Abstract
The production of RanGTP around chromosomes is crucial for spindle microtubule assembly in mitosis. Previous work has shown that hepatoma up-regulated protein (HURP) is a Ran target, required for microtubule stabilization and spindle organization. Here we report a detailed analysis of HURP function in Xenopus laevis mitotic egg extracts. HURP depletion severely impairs bipolar spindle assembly around chromosomes: the few spindles that do form show a significant decrease in microtubule density at the spindle midzone. HURP depletion does not interfere with microtubule growth from purified centrosomes, but completely abolishes microtubule assembly induced by chromatin beads or RanGTP. Simultaneous depletion of the microtubule destabilizer MCAK with HURP does not rescue the phenotype, demonstrating that the effect of HURP is not to antagonize the destabilization activity of MCAK. Although the phenotype of HURP depletion closely resembles that reported for TPX2 depletion, we find no evidence that TPX2 and HURP physically interact or that they influence each other in their effects on spindle microtubules. Our data indicate that HURP and TPX2 have nonredundant functions essential for chromatin-induced microtubule assembly.
INTRODUCTION
The mitotic spindle segregates chromosomes during mitosis. It consists of dynamic microtubules (MTs) whose behavior is regulated by microtubule-associated proteins (MAPs) and motors. In most somatic animal cells, the assembly of a functional mitotic spindle results from the combination of two independent MT assembly processes: a centrosomal and a chromosomal pathway (reviewed in Walczak and Heald, 2008). The chromosomal pathway is crucial in cells lacking centrosomes, like plant cells and some female meiotic cells. The role of chromatin is exerted through the activation of the GTPase Ran (RanGTP) and its targets. RCC1, the exchange factor for Ran, is associated with chromosomes (Ohtsubo et al., 1989). This implies that RanGTP has a higher concentration near chromosomes, even after mitotic nuclear envelope disassembly (Kalab et al., 2002; Caudron et al., 2005; Kalab et al., 2006). MAPs subjected to Ran regulation are proposed to become locally activated, mostly by release from importin α and/or β (Gruss and Vernos, 2004) in the vicinity of chromosomes, i.e., where the spindle forms. In Xenopus egg extracts RanGTP has been shown to stimulate assembly of MTs and their self-organization into first aster-like structures then bipolar arrays, even in the absence of centrosomes and chromosomes (Carazo-Salas et al., 1999; Kalab et al., 1999; Ohba et al., 1999; Wilde and Zheng, 1999). Conversely, in the absence of RanGTP, mitotic spindles do not form in response to chromatin (Carazo-Salas et al., 1999).
One of the best characterized MAPs regulated by RanGTP is TPX2. This protein is required for chromatin-dependent MT assembly (Gruss et al., 2001, 2002; Schatz et al., 2003; Brunet et al., 2004) and is also involved later in spindle pole organization by targeting the motor Xklp2 to MTs (Wittmann et al., 2000). Hepatoma up-regulated protein (HURP), first identified as a cell cycle–regulated protein (Tsou et al., 2003), was recently reported to be a Ran target required for spindle formation based on studies of its expression profile (Wong and Fang, 2006), its identification as a spindle component (Sillje et al., 2006), and its presence in a complex required for the bipolarization of Ran spindles (Koffa et al., 2006). These analyses showed that HURP is critical for kinetochore fiber stabilization in human cells and thus for accurate chromosome alignment and segregation. The activity of HURP in spindle bipolarization in the Xenopus system depends on its assembly into a multicomponent complex, whose formation requires Aurora A kinase activity as well as MT binding (Koffa et al., 2006). HURP's binding to MTs was recently shown to require direct phosphorylation of the protein by Aurora A, an event that releases an inhibitory intra-HURP interaction (Wong et al., 2008). Analysis of HURP in total HeLa cell extracts (Wong et al., 2008) or in complete Xenopus egg extracts in the absence of MTs (see below) indicates that HURP also exists in a free state outside of the Aurora A–dependent complex.
Here, we characterize the free Xenopus HURP and report a novel HURP function. We show that HURP, independently of TPX2, is essential for chromatin- and RanGTP-dependent MT assembly. In contrast, it plays no role in centrosome-dependent MT nucleation and dynamics. Thus, HURP is a MAP required at several stages of Xenopus spindle assembly: first in early MT assembly mediated by RanGTP, then in MT stabilization, and finally, within the HURP-complex, in the organization of MTs into bipolar arrays.
MATERIALS AND METHODS
Expression and Purification of Recombinant Proteins
Full-length Xenopus laevis HURP (xlHURP) cDNA was assembled from two overlapping ESTs obtained from RZPD (Clone ID: IRAKp961L18169Q2 and IMAGp998O1015902Q1), cloned in a pET28 expression vector with an N-terminal 6-histidine tag. Expression in Escherichia coli (Rosetta strain, Novagen, Darmstadt, Germany): bacteria was induced with 0.2 mM IPTG and for 3 h at RT. Purification was performed under native conditions using nickel-NTA agarose beads (Qiagen, Hilden, Germany), according to standard protocols. xlHURP was eluted with 400 mM imidazole, dialyzed against 20 mM Tris-HCl (pH 7.5), 300 mM NaCl, and 8.7% glycerol.
Full-length xlHURP cDNA was also cloned into pFastBAC HTa (Invitrogen, Karlsruhe, Germany), expressed in Sf9 cells, purified according to standard protocols and used for polyclonal antibody production in rabbits.
RanQ69L and GFP-TPX2 expression and purification was described previously (Bischoff et al., 1994; Wittmann et al., 2000).
Antibodies
The C-terminal fragment of Xenopus tropicalis HURP (Koffa et al., 2006) and full-length xlHURP were used for rabbit polyclonal antibody production and purified against recombinant proteins according to standard protocols.
