Abstract
Aberrant Wnt signal transduction is involved in many human diseases such as cancer and neurodegenerative disorders. The key effector protein of the canonical Wnt pathway is β-catenin, which functions with T-cell factor/lymphoid enhancer factor (TCF/LEF) to activate gene transcription that leads to expression of Wnt target genes. In this study we provide results obtained from a novel functional screen of a human brain cDNA library used to identify 63 genes that are putative negative Wnt regulators. These genes were divided into eight functional groups that include known canonical and noncanonical Wnt pathway components and genes that had not yet been assigned to the Wnt pathway. One of the groups, the presenilin-binding proteins, contains the modifier of cell adhesion (MOCA) gene. We show that MOCA is a novel inhibitor of Wnt/β-catenin signaling. MOCA forms a complex with β-catenin and inhibits transcription of known Wnt target genes. Epistasis experiments indicate that MOCA acts to reduce the levels of nuclear β-catenin, increase the levels of membrane-bound β-catenin, and enhances cell–cell adhesion. Therefore, our data indicate that MOCA is a novel Wnt negative regulator and demonstrate that this screening approach can be a rapid means for isolation of new Wnt regulators.
INTRODUCTION
Wnt proteins are a family of secreted glycoprotein ligands that initiate signaling pathways involved in fundamental cellular functions such as cell growth, differentiation, and polarity (Akiyama, 2000; Huelsken and Birchmeier, 2001; Moon et al., 2002; Logan and Nusse, 2004; Nelson and Nusse, 2004). When misregulated, the Wnt signaling pathway is involved in oncogenesis (Peifer and Polakis, 2000; Polakis, 2000; Logan and Nusse, 2004; Barker and Clevers, 2006). This signaling pathway is broadly divided into the canonical and noncanonical Wnt signaling pathways. The canonical pathway, which regulates the ability of the β-catenin protein to drive activation of specific target genes, is initiated after binding of Wnt to the receptor/coreceptor complex comprised of the seven transmembrane Frizzled (Fz) receptor and low-density lipoprotein receptor-related proteins (LRP) 5 or 6. This results in the formation of Dishevelled (Dvl)-Fz complexes, which in turn relocalize Axin and activate Dvl, leading to inactivation of glycogen synthase kinase (GSK)-3β (Akiyama, 2000; Huelsken and Birchmeier, 2001). This process causes β-catenin to accumulate in the cytoplasm and to enter the nucleus, where it interacts with members of the T-cell factor/lymphoid enhancer factor (TCF/LEF) family of transcription factors. The direct interaction of TCF/LEF factors with nuclear β-catenin relieves TCF/LEF from corepressors and converts the TCF proteins into potent transcriptional activators, ensuring expression and activation of Wnt target gene expression (Hurlstone and Clevers, 2002). In the absence of a Wnt signal, β-catenin associates in a “destruction complex” together with Axin, adenomatous polyposis coli (APC), GSK-3β, casein kinase I (CKI), FRAT and others. When this protein complex is assembled, β-catenin is actively degraded by the cellular proteasome pathway (Peifer and Polakis, 2000). The noncanonical, β-catenin-independent Wnt signaling pathway that primarily modulates cell movements is subdivided into at least two additional pathways. One is the planar cell polarity (PCP) pathway, which activates the c-Jun-N-terminal kinase (JNK) and, perhaps, small GTP-binding proteins. Also activated is the Wnt/Ca2+ pathway: Wnt and Frizzled homologues can activate calcium/calmodulin-dependent kinase II (CamKII) and protein kinase C (PKC; Kohn and Moon, 2005; Widelitz, 2005).
The aberrant activation of the Wnt/β-catenin pathway has a well-established role in driving cancer progression (Barker and Clevers, 2006). This signaling pathway also plays a key role in diverse aspects of neurodevelopment by regulating axon guidance, dendritic development, axon remodeling, and synapse formation (reviewed in Wilson and Houart, 2004; Ciani and Salinas, 2005), and aberrant Wnt signaling is associated with neurodegenerative pathological conditions such as autism, schizophrenia, and Alzheimer's disease (AD; De Ferrari and Moon, 2006).
To better understand the correlation between the Wnt signaling pathway and neurodevelopment and neurodegenerative disorders, we have screened a human embryonic brain cDNA library using a novel method based on the ability of cells to survive only when the Wnt pathway is turned off. Here we present evidence for the identification of a novel set of direct and indirect Wnt regulators. We have divided these genes into distinct functional groups and have demonstrated that the modifier of cell adhesion (MOCA) negatively regulates Wnt signaling. MOCA, also called presenilin (PS)-binding protein (PBP) and dedicator of cytokinesis 3 (DOCK3), is a member of the DOCK180 family of proteins known to be involved in regulation of cytoskeletal organization and cell–cell interactions (Kashiwa et al., 2000). MOCA has been linked to AD pathology, activates Rac1, alters NIH3T3 cells morphology, and regulates N-cadherin–dependent adhesion (Namekata et al., 2004; Chen et al., 2005). In this study, we show for the first time that MOCA exists in a complex with β-catenin, represses the canonical Wnt pathway, decreases cell migration, and increases cell adhesion.
MATERIALS AND METHODS
Plasmids and Library Construction
The expression plasmid TCF/HSV-TK, which contains the herpes simplex virus thymidine kinase gene (HSV-TK) under the control of three TCF-binding sites, was constructed using the pEGFP-C2 (Clontech, Palo Alto, CA) plasmid as a backbone. The HSV-TK sequence was cloned using HindIII and BamHI restriction sites downstream of the TCF-binding sites that were inserted using the AseI and HindIII sites. The blasticidin-resistance gene was added using the AseI site. An oligo dT cDNA expression library made from embryonic human brain RNA (Invitrogen, Carlsbad, CA) was size selected and the 1.2–5.0-kb fraction was inserted into the episomal pREP4 vector (Invitrogen). The cDNA expression library used for the screen contained 2 × 107 independent inserts.
The mouse Axin coding region was inserted into the pREP4 using XhoI and BglII. The human wild-type β-catenin coding region was inserted into the pEGFP-C2 (Clontech) plasmid. A short-hairpin RNA construct was generated using the plasmid vector pSuper.retro.puro (OligoEngine, Seattle, WA). The targeting sequence for MOCA was 5′-GCAAGTAGTTGGAGCCTGTAA-3′. The Wnt-responsive, TCF-dependent luciferase constructs, pTOPFLASH and its mutated version pFOPFLASH and FLAG-tagged β-catenin (ΔS45, in which Ser45 is deleted), were kindly provided by Dr. H. Clevers (Utrecht University, Utrecht, The Netherlands) and have been described previously (Morin et al., 1997). Human Frizzled 1 (HFz1) described previously (Gazit et al., 1999) was kindly provided by Dr. S. A. Aaronson (Mount Sinai Medical Center, NY). FLAG-tagged Dvl and FLAG-tagged FRAT were kindly provided by Dr. A. Gazit (Tel Aviv University, Israel). FLAG-tagged GSK3-β was a kind gift from Dr. H. Eldar-Finkelman (Tel Aviv University). Wnt-3a-HA and pCMV/β-galactosidase (β-Gal) expression plasmids, used to evaluate the efficiency of transfection, were purchased from Upstate Biotechnology (Lake Placid, NY) and Clontech, respectively. pCMV-Renilla used to evaluate transfection efficiency was purchased from Promega (Madison, WI). The pCis2/HA-tagged MOCA plasmid, pCis2 empty vector, the siRNA MOCA, and scrambled sequence (Chen et al., 2005) were kindly provided by Dr. D. Schubert (The Salk Institute for Biological Studies, La Jolla, CA), the cyclin D1/Luciferase reporter harboring the β-catenin-TCF response element driving luciferase expression was kindly provided by Drs. R. G. Pestell and C. Albanese (Albert Einstein College of Medicine, New York, NY). MG132 (Calbiochem, San Diego, CA) and hygromycin (Invivogen, San Diego, CA) were used at concentrations as indicated.
Cell Cultures, Transfections, and Luciferase Reporter Assays
Human embryonic kidney cell lines HEK293T, HEK293-EBNA (stably expressing the EBNA-1 gene; Invitrogen), L Wnt-3A cells (ATCC, Manassas, VA; no. CRL-2647), and colon carcinoma SW480 cells were cultured in DMEM supplemented with 10% fetal calf serum (FCS), and 100 U/ml penicillin/streptomycin. Undifferentiated P19 (mouse, C3H.He, teratocarcinoma) cells (ATCC; no. CRL-1825) were cultured in Alpha MEM supplemented with 5% FCS, and 100 U/ml penicillin/streptomycin. For neural differentiation, P19 cells were grown in Alpha MEM supplemented with 5% FCS and 1 μM retinoic acid (Sigma-Aldrich, St. Louis, MO) for 4 d and plated on poly-l-lysine (Sigma-Aldrich)-coated dished. Neural differentiated cells were then cultured in DMEM supplemented with 2.5% FCS and 2 mM glutamine (Biological Industries Israel, Beit Haemek, Israel). Cells were kept in a humidified 5% CO2 atmosphere at 37°C. For establishment of HEK293-EBNA cells stably expressing TCF/HSV-TK, cells were transfected with TCF/HSV-TK and selected by 10 μg/ml blasticidin (Invivogen). Different cell colonies stably expressing the TCF/HSV-TK (293EBNA-TK cells) plasmid were screened for their ability to specifically express the TK gene upon Wnt-3A activation.
