly turned over in the lysosome (Wu et al., 2007; Ushioda et al., 2008).
Depletion of JAMP in C. elegans Causes ER Stress and Impairs Development in Response to ER Stress
C. elegans mutants in ERAD and UPR genes exhibit reduced survival and delayed development under ER stress conditions (Shen et al., 2001; Ye et al., 2004). We thus assessed changes in UPR and the ER-stress response in a C. elegans strain (jamp-1(ok765)) in which the JAMP orthologue is largely deleted, leaving only 41 aa of the N-terminus (see Materials and Methods for details). C. elegans JAMP exhibits ∼30% similarity with human JAMP. Analysis of the jamp-1(ok765) response to ER stress revealed that 65% of mutant worms (n = 479) exhibited growth arrest at or before the L3 stage, compared with 27% (n = 482) of wild-type (WT; N2) worms (Figure 6A). Of note, greater sensitivity was observed in response to low rather than high concentrations of tunicamycin, suggesting that JAMP is required for a physiological threshold response to ER stress, which may utilize different pathways compared with responses to more toxic doses (Shen et al., 2001). Further, expression levels of hsp-4::gfp, an indicator of UPR activation (Harding et al., 2002), were high in jamp-1(ok765) relative to WT worms maintained in normal growth medium (Figure 6B). Although restricted to the intestine, hsp-4:gfp expression was stronger in anterior and posterior intestinal cells (Figure 6B), and high rates of embryonic lethality accompanied by strong GFP expression were observed in a jamp-1(ok756);hsp-4::gfp strain (data not shown). Our findings are consistent with studies of C. elegans mutants in derlin, Sel1, or Abu1, which exhibit similar phenotypes (Ye et al., 2004).
DISCUSSION
Independent groups have reported possible recruitment of proteasome subunits to the ER (Kruger et al., 2001; Lee et al., 2004; Kalies et al., 2005; Okuda-Shimizu and Hendershot, 2007) and suggested that such recruitment may be required for efficient degradation of ER-exported ubiquitinated proteins. Here we demonstrate that JAMP is part of a complex that contains components that facilitate ERAD, in addition to specific proteasome subunits, which is required for efficient clearance of ER-residing misfolded proteins in mammalian cells. JAMP contribution to ERAD may not be global, because initial studies reveal that it does not affect degradation of ER-luminal substrates (ERAD-L) or cytosol-localized misfolded proteins. The effect on clearance of CFTRΔ508, CD3δ, and TCRα shown here implies that JAMP activity may be limited to substrates of ubiquitin ligases/chaperones with which it associates or to proteins positioned proximal to JAMP's location in the ER (i.e., ERAD-M; Carvalho et al., 2006).
The notion that JAMP does not serve all ERAD processes is consistent with the finding that JAMP associates with different channel proteins implicated in ERAD, including Sec61 and Derlin1 (Scott and Schekman, 2008). However, association with Derlin1 but not with Sec61 was seen only after expression of misfolded protein. These observations suggest that JAMP may serve diverse functions in the ER-stress response.
Originally we identified JAMP as an RNF5/RMA1-associated protein. RNF5/RMA1 is an ER membrane-anchored RING finger ubiquitin ligase regulating protein trafficking and degradation (Broday et al., 2004; Didier et al., 2003; our unpublished data). RNF5/RMA1 was also shown, together with the ubiquitin ligases CHIP or gp78, to mediate CFTRΔ508 ubiquitination, resulting in its degradation (Younger et al., 2006; Morito et al., 2008). JAMP associates with both RNF5/RMA1 and gp78 (data not shown), consistent with the cooperation reported for these ligases in ubiquitination of misfolded CFTR protein (Younger et al., 2006; Morito et al., 2008). Association of RNF5 with JAMP results in its ubiquitination but not degradation, suggesting regulation of JAMP's contribution to ERAD by RNF5 (our unpublished data).
We provide important support for the role of JAMP in ERAD by using a C. elegans strain harboring deletion of most of the jamp-1 gene (jamp-1(ok756). Exposure of these mutants to ER stress promotes developmental arrest at low concentrations of tunicamycin, suggesting that JAMP depletion causes endogenous ER stress during normal growth conditions that is exacerbated by drug treatment. The role of JAMP in causing stress and cell death is likely to also depend on cellular factors including type and degree of stress and its effect on proteaseome recruitment and/or control of JNK activity. This finding is further supported by observation of high basal levels of the UPR-inducible hsp-4 promoter::gfp reporter in jamp-1(ok756) mutant, findings consistent with observation of worms depleted of derlin, Sel1, or Ero1 (Ye et al., 2004).