Rabbit polyclonal antibodies against TPX2, MCAK, and EB1 have been described (Niethammer et al., 2007; Yokoyama et al., 2008).
Xenopus Mitotic Extract Preparation, Immunodepletion, and Immunoprecipitation Assays
X. laevis cytostatic factor (CSF) metaphase-arrested extracts were prepared as described (Desai et al., 1999). To visualize MTs Cy3-labeled tubulin was added (0.2 mg/ml): tubulin was purified from pig brain (Castoldi and Popov, 2003) and labeled (Hyman et al., 1991).
For HURP depletion protein A–coated magnetic beads (Dynal, Hamburg, Germany) were used in a 1:1 ratio of beads to extracts. Beads were incubated with 0.3 μg of affinity-purified anti-HURP-C antibody per microliter of beads in PBS, 0.1% Triton X-100 for 1 h at 4°C with rotation. Antibody-coated beads were washed in cytostatic factor extract buffer (CSF-XB), and extracts were applied and incubated on ice for 30 min for three rounds of depletion. TPX2 and MCAK depletion were as described (Wittmann et al., 2000; Hannak and Heald, 2006). IgG from rabbit serum (Sigma, Munich, Germany) were used for control mock depletion.
For immunoprecipitation either total mitotic extracts or nuclear localization signal (NLS) proteins were incubated with antibody-coated beads for 1–2 h at 4°C, and beads were washed in CSF-XB and eluted with SDS-sample buffer.
Sperm spindles and spindles assembled around DNA beads were prepared as described (Hannak and Heald, 2006). Centrosomes were prepared as described (Bornens et al., 1987), and asters were grown from centrosomes as in Peset et al. (2005). Images were acquired using a Leica SP2 FCS confocal microscope or a Leica DMI4000 B inverted microscope (Heidelberg, Germany), with 63×/1.4 NA oil objective lens and laser lines at 405, 488, and 546 nm to excite, respectively, Hoechst 33342, GFP or Alexa 488, and Cy3. The average length of MTs nucleated from centrosomes was quantified using a macro written in MATLAB (MathWorks, Munich, Germany).
NLS Protein Preparation, Fractionation, and Immunoprecipitation
NLS proteins were prepared as described (Yokoyama et al., 2008). NLS proteins were fractionated using a 1-ml Mono S column. The bound proteins were eluted with 30 bed volumes of a 100–500 mM KCl gradient. Fractions containing HURP and TPX2 were pooled, adjusted to 100 mM KCl, and applied to a 100 μl Mono Q column. The bound proteins were eluted with 30 bed volumes of a 100–500 mM KCl gradient. NLS proteins were fractionated by gel filtration, using a Superose 6 column, in 100 mM KCl.
RESULTS
HURP Is Required for Spindle Assembly
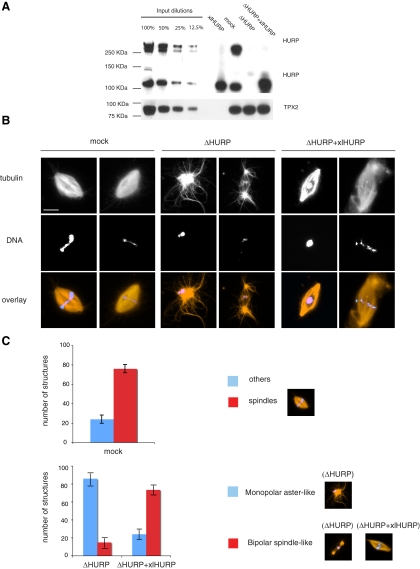
To elucidate the function of free HURP in X. laevis mitotic egg extracts we first performed immunodepletion and addback experiments. Control depleted (mock), HURP depleted (ΔHURP), and HURP-depleted extracts complemented with recombinant xlHURP (ΔHURP+xlHURP) were prepared. HURP depletion did not affect any other MAP tested. In particular, levels of XMAP215, Eg5, TPX2, and Aurora A, previously described as members of the same MT-dependent HURP complex (Koffa et al., 2006), were unperturbed (Figure 1A and Supplemental Figure S1). The HURP complex specifically forms upon binding of its members to MTs, and therefore interactions between HURP, XMAP215, Eg5, TPX2, and Aurora A occur within the MAP fraction but not in total Xenopus egg extracts (Supplemental Figure S3, A and B; Koffa et al., 2006).
Figure 1.
HURP is required for mitotic spindle assembly. (A) Western blot of HURP and TPX2 comparing dilutions of untreated Xenopus mitotic egg extracts with mock-depleted, HURP-depleted (ΔHURP), and HURP-depleted extracts supplemented with recombinant xlHURP (ΔHURP+xlHURP). (B) Sperm spindle assembly: cycled spindles were assembled in mock, ΔHURP, and ΔHURP+xlHURP extracts in the presence of sperm nuclei and Cy3-labeled tubulin (red) and fixed at 45–60 min. DNA was stained with Hoechst 33342 (blue). Scale bar, 15 μm. (C) Quantitation of the structures formed in mock, ΔHURP, and ΔHURP+xlHURP extracts. One hundred structures from three independent experiments were counted. Error bars, SD.
In Xenopus egg extracts HURP is present in two forms of roughly 120 and >250 kDa in size (Figure 1A and Supplemental Figure S2), the latter having a higher affinity for MTs, presumably being the result of a very stable modification (Koffa et al., 2006). Western blotting, performed using various commercial reagents that detect protein phosphorylation, glycosylation, SUMOylation, and ubiquitination did not provide insight into the nature of the modification. Additionally, sequencing of the high-molecular-weight form of HURP by mass spectrometry identified exclusively unmodified HURP peptides (our unpublished data). Bacterially expressed xlHURP was used to estimate the endogenous HURP concentration. The concentration of the 120-kDa HURP form is ca. 370 nM (Supplemental Figure S2), and the total concentration of HURP (the 120- and >250-kDa forms) is ca. 750 nM (Supplemental Figure S2). In complementation assays, xlHURP is added to a final concentration of roughly one or two times the total (i.e., modified and unmodified) endogenous HURP (Supplemental Figure S2).