For establishment of HEK293 cells stably expressing MOCA, cells were transfected with pCis2/HA-MOCA or with pCis2 as described previously (Chen et al., 2005) and selected by 800 μg/ml G418 (AG Scientific, San Diego, CA). SW480, P19, HEK293-EBNA, TCF/HSV-TK, and HEK293/MOCA cells were transfected using JetPEI (PolyPlus Transfection, New York, NY). For HEK293T cells, the standard CaPO4 precipitation method was used. For Wnt signaling reporter assays, HEK293T cells growing in 24-well dishes were transfected at 60–70% confluence with 0.2 μg of pTOPFLASH, 0.2 μg HFz1, 0.02 μg Wnt-3A-HA, 0.02 μg β-Gal and 0.2 μg of isolated plasmid (or Prep4 empty vector as a control), or various DNA concentrations of pCis2/HA-MOCA. Forty-eight hours after transfection, the luciferase levels were measured using a luciferase assay kit (Promega). For Wnt signaling reporter assays with siMOCA, HEK293/MOCA cells grown in 35-mm dishes were transfected with 1 μg of pTOPFLASH, 1 μg HFz1, 0.1 μg Wnt-3A-HA, 0.1 μg β-Gal, and 8 μg of siMOCA or scrMOCA (Chen et al., 2005). Luciferase levels were measured 72 h after transfection. To confirm activation of TCF luciferase in P19 cells, cells grown in 35-mm dishes were transfected with 1 μg FLAG-tagged β-catenin (ΔS45) or empty vector, 1 μg of pTOPFLASH, and 0.1 μg pCMV-Renilla. Luciferase levels were measured 24 h after transfection. For TCF luciferase assay with siMOCA, P19 cells were transfected with 1 μg of pTOPFLASH, 0.1 μg pCMV-renilla, and 4 μg of siMOCA or scrMOCA (Chen et al., 2005) and 6 μg of pSuper/siMOCA or the appropriate pSuper/scrMOCA. Luciferase levels were measured 72 h after transfection of siMOCA constructs and 24 h after transfection of the pTOPFLASH plasmid. In all assays, FOPFLASH activity was measured by replacing the pTOPFLASH with pFOPFLASH under equivalent conditions of Wnt 3A-induced signaling. Data are presented as mean values and SDs for at least three independent experiments done in duplicate.
cDNA Library Amplification
Amplification of the plasmid library was done on selective agar plates to avoid disproportionate amplification. The previously transformed sample was diluted to a density of 2.2 × 104 colony-forming units/plate (in 0.4 ml LB/plate) and plated on 55 × 150-mm 2YT + Amp plates. Plates were incubated at 37°C for 17 h. Cells were harvested and combined from all plates for plasmid DNA isolation.
Functional pREP4/cDNA Library Screening
The cDNA expression library was transfected into 293EBNA-TK cells at 10 times the number of independent clones expressed in the library, a complexity that maximized the potential representation of the library genes. One plate of TCF/HSV-TK cells (3 × 106 cells/9-cm plate) was transfected with 10 μg of the library DNA. Twenty-four hours after transfection, cells were subdivided into 15 pools and plated on 9-cm plates. Twenty-four hours later, the growth medium was replaced with Wnt-3A–conditioned media (CM; Shibamoto et al., 1998) containing 3 μg/ml ganciclovir (Sigma-Aldrich). Two weeks later 350 μg/ml hygromycin B (Invivogen) was added to the growth medium. Plasmid DNA was recovered from cells that survived the selection using the Hirt's method as previously described (Chinsky and Soeiro, 1981). Control transfection colonies were fixed in cold methanol for 10 min followed by 10% Giemsa (Merck, Rahway, NJ) stain for 15 min.
Immunoprecipitation, Western Blot Analysis, and Cell Fractionation
For coimmunoprecipitations, HEK293T cells were cotransfected with 8 μg MOCA-hemagglutinin (HA) or HA-empty vector together with 7 μg β-cateninΔS45-FLAG or FLAG empty vector. Total cell lysates were prepared by solubilization in lysis buffer (150 mM NaCl, 50 mM Tris, pH 7.5, and 0.2% NP-40). Protein concentrations were determined by Coomassie Plus. Cell lysate (1.5 mg) was incubated with anti-FLAG M2-agarose affinity gel (Sigma-Aldrich) for 4 h at 4°C on a rocker platform. For coimmunoprecipitation of endogenous β-catenin, HEK293/MOCA, and HEK293/vector cells were lysed as described above. Precleared cell lysates (2 mg) were immunoprecipitated with 8 μg of HA-probe (Y-11) antibody (Santa Cruz Biotechnology, Santa Cruz, CA) overnight at 4°C. Protein A/G plus agarose beads (Santa Cruz Biotechnology) were added and incubated on a rocker platform at 4°C for 1 h. Beads were collected by centrifugation and washed three times in lysis buffer. Protein samples were analyzed by SDS-PAGE and transferred to nitrocellulose membranes, blocked with 5% low fat milk, and detected with the appropriate antibody.
To prepare membrane fractions, cells were washed in phosphate-buffered saline (PBS) and incubated for 5 min at room temperature in buffer S (0.25 M sucrose, 1 mM imidazole, and 5 mM MgCl2). The medium was aspirated and cells were harvested in buffer S containing 1 mM DTT and a cocktail of protease inhibitors, incubated on ice for 15 min, and then homogenized. Cells were centrifuged at 800 × g at 4°C to pellet nuclei, after which the supernatant was centrifuged at 4°C for 45 min at 100,000 × g in a Beckman TL-120.2 rotor (Hercules, CA). The supernatant (cytosol) was collected, and the remaining pellet (membrane fraction) was resuspended in buffer containing TNE (25 mM Tris, pH 7.4, 150 mM NaCl, 1% NP-40, 4 mM EDTA, 25 mM sodium fluoride, and 1 mM sodium orthovanadate) and sonicated. Protein concentrations from the cell fractions were determined using the Bio-Rad protein assay kit (Hercules, CA). For Western blot analysis cell lysates were prepared as described above for immunoprecipitates samples. Equal protein amounts were subjected to SDS-PAGE and transferred to nitrocellulose membranes. Filters were incubated with anti-HA rat mAb clone (3F10, Roche Diagnostics, Alameda, CA) diluted 1:2500, anti FLAG rabbit polyclonal antibody (Sigma-Aldrich) diluted 1:400, anti-β-catenin (BD Transduction Laboratories, Lexington, KY) diluted 1:5000, and anti-cyclin D1 clone (A-12) diluted 1:500 and anti-LEF 1 (H-70) diluted 1:300 (Santa Cruz Biotechnology). Anti-E-cadherin and anti-N-cadherin (BD Transduction Laboratories) were diluted 1:5000, whereas anti-GFP (green fluorescent protein) and anti-p120 (H-90; Santa Cruz Biotechnology) were diluted 1:500. β-actin (MP Biomedicals, Solon, OH), diluted 1:10,000, was used as a loading control. Horseradish peroxidase (HRP)-conjugated goat anti-rat antibody (Santa Cruz Biotechnology) and HRP goat anti-mouse and anti-rabbit antibodies (Jackson ImmunoResearch Laboratories, West Grove, PA) were used as secondary antibodies. Antibodies were visualized by enhanced chemiluminescence (Amersham Pharmacia, Piscataway, NJ).
RT-PCR
Total RNA was isolated from undifferentiated and neural differentiated P19 cells using Trireagent (Sigma-Aldrich) according to manufacturer's instructions. Total RNA from each sample (0.1–1 μg) was used to obtain the first-strand cDNA using SuperScript First-Strand Synthesis System for PCR (Invitrogen) according to manufacturer's protocol. The cDNA was used as a template for PCR using PCR ready mix (New England Biolabs, Ipswich, MA). The primers used for the PCR reactions were as follows: 5′CTGGATCCGGAAAATGGAG3′ (forward) and 5′ACTCGCTCAGCATCCTCTGT3′ (reverse) for the MOCA gene and 5′AGGCCAGACTTTGTTGGATT3′ (forward) 5′TTTGGCTTTTCCAGTTTCACT3′ (reverse) for HPRT gene. Amplification was performed at 94°C for 30 s, 57°C for 30 s, and 72°C for 1 min for 35 cycles. HPRT was used as an endogenous mRNA control. Data are presented as mean values and SDs for at least three independent experiments.
Immunofluorescence Staining
SW480 cells were transfected with pCis2/HA-tagged MOCA and 48 h later were fixed in 3.7% paraformaldehyde in PBS for 20 min at room temperature, permeabilized (0.1% triton in PBS) for 30 min, and blocked (1% BSA and 0.1% triton in PBS) for 1 h at room temperature. HEK293T cells transfected with pCis2/MOCA-HA were treated for 24 h with 20 mM LiCl (Sigma-Aldrich), and 24 h after transfection the cells were fixed as described above. HEK293/MOCA and HEK293/vector were grown on pre-coated poly-l-lysine coverslips and fixed (see above). Primary antibodies included mouse monoclonal anti-β-catenin, anti-E-cadherin, anti-N-cadherin (BD Transduction Laboratories) diluted 1:500, anti-HA rat mAb clone (3F10, Roche Diagnostics) diluted 1:300, active β-catenin (clone 8E7, Upstate Biotechnology) diluted 1:500, and anti-p120 (H-90, Santa Cruz Biotechnology) diluted 1:300. The cells were washed and exposed for 1 h to FITC-conjugated anti-mouse antibody (Sigma) and rhodamine anti-rat antibody (Molecular Probes, Eugene, OR). 4-6′ diamidino-2 phenylindole (DAPI, Sigma) was used to stain cell nuclei. Fixed cells were imaged in a Leica SP2 confocal microscope (Leica Microsystems, Bannockburn, IL).
Migration Assay
Cell migration was assayed in 24-well, 8-mm pore membrane Transwell cell culture chambers (Costar, Cambridge, MA). Cells (0.75 × 105) were seeded in the upper chamber in DMEM medium. Growing medium or Wnt 3A CM (diluted 1:2) was added to the lower chamber. Twenty hours later the cells were fixed in cold methanol and stained with Giemsa. Cells that had migrated to the lower surface of the membrane were visualized, and the intensity of the Giemsa stained cells was quantified by a computer-assisted densitometer (TINA 2.0c; Fuji BAS, Tokyo, Japan).