Elevated ER stress and induction of UPR markers seen in C. elegans JAMP mutant support the notion that JAMP constitutes an important regulatory component of the ERAD system as part of the UPR. The finding that loss of JAMP increases ER stress and causes accumulation of misfolded and labile proteins seen in both human and worm systems indicates a critical role for JAMP in maintaining efficient clearance of misfolded proteins under normal physiological conditions. Thus, we conclude that JAMP is important to prevent ER stress, a function amplified in response to ER stress.
JNK, a JAMP-associated protein, is also an important regulatory component of ER stress and UPR (Urano et al., 2002). We have demonstrated that JAMP association with JNK prolongs JNK kinase activity (Kadoya et al., 2005). The possibility that JNK may phosphorylate JAMP or associated proteins at the ER is currently being investigated. It is expected that the type and degree of stress will dictate JAMP function in coordinated recruitment of proteasomes and/or effect on JNK-mediated apoptosis.
Given the growing evidence supporting redistribution of mammalian proteasomes during specific phases of cell cycle and also in response to cytokine stimuli (Amsterdam et al., 1993; Wilkinson et al., 1998; Brooks et al., 2000; Isono et al., 2007), it is possible that receptors for proteasome localization exist in mammalian cells and facilitate spatial organization of proteasomes to support timely cellular processes. We propose that JAMP contributes to proteasome localization at the ER as a mechanism for normal maintenance under nonstress conditions, and even more so after ER stress response, when JAMP levels temporarily increase to support the need for proper processing of ERAD.
In summary, this study identifies JAMP as a novel component of the ERAD machinery, which facilitates degradation of ERAD substrates by organizing proteasomes in the vicinity of the ER. JAMP thus emerges as part of the ERAD system, which marks, escorts, and exports ER-resident proteins destined for degradation. Collectively, our findings offer a new paradigm for regulation of proteasome localization and consequently ERAD efficiency.
Supplementary Material
ACKNOWLEDGMENTS
We thank Drs. Keiji Tanaka, Ron Kopito, Allan Weisman, Nico Dantuma (Karolinska Institute, Stockholm, Sweden), Maria Masucci (Karolinska Institute), Gergely Lukacs (The Hospital for Sick Children, Toronto, ON, Canada), Yihong Ye, Mei-Fan Chen (Burnham Institute for Medical Research, La Jolla, CA), H. H. Meyer (Institute of Biochemistry, Swiss Federal Institute of Technology Zurich, Zurich, Switzerland), and John Reed (Burnham Institute for Medical Research) for reagents used in this study. We thank Ed Mosonov for help with confocal microscopy and live cell imaging and Gal Zaks for technical assistance. We also thank the Caenorhabditis Genetic Center (University of Minnesota, Minneapolis, MN) for providing strains used in this study. We thank Drs. Alfred Goldberg, Francesca Marassi, and members of the Ronai lab for advice and discussions. Support from National Cancer Institute Grant CA097105 to Z.R. and Israel Science Foundation Grant ISF 980/06 to L.B. is gratefully acknowledged.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-08-0839) on September 10, 2008.
REFERENCES
- Amsterdam A., Pitzer F., Baumeister W. Changes in intracellular localization of proteasomes in immortalized ovarian granulosa cells during mitosis associated with a role in cell cycle control. Proc. Natl. Acad. Sci. USA. 1993;90:99–103. doi: 10.1073/pnas.90.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broday L., Kolotuev I., Didier C., Bhoumik A., Podbilewicz B., Ronai Z. The LIM domain protein UNC-95 is required for the assembly of muscle attachment structures and is regulated by the RING finger protein RNF-5 in C. elegans. J. Cell Biol. 2004;165:857–867. doi: 10.1083/jcb.200401133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky J. L. The protective and destructive roles played by molecular chaperones during ERAD (endoplasmic-reticulum-associated degradation) Biochem. J. 2007;404:353–363. doi: 10.1042/BJ20061890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks P., Fuertes G., Murray R. Z., Bose S., Knecht E., Rechsteiner M. C., Hendil K. B., Tanaka K., Dyson J., Rivett J. Subcellular localization of proteasomes and their regulatory complexes in mammalian cells. Biochem. J. 2000;346(Pt 1):155–161. [PMC free article] [PubMed] [Google Scholar]