We first examined the effect of immunodepletion and addback on sperm spindle assembly. ΔHURP extracts had a dramatically reduced capacity to organize MTs into bipolar structures. More than 80% of the structures formed were monopolar, often without any clear organization (Figure 1B, ΔHURP, left panel, and C). Only a small percentage of the structures (<20%, Figure 1C) were bipolar, but even these bipolar structures were aberrant and had a very low MT content (Figure 1B, ΔHURP, right panel). Interestingly, spindles formed in ΔHURP extracts contained few MTs in the midzone, but astral MTs were more abundant than in mock-depleted extracts (Figure 1B, ΔHURP, right panel).
The defects observed in ΔHURP extracts were specifically dependent on HURP, because recombinant xlHURP was able to complement many of the effects of HURP depletion: both MT density and MT organization were considerably restored and more than 70% of the structures formed were bipolar (Figure 1, B and C).
HURP Is Not Required for MT Growth from Centrosomes and Is Not a Plus-End MT Stabilizer
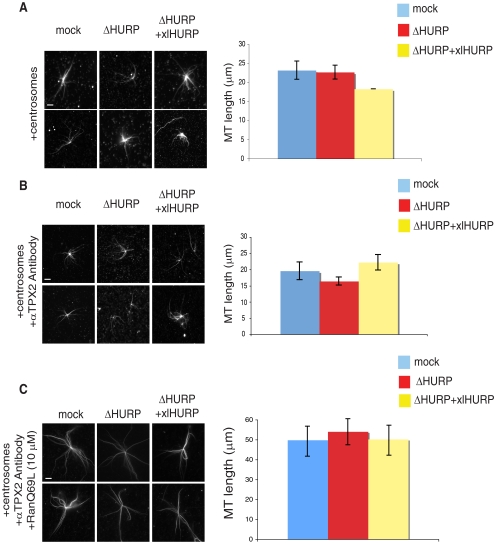
In HeLa cells, as well as in Xenopus extracts and in pure tubulin, HURP has been reported to behave as a MT stabilizer (Koffa et al., 2006; Sillje et al., 2006; Wong and Fang, 2006; Santarella et al., 2007). We therefore examined whether HURP might function in our assays by stabilizing MT plus ends. We analyzed centrosomal aster formation in mock, ΔHURP, and ΔHURP+xlHURP extracts. Samples were fixed and measured after 20 min of extract incubation with purified centrosomes, when aster length had reached a steady state. We observed no effects of HURP depletion on aster MT length (Figure 2A). To exclude any potential contribution of TPX2 to MT assembly, the experiment was repeated in the presence of αTPX2 antibodies, with similar results (Figure 2B). We further examined whether HURP is required for RanGTP-dependent stabilization of MTs nucleated from centrosomes (Carazo-Salas et al., 2001). In mock, ΔHURP, and ΔHURP+xlHURP extracts, aster MTs were twice as long in the presence of RanQ69L (RanGTP) as in the control extracts (Figure 2C; Carazo-Salas et al., 2001).
Figure 2.
HURP is not required for MT growth from centrosomes. (A) Asters nucleated from centrosomes. Mock, ΔHURP, and ΔHURP+xlHURP extracts were incubated with centrosomes and Cy3-labeled tubulin and fixed at 20 min (left panel). MT length was calculated for 40 asters in three independent experiments (right panel). Error bars, SD. Scale bar, 10 μm. (B) Asters nucleated from centrosomes in the absence of active TPX2. Mock, ΔHURP, and ΔHURP+xlHURP extracts were incubated with centrosomes, Cy3-labeled tubulin, and αTPX2 antibodies and fixed at 20 min (left panel). MT length was calculated for 40 asters in three independent experiments (right panel). Error bars, SD. Scale bar, 10 μm. (C) RanQ69L effect on MT stability. Mock, ΔHURP, and ΔHURP+xlHURP extracts were incubated with centrosomes, Cy3-labeled tubulin, αTPX2 antibodies, and 10 μM RanQ69L and fixed at 20 min (left panel). MT length was calculated for 40 asters in three independent experiments (right panel). Error bars, SD. Scale bar, 10 μm.
Together, these results show that HURP is not required for MT nucleation from centrosomes, nor does HURP act as a plus-end MT stabilizer either in the presence or absence of RanQ69L.