Statistical Analysis
Results were presented as mean ± SEM. Statistical analysis among groups was performed using Student's t test. p < 0.05 was regarded as statistically significant.
Database Search
Database homology searches were carried out using the National Center for Biotechnology Information blast server (http://www.ncbi.nlm.nih.gov/).
RESULTS
Functional Screen for Isolation of Wnt Signaling Inhibitors
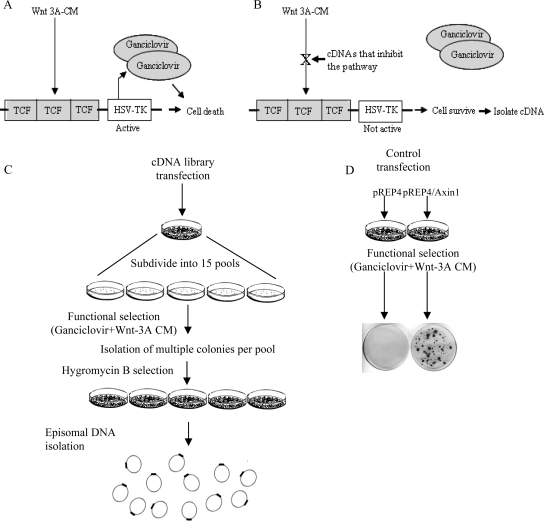
To generate a system for the identification of inhibitors of the Wnt signaling pathway (illustrated in Figure 1, A and B), a reporter plasmid carrying the HSV-TK coding region downstream of three consensus TCF-binding sites (TCF/HSV-TK) was constructed (see Material and Methods) and transfected into HEK293-EBNA cells. Cells that stably expressed this reporter plasmid were isolated. This cell line that expresses the EBNA-1 protein that drives episomal replication of ori-P–containing plasmids was termed 293EBNA-TK. Treatment of 293EBNA-TK cells with Wnt-3A CM resulted in expression of the HSV-TK gene. The HSV-TK enzyme converts ganciclovir to the phosphorylated form that can be incorporated into DNA, thereby blocking DNA synthesis and leading to cell death (Moolten, 1986). Previously it was shown that expression of the TK gene under control of a TCF-responsive promoter selectively kills cells treated with ganciclovir (Kwong et al., 2002). The feasibility of the system was confirmed: 293EBNA-TK cells were treated with Wnt-3A CM in the presence of ganciclovir and hygromycin B (a gene coding for hygromycin B resistance is expressed on the pREP4 vector). All cells died within 1 wk of treatment. In contrast, transfection of cells with a plasmid coding for Axin, a negative regulator of the Wnt pathway, allowed cell survival under the same conditions (Figure 1D). The Axin encoding plasmid was recovered from the cells (data not shown). This result suggests that negative regulators of the Wnt signaling pathway, such as Axin, can be isolated using this method.
Figure 1.
Schematic of the screening strategy. (A and B) Illustration of the basic screening strategy. This screen is based on activation of TCF target gene transcription by Wnt; expression of the TK gene converts ganciclovir into a toxic compound causing cell death. When a library member that inhibits Wnt signaling is expressed, TK gene expression is not induced and cells survive. (C) An episomal embryonic human brain cDNA library was transfected into 293EBNA-TK cells (see Materials and Methods). Forty-eight hours later, the cells were divided into 15 pools and were exposed to Wnt-3A CM and ganciclovir. Three to four weeks later, surviving cell colonies were isolated and grown in the presence of hygromycin B. Plasmid DNA was extracted from confluent plates using the Hirt's method. (D) As a proof of principle, 293EBNA-TK cells were transfected with an empty vector (pREP4) or an Axin1 expression vector (pREP4/Axin1). These cells were subjected to the same conditions as the library-transfected plates but instead of DNA extraction, the cells were fixed and stained with Giemsa.
Next, a human embryonic brain library was cloned into the pREP4 vector that contains an EBV oriP and EBNA-1, which allow for high-copy episomal replication of the plasmids in 293EBNA cells. The library was transfected into 293EBNA-TK cells. Colonies were isolated 3–4 wk after exposure to Wnt-3A CM and selection with ganciclovir and hygromycin B (Figure 1C). The plasmid DNA was extracted from cells that survived the selection using the Hirt's method. Extracted DNA was first analyzed using the Sau3A restriction enzyme. The restriction maps indicated that in most cases, the extracted DNA from a cell colony was uniform. However, in some cases a single cell colony expressed more than one plasmid (not shown). All plasmids were sequenced and the results are shown in Table 1.
Table 1.
Summary of identified genes in functional categories
| Screening ID | Accession no. | Gene | Repeats | Main function |
|---|---|---|---|---|
| Wnt canonical-associated genes | ||||
| 40 (1) | NM_180976 | Protein phosphatase 2, regulatory subunit B (B56) | 1 | Substrate specificity regulator of PP2A |
| 214 (1) | NM_019884 | Glycogen synthase kinase 3 alpha (GSK3A) | 1 | Serine/Threonine kinase |
| 208 | NM_015881 | Dickkopf homolog 3 (DKK3) | 1 | Wnt signaling antagonist |
| 506 | NM_004360 | E-cadherin (CDH1) | 1 | Cell adhesion |
| Wnt Ca2+ pathway–associated genes | ||||
| 50, 309 | NM_015981 | Calcium/calmodulin-dependent protein kinase (CAM Kinase) II | 2 | Ca2+and calmodulin-dependent serine/threonine protein kinase |
| 506 | NM_004360 | E-cadherin (CDH1) | 1 | Cell adhesion |
| Presenilin–associated genes | ||||
| 1 (5) | NM_001831 | Clusterin (CLU) | 1 | Chaparone protein, induces IκBα stabilization |
| 40 (2) | NM_015470 | Rab11 family interacting protein 5 (class 1) (Rab11-FIP5) | 1 | Vesicular transport |
| 223 (3), 231 | NM_001642 | Amyloid beta A4 precursor like protein 2 (APLP2) | 2 | Regulation of hemostasis. May interact with cellular G-protein signaling pathways |
| 215 (1), 252, 400 | NM_004947 | Modifier of cell adhesion (MOCA) or dictator of cytokinesis 3 (DOCK3) | 3 | Cell adhesion |
| 215 (2) | NM_004331 | Bcl2/adenovirus E1B 19-kDa interacting protein 3-like (BNIP3L) | 1 | Induces apoptosis, may function as a tumor suppressor |
| 242 | XM_001129341 | Bcl-2–associated transcription factor 4 (Btf) | 1 | Cell death |
| 269 (1) | NM_130442 | Engulfment and cell motility 1 (ELMO1) | 1 | Interactor of DOCK180 |
| 506 | NM_004360 | E-cadherin (CDH1) | 1 | Cell adhesion |
| Cell adhesion genes | ||||
| 1 (3) | NM_153831 | PTK2 or FAK tyrosin kinase | 1 | Tyrosin Kinase |
| 27, 67 | NM_139159 | Dipeptidyl peptidase 9 (DPP9) | 2 | Tissue remodeling |
| 214 (2) | NM_003385 | Visinin-like 1 (VSNL1) | 1 | Ca2+ sensor, regulation of cell adhesion and migration |
| 244 | NM_004356 | CD81 | 1 | Intergrin signaling |
| 506 | NM_004360 | E-cadherin (CDH1) | 1 | Cell adhesion |
| 247 (2) | NM_004819 | Symplekin (SYMPK) | 1 | A plaque protein |
| 320 | AK226066 | Lysosomal-associated membrane protein 2 (LAMP2) | 1 | Cell adhesion |
| Ca2+-binding proteins | ||||
| 7 (1), 73 | NM_001001331 | ATPase, Ca2+ transporting, plasma membrane 2 (ATP2B2) | 2 | Main activator of PMCA-mediated Ca2+ transport |
| 50, 309 | NM_015981 | Calcium/calmodulin-dependent protein kinase (CAM Kinase) II | 2 | Ca2+ and calmodulin-dependent serine/threonine protein kinase |
| 77 | NM_006888 | Calmodulun 1 (phosphorylase kinase delta) CALM1 | 1 | Developmental processes |
| 502 | NM_001743 | Calmodulin 2 (phosphorylase kinase delta) CALM2 | 1 | Developmental processes |
| 504 (1) | NM_021116 | Adenylate cyclase 1 (brain) (ADCY1) | 1 | Calmodulin-sensitive adenylyl cyclase |
| 19 | NM_014711 | CP110 protein | 1 | Calmodulin interactor, regulates cell cycle |
| 214 (2) | NM_003385 | Visinin-like 1 (VSNL1) | 1 | Ca2+ sensor, regulation of cell adhesion and migration |
| Trafficking proteins | ||||
| 1 (7) | NM_139345 | Bridging integrator 1 (BIN1) | 1 | Regulates synaptic vesicle endocytosis, actin cytoskeletal organization, transcription, and stress responses |
| 18 | NM_006650 | Complexin 2 (CPLX2) | 2 | Ca2+ triggered stabilizes trans-SNARE complex |
| 25 (5), 507 (2) | NM_001788 | Septin 7 (SEPT7) | 2 | Interacts with syntaxin-SNARE complex |
| 40 (2) | NM_015470 | Rab11 family interacting protein 5 (class 1) (Rab11-FIP5) | 1 | Vesicular transport |
| 215 (6) | NM_004311 | ADP-ribosylation factor-like 3 (ARL3) | 1 | Binds GTP, involves in cell signaling or vesicular transport machinery |
| 410 | NM_005639 | Synaptotagmin I (SYT1) | 1 | Ca2+ sensor, acts with complexin in fusion vesicles to membrane |
| Cytoskeleton-associated proteins | ||||
| 1 (1) | NM_020782 | Kelch domain containing 5 (KLHDC5) | 1 | Actin-binding protein |