- Carvalho P., Goder V., Rapoport T. A. Distinct ubiquitin-ligase complexes define convergent pathways for the degradation of ER proteins. Cell. 2006;126:361–373. doi: 10.1016/j.cell.2006.05.043. [DOI] [PubMed] [Google Scholar]
- Chae H. J., et al. BI-1 regulates an apoptosis pathway linked to endoplasmic reticulum stress. Mol. Cell. 2004;15:355–366. doi: 10.1016/j.molcel.2004.06.038. [DOI] [PubMed] [Google Scholar]
- DeLaBarre B., Christianson J. C., Kopito R. R., Brunger A. T. Central pore residues mediate the p97/VCP activity required for ERAD. Mol. Cell. 2006;22:451–462. doi: 10.1016/j.molcel.2006.03.036. [DOI] [PubMed] [Google Scholar]
- Delaunay A., et al. The ER-bound RING finger protein 5 (RNF5/RMA1) causes degenerative myopathy in transgenic mice and is deregulated in inclusion body myositis. PloS ONE. 2008;3:e1609. doi: 10.1371/journal.pone.0001609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didier C., et al. RNF5, a RING finger protein that regulates cell motility by targeting paxillin ubiquitination and altered localization. Mol. Cell. Biol. 2003;23:5331–5345. doi: 10.1128/MCB.23.15.5331-5345.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdjument-Bromage H., Lui M., Lacomis L., Grewal A., Annan R. S., McNulty D. E., Carr S. A., Tempst P. Examination of micro-tip reversed-phase liquid chromatographic extraction of peptide pools for mass spectrometric analysis. J. Chromatogr. A. 1998;826:167–181. doi: 10.1016/s0021-9673(98)00705-5. [DOI] [PubMed] [Google Scholar]
- Fang S., Ferrone M., Yang C., Jensen J. P., Tiwari S., Weissman A. M. The tumor autocrine motility factor receptor, gp78, is a ubiquitin protein ligase implicated in degradation from the endoplasmic reticulum. Proc. Natl. Acad. Sci. USA. 2001;98:14422–14427. doi: 10.1073/pnas.251401598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampton R. Y. ER-associated degradation in protein quality control and cellular regulation. Curr. Opin. Cell Biol. 2002;14:476–482. doi: 10.1016/s0955-0674(02)00358-7. [DOI] [PubMed] [Google Scholar]
- Harding H. P., Calfon M., Urano F., Novoa I., Ron D. Transcriptional and translational control in the Mammalian unfolded protein response. Annu. Rev. Cell Dev. Biol. 2002;18:575–599. doi: 10.1146/annurev.cellbio.18.011402.160624. [DOI] [PubMed] [Google Scholar]
- Isono E., Nishihara K., Saeki Y., Yashiroda H., Kamata N., Ge L., Ueda T., Kikuchi Y., Tanaka K., Nakano A., Toh-e A. The assembly pathway of the 19S regulatory particle of the yeast 26S proteasome. Mol. Biol. Cell. 2007;18:569–580. doi: 10.1091/mbc.E06-07-0635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadoya T., Khurana A., Tcherpakov M., Bromberg K. D., Didier C., Broday L., Asahara T., Bhoumik A., Ronai Z. JAMP, a Jun N-terminal kinase 1 (JNK1)-associated membrane protein, regulates duration of JNK activity. Mol. Cell. Biol. 2005;25:8619–8630. doi: 10.1128/MCB.25.19.8619-8630.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalies K. U., Allan S., Sergeyenko T., Kroger H., Romisch K. The protein translocation channel binds proteasomes to the endoplasmic reticulum membrane. EMBO J. 2005;24:2284–2293. doi: 10.1038/sj.emboj.7600731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh A., Larsson B., von Heijne G., Sonnhammer E. L. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- Kruger E., Kloetzel P. M., Enenkel C. 20S proteasome biogenesis. Biochimie. 2001;83:289–293. doi: 10.1016/s0300-9084(01)01241-x. [DOI] [PubMed] [Google Scholar]
- Lee R. J., Liu C. W., Harty C., McCracken A. A., Latterich M., Romisch K., DeMartino G. N., Thomas P. J., Brodsky J. L. Uncoupling retro-translocation and degradation in the ER-associated degradation of a soluble protein. EMBO J. 2004;23:2206–2215. doi: 10.1038/sj.emboj.7600232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manders E. M., Stap J., Brakenhoff G. J., van Driel R., Aten J. A. Dynamics of three-dimensional replication patterns during the S-phase, analysed by double labelling of DNA and confocal microscopy. J. Cell Sci. 1992;103(Pt 3):857–862. doi: 10.1242/jcs.103.3.857. [DOI] [PubMed] [Google Scholar]