HURP Is Required for RanGTP and Chromatin-dependent Microtubule Assembly
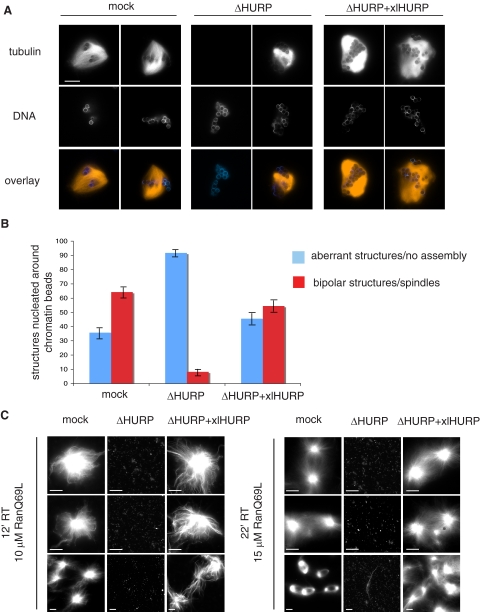
We next tested whether HURP was required for chromatin-dependent MT nucleation, using chromatin beads to promote spindle assembly in egg extracts. In mock-treated extracts, bipolar spindles efficiently formed around chromatin beads, whereas almost no bipolar structures formed in ΔHURP extracts (Figure 3A). Instead, MTs were either absent around the bead clusters (Figure 3A, ΔHURP, left panel) or, if they assembled, they did not form bipolar spindles (Figure 3A, ΔHURP, right panel). This indicated that without HURP, chromatin-dependent MT assembly is severely impaired. Addition of xlHURP to ΔHURP extracts rescued both MT formation and bipolar spindle organization (Figure 3A, ΔHURP+xlHURP). We further quantified these observations as follows: the structures observed around DNA beads were classified as “aberrant structures/no assembly” if MTs assembled without bipolar organization (as in Figure 3A, ΔHURP, left panel) or if MT were completely absent (as in Figure 3A, ΔHURP, right panel) and as “bipolar structures” (as in Figure 3A, mock and ΔHURP+xlHURP, left panel). We found that in the depleted extract, 90% of the observed structures were aberrant, most of them lacking MTs.
Figure 3.
HURP is required for chromatin-dependent microtubule nucleation. (A) Spindle assembly around DNA beads. Mock, ΔHURP, and ΔHURP+xlHURP extracts were incubated with DNA beads and Cy3-labeled tubulin (red) and fixed at 45–60 min. DNA was stained with Hoechst 33342 (blue). Scale bar, 15 μm. (B) Quantitation of structures around 100 DNA bead clusters in three independent experiments. Error bars, SD. (C) Ran-aster and Ran-spindle assembly. Mock, ΔHURP, and ΔHURP+xlHURP extracts were incubated with RanQ69L and Cy3-labeled tubulin and fixed at the indicated time points. Scale bar, 15 μm.
To obtain stronger evidence that HURP is indeed essential for chromatin-dependent MT assembly, we tested how ΔHURP extracts responded to excess RanQ69L. In this case, instead of having chromatin as a focused source of RanGTP, an equally distributed high concentration of RanGTP was present throughout the extract. Both Ran-aster and Ran-spindle formation were analyzed: shorter incubation with lower concentration of RanQ69L allowed generation of Ran-asters (Figure 3C, left panel); longer incubation with higher RanQ69L concentration was necessary for Ran-spindle formation (Figure 3C, right panel).
ΔHURP extracts did not nucleate MTs either at early or at later time points with either low or high RanQ69L (Figure 3C). In contrast, ΔHURP extracts containing xlHURP behaved like the mock extracts: they nucleated MTs, which became organized first into aster structures (Figure 3C, left panel, ΔHURP+xlHURP) and then into bipolar spindle-like structures (Figure 3C, right panel, ΔHURP+xlHURP). After longer incubation times in the presence of both RanQ69L and xlHURP overgrowth of MTs, which tended to form bundles, was observed (as in Figure 3C, right panel, ΔHURP+xlHURP, bottom).
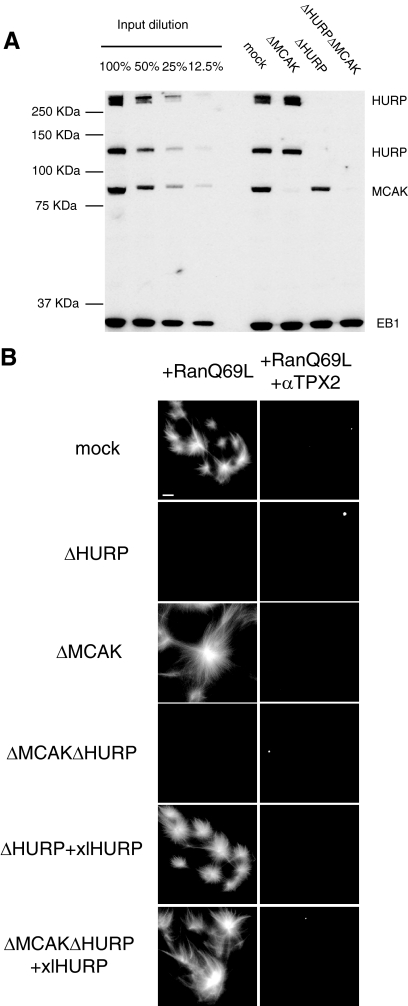
These observations suggested that HURP was either required for MT assembly or for the stabilization of very short-lived MTs against the effect of MT destabilization. To distinguish between these two possibilities, we examined the effect of HURP depletion in extracts from which the predominant MT destabilizer MCAK (Walczak et al., 1996) had been removed. Ran-structure assembly was repeated after codepleting MCAK and HURP (Figure 4A) and assayed in the presence and absence of TPX2 activity. MCAK depletion (ΔMCAK) led to overgrowth of MTs (Figure 4B, ΔMCAK+RanQ69L panel), confirming that without MCAK MTs are strongly stabilized (Walczak et al., 1996). As expected, αTPX2 antibodies added to ΔMCAK extracts prevented nucleation (Figure 4B, ΔMCAK+RanQ69L+αTPX2). The double HURP- and MCAK-depleted extracts (ΔMCAKΔHURP) were completely inactive in MT formation, either in the presence or absence of αTPX2 antibodies (Figure 4B, ΔMCAKΔHURP panels). Addition of xlHURP restored MT assembly to both singly depleted ΔHURP (Figure 4B, ΔHURP+xlHURP+RanQ69L panel) and doubly depleted ΔMCAKΔHURP extracts (Figure 4B, ΔMCAKΔHURP+xlHURP+RanQ69L panel). As expected, asters nucleated in the latter condition had longer MTs, due to the absence of MCAK. In the presence of αTPX2 antibodies, the addition of xlHURP to singly and doubly depleted extracts was not sufficient to rescue MT assembly (Figure 4B, ΔHURP+xlHURP+RanQ69L+αTPX2 and ΔMCAKΔHURP+xlHURP+RanQ69L+αTPX2 panels), suggesting that both HURP and TPX2 activities are required to promote MT assembly in response to RanGTP (see below). Taken together, these experiments indicate that HURP is required for the very early steps of RanGTP and chromatin-dependent MT assembly.