| 1 (7) | NM_139345 | Bridging integrator 1 (BIN1) | 1 | Regulates synaptic vesicle endocytosis, actin cytoskeletal organization, transcription, and stress responses |
| 1 (9), 273, 83 (1), 269 (3), 275 | NM_006082 | Alpha-tubulin (K-TUBA-1) | 5 | Major constituent of microtubules |
| 310 |
NM_000428 NM_032035 |
Latent transforming growth factor beta binding protein 2 (LTBP2) | 1 | Structural role within elastic fibers |
| 319 (4) | NM_001024858 | Spectin beta erythrocytic (includes spherocytosis clinical type I) (SPTB) | 1 | Actin binding, structural constituent of cytoskeleton |
| Transcription regulatory proteins | ||||
| 219 | NM_006300 | Zinc finger protein 230 (ZNF230) | 1 | Putative transcriptional regulator |
| 504 (2) | NM_012102 | Arginine-glutamic acid dipeptide (RE) repeats (RERE) | 1 | Transcriptional repressor during development, overexpression triggers apoptosis |
| Other genes | ||||
| 7 (2) | NT_025741 | Chromosome 6 genomic contig (unknown) | 1 | Unknown function |
| 25 (3) | NM_024293 | Chromosome 2 open reading frame 17 (C2orf17) | 1 | Unknown function |
| 55 | XM_937098 | Endonuclease domain-containing 1 (ENDOD1) | 1 | Putative Dnase or Rnase |
| 25 (4) | NM_001247 | Ectonucleoside triphosphate diphosphohydrolase 6 nucleotidase (ENTPD6) | 1 | Putative NTPase |
| 28 (1) | NM_004171 | Solute carrier family 1, member 2 (SLC1A2) | 2 | Glutamate transport |
| 80 (1) | NM_013267 | Glutaminase (GLS2) | 1 | Regulation of glutamine catabolism |
| 82 | NM_014612 | Family with sequence similarity 120A (FAM120A) | 1 | Nuclease activity |
| 214 (3) | NM_030649 | Centaurin, beta 5 (CENTB5) | 1 | Regulation of GTPase activity |
| 220 (4) | NM_020132 | 1-Acylglycerol-3-phosphate O-acyltransferase 3 (AGPAT3) | 1 | Phospholipid metabolism |
| NM_031487 | ||||
| 220 (5) | NM_020378 | N-acetyltransferase 14 (NAT14) or Klp1 | 1 | Putative cell cycle regulation |
| 223 (1) | NM_013271 | Proprotein convertase subtilisin/kexin type 1 inhibitor (PCSK1N) | 1 | Processing of hormone and other protein precursors (PCSK1), PCSK1 inhibitor (PCSK1N) |
| 223 (2), 226 | NC_001807 | Mitochondrion, complete genome | 2 | |
| 224 (1) | NM_012247 | Selenophosphate synthetase 1 (SEPHS1) | 1 | Synthesizes selenophosphate from selenide and ATP, signal transduction |
| 224 (2) | NM_001025101 | Myelin basic protein (MBP) | 1 | A major constituent of the myelin sheath |
| 248 (1) | XM_939275 | SH2 domain-containing 5 (SH2D5) | 1 | Uncharacterized protein that has similar domain architecture to the Shc family of scaffolding proteins |
| 248 (2) | NM_001042492 | Neurofibromin 1 (NF1) | 1 | Tumor suppressor, GTPase-activating activity (GAP) toward RAS and cAMP-mediated signaling |
| 267 |
NM_174933 XM_372144 |
Phytanoyl-CoA dioxygenase domain-containing 1 (PHYHD1) | 1 | Oxidoreductase activity |
| 303 | NM_006516 | Solute carrier family 2, member 1 (SLC2A1) | 1 | Facilitative glucose transporter |
| 302, 305 | NM_014899 | Rho-related BTB domain-containing 3 (RHOBTB3) | 3 | May play as a scaffold, participating in diverse signal transduction cascades |
| 319 (1) | NM_014603 | Cerebellar degeneration-related protein 2-like (CDR2L) | 1 | Unknown function |
| 319 (3) | XM_941377 | KIAA0748 | 1 | Unknown function |
| 450 (1) | NM_024812 | Brain and acute leukemia, cytoplasmic (BAALC) | 2 | May play a synaptic role at the postsynaptic lipid rafts by interacting with CAMKIIA |
| 450 (3) | NT_025741 | Chromosome 6 genomic contig (unknown*) | 1 | Unknown function |
| 450 (4) | NM_198264 | Chromosome 1 open reading frame 2 (C1orf2) | 1 | Unknown function |
| 501 | DQ834604 | VN-24 NADH dehydrogenase subunit 2 (ND2) | 1 | Potassium transport |
| 503 | NM_002778 | Prosaposin (PSAP) | 1 | Signal transduction and cancer development |
| 505 (1) | NM_002948 | Ribosomal protein L15 (RPL15) | 1 | Component of ribosome. Secondary functions in DNA repair, apoptosis, drug resistance, and proliferation |
| 505 (2) | NM_006818 | Myeloid/lymphoid or mixed-lineage leukemia translocated to, 11 (MLLT11) | 1 | Unknown function |
| 507 (1) | NM_016065 | Mitochondrial ribosomal protein S16 (MRPS16) | 1 | Component of the mitochondrial ribosome small subunit (28S) |
Asterisk indicates gene as describe in Figure 2.
Functional Classification of Putative Wnt Inhibitors
Analysis of the putative Wnt signaling inhibitors revealed that the majority of these genes fall into distinct functional categories. As shown in Table 1 these genes can be divided into known canonical Wnt regulators GSK-3α (Clevers, 2006), PP2A (Li et al., 2001), DKK3 (Byun et al., 2005), E-cadherin (Orsulic et al., 1999), and noncanonical Wnt regulators (CAM kinase II; Montcouquiol et al., 2006). Other categories include PS-associated proteins (eight genes), cell adhesion proteins (seven genes), Ca2+-binding proteins (seven genes), trafficking proteins (six genes), cytoskeletal proteins (five genes), and transcription regulators (two genes). Some of these genes can be classified in more than one functional group. For example, BIN1 is both a trafficking-associated protein as well as an actin cytoskeleton-associated protein.
Secondary Screening of the Putative Wnt Inhibitors
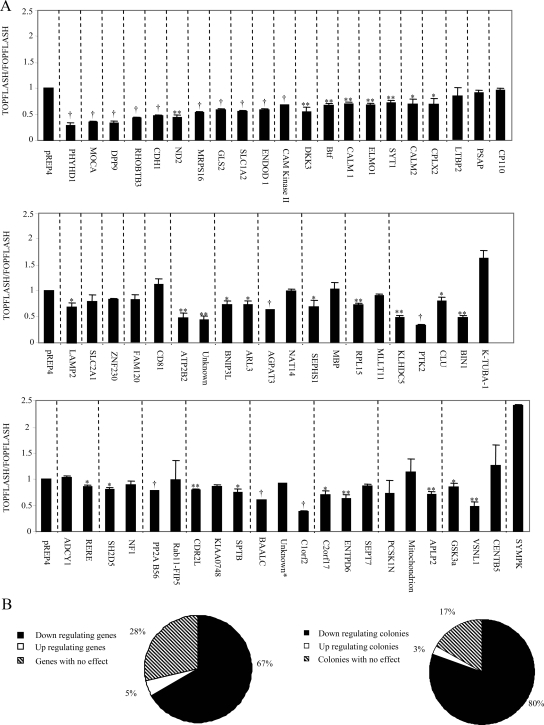
The screen described above was suitable for the first round of functional selection, because in this system a single genetic change was sufficient to yield a weak yet selectable phenotype of increased survival in a population of cells subjected to a long-term selection. However, because this characteristic of the system may also lead to a significant level of nonspecific background, cDNAs obtained in the first round of selection had to be individually tested by a secondary test. The pTOPFLASH plasmid contains wild-type TCF-binding sites fused upstream to a luciferase reporter gene and is widely used for measuring the ability of a given gene to specifically activate or inhibit the Wnt signaling pathway (Morin et al., 1997). The pFOPFLASH plasmid contains mutated TCF-binding sites and serves as a control for Wnt signal specificity. Therefore, this assay was used as a secondary screen to test the isolated cDNAs for their ability to specifically inhibit the Wnt signaling pathway.
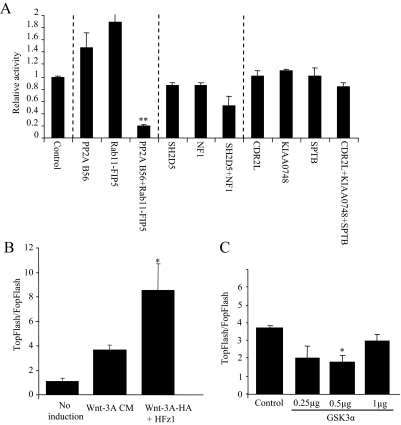
The pTOPFLASH or pFOPFLASH plasmids were cotransfected with plasmids expressing HFz1, HA-Wnt-3A, and individually isolated cDNA plasmids into HEK293T cells. The resulting luciferase levels were measured. As expected, most cDNAs decreased the Wnt signal by at least 20% (Figure 2). Some cDNA clones increased the Wnt signal and the rest did not specifically affect the TOPFLASH reporter plasmid activity (Figure 2). As our primary screen was aimed at identifying Wnt inhibitors, this secondary screen ensured robustness and specificity of the assay. The results from the secondary screen did not correspond exactly to the results from the primary screen. There are a number of reasons for these differences. First, a number of the cell colonies isolated in our primary screen contained more then one cDNA type (Table 1, Figure 2). When these cDNAs were expressed individually they did not decrease TOPFLASH values, but when the cDNAs were cotransfected (as in the primary screen), they synergistically inhibited the Wnt signal and reduced TOPFLASH levels (Figure 3A). Another difference between the screens is that under the primary screening conditions the cells were exposed to lower levels of Wnt signal for a longer time. As can be seen from Figure 3B, cells exposed to Wnt-3A CM (as in the primary screen) showed lower levels of Wnt signal compared with cells transfected with HFZ-1 and HA-Wnt-3A (as in the TOPFLASH screen). Finally, the effect on Wnt signaling may also depend on the gene dosage. For example, GSK-3α, which was isolated in our screen, more strongly inhibited the Wnt signal at the lower DNA concentrations (Figure 3C).