- Morito D., Hirao K., Oda Y., Hosokawa N., Tokunaga F., Cyr D. M., Tanaka K., Iwai K., Nagata A. K. Gp78 cooperates with RMA1 in endoplasmic reticulum-associated degradation of CFTR{Delta}F508. Mol. Biol. Cell. 2008;19:1328–1336. doi: 10.1091/mbc.E07-06-0601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda Y., Okada T., Yoshida H., Kaufman R. J., Nagata K., Mori K. Derlin-2 and Derlin-3 are regulated by the mammalian unfolded protein response and are required for ER-associated degradation. J. Cell Biol. 2006;172:383–393. doi: 10.1083/jcb.200507057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuda-Shimizu Y., Hendershot L. M. Characterization of an ERAD pathway for nonglycosylated BiP substrates, which require Herp. Mol. Cell. 2007;28:544–554. doi: 10.1016/j.molcel.2007.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne A. R., Rapoport T. A., van den Berg B. Protein translocation by the Sec61/SecY channel. Annu. Rev. Cell Dev. Biol. 2005;21:529–550. doi: 10.1146/annurev.cellbio.21.012704.133214. [DOI] [PubMed] [Google Scholar]
- Pulak R., Anderson P. mRNA surveillance by the Caenorhabditis elegans smg genes. Genes Dev. 1993;7:1885–1897. doi: 10.1101/gad.7.10.1885. [DOI] [PubMed] [Google Scholar]
- Ron D., Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat. Rev. Mol. Cell Biol. 2007;8:519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- Rubin D. M., Coux O., Wefes I., Hengartner C., Young R. A., Goldberg A. L., Finley D. Identification of the gal4 suppressor Sug1 as a subunit of the yeast 26S proteasome. Nature. 1996;379:655–657. doi: 10.1038/379655a0. [DOI] [PubMed] [Google Scholar]
- Russell S. J., Sathyanarayana U. G., Johnston S. A. Isolation and characterization of SUG2. A novel ATPase family component of the yeast 26 S proteasome. J. Biol. Chem. 1996;271:32810–32817. doi: 10.1074/jbc.271.51.32810. [DOI] [PubMed] [Google Scholar]
- Shen X., et al. Complementary signaling pathways regulate the unfolded protein response and are required for C. elegans development. Cell. 2001;107:893–903. doi: 10.1016/s0092-8674(01)00612-2. [DOI] [PubMed] [Google Scholar]
- Scott D. C., Schekman R. Role of Sec61p in the ER-associated degradation of short-lived transmembrane proteins. J. Cell Biol. 2008;181:1095–1105. doi: 10.1083/jcb.200804053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urano F., Calfon M., Yoneda T., Yun C., Kiraly M., Clark S. G., Ron D. A survival pathway for Caenorhabditis elegans with a blocked unfolded protein response. J. Cell Biol. 2002;158:639–646. doi: 10.1083/jcb.200203086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ushioda R., Hoseki J., Araki K., Jansen G., Thomas D. Y., Nagata K. ERdj5 is required as a disulfide reductase for degradation of misfolded proteins in the ER. Science. 2008;321:569–572. doi: 10.1126/science.1159293. [DOI] [PubMed] [Google Scholar]
- Wilkinson C. R., Wallace M., Morphew M., Perry P., Allshire R., Javerzat J. P., McIntosh J. R., Gordon C. Localization of the 26S proteasome during mitosis and meiosis in fission yeast. EMBO J. 1998;17:6465–6476. doi: 10.1093/emboj/17.22.6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wójcik C., Rowicka M., Kudlicki A., Nowis D., McConnell E., Kujawa M., DeMartino G. N. Valosin-containing protein (p97) is a regulator of endoplasmic reticulum stress and of the degradation of N-end rule and ubiquitin-fusion degradation pathway substrates in mammalian cells. Mol. Biol. Cell. 2006;17:4606–4618. doi: 10.1091/mbc.E06-05-0432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Kaufman R. J. From acute ER stress to physiological roles of the Unfolded Protein Response. Cell Death Differ. 2006;13:374–384. doi: 10.1038/sj.cdd.4401840. [DOI] [PubMed] [Google Scholar]
- Wu Y., Termine D. J., Swulius M. T., Moremen K. W., Sifers R. N. Human endoplasmic reticulum mannosidase I is subject to regulated proteolysis. J. Biol. Chem. 2007;282:4841–4849. doi: 10.1074/jbc.M607156200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y., Shibata Y., Yun C., Ron D., Rapoport T. A. A membrane protein complex mediates retro-translocation from the ER lumen into the cytosol. Nature. 2004;429:841–847. doi: 10.1038/nature02656. [DOI] [PubMed] [Google Scholar]
- Younger J. M., Chen L., Ren H. Y., Rosser M. F., Turnbull E. L., Fan C. Y., Patterson C., Cyr D. M. Sequential quality-control checkpoints triage misfolded cystic fibrosis transmembrane conductance regulator. Cell. 2006;126:571–582. doi: 10.1016/j.cell.2006.06.041. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.