Figure 4.
HURP is required for Ran-dependent microtubule nucleation in the absence of MCAK. (A) Western blot of HURP, MCAK, and EB1 (as loading control) comparing dilutions of total untreated Xenopus mitotic egg extracts and mock-depleted, HURP-depleted (ΔHURP), MCAK-depleted (ΔMCAK) and double depleted (ΔMCALΔHURP) extracts. (B) Ran-induced nucleation assay. Mock, ΔMCAK, and ΔMCAKΔHURP extracts were incubated with Cy3-labeled tubulin, 10 μM RanQ69L, and, where indicated, αTPX2 antibodies and xlHURP and fixed at 10 min. Scale bar, 15 μm.
HURP and TPX2 Have Independent Functions
TPX2 has been shown to have a functional profile similar to that now described for HURP (Wittmann et al., 2000; Gruss et al., 2001). Our next aim was therefore to determine the relationship between the two MAPs. Although HURP and TPX2 are known to interact with each other when bound to MTs (Supplemental Figure S3A; Koffa et al., 2006), immunoprecipitation experiments performed in total Xenopus mitotic egg extracts provided no evidence that HURP and TPX2 interact detectably in the absence of MTs, either in the absence or in the presence of RanQ69L (Supplemental Figure S3B). We further examined possible interactions in a specific fraction of extracts, the NLS protein fraction, in which both are enriched (Yokoyama et al., 2008). NLS proteins were subjected to fractionation by ion exchange chromatography. After Mono S separation, Western blot analysis revealed a partial cofractionation of HURP and TPX2 (Supplemental Figure S4A). Fractions containing both HURP and TPX2 were further separated on Mono Q. Western blot analysis again revealed partial cofractionation (Supplemental Figure S4A). However, when the fractions eluted from Mono Q containing both HURP and TPX2, or total NLS proteins, were separated by gel filtration, no significant cofractionation was observed (unpublished data and Supplemental Figure S4B). Moreover, in agreement with the immunoprecipitations performed in total extracts (Supplemental Figure S3B), further immunoprecipitation experiments using the NLS fraction showed no evidence of coprecipitation of HURP and TPX2 (Supplemental Figure S3C).
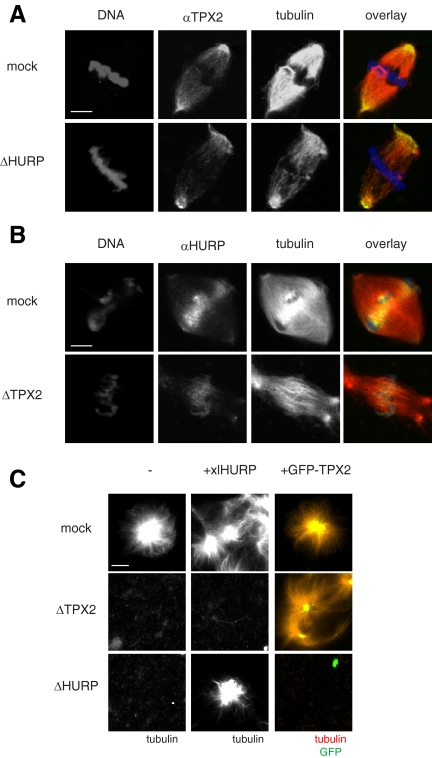
Additional evidence that HURP and TPX2 work independently came from spindle immunolocalization experiments. In mock depleted extracts HURP and TPX2 concentrate at different sites of the mitotic spindle: TPX2 is enriched at spindle poles (Wittmann et al., 2000; Gruss et al., 2002), whereas most HURP is found on MTs in the spindle midzone (Koffa et al., 2006; Sillje et al., 2006; Wong and Fang, 2006; Tedeschi et al., 2007; Wong et al., 2008). Both HURP and TPX2 depletion impair sperm (i.e., chromatin plus centrosome-induced) spindle formation. However, a few bipolar structures still form (Wittmann et al., 2000; Gruss et al., 2001). We examined whether HURP or TPX2 localization was affected by depletion of the other protein. We observed that TPX2 still localizes to spindle poles after HURP depletion (Figure 5A) and that HURP localization to the midzone is maintained upon TPX2 depletion (Figure 5B). These results indicate that HURP and TPX2 localize on the mitotic spindle independently of each other.
Figure 5.
HURP and TPX2 have independent function and distinct localization. (A) Immunolocalization of TPX2. Cycled spindles were assembled in mock or ΔHURP extracts in the presence of sperm nuclei and Cy3-labeled tubulin (red) and fixed at 45–60 min. TPX2 was detected using αTPX2 antibodies (green), and DNA was stained with Hoechst 33342 (blue). Scale bar, 15 μm. (B) Immunolocalization of HURP. Cycled spindles were assembled in mock or ΔTPX2 extracts in the presence of sperm nuclei and Cy3-labeled tubulin (red) and fixed at 45–60 min. HURP was detected using αHURP antibodies (green) and DNA was stained with Hoechst 33342 (blue). Scale bar, 15 μm. (C) Ran-induced nucleation assay. Mock, ΔHURP, and ΔTPX2 extracts were incubated with 10 μM RanQ69L, Cy3-labeled tubulin (red) and buffer (−), 1 μM xlHURP, or 0.5 μM GFP-TPX2 (green) and fixed at 10 min. Scale bar, 15 μm.