Figure 2.
Brain cDNA library secondary screening results. (A) Individual plasmids (as indicated) obtained from the primary screen or pREP4 (control) were cotransfected with HA-Wnt-3A, HFz1, pTOPFLASH, or pFOPFLASH and β-Gal into HEK293T cells. Forty-eight hours later, luciferase and β-Gal activities were analyzed in cell lysates. Data are presented as the pTOPFLASH/pFOPFLASH mean values and with SDs for at least three independent experiments done in duplicate. Dashed lines group the cDNAs isolated from the same cell colony. (B) The affect on the Wnt pathway is also shown as a pie chart of the individual genes and as the genes isolated from the same cell colony. Inhibition is defined as 20% compared with control Strikes indicate significance difference from control empty vector in Wnt signaling levels (*p < 0.05, **p < 0.01, †p < 0.001, Student's t test).
Figure 3.
Differences between first and second screening results. (A) Inhibition of the Wnt signal induced by different genes coexpressed in clones from one cell. HEK293T cells were transfected with pTOPFLASH or pFOPFLASH, β-Gal, HA-Wnt-3A, HFz1, and empty vector or different cDNAs isolated after selection. The cDNAs were individually or cotransfected as indicated. Asterisks indicate significant difference from control (**p < 0.01). (B) Comparison of the Wnt signaling level in both screening methods. HEK293T cells were transiently transfected with pTOPFLASH or pFOPFLASH reporters and β-Gal as an internal control. Wnt signaling was induced by supplementing cells with Wnt-3A CM or, as a comparison, by transiently overexpressing HA-Wnt-3A and HFz1. Asterisks indicate a significant difference between the two methods of Wnt induction control (*p < 0.05). (C) Level of gene expression. HEK293T cells were transfected with the pTOPFLASH or pFOPFLASH reporters, β-Ga,l and increasing DNA concentrations of GSK3α. Wnt signaling was induced by supplementing cells with Wnt-3A CM. Cell lysates were measured for luciferase and β-Gal activities 72 h after transfection. Asterisk indicate a significant difference from control signal (*p < 0.05). The results represent average values of at least three independent experiments. Statistical analysis was performed using Student's t test.
MOCA: A Novel Wnt Regulator, Forms a Complex with β-Catenin
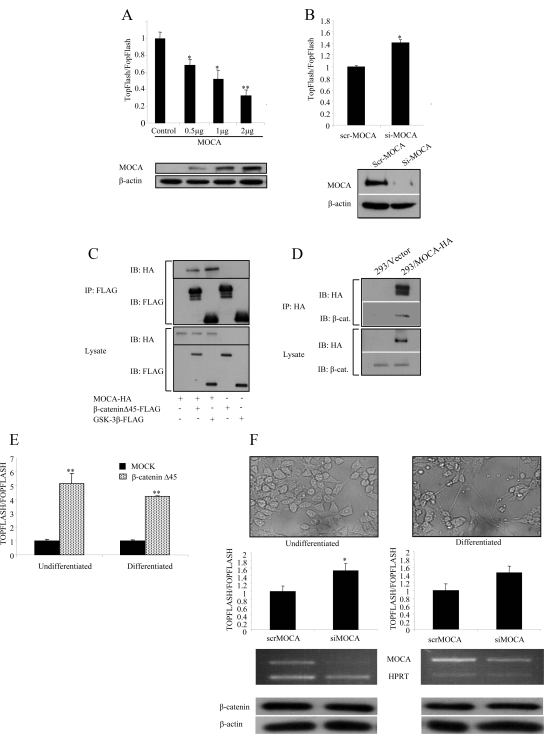
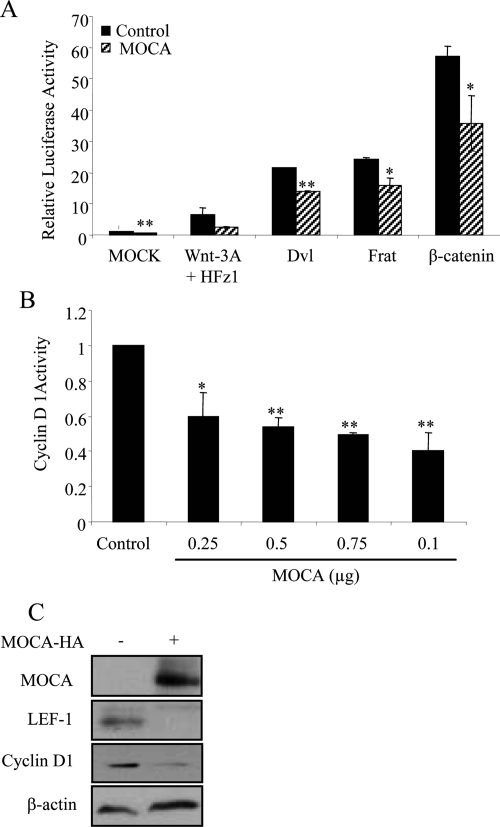
One of the genes identified in our screen was MOCA, a PS-binding protein. MOCA was selected three times. The full-length cDNA of MOCA (a generous gift from Dr. D. Schubert) was tested for its ability to specifically affect the Wnt pathway by using the TOPFLASH assay. The pTOPFLASH reporter plasmid was cotransfected with the plasmids expressing HA-Wnt-3A, HFz1, and HA-tagged MOCA (at various DNA concentrations) into HEK293T cells. HEK293 cells were used to evaluate the effect of MOCA on Wnt signaling, because they show all cell biological features of AD-associated proteins observed in primary neuronal cells (Capell, 1998). As shown in Figure 4A, transfection of MOCA resulted in a concentration-dependent decrease of the TOPFLASH activity. To confirm the specificity of the activity, the cells were transfected with pFOPFLASH. To further verify the specificity of the effect of MOCA on β-catenin signaling, RNA interference was used to inhibit MOCA expression in HEK293 cells that stably express MOCA (HEK293/MOCA). A small interfering RNA (siRNA) that was previously shown to specifically silence MOCA expression was used (Chen et al., 2005). As shown in Figure 4B, down-regulation of MOCA increased Wnt signaling levels compared with a control siRNA. Similar results were obtained using an additional siRNA targeting the MOCA transcript (described in Materials and Methods).
Figure 4.
MOCA forms a complex with β-catenin and inhibits the Wnt pathway. (A) HEK293T cells were transfected with pTOPFLASH or pFOPFLASH reporters, β-Gal, HA-Wnt-3A, HFz1, and with various concentrations of full-length HA-MOCA or empty vector. Luciferase and β-Gal activities were measured 48 h after transfection. The bottom panels show protein levels in transfected HEK293T cells determined using anti-HA antibodies. β-Actin served as a loading control. (B) HEK293/MOCA cells were transfected with the pTOPFLASH or pFOPFLASH reporters, β-Gal, HA-Wnt-3A, HFz1, and with either siRNA targeting MOCA (siMOCA) or scrambled siRNA (scrMOCA). Luciferase and β-Gal activities were measured 72 h after transfection. MOCA protein levels in siMOCA- or scrMOCA–transfected HEK293/MOCA cells were measured 72 h after transfection using an anti-HA antibody. (C) HEK293T cells were cotransfected with MOCA-HA, β-cateninΔS45-FLAG, FLAG-GSK-3β, or empty vector as indicated. Total cellular protein (1.5 μg) was immunoprecipitated with FLAG mAb, and the immunoprecipitate was analyzed by Western blotting using anti-HA and anti-FLAG antibodies. (D) 293/MOCA cells were cotransfected with MOCA-HA or empty vector as indicated. Total cellular protein (2 mg) was immunoprecipitated with an anti-HA antibody, and the immunoprecipitate was analyzed by Western blot using a β-catenin antibody. (E) To ensure that the Wnt signaling pathway can be transmitted in P19 cells, both undifferentiated and differentiated P19 cells were transfected with an active form of β-catenin, and the luciferase levels were measured 24 h later. (F) Using transient transfection of specific MOCA siRNA-encoding plasmids, the endogenous MOCA protein in both undifferentiated and differentiated P19 was silenced. Forty-eight hours later the cells were transfected with the reporter plasmids; 24 h after transfection, the levels of the luciferase activity were measured. The levels of endogenous MOCA and a housekeeping gene, HPRT, were measured using RT-PCR. Bottom panels, Western blot analysis of β-catenin expression levels. Actin served as a loading control. Asterisks indicate a significant difference from control signal (*p < 0.05; **p < 0.01). The results represent average values of at least three independent experiments. Statistical analysis was performed using Student's t test.
Next, we asked whether MOCA interacted in vivo with β-catenin, the core canonical Wnt signaling protein, and with GSK-3β, a member of the β-catenin degradation complex. HEK293T cells were cotransfected with constructs encoding HX-tagged MOCA and FLAG-tagged β-cateninΔS45, FLAG-tagged GSK-3β, or empty vector. As shown in Figure 4C, MOCA-HA specifically coimmunoprecipitated with β-cateninΔS45-FLAG and with FLAG-GSK-3β. Next, HEK293T cells stably expressing MOCA (HEK293/MOCA) were used. Figure 4D shows that immunoprecipitates of MOCA included endogenous β-catenin. These results suggest that MOCA interacts with β-catenin either directly or through another protein in a complex. As MOCA is mainly expressed in neuronal cells (although we have found MOCA to be expressed in other tissue types; data not shown), we examined whether MOCA can affect the Wnt pathway in neuronal cells. For these experiments we used P19 cells, a pluripotent stem cell line of mouse teratocarcinomas that can be induced to differentiate into neuronal cells in the presence of retinoic acid (Kotani et al., 2002). First we confirmed that transfection of a stabilized form of β-catenin activated the Wnt pathway in both undifferentiated and differentiated P19 cells (Figure 4E). Next, we depleted MOCA from both undifferentiated and differentiated P19 cells that were transfected with the TOP/FOPFLASH plasmids.