Finally, we examined whether HURP and TPX2 are redundant with one another. To test this, excess recombinant xlHURP was added to ΔTPX2 extracts and excess recombinant GFP-TPX2 was added to ΔHURP extracts. MT nucleation was induced by RanQ69L addition. Although xlHURP addition complemented ΔHURP extracts and GFP-TPX2 complemented ΔTPX2 extracts, no cross-complementation was seen (Figure 5C). In conclusion, our results reveal a novel function for HURP. Both HURP and TPX2 are independently essential for RanGTP and chromatin-dependent MT assembly in Xenopus extracts.
DISCUSSION
HURP was recently identified as a Ran-target, both in human cells (Sillje et al., 2006) and in Xenopus egg extracts, where it is the essential component of a complex required for the bipolarization of Ran spindles. The HURP complex forms upon binding to MTs (Supplemental Figure S3A and Koffa et al., 2006). However, immunoprecipitations performed either in HeLa cell extracts (Wong et al., 2008) or in complete Xenopus egg extracts in the absence of prepolymerized MTs suggest that HURP also exists in a free state. Consistent with this, in complete extracts, none of the components of the HURP complex are detected by Western blotting in HURP immunoprecipitations (Supplemental Figure S3B and our unpublished data). Sequencing of coimmunoprecipitated proteins by mass spectrometry also failed to identify any members of the HURP complex, whereas importin β, another known HURP interactor (Sillje et al., 2006), was found (our unpublished data). Moreover, although the MAP fraction where HURP is quantitatively present in the complex is active in promoting bipolarization of Ran-induced asters (Koffa et al., 2006), recombinant HURP is unable to do so (our unpublished data). We therefore sought to analyze whether free HURP also has a function by performing immunodepletion and restoration in Xenopus egg extracts, an approach that revealed a novel HURP function during early meiotic spindle assembly.
We first addressed the role of HURP in spindle formation in the presence of both inducers of MT assembly, centrosomes, and chromatin. HURP depletion severely impaired bipolar spindle formation (Figure 1, B and C). The specificity of these observations is proven by restoration of bipolarity on addition of xlHURP (Figure 1, B and C). Interestingly, in the few bipolar structures that could still form upon HURP depletion, we observed a significant decrease in kinetochore and interpolar MT overlaps in the spindle midzone, whereas astral MTs did not seem to be affected (Figure 1B, ΔHURP, right panel).
To elucidate the molecular mechanism of these HURP effects, we examined the role of HURP in either centrosome- or chromatin-dependent MT assembly. We tested whether and how HURP influences MT growth from centrosomes, in the light of HURP's reported function as an MT stabilizer (Koffa et al., 2006; Sillje et al., 2006; Wong and Fang, 2006; Santarella et al., 2007). In particular, we tested whether HURP behaves as a plus-end MT stabilizer. HURP depletion did not affect centrosomal aster length (Figure 2), even in the absence of active TPX2 or the presence of excess RanGTP. These results indicate that HURP does not contribute to centrosome-dependent MT growth or plus-end stabilization, in contrast to XMAP215 or Cdk11 (Niethammer et al., 2007). However, HURP-depleted extracts were almost completely unable to generate bipolar spindles around chromatin beads (Figure 3, A and B). The chromatin-dependent pathway of spindle assembly operates through the production of RanGTP. Interestingly, neither Ran-asters nor Ran-spindles formed in HURP-depleted extracts (Figure 3C). Therefore, HURP depletion results in a much stronger phenotype than that seen after HURP antibody inhibition (Koffa et al., 2006). This argues that previous antibody addition did not completely inactivate HURP and that sufficient amounts of HURP remained free and active to allow the formation of partly organized structures. The fact that HURP has a very early role in chromatin-mediated MT assembly is supported by the fact that MCAK depletion does not rescue HURP depletion (Figure 4B), which clearly shows that HURP activity in MT assembly precedes that of MCAK.
Taken together these results indicate that HURP works as a direct chromatin/RanGTP-target in early MT assembly, similarly to TPX2 (Wittmann et al., 2000; Gruss et al., 2001). We have previously shown that the two proteins belong to the Aurora A kinase-dependent HURP complex that forms on polymerized MTs (Koffa et al., 2006) and that functions in the bipolarization of Xenopus mitotic spindles. In the absence of MT, in total Xenopus extracts, the two MAPs do not detectably interact with each other (Supplemental Figure S3). However, both HURP and TPX2 are very abundant proteins, and it was possible that minor fractions of the two proteins interact. However, even in fractions that were highly enriched for HURP and TPX2, we failed to detect any interaction between the proteins in the absence of MTs. These biochemical conclusions are in line with localization studies showing that HURP and TPX2 are enriched on distinct domains of the mitotic spindle: HURP remains at the spindle midzone, whereas TPX2 moves toward spindle poles (Figure 5, A and B, mock panels). Furthermore, this localization is preserved upon depletion of the other MAP, suggesting that the two proteins work and localize independently of each other (Figure 5, A and B, ΔHURP and ΔTPX2 panels). This reflects the fact that, even though both are required for early steps of meiotic spindle MT assembly, HURP and TPX2 carry out distinct later functions in association with different partners: TPX2 is known to be transported to the spindle poles by the dynein–dynactin complex (Wittmann et al., 2000), whereas HURP has been shown to interact with the plus end motor protein Eg5 (Koffa et al., 2006) that also localizes to the spindle midzone (Sawin and Mitchison, 1995; Lockhart and Cross, 1996; Blangy et al., 1997). At later stages of spindle assembly TPX2 is required, together with Xklp2 and dynein, for MT focusing and spindle pole organization (Wittmann et al., 2000), whereas HURP is crucial for spindle bipolarization, metaphase chromosome alignment, and stabilization of kinetochore fibers (Koffa et al., 2006; Sillje et al., 2006).