Results shown in Figure 4F demonstrate that depletion of MOCA induced TCF/β-catenin–dependent transcription although the total protein levels of β-catenin were not affected. Similar results were obtained using an additional siRNA targeting the MOCA sequence (data not shown).
MOCA Functions Downstream from β-Catenin to Represses Wnt Target Gene Expression
We conducted epistasis experiments, based on TOPFLASH assays, to identify the level within the Wnt signaling cascade at which MOCA acts. First, we examined cells transfected with plasmids encoding Wnt3A and HFz1, which stimulated TOPFLASH approximately eight times (Figure 5A). In cells overexpressing MOCA, the TOPFLASH values were reduced to ∼35% of the control. MOCA expression also reduced the TOPFLASH levels of cells expressing Dvl, FRAT, or β-catenin suggesting that MOCA acts after the stabilization of β-catenin in the pathway. The cyclin D1 gene is regulated by β-catenin and plays an important role in growth of many types of tumors (Donnellan and Chetty, 1998). To determine the effect of MOCA on β-catenin–dependent gene expression, a reporter construct containing the cyclin D1 promoter gene was cotransfected with HA-tagged MOCA into HEK293T cells. The expression of MOCA led to suppressed cyclin D1 promoter activity (Figure 5B). To test whether MOCA reduced expression of Wnt-responsive genes, the levels of endogenous cyclin D1 and LEF1 expression were measured in SW480 cells that overexpressed MOCA. LEF1 was chosen for this assay as previous work has shown that the cyclin D1 promoter contains a consensus LEF-binding site that represents the main contributor to β-catenin transactivation (Soriano et al., 2001). Indeed, Western blot analysis revealed that the MOCA-induced inhibition of the Wnt canonical pathway was accompanied by decreased levels of expression of both cyclin D1 and LEF1 in SW480 cells (Figure 5C). Together, these results suggest that MOCA functions downstream of β-catenin to represses Wnt target gene expression.
Figure 5.
Epistasis analysis of MOCA's function in the Wnt pathway and effect on Wnt target genes. (A) TOPFLASH/FOPFLASH assays in HEK293T cells transfected with MOCA and cotransfected with empty vector, Wnt 3A and HFz1, Dvl, FRAT, or β-catenin. (B) MOCA represses Wnt-3A induced activation of the cyclin D1 promoter. HEK293T cells were transfected with HA-Wnt-3A, HFz1, cyclin D1/luciferase, β-Gal, and with various concentrations of MOCA-HA or with an empty vector. Luciferase and β-Gal levels were measured 48 h after transfection. (C) MOCA represses cyclin D1 and LEF1 endogenous expression in SW480 cells. SW480 cells overexpressing MOCA were analyzed by Western blotting for cyclin D1 and LEF1 expression levels 72 h after transfection. β-Actin was used as a loading control. Asterisks indicate significant difference from control (empty vector) of Wnt signal induction (*p < 0.05, **p < 0.001, Student's t test).
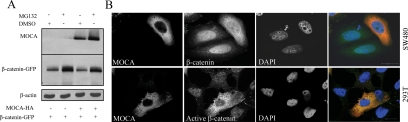
MOCA Does Not Affect the Stability of β-Catenin But Alters Its Subcellular Distribution
Because our results suggested that MOCA acts downstream from the β-catenin destruction complex, we examined whether MOCA alters the amount of the β-catenin protein at the proteasome level. For these experiments the proteasome inhibitor MG132 was used. HEK293T cells transfected with β-catenin and either MOCA or empty vector were treated with 50 μM MG132 for 6 h. Treatment of HEK293T cell transfectants with MG132 led to accumulation of the β-catenin protein; this accumulation of β-catenin was not affected by the expression of MOCA (Figure 6A). Aberrant Wnt signaling is known to be involved in the development of colorectal cancer (CRC). Therefore, to gain insight into the mechanism of the MOCA inhibitory effect on the Wnt canonical pathway, experiments were performed to examine whether the expression of MOCA affected the localization or abundance of endogenous β-catenin protein in the CRC cell line SW480. SW480 cells express a mutant APC gene, resulting in high levels of nuclear β-catenin (Rubinfeld et al., 1993). For this experiment SW480 cells were transfected with MOCA-HA and stained for both the HA-tagged MOCA and endogenous β-catenin. Untransfected SW480 cells showed strong, mostly nuclear staining of endogenous β-catenin, whereas MOCA transfectants expressed lower levels of nuclear β-catenin and higher levels of cytoplasmic β-catenin (Figure 6B). To further examine the effect of MOCA on the localization of β-catenin, HEK293T cells were incubated with LiCl, which leads to nuclear accumulation of active, dephosphorylated β-catenin that can be detected by using an antibody specific for the dephosphorylated form (dephospho-β-catenin antibody anti-ABC, clone 8E7). Dephosphorylated β-catenin transduces the Wnt signal (Staal et al., 2002; van Noort et al., 2002). As in SW480 cells, expression of MOCA in cells that express high levels of active β-catenin resulted in a decrease in nuclear levels of β-catenin and an increase in cytoplasmic levels of active (dephosphorylated) β-catenin. These data argue that MOCA does not affect the total amount of β-catenin but does affect the subcellular distribution of active β-catenin.
Figure 6.
MOCA does not affect stability of β-catenin but alters its subcellular distribution. (A) HEK293T cells transiently transfected with GFP–β-catenin were cotransfected with either HA-MOCA or empty vector. Forty hours later, the cells were treated with DMSO or MG132, a specific proteasome inhibitor. Six hours later, cell lysates were subjected to Western blot analysis using anti-GFP or anti-HA antibodies. β-Actin was used as a loading control. (B) SW480 cells (top panel) or LiCl-treated HEK293T cells (bottom panel) were transiently transfected with MOCA-HA and 48 h after transfection were subjected to immunofluorescence analysis with anti-β-catenin (SW480) or anti-dephospho-β-catenin (HEK293T) and anti-HA antibodies, followed by FITC-conjugated anti-mouse antibody and rhodamine anti-rat antibody, respectively. Green, β-catenin; red, HA. Images viewed represent 90% of MOCA-expressing cells.
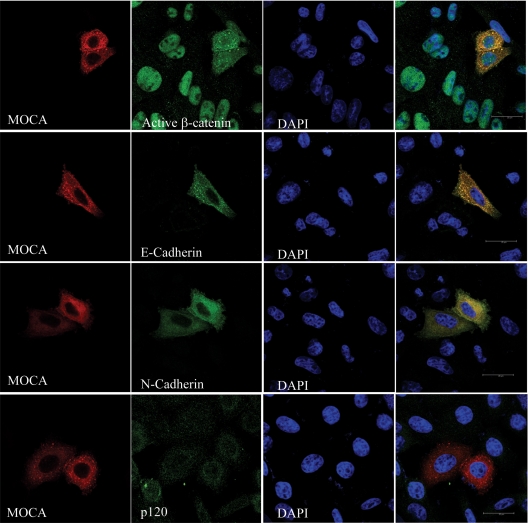
MOCA Increases Expression of Adhesion Proteins in SW480 Cells
As well as playing a central role in the canonical Wnt pathway, β-catenin is an integral component of adherence junctions, and the distribution of β-catenin between these two pools is well coordinated (Gavert and Ben-Ze'ev, 2007). The effect of MOCA on junction proteins was evaluated in SW480 cells. When grown in sparse cultures (subconfluent), SW480 cells do not form adherence junctions and express low levels of adherence proteins (Gavert and Ben-Ze'ev, 2007). Results shown in Figure 7 demonstrate that expression of MOCA in SW480 cells leads to a dramatic expression of both E- and N-cadherin. Expression of MOCA did not change the levels of expression of the p120 protein. As in the LiCl-treated HEK293T cells (Figure 6B), expression of MOCA led to reduced levels of active β-catenin in the nucleus. It is interesting to note that under these experiment conditions, MOCA colocalized with cytoplasmic active β-catenin and with E- and N-cadherin, indicating that these proteins may be part of the same protein complex.
Figure 7.
MOCA increases expression of adhesion proteins in SW480 cells. SW480 cells were transiently transfected with MOCA-HA and 48 h after transfection were subjected to immunofluorescence analysis using indicated antibodies. Secondary antibodies included FITC-conjugated anti-mouse antibody and rhodamine anti-rat antibody. DAPI staining was used to mark the nucleus.
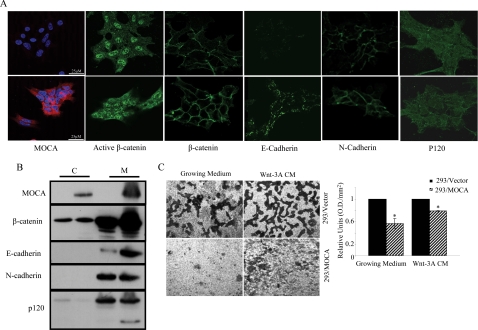
MOCA Induces Translocation of β-Catenin to the Membranes and Increases Cell Adhesion in HEK293T Cells
Our data indicating that MOCA both represses the canonical Wnt pathway and leads to expression of adhesion proteins suggested that MOCA may function by increasing the fraction of β-catenin bound to the membrane. To confirm this notion, we established HEK293T cells that stably express the MOCA protein (Figure 8A, first panel). These cells were stained for endogenous β-catenin, E- and N-cadherin, and p120. In concert with previously shown data (Chen et al., 2005) our results demonstrate that expression of MOCA dramatically increased the levels of E-cadherin and had a more moderate effect on the levels of β-catenin and N-cadherin expression (Figure 8A). Western blot analysis on cells fractionated into membrane and cytoplasmic pools confirmed this observation and showed that expression of MOCA led to elevated levels of membrane bound β-catenin (Figure 8B). It is interesting to note that although the total levels of p120 did not change as a result of the expression of MOCA, an additional, shorter p120 isoform was expressed under these conditions. It has been shown that association of the shorter p120 isoform with E-cadherin is correlated with strong cell–cell adhesion (Seidel et al., 2004). To test whether MOCA can enhance cell adhesion and as a result suppress cell migration, we performed migration assays. Both normal growing medium and Wnt3A-conditioned medium were used in Transwell chambers. We observed that under both of these conditions, cells migrated to the bottom chamber (Figure 8C). The Wnt3A CM had a more pronounced effect, as expected from its chemoattractant properties. This cell migration was inhibited, under both conditions, in cells expressing MOCA relative to untransduced cells.