In conclusion, we show here that HURP fulfills an essential function in the early steps of MT assembly driven by chromatin/RanGTP similar to, but independent of TPX2.
Supplementary Material
ACKNOWLEDGMENTS
We thank F. Nédélec for writing the MATLAB macro used for aster analysis and W. Antonin, J. Ellenberg, S. Kandels-Lewis, and members of the Karsenti and Mattaj laboratories for helpful discussions.
Abbreviations used:
- CSF-XB
cytostatic factor extract buffer
- HURP
hepatoma up-regulated protein
- MAP
microtubule-associated protein
- MT
microtubule.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-06-0624) on September 17, 2008.
REFERENCES
- Bischoff F. R., Klebe C., Kretschmer J., Wittinghofer A., Ponstingl H. RanGAP1 induces GTPase activity of nuclear Ras-related Ran. Proc. Natl. Acad. Sci. USA. 1994;91:2587–2591. doi: 10.1073/pnas.91.7.2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blangy A., Arnaud L., Nigg E. A. Phosphorylation by p34cdc2 protein kinase regulates binding of the kinesin-related motor HsEg5 to the dynactin subunit p150. J. Biol. Chem. 1997;272:19418–19424. doi: 10.1074/jbc.272.31.19418. [DOI] [PubMed] [Google Scholar]
- Bornens M., Paintrand M., Berges J., Marty M. C., Karsenti E. Structural and chemical characterization of isolated centrosomes. Cell Motil. Cytoskelet. 1987;8:238–249. doi: 10.1002/cm.970080305. [DOI] [PubMed] [Google Scholar]
- Brunet S., Sardon T., Zimmerman T., Wittmann T., Pepperkok R., Karsenti E., Vernos I. Characterization of the TPX2 domains involved in microtubule nucleation and spindle assembly in Xenopus egg extracts. Mol. Biol. Cell. 2004;15:5318–5328. doi: 10.1091/mbc.E04-05-0385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carazo-Salas R. E., Gruss O. J., Mattaj I. W., Karsenti E. Ran-GTP coordinates regulation of microtubule nucleation and dynamics during mitotic-spindle assembly. Nat. Cell Biol. 2001;3:228–234. doi: 10.1038/35060009. [DOI] [PubMed] [Google Scholar]
- Carazo-Salas R. E., Guarguaglini G., Gruss O. J., Segref A., Karsenti E., Mattaj I. W. Generation of GTP-bound Ran by RCC1 is required for chromatin-induced mitotic spindle formation. Nature. 1999;400:178–181. doi: 10.1038/22133. [DOI] [PubMed] [Google Scholar]
- Castoldi M., Popov A. V. Purification of brain tubulin through two cycles of polymerization-depolymerization in a high-molarity buffer. Protein Expr. Purif. 2003;32:83–88. doi: 10.1016/S1046-5928(03)00218-3. [DOI] [PubMed] [Google Scholar]
- Caudron M., Bunt G., Bastiaens P., Karsenti E. Spatial coordination of spindle assembly by chromosome-mediated signaling gradients. Science. 2005;309:1373–1376. doi: 10.1126/science.1115964. [DOI] [PubMed] [Google Scholar]
- Desai A., Murray A., Mitchison T. J., Walczak C. E. The use of Xenopus egg extracts to study mitotic spindle assembly and function in vitro. Methods Cell Biol. 1999;61:385–412. doi: 10.1016/s0091-679x(08)61991-3. [DOI] [PubMed] [Google Scholar]
- Gruss O. J., Carazo-Salas R. E., Schatz C. A., Guarguaglini G., Kast J., Wilm M., Le Bot N., Vernos I., Karsenti E., Mattaj I. W. Ran induces spindle assembly by reversing the inhibitory effect of importin alpha on TPX2 activity. Cell. 2001;104:83–93. doi: 10.1016/s0092-8674(01)00193-3. [DOI] [PubMed] [Google Scholar]
- Gruss O. J., Vernos I. The mechanism of spindle assembly: functions of Ran and its target TPX2. J. Cell Biol. 2004;166:949–955. doi: 10.1083/jcb.200312112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruss O. J., Wittmann M., Yokoyama H., Pepperkok R., Kufer T., Sillje H., Karsenti E., Mattaj I. W., Vernos I. Chromosome-induced microtubule assembly mediated by TPX2 is required for spindle formation in HeLa cells. Nat. Cell Biol. 2002;4:871–879. doi: 10.1038/ncb870. [DOI] [PubMed] [Google Scholar]
- Hannak E., Heald R. Investigating mitotic spindle assembly and function in vitro using Xenopus laevis egg extracts. Nat. Protoc. 2006;1:2305–2314. doi: 10.1038/nprot.2006.396. [DOI] [PubMed] [Google Scholar]
- Hyman A., Drechsel D., Kellogg D., Salser S., Sawin K., Steffen P., Wordeman L., Mitchison T. Preparation of modified tubulins. Methods Enzymol. 1991;196:478–485. doi: 10.1016/0076-6879(91)96041-o. [DOI] [PubMed] [Google Scholar]
- Kalab P., Pralle A., Isacoff E. Y., Heald R., Weis K. Analysis of a RanGTP-regulated gradient in mitotic somatic cells. Nature. 2006;440:697–701. doi: 10.1038/nature04589. [DOI] [PubMed] [Google Scholar]
- Kalab P., Pu R. T., Dasso M. The ran GTPase regulates mitotic spindle assembly. Curr. Biol. 1999;9:481–484. doi: 10.1016/s0960-9822(99)80213-9. [DOI] [PubMed] [Google Scholar]
- Kalab P., Weis K., Heald R. Visualization of a Ran-GTP gradient in interphase and mitotic Xenopus egg extracts. Science. 2002;295:2452–2456. doi: 10.1126/science.1068798. [DOI] [PubMed] [Google Scholar]
- Koffa M. D., Casanova C. M., Santarella R., Kocher T., Wilm M., Mattaj I. W. HURP is part of a Ran-dependent complex involved in spindle formation. Curr. Biol. 2006;16:743–754. doi: 10.1016/j.cub.2006.03.056. [DOI] [PubMed] [Google Scholar]
- Lockhart A., Cross R. A. Kinetics and motility of the Eg5 microtubule motor. Biochemistry. 1996;35:2365–2373. doi: 10.1021/bi952318n. [DOI] [PubMed] [Google Scholar]
- Niethammer P., Kronja I., Kandels-Lewis S., Rybina S., Bastiaens P., Karsenti E. Discrete states of a protein interaction network govern interphase and mitotic microtubule dynamics. PLoS Biol. 2007;5:e29. doi: 10.1371/journal.pbio.0050029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohba T., Nakamura M., Nishitani H., Nishimoto T. Self-organization of microtubule asters induced in Xenopus egg extracts by GTP-bound Ran. Science. 1999;284:1356–1358. doi: 10.1126/science.284.5418.1356. [DOI] [PubMed] [Google Scholar]
- Ohtsubo M., Okazaki H., Nishimoto T. The RCC1 protein, a regulator for the onset of chromosome condensation locates in the nucleus and binds to DNA. J. Cell Biol. 1989;109:1389–1397. doi: 10.1083/jcb.109.4.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peset I., Seiler J., Sardon T., Bejarano L. A., Rybina S., Vernos I. Function and regulation of Maskin, a TACC family protein, in microtubule growth during mitosis. J. Cell Biol. 2005;170:1057–1066. doi: 10.1083/jcb.200504037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santarella R. A., Koffa M. D., Tittmann P., Gross H., Hoenger A. HURP wraps microtubule ends with an additional tubulin sheet that has a novel conformation of tubulin. J. Mol. Biol. 2007;365:1587–1595. doi: 10.1016/j.jmb.2006.10.064. [DOI] [PubMed] [Google Scholar]
- Sawin K. E., Mitchison T. J. Mutations in the kinesin-like protein Eg5 disrupting localization to the mitotic spindle. Proc. Natl. Acad. Sci. USA. 1995;92:4289–4293. doi: 10.1073/pnas.92.10.4289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatz C. A., Santarella R., Hoenger A., Karsenti E., Mattaj I. W., Gruss O. J., Carazo-Salas R. E. Importin alpha-regulated nucleation of microtubules by TPX2. EMBO J. 2003;22:2060–2070. doi: 10.1093/emboj/cdg195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sillje H. H., Nagel S., Korner R., Nigg E. A. HURP is a Ran-importin beta-regulated protein that stabilizes kinetochore microtubules in the vicinity of chromosomes. Curr. Biol. 2006;16:731–742. doi: 10.1016/j.cub.2006.02.070. [DOI] [PubMed] [Google Scholar]
- Tedeschi A., Ciciarello M., Mangiacasale R., Roscioli E., Rensen W. M., Lavia P. RANBP1 localizes a subset of mitotic regulatory factors on spindle microtubules and regulates chromosome segregation in human cells. J. Cell Sci. 2007;120:3748–3761. doi: 10.1242/jcs.009308. [DOI] [PubMed] [Google Scholar]
- Tsou A. P., et al. Identification of a novel cell cycle regulated gene, HURP, overexpressed in human hepatocellular carcinoma. Oncogene. 2003;22:298–307. doi: 10.1038/sj.onc.1206129. [DOI] [PubMed] [Google Scholar]
- Walczak C. E., Heald R. Mechanisms of mitotic spindle assembly and function. Int. Rev. Cytol. 2008;265:111–158. doi: 10.1016/S0074-7696(07)65003-7. [DOI] [PubMed] [Google Scholar]
- Walczak C. E., Mitchison T. J., Desai A. XKCM1, a Xenopus kinesin-related protein that regulates microtubule dynamics during mitotic spindle assembly. Cell. 1996;84:37–47. doi: 10.1016/s0092-8674(00)80991-5. [DOI] [PubMed] [Google Scholar]
- Wilde A., Zheng Y. Stimulation of microtubule aster formation and spindle assembly by the small GTPase Ran. Science. 1999;284:1359–1362. doi: 10.1126/science.284.5418.1359. [DOI] [PubMed] [Google Scholar]
- Wittmann T., Wilm M., Karsenti E., Vernos I. TPX2, A novel Xenopus MAP involved in spindle pole organization. J. Cell Biol. 2000;149:1405–1418. doi: 10.1083/jcb.149.7.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong J., Fang G. HURP controls spindle dynamics to promote proper interkinetochore tension and efficient kinetochore capture. J. Cell Biol. 2006;173:879–891. doi: 10.1083/jcb.200511132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong J., Lerrigo R., Jang C. Y., Fang G. Aurora A regulates the activity of HURP by controlling the accessibility of its microtubule-binding domain. Mol. Biol. Cell. 2008;19:2083–2091. doi: 10.1091/mbc.E07-10-1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama H., Gruss O. J., Rybina S., Caudron M., Schelder M., Wilm M., Mattaj I. W., Karsenti E. Cdk11 is a RanGTP-dependent microtubule stabilization factor that regulates spindle assembly rate. J. Cell Biol. 2008;180:867–875. doi: 10.1083/jcb.200706189. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.