Figure 8.
MOCA induces translocation of β-catenin to the membranes and inhibits cell migration in HEK293 cells. (A) HEK293/Vector or HEK293/MOCA cells were subjected to immunofluorescence analysis using indicated antibodies. Secondary antibodies included FITC-conjugated anti-mouse antibody and rhodamine anti-rat antibody. (B) Western blot analysis of cytoplasmic (C) and membrane (M) cell fractions were performed using indicated antibodies. (C) HEK293/Vector or HEK293/MOCA cells were seeded in the upper Transwell chamber. Cell migration to the lower chamber under both normal and Wnt 3A condition medium conditions was inhibited by the expression of MOCA. Quantification of the cell migration is shown in the bar graph. Asterisks indicate significant difference from control (*p < 0.05, Student's t test).
DISCUSSION
In the present study, we have taken advantage of our ability to recapitulate Wnt signaling induction under defined conditions to establish a novel functional screening system in mammalian cells that allows rapid identification of genes whose expression specifically inhibits the Wnt signaling pathway. This screening method was able to detect individual genes, as well as genes that act synergistically, to inhibit the Wnt pathway. We used a brain-derived cDNA library, and the positive clones identified in the screen resulted in a collection of eight gene groups and 63 individual genes that are Wnt inhibitors. The identified putative Wnt regulators were divided into several functional groups, including known Wnt regulators, PS-associated genes, cell adhesion genes, Ca2+-binding proteins, trafficking proteins, cytoskeleton-associated proteins, and transcription regulators.
The PS-binding protein MOCA was isolated independently from three different cell colonies in the screen. Presenilin 1 (PS1) and presenilin 2 (PS2) are polytopic membrane proteins; these genes are mutated in people who have AD (Sherrington et al., 1995), and several lines of evidence support a role for Wnt signaling in AD (De Ferrari and Moon, 2006). We demonstrated that MOCA forms a cellular complex with β-catenin and specifically inhibits the Wnt pathway in both epithelial and neuronal cell lines. Moreover, MOCA functions downstream of the β-catenin degradation complex in the Wnt signaling pathway to suppress Wnt target gene expression. Our results suggest that MOCA does not facilitate degradation of β-catenin, but rather decreases nuclear levels of active β-catenin. Expression of MOCA increased the expression of cell adhesion molecules and as a result enhanced cell adhesion. Moreover, MOCA colocalized with these adhesion molecules in cancer cells and induced the expression of a short p120 isoform that has been associated with enhanced cell adhesion. It has been shown that under conditions that favor cell–cell adhesion, PS1 stabilizes the E-cadherin–catenin adhesion complex (Baki et al., 2001). Combined with the present data, this suggests that MOCA and PS may function synergistically to enhance cell adhesion. Increased levels of β-catenin at adherence junctions results in a decrease in the soluble free cytoplasmic β-catenin that is a key regulator of the Wnt signaling pathway. Because β-catenin must translocate to the nucleus to activate expression of target genes (Polakis, 2000), MOCA may reduce expression of Wnt target genes by increasing the levels of membrane-bound β-catenin.
It is important to note that our results (Figures 6–8) demonstrate that MOCA is a cytoplasmatic and membrane-bound protein. On the other hand our epistasis experiments suggest that MOCA functions downstream or at the level of β-catenin. This could result from cytoplasmic sequestration of β-catenin by MOCA. Another possibility is that MOCA functions to down-regulate the Wnt signal by binding β-catenin in the cytoplasm and relocalizing β-catenin to the membrane. Other proteins that affect the Wnt pathway such as Axin, Dvl, and p120 have been shown to regulate the cellular distribution of β-catenin and in this way affect the signal (Cliffe et al., 2003; Spring et al., 2005; Bilic et al., 2007).
PSs interact with Wnt signaling components such as β-catenin (Tesco et al., 1998) and GSK-3β (Takashima et al., 1998). Both MOCA and β-catenin bind PS1, and the loss of both has been implicated in AD. Because our results do not show direct binding between β-catenin and MOCA, it is possible that this interaction is mediated through PS. It is interesting to note that AD-associated PS1 mutations increase the degradation of β-catenin in the cerebral cortex (Zhang et al., 1998) and that MOCA is expressed almost exclusively in the cerebral cortex and hippocampus, which are brain areas that are prone to nerve degeneration associated with AD (Kashiwa et al., 2000). All together, these data suggest that MOCA and PS may work together to regulate Wnt signaling involved in neurodevelopment and the pathogenesis of AD.
Wnt signaling is essential for many biological functions. Our data suggest that MOCA may be involved in these processes through regulation of Wnt signaling. Previous work (Chen et al., 2005), and this study show that expression of MOCA increased the levels of β-catenin in membranes of HEK293T cells. Moreover, it has been shown that β-catenin is stabilized after transport to the membrane (Chen et al., 2005) and that the binding of β-catenin to cadherins at the membranes leads to β-catenin stabilization (Nelson and Nusse, 2004). The two pools of β-catenin exert different functions: The membrane bound β-catenin is involved in cell–cell adhesion, whereas the cytosolic β-catenin is transported to the nucleus to activate Wnt target genes and promote cell proliferation. Therefore, by elevating the levels of membrane bound-β-catenin, MOCA may lead to a decrease in the levels of free β-catenin available for signaling, which, in turn, results in decreased expression of Wnt target genes. Thus, by regulating the localization of β-catenin and increasing cell–cell adhesion, MOCA plays an important role in determining cell fate.
In summary, we have identified a number of novel Wnt signaling inhibitors using a functional screen in mammalian cells. Although elucidation of the specific molecular connections among all our identified potential Wnt regulators and the Wnt signaling cascade is beyond the scope of this study, we have demonstrated a novel link between the Wnt signaling pathway and the MOCA protein. Because Wnt signaling is essential for normal development and pathogenesis of various cancers and neuronal diseases, the interaction between MOCA and the Wnt signaling pathway provides new insights into the molecular mechanisms of Wnt signaling and its associated cellular functions.
ACKNOWLEDGMENTS
We thank Dr. Arnona Gazit from the Department of Human Microbiology, Sackler School of Medicine, Tel Aviv University, Tel Aviv, Israel, and Qi Chen from the Cellular Neurobiology Laboratory, The Salk Institute for Biological Studies, La Jolla, CA, for critical reading of the manuscript. We are grateful to Drs. H. Clevers, S. A. Aaronson, D. Schubert, R. G. Pestell, and C. Albanese for kindly providing various reagents used in this study. This work was supported by the Israel Cancer Research Fund (ICRF); the Israel Science Foundation (ISF); the Israeli Ministry of Health, the Recanati Foundation, and the Adams Super Center for Brain Studies.
Abbreviations used:
- AD
Alzheimer's disease
- MOCA
modifier of cell adhesion
- PS
presenilin
- TCF/LEF
T-cell factor/lymphoid enhancer factor.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E07-10-1046) on August 20, 2008.
REFERENCES
- Akiyama T. Wnt/beta-catenin signaling. Cytokine Growth Factor Rev. 2000;11:273–282. doi: 10.1016/s1359-6101(00)00011-3. [DOI] [PubMed] [Google Scholar]
- Baki L., et al. Presenilin-1 binds cytoplasmic epithelial cadherin, inhibits cadherin/p120 association, and regulates stability and function of the cadherin/catenin adhesion complex. Proc. Natl. Acad. Sci. USA. 2001;98:2381–2386. doi: 10.1073/pnas.041603398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker N., Clevers H. Mining the Wnt pathway for cancer therapeutics. Nat. Rev. Drug Discov. 2006;5:997–1014. doi: 10.1038/nrd2154. [DOI] [PubMed] [Google Scholar]
- Bilic J., Huang Y. L., Davidson G., Zimmermann T., Cruciat C. M., Bienz M., Niehrs C. Wnt induces LRP6 signalosomes and promotes dishevelled-dependent LRP6 phosphorylation. Science. 2007;316:1619–1622. doi: 10.1126/science.1137065. [DOI] [PubMed] [Google Scholar]
- Byun T., Karimi M., Marsh J. L., Milovanovic T., Lin F., Holcombe R. F. Expression of secreted Wnt antagonists in gastrointestinal tissues: potential role in stem cell homeostasis. J. Clin. Pathol. 2005;58:515–519. doi: 10.1136/jcp.2004.018598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capell A., Grünberg J., Pesold B., Diehlmann A., Cintron M., Nixon R., Beyreuther K., Selkoe D. J., Haass C. The proteolytic fragments of the Alzheimer's disease-associated presenelin-1 form heterodimers and occur as a 100-150-kDa molecular mass complex. J. Biol. Chem. 1998;273:3205–3211. doi: 10.1074/jbc.273.6.3205. [DOI] [PubMed] [Google Scholar]
- Chen Q., Chen T. J., Letourneau P. C., Costa Lda F., Schubert D. Modifier of cell adhesion regulates N-cadherin-mediated cell-cell adhesion and neurite outgrowth. J. Neurosci. 2005;25:281–290. doi: 10.1523/JNEUROSCI.3692-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinsky J., Soeiro R. Fv-1 host restriction of Friend leukemia virus: analysis of unintegrated proviral DNA. J. Virol. 1981;40:45–55. doi: 10.1128/jvi.40.1.45-55.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciani L., Salinas P. C. WNTs in the vertebrate nervous system: from patterning to neuronal connectivity. Nat. Rev. Neurosci. 2005;6:351–362. doi: 10.1038/nrn1665. [DOI] [PubMed] [Google Scholar]
- Clevers H. Wnt/beta-catenin signaling in development and disease. Cell. 2006;127:469–480. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- Cliffe A., Hamada F., Bienz M. A role of Dishevelled in relocating Axin to the plasma membrane during wingless signaling. Curr. Biol. 2003;13:960–966. doi: 10.1016/s0960-9822(03)00370-1. [DOI] [PubMed] [Google Scholar]
- De Ferrari G. V., Moon R. T. The ups and downs of Wnt signaling in prevalent neurological disorders. Oncogene. 2006;25:7545–7553. doi: 10.1038/sj.onc.1210064. [DOI] [PubMed] [Google Scholar]
- Donnellan R., Chetty R. Cyclin D1 and human neoplasia. Mol. Pathol. 1998;51:1–7. doi: 10.1136/mp.51.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavert N., Ben-Ze'ev A. beta-Catenin signaling in biological control and cancer. J. Cell Biochem. 2007;102:820–828. doi: 10.1002/jcb.21505. [DOI] [PubMed] [Google Scholar]
- Gazit A., Yaniv A., Bafico A., Pramila T., Igarashi M., Kitajewski J., Aaronson S. A. Human frizzled 1 interacts with transforming Wnts to transduce a TCF dependent transcriptional response. Oncogene. 1999;18:5959–5966. doi: 10.1038/sj.onc.1202985. [DOI] [PubMed] [Google Scholar]
- Huelsken J., Birchmeier W. New aspects of Wnt signaling pathways in higher vertebrates. Curr. Opin. Genet. Dev. 2001;11:547–553. doi: 10.1016/s0959-437x(00)00231-8. [DOI] [PubMed] [Google Scholar]
- Hurlstone A., Clevers H. T-cell factors: turn-ons and turn-offs. EMBO J. 2002;21:2303–2311. doi: 10.1093/emboj/21.10.2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashiwa A., Yoshida H., Lee S., Paladino T., Liu Y., Chen Q., Dargusch R., Schubert D., Kimura H. Isolation and characterization of novel presenilin binding protein. J. Neurochem. 2000;75:109–116. doi: 10.1046/j.1471-4159.2000.0750109.x. [DOI] [PubMed] [Google Scholar]
- Kohn A. D., Moon R. T. Wnt and calcium signaling: beta-catenin-independent pathways. Cell Calcium. 2005;38:439–446. doi: 10.1016/j.ceca.2005.06.022. [DOI] [PubMed] [Google Scholar]
- Kotani M., Osanai T., Tajima Y., Kato H., Imada M., Kaneda H., Kubo H., Sakuraba H. Identification of neuronal cell lineage-specific molecules in the neuronal differentiation of P19 EC cells and mouse central nervous system. J. Neurosci. Res. 2002;67:595–606. doi: 10.1002/jnr.10150. [DOI] [PubMed] [Google Scholar]
- Kwong K. Y., Zou Y., Day C. P., Hung M. C. The suppression of colon cancer cell growth in nude mice by targeting beta-catenin/TCF pathway. Oncogene. 2002;21:8340–8346. doi: 10.1038/sj.onc.1206050. [DOI] [PubMed] [Google Scholar]
- Li X., Yost H. J., Virshup D. M., Seeling J. M. Protein phosphatase 2A and its B56 regulatory subunit inhibit Wnt signaling in. Xenopus. EMBO J. 2001;20:4122–4131. doi: 10.1093/emboj/20.15.4122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan C. Y., Nusse R. The Wnt signaling pathway in development and disease. Annu. Rev. Cell Dev. Biol. 2004;20:781–810. doi: 10.1146/annurev.cellbio.20.010403.113126. [DOI] [PubMed] [Google Scholar]
- Montcouquiol M., Crenshaw E. B., 3rd, Kelley M. W. Noncanonical Wnt signaling and neural polarity. Annu. Rev. Neurosci. 2006;29:363–386. doi: 10.1146/annurev.neuro.29.051605.112933. [DOI] [PubMed] [Google Scholar]
- Moolten F. L. Tumor chemosensitivity conferred by inserted herpes thymidine kinase genes: paradigm for a prospective cancer control strategy. Cancer Res. 1986;46:5276–5281. [PubMed] [Google Scholar]
- Moon R. T., Bowerman B., Boutros M., Perrimon N. The promise and perils of Wnt signaling through beta-catenin. Science. 2002;296:1644–1646. doi: 10.1126/science.1071549. [DOI] [PubMed] [Google Scholar]
- Morin P. J., Sparks A. B., Korinek V., Barker N., Clevers H., Vogelstein B., Kinzler K. W. Activation of beta-catenin-Tcf signaling in colon cancer by mutations in beta-catenin or APC. Science. 1997;275:1787–1790. doi: 10.1126/science.275.5307.1787. [DOI] [PubMed] [Google Scholar]
- Namekata K., Enokido Y., Iwasawa K., Kimura H. MOCA induces membrane spreading by activating Rac1. J. Biol. Chem. 2004;279:14331–14337. doi: 10.1074/jbc.M311275200. [DOI] [PubMed] [Google Scholar]
- Nelson W. J., Nusse R. Convergence of Wnt, beta-catenin, and cadherin pathways. Science. 2004;303:1483–1487. doi: 10.1126/science.1094291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orsulic S., Huber O., Aberle H., Arnold S., Kemler R. E-cadherin binding prevents beta-catenin nuclear localization and beta-catenin/LEF-1-mediated transactivation. J. Cell Sci. 1999;112(Pt 8):1237–1245. doi: 10.1242/jcs.112.8.1237. [DOI] [PubMed] [Google Scholar]
- Peifer M., Polakis P. Wnt signaling in oncogenesis and embryogenesis—a look outside the nucleus. Science. 2000;287:1606–1609. doi: 10.1126/science.287.5458.1606. [DOI] [PubMed] [Google Scholar]
- Polakis P. Wnt signaling and cancer. Genes Dev. 2000;14:1837–1851. [PubMed] [Google Scholar]
- Rubinfeld B., Souza B., Albert I., Muller O., Chamberlain S. H., Masiarz F. R., Munemitsu S., Polakis P. Association of the APC gene product with beta-catenin. Science. 1993;262:1731–1734. doi: 10.1126/science.8259518. [DOI] [PubMed] [Google Scholar]
- Seidel B., Braeg S., Adler G., Wedlich D., Menke A. E- and N-cadherin differ with respect to their associated p120ctn isoforms and their ability to suppress invasive growth in pancreatic cancer cells. Oncogene. 2004;23:5532–5542. doi: 10.1038/sj.onc.1207718. [DOI] [PubMed] [Google Scholar]
- Sherrington R., et al. Cloning of a gene bearing missense mutations in early-onset familial Alzheimer's disease. Nature. 1995;375:754–760. doi: 10.1038/375754a0. [DOI] [PubMed] [Google Scholar]
- Shibamoto S., Higano K., Takada R., Ito F., Takeichi M., Takada S. Cytoskeletal reorganization by soluble Wnt-3a protein signalling. Genes Cells. 1998;3:659–670. doi: 10.1046/j.1365-2443.1998.00221.x. [DOI] [PubMed] [Google Scholar]
- Soriano S., Kang D. E., Fu M., Pestell R., Chevallier N., Zheng H., Koo E. H. Presenilin 1 negatively regulates beta-catenin/T cell factor/lymphoid enhancer factor-1 signaling independently of beta-amyloid precursor protein and notch processing. J. Cell Biol. 2001;152:785–794. doi: 10.1083/jcb.152.4.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spring C. M., Kelly K. F., O'Kelly I., Graham M., Crawford H. C., Daniel J. M. The catenin p120ctn inhibits Kaiso-mediated transcriptional repression of the beta-catenin/TCF target gene matrilysin. Exp. Cell Res. 2005;305:253–265. doi: 10.1016/j.yexcr.2005.01.007. [DOI] [PubMed] [Google Scholar]
- Staal F. J., Noort Mv M., Strous G. J., Clevers H. C. Wnt signals are transmitted through N-terminally dephosphorylated beta-catenin. EMBO Rep. 2002;3:63–68. doi: 10.1093/embo-reports/kvf002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takashima A., et al. Presenilin 1 associates with glycogen synthase kinase-3beta and its substrate tau. Proc. Natl. Acad. Sci. USA. 1998;95:9637–9641. doi: 10.1073/pnas.95.16.9637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesco G., Kim T. W., Diehlmann A., Beyreuther K., Tanzi R. E. Abrogation of the presenilin 1/beta-catenin interaction and preservation of the heterodimeric presenilin 1 complex following caspase activation. J. Biol. Chem. 1998;273:33909–33914. doi: 10.1074/jbc.273.51.33909. [DOI] [PubMed] [Google Scholar]
- van Noort M., Meeldijk J., van der Zee R., Destree O., Clevers H. Wnt signaling controls the phosphorylation status of beta-catenin. J. Biol. Chem. 2002;277:17901–17905. doi: 10.1074/jbc.M111635200. [DOI] [PubMed] [Google Scholar]
- Widelitz R. Wnt signaling through canonical and non-canonical pathways: recent progress. Growth Factors. 2005;23:111–116. doi: 10.1080/08977190500125746. [DOI] [PubMed] [Google Scholar]
- Wilson S. W., Houart C. Early steps in the development of the forebrain. Dev. Cell. 2004;6:167–181. doi: 10.1016/s1534-5807(04)00027-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., et al. Destabilization of beta-catenin by mutations in presenilin-1 potentiates neuronal apoptosis. Nature. 1998;395:698–702. doi: 10.1038/27208. [DOI] [PubMed] [Google Scholar]